Summary
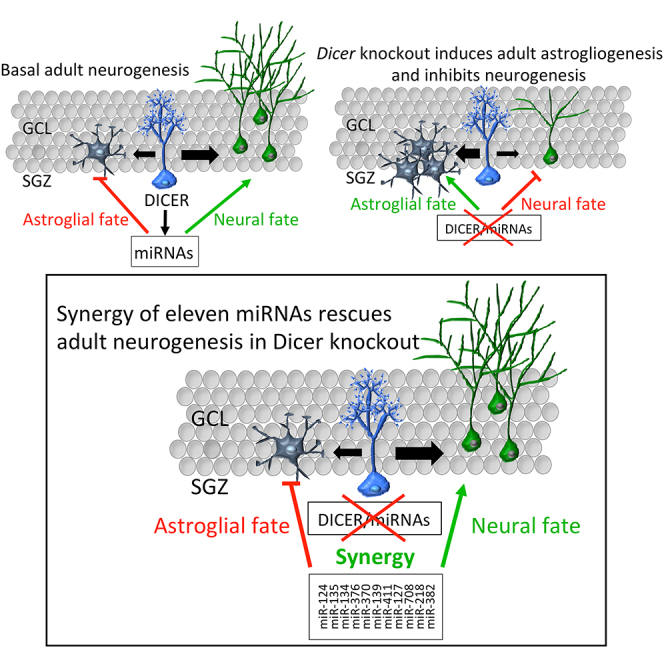
Adult neurogenesis requires the precise control of neuronal versus astrocyte lineage determination in neural stem cells. While microRNAs (miRNAs) are critically involved in this step during development, their actions in adult hippocampal neural stem cells (aNSCs) has been unclear. As entry point to address that question we chose DICER, an endoribonuclease essential for miRNA biogenesis and other RNAi-related processes. By specific ablation of Dicer in aNSCs in vivo and in vitro, we demonstrate that miRNAs are required for the generation of new neurons, but not astrocytes, in the adult murine hippocampus. Moreover, we identify 11 miRNAs, of which 9 have not been previously characterized in neurogenesis, that determine neurogenic lineage fate choice of aNSCs at the expense of astrogliogenesis. Finally, we propose that the 11 miRNAs sustain adult hippocampal neurogenesis through synergistic modulation of 26 putative targets from different pathways.
Keywords: mouse, hippocampus, neural stem cells, fate choice, adult neurogenesis, astrogliogenesis, DICER, microRNAs, synergy
Graphical Abstract
Highlights
-
•
Dicer depletion in aNSCs impairs neurogenesis and stimulates astrogliogenesis
-
•
Synergy of 11 miRNAs sustains neuronal fate of aNSCs
-
•
miRNA converge on multiple targets in different pathways to induce neurogenesis
In this article, the authors demonstrate that Dicer-dependent miRNAs are required for the generation of new neurons, but not astrocytes, in the adult hippocampus in vivo and in vitro. The authors identify a new set of 11 miRNAs that synergistically converge on multiple targets in different pathways to sustain neurogenic lineage fate commitment in aNSCs.
Introduction
Neural stem cells (NSCs) resident in the two main neurogenic niches of the adult mammalian brain (the subventricular zone [SVZ] of the lateral ventricle and the subgranular zone of the hippocampal dentate gyrus [DG]) are two heterogeneous populations of radial glia-like precursor cells that have astrocytic properties, express bona fide stem cell markers and rarely divide. These cells have the capacity to self-renew and differentiate, giving rise to both neurons and glia (reviewed in Bond et al., 2015, Kempermann et al., 2015). The mechanisms of fate determination in adult hippocampal NSC (aNSC) lineage is a highly debated topic (Bonaguidi et al., 2012, Kempermann, 2011) and of fundamental importance. In addition, understanding the molecular mechanisms underlying lineage determination might provide new avenues to prevent age-dependent loss of neurogenesis (Encinas et al., 2011, Marlatt and Lucassen, 2010, Pons-Espinal et al., 2013), or the pathological generation of undesirable cells such as activated glia upon trauma and epilepsy (Dibajnia and Morshead, 2013, Doetsch et al., 2002, Shimada et al., 2012, Sierra et al., 2015).
Regulation of aNSC fate determination is known to be possible at the transcriptional level (Beckervordersandforth et al., 2015), but accumulating evidence indicates that additional control layers, such as epigenetics and non-coding RNAs, are involved in this mechanism (Castel and Martienssen, 2013, Cernilogar et al., 2011, Huang and Li, 2014, Li, 2014, Noguchi et al., 2015, Schouten et al., 2012). MicroRNAs (miRNAs) are small (∼22 nt long) single-stranded non-coding RNAs, which post-transcriptionally repress target mRNAs through imperfect miRNA-mRNA binding (Agarwal et al., 2015, Ha and Kim, 2014, Krol et al., 2010). They exert their regulatory functions in a highly combinatorial way: one miRNA can regulate several mRNAs in parallel (Lim et al., 2005), and different miRNAs can target one mRNA simultaneously, thus repressing its expression more efficiently (Selbach et al., 2008). Based on these observations, miRNAs are predicted to regulate the majority of mammalian mRNAs (Friedman et al., 2009).
Despite the known functions of miRNAs in fate determination of embryonic and adult SVZ NSCs (Barca-Mayo and De Pietri Tonelli, 2014, Cheng et al., 2009, Zhao et al., 2009), as well as survival and dendritic maturation of adult-born neurons in the DG (Konopka et al., 2010, Magill et al., 2010, Schouten et al., 2015, Smrt et al., 2010), it has been unknown whether miRNAs regulate neuronal versus astrocyte lineage fate determination in the adult hippocampus. Indeed, as single miRNAs could have opposite effects depending on the spatiotemporal expression of their targets (Zhu et al., 2011), it is possible to hypothesize that the same miRNAs might exert different functions in various cell types involved in adult hippocampal neurogenesis.
Current approaches to infer miRNA functions in vivo either manipulate single miRNAs/targets or deplete miRNAs by conditional knockouts of genes encoding essential miRNA biogenesis proteins such as DROSHA, DGCR8, or DICER. Although both approaches successfully demonstrated critical functions for specific miRNAs and miRNA biogenesis proteins in neurogenesis (Aksoy-Aksel et al., 2014, Barca-Mayo and De Pietri Tonelli, 2014, Schouten et al., 2012), most of these studies neglected the intrinsic combinatorial nature of miRNA-dependent control (Schmiedel et al., 2015, Siciliano et al., 2013), or left unresolved the question of miRNA-specific versus miRNA-independent functions of miRNA biogenesis proteins (Yang and Lai, 2011).
By conditional ablation of Dicer specifically in bona fide aNSCs of the adult hippocampus in vivo and in vitro and by manipulation of specific miRNAs, here we studied the role of miRNAs for lineage fate choice of aNSCs. Our study identified a set of 11 miRNAs that, by synergistic enforcement of gene-regulatory networks, allows aNSCs to acquire the neurogenic fate at the expense of astrogliogenesis.
Results
Split-Cre Virus-Mediated Dicer Ablation In Vivo Impairs Neurogenesis, but Not Astrogliogenesis, in the Adult Hippocampus
To study the role of DICER in adult hippocampal neurogenesis in vivo, we first crossed a mouse line carrying a conditional allele for Dicer (Dicerflox, Murchison et al., 2005) with a Cre-inducible reporter mouse line (Td-Tomatoflox, Madisen et al., 2010). To achieve conditional ablation of Dicer and expression of Tomato in bona fide type 1 aNSCs, we injected split-Cre viruses (allowing specific expression of an active Cre recombinase in type 1 aNSCs, based on the coincident activity of human glial fibrillary acidic protein [hGFAP] and Prominin1 promoters) (Figures 1A and S1A; Beckervordersandforth et al., 2014) in the DG of 8-week-old Dicerwt/wt Td-Tomatoflox/wt (wild-type; WT), Dicerflox/wt Td-Tomatoflox/wt (Dicer HT), and Dicerflox/flox Td-Tomatoflox/wt (Dicer cKO) mice, and followed the fate of the labeled cells in the subgranular zone (SGZ) and granule cell layer (GCL) of the hippocampus.
