Abstract
Cytoplasmic dynein is a microtubule-associated minus-end-directed motor protein. CaDYN1 encodes the single dynein heavy-chain gene of Candida albicans. The open reading frames of both alleles of CaDYN1 were completely deleted via a PCR-based approach. Cadyn1 mutants are viable but grow more slowly than the wild type. In vivo time-lapse microscopy was used to compare growth of wild-type (SC5314) and dyn1 mutant strains during yeast growth and after hyphal induction. During yeast-like growth, Cadyn1 strains formed chains of cells. Chromosomal TUB1-GFP and HHF1-GFP alleles were used both in wild-type and mutant strains to monitor the orientation of mitotic spindles and nuclear positioning in C. albicans. In vivo fluorescence time-lapse analyses with HHF1-GFP over several generations indicated defects in dyn1 cells in the realignment of spindles with the mother-daughter axis of yeast cells compared to that of the wild type. Mitosis in the dyn1 mutant, in contrast to that of wild-type yeast cells, was very frequently completed in the mother cells. Nevertheless, daughter nuclei were faithfully transported into the daughter cells, resulting in only a small number of multinucleate cells. Cadyn1 mutant strains responded to hypha-inducing media containing l-proline or serum with initial germ tube formation. Elongation of the hyphal tubes eventually came to a halt, and these tubes showed a defect in the tipward localization of nuclei. Using a heterozygous DYN1/dyn1 strain in which the remaining copy was controlled by the regulatable MAL2 promoter, we could switch between wild-type and mutant phenotypes depending on the carbon source, indicating that the observed mutant phenotypes were solely due to deletion of DYN1.
Faithful segregation of nuclei is essential for the proliferation of the eukaryotic cell. Dynamic behavior of nuclei is required at several stages during the cell cycle for correct nuclear positioning, orientation of the mitotic spindle, and distribution of nuclei in the cytoplasm, particularly in filamentous fungi (18). Nuclear distribution has been studied in detail in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, as well as in the filamentous fungi Aspergillus nidulans and Neurospora crassa (30, 31, 32). The spindle pole body (SPB) plays a central role in nuclear dynamics, as it is the organizing center of cytoplasmic microtubules (MTs) that emanate from the SPB outer plaque (4, 5). These cytoplasmic MTs interact with the cell cortex in S. cerevisiae via a search-and-capture mechanism that results in the orientation of the mitotic spindle in the mother-bud axis, in the positioning of the nucleus at the bud neck, and in the translocation of the daughter nucleus into the bud. A set of proteins are involved in this process, including Kar9p, Bim1p, Bud6p, the formin Bni1p, and Myo2p (3, 8, 12, 17, 33, 34). During anaphase, nuclear movement depends on the minus-end-directed microtubule motor protein Dyn1p (the S. cerevisiae dynein heavy-chain homolog), which provides the pulling force (1). Deletion of DYN1 in S. cerevisiae or mutations in the conserved AAA domains of dynein involved in nucleotide binding and hydrolysis resulted in slower growing but viable mutant strains which displayed disruption of normal nuclear movement (13, 20). S. cerevisiae dynein mutants were shown not to be defective in mating and karyogamy (13). In contrast to short-range nuclear migration which delivers the nucleus into the mating projection of S. cerevisiae, in the hyphae of filamentous fungi long-range transport of nuclei and organelles into the hyphal tip is required and a uniform distribution of nuclei is achieved to support growth and development of a mycelium. Dynein heavy-chain mutants in A. nidulans (nudA) and N. crassa (ro-1) revealed severe defects in nuclear distribution (19, 29). Whereas during germination (the process in which a germ tube is formed from a round-shaped germ cell) in A. nidulans nudA mutant nuclei are not transported into the hyphal tube, the dynein heavy-chain mutants, such as dhc1, in the filamentous fungus Ashbya gossypii show a clustering of nuclei in the hyphal tips (2). In the N. crassa ropy class of mutants, with several genes of the dynein/dynactin complex, defects in nuclear positioning were also accompanied by morphological alterations (6, 19, 25, 26). In contrast to the hyphal morphology of the wild type, ropy mutants show distorted morphologies indicative of a loss-of-growth directionality and an out-of-tip-focus Spitzenkörper (21). Cytoplasmic dynein in dimorphic fungi has so far only been studied in the plant pathogenic fungus Ustilago maydis, in which it is encoded by two genes, dyn1 and dyn2 (23). Dynein is essential in U. maydis, in contrast to the cases of S. cerevisiae, and, as we show here, Candida albicans. In this report we present mutational analysis of the DYN1 gene of C. albicans and, by using in vivo time-lapse microscopy, show its involvement in nuclear positioning and spindle movement during yeast-like growth. Slight growth defects in the yeast phase yielded elongated cells, whereas under regimens inducing hyphal growth in C. albicans dyn1 mutants exhibited defects in hyphal formation and in the maintenance of hyphal growth that can be explained by defects in nuclear distribution within the hyphae.
