Abstract
The Achaete-scute homolog 1 (Ascl1) protein regulates a large subset of genes that leads neuronal progenitor cells to distinctive differentiation pathways during human brain development. Although it is well known that Ascl1 binds DNA as a homo- or heterodimer via its basic helix-loop-helix (bHLH) motif, little is known about the conformational sampling properties of the DNA-free full-length protein, and in particular about the bHLH domain-flanking N- and C-terminal segments, which are predicted to be highly disordered in solution. The structural heterogeneity, low solubility, and high aggregation propensity of Ascl1 in aqueous buffer solutions make high-resolution studies of this protein a challenging task. Here, we have adopted a fragment-based strategy that allowed us to obtain high-quality NMR data providing, to our knowledge, the first comprehensive high-resolution information on the structural propensities and conformational dynamics of Ascl1. The emerging picture is that of an overall extended and highly dynamic polypeptide chain comprising three helical segments and lacking persistent long-range interactions. We also show that the C-terminal helix of the bHLH domain is involved in intermolecular interactions, even in the absence of DNA. Our results contribute to a better understanding of the mechanisms of action that govern the regulation of proneural transcription factors.
Introduction
The development of the human brain during fetal and postnatal stages relies on the rapid division and differentiation of billions of cells with different phenotypes. The proneural basic helix-loop-helix (bHLH) transcription factors contribute to the specification of distinct neural cell fates (1, 2). Among these, the Achaete-scute Homolog 1 (Ascl1) protein (also known as Mash1) plays a central role in phenotype differentiation of neuronal progenitor cells by regulating the expression of a large subset of genes. The role of Ascl1 in neuronal differentiation has been extensively reviewed, highlighting the importance of its activity (1, 2, 3, 4, 5). However, little is known so far about its structure and the exact mechanism of regulation.
The Ascl1 sequence comprises 236 amino acid residues and can be divided into three domains: 1) an N-terminal region (residues 1 - 121) containing adjacent polyA and polyQ stretches of 13 and 12 residues in length, respectively; 2) a central bHLH domain (6) (residues 122–179) that is responsible for homo- and heterodimerization of the protein, as well as for DNA binding through interaction of positively charged residues (basic region) with the negatively charged DNA backbone (7); and 3) a shorter C-terminal region (residues 180–236) rich in Ser-Pro pairs that are conserved among bHLH transcription factors. Phosphorylation through proline-directed kinases of these serine residues contributes to the regulation of Ascl1 activity (8). Although the bHLH domain and its interaction with DNA has been studied previously by a variety of biochemical and biophysical methods (9, 10, 11, 12), no high-resolution structural information is available so far for the DNA-free bHLH domain or for the flanking N- and C-terminal regions.
As the structural heterogeneity that characterizes the bHLH protein family has long been known to affect protein solubility, production of stable and soluble samples of Ascl1 is challenging, and a structural description at atomic resolution is still missing. In this work, we present, to our knowledge, the first high-resolution structural data for the N- and C-terminal regions of Ascl1, as well as the bHLH domain in the absence of DNA, based on NMR studies of four different overlapping Ascl1 fragments. Our findings provide important information on the structural features of Ascl1, in particular the presence of three peptide segments with a high propensity for α-helical conformation. Two of these segments are in the bHLH domain and are implicated in DNA binding, whereas the functional role of the third one (polyA/polyQ) remains to be elucidated. The rest of the Ascl1 peptide chain is highly disordered, a feature that favors transient interactions with kinases and other regulatory proteins for fine tuning of the protein’s activity.
Materials and Methods
Ascl1 cloning, bacterial expression, and production of 13C/15N labeled samples
Full-length Ascl1 and Ascl1 fragments were cloned in the expression plasmids pDEST17 (Thermo Fisher Scientific, Waltham, MA) or pET21a (Eurofins, Luxembourg, Luxembourg). All constructs were designed to bear a six-residue His-tag. Initial constructs contained a linker region and a TEV cleavage sequence between the His-tag and the protein coding region (see Supporting Material). In later constructs, the TEV-cleavage site was omitted as the TEV protease is not active in the final buffer conditions.
