The oomycetes are a class of eukaryotes and include ecologically significant animal and plant pathogens. Single-gene and multigene phylogenetic studies of individual oomycete genera and of members of the larger classes have resulted in conflicting conclusions concerning interspecies relationships among these species, particularly for the Phytophthora genus. The onset of next-generation sequencing techniques now means that a wealth of oomycete genomic data is available. For the first time, we have used genome-scale phylogenetic methods to resolve oomycete phylogenetic relationships. We used supertree methods to generate single-gene and multigene species phylogenies. Overall, our supertree analyses utilized phylogenetic data from 8,355 oomycete gene families. We have also complemented our analyses with superalignment phylogenies derived from 131 single-copy ubiquitous gene families. Our results show that a genome-scale approach to oomycete phylogeny resolves oomycete classes and clades. Our analysis represents an important first step in large-scale phylogenomic analysis of the oomycetes.
KEYWORDS: oomycete, phylogeny, Phytophthora, species phylogeny, phylogenomics, supermatrix, supertrees
ABSTRACT
The oomycetes are a class of microscopic, filamentous eukaryotes within the Stramenopiles-Alveolata-Rhizaria (SAR) supergroup which includes ecologically significant animal and plant pathogens, most infamously the causative agent of potato blight Phytophthora infestans. Single-gene and concatenated phylogenetic studies both of individual oomycete genera and of members of the larger class have resulted in conflicting conclusions concerning species phylogenies within the oomycetes, particularly for the large Phytophthora genus. Genome-scale phylogenetic studies have successfully resolved many eukaryotic relationships by using supertree methods, which combine large numbers of potentially disparate trees to determine evolutionary relationships that cannot be inferred from individual phylogenies alone. With a sufficient amount of genomic data now available, we have undertaken the first whole-genome phylogenetic analysis of the oomycetes using data from 37 oomycete species and 6 SAR species. In our analysis, we used established supertree methods to generate phylogenies from 8,355 homologous oomycete and SAR gene families and have complemented those analyses with both phylogenomic network and concatenated supermatrix analyses. Our results show that a genome-scale approach to oomycete phylogeny resolves oomycete classes and individual clades within the problematic Phytophthora genus. Support for the resolution of the inferred relationships between individual Phytophthora clades varies depending on the methodology used. Our analysis represents an important first step in large-scale phylogenomic analysis of the oomycetes.
IMPORTANCE The oomycetes are a class of eukaryotes and include ecologically significant animal and plant pathogens. Single-gene and multigene phylogenetic studies of individual oomycete genera and of members of the larger classes have resulted in conflicting conclusions concerning interspecies relationships among these species, particularly for the Phytophthora genus. The onset of next-generation sequencing techniques now means that a wealth of oomycete genomic data is available. For the first time, we have used genome-scale phylogenetic methods to resolve oomycete phylogenetic relationships. We used supertree methods to generate single-gene and multigene species phylogenies. Overall, our supertree analyses utilized phylogenetic data from 8,355 oomycete gene families. We have also complemented our analyses with superalignment phylogenies derived from 131 single-copy ubiquitous gene families. Our results show that a genome-scale approach to oomycete phylogeny resolves oomycete classes and clades. Our analysis represents an important first step in large-scale phylogenomic analysis of the oomycetes.
INTRODUCTION
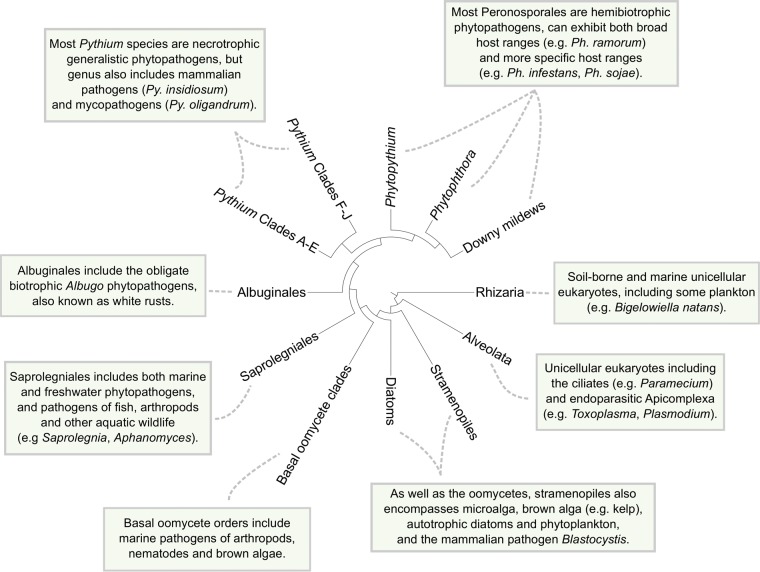
The oomycetes are a class of microscopic eukaryotes which include some of the most ecologically destructive marine and terrestrial eukaryotic species (1). Oomycete species display filamentous morphology and ecological roles very similar to those of fungi and were historically regarded as a basal fungal lineage (2). As morphological and molecular studies have improved since the latter half of the 20th century, the oomycetes have come to be understood as very distant relations of “true” fungi. They have independently evolved similar morphology and lifestyles through convergent evolution and limited interkingdom horizontal gene transfer (HGT) (2–5). Present phylogenomic studies place the oomycetes in the diverse stramenopiles lineage within the Stramenopiles-Alveolata-Rhizaria (SAR) eukaryotic supergroup (6–10) (Fig. 1). The stramenopiles were previously placed within Chromista (11) and then within the “chromalveolates” supergroup (Chromista plus Alveolata) on the basis of a hypothesized last common ancestor on the plastid lineage (12, 13). While early phylogenetic analyses supported the concept of a single origin for the “chromalveolate” plastid (14, 15), later plastome-wide and nuclear phylogenetic and HGT analyses have consistently failed to support a monophyletic chromalevolate grouping (16–21). In contrast, molecular evidence for the monophyly of the current SAR supergroup has been demonstrated in multiple phylogenetic analyses (18, 20, 22–26).
FIG 1 .
Consensus phylogeny of the oomycetes class within the greater SAR grouping, including information pertaining to various taxa. The cladogram was adapted from Judelson (10).
The oomycetes are thought to have diverged from diatoms between the Late Proterozoic and the mid-Paleozoic eras (~0.4 to 0.6 billion years ago [bya]) (27, 28) and have been found to have been present as early as the Devonian period (~400 million years ago [mya]) in the fossil record (29). Though many described species are phytopathogens, oomycete phytopathogenicity is thought to be a derived trait which has evolved independently in many lineages (30). Many species are as yet unsampled, and the class phylogeny of the oomycetes is still subject to revision; with current data, however, the oomycetes can be split into the earliest diverging clades and the later “crown” taxa (31–33) (Fig. 1). With the exception of some species infecting terrestrial nematodes (31), the earliest diverging oomycete clades are otherwise exclusively marine in habitat (1). The remaining “crown” oomycetes can be subdivided into the predominantly marine and freshwater “saprolegnian” branches and the predominantly terrestrial “peronosporalean” branches, which diverged in the Early Mesozoic era (1, 28, 34–36). The “saprolegnian” branches include the fish pathogen Saprolegnia, also known as “cotton mould” (37), and the animal- and plant-pathogenic Aphanomyces genus (34, 38). The “peronosporalean” branches include the best-characterized oomycete taxa, Phytophthora and Pythium, and the more basal Albuginales order (1, 35). The majority of “peronosporalean” oomycetes are phytopathogens, although Pythium includes species capable of infecting animals or acting as mycoparasitic biocontrol agents (39, 40) (Fig. 1).
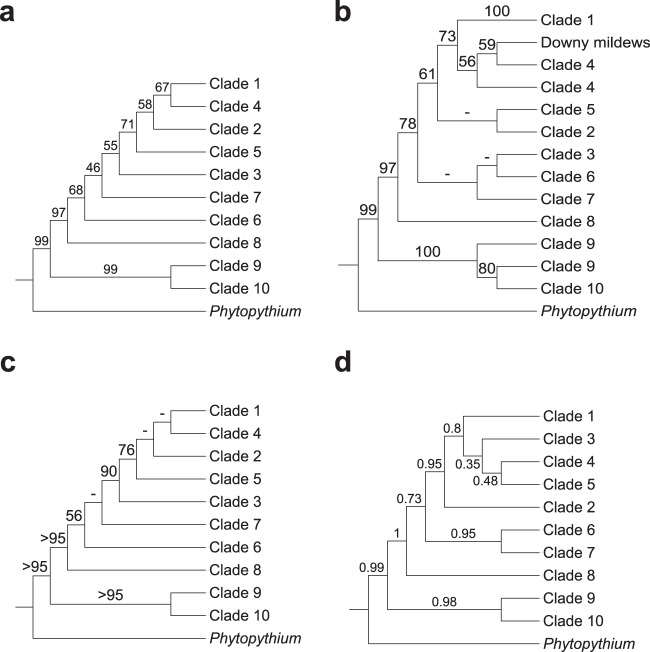
Phytophthora is the largest genus (>120 described species) within the order Peronosporales and was divided into 10 phylogenetic clades on the basis of initial internal transcribed spacer (ITS) analysis and, later, combined nuclear and mitochondrial analyses (41, 42) (Fig. 2a). The largest clades (clades 1, 2, 7, and 8) are further divided into subclades, while the smallest clades (clades 5 and 10) contain fewer than five described species at present (43, 44). Initial ITS phylogeny data reported by Cooke et al. (41) suggested that Phytophthora was paraphyletic with respect to basal clades 9 and 10; however, later multigene and combined nuclear and mitochondrial studies have placed these clades within Phytophthora (42, 44, 45). Generally, species within Phytophthora clades do not share consistent morphological features or reproductive strategies, although clades 6 to 8 form a distinct branch of terrestrial species with predominantly nonpapillate sporangia within the genus tree (44). While many recent phylogenetic analyses have supported the current designation by Blair et al. (42) of 10 distinct phylogenetic clades within Phytophthora, many of the same analyses draw conflicting conclusions as to the relationships among these clades. In their analysis, Blair et al. (42) found strong support by maximum-likelihood, maximum-parsimony, and Bayesian methods for the 10 phylogenetic clades using data from seven highly conserved nuclear loci (including markers from 28S ribosomal DNA [rDNA], Hsp90, and β-tubulin) from 82 Phytophthora species (Fig. 2a). The relationship between the clades reported in Blair et al. (42) was mostly upheld in a follow-up analysis by Runge et al. (46) which included homologous data from an additional 39 Phytophthora species and other Peronosporales species. One noticeable difference was that their analysis placed clades 3, 6, and 7 as sister clades within a monophyletic clade with strong support by the minimum-evolution, maximum-likelihood, and Bayesian methods, while the clades were more distantly related in the analysis by Blair et al. (42) (Fig. 2a and b). The addition of four mitochondrial markers (cox2, nad9, rps10, and secY) in a later 11-locus analysis by Martin et al. (47), while topologically supporting the data from Blair et al. (42), displayed poor resolution for many interclade relationships (particularly for more extensively derived clades such as clades 1 to 5) within Phytophthora by the maximum-likelihood, maximum-parsimony, and Bayesian methods (Fig. 2c). A coalescent approach using a similar data set by the same authors showed improved Bayesian support among some Phytophthora clades (e.g., clades 1 to 5) but weaker support for other clades and a conflicting topology from the 11-locus analysis (47) (Fig. 2d).