MATERIALS AND METHODS
Strains and media.
C. albicans strains used in this study are listed in Table 1. Cells were grown in YPD (1% yeast extract, 2% peptone, 2% glucose) or complete supplement medium (CSM) at 30°C. For hyphal induction in liquid minimal media, 10% serum (calf serum; Sigma) was added to CSM and then was incubated at 37°C. For hyphal induction on plates, CSM with 10 to 20% serum or Spider medium (14) was used. MAL2p-DYN1 cultures were grown on minimal medium containing 2% maltose as the sole carbon source to induce expression. Plates were incubated 4 to 7 days at 37°C prior to photography.
TABLE 1.
Strains used in this study
| Straina | Genotype | Reference |
|---|---|---|
| SC5314 | Candida albicans wild type | 5a |
| BWP17 | ura3::λimm34/ura3::λimm34 his1::hisG/his1::hisG arg4::hisG/arg4/hisG | 28a |
| GC1 | DYN1/dyn1::URA3 | This study |
| GC2 | DYN1/dyn1::HIS1 | This study |
| GC3 | dyn1::URA3/dyn1::HIS1 | This study |
| GC4 | dyn1::HIS1/dyn1::URA3 | This study |
| GC6 | TUB1/TUB1-GFP::URA3 | This study |
| GC8 | dyn1::URA3/dyn1::HIS1 TUB1/TUB1-GFP::ARG4 | This study |
| GC12 | HHF1/HHF1-GFP::URA3 | This study |
| GC17 | dyn1::URA3/dyn1::HIS1 HHF1/HHF1-GFP::ARG4 | This study |
| CAT23 | dyn1::URA3/MAL2p-DYN1:HIS1 | This study |
All GC strains are derivates of BWP17 with the indicated genotypic alterations.
Disruption of CaDYN1.
The C. albicans homolog of the dynein heavy-chain gene DYN1 was identified in the genomic sequence (http://www-sequence.stanford.edu/group/candida). Deletions of the complete open reading frames of both alleles of CaDYN1 were performed by PCR-generated FA-URA3 and FA-HIS1 disruption cassettes (using primers #678 and #679) as described previously (7). Primers used for construction of the cassettes and verification of the deletions are listed in Table 2. Independent transformants were produced, and the disruptions were verified by PCR on whole yeast cells as described previously (27). Identical phenotypes of the independent homozygous transformants were used to indicate that the correct mutant strains were constructed. To place DYN1 under control of the regulatable MAL2 promoter, we used the heterozygous DYN1/dyn1::URA3 (GC1) strain and transformed it with PCR fragments amplified from pFA-HIS1-MAL2p with primers #678 and #1122. Promoter shutdown experiments further verified the correct construction of mutant strains, which therefore were not further analyzed by DNA-hybridization techniques.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequencea |
|---|---|
| #599 U3 | GGAGTTGGATTAGATGATAAAGGTGATGG |
| #600 U2 | GTGTTACGAATCAATGGCACTACAGC |
| #601 H2 | CAACGAAATGGCCTCCCCTACCACAG |
| #602 H3 | GGACGAATTGAAGAAAGCTGGTGCAACCGGCTAACCAGCTGCAATCAAGTTTAATGGGATCAATTAAC |
| #678 S1-DYN1 | TATAGTGTTGTCTGATACACGTCTAATTGTGCAAAAGTACACACACAAACCAAAAGTGTGAGgaagcttcgtacgctgcaggtcAATCTTCAGATAGATCAAGTACAGATTCCACGATTTCTA |
| #679 S2-DYN1 | ATCCAAGTTGTTTTTTCACTAACAGATTGGCATCATCAGACAAGTAAATCCAAGATAATGGtctgatatcatcgatgaattcgag |
| #680 G1-DYN1 | GTGTGACTTCAACCCTTCTTTG |
| #681 G4-DYN1 | GATGCTTCCGGTTCTATCAACAG |
| #692 S1-HHF1-GFP | CTGTTACTTACACTGAACATGCTAAAAGAAAACCGTCACTTCATTGGATGTTGTTTACGCTTTGAAGAGACAAGGTAGAACCTTGTATGGTTTCGGTGGTggtgctggcgcaggtgcttc |
| #693 S2-HHF1-GFP | GATAATGAACTTCAATGAATGACCATTTTATCTGACCAATAAAATCAAAATAGTAAAAAATTGGTGGAAATAAGATACCGAAAATAATTTGCTTGCCTTGtctgatattcatcgaatgaattcgag |
| #694 S1-TUB1-GFP | AGAAGGTGAATTCACTGAAGCTAGAGAAGACTTGGCTGCTTTAGAGAGAGATTATATTGAAGTTGGTACTGATTCTTT |
| CCCTGAAGAAGAAGAAGAATATggtgctggcgcaggtgcttc | |
| #695 S2-TUB1-GFP | CCCTCCTCTTAACCATTTGACACACCAAGAGAGTCAATTCCAAAAGTAAAAATTAAAAATCGGGCTTGGGAGTTCGGGTATATATGGTATATATATATAAGtctgatatcatcgatgaattcgag |
| #1121 G4-DYN1-MAL2p | GCGGTATATCATTCCTAAGTTCACC |