All proteins were expressed in Esherichia coli by isopropyl β-D-1-thiogalactopyranoside induction at OD600 = 0.6 for 18 h at 25°C in minimal growth medium M9, which was supplemented with 15N-labeled NH4Cl for full-length Ascl1 and 15N-labeled NH4Cl, 13C-labeled glucose for fragments A, B, C, and D. The proteins of interest were all found to be expressed as inclusion bodies. To purify inclusion bodies, cell pellets were dissolved in 50 mM Tris, 5 mM imidazole; and 1 mM dithiothreitol (DTT) (pH 8.5), homogenized and sonicated on ice, then ultra-centrifuged. Pellets from ultracentrifugation were dissolved in a buffer of 50 mM Tris, 100 mM NaCl, 1 mM DTT, and 0.2 mM EDTA, supplemented with 25% w/v sucrose (pH 8), then mixed with an equivalent volume of buffer (50 mM Tris, 100 mM NaCl, 1 mM DTT, and 0.2 mM EDTA, supplemented with 2% v/v Triton X-100 (pH 8)). The solution was ultra-centrifuged and the pellet was dissolved in buffer (50 mM Tris, 1 mM DTT; and 0.2 mM EDTA (pH 8)) and ultracentrifuged one last time. The pellet containing the inclusion bodies was finally dissolved in 50 mM Tris, 8 M urea, and 1 mM DTT (pH 8.5)). The presence of the protein of interest was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The samples were subsequently incubated with NiNTA beads in binding buffer (50 mM Tris, 8 M urea, 5 mM imidazole, and 1 mM DTT (pH 8.5)) and successfully separated from impurities by elution using a linear gradient of imidazole (from 50 to 250 mM).
The solubility of Ascl1 fragments was tested for different buffers (see the Supporting Material) to identify optimal conditions for NMR studies. Samples were loaded in a dialysis membrane and dialyzed against a 1:30 ratio volume of test buffer. The dialysis step was repeated three times to obtain a dilution factor of 3 × 104 of the initial buffer concentration. Each dialysis step was performed for 2 h at 4°C with gentle stirring. The presence of the protein of interest in the soluble and insoluble fractions was confirmed by SDS-PAGE.
The purified Ascl1 fragments were dissolved in a buffer of 50 mM TrisHCl, 5 mM DTT, and 2 mM SDS (for fragments C and D) (pH 6.5) at protein concentrations varying from 100 to 400 μM and put into standard (or Shigemi-type) 5 mm NMR glass tubes.
NMR experiments
All NMR experiments were recorded on Bruker Avance spectrometers operating at different magnetic field strengths (600, 700, 850, or 950 MHz 1H frequency) and equipped with cryogenically cooled triple-resonance probeheads (TCI, Bruker, Billerica, MA). A set of three-dimensional (3D) BEST-TROSY HCN and HNN spectra (13) were recorded for sequential resonance assignment, complemented by a sequential HADAMAC experiment for amino acid type identification (14). 15N relaxation measurements were performed using conventional pulse schemes (15) for R1, R2, and heteronuclear nuclear Overhauser effect (HETNOE) experiments. The overall correlation time, related to the size of the particle, was probed by means of a TRACT experiment (16) measuring the relaxation-rate difference of the two doublet components of a 1H-coupled 15N in a series of one-dimensional (1D) experiments. Translational diffusion measurements were made with a 1D 1H-detected, 15N-edited pulsed-field gradient echo sequence (17). Diffusion coefficients, D, were obtained by signal integration over the 1D NMR spectra recorded as a function of the gradient strength, G, and by data fitting to the function , with δ the gradient duration, τ the recovery delay after each gradient, γH the gyromagnetic ratio, and Δ the delay between the two gradient pulses.
NMR data were processed using TopSpin 3.2 software (Bruker BioSpin), and were analyzed with the CcpNmr Analysis software (http://www.ccpn.ac.uk/software/analysis). The neighbor-corrected secondary structural propensities were determined using the ncSPC software (http://nmr.chem.rug.nl/ncSPC/). Tables of assigned chemical shifts corresponding to fragments A, B, C, and D were submitted to the Biological Magnetic Resonance Bank (http://www.bmrb.wisc.edu/) with accession numbers 27023, 27024, 27025, and 27026, respectively.
Results and Discussion
Ascl1 is predicted to be structurally heterogeneous
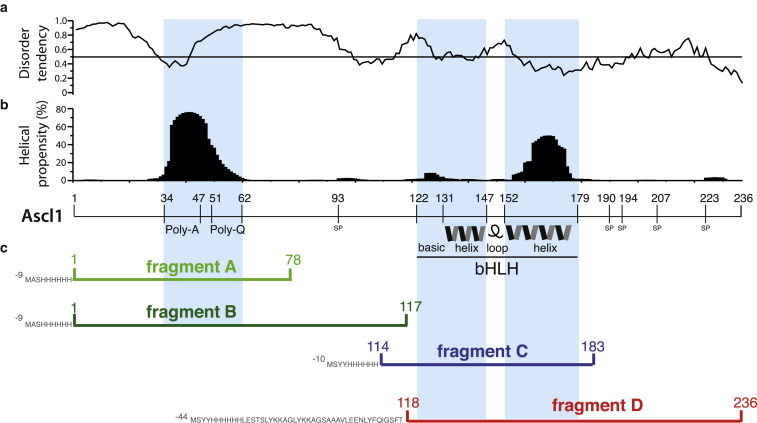
As mentioned before, the primary sequence of Ascl1 can be subdivided into three main regions: the N-terminal region, the bHLH domain, and the C-terminus. A computational analysis of the extent of disorder along the Ascl1 peptide sequence obtained using IUPred (18) predicts that the N-terminal part is highly disordered in solution (Fig. 1 a), except for the polyA stretch, which has a high propensity to adopt helical conformation (Fig. 1 b), as shown by Agadir (19) calculations. The bHLH domain region is also predicted to be (more) structured, although the helical content predicted by Agadir does not match the requirements to form a stable HLH motif. Finally, no helical elements, either well structured or completely disordered, are predicted in the C-terminal part of the protein, which is expected to be structurally heterogeneous.
