Abstract
Intracellular lipid droplets (LDs) are the main cellular site of metabolic energy storage. Their structure is unique inside the cell, with a core of esterified fatty acids and sterols, mainly triglycerides and sterol esters, surrounded by a single monolayer of phospholipids. Numerous peripheral proteins, including several that were previously associated with intracellular compartments surrounded by a lipid bilayer, have been recently shown to target the surface of LDs, but how they are able to selectively target this organelle remains largely unknown. Here, we use atomistic and coarse-grained molecular dynamics simulations to investigate the molecular properties of the LD surface and to characterize how it differs from that of a lipid bilayer. Our data suggest that although several surface properties are remarkably similar between the two structures, key differences originate from the interdigitation between surface phospholipids and core neutral lipids that occurs in LDs. This property is extremely sensitive to membrane undulations, unlike in lipid bilayers, and it strongly affects both lipid-packing defects and the lateral pressure profile. We observed a marked change in overall surface properties for surface tensions >10 mN/m, indicative of a bimodal behavior. Our simulations provide a comprehensive molecular characterization of the unique surface properties of LDs and suggest how the molecular properties of the surface lipid monolayer can be modulated by the underlying neutral lipids.
Introduction
Lipid droplets (LDs) are ubiquitous intracellular organelles that constitute the main storage site of metabolic energy inside the cell (1). As a consequence, they are implicated in many metabolic diseases, including obesity and lipodystrophy, as well as in the infectious cycle of human pathogens (2).
In LDs, energy is stored via esterification of free fatty acids and sterols, which are cytotoxic at elevated concentrations, in neutral lipids, mostly triglycerides (TAGs) and sterol esters. These neutral lipids are highly hydrophobic and can thus be efficiently packed in the absence of water, unlike sugars (1).
To prevent the energetically unfavorable direct exposure of these large fat assemblies to the aqueous environment of the cytosol, LDs are coated by a single monolayer of lipids (3). This molecular arrangement, consisting of a core of neutral lipids surrounded by a lipid monolayer, is unique inside the cell. As a result, LDs can be considered intracellular oil emulsions, and being able to correctly reproduce their interfacial physics, including surface tension (ST) and pressure, is paramount to understanding their molecular properties.
Recent evidence suggests that several proteins previously associated with other intracellular organelles, mainly the endoplasmic reticulum (ER) and the Golgi apparatus, also bind to LDs (4, 5, 6). From a molecular standpoint, these observations raise the question of how these proteins are able to bind membranes with such different molecular structures (a lipid bilayer for the ER or the Golgi versus a lipid monolayer surrounding a core of neutral lipids for LDs).
A common trait of many cytosolic proteins found to bind LDs is the presence of an amphipathic helix (AH) in their sequence (5, 7, 8, 9). Through a combination of cellular, biochemical, and in silico methodologies, it has been shown that binding of AHs to lipid bilayers (10, 11), as well as to ER and Golgi membranes (12), is driven by the presence of lipid-packing defects, i.e., interfacial voids at the membrane-water interface able to accommodate the binding of hydrophobic residues. Based on indirect observations from cellular studies (5, 13), a similar mechanism has been recently proposed for the targeting of peripheral proteins to LDs (14), but this model remains speculative, since no direct measurements are possible in vivo.
A more accurate evaluation of the role of LD properties in protein targeting has been drawn using an in vitro binding assay based on a microfluidic setup (15, 16). There, an increase in interfacial tension in reconstituted LDs was shown to promote the adsorption of the AH of α-synuclein (15). Since increasing interfacial tension entails an increase in area per lipid, the authors proposed that this could, in turn, have a direct effect on lipid-packing defects. However, a direct quantification of lipid-packing defects in model LDs is still lacking, and molecular dynamics (MD) simulations are the only methodology that is currently able to quantify them.
MD simulations of LD-like water/phospholipid/oil emulsions, however, have been relatively scarce so far, especially in comparison with those describing lipid bilayers. All-atom and united-atom (UA) simulations have mainly focused on general properties of LD-like trilayer systems (17, 18, 19), and a few coarse-grained (CG) studies focusing on LD composition (20, 21, 22) or LD formation (22) have been reported. Other CG studies (23, 24) describe the closely related (but of much smaller radius) high- and low-density lipoproteins (HDLs/LDLs). However, with the partial exception of one study investigating the binding of apolipoproteins to HDLs/LDLs (25), all these studies (17, 18, 19, 20, 23, 24) have characterized general properties of LD-mimicking systems without focusing on the LD surface and how its unique properties may drive selective protein adsorption. Of note, all CG studies have used the MARTINI force field, even though shortcomings of this model on interfaces (26), such as the presence of two negative peaks in the lateral pressure profile (LPP) at the oil/water boundary (27) and an unrealistically high interfacial tension in water/phospholipid/oil systems (25), have been reported.
In this work, we investigate the molecular properties of the surfaces of LD-mimicking systems using both UA and CG MD simulations based on the CG lipid model by Klein et al. (28, 29), after developing new parameters for TAGs. Our simulations indicate that the LD surface has various similarities to that of lipid bilayers with a comparable phospholipid composition, thus providing a rationale for why several proteins are able to bind to both LDs and bilayer-like cellular membranes, such as the ER or the Golgi apparatus. At the same time, the ability of TAGs to adopt multiple conformations and to interdigitate with the monolayer phospholipids, especially in response to variations in ST, induces significant differences in the LPP of LDs with respect to that of lipid bilayers. In turn, this has a marked effect on the LD surface, suggesting that the molecular properties of the surface lipid monolayer are strongly influenced by the underlying neutral lipids.
Materials and Methods
Atomistic simulations
Atomistic simulations were performed with GROMACS 4 (30) using the UA Berger force field for palmitoyloleoylphosphatidylcholine (POPC) phospholipids (31) and the simple-point-charge water model (32). For trioleins (TOs, tri-C18:1), the parameters adapted from Berger by Vattulainen et al. (33) were used (starting from a POPC molecule, the sn-3 was replaced by an oleoyl chain). For all oleoyl chains (POPC and TO), the correction on the double bond of (34) was applied.
Several systems were simulated (Table 1): 1) a liquid TO system (pureTO); 2) a POPC lipid bilayer (POPCbil); 3) TO/water (TO/W) and 4) TO/vacuum interfaces; and 5) five LD-like trilayer systems consisting of a mixture of TO, POPC, and water molecules at different STs ranging from 0 to 12.8 mN/m. A detailed description of how the different systems were constructed is given in the Supporting Material.
Table 1.
List of UA Simulations Performed in This Work
| System | Time (ns) | No. of TOs | No. of POPCs | No. of Waters | Acronym |
|---|---|---|---|---|---|
| pureTO | 150 | 108 | – | – | pureTO |
| TO/vacuum | 200 | 108 | – | – | TO/vac |
| TO/water | 200 | 108 | – | 5292 | TO/W |
| POPC bilayer NPT | 300 | – | 200 | 7943 | POPCbil |
| LD NPT | 300 | 204 | 200 | 7941 | LD0 |
| LD NPT 4× surface | 300 | 816 | 800 | 31,764 | LD0-4× |
| LD 1% area increase | 300 | 204 | 200 | 7929 | LD1 |
| LD 2% area increase | 300 | 204 | 200 | 7924 | LD2 |
| LD 5% area increase | 300 | 204 | 200 | 7915 | LD5 |
| LD 10% area increase | 300 | 204 | 200 | 7903 | LD10 |
ST is described here as a percentage of the increase with respect to the area of a bilayer (with the same number of POPC molecules) at equilibrium (thus at 0 ST). The corresponding values of ST in units of millinewtons per meter are given in Table 2. A snapshot of the LD0 system is presented in Fig. 1.
All UA simulations were analyzed with classical observables of lipid systems (area per lipid, order parameter, POPC monolayer thickness, and diffusion). Details of these analyses are given in the Supporting Material. Frames were saved every 100 ps, and trajectory analyses were performed on the last 200 ns for each simulation (the first 100 ns were systematically discarded from the analysis). All simulations were performed twice (or three times for LD10) and divided into three blocks. Except as otherwise stated, the final results are presented as an average over six blocks (three blocks times two trajectories), and the error is the corresponding standard deviation.
In addition, we also performed the following non-standard analyses.
Interdigitation
For all LD systems, interdigitation between TO and POPC molecules was evaluated as in Das et al. (35). Briefly, an overlap parameter, ρov, was evaluated from the density of TO and POPC (ρTO and ρPOPC, respectively):
| (1) |
The amount of interdigitation, λov, was then computed by integrating this quantity:
| (2) |
where L is the box length in the z dimension. λov has a unit of length scale and can be seen as the length over which interdigitation occurs: it equals 0 when no interdigitation is present, and can reach a few nanometers when interdigitation between the two lipid species is important. It is a meaningful quantity to compare interdigitation in the different systems. For the POPCbil system, interdigitation was evaluated between the two leaflets.