| #1122 S2-DYN1-MAL2p | CACACTGTGTAAATTTCGGCACATTTTCATTAAAAGGTAGAAAGCCTATTATACAGTCATATAATTGATTGACTGATAATAAGGGTGTGACCAGTTCTTCCcattgtagttgattattagttaaaccac |
Uppercase sequences correspond to Candida albicans genomic DNA. Lowercase sequences correspond to 3′-terminal annealing regions for the amplification of transformation cassettes. All sequences are written from 5′ to 3′.
GFP tagging of HHF1 and TUB1.
In order to fuse the genes encoding either histone H4 or α-tubulin with green fluorescent protein (GFP), a PCR-based approach was applied. To this end, transformation cassettes were amplified from plasmids pFA-GFP-URA3 (for use in BWP17) or pFA-GFP-ARG4 (for use in GC3) with primers #692 and #693 for HHF1 and primers #694 and #695 for TUB1. With these primers, 100 bp of homology to two positions at the 3′ ends of the target genes were added to PCR-amplified cassettes as described previously (7). The PCR fragments were then used to transform either BWP17 or the homozygous dyn1/dyn1 mutant strain GC3 to generate strains that contain heterozygous HHF1/HHF1-GFP or TUB1/TUB1-GFP alleles.
Transformation of C. albicans.
The lithium-acetate procedure was used to transform C. albicans as described previously (27). Basic features of this protocol include an overnight incubation with lithium-acetate followed by heat shock for 15 min at 44°C.
Staining procedures.
For examination of nuclear positioning in the heterozygote mutants GC1 and GC2 and the TUB1-GFP-labeled strains, cells were stained with 4,6-diamino-2-phenylindol (DAPI; 1 mg/ml; Molecular Probes). For this staining, 200 μl of a cell suspension was fixed with 500 μl of 70% ethanol, and 1 μl of DAPI was added. After incubation for 2 min at room temperature, cells were analyzed by fluorescence microscopy. Chitin staining was done by directly adding 1 μl of calcofluor (1 mg/ml) to a 100-μl cell suspension followed by an incubation of 15 min at room temperature and a subsequent washing step. The positioning of septa is a valuable criterion to distinguish between true hyphae and pseudohyphae and was used to score the cell morphological defects of dyn1 strains (24). Additionally, septation in hyphae occurs without producing constrictions at the septal site, whereas in pseudohyphal cells invaginations at the sites of septation are found.
Time-lapse microscopy.
Strains were grown to exponential phase either in complete or minimal medium, harvested, washed, and resuspended in sterile water. Small aliquots of cells (1.5 μl) were applied on microscopy slides with deep wells. Enrichment of media with oxygen and preparation of the media for microscopy were done as described previously (28). Temperature control was achieved via a heat stage mounted on the microscope table. Microscopy was done on a fully automated motorized Zeiss Axioplan II imaging microscope. Images were acquired by using Metamorph 4.6 software (Universal Imaging Corporation) and a digital imaging system (MicroMax1024; Princeton Instruments). For Hhf1-GFP, acquisition of images was done in 90-s intervals using 0.7-s exposure times and illumination transmission that was reduced to 5% by using appropriate neutral-density filters (Chroma Technology). Image acquisition into stacks was done via custom-designed software journals. Stacks containing brightfield/DIC (differential interference contrast) images were processed separately from images displaying GFP fluorescence. By using these conditions, nuclear dynamics of individual cells could be tracked for more than 10 h. A red look-up table was assigned to the phase-contrast images, and a green look-up table was assigned for the fluorescent images. Stacks were then combined by using overlay tools of the Metamorph software, converted to 8-bit format and processed as video clips with frame rates of 10 images/s.