Figure 1.
Ascl1 domain organization, structure prediction, and fragment design. (a) The disorder tendency was computed with the IUPred software (18, 29), whereas (b) the helical propensity prediction was obtained from Agadir (19). Peculiar sequence features of Ascl1, such as the polyA, polyQ, and Ser-Pro regions, as well as the bHLH domain, are highlighted. (c) The different fragments used in this study are shown at the bottom using a color code that is maintained throughout the manuscript. To see this figure in color, go online.
The Ascl1 sequence is rich in alanine residues, A (36 residues; 15.3% of the full sequence), polar amino acids such as glutamine, Q (30 residues; 12.7%) and serine, S (25 residues; 10.6%), as well as proline, P (18 residues; 8.1%). These four amino acids make up almost half of the protein sequence. This bias in amino-acid-type composition, combined with the presence of many repeats, e.g., QP, SP, SS, PP, polyQ, and polyA, presents a particular challenge for an NMR investigation of full-length Ascl1.
Design of Ascl1 fragments amenable to NMR studies
First attempts to obtain samples of E. Coli overexpressed full-length Ascl1 that are amenable to an NMR study were unsuccessful, as the resulting 1H-15N correlation spectra were of poor quality (Fig. S1). We therefore generated a library of Ascl1 fragments designed on the basis of structure/disorder predictions. The rationale for this fragment-based approach is that a series of partly overlapping constructs that cover the whole sequence can be obtained while retaining the structural features of full-length Ascl1. A first set of fragments was generated (Table S1), but only a subset thereof showed E. coli overexpression levels compatible with NMR requirements. The set of fragments selected for our subsequent NMR study is shown in Fig. 1 c, and the corresponding NMR 1H-15N fingerprint spectra are displayed in Fig. 2. The polyA -and -polyQ-containing N-terminal region of Ascl1 is covered by fragments A and B, with fragment B extending up to the bHLH domain. Fragment C corresponds to the bHLH domain, whereas fragment D comprises both the bHLH and C-terminal regions of the protein. All fragments were subsequently screened for solubility and NMR spectral quality using a variety of different buffer conditions (Table S2). As a result, fragments A and B were found to be soluble in a buffer composed of 50 mM TrisHCl and 5 mM DTT (pH 6.5), whereas for the bHLH-domain containing fragments, C and D, addition of 2 mM SDS was required to prevent extensive protein aggregation and to obtain samples suitable for an NMR investigation. Our observations are in line with previous reports on the highly homologous proneural transcription factor neurogenin-1, which was found to be insoluble in all tested buffer conditions (20). Fragment D has an additional 44-residue N-terminal extension comprising a cleavable His tag that could not be removed due to an inactive TEV enzyme in the SDS-containing NMR buffer.
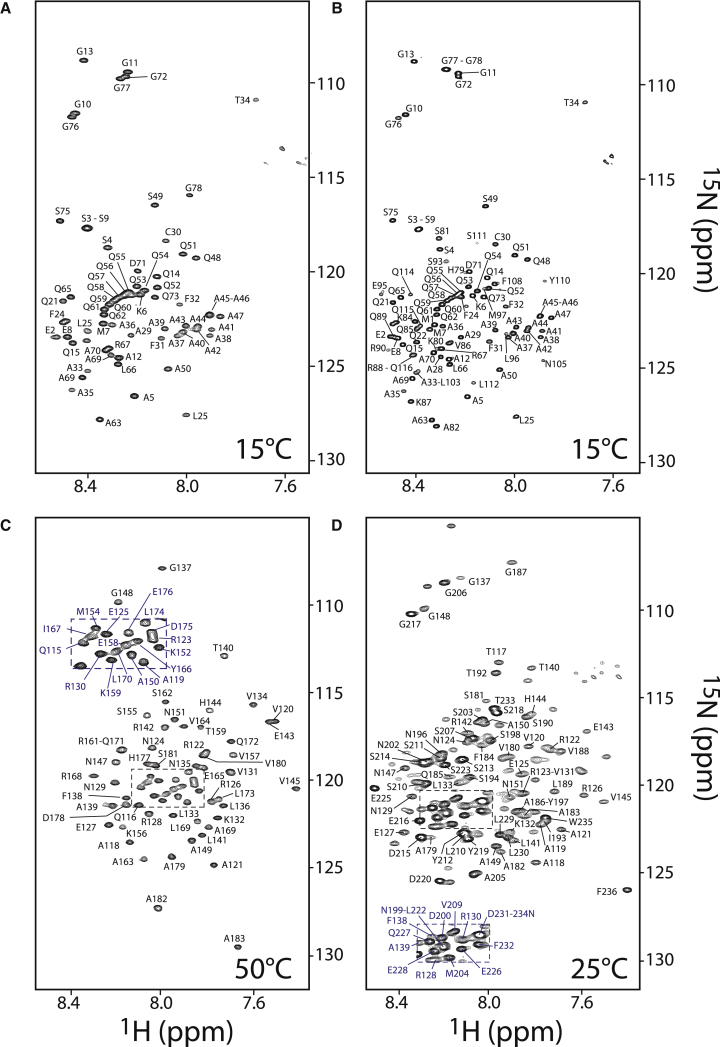
Figure 2.
