Abstract
The study of the bacteriophage λ has been critical to the discipline of molecular biology. It was the source of key discoveries in the mechanisms of, among other processes, gene regulation, recombination, and transcription initiation and termination. We trace here the events surrounding these findings and draw on the recollections of the participants. We show how a particular atmosphere of interactions among creative scientists yielded spectacular insights into how living things work.
INTRODUCTION
The origins of molecular biology are deeply enmeshed with the discovery and characterization of the temperate coliphage λ. In this review, we trace the early days of λ, drawing on the memories of some of the key players. Amazingly, these memories have not perceptibly faded, probably because many of the workers were then near the beginning of their careers, the questions being addressed were well defined, and progress was rapid. Our intention is to try to reconstruct the interactions and the flow of ideas among the lambdologists. This is both of historical interest and an illustration of how imagination and perseverance—and luck—can yield major scientific insights. This review is more of a memoir than a history of λ studies. We limited discussion of work published after the mid-1980s and omitted several important areas of λ research, in particular those in which our personal involvement was tangential. Among these are autonomous phage DNA replication, homologous recombination, and virion assembly. Omission is not meant to denigrate the importance of these areas to molecular biology, but reflects only our desire to complete this review before the effects of old age begin to limit our capacity to comprehend and to write.
We recommend several books and reviews for readers who wish to pursue this subject. Lambda II (89), although dated, gives an extensive picture of the field as it appeared in 1982. Echols' Operators and Promoters (47) covers much of the same ground as we do but goes far beyond us in subject matter and level of detail. It also has sketches of some of the major players. We hope that our article encourages the reader to read this excellent monograph. The most recent edition of A Genetic Switch (160) presents a highly readable view by one of the pioneers in the field of how the lysis-lysogeny decision is made and sustained. Several relevant reviews are worth perusing (27, 32, 65, 152).
THE DISCOVERY OF LAMBDA
“Lysogenicity, or latent virus, is of frequent occurrence in enteric bacteria, but has been little emphasized in recent genetic studies with E. coli. It is of interest, therefore, that E. coli strain K-12 is lysogenic, but the latent phage is demonstrable only with the help of sensitive indicator strains.” Esther Lederberg, Microbial Genetics Bulletin, 1951.
The isolation of λ was first reported in 1951 by Esther Lederberg (119), then a Ph.D. student at the University of Wisconsin, and later was described, in greater detail, in a 1953 Genetics paper by Esther and Joshua Lederberg (120). The discovery was accidental, when a λ-sensitive strain of Escherichia coli K-12 (W518, obtained after UV irradiation) was crossed with its parent. The mixture yielded plaques, and the source of the virus was the K-12 parent. W518 cells that survived infection became stable lysogens which, like the K-12 parent, were immune to superinfection and which released unaltered phage. Although the Lederbergs were initially opposed to the notion, crosses between lysogens and sensitive cells led them to suggest that λ prophage was chromosomal and linked to gal. Joshua Lederberg recalls that he was convinced, from earlier work of Burnet and Lush (21), that lysogeny was a real phenomenon but that he “fully expected lambda to be a [plasmid]—in fact the term lambda was modeled after Sonneborn's kappa [in Paramecium; see reference 156 for a recent review], so it was quite a shock to discover the contrary.” (In fact, years later Hideo Ikeda and Jun-ichi Tomizawa [98] showed that prophage P1, unlike λ, is a plasmid and not part of the host chromosome!)
The idea that an infecting bacteriophage could assume a noninfectious, heritable form, now known as prophage, was controversial for many years. (An amusing account of the controversy can be found in reference 142.) Although persistent phage release by otherwise healthy bacterial cultures was a well-known phenomenon, skeptics supposed that only a minority of the population was infected at any one time or that virus release was an infrequent and nonlethal event. Many of these doubts were laid to rest by André Lwoff and Antoinette Gutmann (129), who observed single cells of a Bacillus megaterium lysogen during incubation in microdroplets. During such incubation, which provided an environment free of extracellular viruses, they saw occasional cell lysis with concomitant release of infectious phage. Their conclusions were confirmed by Giuseppe Bertani (17), who isolated three different temperate phages from a multiply lysogenic strain of E. coli, most notably phages P1 and P2, both of which have been extensively studied. Any remaining doubts about the existence of a heritable prophage were eliminated by the discovery of an experimental treatment, irradiation with UV light, that induced synchronous phage production by most cells of a lysogenic population of B. megaterium (130). UV irradiation was later found to induce phage production by λ lysogens, and this observation probably contributed to adoption of λ as a model system by many researchers (199).
Although λ was not one of the phages used to put the idea of lysogeny on a solid foundation, it rapidly became the experimental object of choice to investigate the many questions that followed, most notably how the lysogenic state is established after infection and how it is disrupted after induction. It is not entirely clear why λ became the chosen phage, since other candidates were available. One reason is probably the early development of bacterial conjugation as a tool for studying the genetics of λ's host, E. coli K-12. This allowed the Lederbergs to show that λ prophage or, conceivably, a gene required for its maintenance (but see below) is located on the bacterial chromosome (120). This important result was independently confirmed by Élie Wollman at the Pasteur Institute in Paris (205). Further work led to the discovery of λ-mediated transduction of specific bacterial genes (specialized transduction) (143). These discoveries suggested an interesting connection between λ lysogeny and bacterial chromosome structure and attracted other workers to the field. Another major attractant was the congruence of the physical and the genetic. For example, a hypothesis based on genetic inferences could rapidly be confirmed by a physical analysis of λ DNA structure. First, that the genetic material of λ is indeed DNA and that the genetic map is colinear with the DNA were shown directly by DNA transformation (see below) (92, 110). The assumption that genetic recombination results from the exchange of DNA between parental phage was established by differential density labeling of parental DNA and fractionation of recombinant phage particles in a density gradient (134). Finally, electron microscopy of annealed DNA strands revealed for even the most skeptical viewer the nonhomologous regions that represent deletions and substitutions (96, 203). These and similar experiments soothed the fears of hardcore biochemists who felt ill at ease with conclusions based on clear or turbid spots in agar or the analytical problems presented by bacteriophages with circularly permuted chromosomes.
GENETIC ANALYSIS
Soon after λ was discovered and the sensitive derivative of K-12 became available, groups at the California Institute of Technology (Caltech), the University of Geneva, and the Pasteur Institute in Paris initiated genetic analysis of the virus and of the virus-host interaction. Jean Weigle, who adopted λ as his sole research material after he retired from the physics department of the University of Geneva and moved to Max Delbrück's lab in the biology division of Caltech in 1948, was an important vector in this intellectual diffusion (see the dedication to reference 89 and reference 197). Among Weigle's students and close associates were Dale Kaiser, Margaret Lieb, and Ray Appleyard at Caltech and Werner Arber and Grete Kellenberger-Gujer at the University of Geneva, all of whom made substantial contributions to the field. Weigle brought a λ-sensitive strain to Paris in the early 1950s, when the Parisian school of lambdology was founded by François Jacob and Élie Wollman in Lwoff's laboratory (F. Jacob, personal communication).
One of the most consequential fruits of the early genetic studies was the discovery by Weigle and Bertani (18, 198), using phages λ and P2, of an interesting epigenetic phenomenon in which the preferred host of the virus changed according to its most recent host strain. The phenomenon, originally called host-controlled variation, later became known as restriction and modification. Arber and his associates showed that modification was a nonheritable alteration of DNA catalyzed by host-specific enzymes and that restriction was the result of the degradation of unmodified DNA by specific nucleases (6-8, 10, 46). This discovery was, of course, an early and important step in the development of the biotechnology industry. Arber recollects that his work was financed by special credits for the peaceful use of atomic energy and, indeed, developed from a study of “the effects of different irradiations on λ and its host bacteria,” a nice example of unintended consequences.
The classical genetics of λ was extensively developed by Wollman and Jacob at the Pasteur Institute and by Kaiser at Caltech, who isolated a large number of mutants with altered plaque morphology (107, 206). Kaiser notes that “In 1953, he (Weigle) found a number of plaque morphology mutants on his plates and gave them to me to study. I mapped the cI, cII, and cIII mutants to a cluster in the middle of the chromosome; then, on my postdoc with Jacob, I found that the cluster defined the region that specified immunity, which distinguished the lambdoid phages from each other.” The mutations are located on a single linear linkage map, which was later shown by Kaiser, David Hogness, and their associates to be colinear with the phage DNA (92, 93, 109, 110, 165). A plaque morphology mutant was used by Appleyard (5) to show that the chromosomal determinant of λ lysogeny was indeed the prophage and not a gene required for prophage maintenance. The most interesting mutants of this type were those that reduced the frequency of lysogenization. Wild-type λ and other temperate phages form turbid plaques because infected cells have a high probability of surviving, and the survivors, which are immune to reinfection by the same phage, grow within the plaques. Most of the descendants of the survivors are lysogens. Some phage mutants that are unable to lysogenize form clear plaques because there are few surviving cells in their centers. Kaiser (108), in addition to determining the map locations of the clear-plaque-forming (or c) mutations, classified them into three functional groups by complementation tests. Mutants in two groups (cII and cIII) were able to form lysogens, albeit at a low frequency, and, aside from the fact that they released mutant phage, these lysogens were indistinguishable from those formed by wild type λ. By contrast, mutants of the third group (cI) were unable to form lysogens by themselves. At about the same time, Myron Levine (121), working in Salvador Luria's laboratory at the University of Illinois, made similar observations on temperate phage P22, which was later shown to be a λ relative (19). Kaiser (108) surmised that the function controlled by gene cI was required for the maintenance of lysogeny and that those controlled by cII and cIII were required for its establishment. This surmise was supported by an elegant experiment in which he showed that the lysogenization defect of a cI mutant could be complemented by coinfection with a cII or cIII mutant provided that the second phage was added at the same time or later than the first, but not if it was added as little as 5 min earlier. Thus, as expected, the putative establishment functions were needed before the putative maintenance function. Additional experiments by Kaiser and Jacob implicated the cI gene in the specificity of immunity and, erroneously, in the determination of the location of the prophage on the bacterial chromosome (178). We consider this work in more detail below.
Analysis of the functions altered by λ mutations was greatly facilitated by the possibility of isolating defective lysogens. Such lysogens are unable to produce active phage after induction because of a prophage mutation in a gene required for plaque formation. However, they offer a means to propagate a λ mutant that cannot be grown lytically. This approach was exploited by Jacob, Wollman, and a Canadian associate, Clarence Fuerst (99, 103). They were able to identify phage genes important for cell lysis, viral DNA synthesis, and the production of virions. Most important of all, this work led to the identification of mutations that had pleiotropic effects; that is, they blocked several phage functions. It was proposed that the expression of λ genes occurred in an ordered sequence and that the primary defect of a pleiotropic mutant was its inability to complete an early step in phage development. This insight, which suggested that phage growth could be blocked by repression of a single early phage gene, was one of the keys in appreciating that the maintenance of λ in the prophage state had similarities to the control of β-galactosidase synthesis. Understanding the relatedness of λ and the lac operon was a major step in the formulation of a remarkably insightful theory of the regulation of gene expression by François Jacob and Jacques Monod (100). A further key insight was inspired by the phenomenon of zygotic induction (104, 154). Transfer by conjugation of either a λ prophage to a nonlysogen or of the lac operon to a cell lacking the lac operon led to increased production of λ phage or β-galactosidase, respectively. Both observations could be understood if the production of phage and enzyme was each blocked by specific repressors that were present in the donor but not the recipient cell at the moment of DNA transfer.
Another line of early λ genetics was inspired by the discovery of λ-mediated specialized transduction of gal and by the curious properties of λ gal transducing phage. The initial discoveries were made by Larry Morse in Lederberg's laboratory (143-145). These discoveries were ultimately to provide important clues about the nature of the association between the prophage and the host chromosome. Unlike generalized transducing phages, such as P1 and P22, λ carried into the host chromosome only the gal genes among those initially tested (hence the name specialized transduction). Since gal is close to the prophage on the bacterial chromosome and transducing phage was found only in lysates produced by induction of a lysogen, not by λ infection (unlike the case for P1 and P22), it appeared that association between the prophage and the bacterial chromosome necessarily preceded the formation of transducing phage. Another unusual feature was that most of the gal+ transductants reverted to gal− with low frequency upon further cultivation, as if the recipient had not lost its original mutant gene but instead had acquired an extra, and rather unstable, gal+ allele. Most striking of all, induction of a lysogenic transductant (nearly all were lysogens) produced lysates containing many orders of magnitude more gal transducing particles than those produced by induction of ordinary lysogens—the so-called high-frequency transducing (HFT) lysates. The production of such lysates can be considered the first example of in vivo cloning of a foreign gene into a viral vector.
Arber and Campbell independently showed that the gal transducing particles in HFT lysates were defective in plaque formation because they lacked essential phage genes, notably those required for the formation of the virion (hence the name λ dg, for defective, galactose transducing) (9, 30). Arber recalls, “In summer 1956 Jean Weigle brought to Geneva gal− and gal+ strains from the Lederbergs (the Morse et al. papers on λ transduction had just been published). Jean suggested to me to include in my electron microscope studies defective gal+ transductant strains. In their lysates nothing was visible, no heads and no tails, but lysis was O.K. This motivated me to start the genetics of λ-gal. At first, Jean did not tell me that he had already set Allan Campbell on this topic before leaving Caltech. Anyhow, we later got aware of the situation and exchanged information in good friendship.” Both groups suggested that λ genes were replaced with host genes following crossing over between the phage and host chromosomes. Campbell analyzed the defective phage lines by crossing them with a new type of λ mutant he had isolated. These mutants, originally called hd, for host defective, or sus, for suppressor sensitive, and later shown to carry nonsense mutations in vital genes (26), formed plaques on one substrain of E. coli K-12 but not another. Thus, wild-type recombinants could easily be scored. Campbell found that independently isolated HFT lysates typically differed in their content of phage genes. He concluded that a contiguous block of phage genes had been replaced by host gal genes and that the differences among different HFT lysates were the result of the variable position of a crossover point between phage and host chromosomes (29). Later work showed that the position of the crossover point varied in both the phage and host chromosomes (112). This conclusion, which seemed incompatible with the then-popular idea of recombination in a region of phage-host homology, was neatly accounted for as a by-product of Campbell's model of prophage insertion (see below).