FIG 2 .
Congruence of the Peronosporales order among recent multilocus phylogenetic analyses. (a) Seven-locus maximum-likelihood (ML)/maximum-parsimony (MP)/Bayesian phylogeny of Phytophthora by Blair et al. (42). (b) Minimum-evolution (ME)/ML/Bayesian phylogeny of Phytophthora and downy mildews by Runge et al. (46). (c) Eleven-locus ML/MP/Bayesian phylogeny of Phytophthora by Martin et al. (47). (d) Six-locus coalescent phylogeny of Phytophthora by Martin et al. (47). Support values, where given, represent maximum-likelihood bootstrap support, except for panel d, where Bayesian posterior probabilities are given instead.
Placement of other taxa within the Peronosporales order, namely, the “downy mildews,” and the phylogeny of Pythium and the Pythiales order have also been difficult to resolve. The inclusion of two downy mildews species (Hyaloperonospora arabidopsidis and Pseudoperonospora cubensis) in an analysis conducted by Runge et al. placed the two species within Phytophthora clade 4 and sister to clade 1 species such as Phytophthora infestans, implying the existence of a paraphyletic Phytophthora genus (46) (Fig. 2b). However, a subsequent tree reconciliation analysis, inferred using a class phylogeny of 189 oomycete clusters of orthologous groups (COGs), placed H. arabidopsidis as sister to members of the Phytophthora genus (48). Another downy mildew species, Plasmopara halstedii, was placed sister to Phytophthora clade 1 in similar phylogenetic analyses (36, 49). Phytopythium, a morphological intermediate between Phytophthora and Pythium, was reclassified from Pythium clade K to its own genus within the Peronosporales order based on a recent multigene phylogenetic analysis which placed the genus sister to Phytophthora (50). Pythium itself is divided into 10 clades, labeled A to J, which were initially circumscribed with its data and consistent with mitochondrial data (51). The main morphological difference between clades within Pythium is the development of the filamentous sporangium in species within clades A to C from the ancestral globose sporangium observed in the basal clades and Phytopythium (51, 52), with an intermediate contiguous sporangium developing in species within clade D (51) and an elongated sporagium in species within clade H (53). Otherwise, as in Phytophthora, phylogenetic clades generally do not correlate with distinct morphological characters in Pythium (51). A number of phylogenetic analyses suggest that Pythium is polyphyletic (36, 49, 52–55), and there has been recent suggestion that it be amended entirely into at least five new genera (53, 56).
Many of the aforementioned phylogenetic analyses of the oomycetes are based upon a small number of highly conserved nuclear and/or mitochondrial markers, either through consensus analysis or concatenated analysis. The selection of such markers, while usually robust, may unintentionally ignore other types of potential phylogenetic markers that might resolve conflicting analyses, such as lineages which include gene duplication events (20). One solution to the possible limitations of single-gene or small-scale gene phylogenies is to assemble a consensus phylogeny for a given set of taxa using many sources of single-gene phylogenies through supertree analysis, which enables the inclusion of phylogenies with missing or duplicated taxa (57). Matrix representation using parsimony (MRP), in which character matrices are generated for each source phylogeny and merged into a single binary character matrix for maximum-parsimony alignment (58, 59), is one of the most commonly used supertree methods and has seen successful application in a number of eukaryotic phylogenomic studies (60–62). Other methods have been developed for inferring species phylogeny from paralogous gene phylogenies, the most successful of which has been gene tree parsimony (GTP) (63). GTP attempts to find the most parsimonious species tree from a set of source phylogenies with the lowest number of events required to explain incongruences (i.e., gene duplication events) between the source phylogenies and has seen application in large-scale phylogenetic analysis (64). Another method of large-scale phylogenetic analysis is the supermatrix approach of concatenating multiple character data sets for simultaneous analysis (65).
Since the publication of the genome sequences of Phytophthora sojae and Phytophthora ramorum in 2006 (66), the quantity of oomycete genomic data has steadily increased; currently, 37 oomycete species now have publicly available genomic data at the assembly level or higher (Table 1). With this in mind, we have conducted the first whole-genome phylogenetic analysis for the oomycetes as a class, using a variety of supertree and supermatrix approaches which have previously been used in fungal whole-genome phylogenetic analysis (60). In our analysis, we utilized protein data from 37 complete oomycete genomes and 6 complete SAR genomes (as outgroups). This represents all extant genomic data from the four “crown” oomycete orders and covers 8 of the 10 phylogenetic clades within Phytophthora and 7 of the 10 phylogenetic clades within Pythium (Table 1). Our whole-genome phylogenetic analysis of the oomycetes supports the four oomycete orders and the placement of Phytopythium within the Peronosporales and individual clades within Phytophthora and Pythium. The resolution of the Peronosporales as an order varied under different methods, probably due to missing data from clades 4 and 9 within Phytophthora. However, the overall order phylogenies are relatively congruent among our different species phylogenies. This analysis will provide a useful backbone to future genome phylogenies of the oomycetes utilizing more taxonomically extensive data sets.
TABLE 1 .
Taxonomic and genomic information for the 43 oomycete and SAR species in this analysisa
| Species name | Clade | Order | Class | Reference | Gene |
|---|---|---|---|---|---|
| Albugo candida | NA | Albuginales | Oomycota | Links et al. 2011 (73) | 13310 |
| Albugo labiachii | NA | Albuginales | Oomycota | Kemen et al. 2011 (74) | 13804 |
| Hyaloperonospora arabidopsidis | NA | Peronosporales | Oomycota | Baxter et al. 2010 (71) | 14321 |
| Phytophthora agathidicida | Clade 5 | Peronosporales | Oomycota | Studholme et al. 2016 (70) | 14110* |
| Phytophthora capsici | Clade 2 | Peronosporales | Oomycota | Lamour et al. 2012 (72) | 19805 |
| Phytophthora cinnamomi | Clade 7 | Peronosporales | Oomycota | Studholme et al. 2016 (70) | 12942* |
| Phytophthora cryptogea | Clade 8 | Peronosporales | Oomycota | Feau et al. 2016 (75) | 11876* |
| Phytophthora fragariae | Clade 7 | Peronosporales | Oomycota | Gao et al. 2015 (76) | 13361* |
| Phytophthora infestans | Clade 1 | Peronosporales | Oomycota | Haas et al. 2009 (69) | 17797 |
| Phytophthora kernoviae | Clade 10 | Peronosporales | Oomycota | Sambles et al. 2015 (77) | 10650 |
| Phytophthora lateralis | Clade 8 | Peronosporales | Oomycota | Quinn et al. 2013 (78) | 11635 |
| Phytophthora multivora | Clade 2 | Peronosporales | Oomycota | Studholme et al. 2016 (70) | 15006* |
| Phytophthora nicotianae | Clade 1 | Peronosporales | Oomycota | Liu et al. 2016 (79) | 10521 |
| Phytophthora parasitica | Clade 1 | Peronosporales | Oomycota | Broad Institute (INRA-310 v. 3) | 27942 |
| Phytophthora pinifolia | Clade 6 | Peronosporales | Oomycota | Feau et al. 2016 (75) | 19533* |
| Phytophthora pluvialis | Clade 3 | Peronosporales | Oomycota | Studholme et al. 2016 (70) | 18426* |
| Phytophthora pisi | Clade 7 | Peronosporales | Oomycota | PRJEB6298 | 15495* |
| Phytophthora ramorum | Clade 8 | Peronosporales | Oomycota | Tyler et al. 2006 (66) | 15743 |
| Phytophthora rubi | Clade 7 | Peronosporales | Oomycota | PRJNA244739 | 15462* |
| Phytophthora sojae | Clade 7 | Peronosporales | Oomycota | Tyler et al. 2006 (66) | 26584 |
| Phytophthora taxon Totara | Clade 3 | Peronosporales | Oomycota | Studholme et al. 2016 (70) | 16691* |
| Plasmopara halstedii | NA | Peronosporales | Oomycota | Sharma et al. 2015 (80) | 15469 |
| Plasmopara viticola | NA | Peronosporales | Oomycota | PRJNA329579 | 12048* |
| Phytopythium vexans | NA | Peronosporales | Oomycota | Adhikari et al. 2013 (67) | 11958 |
| Pilasporangium apinafurcum | NA | Pythiales | Oomycota | PRJDB3797 | 13184* |
| Pythium aphanidermatum | Clade A | Pythiales | Oomycota | Adhikari et al. 2013 (67) | 12312 |
| Pythium arrhenomanes | Clade B | Pythiales | Oomycota | Adhikari et al. 2013 (67) | 13805 |
| Pythium insidiosum | Clade C | Pythiales | Oomycota | Rujirawat et al. 2015 (81) | 19290* |
| Pythium irregulare | Clade F | Pythiales | Oomycota | Adhikari et al. 2013 (67) | 13805 |
| Pythium iwayami | Clade G | Pythiales | Oomycota | Adhikari et al. 2013 (67) | 14875 |
| Pythium oligandrum | Clade D | Pythiales | Oomycota | Berger et al. 2016 (82) | 14292* |
| Pythium ultimum var. sporangiiferum | Clade I | Pythiales | Oomycota | Adhikari et al. 2013 (67) | 14096 |
| Pythium ultimum var. ultimum | Clade I | Pythiales | Oomycota | Lévesque et al. 2010 (68) | 15323 |
| Aphanomyces astaci | NA | Saprolegniales | Oomycota | Broad Institute (APO3 v.2) | 26259 |
| Aphanomyces invadans | NA | Saprolegniales | Oomycota | Broad Institute (9901 v.2) | 20816 |
| Saprolegnia diclina | NA | Saprolegniales | Oomycota | PRJNA168273 | 18229 |
| Saprolegnia parasitica | NA | Saprolegniales | Oomycota | Jiang et al. 2013 (83) | 20121 |
| Aureococcus anophagefferns | NA | Pelagomonadales | Pelagophyceae | Gobler et al. 2011 (84) | 11501 |
| Ectocarpus siliculosus | NA | Ectocarpales | Phaeophyceae | Cock et al. 2010 (87) | 16269 |
| Phaeodactylum tricornutum | NA | Naviculales | Bacillariophyceae | Bowler et al. 2008 (85) | 10402 |
| Thalassiosira psuedonana | NA | Thalassiosirales | Coscinodiscophyceae | Armbrust et al. 2004 (86) | 11776 |
| Paramecium tetraurelia | NA | Peniculida | Oligohymenophorea | Aury et al. 2006 (88) | 39580 |
| Bigelowiella natans | NA | Chlorarachniophyceae | Cercozoa | Curtis et al. 2012 (89) | 21708 |
Protein counts generated in this study from assembly data are highlighted with an asterisk (*). References are to the genome publications where possible and otherwise to the NCBI BioProject identifier or the Broad Institute strain identifier and assembly version. NA, not applicable.