1H-15N BEST-TROSY correlation spectra of the different Ascl1 fragments designed and used for this study. (A) Residues 1–78. (B) Residues 1–117. (C) Residues 114–183 (the bHLH domain). (D) Residues 118–236. The spectra shown were recorded at an optimized sample temperature at which the NMR assignment was performed. Correlation peaks in the different NMR spectra are annotated by the corresponding residue number and type. To see this figure in color, go online.
Two-dimensional (2D) 1H-15N correlation “fingerprint” spectra of all four Ascl1 fragments are plotted in Fig. 2. The poor spectral dispersion observed in these spectra for all protein constructs is indicative of highly flexible molecules that lack a persistent conformation in solution. The NMR spectra of the different fragments exhibited a better signal/noise (S/N) ratio and improved homogeneity in crosspeak intensities with respect to the full-length protein. In addition, the fragment-based approach allowed individual optimization of the sample temperature at which the series of NMR assignment experiments was performed. The fragment-based approach is validated by the fact that only small variations in chemical shifts were observed for residues that are part of different fragments, except for a few terminal residues that showed increased mobility.
NMR assignments and conformational dynamics in Ascl1
NMR backbone resonance assignments for all four Ascl1 fragments were obtained from a series of 3D BEST-TROSY (13, 21) triple resonance and HADAMAC (14) spectra recorded at 15°C (A and B), 50°C (C), and 25°C (D) on different high-field NMR spectrometers (700–950 MHz). Almost complete backbone (1H, 15N, 13CA, 13CB, 13CO) resonance assignment was obtained under these optimized experimental conditions. The 1H-15N assignments were then transferred to 25°C for all fragments for subsequent 15N relaxation experiments by recording a temperature series of 1H-15N BEST-TROSY spectra, helped by additional 3D HNCO and HN(CO)CACB experiments recorded at 25°C.
The N-terminal part of Ascl1 (fragments A and B) contains two homorepeats, the first one comprising 13 alanines in a row (polyA: residues 35–47), and the second comprising 12 glutamines (polyQ: residues 51–62). Despite this high degree of chemical equivalence, both homorepeats could be assigned in a sequence-specific manner. Interestingly, the polyQ segment forms a characteristic pseudodiagonal arrangement of crosspeaks in the two-dimensional (2D) 1H-15N correlation spectrum (Fig. 2, A and B). This feature has been observed before in polyQ-containing proteins (22) and related to the presence of helical propensity in the polyQ segment. Observing such a characteristic crosspeak arrangement therefore provides some structural information even before sequence-specific NMR assignment.
