Abstract
Most of the present knowledge about cell organization and function is based on molecular and genetic methods as well as cytological investigations. While electron microscopy allows identifying cell substructures until a resolution of ∼1 nm, the resolution of fluorescence microscopy is restricted to ∼200 nm due to the diffraction limit of light. However, the advantage of this technique is the possibility to identify and co-localize specifically labeled structures and molecules. The recently developed super-resolution microscopy techniques, such as Structured Illumination Microscopy, Photoactivated Localization Microscopy, Stochastic Optical Reconstruction Microscopy, and Stimulated Emission Depletion microscopy allow analyzing structures and molecules beyond the diffraction limit of light. Recently, there is an increasing application of these techniques in cell biology. This review evaluates and summarizes especially the data achieved until now in analyzing the organization and function of plant cells, chromosomes and interphase nuclei using super-resolution techniques.
Keywords: chromosome, nucleus, PALM, plant cell, STED, SIM, STORM, super-resolution
Introduction
Light microscopy of DNA and proteins specifically fluorescently labelled by FISH and immunostaining, respectively, as well as live cell imaging based on fluorescent recombinant proteins significantly increased our knowledge concerning cell organization and function, and is an important advantage compared to electron microscopy.
However, due to the diffraction limit of light as defined by Abbe (1873) the spatial resolution of light microscopy including conventional fluorescence techniques is restricted, and reaches only ∼200 nm laterally and ∼600 nm in the axial dimension in biological specimens (Pawley, 1995). Thus, this limited resolution did not allow identifying single molecules and structures with the resolution achieved by electron microscopy.
Recently, to overcome this restriction and to bridge the resolution gap between light and electron microscopy the so-called super-resolution (also referred as optical nanoscopy) techniques SIM, PALM, STORM, and STED offering new insights into molecular structures, interactions and functions were developed. These “subdiffraction” methods can be divided into two different principles: (i) localization of individual fluorophores in the specimen with subdiffraction precision (PALM, STORM), and (ii) structuring the illumination light to collect high spatial frequencies in the image that contain high resolution information (SIM, STED) (Rego et al., 2012).
To honor the establishment of these stunning methods as PALM/STORM and STED the Nobel Prize in Chemistry 2014 was awarded to Eric Betzig, William Moerner and Stefan Hell, respectively (Gevaux, 2014; Möckl et al., 2014; Stelzer, 2014). The meanwhile widely applied SIM technique has mainly been developed by the late Mats Gustafsson (Gustafsson, 2005; Gustafsson et al., 2008).
In addition to special super-resolution microscope systems established in different research groups, since 2004 also commercial systems are produced by companies as Leica Microsystems (Leica TCS SP8 STED 3X - STED; Leica SR GSD 3D microscope - STORM), Carl Zeiss (Elyra S.1 - SIM; Elyra P.1 - PALM; Elyra PS.1 - combination of SIM and PALM), Nikon (N-SIM; N-STORM; combination of N-SIM and N-STORM) and GE Healthcare Life Sciences (DeltaVision OMX System - SIM; DeltaVision Localization Microscopy System - STORM; combination of SIM and STORM).
Excellent reviews describing and comparing the different super-resolution light microscopy methods (Schermelleh et al., 2010; Agrawal et al., 2013; Allen et al., 2014; Komis et al., 2015b; Nienhaus and Nienhaus, 2016) already exist. These techniques were applied successfully in cell biology (Rouquette et al., 2010; Han et al., 2013; Fornasiero and Opazo, 2015) at fixed and live specimens from both pro- and eukaryotes and helped to discover new structures.
Compared to animal tissues and due to varying refractive indices of plant cell organelles inducing spherical aberrations and light scattering, plant cell imaging is particularly challenging (Komis et al., 2015b). Nevertheless, the applications in this field are also increasing. Here I summarize and evaluate the recent achievements in plant cell research applying different super-resolution techniques.
Applications in Plant Cell Research
Similar as in other organisms analyzing cell structures of plants is essential to understand biological functions. Thus, increasing efforts are undertaken to apply super-resolution techniques in plant cell research.
Investigations were performed on structures outside and inside of the nucleus, and on condensed chromosomes during cell division (Table 1). Most results were obtained from fixed material, but also live cell imaging based on fluorescently tagged proteins was performed.
Table 1.
Applications of super-resolution methods to analyze plant cell structures.
| Structures/cell stages analyzed | Results | Speciesa | Methods | Reference |
|---|---|---|---|---|
| Cellular components outside of the nucleus/chromosomes | ||||
| Pollen | Analysis of pollen surface texture and shape | 2 | SIM | Sivaguru et al., 2012 |
| Cellulose microfibrils | Analysis of the cellulose microfibril architecture in the cell walls of onion-bulb scale epidermal cells | 3 | STORM | Liesche et al., 2013 |
| Cortical microtubules | Organization and development of cortical microtubules in living hypocotyl epidermal cells | 4 | SIM | Komis et al., 2014 |
| Cortical microtubules | Vizualization of cortical microtubules in living cotyledon, petiole and root cells | 4 | SIM | Komis et al., 2015a |
| Cortical microtubules | Organization and quantitation of cortical microtubules in different root cells | 4 | STORM | Dong et al., 2015 |
| Endosomes | Imaging and quantification of early and late endosomes during root hair formation | 4 | SIM | von Wangenheim et al., 2016 |
| Plasmodesmata | Organization of plasmodesmata in leaf parenchyma cells | 11 | SIM | Fitzgibbon et al., 2010 |
| Plasmodesmata | Arrangement of plasmodesmata and callose in leaf cells | 4 | SIM | Bell and Oparka, 2011 |
| Plasma membrane | Tracking of individual membrane protein motions in living root epidermal cells | 4 | PALM | Hosy et al., 2015 |
| Plasma membrane | Detection of polar-competent PIN protein clusters responsible for auxin transport in the apical plasma membrane of living root cells | 4 | STED | Kleine-Vehn et al., 2011 |
| Perinuclear cell region | Localization of perinuclear actin in living tobacco cells (BY-2) | 11 | PALM | Durst et al., 2014 |
| Plasmodesmata and virus proteins | Identification of callose and viral movement proteins in the central cavities of complex plasmodesmata in leaf epidermal cells | 4 | SIM | Fitzgibbon et al., 2013 |
| Plasmodesmata and virus proteins | Localization of plant virus movement proteins in plasmodesmata | 10 | SIM | Tilsner et al., 2013 |
| Sieve element reticulum and virus proteins | Arrangement of the sieve element reticulum, callose, and cellulose, and identification of potato virus X filaments in phloem cells | 10, 11 | SIM | Bell et al., 2013 |
| Membrane structures and virus filaments | Visualization of membrane structures of pseudoviral replication complexes and individual potato virus X filaments in leaf cells | 10 | SIM | Linnik et al., 2013 |
| Plasmodesmata and virus proteins | Localization of RTNLB proteins in the the central desmotubule of plasmodesmata and their colocalization with tobacco mosaic virus movement proteins | 4 | SIM | Knox et al., 2015 |