RESULTS AND DISCUSSION
Identification of orthologous and paralogous oomycete and SAR gene families.
For our supertree analyses, we constructed a data set containing 43 complete genomes, consisting of 37 from oomycete species and 6 outgroups from other species within the SAR supergroup (Materials and Methods; Table 1). Of these 37 oomycete genomes, 26 were from either Phytophthora species or Pythium species representing the majority of clades within both genera, and the remainder were sampled from all four of the “crown” orders (66–89). We downloaded proteomes for 23 oomycete species which were available from public databases, and we generated corresponding proteomes for the remaining 14 species from publicly available assembly data using bespoke oomycete reference templates with AUGUSTUS and GeneMark-ES (90, 91) (Table S1). In total, our final data set contained 702,132 protein sequences from 37 complete oomycete genomes and 6 complete SAR genomes (Table 1).
Taxonomy, assembly statistics, and protein predictions for 14 oomycete species. AUGUSTUS ab initio protein predictions were carried out using parameters generated from genomic, EST, and protein data from template species. Protein prediction for Pi. apinafurcum was carried out using GeneMark-ES and AUGUSTUS. Protein sets used in whole-genome phylogenetic analyses in this study are highlighted in bold. Download TABLE S1, DOCX file, 0.02 MB (17.6KB, docx) .
Copyright © 2017 McCarthy and Fitzpatrick.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The initial step in determining the phylogeny of the 43 oomycete and SAR genomes in our data set through supertree methods was to identify groups of closely related orthologs or paralogs within our data set, which we termed gene families, and to use these groups to generate gene phylogenies to use as source data for our methods. To identify families of orthologous and paralogous genes in our data set, we set the following criteria:
(1) A single-copy gene family must contain no more than one orthologous gene per species and must be present in four or more species.
(2) A multicopy gene family must contain at least four unique species, and two or more paralogs must be present in at least one of the species.
Using OrthoMCL (92), with an inflation value of 1.5 and a strict BLASTp cutoff value of 10−20 (93) and bespoke Python scripting, we identified over 56,000 homologous oomycete and SAR gene families in our data set. Of these, 2,853 families matched our criterion for single-copy families and 11,158 families matched our criterion for multicopy families. By aligning each of these gene families in MUSCLE (94) and sampling for highly conserved regions using Gblocks (95), both using the default parameters, and then carrying out permutation-tail possibility (PTP) tests for every remaining sampled alignment using PAUP* (96, 97), we were able to remove 576 single-copy gene families and 5,103 multicopy gene families with poor phylogenetic signal from our data. All remaining gene families had their evolutionary model estimated using ProtTest (98) (Table S2), and maximum-likelihood gene phylogenies were generated using PhyML with 100 bootstrap replicates (99). We generated phylogenetic reconstructions for 2,280 orthologous gene families (containing 35,622 genes) and 6,055 paralogous gene families (containing 174,282 genes). In total, from our 43-genome data set, we identified 8,335 individual gene phylogenies, containing 209,904 oomycete and SAR genes.
Selection of evolutionary models (not including additional parameters) for single-copy and multicopy gene phylogenies by ProtTest. Download TABLE S2, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2017 McCarthy and Fitzpatrick.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supetree phylogenies fully resolve oomycete class and order phylogenies.
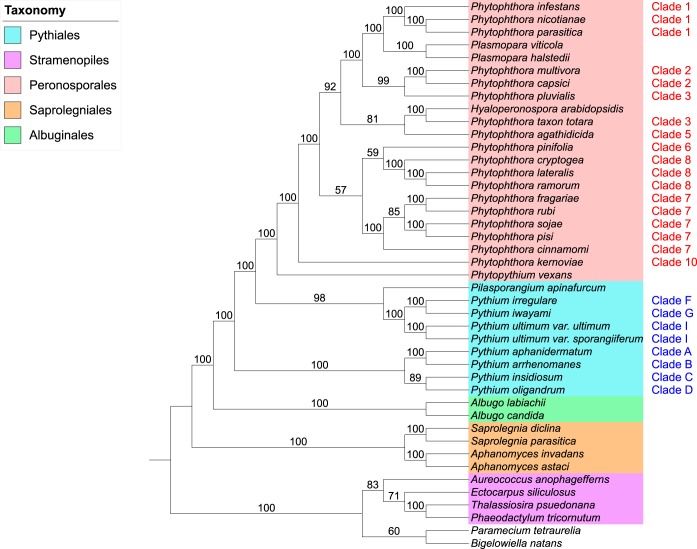
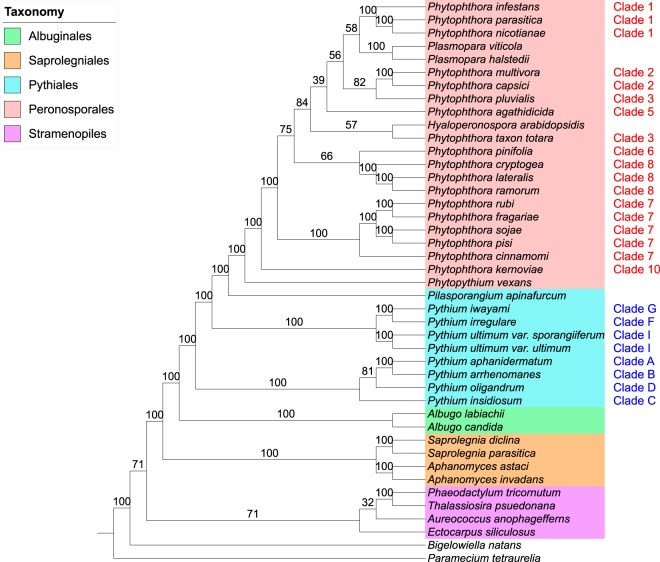
All 2,280 orthologous single-copy gene phylogenies (35,622 genes in total) were used as input for CLANN (100), which implements a matrix representation using parsimony (MRP) method to determine consensus phylogeny for many source phylogenies with overlapping taxa or missing taxa. An MRP supertree phylogeny was generated in CLANN using a heuristic search with 100 bootstrap replicates. The supertree was visualized and annotated within the Interactive Tree of Life (iTOL) website (101) and rooted at the branch containing the SAR outgroups, Paramecium tetraurelia (Alveolata), Bigelowiella natans (Rhizaria), and four stramenopiles species (Fig. 3).
FIG 3 .
Matrix representation with parsimony (MRP) supertree of 37 oomycete species and 6 SAR species (2,280 source phylogenies). The supertree was generated in CLANN. The phylogeny is rooted at the SAR branch. Phytophthora clades as designated by Blair et al. (42) and Pythium clades as designated by de Cock et al. (50) are indicated in red and blue, respectively. No color, P. tetraurelia (Alveolata) and B. natans (Rhizaria).
MRP supertree analysis of 2,280 orthologous single-copy oomycete gene phylogenies supported the four “crown” oomycete orders (Saprolegniales, Albuginales, Pythiales, and Peronosporales), with maximum bootstrap support (BP) (Fig. 3). The MRP supertree reflects the consensus phylogeny of the oomycetes (31–33) (Fig. 1). The Saprolegniales species represent the most basal “crown” order, and the Albuginales is a sister order to the Pythiales and Peronosporales. Within the Pythiales themselves, a highly supported split among Pythium clades A to D (100% BP) and clades F to I (100% BP) was observed, matching similar splits seen in small-scale analyses (51, 52) (Fig. 3). Pilasporangium apinafurcum, a Pythiales species, is placed sister to Pythium clades F to I (98% BP). Phytopythium vexans is placed at the base of the Peronosporales order (Fig. 3), supporting the recent reclassification of the Phytopythium genus from the Pythiales (50). Many individual Phytophthora clades within the Peronosporales are well supported. In addition, the “downy mildews” species in our data set (Hyaloperonospora arabidopsidis and two Plasmopara species) place as derived taxa within the Peronosporales order rather than as basal to Phytophthora (Fig. 3). The overall phylogeny of the Peronosporales in our MRP supertree is summarized in Fig. 4a and discussed in greater detail later in the text. As an additional analysis, a consensus supernetwork of the phylogenetic splits within the 2,280 single-copy gene phylogenies was generated in SplitsTree (102) (see Fig. S1 in the supplemental material). The network further highlights support for the four “crown” oomycete orders and the division of the Pythiales order as in the supertree phylogeny; it also recapitulates many of individual Phytophthora clades and intraorder relationships within the Peronosporales (Fig. 3 and 4a; Fig. S1).
FIG 4 .
Congruence of the Peronosporales order data between our supertree and supermatrix methods. (a) MRP analysis. (b) GTP analysis. (c) Concatenated supermatrix analysis. For full phylogenies, refer to Fig. 3, 5, and 6, respectively.
Consensus network of phylogenetic splits within 2,280 single-copy oomycete and SAR phylogenies. The network was generated in SplitsTree. Clades are colored by taxonomic order as follows: red, Peronosporales; blue, Pythiales; green, Albuginales; orange, Saprolegniales; no color, P. tetraurelia (Alveolata) and B. natans (Rhizara). Download FIG S1, PDF file, 0.1 MB (121.4KB, pdf) .
Copyright © 2017 McCarthy and Fitzpatrick.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Both the 2,280 single-copy phylogenies and the 6,055 multicopy phylogenies (209,904 genes in total) were used as input for DupTree (103), which uses a gene tree parsimony (GTP) method to determine consensus phylogeny for many source phylogenies that may include gene duplication events. The source data were bootstrapped with 100 replicates, and the resultant consensus GTP supertree was rooted at the branch containing Paramecium tetraurelia, Bigelowiella natans, and the other stramenopiles species (Fig. 5). As in the single-gene MRP supertree, all four individual crown oomycete orders and the oomycete class phylogeny are highly supported. The Pythiales order is once again split into highly supported sister branches containing clades A to D (100% BP) and clades F to I (100% BP) (Fig. 5). The Peronosporales order is highly supported again (100% BP), as is the placement of Phytopythium vexans at the base of this order (Fig. 5). As with the single-gene MRP supertree, the downy mildews (P. viticola and P. halstedii) are found as sister taxa to clade 1 Phytophthora species. However, it is worth pointing out that phylogenetic support for this grouping is weaker in the GTP supertree (58% BP) (Fig. 4b and 5) than in the MRP supertree, where support is very strong (100% BP) (Fig. 3). Overall, the phylogeny of the Peronosporales order in our GTP supertree displays weaker bootstrap support at some branches than in the single-gene MRP supertree. However, with the exception of the placement of clade 5, the overall taxonomic congruence between the two supertree approaches for the Peronosporales is high (Fig. 3, 4a and b, and 5).