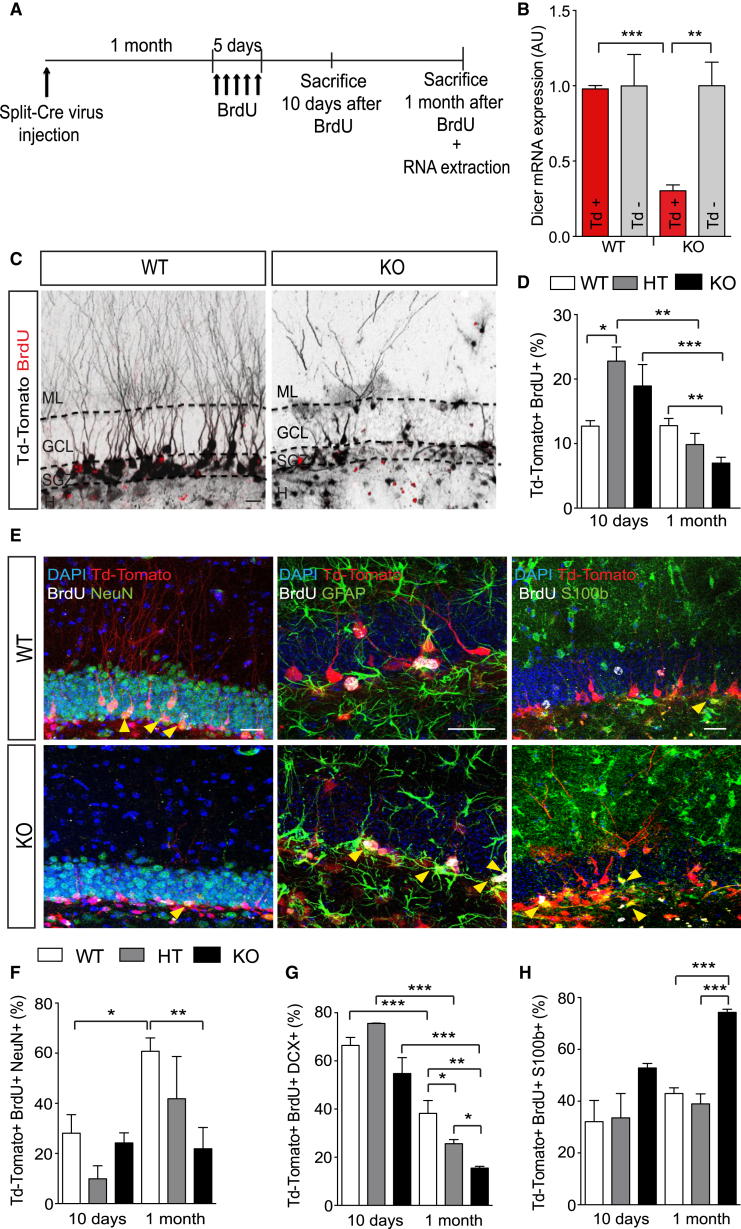
Figure 1.
Split-Cre Virus-Mediated Dicer Ablation In Vivo Impairs Neuronal Differentiation and Survival but Not Astrogliogenesis
(A) Schematic representation of the experiment.
(B) qRT-PCR quantification of Dicer mRNA from FACS-sorted Td-Tomato+ aNSCs 2 months after split-Cre virus injection.
(C and E) Representative micrographs showing recombined Td-Tomato/BrdU double-positive cells from Dicer WT and cKO mice 1 month after BrdU injection (C), co-expressing NeuN (E, left panel), GFAP (E, middle panel) and S100b (E, right panel). Yellow arrowheads show Td-Tomato/BrdU double-positive cells co-expressing NeuN, GFAP, or S100b.
(D) Percentage of Td-Tomato+ cells expressing BrdU after 10 days, or 1 month after BrdU injections.
(F–H) Percentage of Td-Tomato/BrdU double-positive cells co-expressing NeuN (F), DCX (G), or S100b (H) 10 days or 1 month after BrdU injections.
ML, molecular layer; GCL, granular cell layer; SGZ, subgranular zone; H, Hilus. Data are expressed as mean ± SEM, n = 4–6 mice per group. Unpaired t test was used for Dicer mRNA expression analysis. One-way ANOVA Bonferroni as post hoc was used to analyze cell marker quantification. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bars, 20 μm.
To ascertain Dicer ablation in vivo, we sorted Tomato+ cells by fluorescence-activated cell sorting (FACS) and, as internal control non-infected Tomato− cells, from the DG of WT and Dicer cKO mice, and quantified Dicer mRNA levels by quantitative real-time PCR (qRT-PCR). This quantification confirmed a 70% reduction of Dicer mRNA levels in Tomato+ cells from Dicer cKO mice, compared with Tomato+ cells from WT mice (Figure 1B, p = 0.0001) and Tomato− cells from both WT and Dicer cKO mice (Figure 1B, p = 0.003).
To investigate the survival of the progeny originating from the Dicer cKO aNSCs at 1 month after virus injection, we administered bromodeoxyuridine (BrdU) for 5 consecutive days. Ten days or 1 month after BrdU, we quantified the proportion of Tomato/BrdU double-positive cells in the SGZ/GCL of WT, Dicer HT, and cKO mice (Figure 1A). The proportion of Tomato/BrdU double-positive cells in Dicer cKO and Dicer HT mice showed a slight increase at 10 days (Figure 1D), but significantly decreased in Dicer cKO mice at 1 month (Figures 1C and 1D, p = 0.006). This result indicated that Dicer depletion impaired survival of newborn cells in the SGZ/GCL.
Moreover, we also observed a dramatic reduction in the number of processes and arborization of Tomato+ Dicer cKO cells in the GCL and molecular layer (ML) of the hippocampus compared with Tomato+ WT cells (Figure 1C). This finding suggested that Dicer depletion impaired the differentiation and maturation of the surviving cells.
Next, we assessed the role of DICER in neuronal fate choice. We quantified the proportion of newborn cells co-expressing the immature neuronal marker doublecortin (DCX) or postmitotic neuronal marker NeuN in the SGZ/GCL of the adult hippocampus of Dicer WT, HT, and cKO mice (as in Figure 1A). At 10 days we did not find differences in DCX and NeuN expression among the groups (Figures 1F and 1G). However, at 1 month we found that 40% of Dicer WT cells also co-expressed DCX, whereas only 26% of Dicer HT cells and 10% of Dicer cKO cells did so, respectively (Figure 1G, WT versus KO, p = 0.0012; WT versus HT, p = 0.039). Consistently, at the same age, only 20% of Dicer cKO cells co-expressed NeuN, compared with 60% of NeuN+ Dicer WT neurons (Figures 1E and 1F, p = 0.0058). Moreover, although the proportion of newborn NeuN+ neurons increased significantly between 10 days and 1 month in the SGZ/GCL of Dicer WT mice (Figures 1E and 1F, p = 0.0062), this population did not grow over time in Dicer cKO mice (p = 0.72). These results indicate that Dicer depletion impairs neuronal differentiation and maturation in the adult mouse hippocampus in vivo.
We then assessed the role of DICER on adult astrogliogenesis by immunostaining for three different astrocyte markers, GFAP, S100b (Figure 1E), and glutamine synthetase (GS; Figure S1B) and found results complementary to the findings on neurogenesis. Whereas no significant differences were observed for S100b at 10 days between the three Dicer genotypes; at 1 month the proportion of S100b+ (Figure 1H, p = 0.0002) and GS+ (Figure S1C, p = 0.024) was about twice as high in the SGZ/GCL of Dicer cKO mice than in controls. Moreover, upon Dicer ablation we did not observe an increase in progenitor markers such as Nestin or SOX2 (Figures S1B and S1C), largely excluding the possibility that Dicer cKO cells remained in undifferentiated or quiescent state. Thus, these results indicated that Dicer depletion in type 1 aNSCs impaired neurogenesis and favored astrogliogenesis in the adult hippocampus in vivo.