| Membrane domains and fungus structures | Identification of extrahaustorial membrane domains and proteins in living leaf cells after Phytophtora infestans infection | 10 | SIM | Bozkurt et al., 2014 |
| Chromatin and protein organization in interphase nuclei | ||||
| Somatic interphase | Chromatin ring formation of satellite DNA | 14 | SIM | Ribeiro et al., 2017 |
| Interphase nuclei, mitosis | Distribution of histone H3K4me2, H3K9me2 and H3K27me3 in holocentric chromosomes | 9 | SIM | Heckmann et al., 2013 |
| Interphase nuclei | Detection of active and inactive RNAPII in the proximity of B chromosome chromatin | 17 | SIM | Ma et al., 2017 |
| Meristematic nuclei | Determination of the spatio-temporal distribution of rDNA during replication | 4 | SIM | Dvořáčková et al., in press |
| Meristematic and endopolyploid nuclei | Distribution and quantification of active and inactive RNAPII within euchromatin | 4, 17 | SIM | Schubert, 2014 |
| Endopolyploid nuclei | Distribution and quantification of active and inactive RNAPII within euchromatin | 4 | SIM, PALM | Schubert and Weisshart, 2015 |
| Interphase nuclei | Co-localization of the transcript elongation factor SPT5 and RNAPII within euchromatin | 4 | SIM | Dürr et al., 2014 |
| Interphase nuclei | Co-localization between the transcript elongation factors SPT6L, ELF7 and RNAPII | 4 | SIM | Antosz et al., 2017 |
| Differentiated nuclei | Distribution of SMC3 and CAP-D3; eu- and heterochromatin characterization in endopolyploid nuclei | 4 | SIM | Schubert et al., 2013 |
| Chromatin/protein organization along condensed chromosomes | ||||
| Pachytene | Mitochondrial and plastidal DNA localization in B chromosomes | 17 | SIM | Klemme, 2013 |
| Somatic metaphase | Accumulation of chloroplast- and mitochondria-derived sequences in B chromosomes | 1 | SIM | Ruban et al., 2014 |
| Somatic metaphase | Localization of satellite repeats along holocentric chromosomes | 13, 14, 15, 16 | SIM | Ribeiro et al., 2017 |
| Pachytene | Evaluation of the cytosine methylation status of satellite DNAs | 5 | SIM | Zakrzewski et al., 2014 |
| Somatic metaphase | Detection of sister chromatid exchanges | 7 | SIM | Vu et al., 2014 |
| Somatic metaphase | Sister chromatid exchange arrangement in mono- and holocentric chromosomes | 9, 17 | SIM | Schubert et al., 2016b |
| Somatic metaphase | Localization of H2AThr120ph and H3S10ph at chromosome arms | 6 | SIM | Sousa et al., 2016 |
| Meiosis | Analysis of the synaptonemal complex formation and the progression of meiotic chromosome synapsis | 7 | SIM | Phillips et al., 2012 |
| Meiosis | Analysis of the synaptonemal complex organization and interlock formation | 19 | SIM | Wang et al., 2009 |
| Zygotene | Colocalization of AFD1 and ASY1 during the synaptonemal complex formation | 19 | SIM | Gustafsson et al., 2008 |
| Mitosis | Visualization of 3xHMG-box proteins at somatic chromosomes | 4 | SIM | Antosch et al., 2015 |
| Centromeres | ||||
| Somatic metaphase | Deviating centromere chromatin organization in A and B chromosomes | 17 | SIM | Banaei-Moghaddam et al., 2012 |
| Metaphase I | Detection of CENH3 at the centromeres of bi- and univalents | 17 | SIM | Cuacos, 2013 |
| Interphase nuclei, mitosis, meiosis | Chromatin ring formation at centromeres | 1, 4, 7, 17, 18 | SIM | Schubert et al., 2016a |
| Interphase nuclei, mitosis | Co-localization of CENH3 and centromere-specific repeats in holocentromeres | 15 | SIM | Marques et al., 2015 |
| Interphase nuclei, mitosis, meiosis | Co-localization of tubulin, CENH3, CENP-C and centromere-specific repeats in holocentromeres | 15 | SIM | Marques et al., 2016 |
| Somatic inter- and metaphase | Localization of CENH3 and centromeric repeats along holocentric chromosomes | 13, 14, 15, 16 | SIM | Ribeiro et al., 2017 |
| Interphase nuclei, mitosis, meiosis | CENH3 amount measurements based on fluorescence intensities | 17 | SIM | Schubert et al., 2014 |
| Somatic metaphase | Localization of CENH3 along holocentric chromosomes | 15 | SIM | Cabral et al., 2014 |
| Mitosis, meiosis | Co-localization of the two CENH3 variants | 7 | SIM | Karimi-Ashtiyani et al., 2015 |
| Somatic inter- and metaphase | Intermingled co-localizaton of αCENH3 and βCENH3 | 7 | SIM | Ishii et al., 2015 |
| Interphase nuclei | Intermingled colocalization of A. thaliana and Zea mays CENH3s | 4 | SIM | Maheshwari et al., 2016 |
| Somatic metaphase | Intermingled co-localizaton of αCENH3 and H2AThr120ph | 7 | SIM | Demidov et al., 2014 |
| Somatic metaphase | Localization of CENH3 and H2AThr120ph in holokinetic chromosomes | 9 | SIM | Jankowska et al., 2015 |
| Mitosis | Co-localization of α and βCENH3, H2AThr120ph and tubulin at holo- and monocentromeres | 9, 7 | SIM | Wanner et al., 2015 |
| Meiosis | Co-localization of CENH3, H2AThr120ph and tubulin at holocentromeres | 9 | SIM | Heckmann et al., 2014 |
| Somatic metaphase | Co-localization of CENH3 and α-kleisin in mono- and holocentromeres | 9, 7 | SIM | Ma et al., 2016 |
| Meristematic nuclei | Co-localization of CENH3 and KNL2 | 4 | SIM | Lermontova et al., 2013 |
| Root tip nuclei | Co-localization of CENH3 and KNL2 | 4 | SIM | Sandmann et al., 2017 |
| Interphase nuclei, mitosis | Co-localization of CENH3 and GIPs at centromeres | 4 | SIM | Batzenschlager et al., 2015 |
| Somatic metaphase | Co-localization of both CENH3 variants, H2AThr120ph and H3S28ph in polycentric chromosomes | 8, 12 | SIM | Neumann et al., 2016 |
| Somatic metaphase | Localization of H2AThr120ph and H3S10ph at centromeres | 6 | SIM | Sousa et al., 2016 |
| (Sub)telomeres | ||||
| Somatic metaphase | Localization of telomeric repeats in holokinetic chromosomes | 9 | SIM | Jankowska et al., 2015 |
| Interphase nuclei, mitosis, meiosis | Chromatin ring formation at subtelomeres | 1, 4, 7, 17, 18 | SIM | Schubert et al., 2016a |
a(1) Aegilops speltoides (Boiss.) Chennav.; (2) Agropyron repens (L.) P. Beauv.; (3) Allium cepa L.; (4) Arabidopsis thaliana (L.) Heynh.; (5) Beta vulgaris L.; (6) Coccinia grandis (L.) Voigt; (7) Hordeum vulgare L.; (8) Lathyrus sativus L.; (9) Luzula elegans Lowe; (10) Nicotiana benthamiana Domin; (11) Nicotiana tabacum L.; (12) Pisum sativum L.; (13) Rhynchospora ciliata (G. Mey.) Kük.; (14) Rhynchospora globosa (Kunth) Roem. & Schult.; (15) Rhynchospora pubera (Vahl) Boeckeler; (16) Rhynchospora tenuis Link; (17) Secale cereale L.; (18) Triticum aestivum L.; (19) Zea mays L.
Fixed specimens were stained with specific antibodies and/or hybridized with labeled DNA probes. Such experiments were the basis to quantify and colocalize polysaccharides, proteins and DNA.
Live cell imaging has already successfully applied to follow the development of cytoskeleton components, membrane proteins and fungal infection structures.
Meanwhile, in addition to model plants as Arabidopsis and tobacco, also cereals and holocentric wild species were investigated by super-resolution microscopy.
Till now, among the different super-resolution techniques most results were achieved by SIM, whereas fewer applications are based on PALM/STORM and STED (Table 1).
Cellular Components Outside of the Nucleus/Chromosomes
Super-resolved imaging was applied to investigate interactions between different cells, cellular organelles and nuclei.
In this field Oparka and co-workers studied especially by using SIM cell-cell interactions by plasmodesmata imaging (Bell and Oparka, 2011, 2015) and localized plant virus proteins therein. Additionally, by other groups cytoskeleton components and membrane structures were analyzed on fixed specimens (Table 1). STORM was applied to elucidate the cellulose microfibril organization in onion cells (Liesche et al., 2013) and the cortical microtubule arrangement in Arabidopsis roots (Dong et al., 2015).