PROPHAGE INSERTION
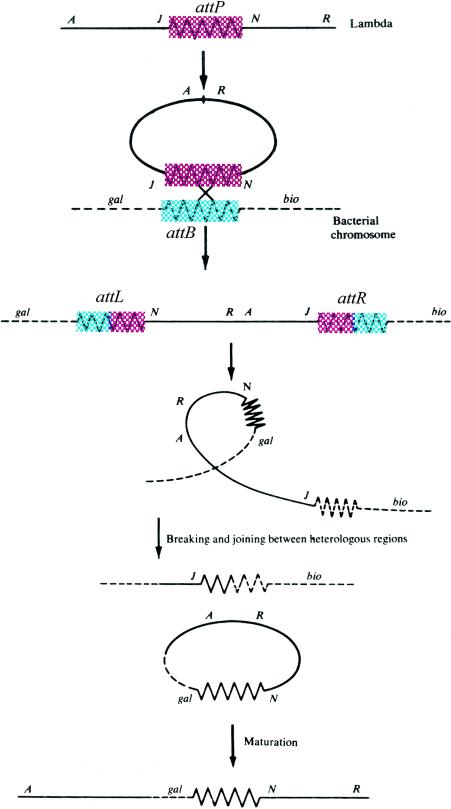
The nature of the linkage between the prophage and the bacterial chromosome was controversial for several years after the chromosomal location of the prophage was established. Although a λ lysogen is stable during normal bacterial growth, “cured” cells, which have lost the entire prophage, can easily be found among the survivors of induction. These cured cells can readily be lysogenized after reinfection. Induction of a lysogen, as already noted, leads to the production of phages that are genetically identical to the one used to establish the lysogen. These observations argued that the prophage-bacterial link can be formed and disrupted without alteration of either partner. Jacob and Wollman (see chapter 15 of reference 102) suggested that this kind of linkage could best be explained by persistent pairing or attachment of two homologous regions, one in the prophage and the other in the bacterial chromosome. Although the model is incorrect for λ and all other phages for which it has been tested, it was popular for some time, and a relic persists in the name of the DNA sequences that are directly involved in λ integration into the host: the “attachment” or att sites. An alternative model, prophage insertion, was proposed by Campbell in a 1962 review (28) and is shown in Fig. 1. Campbell's model was attractive because of its simplicity: it did not postulate a novel type of DNA-DNA linkage, such as persistent pairing, and it relied on a known process for insertion, i.e., recombination of homologous regions within each chromosome. However, the model was incomplete in some important respects and did not immediately sweep the field. First, it did not provide a mechanism to convert the λ chromosome from the linear DNA molecule found in virions to the ring proposed as an intermediate in prophage insertion. Second, the high frequencies of lysogeny after infection and of phage production after induction seemed to require extensive homology between the two chromosomes, and this had not been found (38). Finally, lysogenization was impeded by the presence of a resident λ prophage in the infected cell, although the model predicts, instead, that the increased homology provided by the prophage should favor insertion of the superinfecting phage. Although these problems and others seemed quite serious at the time (see, for example, chapter 17 of reference 88), they were resolved in favor of the insertion model within a few years. Nevertheless, it is clear that when it was published, Campbell's insertion model was a triumph of imagination over some of the presumed facts. Campbell recalls, “I'm sure I had not formulated the idea when I started writing. It came as an effort to put the existing data together. A major datum was a paper by Calef and Licciardello (24), where they mapped a λ prophage onto the E. coli chromosome and came out with an unexpected order. There were also some results of Appleyard's, which I cited in the review. While I was writing, a manuscript arrived in the mail from Frank Stahl suggesting that prokaryotic chromosomes had to be circular (I've forgotten why). I was also pleased with how easily I could interpret my results on genetic content of gal phages using the model.” Understanding the genetic content of specialized gal transducing phages, a major accomplishment of the model, requires that one assumes that they are formed by a process similar to prophage excision but occurring by infrequent recombination at many different points within the prophage and adjacent bacterial DNA (Fig. 1). The formation of transducing phage chromosomes by such abnormal excision is formally similar to the formation of genetic deletions, although the mechanisms need not be identical.
FIG. 1.
Model of prophage insertion according to Campbell (31), modified to show sequences specific to prophage and bacterial attachment sites. The four attachment sites, shown as zigzag lines, are colored differently in the top part of the figure to indicate that they differ in sequence and function (see the text). The phage chromosome is shown as a solid line, and a segment of the bacterial chromosome is shown as a dashed line. The points of genetic exchange between attP and attB are indicated (×). The positions of phage genes A, J, N, and R and bacterial genes gal and bio are included for orientation. Normal prophage excision occurs by recombination between attL and attR. Formation of a λ gal transducing phage chromosome is illustrated in the bottom part of the figure.
One of the problems that delayed acceptance of the insertion model, i.e., how the ring intermediate is formed, had a particularly simple and elegant solution. Alfred Hershey and his associates at Cold Spring Harbor Laboratory discovered that a linear λ chromosome could be converted to a “folded form,” later shown to be a ring, by joining of two distinguishable cohesive sites at or near the molecular ends (91). Joining occurred readily when the DNA was incubated under conditions propitious for annealing of cDNA strands. The cohesive sites are two short cDNA sequences, one at the left and the other at the right chromosome end (91, 207). The left and right cohesive ends join shortly after infection, and the remaining nicks are sealed covalently. The sealing mechanism was shown by Martin Gellert (68) at the National Institutes of Health to be promoted by an E. coli enzyme, DNA ligase. This discovery, of course, marked the beginning of the age of cloning and biotechnology, in which so many of us have made our fortunes. The cohesive ends are regenerated near the end of the lytic cycle, as the phage DNA is packaged within virions (see below).
Explaining the high efficiency of insertion and its inhibition by a preexisting prophage was equally rewarding but more complicated, since two fundamental, related insights were needed. One, due in large part to Ethan Signer, who worked at the Pasteur Institute and then at the Massachusetts Institute of Technology (MIT), was that recombination of the circular phage chromosome with the bacterial chromosome is catalyzed by a site-specific recombinase encoded by the phage (178, 179). The other insight, due in large part to the work of Franco Guerrini at MIT and the International Laboratory of Genetics and Biophysics in Naples (87), was that the phage and bacterial attachment sites are not identical and that recombination between them follows different rules than does recombination between homologous chromosomes, or homologous recombination. The frequency of homologous recombination typically increases with the length of the homologous segment. In contrast to this rule, Guerrini found that recombination between the phage and bacterial attachment sites occurred more frequently than did recombination between two bacterial attachment sites, an observation that was made independently by Kiyoshi Mizuuchi and Toshio Fukasawa at Osaka University (138). One of the principal difficulties in untangling this story was that both λ and E. coli encode enzymes that promote homologous recombination, and the contribution of both of these pathways had to be understood before that of the λ specialized recombination pathway could be clearly seen. Signer recalls, “Lambdoids integrate efficiently by recombination within a very small region of the phage genome, yet dogma had it that efficient recombination required extensive homology. The key, exciting realization was that high efficiency could just as well come from a recombination enzyme(s) that had site specificity and would recognize such a short region as well as a corresponding region in the chromosome—that such an enzyme wasn't known at the time didn't mean one couldn't exist. Subsequently, on moving to a position at MIT shortly thereafter, I learned that this fit perfectly with a similar proposal of Franco Guerrini's, which came from a completely different approach—yet another instance of breakthroughs made independently but simultaneously.” Acceptance of Guerrini's proposal necessitated a rechristening of the attachment sites to avoid confusion. The phage attachment site, the bacterial attachment site, and the two recombinant attachment sites located to the left and right of the prophage became attP, attB, attL, and attR, respectively (Fig. 1).
Acceptance of the “site-specific recombination” hypothesis seemed to obviate Campbell's proposal that phage-bacterial recombination occurs in a region of homology between the two chromosomes. Nevertheless, this proposal also proved to be correct, although the extent of the homologous segment was undoubtedly shorter than Campbell appreciated at the time. Marc Shulman and Max Gottesman (176) isolated attachment site mutations that could be crossed from the phage in which they were isolated into E. coli, a property best explained by assuming that the mutations are located in a region common to attP and attB. Arthur Landy and Wilma Ross at Brown University (118) sequenced the phage, bacterial, and prophage attachment sites. They found a 15-bp segment common to the four sites within which recombination occurred. The attachment site mutations of Shulman and Gottesman are deletions of a single base pair within this segment. Finally, Robert Weisberg and his collaborators confirmed that homology is important for phage-bacterial recombination, since a base substitution mutation within the common region decreased recombination of the mutant with a wild-type partner or with a partner carrying a different mutation but had no effect on identical partners (200).
This existence of the proposed phage-encoded site-specific recombinase, later dubbed λ integrase, was confirmed when several groups isolated int mutants (71, 75, 184, 210). Since such mutants should not be able to form lysogens, it might seem at first view that their phenotype would resemble that of mutants unable to synthesize repressor; i.e., they would form clear plaques. However previous work showed that this view is flawed and that repression can be established without prophage insertion, a phenomenon called abortive lysogeny. Abortive lysogeny was discovered by Margaret Lieb (123), who showed that survivors of λ infection would not become stable lysogens if they were briefly exposed to high temperature shortly after infection. Later work suggests that this is probably the result of heat inactivation of the Int protein (85). Maria Zichichi and Grete Kellenberger-Gujer (113, 209) found that λ b2, a mutant unable to form stable lysogens, could initiate lysogeny after infecting Hfr cells, but the survivors, unlike those produced by wild-type infection, were unable to transfer the repressed prophage to an F− recipient during conjugation. Furthermore, after several cell divisions, the descendants of these abortive lysogens lost the capacity to produce phage upon induction as well as their immunity to reinfection. Zichichi and Kellenberger-Gujer suggested that the repressed but unintegrated prophage is unable to replicate and is therefore lost by dilution when the cell containing it divides. It was later shown that the lysogenization defect of λ b2 is the result of a partial deletion of attP (62).
The insight that establishment of repression and prophage insertion are distinct steps in the establishment of lysogeny led to the development of a method used to isolate new λ mutants that formed abortive lysogens. Michael Yarmolinsky recalls, “My principal contribution to the study of λ, sensu strictu, was the isolation and characterization, with Max Gottesman, of λ int mutants. For their isolation I had developed a selection based on heteroimmune curing (33) of a thermoinducible λ imm434 prophage that I had isolated for the purpose in 1965. Ironically, the selection I had developed would have yielded xis as well as int mutants, as we later recognized, but when Max joined the lab he persuaded me that we should try instead a method used also by James Zissler. By that method we (and Zissler) obtained several int mutants. Lambda mutants specifically affecting excision were subsequently obtained by Guarneros and Echols.” In the method referred to by Yarmolinsky, mutants that established abortive but not stable lysogeny were identified by allowing infected cells to divide in the absence of reinfection before testing their progeny for loss of immunity.
Guerrini (87) was the first to propose that prophage excision is not a simple chemical reversal of prophage insertion. This proposal opened people's minds to the possibility that the enzymatic requirements for insertion and excision differ, a notion that seemed attractive because it could facilitate differential control of the two processes. There was no compelling evidence for such differential control at the time, but that did not stop Gabriel Guarneros, a Mexican student of Harrison (“Hatch”) Echols at the University of Wisconsin, from looking for λ mutants that were unable to excise (84). “I arrived in Hatch's lab in 1968 (this probably saved my life since I left just a few weeks before students were massacred in Tlatelolco Plaza on October 2). I had seen Hatch's papers on int since I had done some reading before leaving for Madison. Of course, I was pretty naive about λ when I arrived, but I proposed the idea of looking for a phage gene which, together with Int, reversed the λ integration reaction. In fact I was so naive that I never doubted its existence; this state of bliss would never come back to me. I thought of calling mex (main excision function) what was eventually named xis; Hatch probably thought my suggestion somewhat nationalistic.”
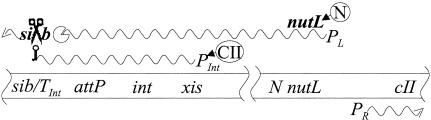
The role of Xis protein in the differential control of insertion and excision has been abundantly documented since Guarneros and Echol's discovery. Indeed, λ goes to extraordinary lengths to ensure that insertion is favored in conditions suitable for stable lysogeny, and excision is favored after induction (49). First, integrase is more stable than Xis protein (201). Because of this, insertion eventually becomes irreversible after infection, provided the cell survives for enough time to allow Xis to decay. Second, after infection Xis is produced from the PL transcript, while Int is produced mainly from the PInt transcript. PInt is activated by the CII protein, and CII activation helps to coordinate the synthesis of Int with that of repressor (35). This favors insertion in cells in which lytic phage growth has been repressed. Finally, after induction, in contrast to infection, Int and Xis are both produced by translation of the PL transcript. Since excision occurs rapidly after induction (127), Xis has time to act before it decays. Gabriel Guarneros, Donald Court, and their associates played the major role in understanding the baroque regulatory network that allows Int to be made from different transcripts in different conditions (Fig. 2) (reviewed in references 49 and 83). The two key regulatory elements are sib, a site of endonucleolytic RNase cleavage that is located promoter distal to int (86), and N, a λ protein that suppresses termination of certain phage transcripts (see The Story of N, below). Transcripts that initiate at the PL promoter contain the sib sequence because termination is suppressed by N. These transcripts are therefore cleaved, and processive exonucleolytic degradation of the upstream RNA destabilizes the int message. Transcripts that initiate at the PInt promoter terminate before sib and are therefore more stable (173, 174). The PL transcript that is made after induction does not contain sib because of the permutation of prophage genes that occurs after insertion (Fig. 1). Court recollects, “With Gabriel's lab, I determined that Int was only expressed from the PInt promoter under CII and CIII control and that it was not expressed from PL despite its strength. Mutants isolated under cII− conditions demonstrated a site beyond int to be responsible, which turned out to be an RNase III site in the RNA that was made only from the PL transcript and was processed by that endonuclease to cause int RNA's exonucleolytic degradation. This was really the first example of a gene regulated from downstream and one of the first examples of regulation by RNA processing.”
