Abstract
Over the past 20 years, more than 78,000 16S rRNA gene sequences have been deposited in GenBank and the Ribosomal Database Project, making the 16S rRNA gene the most widely studied gene for reconstructing bacterial phylogeny. While there is a general appreciation that these sequences are largely unique and derived from diverse species of bacteria, there has not been a quantitative attempt to describe the extent of sequencing efforts to date. We constructed rarefaction curves for each bacterial phylum and for the entire bacterial domain to assess the current state of sampling and the relative taxonomic richness of each phylum. This analysis quantifies the general sense among microbiologists that we are a long way from a complete census of the bacteria on Earth. Moreover, the analysis indicates that current sampling strategies might not be the most effective ones to describe novel diversity because there remain numerous phyla that are globally distributed yet poorly sampled. Based on the current level of sampling, it is not possible to estimate the total number of bacterial species on Earth, but the minimum species richness is 35,498. Considering previous global species richness estimates of 107 to 109, we are certain that this estimate will increase with additional sequencing efforts. The data support previous calls for extensive surveys of multiple chemically disparate environments and of specific phylogenetic groups to advance the census most rapidly.
INTRODUCTION
Estimating the diversity of life is a persistent challenge in biology. In microbiology, the task is complicated by the fact that the subjects of the census are not visible to the naked eye or easily differentiated morphologically, and they are estimated to number over 1030 individual bacteria worldwide (30). The properties of microorganisms necessitate the use of indirect analysis, involving culturing or 16S rRNA gene sequence analysis, to conduct a census of prokaryotes. Previous estimates of the number of bacterial species in the world range from 107 to 109 (6, 7). Although it is well accepted that the number of prokaryotic species in the world is immense and that our efforts to sample them have been inadequate, there has been no systematic analysis to assess how well we have sampled the bacterial world.
Estimating microbial phylogenetic diversity is intrinsically interesting to many microbiologists, but it also plays a crucial role in the functional analysis of microbial communities. Knowledge of the extent of phylogenetic diversity can indicate how many functional groups have not yet been accounted for. For example, 16S rRNA diversity surveys of terrestrial and marine ecosystems revealed that gene sequences belonging to the Acidobacterium phylum (14) and the SAR11 clade of the α-Proteobacteria (8), respectively, represented more than 25% of 16S rRNA sequences. These results have led to the development of improved culturing methods (13, 18). Likewise, Archaea were long thought to exist solely in “extreme” environments, but 16S rRNA gene sequencing analysis indicates that Crenarchaeota live in temperate soils (3, 26) and on the roots of plants (23). Although it is impossible to elucidate function based solely on phylogeny, study of certain groups will be particularly fruitful for the discovery of new examples of certain functions such as antibiotics in the actinobacteria and light-harvesting complexes in the cyanobacteria. It is clear that we are at a relatively early stage in sampling global species richness, only beginning the exploration of ecologically important but unidentified groups of microorganisms.