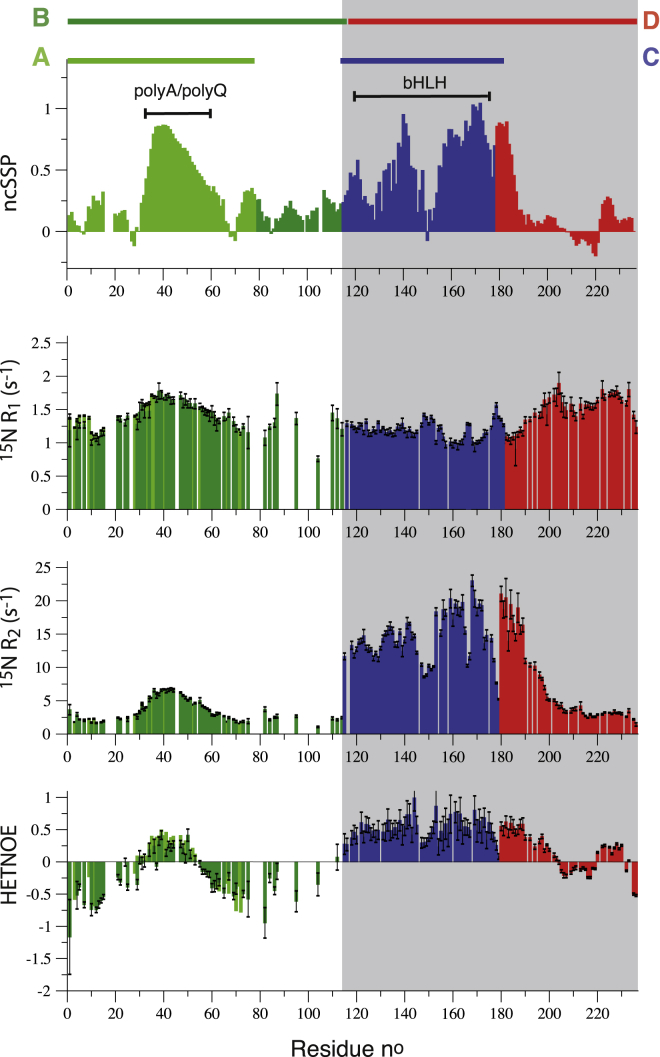
Neighbor-corrected secondary-structural propensities (ncSSP), computed from NMR chemical shifts (23) together with 15N relaxation parameters (R1, R2, HETNOE) are plotted in Fig. 3 as a function of the protein sequence. These data provide a comprehensive picture of the conformational sampling properties of the Ascl1 peptide chain. As predicted (Fig. 1), outside the polyA/polyQ region and the bHLH domain, Ascl1 shows a high degree of conformational disorder, with SSP values <0.3, low order parameters (small or even negative HETNOE values), and decreased R2 rate constants.
Figure 3.
NMR data collected for the four fragments of Ascl1, reporting on conformational features (structure and dynamics) along the polypeptide chain. From top to bottom are shown neighbor-corrected secondary structural propensity (ncSSP) values, calculated from the measured 13C chemical shifts, 15N R1 and R2 relaxation rate constants, and HETNOE intensity ratios. 15N relaxation data were recorded at 25°C, and the magnetic field strength was 600 MHz 1H frequency (700 MHz for fragment D). Data measured for the highly flexible terminal residues at the nonnative fragment ends are not shown. All graphs are color-coded to make clear that the underlying data originate from different NMR samples. White (grey) backgrounds indicate that the data have been measured on sampels without (with) detergent. To see this figure in color, go online.
The N-terminal polyA/polyQ region
The polyA/polyQ segment predominantly populates α-helical conformations that are highly stable at the N-terminal polyA side and become more and more flexible toward the polyQ end. Indeed, the SSP reaches its maximum (>0.8) at residues 37–43, in the central part of the polyA segment, suggesting that this fragment plays a key role in stabilizing the α-helical conformation. The polyA segment acts as a seeding point for α-helix formation in the subsequent polyQ stretch that otherwise would be preferentially found in a random-coil conformation (24). A similar finding has been made recently for the androgen receptor (22), where the polyQ segment in the N-terminal half of the protein adopts a helical conformation because of the presence of a preceding stretch of four leucine residues that act as a helix-inducer in the polyQ segment. The helical conformation is lost when the four preceding leucine residues are removed (22). The gradual (almost linear) decrease in helical propensity observed in the polyQ region most likely also explains the characteristic “pseudo-diagonal” crosspeak pattern observed for these residues. Such a regular pattern is not observed for the polyA α-helical fragment, although crosspeaks are also clustered in a quite narrow spectral region. The 15N relaxation data (R1, R2, and HETNOE) confirm that the polyA region adopts a highly stable structure, with HETNOE values close to 0.5 for the region encompassing residues 38–47, which rapidly decrease, becoming negative at the end of the polyQ tract.