Classification of TO conformations
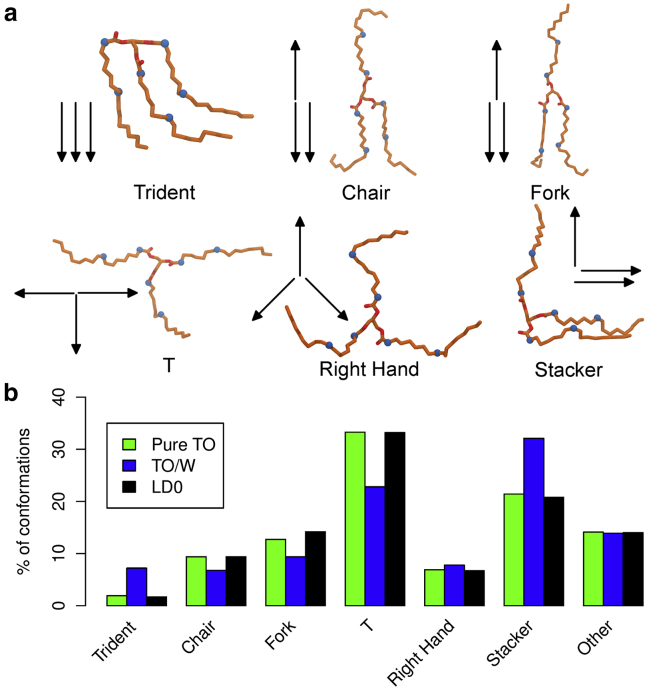
The new, to our knowledge, classification of TO conformations we propose in this study can be implemented as follows. For a given TO molecule, three unit vectors (vsn1, vsn2, vsn3) were defined going from each glycerol carbon up to the first carbon atom of the double bond (we did not go beyond, because of the high flexibility of the last carbon atoms of the tail). Then, the three scalar products, s12 = vsn1.vsn2, s13 = vsn1.vsn3, and s23 = vsn1.vsn3, describing the relative orientation of each chain pair were computed (each of them ranging between −1 and 1). Extreme cases of these scalar products were used to define conformational classes of TO that we called “Trident,” “Fork,” “Chair,” “T,” “Right Hand,” and “Stacker” (see Fig. 3). Note that some of the names from (36) were reused. For example, the ideal “Trident” has the three chains going toward the same direction, so the three scalar products equal 1. To assess the conformation of an arbitrary TO, we computed a Euclidean distance (in the space of the scalar products) to the ideal conformational classes:
| (3) |
where ideal is within {“Trident,” “Fork,” “Chair,” “T,” “Right Hand,” “Stacker”}, and sij(TO) and sij(ideal) are the scalar products of the arbitrary TO and the ideal conformation, respectively. The arbitrary TO was assigned the conformation with the lowest dideal. To avoid getting conformations too far away from these ideal classes, we added the restriction that dideal < 0.7. If all dideal > 0.7, the TO molecule was classified into a seventh class, which we called “Other.” On a typical trilayer simulation (LD0), ∼85–86% of the TO molecules were classified into one of the six well-defined classes {“Trident,” “Fork,” “Chair,” “T,” “Right Hand,” “Stacker”} using these rules. Hence, our classification gave a good overview of TO conformational populations and was a useful tool for comparing UA and CG simulations.
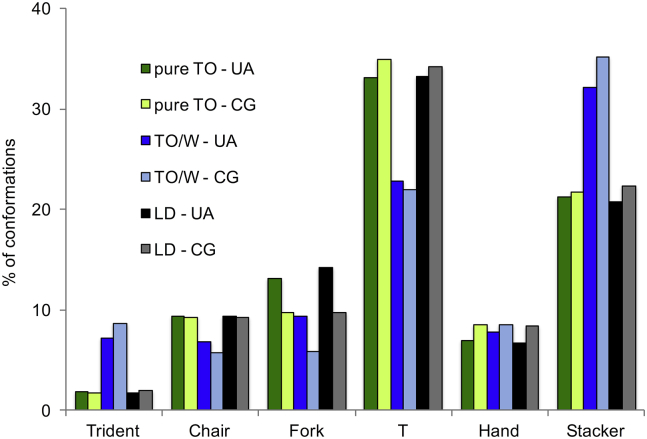
Figure 3.
Conformational analysis of TO molecules in UA simulations. (a) Snapshots of the different ideal conformational classes. Also shown are the names of the different classes and arrow sketches of them. (b) Comparison of the different TO conformational populations between three systems (pureTO, TO/W, and LD0). For all conformational classes and simulations, the error was systematically <0.2%. Numerical values and error are detailed in Table S11. To see this figure in color, go online.
Lipid-packing defects
Lipid-packing defects were computed using a previously described algorithm (10, 12). In brief, the membrane is mapped to a grid (with a granularity of 1 Å) parallel to the plane of the bilayer. Each grid point is scanned vertically starting from outside of the membrane toward its interior (water is excluded from the analysis) up to 1 Å below the sn-2 carbon atom of the nearest glycerol. If only lipid aliphatic atoms are encountered, the grid point is considered as an elementary packing defect of 1 Å2, whereas if any lipid polar atom is encountered, the grid point is not considered as a defect. We then divide elementary defects that are deep (below 1 Å of the glycerol level) or shallow (above the previous threshold). For each type, adjacent elementary defects are then clustered using a connected-component algorithm, and the area of each cluster is calculated (a packing defect is thus a cluster of elementary defects). This analysis is done for each leaflet separately. Statistics are then accumulated over all frames of a trajectory according to the same procedure. The obtained distributions are then fit to a mono-exponential decay:
| (4) |
where p(A) is the probability of finding a defect of area A Å2, πtype is the packing defect constant in units of Å2, type is within (deep, shallow), and b is a constant. The fit is done on defects >15 Å2 and for probabilities ≥10−4. For each type, the higher this constant, the higher the probability of finding large defects.
For LD systems, TO molecules were taken into account only if at least one of their atoms was above the imposed threshold along the normal to the membrane (Fig. S3).
ST, which is a central quantity in this article, is described here in detail. For all simulations, ST was computed from the diagonal values of the pressure tensor (Pxx, Pyy, and Pzz) using the Kirkwood-Irving method (37),
| (5) |
where L is the box length in the z dimension and angled brackets indicate an ensemble average. γ refers interchangeably to ST or interfacial tension in this article.
CG simulations
CG simulations were performed using the MARTINI force field (38) as well as the CG lipid model by Klein et al. (28, 29), which was used as a basis to derive new TO parameters, according to the procedure by Shinoda et al. (39).
MARTINI simulations were performed using the software GROMACS 4 (30), with POPC parameters taken from version 2.0 of the lipid force field (with five beads in the oleoyl tail) and TO parameters taken from (24).
Simulations using the CG lipid model by Klein et al. were generated by converting atomistic snapshots using the CG-it software (https://github.com/CG-it), and simulations were performed with LAMMPS (40). TO parameters were derived according to the procedure by Shinoda et al. (39) (see the Supporting Material for additional information).
For both force fields, several systems (liquid TO, POPC lipid bilayer, TO/W interface, and LD-like trilayer systems) were simulated. All the details (derivation of the model, system construction, simulation details, and analyses) are given in the Supporting Material, together with a list of all the simulated systems (Table S2). Average values and corresponding uncertainties were computed from block averaging over the last 400 ns of each trajectory with a block size of 100 ns and a time resolution of 100 ps.
Results
Structural and dynamic properties of LD systems assessed by UA simulations
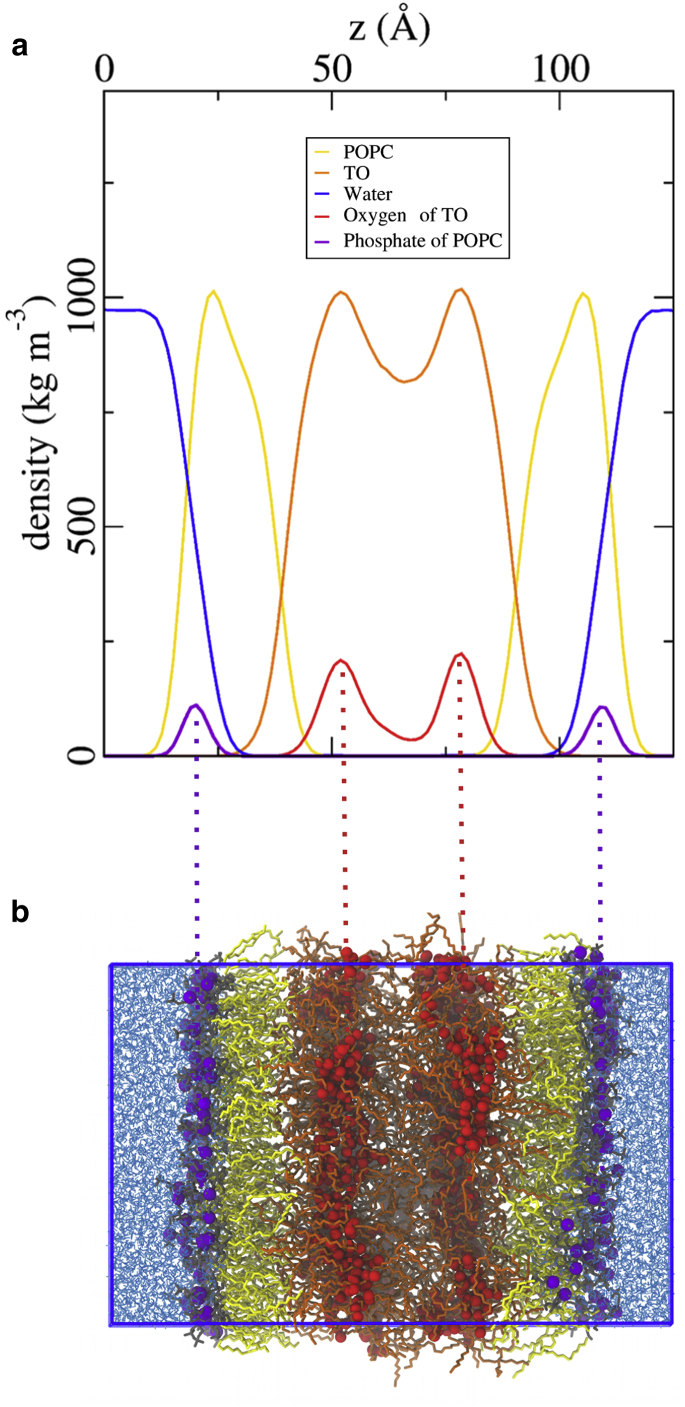
As a first step to obtain molecular insights into the structure of LDs, we built a system that could be tackled by UA simulations. Although LDs are generally spherical, their size is typically within the 100 nm–100 μm range (3, 41) and can thus be considered flat on the scale of proteins binding to it. The typical system for mimicking LDs in UA simulations is a trilayer (17, 18, 19), that is, a layer of neutral lipids sandwiched between two monolayers of phospholipids (Fig. 1). This set-up allows simulation of flat monolayers of phospholipids directly interacting with neutral lipids.