FIG. 2.
Regulation of int expression by RNase III cleavage of sib. Two segments of the λ chromosome with relevant genes and sites are indicated by the two horizontal lines. Transcripts that initiate at PL are antiterminated following binding of N protein to the nutL site (see the text). These transcripts are cleaved at sib by RNase III (indicated by the scissors symbol), and upstream RNA, which includes the Int-coding region, is destabilized by exonucleolytic digestion in a 3′-to-5′ direction (indicated by the “Pacman” symbol). If sufficient CII protein is made, it activates transcription at PInt, a promoter whose start site is within xis. These transcripts terminate at the TInt terminator (indicated by the hairpin symbol), do not contain a complete sib, are not cleaved by RNase III, and are relatively resistant to exonucleolytic digestion.
The genetics of insertion and excision was well developed by the early 1970s. However, the mechanism of recombination could not be fully understood until the reaction was deconstructed in vitro. Michael Syvanen (186) at Stanford University made a first step towards this goal by using extracts that packaged λ DNA molecules that had undergone site-specific recombination into infective phage. Subsequently Howard Nash (148, 149) at the National Institutes of Health developed an in vitro recombination reaction that faithfully reproduced in vivo λ insertion, and Susan Gottesman and Max Gottesman (77) did the same for λ excision. Both kinds of recombination had the expected dependence on Int, and excisive recombination had the additional requirement for Xis. These were the first biologically faithful in vitro recombination reactions. The story is best told by Nash. “I had become fascinated with the mechanism by which λ achieved its specificity of integration. Since I could not think of a plausible mechanism, I was convinced that the only way to find it out was to take the system apart biochemically. Toward this end, I worked for several years to assemble the necessary pieces: a substrate in which the attachment sites were tethered so as to find each other with high frequency (the integrative counterpart to the att2 phage of Shulman and Gottesman [177]), a highly concentrated source of recombination proteins (modeled after the Kornberg lab's procedures for in vitro replication), and an assay for recombinant DNA that was both sensitive and faithful (using the spheroplast transformation protocol of Wackernagel [193] to get the product to package itself into viable phage). I still remember vividly seeing the first plaques on a pyrophosphate-containing plate and realizing that, despite the predictions that recombination was a vital process that required cellular integrity, I had actually reproduced integrative recombination in vitro. In some ways, I regard this advance as my major contribution to lambdology. Certainly, there was a larger intellectual content in subsequent work from my lab that dissected the proteins in the crude extract to figure out how each contributed to the reaction and that studied the topology of the products of in vitro recombination to figure out how the sites came together. But, none of this would have been possible without the establishment of the cell-free system.”
The cell-free system was essential in delineating the details of DNA strand exchange during recombination. Yoshiko Kikuchi and Nash (115) showed that the enzymatic activity used by Int to cleave and reseal DNA strands is that of a type I topoisomerase. Kiyoshi Mizuuchi et al. at the National Institutes of Health, in a technical and intellectual tour de force, showed that strand exchange in vitro occurs at a unique position in each strand (140). The implications of Mizuuchi's experiments agreed with an independent genetic analysis of recombination in vivo between attachment site mutants carried out jointly in the laboratories of Landy and Weisberg (200, 202). Taken together, these results showed that the two points of strand exchange are separated by a 7-bp segment, which was dubbed the overlap region. Earlier genetic studies from the laboratories of Echols and Weisberg (48, 55) suggested that the two reciprocal strand exchanges needed to form a pair of completed recombinant DNA molecules are not always concerted but can occur one at a time. A single reciprocal strand exchange produces a four-armed junction known as a Holliday structure. Consistent with the genetic analysis, Pei Ling Hsu and Landy (97) found that Int could resolve synthetic Holliday structures that contained the att sequences into a pair of nick-free linear molecules.
Among the many other important results to emerge from the development of the cell-free recombination system, we mention only two. One was the discovery that a host component was required for the reaction (150). Purification of the extract revealed that the component was a single, heterodimeric, basic protein that was dubbed integration host factor (IHF) (151). The genes encoding each subunit (ihfA [also called hid/himA] and ihfB [also called hip/himD]) were identified in the laboratories of Daniel Wulff at the State University of New York at Albany, David Friedman at the University of Michigan, and Robert Weisberg (114, 136, 204). IHF, an accessory factor for many bacterial functions, has been co-opted by λ for site-specific recombination, DNA packaging (see below), and other reactions. IHF plays an architectural rather than a catalytic role in site-specific recombination: it bends the DNA substrate so as to allow formation of a recombinogenic Int-attP-IHF complex called the intasome (73).
Investigation of site-specific recombination in vitro also led to the discovery of a new enzyme, DNA gyrase, that couples ATP hydrolysis to the introduction of negative supercoils into DNA. Mizuuchi and Nash (139) found that efficient integrative recombination requires a covalently closed circular substrate and, together with Gellert (69), showed that this requirement reflects the need for negative superhelicity. DNA gyrase, like IHF, was present in the crude extracts initially used for recombination, and gyrase activity supercoiled the relaxed covalently closed substrate molecules.
TERMINASE
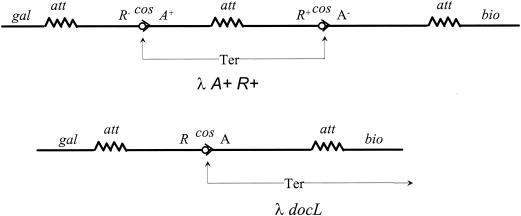
In the virion, λ DNA is a linear, mostly double-stranded DNA molecule with unique, 12-bp single-stranded ends. Many laboratories contributed to the analysis of λ virion DNA structure, including those of Alfred Hershey, Dale Kaiser, David Hogness, Ray Wu, Ross Inman, Norman Davidson, Waclaw Sybalski, and Karol Taylor. Their work is summarized admirably in The Bacteriophage Lambda (90). We have seen that upon infection, λ DNA circularizes by joining its cohesive ends and becomes negatively supercoiled. But how does intracellular λ DNA linearize and regain its cohesive ends? By the mid-1960s it was clear that the site of the joined cohesive ends, or cos (see below), is essential for the formation of infective phage particles. Lambda transducing phage lines that lacked all known phage genes could be propagated and packaged with the help of coinfecting λ, but none that lacked the cohesive ends had ever been found (111, 112). It appeared reasonable that a λ-encoded restriction-like enzyme recognized and cleaved cos during phage development (although type II restriction enzymes had not yet been discovered). The cos-cleaving function was given a name—terminase (abbreviated Ter)—by Suzanne Mousset and René Thomas (147) at the Free University of Brussels, a necessary but insufficient step in understanding the reaction. They substantiated the existence of Ter by superinfecting a lysogen carrying two different genetically marked tandem λ prophages with a phage variant that can grow lytically in an immune host (“heteroimmune superinfection”). Replication of the prophage is severely limited in these conditions (159, 192). The genetic content of prophage DNA appearing in infectious phage particles after superinfection was used to characterize cos cleavage. Mousset and Thomas (146) found that heteroimmune superinfection excised the recombinant expected from Ter cleavage at the two prophage cos sites and packaging of the DNA between them into infectious phage (Fig. 3). The yield was about one recombinant phage per infected lysogen, suggesting that cos cleavage is efficient. Similar infection of a single lysogen yielded much less λ, presumably because an inserted single prophage cannot be packaged into plaque-forming phage particles. This interpretation is supported by the observation that induction of polylysogens carrying tandem, excision-defective prophages gave high yields of active phage (76). By contrast, induction of a lysogen carrying a single excision-defective prophage gave a high yield of a noninfectious particle, λ docL, whose chromosome consists of the left cohesive end, adjacent prophage DNA to the right, and bacterial DNA to the right of attR (125, 185) (Fig. 3) (see below). However, efforts to show that Ter is a simple nuclease were confounded by the connection between cleavage and packaging: cells infected by mutants with mutations in any of the phage genes required for packaging accumulated linear λ concatemers (oligomers of several chromosomes joined head to tail) rather than single λ chromosomes with cohesive ends (45, 183, 194). Elegant studies of DNA packaging in vivo by using phage mutants that contain a short tandem cos duplication ruled out the possibility that Ter acts as a simple nuclease in vivo (54, 56, 57). Such duplication mutants can package an uncut cos, and the uncut copy is more frequently located near the left than near the right chromosome end (see below).
FIG. 3.
Top. Ter action excises a recombinant prophage from a double lysogen (146). The drawing shows a segment of the bacterial chromosome containing two differently marked λ prophages: λ A+ R− and λ A− R+. The attachment sites are indicated by zigzag lines, and the cos sites are indicated by the circle-arrowhead symbol. Heteroimmune superinfection of this dilysogen excises and packages the recombinant expected from Ter cleavage of the two cos sites (see the text). (Bottom) Packaging of λ docL DNA from a single lysogen (125, 185). Induction of a lysogen carrying a single prophage that is unable to excise from the bacterial chromosome leads to abundant production of a noninfectious particle carrying the left cohesive end and prophage and bacterial DNA located to the right of cos.
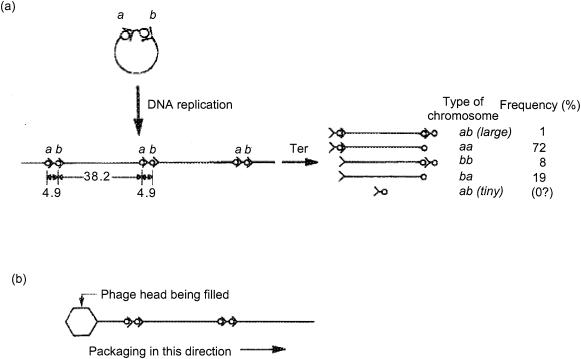
Scott Emmons describes the experiments that led him to a model that links cutting and packaging, and he reveals the origin of the term cos. Emmons, then a student in Robert (“Buzz”) Baldwin's laboratory at Stanford, was studying partial duplications of λ by electron microscopy. The duplications were to be mapped by hybridization with λ DNA that did not carry a duplication. “In heteroduplexes with unduplicated DNA, the duplicated DNA looped out at a range of positions on the chromosomal DNA that indicated the region that was duplicated. One day I found a mutant with the odd property that the added DNA usually didn't make an internal loop but rather a tail at one end of the linear chromosomal DNA molecule. Furthermore, sometimes the tail was at one end of the chromosome and sometimes at the other. I realized that this particular duplication had formed in the concatemeric DNA intermediate that was known to be present during λ replication. Moreover, it contained the joint between the chromosomal ends that formed during intracellular phage growth. When this phage packaged its DNA, it had a choice of two sites for cutting the new ends of a chromosome and used them in various combinations. I studied this phage and deduced some things about the process by which the concatemeric intermediate is packaged, such as that the packaging appeared to be from left to right on the λ chromosome and that the concatemers were two to three chromosomes in length. When I came to write up this work (for the Journal of Molecular Biology), it seemed the joined end site on the concatemer didn't have a proper name. It had been variously named m.m′, after certain minute mutants that mapped to the ends of the λ chromosome, or A.R, after the genes at the two ends. But by then a convention of three-letter gene names had come into use for λ, so I decided the joined site should have one. The logical first choice was ces for cohesive end site, but this reminded me too much of unsanitary waste. cos sounded better and that's how the now-famous λ cos site got its name.”
Emmon's model, described in Fig. 4, neatly explained the efficient packaging of λ docL DNA by a prophage that is unable to excise from the bacterial chromosome after induction. Entry of DNA into the phage head begins at cos and continues rightwards through the prophage into the bacterial chromosome (genes A through bio in Fig. 3). Packaging stops when the head is filled to capacity, because there are no additional copies of cos, as there are on λ DNA concatemers, to signal termination of packaging. The resulting particle is not infective, because unpackaged DNA, continuous with the packaged DNA, protrudes from the head and prevents the attachment of phage tails (185).
FIG. 4.
Model for cos cleavage and DNA packaging (adapted from reference 54 with permission of the publisher). (a) Circular DNA of a cos duplication mutant replicates to produce a concatemer, which is a substrate for Ter cleavage. cos is indicated by the circle-arrowhead symbol (○>). Cleavage separates the left cohesive end (>) from the right (○). The two copies of cos are labeled a and b, and the approximate sizes, in kilobase pairs, of the λ chromosome segments between them are shown. The types of chromosome and their observed frequencies in the progeny phage population were determined by electron microscopy of DNA heteroduplexes and are shown at the right. (b) According to the model, the first Ter cleavage occurs with equal probability at any of the cos copies on a concatemer. Cleavage is linked to DNA packaging, and packaging proceeds rightwards from the first cleaved cos. Several λ chromosomes are cleaved and packaged in a continuous sequence from a typical concatemer. If these assumptions are correct and production of both the tiny and the large ab fragments is rare, aa chromosomes should be more frequent than ba chromosomes, as was observed. This is because only aa chromosomes can follow cleavage of an a cos site in a packaging series on a single concatemer. By contrast, the assumption of leftward packaging predicts an excess of bb over aa chromosomes.