FIG 5 .
Gene tree parsimony (GTP) supertree of 37 oomycete species and 6 SAR species (8,335 source phylogenies). The supertree was generated in DupTree. The phylogeny is rooted at the SAR branch. Phytophthora clades as designated by Blair et al. (42) and Pythium clades as designated by de Cock et al. (50) are indicated in red and blue, respectively. No color, P. tetraurelia (Alveolata) and B. natans (Rhizaria).
The supermatrix approach based on ubiquitous Peronosporales gene phylogenies supports supertree phylogenies.
As a complement to our supertree method phylogenies, we undertook a supermatrix approach to infer the oomycete species phylogeny using oomycete orthologs of known proteins corresponding to clusters of orthologous groups (COG) as phylogenetic markers (104). To identify oomycete COGs, we performed a reciprocal BLASTp analysis of all 458 Saccharomyces cerevisiae COGs against the 37 oomycete proteomes in our full data set (590,896 protein sequences in total) with an E value of 10−10. Overall, 443 oomycete gene families that were reciprocal top hits to S. cerevisiae COGs were retrieved. Of the 443 COG families, 144 families contained an ortholog from all 37 oomycete species and were retained for analysis. A superalignment of 16,934 characters was generated by concatenating the 131 aligned families which retained alignment data after Gblocks sampling with FASconCAT (105). The maximum-likelihood phylogeny of this superalignment was reconstructed in PhyML with 100 bootstrap replicates and an LG+I+G+F amino acid substitution model as selected by ProtTest, and the resultant consensus phylogeny was rooted at the Saprolegniales branch (Fig. S2). This initial supermatrix phylogeny supported the four “crown” orders similarly to our supertree phylogenies; however, poor resolution and inconsistent phylogeny were observed within the Peronosporales, particularly the placement of species from Phytophthora clades 7 and 8; for example, clade 7 species are not monophyletic (Fig. S2). To attempt to tease apart the data corresponding to the poor resolution of the Peronosporales in our maximum-likelihood phylogeny, a neighbor-joining network was generated for the COG superalignment in SplitsTree to visualize the bifurcations within the superalignment (Fig. S3). As can be seen in the network, a significant amount of phylogenetic conflict is obvious and is represented as alternative splits among Peronosporales clades, a phenomenon that is consistent with poor bootstrap support and inconsistent topology (relative to supertrees) throughout the Peronosporales in this class-level supermatrix phylogeny (Fig. S2 and S3).
Maximum-likelihood (ML) supermatrix phylogeny of 37 oomycete species (144 families of Saccharomyces cerevisiae COG homologs, 16,934 characters). Supermatrix phylogeny data were generated in PhyML with an LG+I+G+F amino acid substitution model. The phylogeny is rooted at the Saprolegniales branch. Phytophthora clades as designated by Blair et al. (42) and Pythium clades as designated by de Cock et al. (50) are indicated in red and blue, respectively. No color, P. tetraurelia (Alveolata) and B. natans (Rhizaria). Download FIG S2, PDF file, 0.1 MB (60.2KB, pdf) .
Copyright © 2017 McCarthy and Fitzpatrick.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Neighbor-joining network of phylogenetic splits within oomycete COG supermatrix of 37 oomycete species. The network was generated in SplitsTree. Clades are colored by taxonomic order as follows: red, Peronosporales; blue, Pythiales; green, Albuginales; orange, Saprolegniales. Download FIG S3, PDF file, 0.02 MB (18.3KB, pdf) .
Copyright © 2017 McCarthy and Fitzpatrick.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
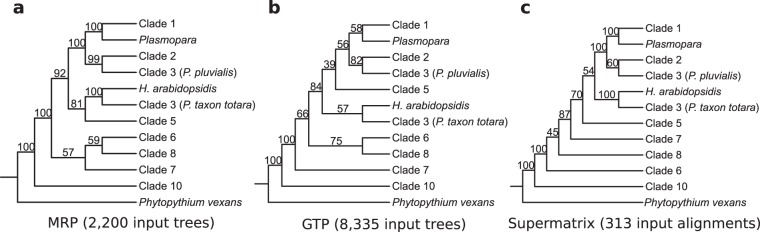
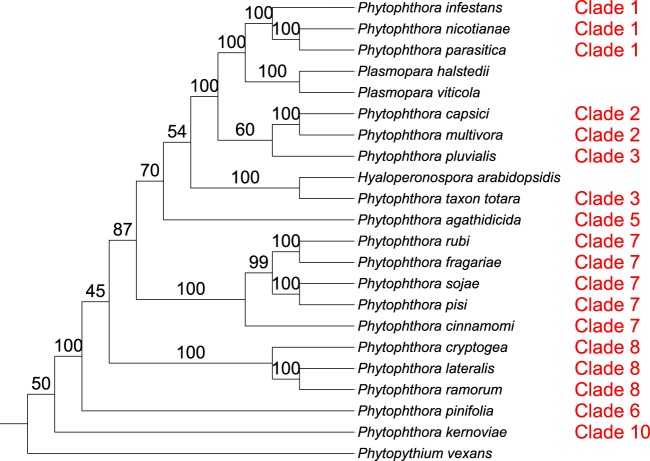
To extend our COG supermatrix phylogeny, we took the approach of generating a supermatrix from ubiquitous gene families within the 22 Peronosporales species in our data set. Using this approach, we hoped to extend the amount of available alignment data for species solely within Peronosporales to improve resolution of the order. We defined a ubiquitous Peronosporales gene family as containing exactly one ortholog from all 22 Peronosporales species in our data set. Using OrthoMCL, with a strict BLASTp E value of 10−20 and an inflation value of 1.5, we identified over 20,000 orthologous gene families in the 22 Peronosporales proteomes in our data set. From these families, we identified 352 ubiquitous gene families within Peronosporales using bespoke Python scripting; each family was then aligned in MUSCLE and sampled in Gblocks. After removing families which did not retain alignment data after Gblocks, we concatenated the remaining 313 gene families into a superalignment that was 47,365 amino acids in length. The maximum-likelihood phylogeny for this superalignment was generated with 100 bootstrap replicates and a JTT+I+G+F evolutionary model. The resultant consensus phylogeny was rooted at Phytopythium vexans (Fig. 6). While resolution of relationships among clades is still weak at some branches, the higher support seen on many other branches and the overall topology of the ubiquitous supermatrix phylogeny represent substantial improvements over the COG supermatrix. Phytophthora clades 1, 2, 7, and 8 are now all monophyletic, with 100% bootstrap support each. The genus is split between the basal lineages (Phytopythium and Phytophthora clades 6 to 10) and the more extensively derived Phytophthora clades (clades 1 to 5) and the downy mildews, which form a monophyletic group (70% BP) (Fig. 4c and 6), an inference that is also observed in our supertree species phylogenies and with the highest degree of congruence to the single-gene MRP supertree (Fig. 4a and b).
FIG 6 .
Maximum-likelihood (ML) supermatrix phylogeny of 22 Peronosporales species (313 ubiquitous Pernosporales gene families, 47,635 characters). The supermatrix phylogeny was generated in PhyML with a JTT+I+G+F amino acid substitution model. The cladogram is rooted at Phytopythium vexans. Phytophthora clades as designated by Blair et al. (2008) are shown in red.
Resolution of the Peronosporales order in phylogenomic analysis.
All three of our whole-genome species phylogenies strongly support the Peronosporales order (Fig. 4) and display a high degree of congruence with one another. Each phylogeny also supports the recent reclassification of Phytopythium from the Pythiales to the Peronosporales as a basal taxon (50). All three phylogenies also show varying but strong bootstrap support (70 to 92% BP) for the divergence of Phytophthora clades 1 to 5 and the downy mildews (Plasmopara spp., H. arabidopsidis) from the remaining Phytophthora clades and Phytopythium at a single point (Fig. 4c). The relationships among these taxa across our phylogenies can be summarized as follows:
(1) The downy mildews species Hyaloperonospora arabidopsidis and Phytophthora taxon Totara (Phytophthora clade 3) are sister taxa, with maximum support in both MRP and supermatrix analysis (Fig. 4a and c). Therefore, Phytophthora clade 3 is not monophyletic in any of our species phylogenies (Fig. 4). Phytophthora taxon Totara has provisionally been assigned to clade 3 based on sequence similarity. Our species phylogenies suggest that it is not actually a clade 3 species.
(2) A close relationship between Phytophthora clades 1 and 2, the clade 3 species Phytophthora pluvialis, and the downy mildew species Plasmopara viticola and Plasmopara halstedii is observed in each phylogeny, with maximum support in both MRP and supermatrix analysis (Fig. 4a and c).
The placement of the clade 5 species Phytophthora agathidcida varies in each phylogeny, but it appears that the species is most closely related to Phytophthora taxon Totara and H. arabidopsidis within the Peronosporales, as is most apparent in the single-gene MRP supertree (81% BP) (Fig. 3 and 4a). As for the more basal clades, both the MRP and GTP phylogenies show support for the idea of clade 6 species Phythophthora pinifolia being sister to Phytophthora clade 8, with highest bootstrap support of 59% and 75%, respectively (Fig. 4a and b).
In our analysis, we set out to resolve relationships within the oomycetes where conflicts have arisen in different analyses, particularly in the Peronosporales order (Fig. 2). With respect to the divergence of Phytophthora clades 1 to 5 and the downy mildews from the remaining basal taxa in the Peronosporales (i.e., Phytophthora clades 6 to 10 and Phytopythium), our results are congruent with the small-scale analyses performed by Blair et al. and Martin et al. (42, 47) (Fig. 2a, c, and d), with closest topological similarity to the latter authors’ 6-locus coalescence method phylogeny (Fig. 2d), despite a lack of data from H. arabidopsidis and Plasmopara species in both analyses and the inclusion of H. arabidopsidis data in the analysis carried out by Runge et al. (46) (Fig. 2b). Our own analysis lacks data from any species in Phytophthora clade 4, which is still unsampled in terms of genome sequencing. In the analysis by Runge et al., H. arabidopsidis branches within paraphyletic Phytophthora clade 4; were there a representative species from clade 4 available, a greater degree of resolution for the relationships among Phytophthora clades 3 to 5 and Hyaloperonospora might be observed. However, it is not clear whether the placement of H. arabidopsidis relative to Phytophthora clade 1 would then recapitulate that described by Runge et al. (46). Similarly, with regard to the basal taxa, our result are relatively congruent with the linearized relationships seen in previous analyses (Fig. 2), although the close relationship of clade 6 species Phytophthora pinifolia to Phytophthora clade 7 seen in our two supertree methods is not reflected in any of the multilocus phylogenies (Fig. 4a and b). The resolution of the relationships among Phytophthora clades 6, 7, and 8 varies both in support and sister group relationships among our analyses (Fig. 4); however, similar variation can be observed between the highlighted multilocus phylogenies (42, 46, 47) (Fig. 2). The lack of available genomic data from Phytophthora clade 9 also prevents any conclusions regards its placement in a whole-genome phylogeny; however, we would expect that it would branch as a sister to clade 10 species such as Phytophthora kernoviae, as the relationship between clades 9 and 10 has been highly supported in multilocus analyses (42, 46, 47).