The DNA-free bHLH domain
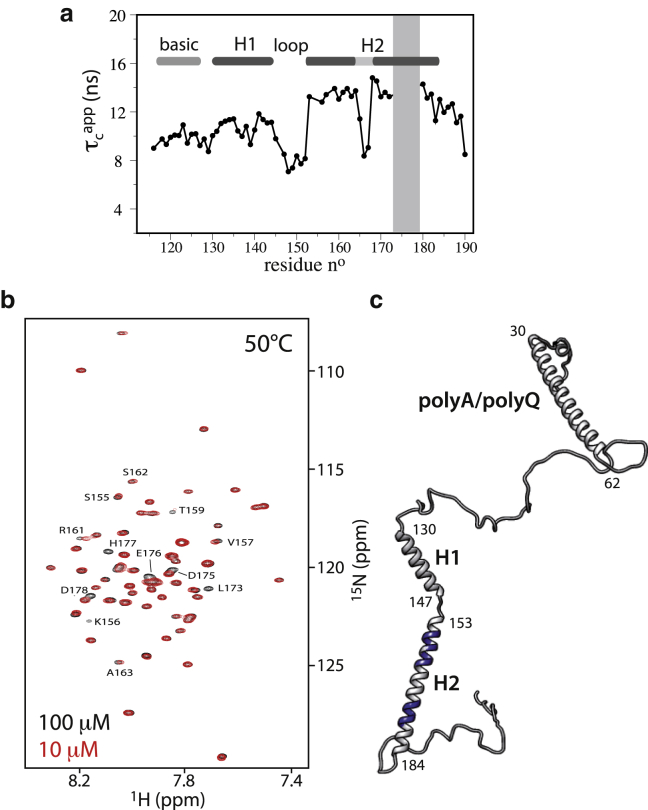
Previous studies of bHLH containing proteins have shown that the bHLH domain in the absence of DNA tends to be structurally disordered and aggregation prone, and that a stable dimeric conformation is only induced upon DNA binding (11, 20). In our hands, the bHLH domain (fragments C and D) of Ascl1 in buffered water solutions either precipitated or yielded soluble protein giving rise to NMR spectra of low quality, most probably due to heterogeneous protein aggregation. Only in the presence of millimolar amounts of detergent (SDS) was it possible to stabilize a homogeneous low-weight conformation that was amenable to NMR investigation. We observe no significant spectral changes when increasing the SDS concentration from 2 mM (below the critical micelle concentration (CMC)) to 40 mM (above the CMC). This indicates that the protein interaction with SDS is mainly of a hydrophobic nature (25). SDS has the combined effect of disrupting long-range intra- and intermolecular interactions, as well as stabilizing native-like helical structure. Although the presence of SDS is certainly not biologically meaningful, our results may give some insights into the physicochemical properties of this bHLH domain. In the presence of detergent, the bHLH domain adopts secondary structure that is similar to what is observed for DNA-bound bHLH domains (7). In particular, the second (C-terminal) helix (H2) is formed and rather stable, as can be deduced from SSP values close to 1 and high HETNOE ratios (∼0.7). This is consistent with AGADIR predictions that indicate a higher probability of helix formation for this part of the bHLH domain. Our data also indicate that the helix formation extends somewhat beyond the classical definition of the bHLH domain up to residue F184. Residue-specific local correlation times (τcapp) that can be computed from the measured R2 and R1 relaxation rates (Fig. 4 a) further indicate the presence of a flexible kink in this helical segment around residues 165–167. The first helix (H1) is structurally less stable, with the highest helical propensity found for residues 138–142. The DNA-binding basic region at the N-terminus also shows some tendency for helical structure, although with significantly lower propensity compared to the other helical elements. The loop region between helices 1 and 2 retains a high degree of local flexibility.
Figure 4.
NMR characterization of the bHLH domain in the absence of DNA. (a) Residue-specific apparent rotational correlation times computed from the 15N relaxation rate constants R2 and R1 as . (b) Overlaid 1H-15N correlation spectra (50°C) of Ascl1 fragment C recorded on samples of 100 μM and 10 μM concentration. Correlation peaks that show concentration-dependent peak shifts or line broadening are annotated by the corresponding residue number and amino acid type. (c) Graphical sketch of full-length Ascl1 representing an example of a transiently occurring α-helix-rich conformer. In addition, residues involved in dimerization, as identified in (b), are highlighted in blue. To see this figure in color, go online.
To obtain some information on the oligomerization state of the bHLH domain under our experimental conditions, we performed additional concentration-dependent NMR measurements. A superposition of 1H-15N correlation spectra of fragment C recorded at sample concentrations of 100 μM and 10 μM is shown in Fig. 4 b. Interestingly, extensive line broadening upon sample dilution is observed for residues 173–178 toward the end of the C-terminal helix, whereas small peak shifts are observed for most residues in the peptide region comprising residues 155–163 at the N-terminal side of the same helical segment (Fig. 4 b), and no significant changes are detected for residues in the central part of the helix. The concentration-dependent line broadening and NMR frequency shifts indicate that the C-terminal helix H2 is involved in an intermolecular exchange process, e.g., monomer-dimer equilibrium. We may speculate that the C-terminal end of the helix is the main interaction side, whereas the N-terminal part is involved in more transient interactions. This model also explains the peculiar motional properties observed for this helical segment (Fig. 4 a). To further characterize this intermolecular exchange process, we measured the rotational and translational diffusion properties of fragment C at 50°C at different sample concentrations (Fig. S2). Our data show that the average particle size is only very little affected by a 10-fold dilution of the protein solution. These findings point toward a model where a dimeric bHLH domain (with helix 2 at the interface) is in exchange with a minor population of a monomeric species. The population of monomeric bHLH increases by a few percent upon dilution, resulting in NMR peak shifts and line broadening without having a measurable effect on the average tumbling correlation time of the molecules.