Figure 1.
Typical structure of an LD system using UA simulations. (a) Density plot corresponding to the snapshot of the LD0 trilayer system in (b). To see this figure in color, go online.
We chose a lipid composition commonly used to study LDs in vitro (42, 43, 44), namely POPC for the phospholipids and TO for the TAGs. The choice of pure TO was further motivated by two other arguments: 1) in regular LDs constituted of mixtures of TAG and sterol esters, TAGs are absolutely required to make the mixture liquid at physiological temperature (pure sterol esters have a melting temperature >37°C) (45); and 2) the TO/W interface is well characterized using biophysical experiments (42, 43).
We first compared the different properties of the POPC monolayer in the LD0 system and in a bilayer membrane (POPCbil) (first two lines of Table 2). Overall, the presence of TO molecules has only a marginal effect on the structural (area, monolayer thickness, and order parameter) and dynamic (46) properties of the POPC monolayer. In contrast, interdigitation between POPC and TO is quite substantial, with a 43% increase compared to the leaflet/leaflet interdigitation in a regular POPC bilayer (Table 2). It must be pointed out, however, that interdigitation is conceptually different between lipid bilayers (interdigitation between the two monolayers) and LD models (interdigitation between the POPC monolayer and TO molecules).
Table 2.
Comparison of Structural and Dynamic Phospholipid Properties between POPCbil and LD Systems in UA Simulations
| Area per Lipid (Å2) | ST (mN/m) | Diffusion (μm2/s) | Monolayer Thickness (Å) | Interdigitation (Å) | Order parameter |
|||
|---|---|---|---|---|---|---|---|---|
| Start | Middle | End | ||||||
| POPCbil | 66.1 ± 0.6 | 0.0 | 4.8 ± 0.6 | 12.7 ± 0.3 | 5.1 ± 0.1 | 0.18 | 0.17 | 0.09 |
| LD0 | 66.4 ± 0.4 | 0.0 | 5.3 ± 0.7 | 13.0 ± 0.4 | 7.3 ± 0.4 | 0.18 | 0.18 | 0.10 |
| LD1 | 67.3 | 2.9 ± 0.6 | 5.2 ± 0.1 | 12.8 ± 0.4 | 7.2 ± 0.4 | 0.18 | 0.17 | 0.10 |
| LD2 | 67.9 | 5.3 ± 0.3 | 5.8 ± 0.4 | 12.7 ± 0.5 | 7.1 ± 0.2 | 0.17 | 0.17 | 0.09 |
| LD5 | 69.9 | 9.2 ± 0.2 | 6.3 | 12.4 ± 0.4 | 7.3 ± 0.2 | 0.16 | 0.15 | 0.09 |
| LD10 | 73.3 | 12.4 ± 0.3 | 9.1 ± 0.5 | 12.4 ± 0.4 | 8.2 ± 0.6 | 0.15 | 0.14 | 0.08 |
MD runs for POPCbil and LD0 are performed in the NPT ensemble (ST = 0), whereas all other simulations (LD1–10) are run at constant xy area. All details on how to calculate these values and the relative uncertainties are described in the Supporting Material. When the uncertainties are not shown, their value is <0.1 mN/m for ST and below 0.1 μm2/s for diffusion.
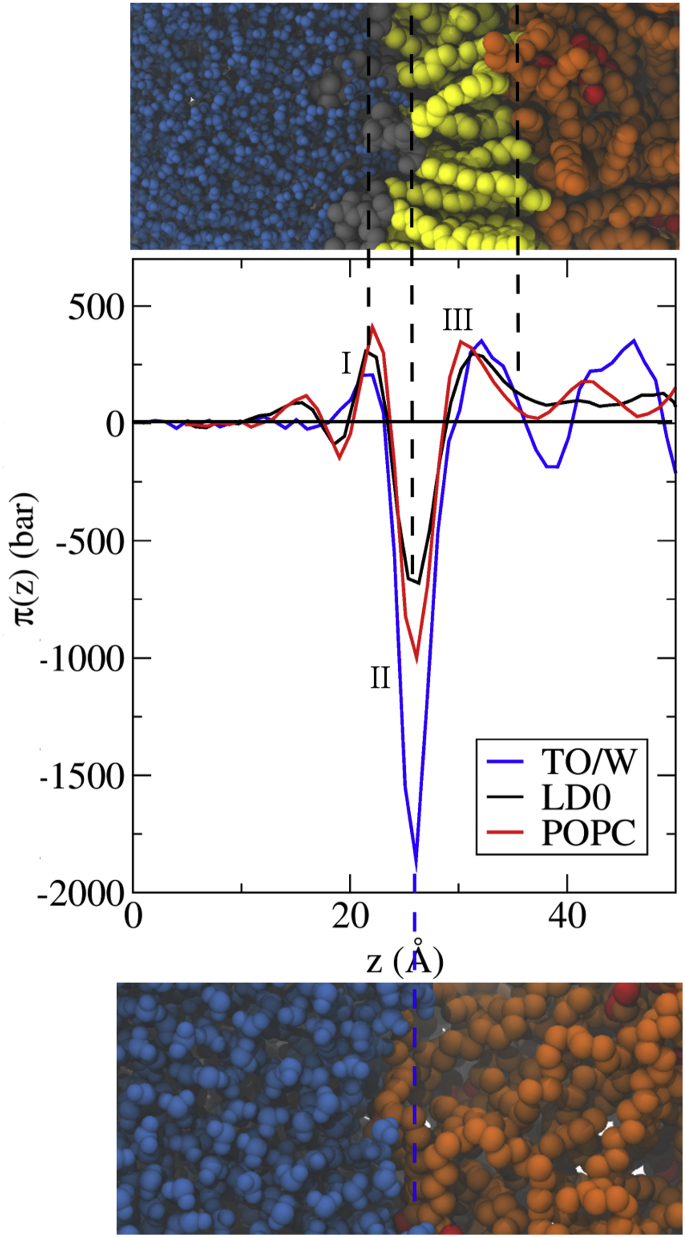
To further investigate the differences between a lipid bilayer and an LD ternary system, we next compared their respective LPPs (Fig. 2). The LPP π(z) is the difference between the lateral and normal components of the pressure tensor (see the Supporting Material), and it describes how the pressure changes along the normal to the interface. When the LPP is positive, the interface wants to expand; when it is negative, the interface wants to shrink.
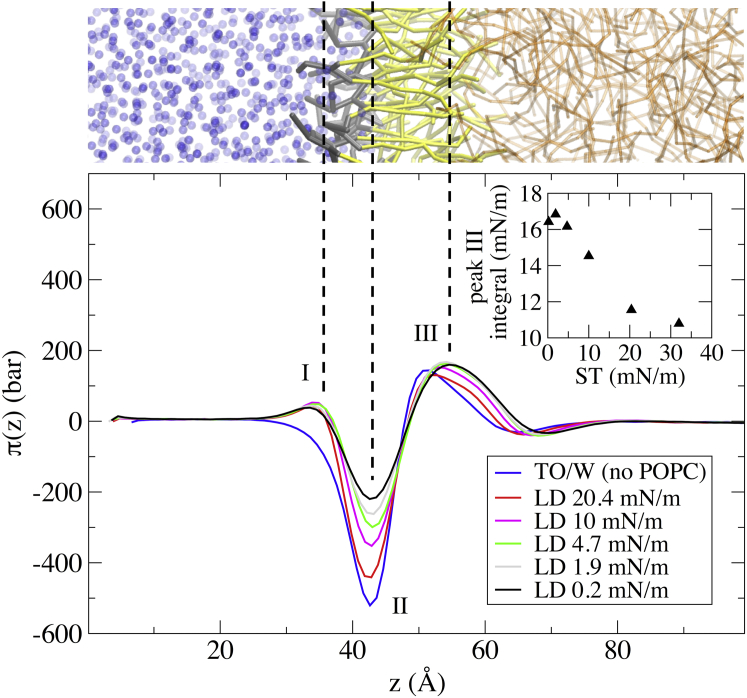
Figure 2.
Comparison between the LPPs of a POPC bilayer, an LD ternary system, and a TO/water interface in UA simulations. (Top) Snapshot of an LD interface. (Middle) LPPs aligned on the negative peak (II). (Bottom) Snapshot of the TO/W system. The two snapshots give an approximate view of where the different peaks occur. The three important peaks are labeled I, II, and III (see text). To see this figure in color, go online.
As can be seen in Fig. 2, the LPPs of LD0 and POPCbil are quite similar, which is consistent with the general properties of Table 2. The negative peak (II) is slightly lower for POPCbil compared to LD0, whereas the positive peak of the polar heads (I) is slightly higher for POPCbil than for LD0. This means that the lateral stresses in the LD system are slightly lower than in a POPC bilayer. We attribute this difference to interdigitation. The positive peak (III) describing the repulsion of the aliphatic chains is also slightly higher for POPCbil than for LD0.
Next, to investigate the spontaneous curvature of the LD monolayer, we computed the first moment of the LPPs (see the Supporting Material). This value, according to the continuum theory of membranes, corresponds to the product of the monolayer bending constant, kc, and the spontaneous curvature, c0. Since kc is positive by definition, the sign of c0 is straightforward to obtain from the first moment of the LPPs. In our MD simulations, the lipid monolayer has a slightly negative curvature in both the POPCbil and the LD0 systems. These results are consistent with the experimental observation that POPC has a moderate negative curvature (47), and they further confirm that the presence of the neutral lipids barely affects most of the monolayer properties (see Table 2). However, further and more detailed investigations will be necessary to properly assess the relevance of monolayer curvature in biological processes involving LDs.
Looking at the TO/W LPP is also instructive: the negative peak (II) is much deeper, and there is no surfactant like POPC that could act to alleviate ST. Instead, we observe a small positive peak (I) that is probably due to water ordering in the vicinity of the interface (this small peak is also present for LD0 and POPCbil, to the left of peak I).