Live cell imaging using fluorescent tags such as GFP and mCherry fused to genetically encoded marker proteins together with SIM has been applied to follow the development of microtubules in Arabidopsis hypocotyl epidermal cells (Komis et al., 2014), but also in cotyledon, petiole and root cells (Komis et al., 2015a). Bozkurt et al. (2014) identified by SIM fungus structures after Phytophtora infection in living tobacco leaves. This indicates that SIM is also a very versatile method with a broad application potential in plant live cell research.
Photoactivated Localization Microscopy on living cells was performed to track Arabidopsis root membrane proteins (Hosy et al., 2015) and to localize perinuclear actin in tobacco (Durst et al., 2014).
To date only one STED application in plant cell research has been published. Kleine-Vehn et al. (2011) detected polar-competent YFP-labeled PIN protein clusters responsible for auxin transport in the apical plasma membrane of living A. thaliana root cells. The high laser power required for STED causing fast bleaching impedes the acquiring of image stacks and longer live cell imaging. In addition, the number of applicable florescence dyes is restricted. The high degree of autofluorescence and the presence of color pigments (e.g., chlorophyll) make plant tissues especially challenging for STED. Obviously, this causes its so far restricted application in plant cell research.
Chromatin and Protein Organization in Interphase Nuclei
To understand such basic cellular functions as transcription, replication, and DNA repair the organization of chromatin, DNA–DNA, DNA–protein and protein-protein interactions have to be investigated in interphase nuclei. For this aim, super-resolution imaging was performed on nuclei in tissue squash preparations. However, especially the imaging of isolated and flow-sorted nuclei delivered excellent resolutions due to the absence of cytoplasm (Table 1).
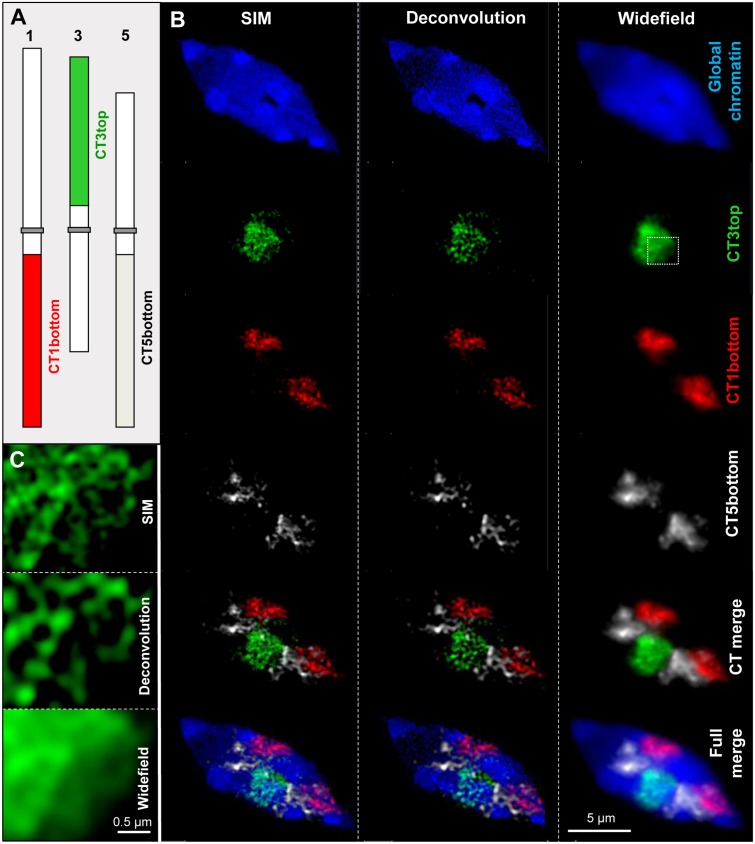
FISH with differently labeled DNA probes allowed investigating the subchromosomal arrangement of chromatin within cell nuclei (Schubert et al., 2013) (Figure 1). Compared to widefield microscopy and deconvolution imaging the increased content of information due to the higher resolution obtained by SIM becomes obvious.
FIGURE 1.
Chromatin organization in a differentiated 8C A. thaliana leaf nucleus. Three chromosome arm territories (CTs) of chromosomes 1, 3, 5 showing similar size were stained in different colors (A). After FISH and SIM the distribution of the euchromatic arm regions becomes visible beyond the diffraction limit of light, in comparison to the resolution achieved by deconvolution and widefield microscopy (B). The enlarged region of CT3top (rectangle) shows clearly the increased resolution compared to widefield and deconvolution imaging (C).
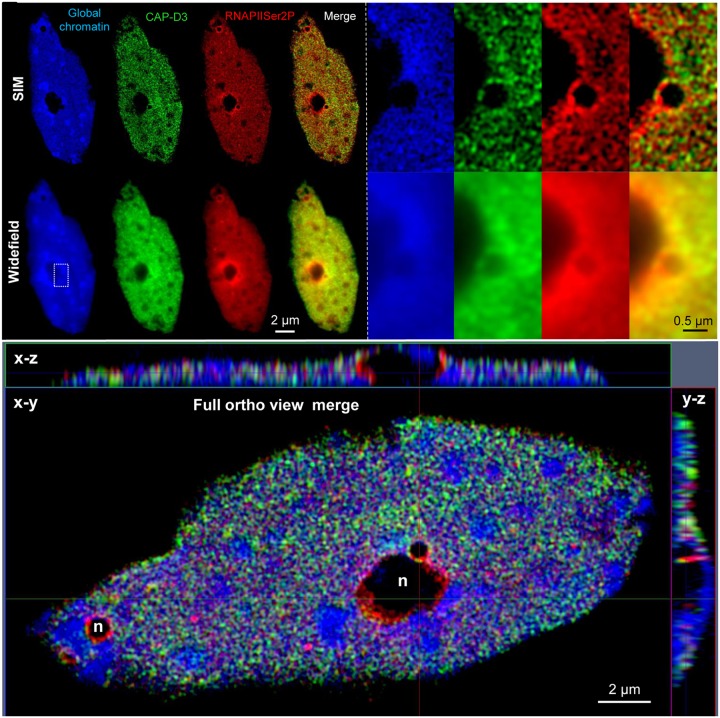
Several publications show the distribution and colocalization of RNA polymerase II (RNAPII) enzymes and interacting factors in Arabidopsis nuclei after labeling with specific antibodies. In addition, the relative and absolute quantification of these molecules in 3D image stacks was performed by SIM and PALM, respectively (Schubert, 2014; Schubert and Weisshart, 2015). Figure 2 demonstrates the colocalization of RNAPII and the structural condensin subunit CAP-D3 within the euchromatin of an isolated 32C Arabidopsis nucleus, and the improved resolution achieved by SIM compared to widefield microscopy.
FIGURE 2.
Distribution and co-localization of active RNAPII (phosphorylated at serine 2) and the condensin subunit CAP-D3 in a flow-sorted differentiated 32C A. thaliana leaf nucleus. After immunostaining with specific antibodies and SIM it becomes visible that both proteins are present within euchromatin, but absent from heterochromatin (dense blue staining) and nucleoli (n) (upper left). Especially the enlarged region (rectangle) shows the increased resolution obtained by SIM compared to widefield microscopy (upper right). The ortho view (below) generated from fully merged SIM image stacks visualizes the nucleus as front (x–y) and side (x–z, y–z) views. Relative voxel intensity measurements based on the SIM image stack using the Imaris 8.0 (Bitplane) software showed that this nucleus contains ∼19% less RNAPII than CAP-D3 molecules, and that ∼81% of these molecules colocalize.