The use of supertree and phylogenomic methods in oomycete systematics.
Our analysis is the first large-scale genome phylogeny of the oomycetes as a class, using all extant genomic data from 37 oomycete species. Our analysis has recapitulated the four crown orders of the oomycetes and many relationships within the two largest-sampled orders, the Pythiales and the Peronosporales. During our analysis, we were conscious of potential characteristics of oomycete genomes that could obfuscate phylogenomic analysis. The role of HGT and its impact on the quality of our analyses were considered; it has been shown that supertree and supermatrix analyses are thought to be susceptible to misleading signal in data sets where a large degree of HGT has occurred, particularly in MRP analysis (106). While HGT from other microbial eukaryotes, fungi, and prokaryotes has been identified within oomycete genomes, the majority of these events are thought to be ancestral or to have not occurred in proportions large enough to impact our results (4, 5, 107). Other factors, such as fast-evolving regions of genomes or ancestral gene loss or duplication events within the oomycetes, are not likely to have affected our analysis, given our genome-wide scale of data acquisition and our strict filtering of gene families with poor phylogenetic signal (10, 48, 96). Intraspecific hybridization within the Phytophthora genus has been increasingly reported in the literature and usually occurs in nature among Phytophthora species within the same phylogenetic clade (108). A number of hybrid species or hybridization events have been described in Phytophthora clades 6 to 8 (108–110); however, none of these species are present in our data set. Also, where hybridization has occurred, it has been between closely related species and, in the case of Phytophthora species, those from the same phylogenetic clade. Taking this into consideration, hybridization should affect intraclade relationships to a greater degree than interclade relationships.
Compared with fungi, particularly in light of the ongoing 1,000 fungal genomes project (http://1000.fungalgenomes.org), there is a relative dearth of genomic data for both the earliest diverging lineages and the “crown” taxa within the oomycetes. With the greater sampling of genomic sequencing of the oomycetes likely to occur in the future, it is our view that subsequent genome phylogenies of the oomycetes will match the success of other eukaryotic genome phylogenies at resolving individual problematic clade and species relationships (60, 62). We suspect that, with a broader sampling of all Phytophthora clades and downy mildew species, we would see better resolution of the Peronosporales within any subsequent oomycete genome phylogenies. Similar approaches with other oomycete taxa, such as Pythium, may disentangle some of the phylogenetic conflicts seen in recent analyses (49, 53). Similarly, sequencing of more Saprolegniales species or basal oomycete species and their inclusion in similar analyses will potentially help uncover further aspects of oomycete evolution, including the evolution of phytopathogenicity. Such analyses, for which ours is a first step, would also provide the benefit of establishing a robust phylogeny for a eukaryotic group with such devastating ecological impact and would hopefully encourage further genomics and phylogenomics research into the oomycetes.
Conclusions.
Using 37 oomycete genomes and 6 SAR genomes, we have carried out the first whole-genome phylogenetic analysis of the oomycetes as a class. The different methods that we used in our analysis support the four “crown” oomycete orders and support many individual phylogenetic clades within genera. Our analysis also generally supports the placement of Phytopythium within the Peronosporales, the placement of the downy mildews within the Phytophthora genus, and the topology of clades within the Pythiales order. However, resolution of the Peronosporales as an order remains weak at some branches, possibly due to a lack of genomic data for some phylogenetic clades within Phytophthora. As the amount of genomic data available for the oomycetes increases, future genome phylogenies of the class should resolve these branches, as well as those within currently unsampled basal lineages or undersampled taxa such as Saprolegnia. Our analysis represents an important backbone for oomycete phylogenetics upon which future analyses can be based.
MATERIALS AND METHODS
Data set assembly.
The predicted proteomes for 29 SAR species (23 oomycete species, 4 other stramenopile species, the alveolate species Paramecium tetraurelia, and the rhizarian species Bigelowiella natans) were obtained from public databases (Table 1). Predicted proteomes for a further 14 oomycete species (10 Phythophthora species, 2 Pythium species, Plasmopara viticola, and Pilasporangium apinafurcum) were generated from publically available assembly data using AUGUSTUS (90). Templates for ab initio protein prediction with AUGUSTUS were generated from assembly and expressed sequence tag (EST) data from a number of reference oomycete species (Phytophthora sojae, Phytophthora capsici, Pythium ultimum var. ultimum, and Plasmopara halstedii) (Table S1). Ph. capsici was used as a reference for Phytophthora species from clades 1 to 5, while Ph. sojae was used as a reference for Phytophthora species from clades 6 to 10. Py. ultimum var. ultimum was used as a reference for two Pythium species and Pi. apinafurcum. P. halstedii was used as a reference for P. viticola. GeneMark-ES (91) was used in conjuction with AUGUSTUS for protein prediction for Pi. apinafurcum. The taxonomy, assembly, and prediction statistics for each of the 14 assemblies included in this study are summarized in Table S1. Our final data set contained 702,132 protein sequences from 37 oomycete genomes and 6 SAR genomes (66–89) (Table 1; Table S1).
Identification and reconstruction of gene phylogenies in oomycete and SAR genomes.
All 702,132 protein sequences in our data set were filtered and clustered into 56,638 orthologous gene families using OrthoMCL (92), with a BLASTp E value cutoff of 10−20 (93) and an inflation value of 1.5. Using bespoke Python scripting, we identified and retrieved two types of gene family containing 200 sequences or fewer from the 56,638 families within our data set as follows:
(1) A total of 2,853 single-copy gene families (single-copy orthologs present in ≥4 species.
(2) A total of 11,158 multicopy gene families (≥1 paralog[s] present in ≥4 species).
Each of these gene families was retrieved and aligned in MUSCLE (94), and highly conserved regions of these alignments were sampled using Gblocks (95) with the default parameters. A total of 266 single-copy gene families and a total of 4,928 multicopy gene families did not retain alignment data after Gblocks sampling and were discarded. Permutation-tail probability (PTP) tests (96) were carried out for every remaining sampled gene family in PAUP* (97), using 100 replicates, to determine whether a given sampled gene family had phylogenetic signal. Those sampled gene families whose PTP test result had a P value of ≤0.05 were considered to have signal and were retained. A total of 2,280 single-copy sampled gene families (containing 35,622 genes in total) and a total of 6,055 multicopy sampled gene families (containing 174,282 genes in total) ultimately satisfied our filtering process. Best-fit amino acid replacement models were selected for every remaining sampled gene family using ProtTest (Table S2), and maximum-likelihood phylogenetic reconstruction was carried out using PhyML with 100 bootstrap replicates.
Supertree analyses of single-copy and paralogous gene phylogenies.
Maximum-parsimony supertree analysis of 2,280 single-copy gene phylogenies (containing 35,622 genes in total) was carried out using CLANN, by performing a subtree prune and regraft (SPR) heuristic search with 100 bootstrap replicates (100). This phylogeny was visualized and annotated as a cladogram using the Interactive Tree of Life (iTOL) website (101) (Fig. 3). As an additional analysis, a consensus supernetwork of phylogenetic multifurcations within the 2,280 individual gene phylogenies was generated in SplitsTree (102) (see Fig. S1 in the supplemental material). Gene tree parsimony (GTP) supertree analyses of all 8,335 gene phylogenies (containing 209,904 genes in total) was carried out using DupTree (103) and a rooted SPR heuristic search of 100 bootstrapped replicates of each phylogeny. A consensus phylogeny was generated from all individual replicates and was visualized and annotated as a cladogram using iTOL (Fig. 5).
Identification and supermatrix analysis of ubiquitous oomycete gene phylogenies.
A reciprocal BLASTp search was carried out with an E value cutoff of 10−10 among all 37 oomycetes proteomes in our data set (590,896 protein sequences in total) and 458 core orthologous genes (COGs) in Saccharomyces cerevisiae from the CEGMA data set (93, 104). A total of 443 oomycete gene families representing oomycete top hits to S. cerevisiae COGs were retrieved, among which 144 families contained an ortholog from all 37 oomycete species in our data set. Each of these 144 families was aligned in MUSCLE and was sampled for highly conserved regions using Gblocks with the default parameters. After 13 families which failed to retain alignment data after Gblocks sampling were removed, the remaining 131 sampled alignments (containing 4,847 genes in total) were concatenated into a superalignment of 16,934 aligned positions. This superalignment was bootstrapped 100 times using Seqboot, and maximum-likelihood phylogenetic trees were generated for each individual replicate using PhyML, with an LG+I+G+F amino acid substitution model as selected by ProtTest. A consensus tree was generated from these replicate trees using Consense, and the consensus tree was visualized and annotated as a cladogram using iTOL (Fig. S2). A neighbor-joining network of phylogenetic splits in the original superalignment was generated in SplitsTree (Fig. S3).
Identification and supermatrix analysis of ubiquitous Peronosporales gene phylogenies.
A total of 347,375 protein sequences from the 22 Peronosporales proteomes in our data set were filtered and clustered into 22,803 orthologous gene families using OrthoMCL, with a BLASTp E value cutoff of 10−20 and an inflation value of 1.5. Using bespoke Python scripting, we identified 352 ubiquitous Peronosporales gene families, which we defined as any family which had exactly one representative ortholog from all 22 Peronosporales species in our data set. Each of these families was aligned in MUSCLE and sampled for highly conserved regions using Gblocks with the default parameters. After 39 gene families which did not retain alignment data after sampling were removed, the remaining 313 sampled alignments (containing 6,886 genes in total) were concatenated into a single superalignment of 47,365 aligned positions. This superalignment was bootstrapped 100 times using Seqboot, and maximum-likelihood phylogenetic trees were generated for each individual replicate using PhyML with a JTT+I+G+F amino acid substitution model, as selected by ProtTest. A consensus tree was generated from these replicate trees using Consense, and the consensus tree was visualized and annotated as a cladogram using iTOL (Fig. 6).
ACKNOWLEDGMENTS
We acknowledge the DJEI/DES/SFI/HEA Irish Centre for High-End Computing (ICHEC) for the provision of computational facilities and support. We also thank Joyce Reilly for generating corroborating results through her undergraduate research project.
C.G.P.M. is funded by Irish Research Council grant number GOIPG/2015/2242.