One other important feature in all LD systems is that the glycerol atoms of TO molecules tend to cluster together regardless of the ST. This is clearly seen in the snapshot of Fig. 1 b and translates to two peaks in the density profile (Fig. 1 a). Since TO molecules are not purely hydrophobic (unlike alkanes), this clustering is thus not entirely surprising. Driven by the hydrophobic effect, the system minimizes the contacts between hydrophobic and hydrophilic moieties. In our trilayer systems, the glycerol moieties of TO molecules form flat layers (Fig. 1). However, in the pureTO system, glycerols also cluster together, but they do not form layers (Fig. S7). This finding suggests that the ordering of TO molecules is likely to be induced by interactions with the neighboring POPC monolayer. Finally, this observation was confirmed by CG simulations: in the presence of a higher amount of TO (Fig. S8), the middle of this layer approaches what could be the core of a real LD, that is, TO molecules cluster together but without any specific order. as in the pure liquid state. In the remainder of this article, we call these TO molecules “bulk” TO. Notably, no appreciable differences for what pertains to LD surface properties, including interdigitation, are observed when varying the number of TO molecules.
In agreement with previous simulations (20, 21), we observed a significant hydration of the TO layer in all LD systems. Although our MD simulations were started without any water in this hydrophobic portion, we observed water molecules going in and out of the TO compartment by crossing both POPC monolayers. Once in the TO compartment, water molecules could reside a few tens of nanoseconds by making hydrogen bonds with the TO glycerol atoms (Fig. S9). The number of water molecules inside the TO was fairly constant (∼10 for 204 TO and 200 POPC) with a turnover of molecules coming in and out (Table S10), and it showed no dependence on ST. This value corresponds to a water content of ∼1 g per kg of oil, and it is consistent with experimental measurements of water content in extra virgin olive oils (in which TO is the major component) (48).
Conformational analysis of TO molecules assessed by UA simulations
To assess whether the conformational flexibilty of TO molecules could play a role in modulating LD surface properties, we next investigated whether they could adopt different conformations depending on the environment: liquid, at a hydrophobic (TO/vacuum) or hydrophilic (TO/W) interface. Since TO molecules can be very flexible at physiological temperature, we expanded the classification of TO conformations first proposed by Brasiello et al. (36) using simple geometric rules to describe the relative orientation of their hydrophobic tails. The results are presented in Fig. 3 for pureTO, TO/W and LD0 (see Table S11 for the other conditions).
The first interesting feature is the relative equality of these populations between pureTO and LD0 systems (of note, increasing ST has a very small effect, if any, on these conformational populations; see Table S11). This observation suggests that the presence of phospholipids near the TO molecules does not change their behavior conformation wise, as if the oil phase of the LD was close to the liquid state (bulk TO). In a sense, this demonstrates that a phospholipid monolayer is a good “container” for oil; the horizontal TO clustering underneath the POPC monolayer (Fig. 1) thus has no effect on TO conformations.
In pureTO and LD systems, the most highly populated conformation is “T” (∼33%), which could represent the main part of bulk TO. Next, we find “Stacker” (∼20%), “Fork” and “Chair” (∼10%), then “Right Hand” and “Trident” (<10%). Of note, because of symmetry, some conformations are more likely to occur because they can arise through multiple ways, whereas others, such as “Tridents,” can only form in one way.
Importantly, the number of “Trident” conformations increases by 350% going from LD0 or pureTO to the TO/W system. “Stacker” conformations also increase by 59%. These increases occur mainly at the expense of “T” and, to a lesser extent, “Chair” and “Fork.” Although the “Trident” should be entropically unfavorable, this conformation is most suited to a hydrophilic interface, since it allows the glycerol moiety to be exposed to water. This explains its abundance in TO/W. The “Stacker” conformation is also favored, for the same reason but to a lesser extent. Exposing TO polar atoms to water is an efficient way to alleviate ST: TO/W ST at 20°C is ∼32 mN/m (42), whereas dodecane/water ST is >50 mN/m (49).
Effect of ST on surface properties of LD systems assessed by UA simulations
Unlike in lipid bilayers, monolayers can undergo extensive stretching (with major variations in area per lipid) at the cost of increasing ST. ST, sometimes called also interfacial tension for liquid/liquid interfaces, is a key parameter to describe emulsions. We thus investigated the effect of increasing ST on our systems (Table 2). The range of ST in our simulations is consistent with that typically used in in vitro experiments (0–20 mN/m) (15, 50).
As expected, POPC monolayer thickness decreases for increasing ST (see Table 2); this behavior can be explained by the larger lateral space accessible to the aliphatic chains at high ST. Interestingly, the other properties (diffusion, interdigitation, and order parameters) do not change much for moderate ST (LD0–LD5 systems translating from 0 to ∼7.7 mN/m) but can substantially vary for LD10 (corresponding to ∼12.8 mN/m). This is especially significant for interdigitation, which remains constant, at ∼7.2 Å, from 0 to 7.7 mN/m, and “jumps” to 8.2 Å for an ST of ∼12.8 mN/m.
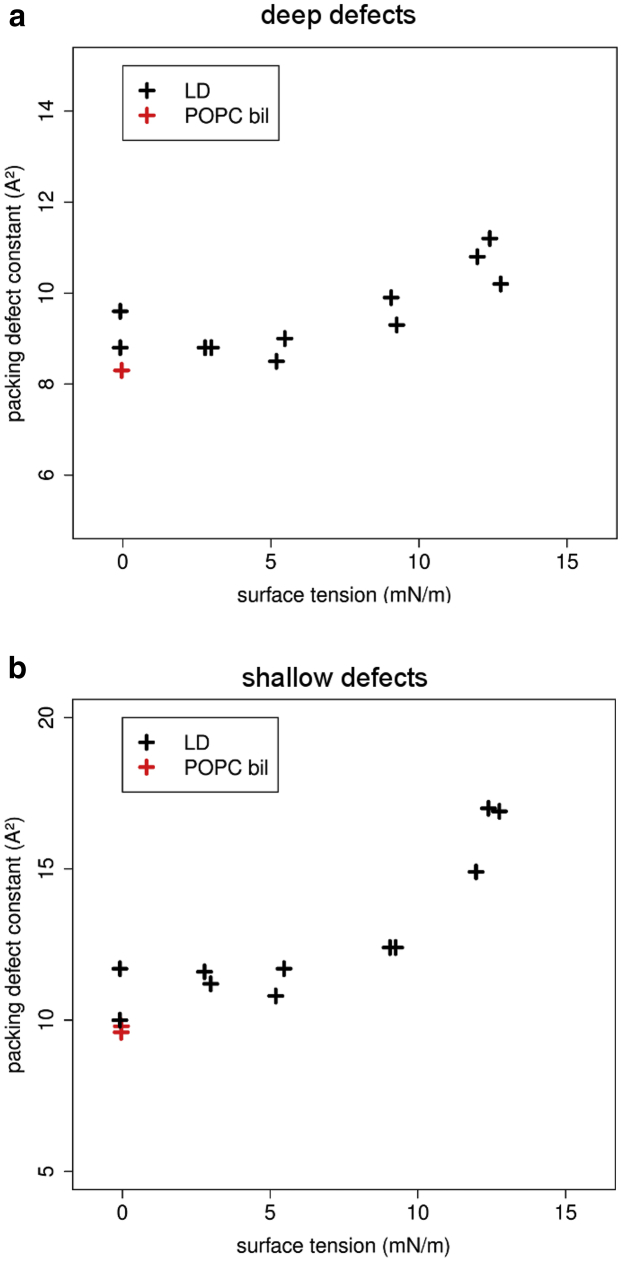
Finally, we determined the presence of interfacial lipid-packing defects in model LDs, also as a function of ST, with the aim of comparing these defects to those evaluated in lipid bilayers of different compositions (12). As previously described (51), we distinguish between two different types of defects: 1) deep packing defects, i.e., those that extend below the glycerol level of the lipid monolayer, and 2) shallow packing defects, i.e., those that emerge at the surface and are above the glycerol level of the lipid monolayer. Previous studies have shown that these defects facilitate the binding of AHs with different chemistries, with AHs with large hydrophobic residues preferentially binding to membranes enriched in deep packing defects and AHs with small hydrophobic residues preferentially binding to membranes enriched in shallow packing defects (51, 52).
The results are presented in Fig. 4. UA simulations show that both deep and shallow defects tend to slightly increase from POPCbil to LD0. This increase probably originates from a higher interdigitation (Table 2) between TO and POPC compared to leaflet/leaflet interdigitation in a POPC bilayer. Lipid-packing defects also increase for increasing ST. This behavior is not unexpected, since area per lipid also increases as a function of ST (Table 2). However, this increase in lipid-packing defects is very modest up to ∼10 mN/m but becomes significant above this value, especially for shallow packing defects. For reference, values >9–10 Å2 for deep defects are indicative of membranes that typically show a high avidity for peripheral proteins (10, 12, 53). Interestingly, this increase in lipid-packing defects at high ST correlates well with the increase in interdigitation.
Figure 4.
Packing-defect constants as a function of ST in UA simulations. (a) Deep packing defects. These defects extend at least 1 Å below the average z-level of glycerol atoms. (b) Shallow packing defects. These defects are above the set threshold of 1 Å below the average z-level of glycerol atoms. The higher the packing-defect constant, the higher the probability of making large defects (10). In this figure, each cross represents an average of both ST and packing-defect constant over one simulation. For better clarity, uncertainties are not represented. Tabulated values and uncertainties for each simulation are detailed in Table S12. To see this figure in color, go online.
Whereas the existence of a bimodal regime would be of marked interest to understanding protein binding to LDs, our UA simulations are still limited in terms of sampling and system size, especially for what concerns the calculation of lipid-packing defects at different STs. We thus moved to CG simulations to tackle these two issues and to simulate larger systems over longer timescales.
Structural properties of LD systems assessed by CG models
To overcome sampling issues, we next investigated model LDs using CG MD simulations. We first used the MARTINI force field (38), since this model has been previously used to characterize model LDs (20, 21, 25) or nascent LDs (22).