Loss of DICER-Dependent miRNAs Does Not Affect aNSC Proliferation and Stemness, but Increases Apoptosis upon Their Differentiation In Vitro
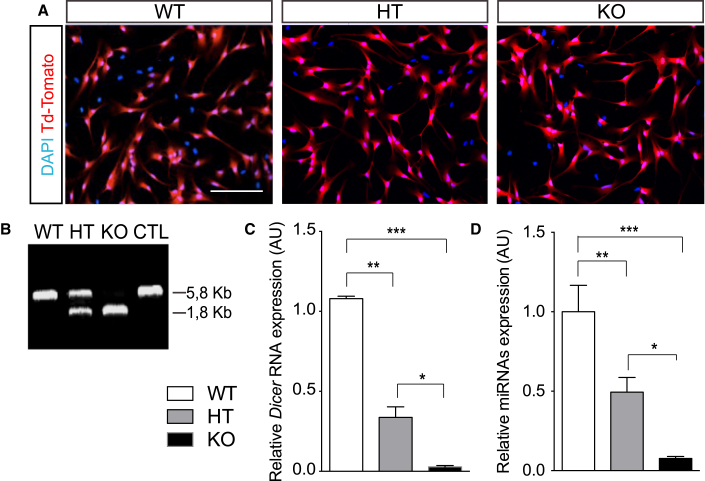
To ascertain the effect of Dicer ablation in vitro, we generated primary aNSCs from the DG of mice WT, HT, and homozygous for Dicerflox allele, which were also heterozygous for the Cre-inducible Td-Tomato allele, by culturing them in monolayers (Figure S1D) as described previously (Babu et al., 2011, Walker and Kempermann, 2014). Upon recombination of Dicerflox locus (Figure 2B) and expression of Tomato protein (Figure 2A), we saw a 50% reduction of Dicer transcript in Dicer HT aNSCs (Figure 2C, p = 0.0082) and almost total loss in Dicer cKO aNSCs (Figure 2C, p = 0.0003) compared with WT. Consequently, mature miRNAs levels were reduced to 50% in Dicer HT aNSCs (Figure 2D, p = 0.01) and almost completely depleted in Dicer cKO aNSCs (Figure 2D, residual miRNA levels 7%; p < 0.0001), compared with WT cells. These results demonstrated that recombination of the Dicerflox allele resulted in efficient depletion of both Dicer transcript and mature miRNAs from hippocampal aNSCs in vitro. Despite efficient loss of DICER-dependent miRNAs, we were able to keep Dicer cKO aNSCs in culture under proliferative conditions for at least 18 days in vitro (DIV). We did not detect major differences in cell morphology or passaging requirements for cells from the three genotypes. Moreover, no differences were observed in the percentage of Tomato+ cells incorporating BrdU after a 2-hr pulse (Figures S2A–S2C and S2G–S2I), or pH3 (Figures S2D and S2E). Consistently, the growth curve was unchanged over several days in culture (Figure S2F). Thus, we concluded that loss of DICER-dependent miRNAs does not affect aNSCs proliferation.
Figure 2.
Dicer and miRNAs Are Depleted after Recombination of Dicerflox Allele in Hippocampal aNSCs In Vitro
(A) Representative micrographs showing Td-Tomato+ aNSCs from Dicer WT, Dicer HT, and Dicer cKO mice after nucleofection with Cre recombinase.
(B) PCR Genotyping of Cre-recombined aNSCs, showing the three Dicer genotypes.
(C) qRT-PCR quantification of Dicer mRNA in Cre-recombined aNSCs.
(D) Average of all miRNAs quantified from recombined aNSCs.
Data are expressed as mean ± SEM, n = 3 independent experiments containing three replicates. One-way ANOVA Bonferroni as post hoc: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bar, 50 μm.
Next, we investigated stem cell markers SOX2, GFAP, and Nestin in aNSCs, and found no change in SOX2 or GFAP expression at both protein and transcript levels in aNSCs from the three genotypes (Figure S3). However, consistent with our in vivo results (Figures S1B and S1C), we detected reduced expression of Nestin in Dicer cKO aNSCs compared with WT aNSCs (Figures S3A–S3C, p = 0.0021; Figure S3D, WT versus cKO p = 0.0053; HT versus cKO p = 0.03). Altogether, these results confirmed that loss of DICER-dependent miRNAs did not primarily affect proliferation, passaging requirements, and expression of stem cell markers of aNSCs. These results are consistent with previous studies on other stem cells types (Murchison et al., 2005) including embryonic NSCs (Andersson et al., 2010, De Pietri Tonelli et al., 2008), reporting that the effects of Dicer ablation are more prominent during cell fate transitions than in self-renewal.
Our data indicated that Dicer ablation impairs survival of newborn neurons in vivo (Figure 1). Thus we investigated the survival of aNSCs from Dicer WT, HT, and cKO upon induction of differentiation. Recombined (Tomato+) Dicer WT, HT, and cKO aNSCs were FACS sorted, and equal numbers of aNSCs were seeded and cultivated under differentiating conditions (Figure S4A). After 6 DIV we found a 30% and 50% reduction in the number of Dicer HT and Dicer cKO aNSCs compared with WT aNSCs (Figures S4B and S4C, WT versus cKO, p = 0.03). Moreover, the reduced survival of Dicer cKO aNSCs was paralleled by a significant increase in the number of pycnotic nuclei (Figures S4B and S4C, p = 0.0051), expression of the apoptotic marker active-caspase-3 (Figures S4B and S4C, p = 0.013), and reduction in the expression of the transcript of anti-apoptotic protein Bcl-2 (Figure S4D, p = 0.04). Thus, consistently with in vivo data (Figure 1), these results indicated that DICER functions are not essential for expansion of aNSCs in vitro, but are required for survival of their progeny.
Loss of DICER-Dependent miRNAs Impairs Neurogenesis but Not Astrogliogenesis In Vitro
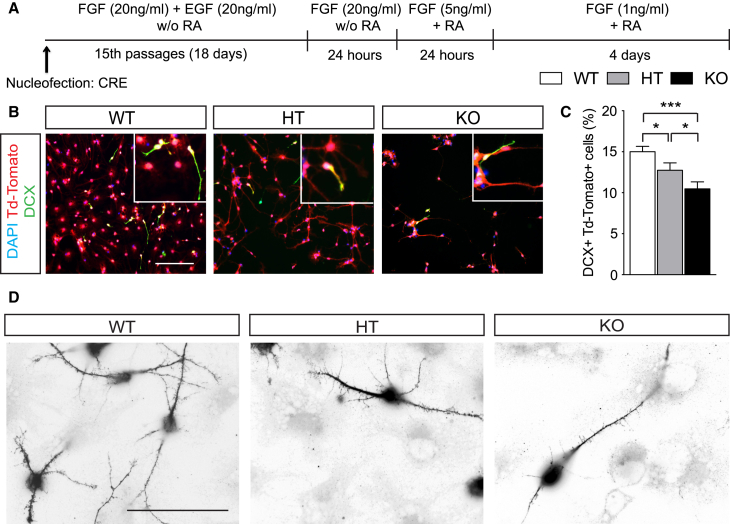
Our in vivo data suggested that Dicer depletion in aNSCs impairs neurogenesis but not astrogliogenesis in the adult mouse hippocampus (Figure 1). Thus, we isolated recombined (Tomato+) Dicer WT, HT, and cKO aNSCs and investigated neurogenesis and neuronal maturation upon differentiation in vitro (Figure 3A). We found that Dicer HT and cKO aNSCs generated significantly fewer DCX+ cells compared with WT aNSCs (Figures 3B and 3C, DCX+ ∼12% in Dicer HT, p = 0.04; ∼9% in Dicer cKO, p = 0.0013). Moreover, we saw a reduction in the number of neurites, branch points, and dendritic spines in DCX+ Dicer cKO cells compared with Dicer WT and HT cells (Figure 3D).
Figure 3.
Loss of Dicer-Dependent miRNA in aNSCs Impairs Neurogenesis and Neuronal Maturation In Vitro
(A) Schematic representation of the protocol and experiment.
(B) Representative micrographs showing recombined Td-Tomato+ aNSCs from Dicer WT, Dicer HT, and Dicer cKO mice after 6 DIV with growth factors titration expressing DCX.
(C) Percentage of Td-Tomato+ cells expressing DCX.
(D) Representative micrographs showing dendritic morphology of immature newly formed neurons expressing DCX.
Data are expressed as mean ± SEM, n = 3 independent experiments containing three replicates. One-way ANOVA Bonferroni as post hoc: ∗p < 0.05, ∗∗∗p < 0.001. Scale bars, 50 μm.
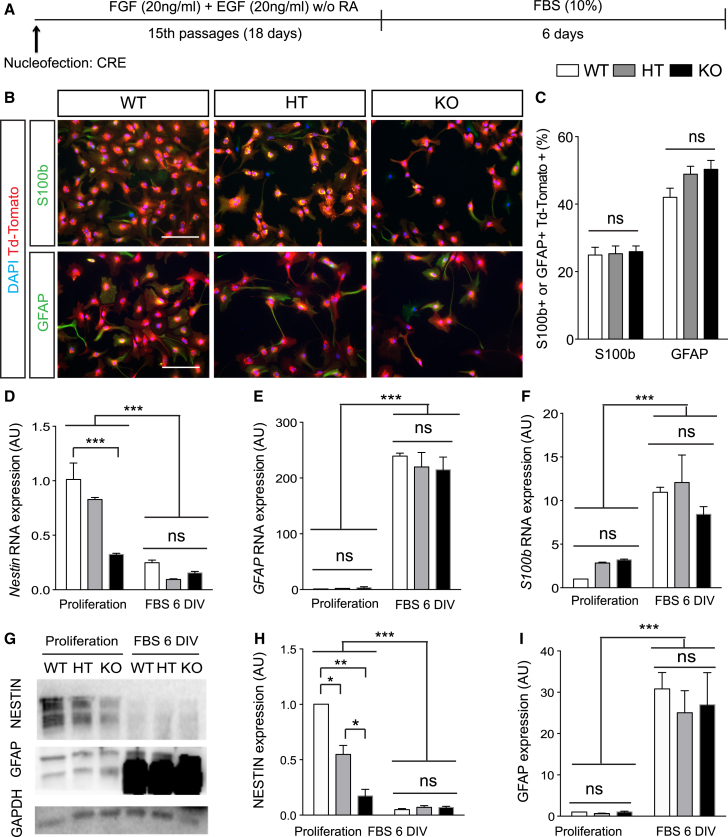
Next, we investigated astrogliogenesis in recombined (Tomato+) Dicer WT, HT, and cKO aNSCs upon differentiation (as in Figure 3A) and found no difference between the three Dicer genotypes (Figure 6B). We then induced astrocyte differentiation with 10% fetal serum for 6 DIV (Figure 4A) and found no difference in the proportion of cells expressing astrocytic markers GFAP and S100b between groups (Figures 4B and 4C). These data were corroborated by a parallel decrease in the expression of the bona fide stem cell marker Nestin, at both mRNA and protein levels (Figures 4D–4H), and by a strong increase in the expression of astrocytic markers GFAP and S100b (Figures 4E, 4F, and 4I) in all genotypes. These results were consistent with our in vivo evidence (Figure 1) and indicated that DICER functions are essential for aNSC differentiation toward neurogenesis and neuronal maturation but not for astrogliogenesis.