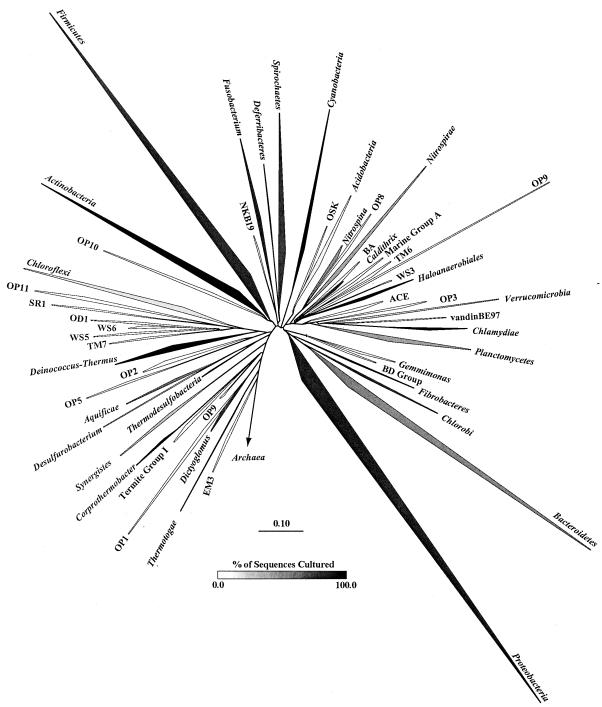
Since Woese and Fox (31) first proposed the 16S rRNA gene as a phylogenetic tool to describe the evolutionary relationships among organisms and Pace et al. (17) described its use for classifying unculturable microorganisms in the environment, over 78,000 16S rRNA gene sequences have been deposited in GenBank (19). These include sequences isolated from cultured bacteria (29) and those amplified directly from environmental samples without prior culturing (17). Sequences obtained by direct amplification from the environment provide the only information available for 99% of the prokaryotes in most natural communities (1). Recent studies have shown that there are at least 50 bacterial phyla, and half of them are composed entirely of uncultured bacteria (9, 10, 19). An additional three phyla contain less than 10% cultured members and six contain more than 90% cultured members (Fig. 1).
FIG. 1.
Phylogenetic tree of the Bacteria showing established phyla (italicized Latinized names) and candidate phyla described previously (9, 10, 19), using the November 2003 ARB database (http://arb-home.de [15]) with 16,964 sequences that are over 1,000 bp. The vertex angle of each wedge indicates the relative abundance of sequences in each phylum, and the length of each side of the wedge indicates the range of branching depth found in that phylum. The density of shading of each wedge corresponds to the proportion of sequences in that phylum obtained from cultured representatives. None of the candidate phyla have cultured representatives.
We sought to answer the exigent question: how complete is the census of prokaryotes as represented by the 16S rRNA sequence database? The answer will indicate which groups have been well sampled and which have not, providing guidance to future studies directed toward discovering new forms of life. We constructed rarefaction curves, which indicate the completeness of sampling for each phylum of Bacteria and for all Bacteria, using the curated 16S rRNA gene accessions in the Ribosomal Database Project-II database (5). We present evidence that argues that the traditional approach of blindly sampling interesting environments is limiting our attempt to census bacterial diversity. Based on the analysis presented here, we suggest complementing blind sampling with a more focused approach predicated on an assessment of which methods, environments, or taxonomic groups are most likely to yield new species in the future.
ANALYZING THE RIBOSOMAL DATABASE PROJECT
Sequence Data Set
The Ribosomal Database Project (RDP-II) is in its second generation of curating 16S rRNA gene sequences. As of the September 2003 release, the RDP-II had assembled a collection of 78,166 partial 16S rRNA gene sequences. The RDP-II collection contains a nondeconvoluted subset of those in GenBank that the RDP-II curators selected based on length and the number of ambiguous bases in each sequence (J. R. Cole, personal communication). The RDP-II assigns sequences to phyla and candidate phyla according to the nomenclature of Bergey's Manual Trust (http://www.cme.msu.edu/bergeys/). Each sequence in the RDP-II is aligned using a stochastic context-free grammar based on 16S rRNA secondary structure (5). We downloaded the 78,166 sequences of the September 2003 release of the RDP-II database as 35 separate files. Twenty-nine of the files contained sequences for individual phyla. The Proteobacteria phylum was divided into separate files for the α, β, γ, δ, ɛ, and unclassified subphyla. One file contained sequences that could not be assigned to a defined phylum. For the purposes of this analysis, we consider each file to contain sequences from one phylum. We selected aligned sequences that overlapped over the first 500 bp, yielding 56,215 sequences.