The connection between cleavage and packaging in vivo, which is still incompletely understood (180), delayed identification of the gene(s) encoding Ter. This problem was resolved by the development of an in vitro cleavage system. Using such a system, James Wang and Dale Kaiser (195) showed that cos cleavage can be uncoupled from DNA packaging in vitro and that the gene A protein but not other known head proteins are required for cleavage. The biochemistry of Ter was first approached successfully by Andrew Becker, Marvin Gold, Helios Murialdo, and their associates at the University of Toronto. Using packaging of concatemeric λ DNA into infectious particles as an assay, they purified an activity that contained the λ A gene product (14-16). The Ter preparation also displayed nonspecific DNA-dependent ATP hydrolytic activity. Cleavage took place in the absence of proheads but did require a factor present in uninfected E. coli. By 1983, they realized that Ter was a hetero-oligomer containing the products of two genes, nu1 and A, which are the two leftmost genes on the lambda chromosome (72). The missing host factor was later shown to be IHF (13, 208).
The structure of cos was solved in the laboratories of Michael Feiss at the University of Iowa (58, 60), Barbara Hohn at the Friedrich Miescher-Institut in Basel (94), and Kenichi Matsubara at Osaka University (137). The sequence required for cos cutting, about 100 bp, is much larger than that of a typical restriction site, consistent with the role of cos as an initiator and terminator of DNA packaging as well as the locus of cleavage. The required sequences lie on both flanks of the points of cleavage and contain Ter and IHF binding sites. More recent results suggest functional specialization: sequences near the left cohesive end are involved in initiating packaging, and sequences near the right cohesive end are involved in terminating packaging (59). The Ter reaction can also be viewed as a recombination event that accounts for the fact that the λ genetic map is linear, although recombination takes place between intracellular DNA circles and concatemers.
IMMUNITY
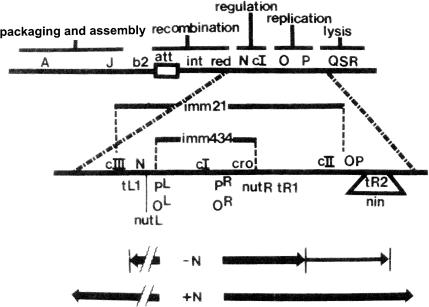
Lambda lysogens are immune, i.e., resistant to infection by another λ. Lambdoid phages with different cI genes and operator sites—heteroimmune phages such as 434 or 21 (Fig. 5) —grow normally on λ lysogens. The λ CI repressor expressed by the prophage prevents the development of the incoming λ but not that of 434 or 21. The basis of this control was elucidated by the work of several groups, most notably those of François Gros at the Pasteur Institute and the Institut de Biologie Physico-Chimique, Harrison Echols and Waclaw Szybalski at the University of Wisconsin, and Anna Marie Skalka at Cold Spring Harbor Laboratory.
FIG. 5.
Patterns of early transcription (adapted from reference 66 with permission of the publisher). The top line is a genetic map of λ (not to scale) showing functional groups of genes. The two lines marked imm21 and imm434 show the extents of the regions of nonhomology between λ imm21, λ imm434, and λ. The arrows at the bottom indicate the extent and intensity of transcription after infection with λ N− or N+. Other symbols are explained in the text.
Repressor prevents the transcription of nearly all λ genes by blocking two early promoters, PL and PR, from which λ lytic development proceeds (Fig. 5) (11, 106, 181, 182, 188). Upon induction, the repressed genes are transcribed according to a developmental program: most genes located on the right segment of the chromosome are expressed early, and genes located on the left segment are expressed late. The accumulation of late transcripts is blocked by mutation in gene N or by inhibition of protein synthesis after induction. Fitting this observation into the Jacob-Monod model of gene control (100), λ repressor directly prevents the synthesis of an early protein(s) whose activity is needed for the expression of other phage genes. Consistent with this model, Thomas and his collaborators showed that some prophage genes could and others could not be “transactivated” by superinfection with a heteroimmune phage (39, 190, 191). The transactivatable class consists principally of genes now known to be expressed late in the infective cycle. The nontransactivatable class consists of early genes N and O, which are on opposite sides of cI. The Szybalski laboratory found that early transcripts, whose syntheses should be directly blocked by repressor, proceeded leftwards and rightwards from PL and PR, respectively (117, 188). All of this work supported the idea that repressor directly blocks the transcription of one or a small number of early genes and that repression of late genes is indirect.
Echols and his coworkers, in a 1968 article (50), demonstrated in vitro that a partially purified λ CI preparation specifically blocked the synthesis of λ RNA, rather than decreasing its stability, and acted in the absence of protein synthesis.
The biochemistry of the cI regulatory system was analyzed in two groundbreaking papers by Mark Ptashne at Harvard University (161, 162). In 1967, Ptashne isolated λ CI repressor and showed that it bound λ DNA in vitro. A 1968 paper with Nancy Hopkins followed, which defined the cis elements recognized by CI, i.e., the operators (163). In 1954, Jacob and Wollman (101) had isolated λ vir as a plaque former on a λ lysogen. They showed that insensitivity to immunity, or virulence, was a result of at least three closely linked mutations, v1, v2, and v3. Jacob and Monod (100) suggested that these mutations reduce the affinity of the operator(s) for repressor. Ptashne and Hopkins confirmed this convincingly in vitro and also demonstrated in vivo that v2 renders the PL operon constitutive, whereas v1v3 affects the PR operon. Thus, cI recognizes two operators, OL and OR. Tom Maniatis and Ptashne went on to demonstrate that OL and OR each consist of three CI binding sites (131, 132). CI bound to these sites as a dimer (105), and the affinities of these subsites for CI dimers were different (133, 135). Furthermore, binding of CI to one subsite was enhanced by binding of CI to another; i.e., binding is cooperative (105). Protein-protein interactions between the C-terminal domains of CI dimers accounted for this cooperativity. At the time, binding of repressor to OL and OR appeared to be independent. More than 30 years were to elapse before Muller-Hill and his collaborators (168) demonstrated cooperativity between CI bound at OL and OR.
Cooperative binding by CI was found to explain the prophage induction by UV. The laboratory of Jeffrey Roberts at Cornell University showed that UV irradiation leads to cleavage of repressor between the N-terminal DNA binding domain and the C-terminal protein interaction domain (170). The major effect of this cleavage was the loss of cooperativity between CI dimers. The organization of the λ operators was proposed by Ptashne to ensure rapid and complete induction after UV irradiation, but this notion was not directly tested until much later by John Little's laboratory at the University of Arizona (126). Ptashne's surmise turned out to be correct.
The isolation of three classes of clear mutants indicated that cI expression was quite different from that of lacI, the gene encoding the Lac operon repressor. If cI protein was made constitutively, like the lac repressor, why would two phage functions, CII and CIII, be required to turn on cI and establish immunity? The regulation of cI expression was clearly a topic of some interest. The control of repressor synthesis in lysogens was studied by two groups: Harvey Eisen and Luiz Pereira da Silva in François Jacob's laboratory in Paris and Enrico Calef and Zdenek Neubauer in Naples. The Paris group, working under the most trying of circumstances (a leaking roof, the odor of tear gas filtering in from the riot-torn streets, and incubators inherited from Pasteur himself), characterized the prophage in bacterial survivors of thermal induction of a λ cIts Nam lysogen (52). Survivors that retained the cIts gene fell into two classes. One carried a mutation that inactivated the DNA replication genes, O or P. The second class carried a mutation in the λ PR promoter that blocked O and P synthesis. The two classes behaved quite differently with respect to λ immunity. At 40°C, both classes were nonimmune. However, when returned to 30°C, O and P mutants remained nonimmune for at least nine generations. In contrast, promoter mutants regained immunity in 2 h.
Based on these observations, Eisen et al. made several conclusions, some quite insightful and others less so. Luiz Pereira da Silva recalls the moment when the import of their data was first understood. “I had observed that some ‘defective lysogens’ had lost immunity, and thinking, as shown by Enrico Calef, that we could be in the presence of a defective lysogen that had lost the repressor or a part of it, I characterized by marker rescue the putative deleted region. To my surprise, all the cI markers could be recovered, and even better, I could recover lambda expressing active CI repressor by crossing the lysogens with defective 434 phage. I was then convinced that something was responsible for the regulation of CI synthesis. Harvey Eisen, working at my side, insisted that I was losing my mind. Jacob told me that the successive cascades might be stopped at some level. Then one day, when Sydney Brenner was in that lab and I was telling him the story, the LIGHT arrived—through Jehovah's inspiration—directly into the brain of Harvey. The defective phage that failed to express CI was not defective in cI but expressed something (that we finally named CRO) that determines whether or not λ can initiate CI synthesis (the flip-flop idea). By comparing the behavior of defective phage mutants in the expression of right and left operon functions, we would be able to confirm this idea. We did the experiment that same night, and wrote the communication to the Comptes Rendus de l'Académie des Sciences the next day. Then we left the hard and dirty work to be finished by Mark Ptashne.”
In other words, the data confirmed that cI expression from a prophage is not constitutive but is, instead, regulated. Since cI (and rex) was the only prophage function whose expression was influenced by the temperature of the medium, they concluded that CI (or a product of a gene in the same operon) positively regulated cI expression. Eureka, CI was its own inducer! However, time plays tricks on one's memory. In fact, the Paris group failed to explain the difference between the two classes of survivors. They proposed that the x mutants expressed CIII, which restored immunity. The possibility of a function, under λ PR control, that negatively regulated cI expression was considered and dismissed (until 1970). Such a function should block repressor synthesis in trans; i.e., a superinfecting λ should form clear plaques on a mutant lysogen that failed to regain immunity at 30°C. Eisen et al. found no evidence of a trans function, possibly because of the plating conditions or media that they used.
However, Enrico Calef and Zdenek Neubauer, also in 1968, working under similarly dismal conditions in Naples, concluded that a trans-acting negative regulator of cI expression must exist (25). Like the Paris group, they isolated and characterized survivors of λ cIts lysogens, and again, as in the work of Eisen et al. (52), two classes were obtained. One class could be shifted readily between immune and nonimmune states by growth at low or high temperature, respectively. The other remained nonimmune at low temperature and, furthermore, prevented repressor expression and lysogenization by superinfecting λ. Calef and Neubauer concluded, correctly, that the prophage in this second class expressed a function that negatively regulated cI expression, both from its own genome and from that of a superinfecting phage.
In 1970, Eisen et al. identified this function (51). They performed a complementation test, introducing both an O mutant and a λ PR-defective mutant prophage into the same host. The O mutant prophage was clearly epistatic; i.e., immunity did not return on a shift down to low temperature. The gene function responsible for preventing repressor synthesis, called cro (also called tof, fed, or anti-immunity), was first mapped and then defined by a mutation, cro27. This mutation was isolated by selecting a cIts N− O− strain that could regain immunity at low temperature. Lambda cIts cro27 failed to grow lytically at low temperature on sensitive cells, because repressor overproduction directed the phage exclusively into the lysogenic pathway. The mutant formed plaques at 40°C, where CIts has reduced activity, or at 33°C if the phage carried an additional cII mutation. Lambda cIts cro27, like the λ PR-defective mutant and a λ imm434 PR-defective mutant isolated by Pero (155), overexpressed a PL operon function, exonuclease, indicating that Cro represses the λ PL operon as well. Both Cro and CI bind OL and OR to repress PL and PR, respectively. At 42°C, a temperature at which CIts is entirely denatured, λ cIts cro27 fails to grow. In the absence of both the CI and Cro repressors, overexpression of PL and PR operon functions inhibits phage development.
The roles of CI, CII, and CIII were defined in two classic papers by Lou Reichardt and Dale Kaiser (166, 167). By measuring the intracellular levels of CI, they concluded that cI transcription could initiate at one of two promoters, PRM and PRE, the promoter for repressor maintenance and the promoter for repressor establishment, respectively. The PRM promoter is activated in lysogens by the binding of CI to the nearby operator, OR. The PRE promoter is located between cro and cII and is activated after infection by CII and CIII. CIII, we now know, acts indirectly on Pre by stabilizing CII. Reichardt went on to climb Mount Everest.
THE STORY OF N
The N protein of phage λ is a transcriptional regulator with remarkable properties. It recognizes sites found exclusively in phage λ and specifically stimulates transcription arising from λ promoters PL and PR. Unlike most transcription activators, it suppresses transcription termination rather than enhancing transcription initiation. These N recognition sites (nut) are distinct from the sites of N action, which are the intrinsic and Rho-dependent terminators in the λ PL and PR operons. The functional nut sites lie in the nascent λ transcripts, rather than in λ DNA, and nut DNA is dispensable for antitermination once it is transcribed. Four host factors, NusA, NusB, NusE, and NusG, stimulate N antitermination in vitro and are required in vivo. Finally, N, and possibly the nascent nut transcript as well, associates with elongating RNA polymerase (RNAP). The resultant antitermination complex persists during translocation. N modification speeds RNAP through pause sites and terminators located more than 10 kb beyond the phage promoters.
The properties of this eccentric system were defined by workers in several laboratories and emerged slowly over many years. In 1957, Jacob, Fuerst, and Wollman (99) reported the isolation of a defective lysogen that was later shown to carry a mutation in the prophage N gene (53). The prophage had a pleiotropic phenotype; upon induction it was unable to replicate, to produce phage endolysin (the product of the R gene), or to synthesize an antigen corresponding to the tail tip protein (the product of the J gene). Working with the Campbell collection of λ amber mutants, Charles Radding, in 1964, noticed that λ N− phage failed to express λ exonuclease and made the natural but incorrect conclusion that N was the exonuclease structural gene (now called exo) (164). In 1966, however, several laboratories verified that N mutants were pleiotropic and added more phenotypes to the list: N mutants are unable to excise themselves from the bacterial chromosome (53), to replicate normally (20, 45), to produce early mRNA (106), or to synthesize phage endolysin (53, 158). Protass and Korn (157) concluded that “N may be a regulatory cistron, the protein product of which ‘turns on’ functions by allowing the initiation of transcription of early λ cistrons.” Well, we were getting closer.