RESULTS
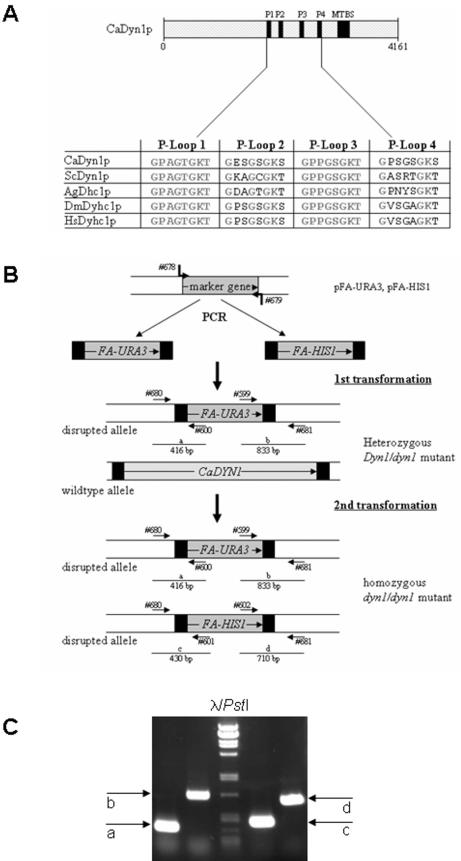
The unique gene DYN1 encodes the C. albicans dynein heavy chain. A search of the C. albicans genome database at Stanford University was performed to identify a sequence homologous to that of the dynein heavy-chain gene DYN1 of S. cerevisiae and DHC1 of A. gossypii. One open reading frame, corresponding to orf19.5999, was found to encode the single dynein heavy-chain gene, designated DYN1 in C. albicans. DYN1 (located on chromosome 3 based on the Stanford data) encodes a protein of 4,161 amino acids, with strong conservation relative to other fungal as well as mammalian dynein homologs, which range from 35% identity to 57% similarity on the protein level. Particularly well conserved are the microtubule-binding domain and the four P loops involved in ATP binding (Fig. 1A).
FIG.1.
The C. albicans dynein heavy chain. (A) Schematic representation of the Dyn1 protein. The positions of the four ATP-binding sites (P-loops; P1, amino acids 1824 to 1832; P2, 2112 to 2119; P3, 2467 to 2474; and P4, 2809 to 2816) and the microtubule binding site (MTBS; amino acids 3151 to 3276) are indicated. Sequence alignment of the four P loops indicates identical P loops in P1 and P3 in the analyzed species, with greater divergence of P loops P2 and P4. Accession numbers of the protein sequences are the following: C. albicans CaDyn1p, orf19.5999.prot; S. cerevisiae ScDyn1p, NP_012980; A. gossypii AgDhc1p, AAK20175; Drosophila melanogaster DmDyhc1p, P37276; human HsDyhc1p, Q14204. (B) Deletion of DYN1 via PCR-based gene targeting. Successive transformation of C. albicans strain BWP17 with PCR-amplified marker genes (FA-URA3 and FA-HIS1) that provide 100 bp of terminal target homology regions. In a first transformation independent, heterozygous DYN1/dyn1 strains were generated. From these strains independent homozygous dyn1/dyn1 mutant strains were derived in a second transformation event. Primer numbers (see Table 2) correspond to specific primers used for cassette amplification or PCR verification as indicated. (C) Ethidium-bromide-stained agarose gel image showing the result of diagnostic PCR that was used to verify the correct insertion of the marker genes at the target locus as described (7). The expected fragment lengths for fragments a to d were as shown in panel B.
DYN1 is not essential for cell viability in C. albicans.
To be able to study the role of dynein on nuclear migration during the different growth stages of C. albicans, we generated homozygous deletion strains via PCR-based gene targeting, starting with independent heterozygous strains (Fig. 1B). Strain BWP17 was chosen as a progenitor strain, because its auxotrophies enabled the sequential disruption of both alleles with the FA-HIS1 and FA-URA3 marker genes according to Gola et al. (7). Independent homozygous dyn1::HIS1/dyn1::URA3 mutant strains were identical in their phenotypes (see below), indicating that correct gene targeting had occurred, as was verified by analytical PCR (Fig. 1B and C). Strains bearing complete open reading frame deletions in the DYN1 genes are viable, demonstrating that C. albicans DYN1 is not an essential gene.
Defects of dyn1 mutant strains during yeast growth phase.