Construction of Rarefaction Curves
One method of comparing 16S rRNA sequences is to calculate distances between known and unknown sequences. Although these comparisons are approximations, distance values of 0.03 are thought to differentiate at the species level, 0.05 at the genus level, 0.10 at the family/class level, and 0.20 at the phylum level (10, 21, 24). While we appreciate that these distinctions are largely arbitrary and continue to be controversial (20, 28), they are useful for purposes of communication and comparison and are widely used (12). To simplify the text, we describe a species as a group of sequences that are all within a distance of 0.03 of each other. Using the supercomputer at the University of Wisconsin—Madison Genome Center and a desktop computer, we constructed distance matrices by using DNADIST from the PHYLIP package with the Jukes-Cantor correction for multiple substitutions (http://evolution.genetics.washington.edu/phylip.html).
We developed a computer program, DOTUR (Distance-based OTU and Richness) that uses a furthest-neighbor (complete-linkage) algorithm to assign sequences into operational taxonomic units (OTUs) and then constructs rarefaction curves for each distance level (http://www.plantpath.wisc.edu/fac/joh/dotur.html [22]). We used the distance matrices from DNADIST as input files for DOTUR.
INTERPRETING THE RICHNESS WITHIN THE RIBOSOMAL DATABASE PROJECT
Interphylum Comparisons
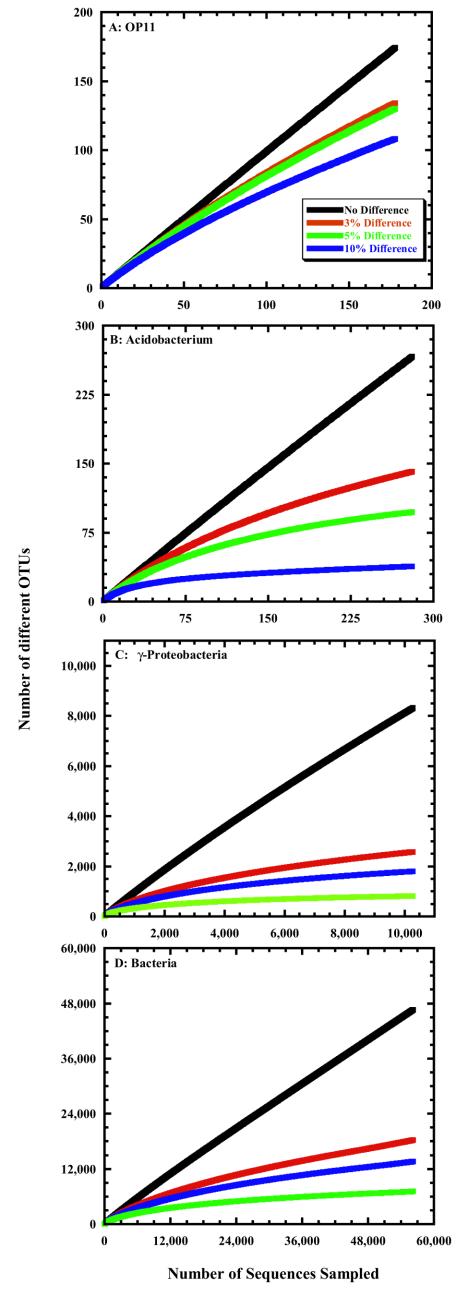
Construction of rarefaction curves for each phylum enabled us to compare the extent of sampling of each phylum at various taxonomic levels and their relative richness. For example, we compared rarefaction curves from OP11 (Fig. 2A), which currently has no cultured representatives (11), Acidobacterium (Fig. 2B), which is one of the most abundant phyla in soil but is difficult to culture (13), and γ-Proteobacteria (Fig. 2C), which is the most well-sampled and well-studied phylum, whose members include Pseudomonas spp. and E. coli (10). Each of these rarefaction curves indicates that the rate of discovering new sequences remains high for all phyla considered, although we are much further along in sampling the γ-Proteobacteria than any other phylum. As the likelihood of finding new sequences decreases, new methods of isolating sequences will be needed or new environments must be sampled to determine the completeness of the census.