In 1968, François Gros's laboratory was devoting itself to a detailed study of λ transcription. Gros recalls, “Our laboratory, at the Institut de Biologie Physico-Chimique, was among the first to attempt dissecting the cartography of λ transcription units at a molecular level, by combining size measurements and genetic mapping, using differential RNA-DNA hybridization with λ DNA preparations carrying various deletions.” The pleiotropy of N mutants was clearly the result of failure to transcribe the λ chromosome. But the transcription pattern of these mutants was odd. They expressed short transcripts from the PL and PR operons, carrying N and O, respectively (116). However, this could still be interpreted to mean that N activated transcription initiation at promoters distal to the 3′ ends of these transcripts.
But that is not how N acts. That N regulates transcription elongation rather than initiation was suggested by the experiments of Denise Luzzati (128). She asked whether N expressed from an exo− heteroimmune superinfecting phage could elicit exonuclease synthesis from a λ N− prophage. The answer was definitive. N could activate exonuclease synthesis only if the prophage was induced, i.e., only after transcription had initiated at PL and PR. This result was clearly incompatible with a role for N in turning on new promoters. Luzzati concluded, “The requirement of repressor inactivation for allowing exonuclease synthesis by an N− prophage, indicates that transcription in the cI to int direction has to start before N protein can produce its effect, suggesting that the function of the N product would be to allow transcription to proceed past some sort of stop signal located, in this case at the end of gene N.” The year before, Court and Sato (37) had isolated deletion mutations that allowed λ N− phage to form plaques. They suspected that these deletions (called nin for N independent) removed stop signals in the pR operon. This was entirely consistent with the model proposed by Luzzati. Without transcription terminators, of course, N would not be required for phage growth.
In 1969 Jeffrey Roberts published work done for his doctoral thesis in the Harvard laboratory of Walter Gilbert (169). In it, he described the isolation of Rho termination factor from E. coli extracts. This work provided the first in vitro support for the notion that N was an antitermination factor. That N might have such a function was known to Roberts from the work of Luzzati and others, but the extensive transcription of the λ chromosome in a purified in vitro system that lacked N argued against the need for an antitermination factor. Why did one need an antitermination function when there was no termination at the relevant sites, TL1 and TR1 (Fig. 5)? Roberts recalls the atmosphere in the Gilbert lab in the late 1960s. “Rho was discovered through a search in uninfected cell extracts for influences that increased the apparent fidelity of transcription. RNA polymerase was a major subject of interest in the Watson-Gilbert laboratory, and Wally was particularly interested in showing that RNA polymerase could work accurately.”
Roberts began his work on transcription by looking for an effect in vitro of the λ c17 mutant, which behaved like a new promoter. In fact, DNA of the c17 mutant supported a modest but distinct increase in overall transcription in vitro rightward from the immunity half of phage DNA. Although this result showed clearly the function of a genetically defined promoter in vitro, there should have been little other transcription in the absence of the N protein regulator, whose mechanism was unknown at the time. Addition of cell extract did in fact reduce background transcription; “… the activity responsible for the effect turned out to be purifiable as a single protein that eventually was named Rho. Rho was purified to homogeneity before its actual activity was known.
“Unexpectedly, the factor had no effect on initiation and in fact depressed total RNA synthesis about threefold. This was consistent with sucrose gradient analysis that showed that the factor decreased the size of RNA products severalfold. [I] identified the slower- and faster-sedimenting peaks of Rho-mediated RNA products as the products of rightward and leftward transcription. The simplest interpretation was that the factor (soon named Rho, for release) terminated RNA synthesis.
“It was known that mRNA synthesis in the λ life cycle starts from the immunity region and goes in both directions and that expression of genes outside the immunity region and gene N required (or mostly required) the activity of the N gene product itself. However, N is not present in the in vitro reaction, so that accurate transcription should stop at these boundaries. Thus, the Rho termination factor might establish the accurate condition by terminating synthesis at the boundaries, and this was shown to be the case.”
Since it was known that N influences transcription from promoters in the immunity region, “The simplest model for the function of the N protein was antitermination. In fact, the size of the in vitro-synthesized RNA was about the same as that identified in vivo for N, and the rightward RNA could correspond to Cro.” The strongest terminator in the PR operon was not, in fact, TR1, which is only about 80% efficient in vivo (36). Stronger terminators are located further downstream, between the P and Q genes. nin5, referred to earlier, was shown to be a deletion in this region by Szybalski and his coworkers in 1971 (61), at a site where he had previously supposed that strong terminators to rightward transcription were located (187). Other N-independent mutants were identified as new promoters in the P-Q region, promoter distal to these terminators (22, 34, 95).
At the end of 1972 and the beginning of 1973, several Canadians reported an in vitro assay for N activity. Using an S30 transcription-translation system, Jack Greenblatt (working at Harvard at the time) demonstrated a requirement for N to synthesize endolysin from a λ DNA template (78). Using this assay, he showed that CI and Cro downregulated N expression (79). Furthermore, N itself was highly unstable (175). Robert Dottin and Mark Pearson (44), using a similar system, reported that the expression of anthranilate synthetase from a template carrying a PL-trpE fusion required extract from induced N+ lysogens. The activity was temperature sensitive if the lysogen carried an Nts prophage. Finally, extracts from induced lysogens of λ imm21 could not support anthranilate synthetase synthesis. Phage 21 cannot complement a λ N mutant, indicating that N21 cannot substitute for Nλ.
The nonexchangeability of Nλ and N21 indicated another remarkable feature of the N reaction. Friedman and his colleagues pointed out that λ differs from λ imm21 only in the immunity region, and the terminators are in a part of the chromosome common to both phage (67). Friedman surmised that the sites of N recognition must differ in λ and λ imm21 and therefore must lie in the immunity region. The sites of N action, however, lay outside this region. Ergo, the N recognition and action sites are distinct.
Where exactly were the sites of N recognition? In 1974 two laboratories reported that N could suppress polarity in bacterial operons (1, 63). Polarity is the ability of a translation-terminating mutation to block expression of downstream genes in the same operon (4). The effect is due to premature transcription termination by Rho, which binds to untranslated mRNA distal to the chain-terminating mutation (2, 41). In certain operon fusions, trp or gal could be expressed from λ PL (1, 23, 63). Polar mutations were suppressed if the genes containing them were transcribed from PL in the presence of N. Although N blocked termination in the fusions, other termination events in the cell were perfectly normal. Indeed, polar mutations in gal were suppressed by N when transcription originated at PL, but not when transcription originated at Pgal. These experiments indicated that N did not act as a general anti-Rho factor but required a cis-acting site in the PL operon. Deletion mapping showed that this N recognition site lies somewhere between the promoter and the end of N. Lambda and λ imm434 share the same N gene, but that of λ imm21 has a different specificity. Therefore, the sites at which N acts, called nut for N utilization, must lie in a region that is identical in the first two phages and differs in the third. The nutL site was defined by John Salstrom and Waclaw Szybalski (172), who selected a cis-acting mutant defective in two PL operon functions, red and gam. The mutation lay in a 17-bp sequence that, with only a single nucleotide difference, appeared at the predicted locations in both PL and PR operons (171). That nut was both necessary and sufficient for N action was shown by de Crombrugghe et al. (42). They cloned a sequence that contained both nutR and tR1 and inserted it between Pgal and tet. In contrast to the case for the fusions described above, transcription arising at Pgal was now modified by N and passed through TR1 to express tet.
But what exactly did N recognize at nut? Among other possibilities, Sankar Adhya and Max Gottesman (2) suggested that N might bind to nascent nut mRNA. This outré notion was supported by the properties of mutations that inhibited N antitermination in the PR operon. The mutations were frameshifts in cro that extended translation across nutR. Realizing that both nutL and nutR lay in untranslated regions, Olson et al. (153) proposed that nut was recognized as RNA and that ribosomes translating this RNA could block recognition.
Whereas N recognition was highly specific, the sites of N action were not. Since N was still required in rho mutants, clearly both Rho-dependent and intrinsic terminators were suppressed by N (74). Indeed, insertion of the classic intrinsic terminator, λ 6S, between nutL and gal did not block expression of the gal operon from PL.
The involvement of host functions in N activity was shown by the isolation of mutants unable to support λ growth by Friedman at the National Institutes of Health and Costa Georgopoulos at Stanford University (64, 70). Friedman's mutant was altered in nusA, and Georgopoulos's mutant was altered in nusB. The role of serendipity in the isolation of the mutants and the etymology of nus is recalled by Friedman: “I was studying the λ N gene as a postdoc in Michael Yarmolinsky's lab (at NIH). Max Gottesman, who had graduated from that lab a year earlier and had moved to a closet down the hall, and I decided to look for E. coli mutants that suppress λ's requirement for N, suppressors of N or, as we referred to them, sun mutants. We were unable to isolate such mutants. So we decided to isolate the opposite type of mutants, those that fail to support N action. I won't go into the details of the selection, but because Max went on one of his many trips to the Pasteur Institute, or so we were told, I was left with the project. Indeed the selection worked, and I showed Michael the results. In his inimical style, he carefully studied the results and finally said, to the best of my memory, ‘You fellows have not isolated a sun mutant but you have isolated the opposite, a nus mutant.’ This explains the etymology of the name nus. Later, after I had moved to Michigan, we decided the acronym stood for N-under supplied. Not great, but the best we could come up with. The initial selection yielded the nusA1 mutation, and then, as in a Hershey heaven,' I continued to use the selection to isolate the nusB5 and nusE71 mutations and, with Lou Baron's help, identified their map locations. Around the same time Costa Georgopoulos was isolating his gro mutations, identifying, among others, the groNB mutation, a nusB allele. I suppose the nusB name was adopted because it was simpler. But in any event we never became competitors; rather, Costa and I became great friends. Max came back to the nus mutations, showing that they affected termination, and later selected suppressors of the Nus phenotype. This search led to the isolation of a mutation that identified the nusG gene. Closing the circle, Max and I were recently on a paper showing that, as [shown by Greenblatt] in vitro, NusG is required for full N action in vivo.” No discussion of Nus factors would be complete without mentioning the elegant in vitro studies of the Greenblatt and Das labs, which not only showed that the Nus proteins are indeed involved in N-mediated antitermination but also identified their roles in the N antitermination complex, a complex sometimes referred to by Sankar Adhya as the “nusosome” and, more colorfully, as the “juggernaut” (3), crashing through obstacles in its path.
Study of the biochemistry of N now moved quickly. In 1980, Greenblatt et al. (81) demonstrated that NusA was identical to L factor, which is required for β-galactosidase synthesis in an S30 transcription-translation extract. The molecular basis of NusA action in this system was puzzling, since Greenblatt et al. found shortly afterward that NusA induced transcription pausing at specific sites (82). On the other hand, NusA bound N and was required for N antitermination (80). This enigma is still unresolved, although recent experiments suggest that N sequesters NusA from its normal role in a transcription elongation complex, thus suppressing transcription termination (152).
In 1984, Das and Wolska (40) reported N-dependent expression of galactokinase, the product of the galK gene, in an S30 system. In addition to N, the reaction required nutL, NusA, NusB, and NusE (ribosomal protein S10). The requirement for a fourth host factor, NusG, was shown much later (122, 124). In a similar system using nutR, Warren and Das (196) showed that translation of the upstream cro gene was not essential for antitermination and that, therefore, the roles of NusE in the N reaction and in translation were distinct. In an utterly brilliant 1987 paper, Das and his colleagues (12) demonstrated that N formed a persistent complex with elongating RNAP at the nut sites. Later, DeVito and Das (43) would show that the complex was stabilized by the Nus proteins, and Greenblatt and his colleagues (141) would demonstrate how N, the Nus proteins, and nut RNA interacted. To follow the still developing story of N, the reader is directed to the recent review in reference 152.
It is striking how many of the key insights in the molecular biology of recombination and gene regulation were made with λ as an experimental tool. It is also striking how many young molecular biologists implicitly assume that the history of their subject, with a few notable exceptions, such as the understanding of DNA structure, began only a short time ago. Many fundamental ideas of molecular biology had an extended gestation period and a difficult birth, and we have tried to make this clear in this review. Nevertheless, it is certain that the development of molecular biology was accelerated because λ drew together an international cohort of researchers who, for the most part, freely exchanged ideas, strains, and students. One is hard put to think of an analogous discipline at present.
Acknowledgments
We are greatly indebted to our many colleagues who helped our failing memories with their own recollections. Sankar Adhya, Don Court, Howard Nash, Amos Oppenheim, Waclaw Szybalski, and Michael Yarmolinsky read the manuscript and made many helpful suggestions. Josette Rouvière-Yaniv helped us to check facts. We apologize to all those whose reminiscences and personal notes we did not directly cite: Sankar Adhya, Paolo Amati, Doug Berg, Giuseppe Bertani, Dick D'Ari, Roger Hendrix, Eduard Kellenberger, Peggy Lieb, Helios Murialdo, Amos Oppenheim, Sandy Parkinson, Mark Ptashne, Frank Stahl, Waclaw Szybalski, René Thomas, and Dan Wulff.
Footnotes
With apologies to William Blake.