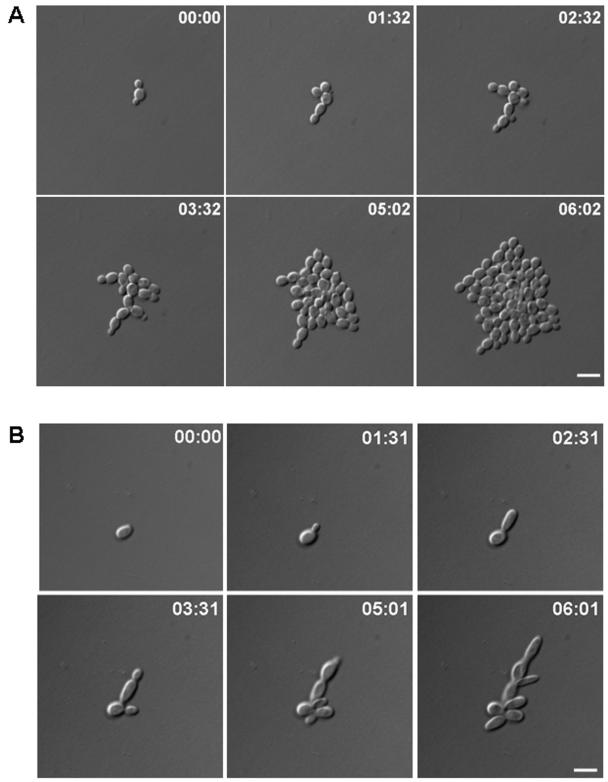
By using in vivo time-lapse microscopy, we monitored growth of the wild-type and dyn1 mutant strains over several generations (Fig. 2). Defects of the dyn1 strain were quantified by using movie data, and additional measurements were taken from yeast cells grown in complete medium (Table 3). One of the major defects is the prolonged cell cycle time of dyn1 mutants compared to that of the wild type, which increased by about 40 min. Cells of the wild type are ellipsoidal in shape, roughly 6 μm in length, and 4.5 μm in width. Cells of the heterozygous DYN1/dyn1 mutant are slightly elongated, whereas the dyn1 mutant cells are considerably longer than wild-type cells (Table 3). In addition, cell aggregates were formed in the homozygous dyn1 mutant strains, indicating cell separation defects. Because dynein is involved in nuclear distribution, we aimed at the visualization of nuclear movement in C. albicans in vivo. To this end we used strain BWP17 and generated a fusion of the histone H4 gene with GFP (see Materials and Methods). As expected, Hhf1p-GFP-derived fluorescence marked the nucleus (Fig. 3). With the strong signal intensity of the GFP label we were able to record time-lapse series to monitor nuclear movement in the progenitor strain (i.e., corresponding to the wild type) (Fig. 3).
FIG. 2.
In vivo time-lapse analyses of yeast phase growth of the wild-type (SC5314) and dyn1 (GC3) strains. Representative frames of movies of the wild-type (A) and dyn1 cells (B) are shown over a growth period of 6 h. Cell cycle times in the dyn1 mutant are longer than in the wild type, resulting in slower growth. Also, cell morphology of dyn1 yeast cells is more elongated than that in the wild type (see Table 3). Time is in hours:minutes. Bars, 10 μm. Movies are available at http://pinguin.biologie.uni-jena.de/phytopathologie/pathogenepilze/index.html.
TABLE 3.
Comparison of wild-type and dyn1 yeast cells
| Parameter | Wild type | GC1 (DYN1/dyn1) | GC3 (dyn1/dyn1) |
|---|---|---|---|
| Cell cycle duration (min) | 71 ± 5 | 83 ± 3 | 114 ± 6 |
| Cell length (n = 500) (μm) | 5.9 ± 0.3 | 7.0 ± 0.4 | 11.4 ± 0.6 |
| Cell width (n = 500) (μm) | 4.5 ± 0.2 | 4.0 ± 0.2 | 5.0 ± 0.3 |
| Single (or budded) cells versus cell aggregates (n = 500) (%) | 87 versus 13 | 81 versus 19 | 28 versus 72 |
FIG. 3.
In vivo fluorescence time-lapse analysis of Hhf1p-GFP in wild-type C. albicans. Representative frames of a movie with strain GC12. Cells were pregrown to exponential phase and were mounted on microscopy slides. Note the realignment of the elongated spindle in the interval between 2 h 13 min and 2 h 21 min, as indicated by arrows. Time is in hours:minutes. Bar, 10 μm. The movie is available at http://pinguin.biologie.uni-jena.de/phytopathologie/pathogenepilze/index.html.
Nuclear positioning in the C. albicans wild type.