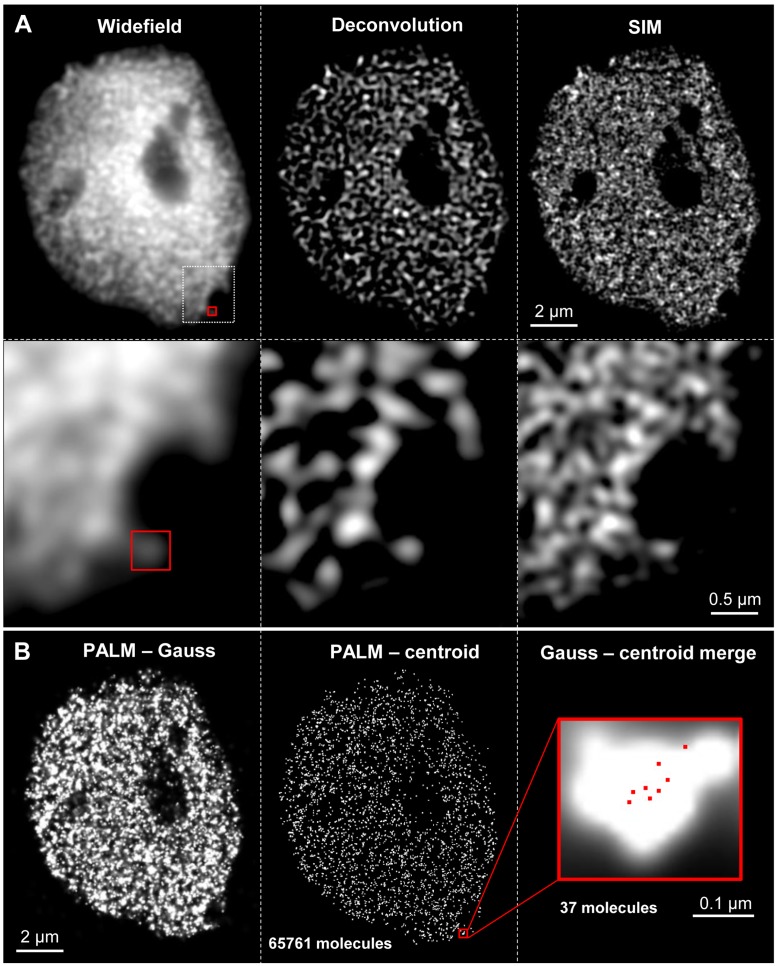
The combination of SIM and PALM feasible with systems such as the Elyra PS.1 from Zeiss enables acquiring image stacks by both techniques subsequently. Then, the combination of these stacks allows counting and localizing single molecules within the structures identified by SIM (Schubert and Weisshart, 2015; Weisshart et al., 2016) (Figure 3).
FIGURE 3.
Distribution and quantification of active RNAPII (phosphorylated at serine 2) molecules in a differentiated 16C A. thaliana leaf nucleus. After immunostaining with specific antibodies SIM (A) and PALM (B) were performed consecutively. (A) Compared to widefield and deconvolution microscopy SIM clearly delivers an increased resolution especially visible in the enlarged region (white rectangle) below. This indicates that RNAPII is distributed network-like within the nucleus. (B) After 3D-PALM the resolution becomes further increased when visualized as Gauss distribution (left). Centroid visualization allows the exact localization and counting of single molecules (center). The nucleus with a z extension of 1.2 μm contains in total 65761 molecules which were counted in 30 slices of 40 nm. The merged Gauss-centroid view (right) represents the localization of eight single molecules in a single slice of a RNAPII cluster (red rectangle) containing in total 37 molecules in five slices with a total z range of 0.2 μm.
Chromatin and Protein Organization Along Condensed Chromosomes
All organisms perform cell divisions, at which for proper segregation during mitosis and meiosis chromosomes have to be condensed. To better understand this process super-resolution microscopy has been used to analyze the distribution and organization of DNA in cereal supernumerary B chromosomes (Ruban et al., 2014), and of satellite DNA along holocentric chromosomes (Ribeiro et al., 2017) (Table 1).
Additionally, the distribution of specific DNA (Zakrzewski et al., 2014) and histone modifications (Sousa et al., 2016) was investigated along condensed chromosomes.
The arrangement of sister chromatids in holocentric chromosomes could be clarified by differential labeling via base analog incorporation during replication followed by SIM (Schubert et al., 2016b).
SIM was also helpful to analyze the synaptonemal complex formation during homologous chromosome paring in prophase I of maize (Gustafsson et al., 2008; Wang et al., 2009) and barley (Phillips et al., 2012).
Centromeres and (Sub)Telomeres
Centromeres/kinetochores as spindle fiber attachment sites are required for proper chromosome segregation during cell division. Telomeres protect chromosome termini from degradation and fusion. Thus, both are essential to maintain genome stability of organisms.
SIM investigations were helpful to get new insight into the centromere organization during the cell cycle via specifically labeling and co-localizing centromere components such as centromere-specific DNA repeats, kinetochore proteins and histone modifications at centromeric chromatin (Table 1).
Especially the finding that phosphorylated histone H2A positive chromatin and different CENH3 variant containing chromatin clusters intermingle to form centromeres has been attained (Demidov et al., 2014; Ishii et al., 2015; Karimi-Ashtiyani et al., 2015), an observation not achievable by refraction-limited widefield resolution. Similar comparative investigations in mono-, poly-, and holocentric plants species provided also new insights into the evolution of centromeres (Wanner et al., 2015; Neumann et al., 2016).
Regarding telomere investigations until now only two publications appeared. One was published about the localization of telomeric repeats in holokinetic Luzula chromosomes (Jankowska et al., 2015), a second about chromatin ring formation at subtelomeres in barley (Schubert et al., 2016a).
Specimen Preparation and the Super-Resolution Methods
All super-resolution techniques are based on imaging fluorescent molecules. Consequently, they are used to label structures and molecules of interest. After fixation of a specimen, which should alter the native structures as less as possible, specific fluorescent affinity probes of preferably small size (FAB fragments of antibodies, nanobodies, snap-tags) may be used for labeling (Fornasiero and Opazo, 2015).
The advantage of genetically encoded and expressed fluorescent proteins (Hedde and Nienhaus, 2014) is that they allow investigating dynamic processes in living cells without fixation artifacts. However, due to phototoxicity live cell nanoscopy is much more challenging than imaging fixed specimens (Fornasiero and Opazo, 2015). Hence, fewer live cell imaging results have been published so far in plant cell research (Table 1).
Both structured illumination and localization microscopy may be used for fixed material but also for imaging living cells. Depending on the different super-resolution techniques and the imaging tasks to be performed (e.g., quantification and colocalization of molecules) the specimen preparations have to be adapted accordingly. Staudt et al. (2007) developed a new embedding medium especially useful for STED microscopy to avoid spherical aberrations induced by the refractive index mismatch between the immersion system and the embedding medium of the sample.
Imaging of Fixed Specimens
The major advantage of SIM is that most preparation and labeling protocols and fluorophores used for widefield fluorescence microscopy are applicable without modification, thus allowing high-throughput experiments. Despite a reliable tissue fixation, the use of high-quality glass slides and especially coverslips (e.g., Marienfeld high precision cover glasses) are important to reach the possible resolution of up to ∼120 nm by a 488 nm excitation. It is quite important to notice that during SIM raw data acquisition an overexposure must be avoided perfectly. Otherwise, artificial not existing structures and shapes can be generated during the SIM calculation.
Using a Zeiss ELYRA PS.1 microscope system the acquiring of image stacks of up to 30 slices at a distance of ∼100 nm at full resolution (∼1024 × 1024 pixel, 80 μm × 80 μm FOV, 100 ms exposure time), which takes ∼4–5 min in a sequential 3-color experiment (see Figure 1) are the basis to calculate 3D-SIM image stacks within ∼10-20 min (Weisshart et al., 2016). These stacks may be used for 3D-movie rendering by, e.g., the commercial ZEN (Zeiss) (e.g., Schubert et al., 2013, 2016a) or Imaris (Bitplane) (Neumann et al., 2016; Schubert et al., 2016b) softwares.