REFERENCES
- 1.Adhya, S., M. Gottesman, and B. de Crombrugghe. 1974. Release of polarity in Escherichia coli by gene N of phage lambda: termination and antitermination of transcription. Proc. Natl. Acad. Sci. USA 71:2534-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adhya, S., and M. E. Gottesman. 1978. Control of transcription termination. Annu. Rev. Biochem. 47:967-996. [DOI] [PubMed] [Google Scholar]
- 3.Adhya, S., M. E. Gottesman, and B. de Crombrugghe. 1974. Termination and antitermination in transcription: control of gene expression. Basic. Life Sci. 3:213-221. [DOI] [PubMed] [Google Scholar]
- 4.Ames, B. N., and P. E. Hartman. 1963. The histidine operon. Cold Spring Harbor Symp. Quant. Biol. 28:349-365. [DOI] [PubMed] [Google Scholar]
- 5.Appleyard, R. K. 1953. Segregation of lambda lysogenicity during bacterial recombination in E. coli K-12. Cold Spring Harbor Symp. Quant. Biol. 18:95-97. [DOI] [PubMed] [Google Scholar]
- 6.Arber, W. 1974. DNA modification and restriction. Prog. Nucleic Acid Res. Mol. Biol. 14:1-37. [DOI] [PubMed] [Google Scholar]
- 7.Arber, W., and D. Dussoix. 1962. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J. Mol. Biol. 5:18-36. [DOI] [PubMed] [Google Scholar]
- 8.Arber, W., S. Hattman, and D. Dussoix. 1963. On the host-controlled modification of bacteriophage lambda. Virology 21:30-35. [DOI] [PubMed] [Google Scholar]
- 9.Arber, W., G. Kellenberger, and J. Weigle. 1957. La défectuosité du phage lambda transducteur. Schweiz. Z. Allg. Pathol. Bakteriol. 20:659-665. [PubMed] [Google Scholar]
- 10.Arber, W., and S. Linn. 1969. DNA modification and restriction. Annu. Rev. Biochem. 38:467-500. [DOI] [PubMed] [Google Scholar]
- 11.Attardi, G., S. Naono, F. Gros, G. Buttin, and F. Jacob. 1963. Régulation de la transcription DNA-RNA messager intervenant dans la biosynthèse d'enzymes bactériens et l'expression des fonctions virales du phage. C. R. Acad. Sci. 256:805-807. [PubMed] [Google Scholar]
- 12.Barik, S., B. Ghosh, W. A. Whalen, D. Lazinski, and A. Das. 1987. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell 50:885-899. [DOI] [PubMed] [Google Scholar]
- 13.Bear, S. E., D. L. Court, and D. I. Friedman. 1984. An accessory role for Escherichia coli integration host factor: characterization of a lambda mutant dependent upon integration host factor for DNA packaging. J. Virol. 52:966-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker, A., and M. Gold. 1978. Enzymatic breakage of the cohesive end site of phage lambda DNA: terminase (ter) reaction. Proc. Natl. Acad. Sci. USA 75:4199-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker, A., and M. Gold. 1975. Isolation of the bacteriophage lambda A-gene protein. Proc. Natl. Acad. Sci. USA 72:581-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker, A., H. Murialdo, and M. Gold. 1977. Studies on an in vitro system for the packaging and maturation of phage lambda DNA. Virology 78:277-290. [DOI] [PubMed] [Google Scholar]
- 17.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertani, G., and J. J. Weigle. 1953. Host controlled variation in bacterial viruses. J. Bacteriol. 65:113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Botstein, D., and I. Herskowitz. 1974. Properties of hybrids between Salmonella phage P22 and coliphage lambda. Nature 251:584-589. [DOI] [PubMed] [Google Scholar]
- 20.Brooks, K. 1965. Studies in the physiological genetics of some suppressor-sensitive mutants of bacteriophage lambda. Virology 26:489-499. [DOI] [PubMed] [Google Scholar]
- 21.Burnet, F. M., and D. Lush. 1936. Induced lysogenicity and mutation of bacteriophage within lysogenic bacteria. Aust. J. Exp. Biol. Med. Sci. 14:27-38. [Google Scholar]
- 22.Butler, B., and H. Echols. 1970. Regulation of bacteriophage lambda development by gene N: properties of a mutation that bypasses N control of late protein synthesis. Virology 40:212-222. [DOI] [PubMed] [Google Scholar]
- 23.Buttin, G., F. Jacob, and J. Monod. 1960. Synthèse constitutive de galactokinase consécutive au développement des bactériophages λ chez Escherichia coli K 12. C. R. Acad. Sci. 250:2471-2473. [PubMed] [Google Scholar]
- 24.Calef, E., and G. Licciardello. 1960. Recombination experiments on prophge host relationships. Virology 12:81-103. [Google Scholar]
- 25.Calef, E., and Z. Neubauer. 1968. Active and inactive states of the CI gene in some lambda defective phages. Cold Spring Harbor Symp. Quant. Biol. 33:765-767. [DOI] [PubMed] [Google Scholar]
- 26.Campbell, A. 1961. Sensitive mutants of bacteriophage lambda. Virology 14:22-32. [DOI] [PubMed] [Google Scholar]
- 27.Campbell, A. 2003. The future of bacteriophage biology. Nat. Rev. Genet. 4:471-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell, A. 1962. Episomes. Adv. Genet. 11:101-145. [Google Scholar]
- 29.Campbell, A. 1963. Distribution of genetic types of transducing lambda phages. Genetics 48:409-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell, A. 1957. Transduction and segregation in Escherichia coli. Virology 4:366-384. [DOI] [PubMed] [Google Scholar]
- 31.Campbell, A. M. 1969. Episomes. Harper & Row, New York, N.Y.
- 32.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen, D. 1959. A variant of phage P2 originating in Escherichia coli strain B. Virology 7:112-126. [DOI] [PubMed] [Google Scholar]
- 34.Costantino, N., M. Zuber, and D. L. Court. 1990. Analysis of mutations in the ninR region of bacteriophage lambda that bypass a requirement for lambda N antitermination. J. Bacteriol. 172:4610-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Court, D. L., S. Adhya, H. A. Nash, and L. W. Enquist. 1977. The phage lambda integration protein (int) is subject to control by the cII and cIII gene products, p. 389-394. In A. I. Bukhari, J. A. Shapiro, and S. Adhya (ed.), DNA insertion elements, plasmids, and episomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Court, D. L., C. Brady, M. Rosenberg, D. L. Wulff, M. Behr, M. Mahoney, and S. U. Izumi. 1980. Control of transcription termination: a rho-dependent termination site in bacteriophage lambda. J. Mol. Biol. 138:231-254. [DOI] [PubMed] [Google Scholar]
- 37.Court, D. L., and K. Sato. 1969. Studies of novel transducing variants of lambda: dispensability of genes N and Q. Virology 39:348-352. [DOI] [PubMed] [Google Scholar]
- 38.Cowie, D. B., and A. D. Hershey. 1965. Multiple sites of interaction with host-cell DNA in the DNA of phage lambda. Proc. Natl. Acad. Sci. USA 53:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dambly, C., M. Couturier, and R. Thomas. 1968. Control of development in temperate bacteriophages. II. Control of lysozyme synthesis. J. Mol. Biol. 32:67-81. [DOI] [PubMed] [Google Scholar]
- 40.Das, A., and K. Wolska. 1984. Transcription antitermination in vitro by lambda N gene product: requirement for a phage nut site and the products of host nusA, nusB, and nusE genes. Cell 38:165-173. [DOI] [PubMed] [Google Scholar]
- 41.de Crombrugghe, B., S. Adhya, M. Gottesman, and I. Pastan. 1973. Effect of Rho on transcription of bacterial operons. Nat. New Biol. 241:260-264. [DOI] [PubMed] [Google Scholar]
- 42.de Crombrugghe, B., M. Mudryj, R. DiLauro, and M. Gottesman. 1979. Specificity of the bacteriophage lambda N gene product (pN): nut sequences are necessary and sufficient for antitermination by pN. Cell 18:1145-1151. [DOI] [PubMed] [Google Scholar]
- 43.DeVito, J., and A. Das. 1994. Control of transcription processivity in phage lambda: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc. Natl. Acad. Sci. USA 91:8660-8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dottin, R. P., and M. L. Pearson. 1973. Regulation by N gene protein of phage lambda of anthranilate synthetase synthesis in vitro. Proc. Natl. Acad. Sci. USA 70:1078-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dove, W. F. 1966. Action of the lambda chromosome. I. Control of functions late in bacteriophage development. J. Mol. Biol. 19:187-201. [DOI] [PubMed] [Google Scholar]
- 46.Dussoix, D., and W. Arber. 1962. Host specificity of DNA produced by Escherichia coli. II. Control over acceptance of DNA from infecting phage lambda. J. Mol. Biol. 5:37-49. [DOI] [PubMed] [Google Scholar]
- 47.Echols, H. 2001. Operators and promoters. The story of molecular biology and its creators. University of California Press, Berkeley.
- 48.Echols, H., and L. Green. 1979. Some properties of site-specific and general recombination inferred from int-initiated exchanges by bacteriophage lambda. Genetics 93:297-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Echols, H., and G. Guarneros. 1983. Control of integration and excision, p. 75-92. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 50.Echols, H., L. Pilarski, and P. Y. Xcheng. 1968. In vitro repression of phage lambda DNA transcription by a partially purified repressor from lysogenic cells. Proc. Natl. Acad. Sci. USA 59:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisen, H., P. Brachet, L. Pereira da Silva, and F. Jacob. 1970. Regulation of repressor expression in lambda. Proc. Natl. Acad. Sci. USA 66:855-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eisen, H., L. Pereira da Silva, and F. Jacob. 1968. Sur la régulation précoce du bactériophage λ. C. R. Acad. Sci. Acad. Sci. Ser. D 266:1176-1178. [PubMed] [Google Scholar]
- 53.Eisen, H. A., C. R. Fuerst, L. Siminovitch, R. Thomas, L. Lambert, L. Pereira da Silva, and F. Jacob. 1966. Genetics and physiology of defective lysogeny in K12 (lambda): studies of early mutants. Virology 30:224-241. [DOI] [PubMed] [Google Scholar]
- 54.Emmons, S. W. 1974. Bacteriophage lambda derivatives carrying two copies of the cohesive end site. J. Mol. Biol. 83:511-525. [DOI] [PubMed] [Google Scholar]
- 55.Enquist, L. W., H. A. Nash, and R. A. Weisberg. 1979. Strand exchange in site-specific recombination. Proc. Natl. Acad. Sci. USA 76:1363-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feiss, M., and A. Bublitz. 1975. Polarized packaging of bacteriophage lambda chromosomes. J. Mol. Biol. 94:583-594. [DOI] [PubMed] [Google Scholar]
- 57.Feiss, M., and A. Campbell. 1974. Duplication of the bacteriophage lambda cohesive end site: genetic studies. J. Mol. Biol. 83:527-540. [DOI] [PubMed] [Google Scholar]
- 58.Feiss, M., I. Kobayashi, and W. Widner. 1983. Separate sites for binding and nicking of bacteriophage lambda DNA by terminase. Proc. Natl. Acad. Sci. USA 80:955-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feiss, M., J. Sippy, and G. Miller. 1985. Processive action of terminase during sequential packaging of bacteriophage lambda chromosomes. J. Mol. Biol. 186:759-771. [DOI] [PubMed] [Google Scholar]
- 60.Feiss, M., W. Widner, G. Miller, G. Johnson, and S. Christiansen. 1983. Structure of the bacteriophage lambda cohesive end site: location of the sites of terminase binding (cosB) and nicking (cosN). Gene 24:207-218. [DOI] [PubMed] [Google Scholar]
- 61.Fiandt, M., Z. Hradecna, H. A. Lozeron, and W. Szybalski. 1971. Electron micrographic mapping of deletions, insertions, inversions, and homologies in the DNAs of coliphages lambda and phi 80, p. 329-354. In A. D. Hershey (ed.), The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 62.Fischer-Fantuzzi, L. 1967. Integration of lambda and lambda-b2 genomes in nonimmune host bacteria carrying a lambda cryptic prophage. Virology 32:18-32. [DOI] [PubMed] [Google Scholar]
- 63.Franklin, N. C. 1974. Altered reading of genetic signals fused to the N operon of bacteriophage lambda: genetic evidence for modification of polymerase by the protein product of the N gene. J. Mol. Biol. 89:33-48. [DOI] [PubMed] [Google Scholar]
- 64.Friedman, D. I. 1971. A bacterial mutant affecting lambda development, p. 733-738. In A. D. Hershey (ed.), The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 65.Friedman, D. I., and D. L. Court. 1995. Transcription antitermination: the lambda paradigm updated. Mol. Microbiol. 18:191-200. [DOI] [PubMed] [Google Scholar]
- 66.Friedman, D. I., and M. E. Gottesman. 1983. Lytic mode of lambda development, p. 21-52. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 67.Friedman, D. I., G. S. Wilgus, and R. J. Mural. 1973. Gene N regulator function of phage lambda immun21: evidence that a site of N action differs from a site of N recognition. J. Mol. Biol. 81:505-516. [DOI] [PubMed] [Google Scholar]
- 68.Gellert, M. 1967. Formation of covalent circles of lambda DNA by E. coli extracts. Proc. Natl. Acad. Sci. USA 57:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gellert, M., K. Mizuuchi, M. H. O'Dea, and H. A. Nash. 1976. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. USA 73:3872-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Georgopoulos, C. P. 1971. Bacterial mutants in which the gene N function of bacteriophage lambda is blocked have an altered RNA polymerase. Proc. Natl. Acad. Sci. USA 68:2977-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gingery, R., and H. Echols. 1967. Mutants of bacteriophage lambda unable to integrate into the host chromosome. Proc. Natl. Acad. Sci. USA 58:1507-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gold, M., and A. Becker. 1983. The bacteriophage lambda terminase. Partial purification and preliminary characterization of properties. J. Biol. Chem. 258:14619-14625. [PubMed] [Google Scholar]
- 73.Goodman, S. D., and H. A. Nash. 1989. Functional replacement of a protein-induced bend in a DNA recombination site. Nature 341:251-254. [DOI] [PubMed] [Google Scholar]
- 74.Gottesman, M. E., S. Adhya, and A. Das. 1980. Transcription antitermination by bacteriophage lambda N gene product. J. Mol. Biol. 140:57-75. [DOI] [PubMed] [Google Scholar]
- 75.Gottesman, M. E., and M. B. Yarmolinsky. 1968. Integration-negative mutants of bacteriophage lambda. J. Mol. Biol. 1:487-505. [DOI] [PubMed] [Google Scholar]
- 76.Gottesman, M. E., and M. B. Yarmolinsky. 1968. The integration and excision of the bacteriophage lambda genome. Cold Spring Harbor Symp. Quant. Biol. 33:735-747. [DOI] [PubMed] [Google Scholar]
- 77.Gottesman, S., and M. Gottesman. 1975. Excision of prophage lambda in a cell-free system. Proc. Natl. Acad. Sci. USA 72:2188-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Greenblatt, J. 1972. Positive control of endolysin synthesis in vitro by the gene N protein of phage lambda. Proc. Natl. Acad. Sci. USA 69:3606-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Greenblatt, J. 1973. Regulation of the expression of the N gene of bacteriophage lambda. Proc. Natl. Acad. Sci. USA 70:421-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Greenblatt, J., and J. Li. 1981. The nusA gene protein of Escherichia coli. Its identification and a demonstration that it interacts with the gene N transcription anti-termination protein of bacteriophage lambda. J. Mol. Biol. 147:11-23. [DOI] [PubMed] [Google Scholar]
- 81.Greenblatt, J., J. Li, S. Adhya, D. I. Friedman, L. S. Baron, B. Redfield, H. F. Kung, and H. Weissbach. 1980. L factor that is required for beta-galactosidase synthesis is the nusA gene product involved in transcription termination. Proc. Natl. Acad. Sci. USA 77:1991-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Greenblatt, J., M. McLimont, and S. Hanly. 1981. Termination of transcription by nusA gene protein of Escherichia coli. Nature 292:215-220. [DOI] [PubMed] [Google Scholar]
- 83.Guarneros, G. 1988. Retroregulation of bacteriophage lambda int gene expression. Curr. Top. Microbiol. Immunol. 136:1-19. [DOI] [PubMed] [Google Scholar]
- 84.Guarneros, G., and H. Echols. 1970. New mutants of bacteriophage lambda with a specific defect in excision from the host chromosome. J. Mol. Biol. 47:565-574. [DOI] [PubMed] [Google Scholar]
- 85.Guarneros, G., and H. Echols. 1973. Thermal asymmetry of site-specific recombination by bacteriophage lambda. Virology 52:30-38. [DOI] [PubMed] [Google Scholar]
- 86.Guarneros, G., C. Montanez, T. Hernandez, and D. L. Court. 1982. Posttranscriptional control of bacteriophage lambda gene expression from a site distal to the gene. Proc. Natl. Acad. Sci. USA 79:238-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guerrini, F. 1969. On the asymmetry of lambda integration sites. J. Mol. Biol. 46:523-542. [DOI] [PubMed] [Google Scholar]
- 88.Hayes, W. 1964. The genetics of bacteria and their viruses. Studies in basic genetics and molecular biology. Blackwell Scientific Publications, Oxford, United Kingdom.