Different phases of nuclear movement were described for S. cerevisiae. They include nuclear positioning in the bud-neck region, elongation of the spindle along the mother-bud axis during anaphase, accompanied by the insertion of the anaphase nucleus into the bud-neck region, and nuclear division (10, 32). Our time-lapse data revealed differences between nuclear positioning in C. albicans and that in S. cerevisiae. These changes are particularly obvious in premitotic nuclei. Here, only a quarter of the observed C. albicans nuclei were positioned directly at the mother-bud neck. Lack of early attachment to the mother-bud neck resulted in about 70% of the observed cases in anaphase spindle elongation that were not coordinated with the mother-bud axis. In the wild type, this lack of coordination was compensated for by the realignment of anaphase spindles, with the mother-bud neck requiring rotational movement of the spindle. Spindle alignment was accomplished prior to nuclear division, which was then followed by nuclear migration of the daughter nucleus into the bud (Table 4). To rule out the chance that the observed nuclear migration phenotypes in C. albicans were due to the strain background or the GFP label employed, we used DAPI staining of fixed cells of the C. albicans wild-type strain SC5314 as well as the CAI-4 and BWP17 strains. Data acquired indicated that 30% of the observed anaphase spindles (n > 200 for each strain) were not aligned in the mother-bud axis in these strains (data not shown). Because the data were derived from fixed cells, those spindles that already were realigned with the mother-bud axis could not be scored. Furthermore, in the C. albicans wild-type strain SC5314 as well as in the HHF1-GFP strain, misalignment of anaphase spindles with respect to the mother-bud axis did not result in the formation of binucleate cells. We conclude that in C. albicans, in contrast to S. cerevisiae, movement of the anaphase spindle to position the spindle in the mother-bud axis is a regular cell cycle event.
TABLE 4.
Nuclear migration in C. albicansa
| Step | GC12 (%) | GC17 (%) |
|---|---|---|
| Nuclear movement to bud site | 28.6 | 12.2 |
| Spindle elongation in mother-daughter axis | 28.6 | 12.2 |
| Spindle realignment after elongation | 71.4 | 0 |
| Completion of mitosis on mother cell | 0 | 87.8 |
| No. of binucleate cells | 0 | 4.3 |
Data was acquired from time-lapse microscopy of HHF1-GFP-tagged strains (n > 100).
Nuclear positioning in the C. albicans dyn1 strain.
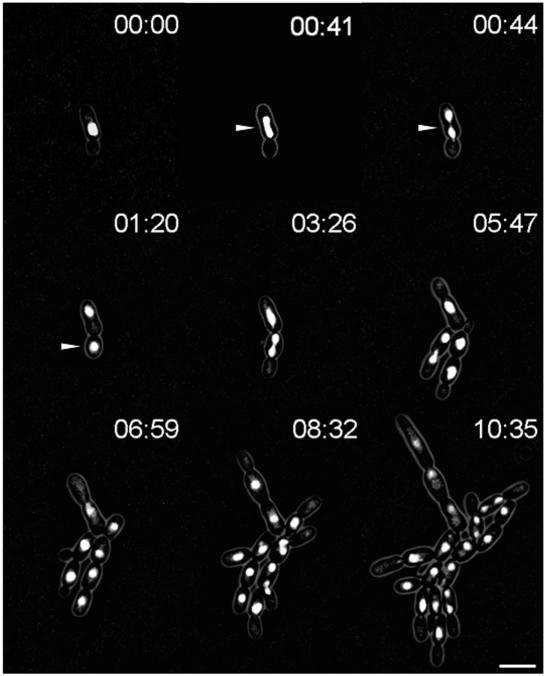
Based on the homozygous dyn1 mutant strain (GC3), we constructed an HHF1-GFP fusion in this background by using the same PCR-based approach described above. This strain appeared, based on morphological criteria, phenotypically unaltered from the GC3 strain and was used for time-lapse analysis to monitor nuclear distribution (Fig. 4). Some striking differences were observed that distinguish nuclear dynamics in the dyn1 mutant from those in the wild type. Movement of preanaphase nuclei towards the bud-neck region occurred less efficiently in the mutant than in the wild type. Spindle elongation of anaphase nuclei was less pronounced in the dyn1 mutant, resulting in spindles shorter (maximum nuclear elongation, 6.2 μm; n = 13) than those of the wild type (maximum nuclear elongation, 8.7 μm; n = 13). Spindle realignment was not observed at all in the dyn1 mutant, which, most notably, resulted in preferential completion of mitosis in the mother cell in the dyn1 mutant strain (87.8% in the dyn1 strain compared to only 7.1% in the wild type) (Table 4). Strikingly, however, even though the majority of mitotic events were completed in dyn1 mother cells, postmitotic nuclear migration ensured the correct segregation of nuclei in the dyn1 mutant, resulting in only a very small number of bi- or multinucleate cells.