To reveal the spatial ultrastructure of cellular components SIM delivers best results after applying FISH and/or immunolabelling at relatively flat (up to ∼10 μm) tissue squashes and spreadings. But especially useful are isolated and flow-sorted cell nuclei free of disturbing cytoplasm (Dürr et al., 2014; Weisshart et al., 2016; Antosz et al., 2017) (Figure 2).
SIM image stacks are also useful to determine relative molecule amounts via pixel intensity measurements in organelles like nuclei, and to measure the degree of colocalization of differently labeled molecules (Dürr et al., 2014; Schubert, 2014; Antosz et al., 2017) (Figure 2).
Bell and Oparka (2015) developed preparative methods especially for imaging plasmodesmata by SIM on fixed plant tissues.
Practically localization microscopy (PALM, STORM) reaches a circa five-fold higher resolution than SIM. Thus, single molecules may be identified, counted and colocalized in single- and even two-color experiments using, e.g., the fluorescence dyes Alexa488 and Cy5 (Schubert and Weisshart, 2015; Weisshart et al., 2016). However, compared to SIM specimen preparation is more challenging and raw data acquisition and calculations are more time-consuming. To achieve reliable results a high labeling density and efficient photoactivation are required (Fernández-Suárez and Ting, 2008).
2D-PALM may be performed under HILO (Tokunaga et al., 2008), Epi and TIRF illumination (Hedde and Nienhaus, 2014), whereby TIRF will give the best signal, followed by HILO and last Epi. 3D-PALM features dependent on the 3D-technology used to capture ranges between 1.2 and 2.0 μm. This range might be extended by using classical z-scans. For a better signal HILO instead of Epi illumination is recommended (van de Linde et al., 2011). TIRF and for best performance also HILO illumination need the positioning of the specimen very close to the coverslip surface. Thus, the preparation of specimens directly onto coverslips has to be performed. These coverslips may be fixed onto slides by rubber cement and then be handled as usual during the staining and washing procedures. After placing them into coverslip chambers, adding a redox reagent and adjusting the pH to the needs of the dye to achieve efficient photoactivation, PALM can be performed (Weisshart et al., 2016). The redox reagent, e.g., 1% 2-mercaptoethanol in 1x PBS can be used if the fluorescence dyes in the specimens are easily accessible. Otherwise, adding of glucose is required (Olivier et al., 2013; Schäfer et al., 2013).
PALM using fluorescent proteins can be performed in buffers like PBS or Hepes. Acquiring raw data sets takes ∼15 min, followed by a calculation procedure of also ∼15 min if, e.g., isolated nuclei and the Elyra PS.1 is used.
Non-commercial super-resolution microscope setups (Hamel et al., 2014) and commercial systems as the Elyra PS.1 (Weisshart et al., 2016) allow producing SIM and PALM/STORM data subsequently. Thereby, the observed SIM structures can be combined with precise PALM/STORM single molecule localization and counting. This clearly increases the information obtained from the specimens under investigation (Figure 3).
Life Cell Imaging
Expressed fluorescent reporter proteins allow visualizing proteins and structures inside living organisms (Hedde and Nienhaus, 2014). Because the implementation of super-resolution microscopy in live cell microscopy is very challenging and both structured illumination and localization microscopy have different advantages and disadvantages (Schermelleh et al., 2010), to date only few publications describing the dynamics of plant cell components appeared (see above).
Propagating seedlings in coverslip chambers under sterile conditions allow live cell imaging by SIM on roots growing closely along the coverslip. However, due to the fast root elongation via cell extension the imaging time (e.g., in Arabidopsis) is limited, because the roots quickly leave (within ∼30 min) the field of view.
The application of hypocotyls circumvents this problem. Samaj and co-workers produced excellent super-resolved movies via SIM by analyzing the microtubule development in Arabidopsis hypocotyl epidermal cells by adapting the settings accordingly to acquire the raw data. For tissue etiolation the seedlings were grown in darkness which induces the thinning of the outer epidermal wall and reduces the thickness of the cuticular surface. Then, after mounting the seedlings into aqueous growth medium, effects causing refractive index mismatches can be reduced (Komis et al., 2014, 2015a).
PALM in living cells was performed on Nicotiana BY-2 cell cultures after transferring the cells into coverslip chambers (Chamber SlidesTM, Thermo Scientific) to localize perinuclear actin (Durst et al., 2014).
Single particle tracking by PALM was used for the first time on living plants by Hosy et al. (2015). They mounted Arabidopsis seedlings between two coverslips to track plasma membrane proteins.
Conclusion and Perspective
Currently, most results obtained by super-resolution microscopy in plant cell research are concentrated in the fields of research groups with access to super-resolution microscopes. But it is expected that the applications will extent in future significantly due to the general applicability of super-resolution to analyze biological specimens, so that super-resolution microscopy will become a standard technique also in plant cell research.
This development will be further accelerated by improving and combining the existing super-resolution methods. Especially SIM has the potential for extended applications in the field of live cell imaging. Additionally, SIM methods are under development to excel its to date achieved two-fold increased resolution.
To image also thick fluorescent samples (Calliphora salivary glands) SIM was combined with line-scanning to remove disturbing out-of-focus fluorescence background deteriorating the illumination pattern (Mandula et al., 2012). Rego et al. (2012) developed nonlinear SIM, and thus were able to visualize with a ∼ 40 nm resolution purified microtubules, mammalian nuclear pores and the actin cytoskeleton by applying the fluorescent photoswitchable protein Dronpa.
SIM can be applied for live cell imaging in multiple colors by using conventional fluorescent dyes as fast as 11 frames/s (Kner et al., 2009) at intensities of only 1 to 100 W/cm2 preventing phototoxicity (Li et al., 2015). Betzig and co-workers extended the resolution of live cell SIM by using an ultrahigh numerical aperture TIRF-SIM and achieved up to 84 nm, and by patterned nonlinear SIM they obtained up to 45-62 nm. By this approach the dynamics of plasma membranes components, mitochondria, actin and the Golgi apparatus in cultured mammalian cells has been imaged (Li et al., 2015). In addition, patterned nonlinear SIM and lattice light sheet microscopy (Chen et al., 2014) were combined to perform 3D live cell imaging beyond the diffraction limit (Li et al., 2015).
Gao et al. (2012) performed in vivo karyotyping of somatic chromosomes and identified the dynamics of the cytoskeleton of fibroblasts by combining an ultrathin planar illumination (produced by scanned Bessel beams) with SIM at thick animal specimens. Similarly, based on this technique the dynamics of mitochondria, filopodia, membrane ruffles, intracellular vesicles, and mitotic chromosomes in living cultured cells were investigated (Planchon et al., 2011).
It is expected that also the use of localization microscopy for absolute molecule quantification will be intensified in future, and that the combination with SIM will be increased to employ the advantages of these different nanoscopical methods. Standing-wave microscopy has the potential for parallel super-resolution imaging as it simultaneously draws on SIM, PALM, and STED technologies Chen and Xi (2015).
Recently, methods were developed to expand biological specimens physically by synthesizing a swellable polymer network within the specimen. This process called expansion microscopy allows to separate labels spaced closer than the optical diffraction limit isotropically. Thus, super-resolution with diffraction-limited microscopes may be achieved (Chen et al., 2015, 2016; Engerer et al., 2016). However, it remains to be tested whether the swelling process is applicable to all organisms and tissues, amongst others those from plants, without disturbing the native structures. Additionally, the technique is not applicable for live cell imaging. Thus, it is not expected that expansion microscopy has the potential to replace optical nanoscopy. But both have the potential to be combined in some special applications.
Furthermore, with the development of genetically encoded markers for electron microscopy (Shu et al., 2011; Martell et al., 2012) correlative approaches with super-resolution techniques will become more powerful in near future. However, due to the harsher fixing conditions and the lack in high specificity electron microscopy will not have the potential to replace completely the super-resolution techniques as live cell imaging is not possible and multi-color labeling is a challenge.