FIG.2.
Rarefaction curves constructed with accessions from the RDP-II in the OP11 (A), Acidobacterium (B), and γ-Proteobacteria (C) phyla and for all 16S rRNA genes (D) at various distances. Each distance represents the maximum difference allowed for DOTUR to consider a group of sequences to be in the same OTU.
Although the bacterial phyla have been sampled to various extents and contain different numbers of sequences, it is possible to use rarefaction to determine differences in relative richness between phyla that would be observed if current sampling practices continued. OP11, which was recently shown to have a patchy distribution in various environments and is relatively sparse within those samples (9), has a relative species richness higher than that of the Acidobacteria but lower than that of the γ-Proteobacteria. We base this on the observation that the 178 sequences in the OP11 phylum contained 134 different species, and after the same sampling effort, the Acidobacteria sequences represented between 100 and 114 species (95% confidence interval) and the γ-Proteobacteria sequences contained between 143 and 162 species (P < 0.05). The relative species richness of the OP11 phylum is not significantly different from that of the β-Proteobacteria (95% confidence interval = 112 to 135 OTUs) and Planctomyces (95% confidence interval = 125 to 144 OTUs) phyla (P > 0.05) for the same sampling effort. Using similar reasoning, we found the relative species richness among the Acidobacteria and Cyanobacteria phyla to be similar. Finally, although the γ-Proteobacteria phylum contains the largest number of sequences, the Firmicutes, Verrucomicrobia, Bacteroidetes, and sequences that were not classified into a phylum each contain greater relative species richness. Rarefaction curves and data files for all of the bacterial phyla are available (http://plantpath.wisc.edu/∼pds/rdpproject.html).
Overall Rarefaction Curves
To construct a rarefaction curve by using the 56,215 partial 16S rRNA gene sequences in a single analysis, we combined the OTU data for distances between 0.00 and 0.10 from each phylum. The time and resources required to construct a single distance matrix for all of the sequences in the analysis would have been prohibitive. Therefore, we assumed that at a distance of 0.10, sequences from different phyla would not be similar, since it is thought that sequences from different phyla have a distance greater than 0.20 between them (10, 12, 21, 24). The merged OTU data were used in a modified form of DOTUR to construct rarefaction curves for various distance levels, using the entire database. Of the 56,215 16S rRNA gene sequences included in the analysis, 35,280 were identical to at least one other sequence.
As expected, the steep slope of the rarefaction curves for the entire data set (Fig. 2D) demonstrated that the census is far from complete. However, considering previous estimates suggesting that there are between 107 and 109 different species of bacteria (6, 7) and that the database contained only 56,215 sequences, we predicted that the species rarefaction curve (3% difference) would be steeper than we observed (Fig. 2D). If we assume that sampling strategies will continue to rely on the same strategies, it does not appear that the species-level curve will reach these estimates of global richness.
Statistical Census of Global Bacterial Richness
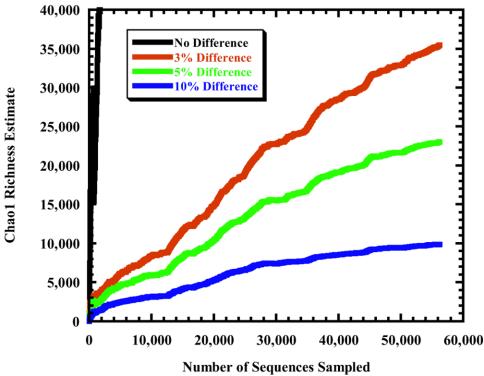
Nonparametric richness estimators permit a mathematical estimate of richness without requiring that each OTU be sampled (12). Assuming that RDP-II accession numbers reflect the order of sampling, we calculated the Chao1 richness estimator (4) for OTUs defined by no difference or no more than 3, 5, and 10% difference between sequences as a function of sampling effort (Fig. 3). The terminal Chao1 richness estimates for each OTU definition were 325,040, 35,498, 23,034, and 9,867 OTUs. Considering the steady rise in the richness estimate with sampling, these estimates are clearly minimum values of richness and should rise with increased sampling. Interestingly, after 26,000 sequences, the rate of change for the estimator at all levels decreased. This indicates that we may be getting closer to obtaining reasonable estimates of richness for OTU definitions near 10%. For the final 10,000 sequences collected, the rate of change for the Chao1 richness estimator was 5.1, 0.36, 0.20, and 0.06 new estimated OTUs per additional sequence sampled for each of the four OTU definitions, respectively.