REFERENCES
- 1.Beakes GW, Glockling SL, Sekimoto S. 2012. The evolutionary phylogeny of the oomycete “fungi.” Protoplasma 249:3–19. doi: 10.1007/s00709-011-0269-2. [DOI] [PubMed] [Google Scholar]
- 2.Lévesque CA. 2011. Fifty years of oomycetes—from consolidation to evolutionary and genomic exploration. Fungal Divers 50:35–46. doi: 10.1007/s13225-011-0128-7. [DOI] [Google Scholar]
- 3.Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ. 2006. Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Curr Biol 16:1857–1864. doi: 10.1016/j.cub.2006.07.052. [DOI] [PubMed] [Google Scholar]
- 4.Richards TA, Soanes DM, Jones MDM, Vasieva O, Leonard G, Paszkiewicz K, Foster PG, Hall N, Talbot NJ. 2011. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci U S A 108:15258–15263. doi: 10.1073/pnas.1105100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savory F, Leonard G, Richards TA. 2015. The role of horizontal gene transfer in the evolution of the oomycetes. PLoS Pathog 11:e1004805. doi: 10.1371/journal.ppat.1004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burki F. 2014. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol 6:a016147. doi: 10.1101/cshperspect.a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsui CKM, Marshall W, Yokoyama R, Honda D, Lippmeier JC, Craven KD, Peterson PD, Berbee ML. 2009. Labyrinthulomycetes phylogeny and its implications for the evolutionary loss of chloroplasts and gain of ectoplasmic gliding. Mol Phylogenet Evol 50:129–140. doi: 10.1016/j.ympev.2008.09.027. [DOI] [PubMed] [Google Scholar]
- 8.Cavalier-Smith T, Chao EEY. 2006. Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista). J Mol Evol 62:388–420. doi: 10.1007/s00239-004-0353-8. [DOI] [PubMed] [Google Scholar]
- 9.Riisberg I, Orr RJS, Kluge R, Shalchian-Tabrizi K, Bowers HA, Patil V, Edvardsen B, Jakobsen KS. 2009. Seven Gene phylogeny of heterokonts. Protist 160:191–204. doi: 10.1016/j.protis.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Judelson HS. 2012. Dynamics and innovations within oomycete genomes: insights into biology, pathology, and evolution. Eukaryot Cell 11:1304–1312. doi: 10.1128/EC.00155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavalier-Smith T. 1981. Eukaryote kingdoms: seven or nine? Biosystems 14:461–481. doi: 10.1016/0303-2647(81)90050-2. [DOI] [PubMed] [Google Scholar]
- 12.Cavalier-Smith T. 1999. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol 46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- 13.Keeling PJ. 2009. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J Eukaryot Microbiol 56:1–8. doi: 10.1111/j.1550-7408.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- 14.Bachvaroff TR, Sanchez Puerta MV, Delwiche CF. 2005. Chlorophyll c-containing plastid relationships based on analyses of a multigene data set with all four chromalveolate lineages. Mol Biol Evol 22:1772–1782. doi: 10.1093/molbev/msi172. [DOI] [PubMed] [Google Scholar]
- 15.Yoon HS, Hackett JD, Pinto G, Bhattacharya D. 2002. The single, ancient origin of chromist plastids. Proc Natl Acad Sci U S A 99:15507–15512. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janouskovec J, Horák A, Oborník M, Lukes J, Keeling PJ. 2010. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A 107:10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice DW, Palmer JD. 2006. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol 4:31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaston D, Roger AJ. 2013. Functional divergence and convergent evolution in the plastid-targeted glyceraldehyde-3-phosphate dehydrogenases of diverse eukaryotic algae. PLoS One 8:e70396. doi: 10.1371/journal.pone.0070396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harper JT, Waanders E, Keeling PJ. 2005. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int J Syst Evol Microbiol 55:487–496. doi: 10.1099/ijs.0.63216-0. [DOI] [PubMed] [Google Scholar]
- 20.Hackett JD, Yoon HS, Li S, Reyes-Prieto A, Rümmele SE, Bhattacharya D. 2007. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of Rhizaria with chromalveolates. Mol Biol Evol 24:1702–1713. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- 21.Keeling PJ. 2001. Foraminifera and Cercozoa are related in actin phylogeny: two orphans find a home? Mol Biol Evol 18:1551–1557. doi: 10.1093/oxfordjournals.molbev.a003941. [DOI] [PubMed] [Google Scholar]
- 22.Baldauf SL. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 23.Burki F, Shalchian-Tabrizi K, Minge M, Skjaeveland A, Nikolaev SI, Jakobsen KS, Pawlowski J. 2007. Phylogenomics reshuffles the eukaryotic supergroups. PLoS One 2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, Simpson AGB, Roger AJ. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci U S A 106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shalchian-Tabrizi K, Kauserud H, Massana R, Klaveness D, Jakobsen KS. 2007. Analysis of environmental 18S ribosomal RNA sequences reveals unknown diversity of the Cosmopolitan phylum Telonemia. Protist 158:173–180. doi: 10.1016/j.protis.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Moreira D, von der Heyden S, Bass D, López-García P, Chao E, Cavalier-Smith T. 2007. Global eukaryote phylogeny: combined small- and large-subunit ribosomal DNA trees support monophyly of Rhizaria, Retaria and Excavata. Mol Phylogenet Evol 44:255–266. doi: 10.1016/j.ympev.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Dick MW. 2001. Straminipilous fungi. Kluwer Academic Publishers, Amsterdam, The Netherlands. [Google Scholar]
- 28.Matari NH, Blair JE. 2014. A multilocus timescale for oomycete evolution estimated under three distinct molecular clock models. BMC Evol Biol 14:101. doi: 10.1186/1471-2148-14-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor TN, Krings M, Kerp H. 2006. Hassiella monospora gen. et sp. nov., a microfungus from the 400 million year old Rhynie chert. Mycol Res 110:628–632. doi: 10.1016/j.mycres.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Thines M, Kamoun S. 2010. Oomycete–plant coevolution: recent advances and future prospects. Curr Opin Plant Biol 13:427–433. doi: 10.1016/j.pbi.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Hakariya M, Hirose D, Tokumasu S. 2007. A molecular phylogeny of Haptoglossa species, terrestrial Peronosporomycetes (oomycetes) endoparasitic on nematodes. Mycoscience 48:169–175. doi: 10.1007/S10267-007-0355-7. [DOI] [Google Scholar]
- 32.Beakes D, Honda D, Thines M, Beakes GW, Honda D, Thines M. 2014. Systematics of the Straminipila: Labyrinthulomycota, Hyphochytriomycota, and Oomycota, p 39–97. In The mycota. Springer, Berlin, Germany. [Google Scholar]
- 33.Sekimoto S, Yokoo K, Kawamura Y, Honda D. 2008. Taxonomy, molecular phylogeny, and ultrastructural morphology of Olpidiopsis porphyrae sp. nov. (oomycetes, straminipiles), a unicellular obligate endoparasite of Bangia and Porphyra spp. (Bangiales, Rhodophyta). Mycol Res 112:361–374. doi: 10.1016/j.mycres.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Jiang RHY, Tyler BM. 2012. Mechanisms and evolution of virulence in oomycetes. Annu Rev Phytopathol 50:295–318. doi: 10.1146/annurev-phyto-081211-172912. [DOI] [PubMed] [Google Scholar]
- 35.Thines M. 2014. Phylogeny and evolution of plant pathogenic oomycetes-a global overview. Eur J Plant Pathol 138:431–447. doi: 10.1007/s10658-013-0366-5. [DOI] [Google Scholar]
- 36.Riethmüller A, Voglmayr H, Göker M, Weiß M, Oberwinkler F. 2002. Phylogenetic relationships of the downy mildews (Peronosporales) and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia 94:834–849. doi: 10.2307/3761698. [DOI] [PubMed] [Google Scholar]
- 37.Hulvey JP, Padgett DE, Bailey JC. 2007. Species boundaries within Saprolegnia (Saprolegniales, Oomycota) based on morphological and DNA sequence data. Mycologia 99:421–429. doi: 10.3852/mycologia.99.3.421. [DOI] [PubMed] [Google Scholar]
- 38.Kamoun S. 2003. Molecular genetics of pathogenic oomycetes. Eukaryot Cell 2:191–199. doi: 10.1128/EC.2.2.191-199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaastra W, Lipman LJA, De Cock AWAM, Exel TK, Pegge RBG, Scheurwater J, Vilela R, Mendoza L. 2010. Pythium insidiosum: an overview. Vet Microbiol 146:1–16. doi: 10.1016/j.vetmic.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 40.Benhamou N, le Floch G, Vallance J, Gerbore J, Grizard D, Rey P. 2012. Pythium oligandrum: an example of opportunistic success. Microbiology 158:2679–2694. doi: 10.1099/mic.0.061457-0. [DOI] [PubMed] [Google Scholar]
- 41.Cooke DE, Drenth A, Duncan JM, Wagels G, Brasier CM. 2000. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet Biol 30:17–32. doi: 10.1006/fgbi.2000.1202. [DOI] [PubMed] [Google Scholar]
- 42.Blair JE, Coffey MD, Park SY, Geiser DM, Kang S. 2008. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet Biol 45:266–277. doi: 10.1016/j.fgb.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 43.Weir BS, Paderes EP, Anand N, Uchida JY, Pennycook SR, Bellgard SE, Beever RE. 2015. A taxonomic revision of Phytophthora clade 5 including two new species, Phytophthora agathidicida and P. Cocois. Phytotaxa 205:21–38. doi: 10.11646/phytotaxa.205.1.2. [DOI] [Google Scholar]
- 44.Kroon LPNM, Bakker FT, Van Den Bosch GBM, Bonants PJM, Flier WG. 2004. Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genet Biol 41:766–782. doi: 10.1016/j.fgb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 45.Martin FN, Tooley PW. 2003. Phylogenetic relationships of Phytophthora ramorum, P. nemorosa, and P. pseudosyringae, three species recovered from areas in California with sudden oak death. Mycol Res 107:1379–1391. doi: 10.1017/S0953756203008785. [DOI] [PubMed] [Google Scholar]