- 89.Hendrix, R. W., J. W. Roberts, F. Stahl, and R. A. Weisberg (ed.). 1983. Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 90.Hershey, A. D. 1971. The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 91.Hershey, A. D., E. Burgi, and L. Ingraham. 1963. Cohesion of DNA molecules isolated from phage lambda. Proc. Natl. Acad. Sci. USA 49:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hogness, D. S., W. Doerfler, J. B. Egan, and L. W. Black. 1966. The position and orientation of genes in lambda and lambda dg DNA. Cold Spring Harbor Symp. Quant. Biol. 31:129-138. [DOI] [PubMed] [Google Scholar]
- 93.Hogness, D. S., and J. R. Simmons. 1964. Breakage of lambda-dg DNA: chemical and genetic characterization of each isolated half-molecule. J. Mol. Biol. 93:411-438. [DOI] [PubMed] [Google Scholar]
- 94.Hohn, B. 1983. DNA sequences necessary for packaging of bacteriophage lambda DNA. Proc. Natl. Acad. Sci. USA 80:7456-7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hopkins, N. 1970. Bypassing a positive regulator: isolation of a lambda mutant that does not require N product to grow. Virology 40:223-229. [DOI] [PubMed] [Google Scholar]
- 96.Hradecna, Z., and W. Szybalski. 1969. Electron micrographic maps of deletions and substitutions in the genomes of transducing coliphages lambda dg and lambda bio. Virology 38:473-477. [DOI] [PubMed] [Google Scholar]
- 97.Hsu, P. L., and A. Landy. 1984. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature 311:721-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ikeda, H., and J. Tomizawa. 1968. Prophage P1, an extrachromosomal replication unit. Cold Spring Harbor Symp. Quant. Biol. 33:791-798. [DOI] [PubMed] [Google Scholar]
- 99.Jacob, F., C. R. Fuerst, and E. L. Wollman. 1957. Recherches sur les bactéries lysogènes défectives. II. Les types physiologiques liés aux mutations du prophage. Ann. Inst. Pasteur (Paris) 93:724-753. [PubMed] [Google Scholar]
- 100.Jacob, F., and J. Monod. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3:318-356. [DOI] [PubMed] [Google Scholar]
- 101.Jacob, F., and E. L. Wollman. 1954. Etude génétique d'un bactériophage tempéré d'Escherichia coli. I. Le système génétique du bactériophage λ. Ann. Inst. Pasteur (Paris) 87:653-673. [PubMed] [Google Scholar]
- 102.Jacob, F., and E. L. Wollman. 1961. Sexuality and the genetics of bacteria. Academic Press, New York, N.Y.
- 103.Jacob, F., and E. L. Wollman. 1956. Recherches sur les bactéries lysogènes défectives. I. Déterminisme génétique de la morphogenèse chez un bactériophage tempéré. Ann. Inst. Pasteur (Paris) 90:282-302. [PubMed] [Google Scholar]
- 104.Jacob, F., and E. L. Wollman. 1956. Les processus de la conjugaison et recombinaison chez Escherichia coli. I. Induction par conjugaison ou induction zygotique. Ann. Inst. Pasteur (Paris) 91:486-510. [PubMed] [Google Scholar]
- 105.Johnson, A. D., B. J. Meyer, and M. Ptashne. 1979. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc. Natl. Acad. Sci. USA 76:5061-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Joyner, A., L. N. Isaacs, H. Echols, and W. S. Sly. 1966. DNA replication and messenger RNA production after induction of wild-type lambda bacteriophage and lambda mutants. J. Mol. Biol. 19:174-186. [DOI] [PubMed] [Google Scholar]
- 107.Kaiser, A. D. 1955. A genetic study of the temperate coliphage. Virology 1:424-443. [DOI] [PubMed] [Google Scholar]
- 108.Kaiser, A. D. 1957. Mutations in a temperate bacteriophage affecting its ability to lysogenize Escherichia coli. Virology 3:42-61. [DOI] [PubMed] [Google Scholar]
- 109.Kaiser, A. D. 1962. The production of phage chromosome fragments and their capacity for genetic transfer. J. Mol. Biol. 4:275-287. [DOI] [PubMed] [Google Scholar]
- 110.Kaiser, A. D., and D. S. Hogness. 1960. The transformation of Escherichia coli with deoxyribonucleic acid isolated from bacteriophage lambda-dg. J. Mol. Biol. 2:392-415. [DOI] [PubMed] [Google Scholar]
- 111.Kayajanian, G. 1968. Studies on the genetics of biotin-transducing, defective variants of bacteriophage lambda. Virology 36:30-41. [DOI] [PubMed] [Google Scholar]
- 112.Kayajanian, G., and A. Campbell. 1966. The relationship between heritable physical and genetic properties of selected gal− and gal+ transducing lambda dg. Virology 30:482-492. [DOI] [PubMed] [Google Scholar]
- 113.Kellenberger, G., M. L. Zichichi, and J. Weigle. 1961. A mutation affecting the DNA content of bacteriophage lambda and its lysogenizing properties. J. Mol. Biol. 3:399-408. [DOI] [PubMed] [Google Scholar]
- 114.Kikuchi, A., E. L. Flamm, and R. A. Weisberg. 1985. An Escherichia coli mutant unable to support site-specific recombination of bacteriophage lambda. J. Mol. Biol. 183:129-140. [DOI] [PubMed] [Google Scholar]
- 115.Kikuchi, Y., and H. A. Nash. 1979. Nicking-closing activity associated with bacteriophage lambda int gene product. Proc. Natl. Acad. Sci. USA 76:3760-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kourilsky, P., L. Marcaud, P. Sheldrick, D. Luzzati, and F. Gros. 1968. Studies of the messenger RNA of bacteriophage lambda. I. Various species synthesized early after induction of the prophage. Proc. Natl. Acad. Sci. USA 61:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kumar, S., K. Bovre, A. Guha, Z. Hradecna, V. R. Maher, and W. Szybalski. 1969. Orientation and control of transcription in E. coli phage lambda. Nature 221:823-825. [DOI] [PubMed] [Google Scholar]
- 118.Landy, A., and W. Ross. 1977. Viral integration and excision: structure of the lambda att sites. Science 197:1147-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lederberg, E. M. 1951. Lysogenicity in E. coli K-12. Genetics 36:560. [Google Scholar]
- 120.Lederberg, E. M., and J. Lederberg. 1953. Genetic studies of lysogenicity in Escherichia coli. Genetics 38:51-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Levine, M. 1957. Mutations in the temperate phage P22 and lysogeny in Salmonella. Virology 3:22-41. [DOI] [PubMed] [Google Scholar]
- 122.Li, J., R. Horwitz, S. McCracken, and J. Greenblatt. 1992. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J. Biol. Chem. 267:6012-6019. [PubMed] [Google Scholar]
- 123.Lieb, M. 1953. The establishment of lysogenicity in Escherichia coli. J. Bacteriol. 65:642-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Linn, T., and J. Greenblatt. 1992. The NusA and NusG proteins of Escherichia coli increase the in vitro readthrough frequency of a transcriptional attenuator preceding the gene for the beta subunit of RNA polymerase. J. Biol. Chem. 267:1449-1454. [PubMed] [Google Scholar]
- 125.Little, J. W., and M. E. Gottesman. 1971. Defective lambda particles whose DNA carries only a single cohesive end, p. 371-394. In A. D. Hershey (ed.), The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 126.Liu, Z., and J. W. Little. 1998. The spacing between binding sites controls the mode of cooperative DNA-protein interactions: implications for evolution of regulatory circuitry. J. Mol. Biol. 278:331-338. [DOI] [PubMed] [Google Scholar]
- 127.Ljungquist, E., and A. I. Bukhari. 1977. State of prophage Mu DNA upon induction. Proc. Natl. Acad. Sci. USA 74:3143-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Luzzati, D. 1970. Regulation of lambda exonuclease synthesis: role of the N gene product and lambda repressor. J. Mol. Biol. 49:515-519. [DOI] [PubMed] [Google Scholar]
- 129.Lwoff, A., and A. Gutmann. 1950. Recherches sur un Bacillus megatherium lysogène. Ann. Inst. Pasteur (Paris) 78:711-739. [PubMed] [Google Scholar]
- 130.Lwoff, A., L. Siminovitch, and N. O. Kjeldgaard. 1950. Induction de la production de bactériophages chez une bactérie lysogène. Ann. Inst. Pasteur (Paris) 79:815-858. [PubMed] [Google Scholar]
- 131.Maniatis, T., and M. Ptashne. 1973. Multiple repressor binding at the operators in bacteriophage lambda. Proc. Natl. Acad. Sci. USA 70:1531-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Maniatis, T., and M. Ptashne. 1973. Structure of the lambda operators. Nature 246:133-136. [DOI] [PubMed] [Google Scholar]
- 133.Maurer, R., B. Meyer, and M. Ptashne. 1980. Gene regulation at the right operator (OR) bacteriophage lambda. I. OR3 and autogenous negative control by repressor. J. Mol. Biol. 139:147-161. [DOI] [PubMed] [Google Scholar]
- 134.Meselson, M., and J. J. Weigle. 1961. Chromosome breakage accompanying genetic recombination in bacteriophage. Proc. Natl. Acad. Sci. USA 47:857-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Meyer, B. J., R. Maurer, and M. Ptashne. 1980. Gene regulation at the right operator (OR) of bacteriophage lambda. II. OR1, OR2, and OR3: their roles in mediating the effects of repressor and cro. J. Mol. Biol. 139:163-194. [DOI] [PubMed] [Google Scholar]
- 136.Miller, H. I., and D. I. Friedman. 1977. Isolation of Escherichia coli mutants unable to support lambda integrative recombination, p. 349-356. In A. I. Bukhari, J. A. Shapiro, and S. Adhya (ed.), DNA insertion elements, plasmids and episomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 137.Miwa, T., and K. Matsubara. 1982. Identification of sequences necessary for packaging DNA into lambda phage heads. Gene 20:267-279. [DOI] [PubMed] [Google Scholar]
- 138.Mizuuchi, K., and T. Fukasawa. 1969. Chromosome mobilization in rec-merodiploids of Escherichia coli K12 following infection with bacteriophage λ. Virology 39:467-481. [DOI] [PubMed] [Google Scholar]
- 139.Mizuuchi, K., and H. A. Nash. 1976. Restriction assay for integrative recombination of bacteriophage lambda DNA in vitro: requirement for closed circular DNA substrate. Proc. Natl. Acad. Sci. USA 73:3524-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mizuuchi, K., R. A. Weisberg, L. W. Enquist, M. Mizuuchi, M. Buraczynska, C. Foeller, P. L. Hsu, W. Ross, and A. Landy. 1981. Structure and function of the phage lambda att site: size, int-binding sites, and location of the crossover point. Cold Spring Harbor Symp. Quant. Biol. 45:429-437. [DOI] [PubMed] [Google Scholar]
- 141.Mogridge, J., T. F. Mah, and J. Greenblatt. 1995. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev. 9:2831-2845. [DOI] [PubMed] [Google Scholar]
- 142.Monod, J. 1971. Du microbe à l'homme, p. 1-9. In J. Monod and E. Borek (ed.), Of microbes and life. Columbia University Press, New York, N.Y.