To assess the extent to which this model is able to describe the complex chemical composition of model LDs, we first computed a number of structural properties and compared them with the results obtained for analogous simulations at atomistic resolution, as described in the previous sections.
As shown in Table 3, several key properties (area per lipid, deep lipid-packing defects, hydration, etc.) are faithfully reproduced by this model. Also, MARTINI appears to be able to correctly describe the preferential conformation of TAGs inside the model LDs, as well as in pure TO systems devoid of the phospholipid monolayers (Table S13).
Table 3.
Comparison of LD Properties between Atomistic and CG Simulations
| Area per Lipid (Å2) | Monolayer Thickness (Å) | Interdigitation (Å) | Deep Lipid-Packing Defects (Å2) | Shallow Lipid-Packing Defects (Å2) | Water/Oil Content (g/kg) | |
|---|---|---|---|---|---|---|
| UA model | 66.4 ± 0.4 | 13.0 ± 0.4 | 7.3 ± 0.4 | 9.2 ± 1.4 | 10.9 ± 1.1 | 0.9 ± 0.2 |
| MARTINI | 65.4 ± 0.4 | 15.2 ± 0.6 | 14.8 ± 0.4 | 9.3 ± 0.2 | 16.1 ± 0.2 | 0.8 ± 0.8 |
| CG model (SDK) | 66.2 ± 0.5 | 13.4 ± 0.5 | 8.1 ± 0.1 | 9.2 ± 0.2 | 12.5 ± 0.2 | 1.5 ± 0.6 |
However, both interdigitation and shallow lipid-packing defects are substantially overestimated using this model (Table 3). These findings, together with previously reported shortcomings of MARTINI when dealing with monolayers at both air/water (26, 27) and oil/water (25, 27) interfaces, suggest that this model may not be optimally suited for investigating surface molecular properties of ternary LD-like systems.
We next tested the CG model based on the parameterization method by Klein et al. (28). The method has been put forward to study lipids and surfactants (54, 55) and it is based on a mixed 3:1 and 2:1 heavy atoms/CG beads mapping, so it features a slightly higher resolution than the MARTINI model, which mostly uses a 4:1 mapping. In addition, the Shinoda-DeVane-Klein (SDK) parameterization method (39) fits non-bonded force-field parameters against interfacial tension measurements and is therefore particularly well suited to study emulsions (28).
To simulate LD model systems, we developed CG parameters for TO molecules according to the general procedure of the SDK model (28) (see the Supporting Material text and Fig. S4). In particular, bonded parameters were derived from the UA simulations of pure TO systems, whereas non-bonded parameters (σij, εij) were taken from the analogous groups in phospholipids, with the exception of interactions between TO ester groups and water molecules, which were tuned to reproduce the interfacial tension of TO molecules with water. Using these parameters, we could correctly reproduce both available experimental properties (Table 4), as well as the conformation of TO molecules in pure TO and TO/W systems, as observed in UA simulations (Fig. 5; Table S14).
Table 4.
Comparison between Computed and Experimental Values Using Our CG Model Based on SDK Parameterization
Figure 5.
Conformational analysis of TO molecules in CG-SDK simulations. Shown is a comparison between the CG and UA analyses of TO conformational populations in the three main systems discussed in the text: pureTO, TO/W, and LD. With the sole exception of “Fork” conformations, the CG model reproduces accurately all of the UA populations. For all conformational classes and simulations, the uncertainty was systematically <0.2%. Numerical values are detailed in Table S14. To see this figure in color, go online.
Next, we prepared LD-mimicking TO-POPC-water systems. Again, we could reproduce both global and structural properties of LD systems computed using UA simulations (Table S14). The only substantial difference between the SDK CG model and its UA counterpart is a somewhat less ordered distribution of TO molecules inside the LD core, as can be appreciated from the lateral density profiles (Fig. S8) and from the LPPs of the pure TO/W systems (Figs. 2 and 8, blue lines). This discrepancy may originate from excessively low entropic barriers in the CG model, as well as from an inadequate TO depth along the membrane normal (z) in the UA simulations.
Figure 8.
Effect of ST on the LPPs of model LDs in CG-SDK simulations. (Top) Snapshot of an LD interface. (Bottom) Lateral pressure as a function of the ST. The positive peak between the polar heads of phospholipids (I) is not affected by ST, whereas major changes can be observed in the negative attractive peak at the hydrophilic/hydrophobic interface of POPC molecules (II). Also, variations in height and shape can be observed in the positive peak between the POPC acyl chains and the TO molecules (III), in particular for values >10 mN/m (purple and red lines). The integral of this peak (inset) is quite constant for low ST but shows a substantial decrease from 10 mN/m to higher STs. The LPP of a TO/W interface using the CG model is shown for comparison. To see this figure in color, go online.
On the other hand, with the sole exception of “Fork” conformations, which are slightly (22%) underestimated by the CG model, all other TO conformations, as well as their variations depending on the environment (pure TO system, TO/W interface, or LD system), are quantitatively reproduced with minimal errors (Fig. 5).
Surface properties of LD systems assessed by CG simulations
We next focused on the surface properties of LDs, notably the characterization of lipid-packing defects. As a first step in investigating whether this CG model is able to correctly reproduce this property in ternary systems, we computed lipid-packing defects in a configuration that was directly derived from the UA LD simulations described above. Notably, we find that this CG model is able to faithfully reproduce both deep and shallow defects from UA simulations (Table 3).
However, when doubling the lateral dimensions of the simulation box (increasing the number of POPC molecules per leaflet from 100 to 400), we observed that deep defects in LDs decreased (from 9.2 ± 0.2 Å2 to 8.1 ± 0.1 Å2), whereas shallow defects increased (from 12.5 ± 0.2 Å2 to 14.1 ± 0.1 Å2). To rule out the possibility that this effect was an artifact of the CG force field, we performed an analogous UA simulation in which we also increased by four times the interfacial surface of our LD model. In agreement with the CG model results, in UA simulations, we also observed a decrease in deep lipid packing defects (from 9.2 ± 1.4 Å2 to 7.5 ± 0.1 Å2) and an increase in shallow ones (from 10.9 ± 1.1 Å2 to 11.8 ± 0.3 Å2). Remarkably, this effect was not observed in lipid bilayers, where increasing the lateral dimension had no effect on lipid-packing defects in either UA or CG simulations.
We hypothesized that these size-dependent effects may originate from a different response of the ternary LD systems, compared to those of a lipid bilayer, to the suppression of long-wavelength undulations that occurs with decreasing system size. To test this hypothesis, we first confirmed that decreasing the lateral (x,y) dimensions decreases undulations (Fig. S15) by measuring the fluctuations of the POPC phosphate groups around their equilibrium z coordinate (normal to the interfacial plane). Next, we computed the interdigitation between TO molecules and surface POPC lipids, and we found that increasing the lateral dimensions substantially increases interdigitation in LD systems (from 8.1 ± 0.1 Å to 10.4 ± 0.3 Å in CG and from 7.3 ± 0.4 Å to 9.3 ± 0.4 Å in UA simulations). This increase of interdigitation should also be facilitated in LD systems, because the oil could act as a “reservoir” and adapt to the shape of the POPC leaflet, unlike in a lipid bilayer, where the two leaflets are coupled.
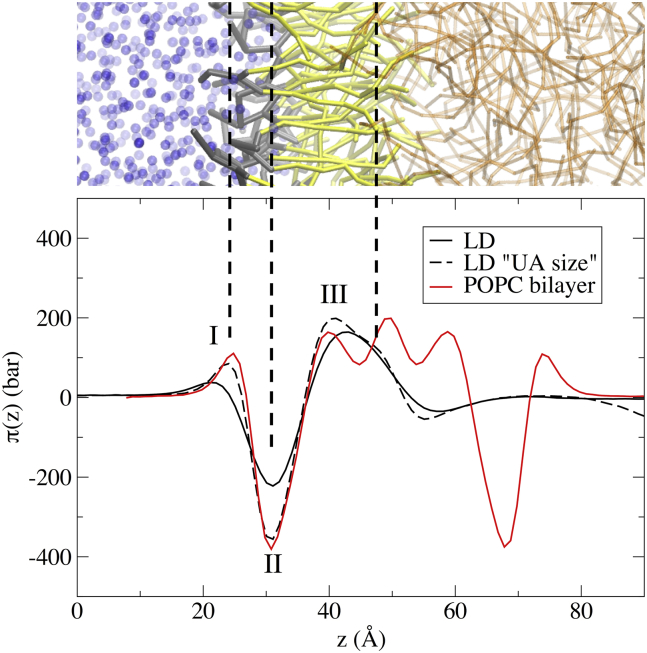
Finally, we computed the LPP for the two LD systems of different sizes and compared them to that of a pure POPC bilayer. As shown in Fig. 6, the LPPs were substantially different when the lateral dimensions of the simulation box were increased, with the smaller system (Fig. 6, black dotted line) showing a pressure profile much more similar to that of a POPC bilayer (Fig. 6, red line). In particular, increasing the lateral dimensions of the box led to a decrease in all the prominent pressure-profile peaks, both repulsive and attractive, despite the fact that there were no changes in the overall ST (which remains negligible in all cases). Regardless of system size, however, the spontaneous curvature of the lipid monolayer (as assessed from the first moment of the LPPs), remained negative both in the POPC bilayer and in the LD simulations, in agreement with the values obtained in the UA simulations. However, the capability of the SDK CG model to reproduce lipid spontaneous curvature has never been comprehensively established, and further studies will be needed to address this aspect.
Figure 6.
Size dependence of LPPs in LD systems in CG-SDK simulations. (Top) Snapshot of an LD interface. (Bottom) Lateral pressure along the membrane normal. Two positive peaks (repulsion between the polar heads of phospholipids (I) and between the POPC acyl chains and the TO molecules (III)) and one negative peak (attraction between the POPC molecules at their hydrophilic/hydrophobic interface (II)) can be observed. The xy surface area of the LD “UA size” model (black dotted line) is one-fourth that of the LD system (black solid line). The LPP of a pure POPC bilayer using the CG force field is shown for comparison. To see this figure in color, go online.