FIG. 4.
In vivo fluorescence time-lapse analysis of Hhf1p-GFP in C. albicans dyn1. Representative frames of a movie with strain GC17. Cells were pregrown to exponential phase and were mounted on microscopy slides using the same conditions as those used for the time-lapse movie with strain GC12 (Fig. 3). Note the completion of mitosis in a mother cell and postmitotic nuclear migration between 41 and 44 min, as marked by the arrowheads. Time is in hours:minutes. Bar, 10 μm. The movie is available at http://pinguin.biologie.uni-jena.de/phytopathologie/pathogenepilze/index.html.
Analysis of spindle positioning by Tub1-GFP fluorescence.
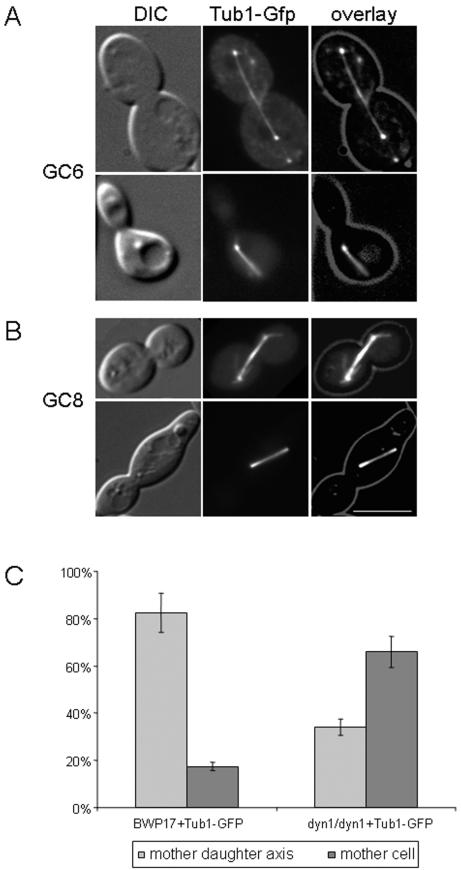
Because the data acquired by monitoring Hhf1-Gfp fluorescence only indirectly revealed spindle positioning, we went on to construct C. albicans strains that carry TUB1-GFP fusions both in the wild-type and dyn1 mutant background (see Materials and Methods). The Tub1p-GFP signal was found to correctly label the microtubules but was too weak to be used for in vivo fluorescent time-lapse recordings. Therefore, we used single time point images to quantify spindle positioning (Fig. 5). These data indicated that in the wild-type background, about 80% of the spindles were found to be in the mother-bud axis (corresponding to DAPI staining of fixed cells; see above), whereas in the dyn1 mutant only 30% of the spindles were correctly positioned. The difference between the in vivo time-lapse data set with the Tub1p-GFP fluorescence data set in the wild type may be due to the fact that spindle realignment is a very fast process (requiring only 6 min). Thus, many spindles that were found to be positioned in the mother-bud axis may in fact have been realigned prior to their observation.
FIG.5.
Orientation of mitotic spindles in C. albicans. Spindle positions in strains GC6 (DYN1/DYN1 TUB1/TUB1-GFP) and GC8 (dyn1/dyn1 TUB1/TUB1-GFP) were analyzed by fluorescence microscopy. (A) Representative images of wild-type spindle positions in which the spindle is either aligned in the mother daughter axis and extends into the daughter (top row) or is elongated in the mother cell (bottom row). (B) Images of the spindle positions in dyn1 cells in which the spindle is either aligned in the mother daughter axis or is misaligned and elongated only in the mother cell. DIC images of cells were merged into an overlay with the images showing the GFP fluorescence. Bar, 10 μm. (C) Quantification (bars indicate the standard deviation of the mean) of spindle positioning in GC6 and GC8 (n = 130 for each strain) corresponding to the observed spindle positions in panels A and B.
Loss of DYN1 results in aberrant hyphal growth in C. albicans.