- 46.Runge F, Telle S, Ploch S, Savory E, Day B, Sharma R, Thines M. 2011. The inclusion of downy mildews in a multi-locus-dataset and its reanalysis reveals a high degree of paraphily in Phytophthora. IMA Fungus 2:163–171. doi: 10.5598/imafungus.2011.02.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin FN, Blair JE, Coffey MD. 2014. A combined mitochondrial and nuclear multilocus phylogeny of the genus Phytophthora. Fungal Genet Biol 66:19–32. doi: 10.1016/j.fgb.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Seidl MF, Van den Ackerveken G, Govers F, Snel B. 2012. Reconstruction of oomycete genome evolution identifies differences in evolutionary trajectories leading to present-day large gene families. Genome Biol Evol 4:199–211. doi: 10.1093/gbe/evs003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robideau GP, Rodrigue N, André Lévesque C. 2014. Codon-based phylogenetics introduces novel flagellar gene markers to oomycete systematics. Mol Phylogenet Evol 79:279–291. doi: 10.1016/j.ympev.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 50.de Cock AWAM, Lodhi AM, Rintoul TL, Bala K, Robideau GP, Abad ZG, Coffey MD, Shahzad S, Lévesque CA. 2015. Phytopythium: molecular phylogeny and systematics. Persoonia Mol Phylogeny Evol Fungi 34:25–39. doi: 10.3767/003158515X685382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lévesque CA, de Cock AW. 2004. Molecular phylogeny and taxonomy of the genus Pythium. Mycol Res 108:1363–1383. doi: 10.1017/s0953756204001431. [DOI] [PubMed] [Google Scholar]
- 52.Villa NO, Kageyama K, Asano T, Suga H. 2006. Phylogenetic relationships of pythium and Phytophthora species based on ITS rDNA, cytochrome oxidase II and beta-tubulin gene sequences. Mycologia 98:410–422. doi: 10.3852/mycologia.98.3.410. [DOI] [PubMed] [Google Scholar]
- 53.Uzuhashi S, Tojo M, Kakishima M. 2010. Phylogeny of the genus Pythium and description of new genera. Mycoscience 51:337–365. doi: 10.1007/S10267-010-0046-7. [DOI] [Google Scholar]
- 54.Hyde KD, Nilsson RH, Alias SA, Ariyawansa HA, Blair JE, Cai L, de Cock AWAM, Dissanayake AJ, Glockling SL, Goonasekara ID, Gorczak M, Hahn M, Jayawardena RS, van Kan JAL, Laurence MH, Lévesque CA, Li X, Liu JK, Maharachchikumbura SSN, Manamgoda DS, Martin FN, McKenzie EHC, McTaggart AR, Mortimer PE, Nair PVR, Pawłowska J, Rintoul TL, Shivas RG, Spies CFJ, Summerell BA, Taylor PWJ, Terhem RB, Udayanga D, Vaghefi N, Walther G, Wilk M, Wrzosek M, Xu JC, Yan JY, Zhou N. 2014. One stop shop: backbones trees for important phytopathogenic genera I (2014). Fungal Divers 67 21–125. doi: 10.1007/s13225-014-0298-1. [DOI] [Google Scholar]
- 55.Rahman AK, Sugitani N, Hatsu M, Takamizawa K. 2003. A role of xylanase, alpha-L-arabinofuranosidase, and xylosidase in xylan degradation. Can J Microbiol 49:58–64. doi: 10.1139/w02-114. [DOI] [PubMed] [Google Scholar]
- 56.Huang JH, Chen CY, Lin YS, Ann PJ, Huang HC, Chung WH. 2013. Six new species of Pythiogeton in Taiwan, with an account of the molecular phylogeny of this genus. Mycoscience 54:130–147. doi: 10.1016/j.myc.2012.09.007. [DOI] [Google Scholar]
- 57.Bininda-Emonds ORP. 2004. The evolution of supertrees. Trends Ecol Evol 19:315–322. doi: 10.1016/j.tree.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 58.Baum BR. 1992. Combining trees as a way of combining data sets for phylogenetic inference, and the desirability of combining gene trees. Taxon 41:3–10. doi: 10.2307/1222480. [DOI] [Google Scholar]
- 59.Ragan MA. 1992. Phylogenetic inference based on matrix representation of trees. Mol Phylogenet Evol 1:53–58. doi: 10.1016/1055-7903(92)90035-F. [DOI] [PubMed] [Google Scholar]
- 60.Fitzpatrick DA, Logue ME, Stajich JE, Butler G. 2006. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol 6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pisani D, Cotton JA, McInerney JO. 2007. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol 24:1752–1760. doi: 10.1093/molbev/msm095. [DOI] [PubMed] [Google Scholar]
- 62.Beck RMD, Bininda-Emonds ORP, Cardillo M, Liu FG, Purvis A. 2006. A higher-level MRP supertree of placental mammals. BMC Evol Biol 6:93. doi: 10.1186/1471-2148-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cotton JA, Page RDM. 2003. Gene tree parsimony vs. uninode coding for phylogenetic reconstruction. Mol Phylogenet Evol 29:298–308. doi: 10.1016/S1055-7903(03)00109-X. [DOI] [PubMed] [Google Scholar]
- 64.Casewell NR, Wagstaff SC, Harrison RA, Wüster W. 2011. Gene tree parsimony of multilocus snake venom protein families reveals species tree conflict as a result of multiple parallel gene loss. Mol Biol Evol 28:1157–1172. doi: 10.1093/molbev/msq302. [DOI] [PubMed] [Google Scholar]
- 65.de Queiroz A, Gatesy J. 2007. The supermatrix approach to systematics. Trends Ecol Evol 22:34–41. doi: 10.1016/j.tree.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 66.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, Chapman J, Damasceno CM, Dorrance AE, Dou D, Dickerman AW, Dubchak IL, Garbelotto M, Gijzen M, Gordon SG, Govers F, Grunwald NJ, Huang W, Ivors KL, Jones RW, Kamoun S, Krampis K, Lamour KH, Lee MK, McDonald WH, Medina M, Meijer HJ, Nordberg EK, Maclean DJ, Ospina-Giraldo MD, Morris PF, Phuntumart V, Putnam NH, Rash S, Rose JK, Sakihama Y, Salamov AA, Savidor A, Scheuring CF, Smith BM, Sobral BW, Terry A, Torto-Alalibo TA, Win J, Xu Z, Zhang H, et al. . 2006. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 67.Adhikari BN, Hamilton JP, Zerillo MM, Tisserat N, Lévesque CA, Buell CR. 2013. Comparative genomics reveals insight into virulence strategies of plant pathogenic oomycetes. PLoS One 8:e75072. doi: 10.1371/journal.pone.0075072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lévesque CA, Brouwer H, Cano L, Hamilton JP, Holt C, Huitema E, Raffaele S, Robideau GP, Thines M, Win J, Zerillo MM, Beakes GW, Boore JL, Busam D, Dumas B, Ferriera S, Fuerstenberg SI, Gachon CMM, Gaulin E, Govers F, Grenville-Briggs L, Horner N, Hostetler J, Jiang RHY, Johnson J, Krajaejun T, Lin H, Meijer HJG, Moore B, Morris P, Phuntmart V, Puiu D, Shetty J, Stajich JE, Tripathy S, Wawra S, van West P, Whitty BR, Coutinho PM, Henrissat B, Martin F, Thomas PD, Tyler BM, De Vries RP, Kamoun S, Yandell M, Tisserat N, Buell CR. 2010. Genome sequence of the necrotrophic plant pathogen Pythium ultimum reveals original pathogenicity mechanisms and effector repertoire. Genome Biol 11:R73. doi: 10.1186/gb-2010-11-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, Bozkurt TO, Ah-Fong AMV, Alvarado L, Anderson VL, Armstrong MR, Avrova A, Baxter L, Beynon J, Boevink PC, Bollmann SR, Bos JIB, Bulone V, Cai G, Cakir C, Carrington JC, Chawner M, Conti L, Costanzo S, Ewan R, Fahlgren N, Fischbach MA, Fugelstad J, Gilroy EM, Gnerre S, Green PJ, Grenville-Briggs LJ, Griffith J, Grünwald NJ, Horn K, Horner NR, Hu C-H, Huitema E, Jeong D-H, Jones AME, Jones JDG, Jones RW, Karlsson EK, Kunjeti SG, Lamour K, Liu Z, et al. . 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 70.Studholme DJ, McDougal RL, Sambles C, Hansen E, Hardy G, Grant M, Ganley RJ, Williams NM. 2016. Genome sequences of six Phytophthora species associated with forests in New Zealand. Genomics Data 7:54–56. doi: 10.1016/j.gdata.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, Kemen E, Thines M, Ah-Fong A, Anderson R, Badejoko W, Bittner-Eddy P, Boore JL, Chibucos MC, Coates M, Dehal P, Delehaunty K, Dong S, Downton P, Dumas B, Fabro G, Fronick C, Fuerstenberg SI, Fulton L, Gaulin E, Govers F, Hughes L, Humphray S, Jiang RHY, Judelson H, Kamoun S, Kyung K, Meijer H, Minx P, Morris P, Nelson J, Phuntumart V, Qutob D, Rehmany A, Rougon-Cardoso A, Ryden P, Torto-Alalibo T, Studholme D, Wang Y, Win J, Wood J, Clifton SW, Rogers J, Van den Ackerveken G, Jones JDG, et al. . 2010. Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330:1549–1551. doi: 10.1126/science.1195203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lamour KH, Stam R, Jupe J, Huitema E. 2012. The oomycete broad-host-range pathogen Phytophthora capsici. Mol Plant Pathol 13:329–337. doi: 10.1111/j.1364-3703.2011.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Links MG, Holub E, Jiang RHY, Sharpe AG, Hegedus D, Beynon E, Sillito D, Clarke WE, Uzuhashi S, Borhan MH. 2011. De novo sequence assembly of Albugo candida reveals a small genome relative to other biotrophic oomycetes. BMC Genomics 12:503. doi: 10.1186/1471-2164-12-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kemen E, Gardiner A, Schultz-Larsen T, Kemen AC, Balmuth AL, Robert-Seilaniantz A, Bailey K, Holub E, Studholme DJ, Maclean D, Jones JDG. 2011. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol 9:e1001094. doi: 10.1371/journal.pbio.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feau N, Taylor G, Dale AL, Dhillon B, Bilodeau GJ, Birol I, Jones SJM, Hamelin RC. 2016. Genome sequences of six Phytophthora species threatening forest ecosystems. Genomics Data 10:85–88. doi: 10.1016/j.gdata.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao R, Cheng Y, Wang Y, Wang Y, Guo L, Zhang G. 2015. Genome sequence of Phytophthora fragariae var. fragariae, a quarantine plant-pathogenic fungus. Genome Announc 3:e00034-15. doi: 10.1128/genomeA.00034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sambles C, Schlenzig A, O’Neill P, Grant M, Studholme DJ. 2015. Draft genome sequences of Phytophthora kernoviae and Phytophthora ramorum lineage EU2 from Scotland. Genomics Data 6:193–194. doi: 10.1016/j.gdata.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quinn L, O’Neill PA, Harrison J, Paskiewicz KH, McCracken AR, Cooke LR, Grant MR, Studholme DJ. 2013. Genome-wide sequencing of Phytophthora lateralis reveals genetic variation among isolates from Lawson cypress (Chamaecyparis lawsoniana) in Northern Ireland. FEMS Microbiol Lett 344:179–185. doi: 10.1111/1574-6968.12179. [DOI] [PubMed] [Google Scholar]