In summary, interdigitation between TAGs and phospholipids leads to a modest decrease in deep lipid-packing defects and to an increase in shallow ones with respect to lipid bilayers. These changes reflect variations in the LPP of the monolayer interface, where all pressure-profile peaks (both positive and negative) are less pronounced (Fig. 6, black line) than in lipid bilayers of analogous phospholipid composition (Fig. 6, red line). Remarkably, these differences cannot be observed when long-wavelength undulations are suppressed, e.g., in the case of a small simulation box, whereas this is not the case for lipid bilayers.
Effect of ST on LD systems assessed by CG simulations
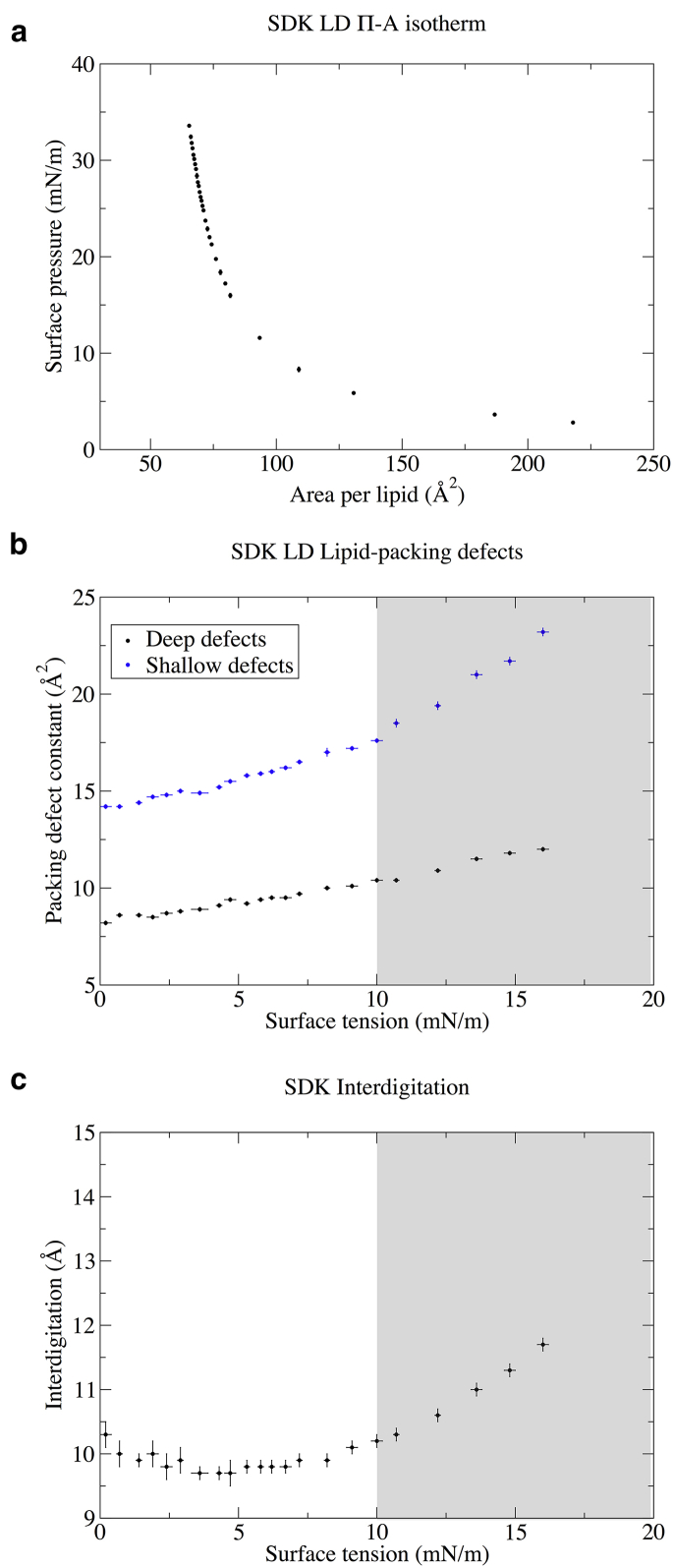
Next, we focused on the effect of ST on surface properties. First of all, our CG model can reproduce the expected decay of the Π-A isotherm of the system over a large range of surface pressures, in qualitative agreement with experimental measurements (50) (Fig. 7 a).
Figure 7.
Effect of ST on LD surface properties in CG-SDK simulations. (a) Pressure-area (Π-A) isotherm of LDs. The surface pressure is defined as Π = γ − γ0, where γ0 = 32 ± 0.1 mN/m is the ST of TO at the TO/W interface (see Table 4). (b) Deep and shallow lipid-packing defects as a function of ST in model LDs. (c) Interdigitation as a function of ST in model LDs. To see this figure in color, go online.
We then computed the variation in both deep and shallow lipid-packing defects as a function of interfacial tension. The results are shown in Fig. 7 b. In agreement with UA simulations, we observed an increase in lipid-packing defects as a function of ST, and a possible two-regime behavior is observed starting at ∼10 mN/m. This effect is especially prominent for shallow packing defects. Interestingly, the increase in lipid-packing defects for increasing STs is modest up to 10 mN/m. As a consequence, within this range of STs, lipid-packing defects in model LDs are remarkably similar to those of the most common phospholipid bilayers (POPC and dioleoylphosphatidylcholine) and very likely similar to those of membranes of the early secretory pathway (56), of which LDs are a part.
Finally, we focused on the molecular basis of the observed increase in packing defects as a function of ST. In particular, we wondered whether, besides the obvious increase in area per lipid when increasing ST, the presence of TO molecules interacting with the lipid monolayer could play a prominent role.
As a first test, we analyzed the change in the conformations of the TO molecules for increasing STs. Only marginal changes were observed for a large range of STs, even when focusing solely on interfacial TO molecules in close contact with the POPC monolayer (Table S16). Next, we computed the LPPs at different STs and we compared them to that of a pure TO/W system (Fig. 8).
Interestingly, although the repulsive peak between polar heads (I) does not vary as a function of ST (with the obvious exception of the TO/W system that lacks phospholipids), a significant increase in the attractive peak in the interfacial layer between the polar head and the acyl chains of POPC molecules (II) can be observed. This peak corresponds to the main peak between TO and water in the TO/W system.
Although variations in this peak seem to be continuous in the range 0–20 mN/m, the two-regime behavior observed for lipid-packing defects can be observed in the subsequent positive peak (III, Fig. 8, inset), which corresponds to the interface between the phospholipid chains and the TO molecules. Although this peak is essentially unchanged in all simulations, with ST <10 mN/m, significant changes can be observed above this value, with an important decrease in this peak indicating a better mixing between POPC chains and TO molecules at high ST. This behavior is accompanied by a marked increase in the interdigitation between TO and POPC molecules (Fig. 7 c) to >10 mN/m and is consistent with the results of the UA simulations (Table 2).
Discussion
Intracellular LDs have been attracting considerable interest in the last few years and are now regarded as a bona fide intracellular organelle that is crucially involved in many lipid trafficking pathways (1, 14, 57). However, obtaining a molecular understanding of cellular processes that take place at the surface of LDs has proven particularly difficult. In vitro reconstitution of ternary systems mimicking LDs is even trickier than the corresponding reconstitution for lipid bilayers, given the unique challenges that oil presents for manipulation, and because of its light-scattering properties (3).
Similarly, MD simulations of ternary systems mimicking LDs are much more challenging than those for lipid bilayers and, as such, are much less frequent in the literature. Some of the technical difficulties are quite obvious: because of the oil layer that separates the two phospholipid monolayers, the required system size is larger, and more computational resources are needed to achieve comparable sampling. Also, system equilibration is not limited to the membrane plane, but is also required along the membrane normal and between neutral and surface lipids. Other challenges are more subtle: due to the natural friction and interdigitation between the surface layer of phospholipids and the underlying oil core, we observed important size effects on both LPPs and lipid-packing defects. These effects are unique to LD-mimicking ternary systems, being absent in MD simulations of lipid bilayers, and they underline the importance of investigating these systems at different levels of resolution.
We have demonstrated how ST crucially modulates the properties of the LD surface. Unlike in lipid bilayers, where the range of accessible tensions is very small (10−3–10−1 mN/m), LDs can access a large spectrum of tensions (0–20 mN/m), and it has been suggested that interactions between LDs and proteins take place within this range in vitro (15, 42). In addition to the expected increase in lipid-packing defects upon ST increase, our simulations indicate the presence of a bimodal behavior, with a modest increase of overall defects below ≈10 mN/m and a sharper increase above this value, especially for shallow lipid-packing defects. This behavior originates from changes in the LPP at the interface between surface lipids and TAGs, leading to a substantial increase in interdigitation between the two species above this value. Furthermore, the main contribution to the ST in the LPP of the LD monolayer originates at the hydrophobic/hydrophilic interface located approximately around the glycerol level of the phospholipids. This peak is markedly lower in LDs in the absence of ST, compared to the corresponding peak for a lipid bilayer, and it becomes comparable at ∼10 mN/m.
Our findings provide interesting insights into numerous processes involving intracellular LDs. First, our observation that lipid-packing defects in model LDs resemble those of model ER and Golgi membranes gives a structural explanation to the puzzling observation that several proteins that are traditionally localized to the ER and/or the Golgi apparatus can also bind to LDs (4, 5, 6). Since all these proteins possess an AH motif, these results are consistent with the model we recently proposed, suggesting that membrane packing defects, as measured in MD simulations, can be used as a proxy to predict the binding of AH-containing peripheral proteins in vitro and in vivo (10, 12, 53, 56). At the same time, we could observe significant differences in the LPPs of LDs when compared to pure lipid bilayers (Fig. 6). Since it has been suggested that AH-containing peripheral proteins sense membrane stresses (58), it is tempting to speculate that proteins that target or interact specifically with LDs might be able to sense these differences, unlike proteins that target both LDs and other membranes.