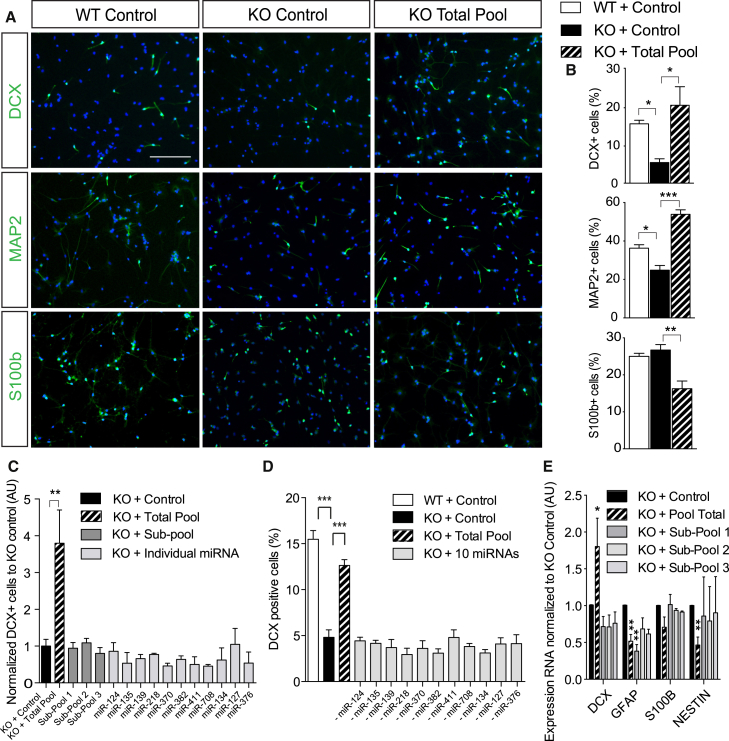
Figure 6.
A Pool of 11 miRNAs Synergistically Rescues Dicer cKO Impairment of Adult Neurogenesis, at the Expense of Astrogliogenesis, In Vitro
(A) Representative micrographs showing aNSCs from Dicer WT and Dicer cKO mice transfected with 250 nM scrambled RNA or total pool (25 nM of each miRNA) after 6 DIV of growth factors withdrawal expressing DCX (upper panel), MAP2 (middle panel), and S100b (bottom panel). Scale bar, 50 μm.
(B) Percentage of DCX-, MAP2-, and S100b-positive aNSCs with respect to DAPI in WT and KO aNSCs transfected with scrambled RNA or total pool.
(C) Proportion of KO aNSCs expressing DCX upon transfection of 250 nM subpool 1 (mir-124-3p + mir-135a-5p), subpool 2 (mir-139-5p + mir-218-5p + mir-411-5p + mir-134-5p + mir-370-3p + mir-382-5p + mir-708-5p), subpool 3 (mir-127-3p + miR-376b-3p), or each miRNA alone with respect to KO control.
(D) Percentage of DCX-positive aNSCs with respect to DAPI after 6 DIV from WT or KO mice transfected with scrambled RNA, total pool, or a pool with ten miRNAs by the withdrawal of individual miRNAs.
(E) mRNA quantification with qRT-PCR from recombined KO aNSCs after 6 DIV.
Data are expressed as mean ± SEM, n ≥ 3 independent experiments containing three replicates. One-way ANOVA with Bonferroni as post hoc test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Figure 4.
Loss of Dicer-Dependent miRNAs Does Not Affect Astrogliogenesis of aNSCs In Vitro
(A) Schematic representation of the protocol and experiment.
(B) Representative micrographs showing recombined Td-Tomato+ aNSCs from Dicer WT, Dicer HT, and Dicer cKO mice after 6 DIV with 10% fetal bovine serum expressing S100b (upper panels) and GFAP (bottom panels).
(C) Percentage of Td-Tomato+ cells expressing astrocyte markers (GFAP and S100b).
(D–F) Relative Nestin (D), GFAP (E), and S100b (F) mRNA quantification with qRT-PCR.
(G–I) Protein quantification of NESTIN (H) and GFAP (I).
Data are expressed as mean ± SEM, n = 3 independent experiments containing three replicates. One-way ANOVA Bonferroni as post hoc: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Scale bar, 50 μm.
A Pool of 11 miRNAs Determines aNSC Neurogenic Fate at the Expense of Astrogliogenesis
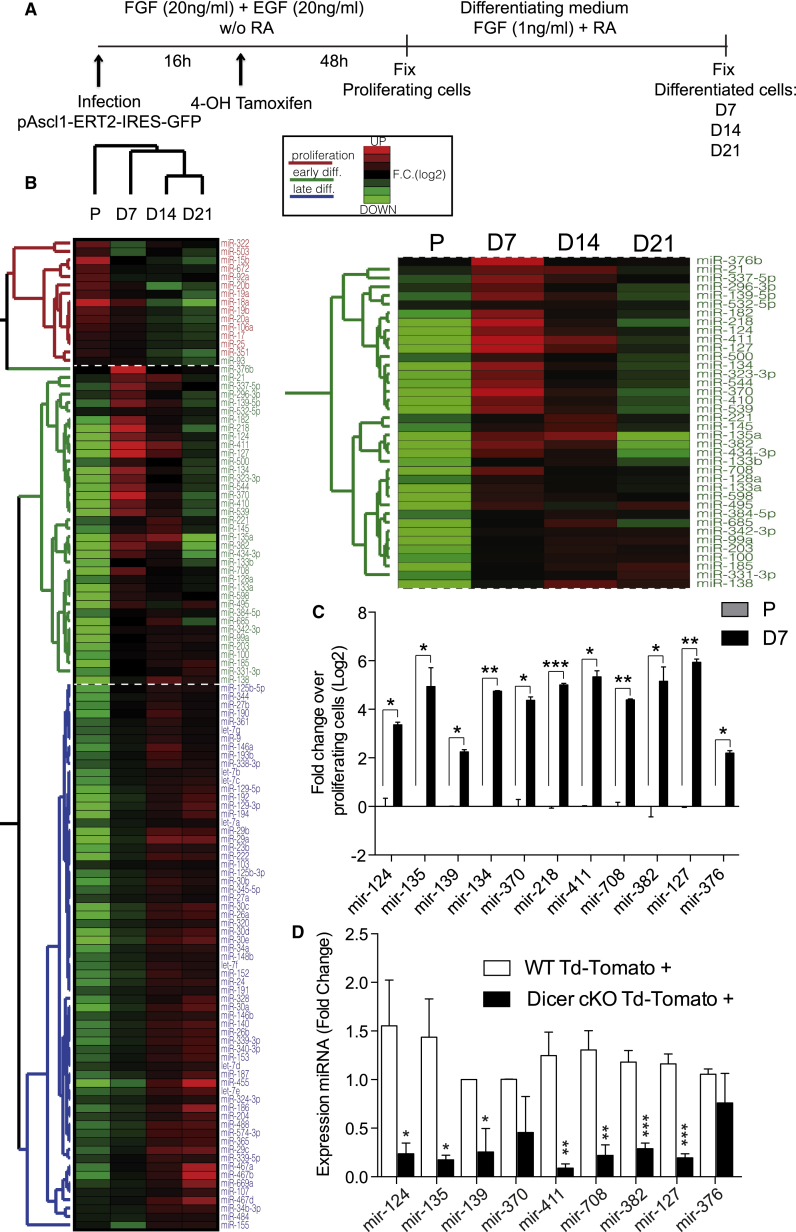
To clarify whether DICER-dependent miRNAs or additional RNAi-related functions of DICER are involved in the control of adult hippocampal neurogenesis, we first analyzed the dynamics of miRNA expression in WT aNSCs under proliferating conditions, as well as upon the induction of neuronal differentiation with virally transmitted ASCL1 expression at 7, 14, and 21 DIV (Figure 5A; Braun et al., 2013). This approach increased the proportion of MAP2+ neurons to 95% (Figures S5A and S5B). By qRT-PCR we detected 335 mature miRNAs in these cells. These miRNAs could be classified into three groups according to the levels and dynamics of their expression during proliferation, early neuronal differentiation (7 DIV), and late neuronal differentiation (14 and 21 DIV) (Figure 5B). As expected, miRNAs known to be involved in proliferation or in neuronal differentiation were dynamically regulated (Figures S5C and S5D; Schouten et al., 2012), thus supporting the validity of our approach.