We can thus conclude that the main structural features present in the DNA-bound form are already transiently present in the DNA-free bHLH domain: 1) two helical segments that are loosely coupled by a flexible linker; and 2) intermolecular interactions involving the C-terminal “dimerization” helix.
The C-terminal serine-and-proline-rich segment
The C-terminal part of Ascl1, which is part of the D fragment, contains a significant number of hydrophobic amino acids, including seven leucine residues, as well as half of the serine residues of full-length Ascl1 (12 out of a total of 25). Among these, five serines are neighbored by proline residues and are likely to be phosphorylation sites for proline-directed kinases (8). All of these putative phosphorylation sites are located in protein regions that are largely unstructured, thus facilitating interaction with the kinase. Not surprisingly, residues 180–184, which have been shown to adopt α-helical structure as part of the second helix of the bHLH domain (Fig. 3), lack such S-P motifs. Furthermore, the peptide region comprising residues 205–220, which shows some elongated average conformation in the nsSSP data (Fig. 3), contains five additional serine residues, which may be involved in posttranslational modifications according to predictions obtained with the NetPhos 3.0 software (26). It has been demonstrated recently for neurogenin-2 that multisite serine phosphorylation in the intrinsically disordered protein regions inhibits the neuronal differentiation activity of this bHLH-containing transcription factor, and that the number, and not the exact location, of phosphorylated residues is relevant for regulation (27). Our findings that the S-P sites and other PTM motifs are located in highly disordered regions suggest a similar role of these PTMs also in Ascl1. Interestingly, the second helix of the bHLH domain, extending up to residue 184, is followed by several phosphorylation sites. Given the sizeable dipole moment of a typical α-helical element, phosphorylation at these sites could result in destabilization of this helix and thus provide a mechanism for functional modulation (28).
Conclusions
Our NMR study, based on the NMR investigation of four overlapping protein fragments, presents, to our knowledge, the first high-resolution characterization of a proneuronal transcription factor. The overall picture emerging from our NMR data is that Ascl1 forms an extended polypeptide chain lacking persistent tertiary interactions, similar to a string of several loosely coupled peptide segments with a high propensity to adopt α-helical conformation. An example of an Ascl1 conformer with a high degree of secondary structure is depicted in Fig. 4 c. Such a conformer interconverts with many other less structured conformations on a subnanosecond timescale. In the absence of DNA as a binding partner, the helices of the bHLH domain, and in particular the N-terminal helix H1, are only transiently formed, and helix H2 undergoes intermolecular interactions, a feature that influences the binding affinity to the DNA targets. The exact role of the polyA/polyQ helical structure for transcriptional regulation and cell differentiation remains to be established.
Author Contributions
H.R.-A., A.D., I.C.F., R.P., and B.B. conceived the research; I.C.F., R.P., and B.B. directed the research; H.R.-A. designed the plasmid library of Ascl1 fragments; L.B., H.R.-A., E.O.C., and I.A. expressed the protein and prepared the samples; L.B., T.H., and S.G.-C. performed the NMR experiments and analyzed the data; L.B., I.C.F., R.P., and B.B. wrote the article. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
This work has been supported in part by the European Commission Projects IDPbyNMR (contract no. 264257), BioNMR (contract no. 261863), and INSTRUCT (contract no. 211252). We also acknowledge access to the high-field NMR and isotope-labeling platforms of the Grenoble Instruct Centre (Integrated Structural Biology Grenoble: UMS 3518 CNRS-CEA-UJF-EMBL) with support from the French Infrastructure for Integrated Structural Biology (FRISBI) (ANR-10-INSB-05-02) and the Grenoble Alliance for Integrated Structural Cell Biology (GRAL) (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB).
Editor: David Eliezer.
Footnotes
Two figures and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30239-4.
Contributor Information
Isabella C. Felli, Email: felli@cerm.unifi.it.
Roberta Pierattelli, Email: pierattelli@cerm.unifi.it.
Bernhard Brutscher, Email: bernhard.brutscher@ibs.fr.