Author Contributions
VS conceived and designed the study, performed the experiments and wrote the manuscript.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer FH and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
I thank Jörg Fuchs for flow sorting of nuclei, Martina Kühne and Andrea Kunze for slide preparation, Andreas Houben and Klaus Weisshart for critical reading of the manuscript.
Abbreviations
- CENH3
centromeric histone H3
- FISH
fluorescence in situ hybridization
- HILO
highly inclined and laminated optical sheet
- PALM
Photoactivated Localization Microscopy
- RNAPII
RNA polymerase II
- SIM
structured illumination microscopy
- STORM
stochastic optical reconstruction microscopy
- STED
stimulated emission depletion
- TIRF
total internal reflection fluorescence
References
- Abbe E. (1873). Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung. Arch. Mikrosk. Anat. 9 413–468. 10.1007/BF02956173 [DOI] [Google Scholar]
- Agrawal U., Reilly D. T., Schroeder C. M. (2013). Zooming in on biological processes with fluorescence nanoscopy. Curr. Opin. Biotechnol. 24 646–653. 10.1016/j.copbio.2013.02.016 [DOI] [PubMed] [Google Scholar]
- Allen J. R., Ross S. T., Davidson M. W. (2014). Structured illumination microscopy for superresolution. Chemphyschem 15 566–576. 10.1002/cphc.201301086 [DOI] [PubMed] [Google Scholar]
- Antosch M., Schubert V., Holzinger P., Houben A., Grasser K. D. (2015). Mitotic lifecycle of chromosomal 3xHMG-box proteins and the role of their N-terminal domain in the association with rDNA loci and proteolysis. New Phytol. 208 1067–1077. 10.1111/nph.13575 [DOI] [PubMed] [Google Scholar]
- Antosz W., Pfab A., Ehrnsberger H. F., Holzinger H., Köllen K., Mortensen S. A., et al. (2017). The composition of the Arabidopsis RNA polymerase II transcript elongation complex reveals the interplay between elongation and mRNA processing factors. Plant Cell. 10.1105/tpc.16.00735 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaei-Moghaddam A. M., Schubert V., Kumke K., Weibeta O., Klemme S., Nagaki K., et al. (2012). Nondisjunction in favor of a chromosome: the mechanism of rye B chromosome drive during pollen mitosis. Plant Cell 24 4124–4134. 10.1105/tpc.112.105270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzenschlager M., Lermontova I., Schubert V., Fuchs J., Berr A., Koini M. A., et al. (2015). Arabidopsis MZT1 homologs GIP1 and GIP2 are essential for centromere architecture. Proc. Natl. Acad. Sci. U.S.A. 112 8656–8660. 10.1073/pnas.1506351112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell K., Mitchell S., Paultre D., Posch M., Oparka K. (2013). Correlative imaging of fluorescent proteins in resin-embedded plant material. Plant Physiol. 161 1595–1603. 10.1104/pp.112.212365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell K., Oparka K. (2011). Imaging plasmodesmata. Protoplasma 248 9–25. 10.1007/s00709-010-0233-6 [DOI] [PubMed] [Google Scholar]
- Bell K., Oparka K. (2015). Preparative methods for imaging plasmodesmata at super-resolution. Methods Mol. Biol. 1217 67–79. 10.1007/978-1-4939-1523-1_4 [DOI] [PubMed] [Google Scholar]
- Bozkurt T. O., Richardson A., Dagdas Y. F., Mongrand S., Kamoun S., Raffaele S. (2014). The plant membrane-associated REMORIN1.3 accumulates in discrete perihaustorial domains and enhances susceptibility to Phytophthora infestans. Plant Physiol. 165 1005–1018. 10.1104/pp.114.235804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral G., Marques A., Schubert V., Pedrosa-Harand A., Schlögelhofer P. (2014). Chiasmatic and achiasmatic inverted meiosis of plants with holocentric chromosomes. Nat. Commun. 5:5070 10.1038/ncomms6070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. C., Legant W. R., Wang K., Shao L., Milkie D. E., Davidson M. W., et al. (2014). Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346:1257998 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Tillberg P. W., Boyden E. S. (2015). Optical imaging. Expansion microscopy. Science 347 543–548. 10.1126/science.1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Wassie A. T., Cote A. J., Sinha A., Alon S., Asano S., et al. (2016). Nanoscale imaging of RNA with expansion microscopy. Nat. Methods 13 679–684. 10.1038/nmeth.3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xi P. (2015). Hundred-thousand light holes push nanoscopy to go parallel. Microsc. Res. Tech. 78 8–10. 10.1002/jemt.22434 [DOI] [PubMed] [Google Scholar]
- Cuacos M. (2013). Analysis of a Secondary Constriction in Chromosome 5R of Rye and Its Neocentromeric Activity. Doctoral thesis, Facultad de Biologia Departamento de Genetica Universidad Complutense de Madrid, Madrid, 98. [Google Scholar]
- Demidov D., Schubert V., Kumke K., Weiss O., Karimi-Ashtiyani R., Buttlar J., et al. (2014). Anti-phosphorylated histone H2AThr120: a universal microscopic marker for centromeric chromatin of mono- and holocentric plant species. Cytogenet. Genome Res. 143 150–156. 10.1159/000360018 [DOI] [PubMed] [Google Scholar]
- Dong B., Yang X., Zhu S., Bassham D. C., Fang N. (2015). Stochastic optical reconstruction microscopy imaging of microtubule arrays in intact Arabidopsis thaliana seedling roots. Sci. Rep. 5:15694 10.1038/srep15694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr J., Lolas I. B., Sorensen B. B., Schubert V., Houben A., Melzer M., et al. (2014). The transcript elongation factor SPT4/SPT5 is involved in auxin-related gene expression in Arabidopsis. Nucleic Acids Res. 42 4332–4347. 10.1093/nar/gku096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durst S., Hedde P. N., Brochhausen L., Nick P., Nienhaus G. U., Maisch J. (2014). Organization of perinuclear actin in live tobacco cells observed by PALM with optical sectioning. J. Plant Physiol. 171 97–108. 10.1016/j.jplph.2013.10.007 [DOI] [PubMed] [Google Scholar]
- Dvořáčková M., Raposo B., Matula P., Fuchs J., Schubert V., Peška V., et al. (in press). Replication of ribosomal DNA in Arabidopsis occurs both inside and outside of the nucleolus during S-phase progression. J. Cell Sci. 10.1242/jcs.202416 [DOI] [PubMed] [Google Scholar]
- Engerer P., Fecher C., Misgeld T. (2016). Super-resolution microscopy writ large. Nat. Biotechnol. 34 928–930. 10.1038/nbt.3669 [DOI] [PubMed] [Google Scholar]
- Fernández-Suárez M., Ting A. Y. (2008). Fluorescent probes for super-resolution imaging in living cells. Nat. Rev. Mol. Cell Biol. 9 929–943. 10.1038/nrm2531 [DOI] [PubMed] [Google Scholar]
- Fitzgibbon J., Beck M., Zhou J., Faulkner C., Robatzek S., Oparka K. (2013). A developmental framework for complex plasmodesmata formation revealed by large-scale imaging of the Arabidopsis leaf epidermis. Plant Cell 25 57–70. 10.1105/tpc.112.105890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon J., Bell K., King E., Oparka K. (2010). Super-resolution imaging of plasmodesmata using three-dimensional structured illumination microscopy. Plant Physiol. 153 1453–1463. 10.1104/pp.110.157941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornasiero E. F., Opazo F. (2015). Super-resolution imaging for cell biologists: concepts, applications, current challenges and developments. Bioessays 37 436–451. 10.1002/bies.201400170 [DOI] [PubMed] [Google Scholar]
- Gao L., Shao L., Higgins C. D., Poulton J. S., Peifer M., Davidson M. W., et al. (2012). Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell 151 1370–1385. 10.1016/j.cell.2012.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaux D. (2014). Nobel prize in chemistry: seeing the nanoscale. Nat. Nanotechnol. 9 878 10.1038/nnano.2014.267 [DOI] [PubMed] [Google Scholar]