Figure 5.
Profiling of miRNA Expression during Neuronal Differentiation of aNSCs In Vitro
(A) Schematic representation of the neuronal differentiation protocol with inducible retrovirus expressing ASCL1 (Ascl1-ERT2-IRES-GFP) and the experiment. Cells were collected during proliferation (P) or differentiation after 7 (D7), 14 (D14), and 21 (D21) DIV.
(B) Heatmap representing the set of miRNAs dynamically regulated upon neuronal differentiation at 7, 14, and 21 DIV. Red indicates high expression and green, low expression.
(C) Fold change of selected miRNAs during differentiation over proliferating cells.
(D) Fold change expression of miRNAs in vivo in FACS-sorted Td-Tomato+ cells from ten adult Dicer WT and ten Dicer cKO mice 2 months after split-Cre recombinase virus injection into the DG. n = 3 independent experiments containing three replicates.
Data are expressed as mean ± SEM. Paired t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We hypothesized that miRNAs, whose expression is associated with early steps of neuronal fate choice, might rescue a DICER-dependent impairment on neurogenesis. We focused on a group of 11 miRNAs that showed a preferential enrichment (fold change log >2) and high levels of expression (<25 Ct values) during early neuronal differentiation (7 DIV): miR-376b-3p, 139-5p, 218-5p, 411-5p, 127-3p, 134-5p, 370-3p, 135a-5p, 382-5p, 708-5p, and 124-3p (Figure 5B, higher magnification; Figures 5C and S5E). To validate these miRNAs in vivo, we performed qRT-PCR analysis of total RNA from Tomato+ cells that were FACS sorted 2 months after split-Cre infection of adult WT or Dicer cKO mice. We could detect most of these miRNAs in Tomato+ WT cells as well as their significant reduction in Dicer cKO (Figure 5D). Next, we transfected in vitro WT and Dicer cKO aNSCs with control miRNAs, or Dicer cKO cells with a pool containing the 11 miRNAs (here referred as “total pool”). Six days after transfection, we found that the total pool, but not control miRNAs, rescued DICER-dependent impairment of neurogenesis to WT levels, as indicated by the expression of neuronal markers DCX (Figures 6A and 6B, p = 0.012; Figure 6E for mRNA quantification, p = 0.03) and MAP2 (Figures 6A and 6B, p = 0.0001). Moreover, we found a concomitant decrease in astrocytic differentiation from Dicer cKO aNSCs 6 days after transfection with the total pool, as revealed by a reduced expression of S100b (Figures 6A and 6B, p = 0.003) and GFAP mRNA (Figure 6E, p = 0.0019), indicating that these miRNAs were sufficient to control the switch between neuronal and astrocyte fates. Remarkably, when administered individually, or in subpools (chosen by the number of shared predicted targets involved in astrogliogenesis or neurogenesis, Table S2 and see below), we did not rescue neurogenesis impairment in Dicer cKO aNSCs (Figures 6C, 6E, and S6A). To further trim down the pool, we tested the withdrawal of individual miRNAs from the total pool in Dicer cKO aNSCs and found that none of them rescued neurogenesis impairment (Figure 6D). These experiments indicate that all of the 11 miRNAs are sufficient to rescue neuronal commitment in miRNA-depleted aNSCs.
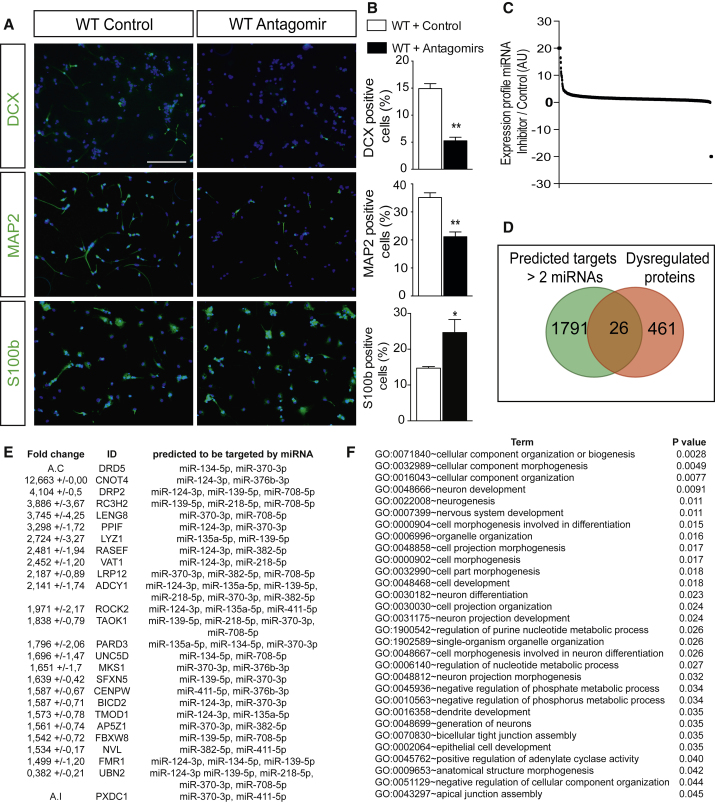
To further ascertain whether these miRNAs are required, we conducted the opposite experiment by transfecting 11 miRNA inhibitors into WT aNSCs. We found a significant reduction in DCX+ cells (Figures 7A and 7B, p = 0.003) and MAP2+ cells (Figures 7A and 7B, p = 0.0025), but an increase in S100b+ cells (Figures 7A and 7B, p = 0.05) compared with control WT cells. Taken together, these results demonstrate that 11 miRNAs are sufficient and required to sustain neuronal fate determination in hippocampal aNSCs, at the expense of astrogliogenesis, presumably through a synergic action.
Figure 7.
Convergence of the 11 miRNAs on Neurogenesis-Related Biological Processes
(A) Representative micrographs showing aNSCs from Dicer WT mice transfected with 250 nM scrambled RNA or a pool of miRNA inhibitors (antagomir) (25 nM of each miRNA inhibitor) after 6 DIV of growth factor withdrawal expressing DCX (upper panel), MAP2 (middle panel), and S100b (bottom panel).
(B) Percentage of DCX-, MAP2-, and S100b-positive aNSCs with respect to DAPI in WT aNSCs transfected with scrambled RNA or pool inhibitor.
(C) Expression proteomics profile of miRNA inhibitor with respect to control group at 6 DIV.
(D) Venn diagram showing the putative predicted targets by at least two miRNAs from the in silico analysis that are significantly dysregulated in the proteomics analysis upon miRNA inhibitor administration.
(E) Dysregulated proteins that are putatively targeted by at least two miRNAs. Fold change and SD for WT aNSCs transfected with a pool of inhibitors versus control RNA for each protein expression and predicted targeting miRNAs are shown. A.C, absent in control samples; A.I, absent in miRNA inhibitor samples.
(F) Gene ontology (GO) analysis significantly represented for the 26 predicted common targets dysregulated upon miRNA inhibition.
Data are expressed as mean ± SEM, n = 3 independent experiments containing three replicates. Unpaired t test: ∗p < 0.05, ∗∗p < 0.01. Scale bar, 50 μm.
Dissecting Pathways and Potential Targets Modulated by the 11 miRNAs in aNSCs
We hypothesized that the 11 miRNAs might synergize to simultaneously suppress pro-gliogenic and anti-neurogenic target genes. Consistently with this hypothesis, upon transfection of total miRNA pool (but not subpools), we found a significant reduction of the expression of negative regulators of neuronal differentiation, such as SP1 (p = 0.048), or astrocyte differentiation such as Tgfbr1 (p = 0.0002) (Figure S6B).