Supporting Material
References
- 1.Imayoshi I., Kageyama R. Review and fate choice of neural progenitor cells. Neuron. 2014;82:9–23. doi: 10.1016/j.neuron.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 2.Wilkinson G., Dennis D., Schuurmans C. Proneural genes in neocortical development. Neuroscience. 2013;253:256–273. doi: 10.1016/j.neuroscience.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 3.Castro D.S., Guillemot F. Old and new functions of proneural factors revealed by the genome-wide characterization of their transcriptional targets. Cell Cycle. 2011;10:4026–4031. doi: 10.4161/cc.10.23.18578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y., Cai S., Chen Z. Intermolecular single-quantum coherence sequences for high-resolution NMR spectra in inhomogeneous fields. J. Magn. Reson. 2010;203:100–107. doi: 10.1016/j.jmr.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Vasconcelos F.F., Castro D.S. Transcriptional control of vertebrate neurogenesis by the proneural factor Ascl1. Front. Cell. Neurosci. 2014;8:412. doi: 10.3389/fncel.2014.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murre C., McCaw P.S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 7.Ma P.C.M., Rould M.A., Pabo C.O. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 8.Ali F.R., Cheng K., Philpott A. The phosphorylation status of Ascl1 is a key determinant of neuronal differentiation and maturation in vivo and in vitro. Development. 2014;141:2216–2224. doi: 10.1242/dev.106377. [DOI] [PubMed] [Google Scholar]
- 9.Künne A.G., Sieber M., Allemann R.K. Thermodynamics of the DNA binding reaction of transcription factor MASH-1. Biochemistry. 1998;37:4217–4223. doi: 10.1021/bi9725374. [DOI] [PubMed] [Google Scholar]
- 10.Sieber M., Allemann R.K. Single chain dimers of MASH-1 bind DNA with enhanced affinity. Nucleic Acids Res. 1998;26:1408–1413. doi: 10.1093/nar/26.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meierhan D., el-Ariss C., Allemann R.K. DNA binding specificity of the basic-helix-loop-helix protein MASH-1. Biochemistry. 1995;34:11026–11036. doi: 10.1021/bi00035a008. [DOI] [PubMed] [Google Scholar]
- 12.Künne A.G.E., Allemann R.K. Covalently linking BHLH subunits of MASH-1 increases specificity of DNA binding. Biochemistry. 1997;36:1085–1091. doi: 10.1021/bi962185l. [DOI] [PubMed] [Google Scholar]
- 13.Solyom Z., Schwarten M., Brutscher B. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J. Biomol. NMR. 2013;55:311–321. doi: 10.1007/s10858-013-9715-0. [DOI] [PubMed] [Google Scholar]
- 14.Lescop E., Rasia R., Brutscher B. Hadamard amino-acid-type edited NMR experiment for fast protein resonance assignment. J. Am. Chem. Soc. 2008;130:5014–5015. doi: 10.1021/ja800914h. [DOI] [PubMed] [Google Scholar]
- 15.Farrow N.A., Muhandiram R., Kay L.E. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 16.Lee D., Hilty C., Wüthrich K. Effective rotational correlation times of proteins from NMR relaxation interference. J. Magn. Reson. 2006;178:72–76. doi: 10.1016/j.jmr.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Morris G.A. Diffusion-ordered spectroscopy. Enzyclopedia NMR. 2009;9:35–44. [Google Scholar]
- 18.Dosztányi Z., Csizmók V., Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 19.Lacroix E., Viguera A.R., Serrano L. Elucidating the folding problem of α-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters. J. Mol. Biol. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- 20.Aguado-Llera D., Goormaghtigh E., Neira J.L. The basic helix-loop-helix region of human neurogenin 1 is a monomeric natively unfolded protein which forms a “fuzzy” complex upon DNA binding. Biochemistry. 2010;49:1577–1589. doi: 10.1021/bi901616z. [DOI] [PubMed] [Google Scholar]
- 21.Favier A., Brutscher B. Recovering lost magnetization: polarization enhancement in biomolecular NMR. J. Biomol. NMR. 2011;49:9–15. doi: 10.1007/s10858-010-9461-5. [DOI] [PubMed] [Google Scholar]
- 22.Eftekharzadeh B., Piai A., Salvatella X. Sequence context influences the structure and aggregation behavior of a PolyQ tract. Biophys. J. 2016;110:2361–2366. doi: 10.1016/j.bpj.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamiola K., Acar B., Mulder F.A. Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J. Am. Chem. Soc. 2010;132:18000–18003. doi: 10.1021/ja105656t. [DOI] [PubMed] [Google Scholar]
- 24.Masino L., Kelly G., Pastore A. Solution structure of polyglutamine tracts in GST-polyglutamine fusion proteins. FEBS Lett. 2002;513:267–272. doi: 10.1016/s0014-5793(02)02335-9. [DOI] [PubMed] [Google Scholar]
- 25.Bhuyan A.K. On the mechanism of SDS-induced protein denaturation. Biopolymers. 2010;93:186–199. doi: 10.1002/bip.21318. [DOI] [PubMed] [Google Scholar]
- 26.Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 27.McDowell G.S., Hindley C.J., Philpott A. Phosphorylation in intrinsically disordered regions regulates the activity of Neurogenin2. BMC Biochem. 2014;15:24. doi: 10.1186/s12858-014-0024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forman-Kay J.D., Mittag T. From sequence and forces to structure, function, and evolution of intrinsically disordered proteins. Structure. 2013;21:1492–1499. doi: 10.1016/j.str.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dosztányi Z., Csizmok V., Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.