FIG. 3.
Collector's curve of the Chao1 nonparametric richness estimator for sequences in the RDP-II. Accession numbers were used to determine the order in which sequences have been sampled. OTUs defined by a collection of identical sequences reached an estimate of 325,040 different OTUs.
Because the Chao1 collector's curves do not level off, it is difficult to determine how many more sequences would need to be sampled to obtain an accurate estimate of global richness. Although others have estimated global richness values ranging between 107 and 109 by using extrapolations from results of individual samples involving DNA-DNA hybridization (7) and theoretical models (6), this is the first attempt to calculate and assess the accuracy of a global statistical census of the number of bacterial taxa from real data collected from a large number of samples.
Caveat Emptor
It is important to be aware of several limitations in the analysis. First, investigators use different criteria when deciding which sequences to submit to the databases. Some researchers submit only the sequences that are different at a defined level of similarity compared to GenBank and RDP-II, others submit only the unique sequences within their libraries, and others deposit all the sequences they obtain. Although the RDP-II is not a systematic sampling effort, it is, in effect, an international collaborative attempt to catalog the Earth's microbial biodiversity. If each of the researchers who deposit sequences into the public databases viewed themselves as contributing to a global census, then perhaps we could achieve better standardization of the criteria used to deposit sequences (e.g., primers, sequencing coverage, chimera testing, and recording sample characteristics). Second, there is no standard set of PCR primers for amplifying 16S rRNA genes from the environment, and so there is not a common basis for comparison. Primer selection could be an important source of bias. The primers typically used to obtain full-length sequences were based on rRNA sequences from cultured organisms and therefore may bias environmental sampling toward sequences that are similar to those already in the database, creating a systematic bias in our sampling of genes that we can clone. Initial analysis of metagenomic sequencing efforts (25, 27) has suggested that standard primers used for amplifying archaeal 16S rRNA genes will not capture all archaeal sequences found by PCR-independent routes (2). The impact of these problems is not clear, and it is not obvious whether similar issues will emerge for bacterial primers. Finally, while the RDP-II screens sequences for length and quality, many of the sequences still have a considerable number of ambiguous bases, which reduce the observed diversity.
A final source of uncertainty in our analysis is the paucity of sequences in many of the phyla that lack cultured representatives. For example, there are only 148 OP11 and 197 Acidobacterium sequences in the RDP-II. Our present analysis predicts that OP11 species are as numerous as the γ-Proteobacteria and that the Acidobacteria phylum contains fewer species than the other two phyla, but additional sampling is necessary to increase our confidence in this prediction.
CONCLUSIONS
Based on the unexpected relatively flat slope of the bacterial rarefaction curves (Fig. 2D), we contend that either current sampling methods are not adequate to identify 107 to 109 different species of bacteria or these estimates are high. Periodic evaluation of sampling progress along a global collector's curve or Chao1 richness estimate curve will enable us to obtain a richness estimate that is based on this informal census effort. Furthermore, the relative paucity of sequences from candidate phyla such as OP11 limits the ability to measure their relative richness and potential biogeographical distribution. Clearly, sampling strategies must change and resources committed to this endeavor must vastly increase to describe the full diversity of bacteria on Earth and better appreciate the potential for discovering novel functional diversity.