To further dissect potential targets and pathways modulated by the 11 miRNAs in aNSCs, we performed shotgun proteomics analysis in WT aNSCs transfected with the 11 miRNA inhibitors or control RNA and cultured them for 6 days in differentiating conditions. As expected, upon inhibition of 11 miRNAs we observed more upregulated proteins (>1.5-fold = 419; Figure 7C) than downregulated (<0.5-fold = 63; Figure 7C), compared with control inhibitors. Of interest, proteins dysregulated upon miRNA inhibition in aNSCs were enriched for metabolism (gene ontology [GO]: 44237; 59.8%, p = 2.5e-4) or cell cycle (GO: 7049; 16%, p=1.5e-5) processes, several of them, such as DRP2 (4.1 ± 0.5), NVL (1.53 ± 0.17), CNOT4 (12.66 ± 0.00), PTBP1 (1.63 ± 0.6), TLE1 (1.83 ± 0.6), and SFXN5 (1.64 ± 0.42) being highly expressed in astrocytes (Cahoy et al., 2008, Zhang et al., 2014) and predicted to be targeted by at least one of the 11 miRNA subjects of our study.
Next, we performed an in silico analysis to identify potential targets for the 11 miRNAs (Tables S1 and S2). We used miRWalk 2.0 with highly restrictive parameters, and found that a substantial fraction of the predicted targets (37%, i.e., 1,817 out of 4,929) were shared by at least two of the 11 miRNAs (Table S1) and had previously been shown to be expressed in developing astrocytes or neurons (Cahoy et al., 2008) (Figure S7).
To provide a molecular mechanism supporting the idea that synergic action of the 11 miRNAs is both necessary and sufficient to sustain adult neurogenesis, we compared the dysregulated proteins upon miRNA inhibition (Figure 7C) with the predicted targets (Table S1) and found 26 proteins of interest (Figure 7D). Remarkably, none of them was a predicted target of all miRNAs. Instead, they were predicted targets of different combinations of the selected miRNAs (Figure 7E). Moreover, based on analyses with DAVID (Jiao et al., 2012) or Reactome (Fabregat et al., 2016) software, we did not find any pathway that was shared between the 26 candidates. In contrast, GO analysis revealed that the 26 proteins shared similar biological processes (Figure 7F), such as nervous system development (GO: 7399; 30%, p = 0.011), neurogenesis (GO: 22008; 27%, p = 0.011), and neuron differentiation (GO: 30182; 24%, p = 0.023).
These results suggested that the 11 miRNAs cooperate by acting upon several targets within different pathways in parallel to determine adult neurogenesis.
Discussion
Our study demonstrates that a set of 11 miRNAs (of which nine were not previously characterized in adult neurogenesis) is essential for neurogenic lineage fate determination of aNSCs in the adult hippocampus, and does so at the expense of astrogliogenesis. Remarkably, these miRNAs could rescue impaired neurogenesis in Dicer cKO aNSCs to WT levels only when administered as a pool, not individually. Thus our study provides evidence for the emerging notion of miRNA “convergence” (or “cooperativity”) that, by synergistic enforcement of gene-regulatory networks, allows the acquisition of neurogenic fate programming in aNSCs.
Adult neurogenesis is a highly conserved process among vertebrates (Gage and Temple, 2013). However, the mechanisms underlying the control of a proper acquisition of the neurogenic versus astrogliogenic fate remains a fundamental question in the field (Bonaguidi et al., 2012, Kempermann, 2011). Here, by targeting bona fide type I aNSCs in vivo and in vitro, we show that loss of DICER-dependent miRNAs in aNSCs impaired neurogenesis but not astrogliogenesis. Thus our results uncover miRNAs as a regulatory level necessary to sustain neurogenic lineage and prevent astrogliogenesis in the adult hippocampal niche. This evidence reinforces the emerging idea that multiple layers of control are required to allow adult neurogenesis to occur properly. This has recently been demonstrated for other epigenetic mechanisms in a similar way (Lv et al., 2013, Noguchi et al., 2015).
An interesting question is why, given these results, DICER/miRNA-depleted aNSCs can still undergo astrocytic differentiation at all? Based on our results, and given the proposed glial nature of aNSCs (Brunne et al., 2010, Kriegstein and Alvarez-Buylla, 2009, Nicola et al., 2015), which share common molecular pathways with non-neurogenic astrocytes (Beckervordersandforth et al., 2014, Buffo et al., 2008, Coskun et al., 2008), we postulate that the gliogenic program in aNSCs might represent rather a “default” developmental path than a fate change, and thus be less dependent on miRNAs (Encinas et al., 2011). Despite we cannot rule out that “immature or intermediate” astrocytes are generated by Dicer-depleted aNSCs in vivo, these newborn cells (as revealed by Td-Tomato and BrdU) were also positive for different astrocytic markers, such as S100b (Figures 1E and 1H) and GS (Figures S1B and S1C) 2 months after Dicer deletion. Moreover, since upon Dicer ablation we did not find increased expression of progenitor markers such as Nestin or SOX2 (Figures S1B and S1C), we postulate that these cells might be bona fide astrocytes, rather than aNSCs remaining in undifferentiated (or returning) or quiescent state. Another possibility is that miRNA-depleted newborn neurons could be more susceptible to apoptosis compared with astrocytes. However, differentiating aNSC Dicer HT (Figure 2) were not more susceptible than WT cells to apoptosis (Figure S4), but still gave rise to fewer neurons (Figure 3). This suggests that miRNA loss in aNSCs can affect the switch toward neurogenesis independently from cell death. Finally, it is still possible that different subtypes of neural and glial progenitor cells exist in the adult hippocampal niche that responds differently to miRNA depletion; hence in the absence of Dicer/miRNAs neurogenesis fails while astrogliogenesis is proportionally increased. This scenario would be consistent with the known heterogeneity of aNSCs (Shin et al., 2015). However, we can still conclude that loss of Dicer and DICER-dependent miRNAs does not impair astrogliogenesis in the adult hippocampal stem cell niche.
Although, due to limitations of the current tools and technical challenges, we could not perform direct manipulation of the 11 miRNAs to rescue/inhibit neurogenesis in vivo, we demonstrated that most of these miRNAs are expressed in adult-born hippocampal neurons, and consequently depleted upon the loss of Dicer in vivo. Moreover, our in vitro experiments identify that all of the 11 miRNAs are sufficient to rescue DICER-dependent impairment of neurogenesis (Figure 6) and are required to sustain neurogenesis in WT aNSCs (Figure 7). Together, these results strongly indicate that miRNAs, rather than other additional DICER functions, determine neuronal fate of aNSCs. In perspective, our approach might be a useful paradigm to functionally investigate other miRNAs and targets. For example, 6 of the 11 miRNAs of our study are encoded by the Dlk1-Dio3 imprinted genomic region, containing the mirG locus that is highly enriched with miRNAs and deregulated in neurodevelopmental disorders and brain tumors (Gardiner et al., 2012, Henriksen et al., 2014). This locus also encodes miR-134, which is important for neuronal synaptogenesis and plasticity (Schratt et al., 2006).
The proposed synergy of miRNA actions refers to the “convergence” or “cooperativity” of miRNAs as a rapidly emerging theme in neurobiology, and has recently been proposed for embryonic neurogenesis (Barca-Mayo and De Pietri Tonelli, 2014), the adult SVZ (Santos et al., 2016), and apoptosis in the adult DG (Schouten et al., 2015). Consistent with this idea, we identified 26 putative targets of the 11 miRNAs that did not share immediate involvement in any pathway, but synergistically regulate biological processes such as neurogenesis, nervous system development, and neuronal differentiation. These results are consistent with our model whereby miRNAs “converge on function” (Barca-Mayo and De Pietri Tonelli, 2014). Given that each of the 11 miRNAs individually did not significantly induce neurogenesis, the synergic action on several targets from different pathways in parallel might compensate for the mild degree of miRNA-dependent regulation of individual mRNA targets. Further studies will be essential to experimentally validate the miRNAs and targets that are, in combination, key in regulating aNSCs neurogenesis.
Finally, the identification of a set of miRNAs that determines neuronal fate of aNSCs raises interesting perspectives with regard to age-dependent loss of hippocampal neurogenesis (Marlatt and Lucassen, 2010) or the generation of undesirable cells upon insults or cell transplantation (Dibajnia and Morshead, 2013, Doetsch et al., 2002, Shimada et al., 2012, Sierra et al., 2015). Perhaps administration of the miRNAs that were the subject of this study will increase our repertoire of approaches to sustain neurogenesis in the aging brain, or to improve efficiency of NSC-based regenerative therapies.