- 79.Liu H, Ma X, Yu H, Fang D, Li Y, Wang X, Wang W, Dong Y, Xiao B. 2016. Genomes and virulence difference between two physiological races of Phytophthora nicotianae. GigaScience 5:3. doi: 10.1186/s13742-016-0108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharma R, Xia X, Cano LM, Evangelisti E, Kemen E, Judelson H, Oome S, Sambles C, van den Hoogen DJ, Kitner M, Klein J, Meijer HJG, Spring O, Win J, Zipper R, Bode HB, Govers F, Kamoun S, Schornack S, Studholme DJ, Van den Ackerveken G, Thines M. 2015. Genome analyses of the sunflower pathogen Plasmopara halstedii provide insights into effector evolution in downy mildews and Phytophthora. BMC Genomics 16:741. doi: 10.1186/s12864-015-1904-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rujirawat T, Patumcharoenpol P, Lohnoo T, Yingyong W, Lerksuthirat T, Tangphatsornruang S, Suriyaphol P, Grenville-Briggs LJ, Garg G, Kittichotirat W, Krajaejun T. 2015. Draft genome sequence of the pathogenic oomycete Pythium insidiosum strain Pi-S, isolated from a patient with pythiosis. Genome Announc 3:e00574-15. doi: 10.1128/genomeA.00574-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berger H, Yacoub A, Gerbore J, Grizard D, Rey P, Sessitsch A, Compant S. 2016. Draft genome sequence of biocontrol agent Pythium oligandrum strain Po37, an oomycota. Genome Announc 4:e00215-16. doi: 10.1128/genomeA.00215-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jiang RHY, de Bruijn I, Haas BJ, Belmonte R, Löbach L, Christie J, van den Ackerveken G, Bottin A, Bulone V, Díaz-Moreno SM, Dumas B, Fan L, Gaulin E, Govers F, Grenville-Briggs LJ, Horner NR, Levin JZ, Mammella M, Meijer HJG, Morris P, Nusbaum C, Oome S, Phillips AJ, van Rooyen D, Rzeszutek E, Saraiva M, Secombes CJ, Seidl MF, Snel B, Stassen JHM, Sykes S, Tripathy S, van den Berg H, Vega-Arreguin JC, Wawra S, Young SK, Zeng Q, Dieguez-Uribeondo J, Russ C, Tyler BM, van West P. 2013. Distinctive expansion of potential virulence genes in the genome of the oomycete fish pathogen Saprolegnia parasitica. PLoS Genet 9:e1003272. doi: 10.1371/journal.pgen.1003272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gobler CJ, Berry DL, Dyhrman ST, Wilhelm SW, Salamov A, Lobanov AV, Zhang Y, Collier JL, Wurch LL, Kustka AB, Dill BD, Shah M, VerBerkmoes NC, Kuo A, Terry A, Pangilinan J, Lindquist EA, Lucas S, Paulsen IT, Hattenrath-Lehmann TK, Talmage SC, Walker EA, Koch F, Burson AM, Marcoval MA, Tang YZ, Lecleir GR, Coyne KJ, Berg GM, Bertrand EM, Saito MA, Gladyshev VN, Grigoriev IV. 2011. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. Proc Natl Acad Sci U S A 108:4352–4357. doi: 10.1073/pnas.1016106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, Maheswari U, Martens C, Maumus F, Otillar RP, Rayko E, Salamov A, Vandepoele K, Beszteri B, Gruber A, Heijde M, Katinka M, Mock T, Valentin K, Verret F, Berges JA, Brownlee C, Cadoret JP, Chiovitti A, Choi CJ, Coesel S, De Martino A, Detter JC, Durkin C, Falciatore A, Fournet J, Haruta M, Huysman MJJ, Jenkins BD, Jiroutova K, Jorgensen RE, Joubert Y, Kaplan A, Kröger N, Kroth PG, La Roche J, Lindquist E, Lommer M, Martin-Jézéquel V, Lopez PJ, Lucas S, Mangogna M, McGinnis K, Medlin LK, et al. . 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 86.Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kröger N, Lau WWY, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 87.Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, Beszteri B, Billiau K, Bonnet E, Bothwell JH, Bowler C, Boyen C, Brownlee C, Carrano CJ, Charrier B, Cho GY, Coelho SM, Collén J, Corre E, Da Silva C, Delage L, Delaroque N, Dittami SM, Doulbeau S, Elias M, Farnham G, Gachon CMM, Gschloessl B, Heesch S, Jabbari K, Jubin C, Kawai H, Kimura K, Kloareg B, Küpper FC, Lang D, Wincker P. 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- 88.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Ségurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, Câmara F, Duharcourt S, Guigo R, Gogendeau D, Katinka M, Keller AM, Kissmehl R, Klotz C, Koll F, Le Mouël A, Lepère G, Malinsky S, Nowacki M, Nowak JK, Plattner H, Poulain J, Ruiz F, Serrano V, Zagulski M, Dessen P, Bétermier M, Weissenbach J, Scarpelli C, Schächter V, Sperling L, Meyer E, Cohen J, Wincker P. 2006. Global trends of whole-genome duplications revealed by the ciliate paramecium tetraurelia. Nature 444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 89.Curtis BA, Tanifuji G, Burki F, Gruber A, Irimia M, Maruyama S, Arias MC, Ball SG, Gile GH, Hirakawa Y, Hopkins JF, Kuo A, Rensing SA, Schmutz J, Symeonidi A, Elias M, Eveleigh RJM, Herman EK, Klute MJ, Nakayama T, Oborník M, Reyes-Prieto A, Armbrust EV, Aves SJ, Beiko RG, Coutinho P, Dacks JB, Durnford DG, Fast NM, Green BR, Grisdale CJ, Hempel F, Henrissat B, Höppner MP, Ishida K-I, Kim E, Kořený L, Kroth PG, Liu Y, Malik S-B, Maier UG, McRose D, Mock T, Neilson JAD, Onodera NT, Poole AM, Pritham EJ, Richards TA, Rocap G, Roy SW, et al. . 2012. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492:59–65. doi: 10.1038/nature11681. [DOI] [PubMed] [Google Scholar]
- 90.Stanke M, Steinkamp R, Waack S, Morgenstern B. 2004. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res 32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. 2008. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res 18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ramsay L, Macaulay M, Degli Ivanissevich S, MacLean K, Cardle L, Fuller J, Edwards KJ, Tuvesson S, Morgante M, Massari A, Maestri E, Marmiroli N, Sjakste T, Ganal M, Powell W, Waugh R. 2000. A simple sequence repeat-based linkage map of barley. Genetics 156:1997–2005. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 96.Faith DP, Cranston PS. 1991. Could a cladogram this short have arisen by chance alone?: on permutation tests for cladistic structure. Cladistics 7:1–28. doi: 10.1111/j.1096-0031.1991.tb00020.x. [DOI] [Google Scholar]
- 97.Swofford LD. 2002. PAUP*: phylogenetic analysis using parsimony (* and other methods) version 4.0 beta. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 98.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 100.Creevey CJ, McInerney JO. 2005. Clann: investigating phylogenetic information through supertree analyses. Bioinformatics 21:390–392. doi: 10.1093/bioinformatics/bti020. [DOI] [PubMed] [Google Scholar]
- 101.Letunic I, Bork P. 2007. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 102.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 103.Wehe A, Bansal MS, Burleigh JG, Eulenstein O. 2008. DupTree: A program for large-scale phylogenetic analyses using gene tree parsimony. Bioinformatics 24:1540–1541. doi: 10.1093/bioinformatics/btn230. [DOI] [PubMed] [Google Scholar]
- 104.Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 105.Kück P, Meusemann K. 2010. FASconCAT: convenient handling of data matrices. Mol Phylogenet Evol 56:1115–1118. doi: 10.1016/j.ympev.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 106.Lapierre P, Lasek-Nesselquist E, Gogarten JP. 2014. The impact of HGT on phylogenomic reconstruction methods. Brief Bioinform 15:79–90. doi: 10.1093/bib/bbs050. [DOI] [PubMed] [Google Scholar]
- 107.McCarthy CGP, Fitzpatrick DA. 2016. Systematic search for evidence of interdomain horizontal gene transfer from prokaryotes to oomycete lineages. mSphere 1:e00195-16. doi: 10.1128/mSphere.00195-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Burgess TI. 2015. Molecular characterization of natural hybrids formed between five related indigenous clade 6 Phytophthora species. PLoS One 10:e0134225. doi: 10.1371/journal.pone.0134225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bertier L, Leus L, D’hondt L, de Cock AW, Höfte M. 2013. Host adaptation and speciation through hybridization and polyploidy in Phytophthora. PLoS One 8:e85385. doi: 10.1371/journal.pone.0085385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Husson C, Aguayo J, Revellin C, Frey P, Ioos R, Marçais B. 2015. Evidence for homoploid speciation in Phytophthora alni supports taxonomic reclassification in this species complex. Fungal Genet Biol 77:12–21. doi: 10.1016/j.fgb.2015.02.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taxonomy, assembly statistics, and protein predictions for 14 oomycete species. AUGUSTUS ab initio protein predictions were carried out using parameters generated from genomic, EST, and protein data from template species. Protein prediction for Pi. apinafurcum was carried out using GeneMark-ES and AUGUSTUS. Protein sets used in whole-genome phylogenetic analyses in this study are highlighted in bold. Download TABLE S1, DOCX file, 0.02 MB (17.6KB, docx) .
Copyright © 2017 McCarthy and Fitzpatrick.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Selection of evolutionary models (not including additional parameters) for single-copy and multicopy gene phylogenies by ProtTest. Download TABLE S2, DOCX file, 0.01 MB (14.4KB, docx) .
Copyright © 2017 McCarthy and Fitzpatrick.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Consensus network of phylogenetic splits within 2,280 single-copy oomycete and SAR phylogenies. The network was generated in SplitsTree. Clades are colored by taxonomic order as follows: red, Peronosporales; blue, Pythiales; green, Albuginales; orange, Saprolegniales; no color, P. tetraurelia (Alveolata) and B. natans (Rhizara). Download FIG S1, PDF file, 0.1 MB (121.4KB, pdf) .
Copyright © 2017 McCarthy and Fitzpatrick.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Maximum-likelihood (ML) supermatrix phylogeny of 37 oomycete species (144 families of Saccharomyces cerevisiae COG homologs, 16,934 characters). Supermatrix phylogeny data were generated in PhyML with an LG+I+G+F amino acid substitution model. The phylogeny is rooted at the Saprolegniales branch. Phytophthora clades as designated by Blair et al. (42) and Pythium clades as designated by de Cock et al. (50) are indicated in red and blue, respectively. No color, P. tetraurelia (Alveolata) and B. natans (Rhizaria). Download FIG S2, PDF file, 0.1 MB (60.2KB, pdf) .
Copyright © 2017 McCarthy and Fitzpatrick.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Neighbor-joining network of phylogenetic splits within oomycete COG supermatrix of 37 oomycete species. The network was generated in SplitsTree. Clades are colored by taxonomic order as follows: red, Peronosporales; blue, Pythiales; green, Albuginales; orange, Saprolegniales. Download FIG S3, PDF file, 0.02 MB (18.3KB, pdf) .
Copyright © 2017 McCarthy and Fitzpatrick.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.