- Gustafsson M. G. (2005). Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc. Natl. Acad. Sci. U.S.A. 102 13081–13086. 10.1073/pnas.0406877102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M. G., Shao L., Carlton P. M., Wang C. J., Golubovskaya I. N., Cande W. Z., et al. (2008). Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94 4957–4970. 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel V., Guichard P., Fournier M., Guiet R., Fluckiger I., Seitz A., et al. (2014). Correlative multicolor 3D SIM and STORM microscopy. Biomed. Opt. Express 5 3326–3336. 10.1364/BOE.5.003326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R., Li Z., Fan Y., Jiang Y. (2013). Recent advances in super-resolution fluorescence imaging and its applications in biology. J. Genet. Genomics 40 583–595. 10.1016/j.jgg.2013.11.003 [DOI] [PubMed] [Google Scholar]
- Heckmann S., Jankowska M., Schubert V., Kumke K., Ma W., Houben A. (2014). Alternative meiotic chromatid segregation in the holocentric plant Luzula elegans. Nat. Commun. 5:4979 10.1038/ncomms5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann S., Macas J., Kumke K., Fuchs J., Schubert V., Ma L., et al. (2013). The holocentric species Luzula elegans shows interplay between centromere and large-scale genome organization. Plant J. 73 555–565. 10.1111/tpj.12054 [DOI] [PubMed] [Google Scholar]
- Hedde P. N., Nienhaus G. U. (2014). Super-resolution localization microscopy with photoactivatable fluorescent marker proteins. Protoplasma 251 349–362. 10.1007/s00709-013-0566-z [DOI] [PubMed] [Google Scholar]
- Hosy E., Martiniere A., Choquet D., Maurel C., Luu D. T. (2015). Super-resolved and dynamic imaging of membrane proteins in plant cells reveal contrasting kinetic profiles and multiple confinement mechanisms. Mol. Plant 8 339–342. 10.1016/j.molp.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Ishii T., Karimi-Ashtiyani R., Banaei-Moghaddam A. M., Schubert V., Fuchs J., Houben A. (2015). The differential loading of two barley CENH3 variants into distinct centromeric substructures is cell type- and development-specific. Chromosome Res. 23 277–284. 10.1007/s10577-015-9466-8 [DOI] [PubMed] [Google Scholar]
- Jankowska M., Fuchs J., Klocke E., Fojtova M., Polanska P., Fajkus J., et al. (2015). Holokinetic centromeres and efficient telomere healing enable rapid karyotype evolution. Chromosoma 124 519–528. 10.1007/s00412-015-0524-y [DOI] [PubMed] [Google Scholar]
- Karimi-Ashtiyani R., Ishii T., Niessen M., Stein N., Heckmann S., Gurushidze M., et al. (2015). Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc. Natl. Acad. Sci. U.S.A. 112 11211–11216. 10.1073/pnas.1504333112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Wabnik K., Martiniere A., Langowski L., Willig K., Naramoto S., et al. (2011). Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol. Syst. Biol. 7 540 10.1038/msb.2011.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemme S. (2013). High Copy Sequences Reveal the Unique Composition and Evolution of the Rye B Chromosome. Doctoral thesis, Martin-Luther-University Halle-Wittenberg, Halle, 95. [Google Scholar]
- Kner P., Chhun B. B., Griffis E. R., Winoto L., Gustafsson M. G. (2009). Super-resolution video microscopy of live cells by structured illumination. Nat. Methods 6 339–342. 10.1038/nmeth.1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K., Wang P. W., Kriechbaumer V., Tilsner J., Frigerio L., Sparkes I., et al. (2015). Putting the squeeze on plasmodesmata: a role for reticulons in primary plasmodesmata formation. Plant Physiol. 168 1563–1572. 10.1104/pp.15.00668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G., Mistrik M., Samajova O., Doskocilova A., Ovecka M., Illes P., et al. (2014). Dynamics and organization of cortical microtubules as revealed by superresolution structured illumination microscopy. Plant Physiol. 165 129–148. 10.1104/pp.114.238477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G., Mistrik M., Samajova O., Ovecka M., Bartek J., Samaj J. (2015a). Superresolution live imaging of plant cells using structured illumination microscopy. Nat. Protocols 10 1248–1263. 10.1038/nprot.2015.083 [DOI] [PubMed] [Google Scholar]
- Komis G., Samajova O., Ovecka M., Samaj J. (2015b). Super-resolution microscopy in plant cell imaging. Trends Plant Sci. 20 834–843. 10.1016/j.tplants.2015.08.013 [DOI] [PubMed] [Google Scholar]
- Lermontova I., Kuhlmann M., Friedel S., Rutten T., Heckmann S., Sandmann M., et al. (2013). Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres. Plant Cell 25 3389–3404. 10.1105/tpc.113.114736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Shao L., Chen B. C., Zhang X., Zhang M., Moses B., et al. (2015). Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 349 aab3500 10.1126/science.aab3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesche J., Ziomkiewicz I., Schulz A. (2013). Super-resolution imaging with Pontamine Fast Scarlet 4BS enables direct visualization of cellulose orientation and cell connection architecture in onion epidermis cells. BMC Plant Biol. 13:226 10.1186/1471-2229-13-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnik O., Liesche J., Tilsner J., Oparka K. J. (2013). Unraveling the structure of viral replication complexes at super-resolution. Front. Plant Sci. 4:6 10.3389/fpls.2013.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Gabriel T. S., Martis M. M., Gursinsky T., Schubert V., Vrana J., et al. (2017). Rye B chromosomes encode a functional Argonaute-like protein with in vitro slicer activities similar to its A chromosome paralog. New Phytol. 213 916–928. 10.1111/nph.14110 [DOI] [PubMed] [Google Scholar]
- Ma W., Schubert V., Martis M. M., Hause G., Liu Z., Shen Y., et al. (2016). The distribution of alpha-kleisin during meiosis in the holocentromeric plant Luzula elegans. Chromosome Res. 24 393–405. 10.1007/s10577-016-9529-5 [DOI] [PubMed] [Google Scholar]
- Maheshwari S., Ishii T., Brown C. T., Houben A., Comai L. (2016). Centromere location in Arabidopsis is unaltered by drastic divergence in CENH3 protein sequence. Genome Res. 27 471–478. 10.1101/gr.214619.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandula O., Kielhorn M., Wicker K., Krampert G., Kleppe I., Heintzmann R. (2012). Line scan–structured illumination microscopy super-resolution imaging in thick fluorescent samples. Opt. Express 20 24167–24174. 10.1364/OE.20.024167 [DOI] [PubMed] [Google Scholar]
- Marques A., Ribeiro T., Neumann P., Macas J., Novak P., Schubert V., et al. (2015). Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proc. Natl. Acad. Sci. U.S.A. 112 13633–13638. 10.1073/pnas.1512255112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A., Schubert V., Houben A., Pedrosa-Harand A. (2016). Restructuring of holocentric centromeres during meiosis in the plant Rhynchospora pubera. Genetics 204 555–568. 10.1534/genetics.116.191213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell J. D., Deerinck T. J., Sancak Y., Poulos T. L., Mootha V. K., Sosinsky G. E., et al. (2012). Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 30 1143–1148. 10.1038/nbt.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möckl L., Lamb D. C., Bräuchle C. (2014). Super-resolved fluorescence microscopy: nobel Prize in Chemistry 2014 for Eric Betzig, Stefan Hell, and William E. Moerner. Angew. Chem. Int. Ed. Engl. 53 13972–13977. 10.1002/anie.201410265 [DOI] [PubMed] [Google Scholar]