Experimental Procedures
Animals
Mice were housed under standard laboratory conditions at Istituto Italiano di Tecnologia (IIT). All experiments and procedures were approved by the Italian authorities (permit nos. 056/2013 and 214/2015-PR) and were conducted in accordance with the Guide for the Care and Use of Laboratory Animals of the European Community Council Directives. Dicerflox/flox mice (Murchison et al., 2005) were crossed with Td-Tomatoflox/wt knockin reporter mice (Jackson Laboratory stock number 007908; Madisen et al., 2010). Dicerwt/wt Td-Tomatoflox/wt (Dicer WT), Dicerflox/wt Td-Tomatoflox/wt (Dicer HT), and Dicerflox/flox Td-Tomatoflox/wt (Dicer cKO) were used for experiments.
aNSC Preparation, Culture Conditions, and miRNA Administration
Hippocampal NSCs were prepared and expanded as described previously (Babu et al., 2011, Walker and Kempermann, 2014). Dicer ablation was obtained in proliferating aNSCs by nucleofection (Amaxa, Lonza) of 5 μg of Cre recombinase-expressing vector under the control of constitutive cytomegalovirus enhancer/chicken β-actin (CAG) promoter (pCAGGS-CRE). Detailed cell-culture protocols are described in Supplemental Experimental Procedures. For miRNA administration, proliferating aNSCs were nucleofected (Amaxa, Lonza) with a pool of mimics or antagomirs at equimolar concentration to a final concentration 250 nM (Dharmacon, negative control, CN-001000-01-05); or with a mix of individual miRNA mimics each at 25 nM, plus negative control to a final concentration of 250 nM. Twenty-four hours after nucleofection, cells were plated in differentiation medium and harvested after 6 days.
Immunofluorescence
Immunofluorescence staining on brain slices was performed in one of every six sections of the hippocampus. A list of primary antibodies and detailed protocol are provided in Supplemental Experimental Procedures. Images were obtained with the Confocal A1 Nikon Inverted SFC with 40× objective. Quantification and analysis in the DG was performed using NIS-Elements software (Nikon). Immunofluorescence on cell cultures was performed as previously described (Babu et al., 2011). Images were obtained using the microscope Nikon Eclipse at 20× or 40× magnification, and cell-counter plugin in ImageJ software (Macbiophotonics) was used to keep track of counted cells.
RNA/Protein Extraction, Analysis, and Proteomics
For RNA extraction and cDNA preparation, six to ten mice (each Dicer genotype) were euthanized at the indicated time points. DG cells were dissociated with the Neural Tissue Dissociation Kit P (Miltenyi Biotec), and FACS-sorted cells were immediately processed for RNA extraction. Cre-nucleofected aNSCs in culture were harvested at the indicated time points. Total RNA was extracted with QIAzol protocol (Qiagen) and RNA purified with RNeasy Mini Kit, or miRNeasy Mini Kit (Qiagen) following the manufacturer's instructions. cDNA (for mRNAs) synthesis was obtained with ImProm-II reverse transcriptase (Promega); cDNA (from miRNA) was prepared with an miScript II RT kit using the HiSpec Buffer (Qiagen) according to the manufacturer's instructions. mRNA was quantified with a QuantiFast SYBR Green PCR Kit (Qiagen) on a ABI-7500 Real-Time PCR System (Applied Biosystems). Each sample was normalized to GAPDH or Actin levels. Specific primers used for gene expression analysis are listed in Supplemental Experimental Procedures. miRNAs were quantified with the Mouse Cell Differentiation & Development miScript miRNA PCR Array (Qiagen) and miScript SYBR Green PCR kit (Qiagen) following the manufacturer's recommendations on an ABI-7500 Real-Time PCR System (Applied Biosystems) or with TaqMan Array Rodent MicroRNA A Cards Set v3.0 (Thermo Fisher) following the manufacturer's recommendations with a ViiA 7 Real-Time PCR system (Thermo Fisher), for which original Ct values are available on request. For western blotting, proteins were extracted from aNSCs by RIPA buffer containing protease inhibitors (Complete mini EDTA-free, Roche), separated by SDS-PAGE on a 10% Tris gradient gel and transferred to a nitrocellulose membrane (Bio-Rad). Membranes were probed overnight using primary antibodies: rabbit GFAP (1:5,000; catalog no. Z-0334, Dako), rat anti-NESTIN (1:1,000; 556309, BD-Pharmingen), rabbit GAPDH (1:4500; AM4300, Applied Biosystems), and secondary horseradish peroxidase-conjugated antibodies (1:2,500; anti-rabbit immunoglobulin G [IgG], A16074, Life Technologies; anti-rat IgG, 31470, Thermo Fisher). Bands were detected by ECL (Millipore) using ImageQuant LAS 4000 Mini (GE Healthcare) and quantified using ImageJ software.
For proteomics, aNSCs (three independent experiments) were lysed with RIPA buffer and 60 μg of proteins was collected from all the samples to isobarically label them using TMT Sixplex kits (Thermo Fisher Scientific). Protein pools were processed for liquid chromatography-tandem mass spectrometry analysis (see Supplemental Experimental Procedures).
Statistical Analysis
Data are presented as mean ± SEM and were analyzed using Prism 6 (GraphPad). Statistical significance was assessed with a two-tailed unpaired t test for two experimental groups. For experiments with three or more groups, one-way ANOVA with Bonferroni's multiple comparison test as post hoc was used. Results were considered significant when p < 0.05.
Author Contributions
M.P.E. performed all experiments and analyses, and co-wrote the manuscript. E.d.L. performed some in vitro experiments and analysis. Initial preparation of in vitro hippocampal aNSC cultures was performed in the G.K. laboratory under the supervision of K.F. M.J.M. performed analysis of miRNA qRT-PCR in the F.N. laboratory, who supervised this part of the work. R.B. produced initial stocks and helped to set up split-Cre virus. A.A. performed and analyzed the proteomics expression experiments. D.D.P.T conceived and coordinated the project and co-wrote the manuscript. All authors approved the final version of the manuscript.
Acknowledgments
We thank Dr. G. Hannon (Cold Spring Harbor Laboratory, USA) for kindly providing Dicer-Flox mouse line; Dr. S. Jessberger (University of Zurich, Switzerland) for Ascl1-ERT2-IRES-GFP viral construct; and Drs M. Götz and J. Ninkovic (Helmholtz Zentrum, München, Germany) for the split-Cre viral constructs. We thank Drs. P. Oloth (DZNE), T. Walker (CRTD), A. Simi, and A Contestabile (IIT) for advice on aNSC preparation and differentiation. We thank R. Pelizzoli and IIT-NBT technical staff (M. Pesce, F. Succol, and M. Nanni) for excellent help. We also thank the Animal Facility of IIT Genoa (F. Piccardi; D. Cantatore; R. Navone, and M. Morini) for assistance in animal experiments. D.D.P.T. was supported by intramural funds of Fondazione Istituto Italiano di Tecnologia. This research was supported by intramural funds of Fondazione Istituto Italiano di Tecnologia and by Fondazione Cariplo grant no. 2015-0590 to D.D.P.T. and F.N.
Published: March 16, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.02.012.