- 143.Morse, M. L. 1954. Transduction of certain loci in Escherichia coli K12. Genetics 39:984. [Google Scholar]
- 144.Morse, M. L., E. M. Lederberg, and J. Lederberg. 1956. Transductional heterogenotes in Escherichia coli. Genetics 41:758-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morse, M. L., E. M. Lederberg, and J. Lederberg. 1956. Transduction in Escherichia coli K-12. Genetics 41:142-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mousset, S., and R. Thomas. 1969. Ter, a function which generates the ends of the mature lambda chromosome. Nature 221:242-244. [DOI] [PubMed] [Google Scholar]
- 147.Mousset, S., and R. Thomas. 1968. L'excision du prophage des bactéries dilysogènes par surinfection hétéroimmune. C. R. Acad. Sci. Acad. Sci. Ser. D. 266:1904-1907. [PubMed] [Google Scholar]
- 148.Nash, H. A. 1975. Integrative recombination in bacteriophage lambda: analysis of recombinant DNA. J. Mol. Biol. 91:501-514. [DOI] [PubMed] [Google Scholar]
- 149.Nash, H. A. 1975. Integrative recombination of bacteriophage lambda DNA in vitro. Proc. Natl. Acad. Sci. USA 72:1072-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nash, H. A., K. Mizuuchi, R. A. Weisberg, Y. Kikuchi, and M. Gellert. 1977. Integrative recombination of bacteriophage lambda—the biochemical approach to DNA insertions, p. 363-373. In A. I. Bukhari, J. A. Shapiro, and S. L. Adhya (ed.), DNA insertion elements, plasmids, and episomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 151.Nash, H. A., and C. A. Robertson. 1981. Purification and properties of the Escherichia coli protein factor required for lambda integrative recombination. J. Biol. Chem. 256:9246-9253. [PubMed] [Google Scholar]
- 152.Nudler, E., and M. E. Gottesman. 2002. Transcription termination and anti-termination in E. coli. Genes Cells 7:755-768. [DOI] [PubMed] [Google Scholar]
- 153.Olson, E. R., E. L. Flamm, and D. I. Friedman. 1982. Analysis of nutR: a region of phage lambda required for antitermination of transcription. Cell 31:61-70. [DOI] [PubMed] [Google Scholar]
- 154.Pardee, A. B., F. Jacob, and J. Monod. 1958. Sur l'expression et le rôle des allèles “inductible” et “constitutive” dans la synthèse de la β-galactosidase chez des zygotes d'Escherichia coli. C. R. Acad. Sci. 246:3125-3128. [PubMed] [Google Scholar]
- 155.Pero, J. 1970. Location of the phage lambda gene responsible for turning off lambda-exonuclease synthesis. Virology 40:65-71. [DOI] [PubMed] [Google Scholar]
- 156.Pond, F. R., I. Gibson, J. Lalucat, and R. L. Quackenbush. 1989. R-body-producing bacteria. Microbiol. Rev. 53:25-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Protass, J. J., and D. Korn. 1966. Function of the N cistron of bacteriophage lambda. Proc. Natl. Acad. Sci. USA 55:1089-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Protass, J. J., and D. Korn. 1966. Control of exonuclease and endolysin synthesis during development of bacteriophage lambda. J. Biol. Chem. 241:4175-4179. [PubMed] [Google Scholar]
- 159.Ptashne, M. 1965. The detachment and maturation of conserved lambda prophage DNA. J. Mol. Biol. 11:90-96. [DOI] [PubMed] [Google Scholar]
- 160.Ptashne, M. 2004. A genetic switch. Phage lambda revisited. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 161.Ptashne, M. 1967. Isolation of the λ phage repressor. Proc. Natl. Acad. Sci. USA 57:306-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ptashne, M. 1967. Specific binding of the lambda phage repressor to lambda DNA. Nature 214:232-234. [DOI] [PubMed] [Google Scholar]
- 163.Ptashne, M., and N. Hopkins. 1968. The operators controlled by the lambda phage repressor. Proc. Natl. Acad. Sci. USA 60:1282-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Radding, C. M. 1964. Nuclease activity in defective lysogens of phage lambda. Biochem. Biophys. Res. Commun. 15:8-12. [DOI] [PubMed] [Google Scholar]
- 165.Radding, C. M., and A. D. Kaiser. 1963. Gene transfer by broken molecules of lambda-dna: activity of the left half-molecule. J. Mol. Biol. 16:225-233. [DOI] [PubMed] [Google Scholar]
- 166.Reichardt, L., and A. D. Kaiser. 1971. Control of lambda repressor synthesis. Proc. Natl. Acad. Sci. USA 68:2185-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Reichardt, L. F. 1975. Control of bacteriophage lambda repressor synthesis: regulation of the maintenance pathway of the cro and cI products. J. Mol. Biol. 93:289-309. [DOI] [PubMed] [Google Scholar]
- 168.Revet, B., B. Wilcken-Bergmann, H. Bessert, A. Barker, and B. Muller-Hill. 1999. Four dimers of lambda repressor bound to two suitably spaced pairs of lambda operators form octamers and DNA loops over large distances. Curr. Biol. 9:151-154. [DOI] [PubMed] [Google Scholar]
- 169.Roberts, J. W. 1969. Termination factor for RNA synthesis. Nature 224:1168-1174. [DOI] [PubMed] [Google Scholar]
- 170.Roberts, J. W., and C. W. Roberts. 1975. Proteolytic cleavage of bacteriophage lambda repressor in induction. Proc. Natl. Acad. Sci. USA 72:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rosenberg, M., D. L. Court, H. Shimatake, C. Brady, and D. L. Wulff. 1978. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature 272:414-423. [DOI] [PubMed] [Google Scholar]
- 172.Salstrom, J. S., and W. Szybalski. 1978. Coliphage lambda nutL−: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J. Mol. Biol. 124:195-221. [DOI] [PubMed] [Google Scholar]
- 173.Schmeissner, U., K. McKenney, M. Rosenberg, and D. L. Court. 1984. Removal of a terminator structure by RNA processing regulates int gene expression. J. Mol. Biol. 176:39-53. [DOI] [PubMed] [Google Scholar]
- 174.Schmeissner, U., K. McKenney, M. Rosenberg, and D. L. Court. 1984. Transcription terminator involved in the expression of the int gene of phage lambda. Gene 28:343-350. [DOI] [PubMed] [Google Scholar]
- 175.Schwartz, M. 1970. On the function of the N cistron in phage lambda. Virology 40:23-33. [DOI] [PubMed] [Google Scholar]
- 176.Shulman, M., and M. E. Gottesman. 1973. Attachment site mutants of bacteriophage lambda. J. Mol. Biol. 81:461-482. [DOI] [PubMed] [Google Scholar]
- 177.Shulman, M., and M. E. Gottesman. 1971. Lambda att2: a transducing phage capable of intramolecular int-xis promoted recombination, p. 477-487. In A. D. Hershey (ed.), The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 178.Signer, E. R. 1968. Lysogeny: the integration problem. Annu. Rev. Microbiol. 22:451-488. [DOI] [PubMed] [Google Scholar]
- 179.Signer, E. R., and J. Weil. 1968. Site-specific recombination in bacteriophage lambda. Cold Spring Harbor Symp. Quant. Biol. 33:715-719. [DOI] [PubMed] [Google Scholar]
- 180.Sippy, J., and M. Feiss. 2004. Initial cos cleavage of bacteriophage lambda concatemers requires proheads and gpFI in vivo. Mol. Microbiol. 52:501-513. [DOI] [PubMed] [Google Scholar]
- 181.Skalka, A. 1966. Regional and temporal control of genetic transcription in phage lambda. Proc. Natl. Acad. Sci. USA 55:1190-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Skalka, A., B. Butler, and H. Echols. 1967. Genetic control of transcription during development of phage lambda. Proc. Natl. Acad. Sci. USA 58:576-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Skalka, A., M. Poonian, and P. Bartl. 1972. Concatemers in DNA replication: electron microscopic studies of partially denatured intracellular lambda DNA. J. Mol. Biol. 64:541-550. [DOI] [PubMed] [Google Scholar]
- 184.Smith, H. O., and M. Levine. 1967. A phage P22 gene controlling integration of prophage. Virology 31:207-216. [DOI] [PubMed] [Google Scholar]
- 185.Sternberg, N., and R. A. Weisberg. 1975. Packaging of prophage and host DNA by coliphage lambda. Nature 256:97-103. [DOI] [PubMed] [Google Scholar]
- 186.Syvanen, M. 1974. In vitro genetic recombination of bacteriophage lambda. Proc. Natl. Acad. Sci. USA 71:2496-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Szybalski, W., K. Bovre, M. Fiandt, S. Hayes, Z. Hradecna, S. Kumar, H. A. Lozeron, H. J. Nijkamp, and W. Stevens. 1970. Transcriptional units and their controls in Escherichia coli phage λ: operons and scriptons. Cold Spring Harbor Symp. Quant. Biol. 35:341. [Google Scholar]
- 188.Taylor, K., Z. Hradecna, and W. Szybalski. 1967. Asymmetric distribution of the transcribing regions on the complementary strands of coliphage lambda DNA. Proc. Natl. Acad. Sci. USA 57:1618-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Thomas, R. 1966. Control of development in temperate bacteriophages. I. Induction of prophage genes following hetero-immune superinfection. J. Mol. Biol. 22:79-95. [Google Scholar]
- 191.Thomas, R. 1970. Control of development in temperate bacteriophages. 3. Which prophage genes are and which are not trans-activable in the presence of immunity? J. Mol. Biol. 49:393-404. [DOI] [PubMed] [Google Scholar]
- 192.Thomas, R., and L. E. Bertani. 1964. On the control of the replication of temperate bacteriophages superinfecting immune hosts. Virology 24:241-253. [DOI] [PubMed] [Google Scholar]
- 193.Wackernagel, W. 1972. An improved spheroplast assay for lambda-DNA and the influence of the bacterial genotype on the transfection rate. Virology 48:94-103. [DOI] [PubMed] [Google Scholar]
- 194.Wake, R. G., A. D. Kaiser, and R. B. Inman. 1972. Isolation and structure of phage lambda head-mutant DNA. J. Mol. Biol. 64:519-540. [DOI] [PubMed] [Google Scholar]
- 195.Wang, J. C., and A. D. Kaiser. 1973. Evidence that the cohesive ends of mature lambda DNA are generated by the gene A product. Nat. New Biol. 241:16-17. [DOI] [PubMed] [Google Scholar]
- 196.Warren, F., and A. Das. 1984. Formation of termination-resistant transcription complex at phage lambda nut locus: effects of altered translation and a ribosomal mutation. Proc. Natl. Acad. Sci. USA 81:3612-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Weigle, J. J. 1966. Story and structure of the λ transducing phage, p. 226-235. In J. Cairns, G. S. Stent, and J. D. Watson (ed.), Phage and the origins of molecular biology. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 198.Weigle, J. J., and G. Bertani. 1953. Variations des bactériophages conditionnées par les bactéries hôtes. Ann. Inst. Pasteur (Paris) 84:175-179. [PubMed] [Google Scholar]
- 199.Weigle, J. J., and M. Delbrück. 1951. Mutual exclusion between an infecting phage and a carried phage. J. Bacteriol. 62:301-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Weisberg, R. A., L. W. Enquist, C. Foeller, and A. Landy. 1983. Role for DNA homology in site-specific recombination. The isolation and characterization of a site affinity mutant of coliphage lambda. J. Mol. Biol. 170:319-342. [DOI] [PubMed] [Google Scholar]
- 201.Weisberg, R. A., and M. E. Gottesman. 1971. The stability of Int and Xis functions, p. 489-500. In A. D. Hershey (ed.), The bacteriophage lambda. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 202.Weisberg, R. A., and A. Landy. 1983. Site-specific recombination in phage lambda, p. 211-250. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 203.Westmoreland, B. C., W. Szybalski, and H. Ris. 1969. Mapping of deletions and substitutions in heteroduplex DNA molecules of bacteriophage lambda by electron microscopy. Science 163:1343-1348. [DOI] [PubMed] [Google Scholar]
- 204.Williams, J. G. K., D. L. Wulff, and H. A. Nash. 1977. A mutant of Eschrichia coli deficient in a host function required for phage lambda integration and excision, p. 357-362. In A. I. Bukhari, J. A. Shapiro, and S. Adhya (ed.), DNA insertion elements, plasmids, and episomes. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 205.Wollman, E. L. 1953. Sur le déterminisme génétique de la lysogénie. Ann. Inst. Pasteur (Paris) 84:281-293. [PubMed] [Google Scholar]
- 206.Wollman, E. L., and F. Jacob. 1954. Etude génétique d'un bactériophage tempéré. II. Mécanisme de la recombinaison génétique. Ann. Inst. Pasteur (Paris) 87:674-690. [PubMed] [Google Scholar]
- 207.Wu, R., and A. D. Kaiser. 1967. Mapping the 5′-terminal nucleotides of the DNA of bacteriophage lambda and related phages. Proc. Natl. Acad. Sci. USA 57:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Xin, W. N., and M. Feiss. 1988. The interaction of Escherichia coli integration host factor with the cohesive end sites of phages lambda and 21. Nucleic Acids Res. 16:2015-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zichichi, M. L., and G. Kellenberger. 1963. Two distinct functions in the lysogenization process: the repression of phage multiplication and incorporation of the prophage in the bacterial genome. Virology 19:450-460. [DOI] [PubMed] [Google Scholar]
- 210.Zissler, J. 1967. Integration-negative (int) mutants of phage lambda. Virology 31:189. [DOI] [PubMed] [Google Scholar]