In addition, the behavior observed for LD surface properties as a function of ST suggests an intriguing scenario: when ST is <10 mN/m, only proteins that possess AHs “well-adapted” to LDs and to membrane binding can selectively target LDs; when ST goes above 10 mN/m, several proteins, including those that should not bind to LDs physiologically, may be recruited to LDs. This dual-mode behavior could be potentially tested in vitro by controlling LD ST with microfluidic devices (15) or micromanipulation techniques (50). Also, a direct consequence of the previous observation is that we can speculate that the ST of intracellular LDs is unlikely to increase beyond 10 mN/m in non-pathological conditions; otherwise, the consequences to intracellular trafficking would be significant.
The observation that interdigitation remains constant for ST of <10 mN/m, whereas it increases dramatically for ST above this threshold, is also surprising, and it has potentially important implications in TAG metabolism. Lipid remodeling enzymes, and specifically those that break down TAGs (such as adipose TAG lipase, for example) are found at the surface of LDs (59). Even though their exact mechanism of action remains unknown, increasing substrate availability, as in the case of increased interdigitation, should probably enhance their activity and hence TAG breakdown. Thus, our simulations suggest only marginal increased activity of adipose TAG lipase as a function of ST for values <10 mN/m and a substantial increase above this threshold. Of note, breakdown of TAGs would not only decrease the number of oil molecules in the core but also would increase the number of surfactant molecules (breakdown of one TAG results in the formation of one free fatty acid and one diacylglycerol). Since those molecules would reduce ST, our observations suggest the possible presence of a feedback mechanism whereby upon reaching values of ST of ≈10 mN/m, lipase activity would substantially increase, or, conversely, a ST increase above 10 mN/m would be strongly prevented in the presence of lipases.
Our simulations also have interesting ramifications for what pertains to the mechanism of LD biogenesis. Even though this process is largely unknown, it has been convincingly proposed that LD formation may start from the accumulation of TAG molecules between the two leaflets of the ER (22). In such a scenario, a transition from a lipid bilayer devoid of TAGs to a fully formed LD would involve structural changes similar to those we observed when we increased the lateral size of our simulation box. Our MD simulations suggest that an increase in LD size is coupled to a decrease in both lipid-packing defects and the main peaks of the LPP (Fig. 6). According to previous theories (12, 58), this may lead to a decrease in the free energy of binding for AH-containing peripheral proteins and hence to their possible unbinding from the membrane surface. Thus, our simulations suggest an interesting molecular mechanism whereby upon LD formation, proteins that were initially recruited to the ER membrane are then expelled (possibly to make room for LD-specific proteins) precisely at the site of LD formation and simply as a consequence of changes in the underlying membrane properties.
In conclusion, understanding how the unique lipid composition of LDs affects their interactions with proteins is a key aspect in LD research. Our observation that not only surface lipids, but also neutral lipids forming the core of LDs, may play a role in shaping surface properties of LDs, and hence in protein targeting, is intriguing. Neutral lipids with different shapes and chemical properties may interact differently with surface phospholipids, due to either entropic or enthalpic contributions. Interestingly, the ratio between TAGs and cholesteryl esters has been proposed as a key factor for protein binding to LDs in cellular experiments (60), although results conflicting with this finding have also been reported (61, 62). We foresee that future MD simulations addressing the lipid complexity of LDs may help to shed light on these questions.
Conclusions
In this work, we used MD simulations at different resolutions (UA and CG, for which a new model, to our knowledge, for TAGs is developed and validated) to extensively investigate the structural and dynamical properties of ternary systems composed of TAGs, phospholipids, and water, mimicking intracellular LDs.
Our simulations indicate that the LD surface has both similarities to (area per lipid, order parameter, POPC thickness, and lipid diffusion) and differences from (interdigitation and LPPs) that of lipid bilayers, providing a first clue of how the uniqueness of the LD membrane may play a role in modulating several cellular processes taking place on the surface of this organelle.
Although this work is only a step toward unveiling the complex cellular mechanisms involving intracellular LDs, our results indicate that MD simulations may become a powerful tool to investigate these processes at the molecular level, especially in combination with cell biology experiments and in vitro reconstitutions.
Author Contributions
S.V., P.F.J.F., and C.L.J. designed research; A.B., R.G., S.V., and P.F.J.F. performed research; A.B., S.V., and P.F.J.F. analyzed the data; P.F.J.F. and S.V. wrote the manuscript.
Acknowledgments
We thank Ilpo Vattulainen for sharing his Berger topology of TO. We thank Juan Vanegas for help on using GROMACS-LS for computing LPPs. We also thank Bruno Antonny, Alenka Čopič, Christopher MacDermaid and Giacomo Fiorin for useful discussions and careful reading of the manuscript.
This work was supported by the French National Research Agency grant LDsurfDynamics, Fondation pour la Recherche Médicale grant no. DEQ20150934717 (to C.L.J.), and granted access to the high-performance computing resources of the National Computing Center for Higher Education (CINES) under allocation c2016077362 made by Grand Equipement National de Calcul Intensif (GENCI).
Editor: Emad Tajkhorshid
Footnotes
Patrick F.J. Fuchs and Stefano Vanni contributed equally to this work.
Stefano Vanni’s present address is Department of Biology, University of Fribourg, Fribourg, Switzerland.
Supporting Materials and Methods, seven figures, and nine tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30246-1.
Contributor Information
Patrick F.J. Fuchs, Email: patrick.fuchs@univ-paris-diderot.fr.
Stefano Vanni, Email: stefano.vanni@unifr.ch.
Supporting Citations
References (63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76) appear in the Supporting Material.
Supporting Material
References
- 1.Walther T.C., Farese R.V., Jr. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herker E., Ott M. Emerging role of lipid droplets in host/pathogen interactions. J. Biol. Chem. 2012;287:2280–2287. doi: 10.1074/jbc.R111.300202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiam A.R., Farese R.V., Jr., Walther T.C. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 2013;14:775–786. doi: 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soni K.G., Mardones G.A., Bonifacino J.S. Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 2009;122:1834–1841. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krahmer N., Guo Y., Walther T.C. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilfling F., Thiam A.R., Walther T.C. Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife. 2014;3:e01607. doi: 10.7554/eLife.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drin G., Antonny B. Amphipathic helices and membrane curvature. FEBS Lett. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 8.Rowe E.R., Mimmack M.L., Savage D.B. Conserved amphipathic helices mediate lipid droplet targeting of perilipins 1–3. J. Biol. Chem. 2016;291:6664–6678. doi: 10.1074/jbc.M115.691048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouvet S., Golinelli-Cohen M.-P., Jackson C.L. Targeting of the Arf-GEF GBF1 to lipid droplets and Golgi membranes. J. Cell Sci. 2013;126:4794–4805. doi: 10.1242/jcs.134254. [DOI] [PubMed] [Google Scholar]
- 10.Vamparys L., Gautier R., Fuchs P.F. Conical lipids in flat bilayers induce packing defects similar to that induced by positive curvature. Biophys. J. 2013;104:585–593. doi: 10.1016/j.bpj.2012.11.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui H., Lyman E., Voth G.A. Mechanism of membrane curvature sensing by amphipathic helix containing proteins. Biophys. J. 2011;100:1271–1279. doi: 10.1016/j.bpj.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanni S., Hirose H., Gautier R. A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat. Commun. 2014;5:4916. doi: 10.1038/ncomms5916. [DOI] [PubMed] [Google Scholar]
- 13.Grippa A., Buxó L., Carvalho P. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 2015;211:829–844. doi: 10.1083/jcb.201502070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kory N., Farese R.V., Jr., Walther T.C. Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol. 2016;26:535–546. doi: 10.1016/j.tcb.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thiam A.R., Antonny B., Pincet F. COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc. Natl. Acad. Sci. USA. 2013;110:13244–13249. doi: 10.1073/pnas.1307685110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kory N., Thiam A.R., Walther T.C. Protein crowding is a determinant of lipid droplet protein composition. Dev. Cell. 2015;34:351–363. doi: 10.1016/j.devcel.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henneré G., Prognon P., Nicolis I. Molecular dynamics study of a phospholipid monolayer at a water/triglyceride interface: towards lipid emulsion modelling. Chem. Phys. Lipids. 2009;157:86–93. doi: 10.1016/j.chemphyslip.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Henneré G., Prognon P., Nicolis I. Molecular dynamics simulation of a mixed lipid emulsion model: influence of the triglycerides on interfacial phospholipid organization. J. Mol. Struct. THEOCHEM. 2009;901:174–185. [Google Scholar]
- 19.Koivuniemi A., Heikelä M., Hyvönen M.T. Atomistic simulations of phosphatidylcholines and cholesteryl esters in high-density lipoprotein-sized lipid droplet and trilayer: clues to cholesteryl ester transport and storage. Biophys. J. 2009;96:4099–4108. doi: 10.1016/j.bpj.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaban V.V., Khandelia H. Lipid structure in triolein lipid droplets. J. Phys. Chem. B. 2014;118:10335–10340. doi: 10.1021/jp503223z. [DOI] [PubMed] [Google Scholar]