Supplemental Information
References
- Agarwal V., Bell G.W., Nam J.-W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy-Aksel A., Zampa F., Schratt G. MicroRNAs and synaptic plasticity—a mutual relationship. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson T., Rahman S., Sansom S.N., Alsiö J.M., Kaneda M., Smith J., O’Carroll D., Tarakhovsky A., Livesey F.J. Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS One. 2010;5:e13453. doi: 10.1371/journal.pone.0013453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu H., Claasen J.-H., Kannan S., Rünker A.E., Palmer T., Kempermann G. A protocol for isolation and enriched monolayer cultivation of neural precursor cells from mouse dentate gyrus. Front. Neurosci. 2011;5:89. doi: 10.3389/fnins.2011.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barca-Mayo O., De Pietri Tonelli D. Convergent microRNA actions coordinate neocortical development. Cell. Mol. Life Sci. 2014;71:2975–2995. doi: 10.1007/s00018-014-1576-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R., Deshpande A., Schäffner I., Huttner H.B., Lepier A., Lie D.C., Götz M. In vivo targeting of adult neural stem cells in the dentate gyrus by a split-cre approach. Stem Cell Rep. 2014;2:153–162. doi: 10.1016/j.stemcr.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R., Zhang C.-L., Lie D.C. Transcription-factor-dependent control of adult hippocampal neurogenesis. Cold Spring Harb. Perspect. Biol. 2015;7:a018879. doi: 10.1101/cshperspect.a018879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi M.A., Song J., Ming G., Song H. A unifying hypothesis on mammalian neural stem cell properties in the adult hippocampus. Curr. Opin. Neurobiol. 2012;22:754–761. doi: 10.1016/j.conb.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond A.M., Ming G., Song H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell. 2015;17:385–395. doi: 10.1016/j.stem.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S.M.G., Machado R.A.C., Jessberger S. Temporal control of retroviral transgene expression in newborn cells in the adult brain. Stem Cell Rep. 2013;1:114–122. doi: 10.1016/j.stemcr.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunne B., Zhao S., Derouiche A., Herz J., May P., Frotscher M., Bock H.H. Origin, maturation, and astroglial transformation of secondary radial glial cells in the developing dentate gyrus. Glia. 2010;58:1553–1569. doi: 10.1002/glia.21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffo A., Rite I., Tripathi P., Lepier A., Colak D., Horn A.-P., Mori T., Götz M. Origin and progeny of reactive gliosis: a source of multipotent cells in the injured brain. Proc. Natl. Acad. Sci. USA. 2008;105:3581–3586. doi: 10.1073/pnas.0709002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel S.E., Martienssen R.A. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernilogar F.M., Onorati M.C., Kothe G.O., Burroughs A.M., Parsi K.M., Breiling A., Lo Sardo F., Saxena A., Miyoshi K., Siomi H. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L.-C., Pastrana E., Tavazoie M., Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun V., Wu H., Blanchi B., Tsao S., Kim K., Zhao J., Biancotti J.C., Hutnick L., Krueger R.C., Fan G. CD133+ neural stem cells in the ependyma of mammalian postnatal forebrain. Proc. Natl. Acad. Sci. USA. 2008;105:1026–1031. doi: 10.1073/pnas.0710000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D., Pulvers J.N., Haffner C., Murchison E.P., Hannon G.J., Huttner W.B. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibajnia P., Morshead C.M. Role of neural precursor cells in promoting repair following stroke. Acta Pharmacol. Sin. 2013;34:78–90. doi: 10.1038/aps.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F., Petreanu L., Caille I., Garcia-Verdugo J.-M., Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36:1021–1034. doi: 10.1016/s0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Encinas J.M., Michurina T.V., Peunova N., Park J.-H., Tordo J., Peterson D.A., Fishell G., Koulakov A., Enikolopov G. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8:566–579. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A., Sidiropoulos K., Garapati P., Gillespie M., Hausmann K., Haw R., Jassal B., Jupe S., Korninger F., McKay S. The reactome pathway knowledge base. Nucleic Acids Res. 2016;44:D481–D487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R.C., Farh K.K.-H., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage F.H., Temple S. Neural stem cells: generating and regenerating the brain. Neuron. 2013;80:588–601. doi: 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- Gardiner E., Beveridge N.J., Wu J.Q., Carr V., Scott R.J., Tooney P.A., Cairns M.J. Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells. Mol. Psychiatry. 2012;17:827–840. doi: 10.1038/mp.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Henriksen M., Johnsen K.B., Olesen P., Pilgaard L., Duroux M. MicroRNA expression signatures and their correlation with clinicopathological features in glioblastoma multiforme. Neuromolecular Med. 2014;16:565–577. doi: 10.1007/s12017-014-8309-7. [DOI] [PubMed] [Google Scholar]
- Huang V., Li L.-C. Demystifying the nuclear function of Argonaute proteins. RNA Biol. 2014;11:18–24. doi: 10.4161/rna.27604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X., Sherman B.T., Huang D.W., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28:1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G. The pessimist’s and optimist’s views of adult neurogenesis. Cell. 2011;145:1009–1011. doi: 10.1016/j.cell.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Kempermann G., Song H., Gage F.H. Neurogenesis in the adult Hippocampus. Cold Spring Harb. Perspect. Med. 2015;5:a018812. doi: 10.1101/cshperspect.a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka W., Kiryk A., Novak M., Herwerth M., Parkitna J.R., Wawrzyniak M., Kowarsch A., Michaluk P., Dzwonek J., Arnsperger T. MicroRNA loss enhances learning and memory in mice. J. Neurosci. 2010;30:14835–14842. doi: 10.1523/JNEUROSCI.3030-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A., Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Li L.-C. Chromatin remodeling by the small RNA machinery in mammalian cells. Epigenetics. 2014;9:45–52. doi: 10.4161/epi.26830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lv J., Xin Y., Zhou W., Qiu Z. The epigenetic switches for neural development and psychiatric disorders. J. Genet. Genomics. 2013;40:339–346. doi: 10.1016/j.jgg.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill S.T., Cambronne X.A., Luikart B.W., Lioy D.T., Leighton B.H., Westbrook G.L., Mandel G., Goodman R.H. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. USA. 2010;107:20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlatt M.W., Lucassen P.J. Neurogenesis and Alzheimer’s disease: biology and pathophysiology in mice and men. Curr. Alzheimer Res. 2010;7:113–125. doi: 10.2174/156720510790691362. [DOI] [PubMed] [Google Scholar]
- Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola Z., Fabel K., Kempermann G. Development of the adult neurogenic niche in the hippocampus of mice. Front. Neuroanat. 2015;9:53. doi: 10.3389/fnana.2015.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi H., Kimura A., Murao N., Matsuda T., Namihira M., Nakashima K. Expression of DNMT1 in neural stem/precursor cells is critical for survival of newly generated neurons in the adult hippocampus. Neurosci. Res. 2015;95:1–11. doi: 10.1016/j.neures.2015.01.014. [DOI] [PubMed] [Google Scholar]
- Pons-Espinal M., de Lagran M.M., Dierssen M. Functional implications of hippocampal adult neurogenesis in intellectual disabilities. Amino Acids. 2013;45:113–131. doi: 10.1007/s00726-013-1489-x. [DOI] [PubMed] [Google Scholar]
- Santos M.C.T., Tegge A.N., Correa B.R., Mahesula S., Kohnke L.Q., Qiao M., Ferreira M.A.R., Kokovay E., Penalva L.O.F. miR-124, -128, and -137 orchestrate neural differentiation by acting on overlapping gene sets containing a highly connected transcription factor network. Stem Cells. 2016;34:220–232. doi: 10.1002/stem.2204. [DOI] [PubMed] [Google Scholar]
- Schmiedel J.M., Klemm S.L., Zheng Y., Sahay A., Blüthgen N., Marks D.S., van Oudenaarden A. Gene expression. MicroRNA control of protein expression noise. Science. 2015;348:128–132. doi: 10.1126/science.aaa1738. [DOI] [PubMed] [Google Scholar]
- Schouten M., Buijink M.R., Lucassen P.J., Fitzsimons C.P. New neurons in aging brains: molecular control by small non-coding RNAs. Front. Neurosci. 2012;6:25. doi: 10.3389/fnins.2012.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten M., Fratantoni S.A., Hubens C.J., Piersma S.R., Pham T.V., Bielefeld P., Voskuyl R.A., Lucassen P.J., Jimenez C.R., Fitzsimons C.P. MicroRNA-124 and -137 cooperativity controls caspase-3 activity through BCL2L13 in hippocampal neural stem cells. Sci. Rep. 2015;5:12448. doi: 10.1038/srep12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G.M., Tuebing F., Nigh E.A., Kane C.G., Sabatini M.E., Kiebler M., Greenberg M.E. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Shimada I.S., LeComte M.D., Granger J.C., Quinlan N.J., Spees J.L. Self-renewal and differentiation of reactive astrocyte-derived neural stem/progenitor cells isolated from the cortical peri-infarct area after stroke. J. Neurosci. 2012;32:7926–7940. doi: 10.1523/JNEUROSCI.4303-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Berg D.A., Zhu Y., Shin J.Y., Song J., Bonaguidi M.A., Enikolopov G., Nauen D.W., Christian K.M., Ming G. Single-cell RNA-Seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano V., Garzilli I., Fracassi C., Criscuolo S., Ventre S., di Bernardo D. miRNAs confer phenotypic robustness to gene networks by suppressing biological noise. Nat. Commun. 2013;4:2364. doi: 10.1038/ncomms3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A., Martín-Suárez S., Valcárcel-Martín R., Pascual-Brazo J., Aelvoet S.-A., Abiega O., Deudero J.J., Brewster A.L., Bernales I., Anderson A.E. Neuronal hyperactivity accelerates depletion of neural stem cells and impairs hippocampal neurogenesis. Cell Stem Cell. 2015;16:488–503. doi: 10.1016/j.stem.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt R.D., Szulwach K.E., Pfeiffer R.L., Li X., Guo W., Pathania M., Teng Z.-Q., Luo Y., Peng J., Bordey A. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase Mind Bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T.L., Kempermann G. One mouse, two cultures: isolation and culture of adult neural stem cells from the two neurogenic zones of individual mice. J. Vis. Exp. 2014:e51225. doi: 10.3791/51225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.-S., Lai E.C. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S., Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Yang L., Du Z. MicroRNA regulation and tissue-specific protein interaction network. PLoS One. 2011;6:e25394. doi: 10.1371/journal.pone.0025394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.