Pace (16) issued a call to sequence 1,000 16S rRNA genes from each of 100 chemically disparate environments. Such a large-scale, intensive sequencing effort is essential to advance our progress along the rarefaction curve. These intensive sequencing efforts will certainly reveal novel phyla that make up a small proportion of communities and are therefore unlikely to be detected until many clones are sequenced. Intensive surveys of specific phyla will enhance our understanding of the biogeography and diversity of each phylum, as was reported by Harris et al. (9) for the OP11 phylum. It is clear from Fig. 1 that although there are many phyla that contain no cultured representatives, there are also poorly sampled phyla dominated by cultured representatives (e.g., Haloanaerobiales, Deferribacteres, and Coprothermobacter). Targeting poorly characterized phyla by using specific PCR primers should improve the efficiency of identifying 16S rRNA genes from novel species.
The National Science Foundation Microbial Observatories Program was launched in 1999 to “support research to discover and characterize novel microorganisms, microbial consortia, communities, activities and other novel properties, and to study their roles in diverse environments” (http://www.nsf.gov/pubs/2004/nsf04586/nsf04586.pdf). This program provides a means of substantially augmenting the microbial census and has accelerated the pace of discovery of new microbial species. To monitor progress toward a complete bacterial census, periodic analyses such as the one presented here should be conducted, and we suggest that an annual report on the “Status of the Microbial Census” would provide a guidepost for the field of microbial diversity.
Acknowledgments
We thank John Holt of the University of Wisconsin—Madison Genome Center Supercomputer facility for his assistance.
This work was supported by the NSF Microbial Observatories program (MCB-0132085), the Howard Hughes Medical Institute, the University of Wisconsin—Madison College of Agricultural and Life Sciences, and a USDA Soil Biology Postdoctoral Fellowship for P.D.S.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, B. J., G. W. Tyson, P. Hugenholtz, and J. F. Banfield. 2004. Analysis of genomic shotgun sequence data from an acid mine drainage biofilm community reveals a novel Euryarchaeota. Abstr. 104th Gen. Meet. Am. Soc. Microbiol. 2004, poster 161.
- 3.Bintrim, S. B., T. J. Donohue, J. Handelsman, G. P. Roberts, and R. M. Goodman. 1997. Molecular phylogeny of archaea from soil. Proc. Natl. Acad. Sci. USA 94:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 5.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dykhuizen, D. E. 1998. Santa Rosalia revisited: Why are there so many species of bacteria? Antonie Leeuwenhaek 73:25-33. [DOI] [PubMed] [Google Scholar]
- 8.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 9.Harris, J. K., S. T. Kelley, and N. R. Pace. 2004. New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 15.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 17.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1985. Analyzing natural microbial populations by rRNA sequences. ASM News 51:4-12. [Google Scholar]
- 18.Rappe, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 19.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 20.Rossello-Mora, R. 2003. Opinion: the species problem, can we achieve a universal concept? Syst. Appl. Microbiol. 26:323-326. [DOI] [PubMed] [Google Scholar]
- 21.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation- independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 22.Schloss, P. D., and J. Handelsman. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 23.Simon, H. M., J. A. Dodsworth, and R. M. Goodman. 2000. Crenarchaeota colonize terrestrial plant roots. Environ. Microbiol. 2:495-505. [DOI] [PubMed] [Google Scholar]
- 24.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence-analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 25.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 26.Ueda, T., Y. Suga, and T. Matsuguchi. 1995. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur. J. Soil Sci. 46:415-421. [Google Scholar]
- 27.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 28.Ward, D. M. 1998. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1:271-277. [DOI] [PubMed] [Google Scholar]
- 29.Ward, D. M., M. M. Bateson, R. Weller, and A. L. Ruff-Roberts. 1992. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv. Microb. Ecol. 12:219-286. [Google Scholar]
- 30.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woese, C. R., and G. E. Fox. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74:5088-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]