- Neumann P., Schubert V., Fukova I., Manning J. E., Houben A., Macas J. (2016). Epigenetic histone marks of extended meta-polycentric centromeres of Lathyrus and Pisum chromosomes. Front. Plant Sci. 7:234 10.3389/fpls.2016.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nienhaus K., Nienhaus G. U. (2016). Where do we stand with super-resolution optical microscopy? J. Mol. Biol. 428 308–322. 10.1016/j.jmb.2015.12.020 [DOI] [PubMed] [Google Scholar]
- Olivier N., Keller D., Rajan V. S., Gonczy P., Manley S. (2013). Simple buffers for 3D STORM microscopy. Biomed. Opt. Express 4 885–899. 10.1364/BOE.4.000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawley J. B. (1995). Handbook of Biological Confocal Microscopy. New York, NY: Plenum Press. [Google Scholar]
- Phillips D., Nibau C., Wnetrzak J., Jenkins G. (2012). High resolution analysis of meiotic chromosome structure and behaviour in barley (Hordeum vulgare L.). PLoS ONE 7:e39539 10.1371/journal.pone.0039539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchon T. A., Gao L., Milkie D. E., Davidson M. W., Galbraith J. A., Galbraith C. G., et al. (2011). Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods 8 417–423. 10.1038/nmeth.1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rego E. H., Shao L., Macklin J. J., Winoto L., Johansson G. A., Kamps-Hughes N., et al. (2012). Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proc. Natl. Acad. Sci. U.S.A. 109 E135–E143. 10.1073/pnas.1107547108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro T., Marques A., Novák P., Schubert V., Vanzela A. L. L., Macas J., et al. (2017). Centromeric and non-centromeric satellite DNA organisation differs in holocentric Rhynchospora species. Chromosoma 126 325–335. 10.1007/s00412-016-0616-3 [DOI] [PubMed] [Google Scholar]
- Rouquette J., Cremer C., Cremer T., Fakan S. (2010). Functional nuclear architecture studied by microscopy: present and future. Intern. Rev. Cell Mol. Biol. 282 1–90. 10.1016/S1937-6448(10)82001-5 [DOI] [PubMed] [Google Scholar]
- Ruban A., Fuchs J., Marques A., Schubert V., Soloviev A., Raskina O., et al. (2014). B chromosomes of Aegilops speltoides are enriched in organelle genome-derived sequences. PLoS ONE 9:e90214 10.1371/journal.pone.0090214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann M., Talbert P., Demidov D., Kuhlmann M., Rutten T., Conrad U., et al. (2017). Targeting of A. thaliana KNL2 to centromeres depends on the conserved CENPC-k motif in its C-terminus. Plant Cell 29 144–155. 10.1105/tpc.16.00720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer P., van de Linde S., Lehmann J., Sauer M., Doose S. (2013). Methylene blue- and thiol-based oxygen depletion for super-resolution imaging. Anal. Chem. 85 3393–3400. 10.1021/ac400035k [DOI] [PubMed] [Google Scholar]
- Schermelleh L., Heintzmann R., Leonhardt H. (2010). A guide to super-resolution fluorescence microscopy. J. Cell Biol. 190 165–175. 10.1083/jcb.201002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V. (2014). RNA polymerase II forms transcription networks in rye and Arabidopsis nuclei and its amount increases with endopolyploidy. Cytogenet. Genome Res. 143 69–77. 10.1159/000365233 [DOI] [PubMed] [Google Scholar]
- Schubert V., Lermontova I., Schubert I. (2013). The Arabidopsis CAP-D proteins are required for correct chromatin organisation, growth and fertility. Chromosoma 122 517–533. 10.1007/s00412-013-0424-y [DOI] [PubMed] [Google Scholar]
- Schubert V., Lermontova I., Schubert I. (2014). Loading of the centromeric histone H3 variant during meiosis – how does it differ from mitosis? Chromosoma 123 491–497. 10.1007/s00412-014-0466-9 [DOI] [PubMed] [Google Scholar]
- Schubert V., Ruban A., Houben A. (2016a). Chromatin ring formation at plant centromeres. Front. Plant Sci. 7:28 10.3389/fpls.2016.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V., Zelkowski M., Klemme S., Houben A. (2016b). Similar sister chromatid arrangement in mono- and holocentric plant chromosomes. Cytogenet. Genome Res. 149 218–225. 10.1159/000447681 [DOI] [PubMed] [Google Scholar]
- Schubert V., Weisshart K. (2015). Abundance and distribution of RNA polymerase II in Arabidopsis interphase nuclei. J. Exp. Bot. 66 1687–1698. 10.1093/jxb/erv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X. K., Lev-Ram V., Deerinck T. J., Qi Y. C., Ramko E. B., Davidson M. W., et al. (2011). A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9:e1001041 10.1371/journal.pbio.1001041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M., Mander L., Fried G., Punyasena S. W. (2012). Capturing the surface texture and shape of pollen: a comparison of microscopy techniques. PLoS ONE 7:e39129 10.1371/journal.pone.0039129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A., Bellot S., Fuchs J., Houben A., Renner S. (2016). Analysis of transposable elements and organellar DNA in male and female genomes of a species with a huge Y-chromosome reveals distinct Y-centromeres. Plant J. 88 387–396. 10.1111/tpj.13254 [DOI] [PubMed] [Google Scholar]
- Staudt T., Lang M. C., Medda R., Engelhardt J., Hell S. W. (2007). 2,2′-thiodiethanol: a new water soluble mounting medium for high resolution optical microscopy. Microsc. Res. Tech. 70 1–9. 10.1002/jemt.20396 [DOI] [PubMed] [Google Scholar]
- Stelzer E. H. (2014). Better imaging through chemistry. Cell 159 1243–1246. 10.1016/j.cell.2014.11.032 [DOI] [PubMed] [Google Scholar]
- Tilsner J., Linnik O., Louveaux M., Roberts I. M., Chapman S. N., Oparka K. J. (2013). Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 201 981–995. 10.1083/jcb.201304003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M., Imamoto N., Sakata-Sogawa K. (2008). Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5 159–161. 10.1038/Nmeth.1171 [DOI] [PubMed] [Google Scholar]
- van de Linde S., Löschberger A., Klein T., Heidbreder M., Wolter S., Heilemann M., et al. (2011). Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6 991–1009. 10.1038/nprot.2011.336 [DOI] [PubMed] [Google Scholar]
- von Wangenheim D., Rosero A., Komis G., Samajova O., Ovecka M., Voigt B., et al. (2016). Endosomal interactions during root hair growth. Front. Plant Sci. 6:1262 10.3389/fpls.2016.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu G. T., Cao H. X., Watanabe K., Hensel G., Blattner F. R., Kumlehn J., et al. (2014). Repair of site-specific DNA double-strand breaks in barley occurs via diverse pathways primarily involving the sister chromatid. Plant Cell 26 2156–2167. 10.1105/tpc.114.126607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. J., Carlton P. M., Golubovskaya I. N., Cande W. Z. (2009). Interlock formation and coiling of meiotic chromosome axes during synapsis. Genetics 183 905–915. 10.1534/genetics.109.108688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner G., Schroeder-Reiter E., Ma W., Houben A., Schubert V. (2015). The ultrastructure of mono- and holocentric plant centromeres: an immunological investigation by structured illumination microscopy and scanning electron microscopy. Chromosoma 124 503–517. 10.1007/s00412-015-0521-1 [DOI] [PubMed] [Google Scholar]
- Weisshart K., Fuchs J., Schubert V. (2016). Structured illumination microscopy (SIM) and photoactivated localization microscopy (PALM) to analyze the abundance and distribution of RNA polymerase II molecules in flow-sorted Arabidopsis nuclei. Bio Protocol. 6 e1725 10.21769/BioProtoc.1725 [DOI] [Google Scholar]
- Zakrzewski F., Schubert V., Viehoever P., Minoche A. E., Dohm J. C., Himmelbauer H., et al. (2014). The CHH motif in sugar beet satellite DNA: a modulator for cytosine methylation. Plant J. 78 937–950. 10.1111/tpj.12519 [DOI] [PubMed] [Google Scholar]