- 21.Chaban V.V., Khandelia H. Distribution of neutral lipids in the lipid droplet core. J. Phys. Chem. B. 2014;118:11145–11151. doi: 10.1021/jp506693d. [DOI] [PubMed] [Google Scholar]
- 22.Khandelia H., Duelund L., Ipsen J.H. Triglyceride blisters in lipid bilayers: implications for lipid droplet biogenesis and the mobile lipid signal in cancer cell membranes. PLoS One. 2010;5:e12811. doi: 10.1371/journal.pone.0012811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koivuniemi A., Sysi-Aho M., Ollila S. Interfacial properties of high-density lipoprotein-like lipid droplets with different lipid and apolipoprotein A-I compositions. Biophys. J. 2013;104:2193–2201. doi: 10.1016/j.bpj.2013.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vuorela T., Catte A., Vattulainen I. Role of lipids in spheroidal high density lipoproteins. PLoS Comput. Biol. 2010;6:e1000964. doi: 10.1371/journal.pcbi.1000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ollila O.H., Lamberg A., Vattulainen I. Interfacial tension and surface pressure of high density lipoprotein, low density lipoprotein, and related lipid droplets. Biophys. J. 2012;103:1236–1244. doi: 10.1016/j.bpj.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baoukina S., Monticelli L., Tieleman D.P. Pressure-area isotherm of a lipid monolayer from molecular dynamics simulations. Langmuir. 2007;23:12617–12623. doi: 10.1021/la702286h. [DOI] [PubMed] [Google Scholar]
- 27.Baoukina S., Marrink S.J., Tieleman D.P. Lateral pressure profiles in lipid monolayers. Faraday Discuss. 2010;144:393–409. doi: 10.1039/b905647e. discussion 445–481. [DOI] [PubMed] [Google Scholar]
- 28.Shinoda W., DeVane R., Klein M.L. Zwitterionic lipid assemblies: molecular dynamics studies of monolayers, bilayers, and vesicles using a new coarse grain force field. J. Phys. Chem. B. 2010;114:6836–6849. doi: 10.1021/jp9107206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDermaid C.M., Kashyap H.K., Fiorin G. Molecular dynamics simulations of cholesterol-rich membranes using a coarse-grained force field for cyclic alkanes. J. Chem. Phys. 2015;143:243144. doi: 10.1063/1.4937153. [DOI] [PubMed] [Google Scholar]
- 30.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 31.Berger O., Edholm O., Jähnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berendsen H.J.C., Postma J.P.M., Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B., editor. Intermolecular Forces. Reidel Publishing; Dordrecht, The Netherlands: 1981. pp. 331–342. [Google Scholar]
- 33.Hall A., Repakova J., Vattulainen I. Modeling of the triglyceride-rich core in lipoprotein particles. J. Phys. Chem. B. 2008;112:13772–13782. doi: 10.1021/jp803950w. [DOI] [PubMed] [Google Scholar]
- 34.Bachar M., Brunelle P., Rauk A. Molecular dynamics simulation of a polyunsaturated lipid bilayer susceptible to lipid peroxidation. J. Phys. Chem. B. 2004;108:7170–7179. [Google Scholar]
- 35.Das C., Noro M.G., Olmsted P.D. Simulation studies of stratum corneum lipid mixtures. Biophys. J. 2009;97:1941–1951. doi: 10.1016/j.bpj.2009.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brasiello A., Crescitelli S., Milano G. Development of a coarse-grained model for simulations of tridecanoin liquid-solid phase transitions. Phys. Chem. Chem. Phys. 2011;13:16618–16628. doi: 10.1039/c1cp20604d. [DOI] [PubMed] [Google Scholar]
- 37.Irving J.H., Kirkwood J.G. The statistical mechanical theory of transport processes. IV. The equations of hydrodynamics. J. Chem. Phys. 1950;18:817–829. [Google Scholar]
- 38.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 39.Shinoda W., DeVane R., Klein M.L. Multi-property fitting and parameterization of a coarse grained model for aqueous surfactants. Mol. Simul. 2007;33:27–36. [Google Scholar]
- 40.Plimpton S. Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 1995;117:1–19. [Google Scholar]
- 41.Farese R.V., Jr., Walther T.C. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitsche M.A., Small D.M. C-terminus of apolipoprotein A-I removes phospholipids from a triolein/phospholipids/water interface, but the N-terminus does not: a possible mechanism for nascent HDL assembly. Biophys. J. 2011;101:353–361. doi: 10.1016/j.bpj.2011.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitsche M.A., Wang L., Small D.M. Adsorption of egg phosphatidylcholine to an air/water and triolein/water bubble interface: use of the 2-dimensional phase rule to estimate the surface composition of a phospholipid/triolein/water surface as a function of surface pressure. J. Phys. Chem. B. 2010;114:3276–3284. doi: 10.1021/jp908730t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duelund L., Jensen G.V., Ipsen J.H. Composition, structure and properties of POPC-triolein mixtures. Evidence of triglyceride domains in phospholipid bilayers. Biochim. Biophys. Acta. 2013;1828:1909–1917. doi: 10.1016/j.bbamem.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 45.Croll D.H., Small D.M., Hamilton J.A. Molecular motions and thermotropic phase behavior of cholesteryl esters with triolein. Biochemistry. 1985;24:7971–7980. doi: 10.1021/bi00348a020. [DOI] [PubMed] [Google Scholar]
- 46.Javanainen M., Monticelli L., Vattulainen I. Free volume theory applied to lateral diffusion in Langmuir monolayers: atomistic simulations for a protein-free model of lung surfactant. Langmuir. 2010;26:15436–15444. doi: 10.1021/la102454m. [DOI] [PubMed] [Google Scholar]
- 47.Kollmitzer B., Heftberger P., Pabst G. Monolayer spontaneous curvature of raft-forming membrane lipids. Soft Matter. 2013;9:10877–10884. doi: 10.1039/C3SM51829A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ragni L., Berardinelli A., Valli E. Assessment of the water content in extra virgin olive oils by time domain reflectometry (TDR) and partial least squares (PLS) regression methods. J. Food Eng. 2012;111:66–72. [Google Scholar]
- 49.Zeppieri S., Rodriguez J., Lopez de Ramos A.L. Interfacial tension of alkane + water systems. J. Chem. Eng. Data. 2001;46:1086–1088. [Google Scholar]
- 50.Delacotte J., Gourier C., Pincet F. Interfacial pressure and phospholipid density at emulsion droplet interface using fluorescence microscopy. Colloids Surf. B Biointerfaces. 2014;117:545–548. doi: 10.1016/j.colsurfb.2013.11.033. [DOI] [PubMed] [Google Scholar]
- 51.Pinot M., Vanni S., Barelli H. Lipid cell biology. Polyunsaturated phospholipids facilitate membrane deformation and fission by endocytic proteins. Science. 2014;345:693–697. doi: 10.1126/science.1255288. [DOI] [PubMed] [Google Scholar]
- 52.Garten M., Prévost C., Vanni S. Methyl-branched lipids promote the membrane adsorption of α-synuclein by enhancing shallow lipid-packing defects. Phys. Chem. Chem. Phys. 2015;17:15589–15597. doi: 10.1039/c5cp00244c. [DOI] [PubMed] [Google Scholar]
- 53.Vanni S., Vamparys L., Antonny B. Amphipathic lipid packing sensor motifs: probing bilayer defects with hydrophobic residues. Biophys. J. 2013;104:575–584. doi: 10.1016/j.bpj.2012.11.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shelley J.C., Shelley M.Y., Klein M.L. A coarse grain model for phospholipid simulations. J. Phys. Chem. B. 2001;105:4464–4470. [Google Scholar]
- 55.Klein M.L., Shinoda W. Large-scale molecular dynamics simulations of self-assembling systems. Science. 2008;321:798–800. doi: 10.1126/science.1157834. [DOI] [PubMed] [Google Scholar]
- 56.Magdeleine M., Gautier R., Antonny B. A filter at the entrance of the Golgi that selects vesicles according to size and bulk lipid composition. eLife. 2016;5:e16988. doi: 10.7554/eLife.16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hashemi H.F., Goodman J.M. The life cycle of lipid droplets. Curr. Opin. Cell Biol. 2015;33:119–124. doi: 10.1016/j.ceb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campelo F., Kozlov M.M. Sensing membrane stresses by protein insertions. PLoS Comput. Biol. 2014;10:e1003556. doi: 10.1371/journal.pcbi.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmermann R., Strauss J.G., Zechner R. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
- 60.Hsieh K., Lee Y.K., Kimmel A.R. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 2012;125:4067–4076. doi: 10.1242/jcs.104943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu D., Yu Y., Wang M.C. In vivo metabolic fingerprinting of neutral lipids with hyperspectral stimulated Raman scattering microscopy. J. Am. Chem. Soc. 2014;136:8820–8828. doi: 10.1021/ja504199s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mishra S., Schneiter R. Expression of perilipin 5 promotes lipid droplet formation in yeast. Commun. Integr. Biol. 2015;8:e1071728. doi: 10.1080/19420889.2015.1071728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
- 64.Botan A., Favela-Rosales F., Tynkkynen J. Toward atomistic resolution structure of phosphatidylcholine headgroup and glycerol backbone at different ambient conditions. J. Phys. Chem. B. 2015;119:15075–15088. doi: 10.1021/acs.jpcb.5b04878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vanegas J.M., Torres-Sánchez A., Arroyo M. Importance of force decomposition for local stress calculations in biomembrane molecular simulations. J. Chem. Theory Comput. 2014;10:691–702. doi: 10.1021/ct4008926. [DOI] [PubMed] [Google Scholar]
- 66.Sodt A.J., Pastor R.W. Molecular modeling of lipid membrane curvature induction by a peptide: more than simply shape. Biophys. J. 2014;106:1958–1969. doi: 10.1016/j.bpj.2014.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Naturforsch. C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 68.Sodt A.J., Venable R.M., Pastor R.W. Nonadditive compositional curvature energetics of lipid bilayers. Phys. Rev. Lett. 2016;117:138104. doi: 10.1103/PhysRevLett.117.138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101–014107. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 70.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 71.Jo S., Lim J.B., Im W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984;52:255–268. [Google Scholar]
- 73.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 74.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 75.Eastwood J.W., Hockney R.W., Lawrence D.N. P3M3DP—The 3-dimensional periodic particle-particle-particle-mesh program. Comput. Phys. Commun. 1980;19:215–261. [Google Scholar]
- 76.Ding W., Palaiokostas M., Orsi M. Effects of lipid composition on bilayer membranes quantified by all-atom molecular dynamics. J. Phys. Chem. B. 2015;119:15263–15274. doi: 10.1021/acs.jpcb.5b06604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.