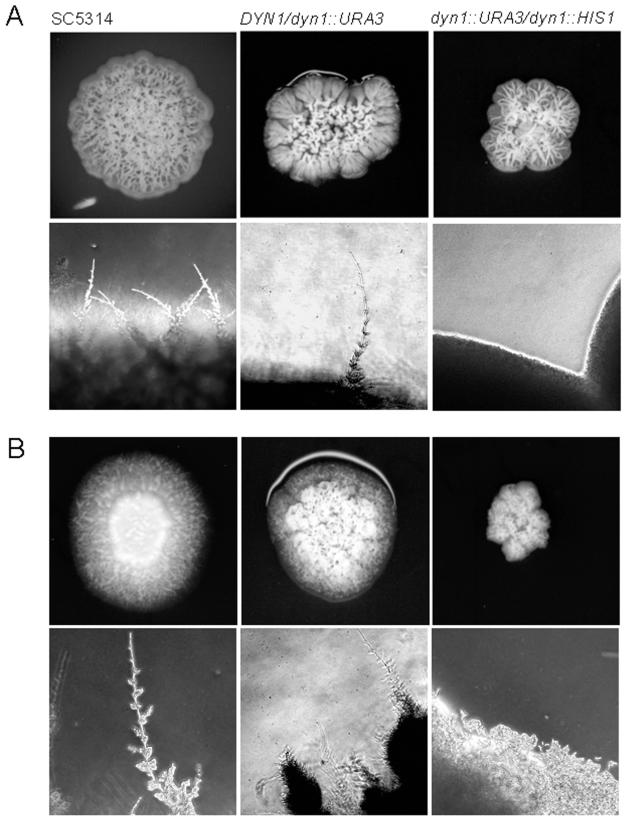
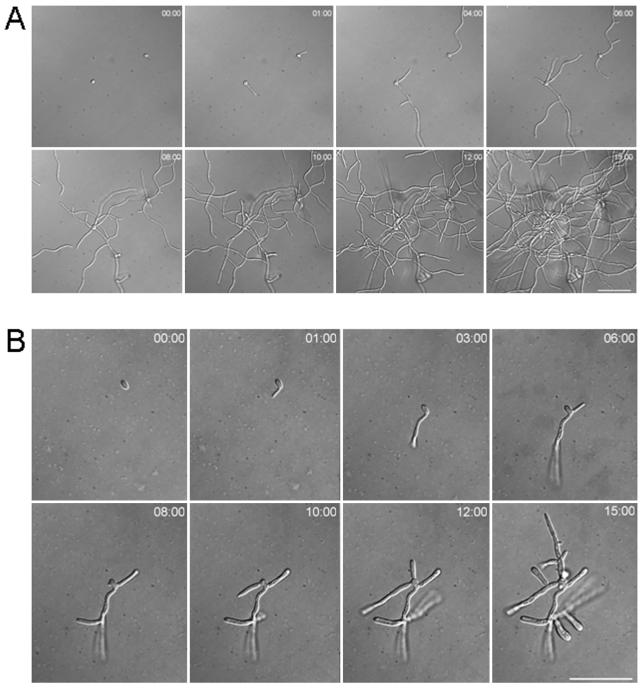
Defects in the dynein/dynactin complex in filamentous fungi have been shown to severely cripple hyphal growth (2, 19, 29). In contrast to the yeast S. cerevisiae, C. albicans is a dimorphic fungus that can be induced to form true hyphae. We used the addition of serum to the medium as well as Spider medium to induce hyphal growth in the dyn1 mutant strains (Fig. 6). While the wild type and the heterozygous DYN1/dyn1 mutant readily responded to hypha-inducing conditions, the homozygous dyn1 mutant showed defects in hypha formation. On these plates, dyn1 colonies showed a less wrinkled appearance, and defects in hyphal growth were particularly evident by the lack of filaments at the colony edges (Fig. 6). To analyze this phenotype in more detail, we applied in vivo time-lapse microscopy to follow growth of the wild-type and dyn1 mutant strains upon hyphal induction (Fig. 7A and B). As described previously (28), the wild type almost immediately responded to the external stimuli (37°C in the presence of serum) with hyphal induction. Hyphal growth was maintained for the 15-h duration of the time-lapse, lateral branches were formed, and a mycelium was generated (Fig. 7A). Hyphal induction of the dyn1 strain resulted in the formation of germ tubes, although the polar growth rate was much slower than that in the wild type (Fig. 7B). However, growth of the filaments came to a halt after a short time, and once stopped it was not resumed at later stages. New filaments were formed from the initial yeast cell and lateral branching was also induced, but mycelium formation was inhibited in dyn1 mutants, corresponding to the results of the plate assays. To quantify these time-lapse data, hyphal induction was done in liquid media, and germ tube and filament formation was monitored after 3 and 6 h (Table 5). These data indicated that initiation of germ tube formation occurs in the dyn1 mutant with frequencies similar to those of the wild type. The appearance of septum formation within germ tubes is a hallmark of hyphal induction (24). We therefore compared septum positioning between the wild type and dyn1 mutant 3 h after hyphal induction (Table 6). At this stage of hyphal induction, the differences between the wild type and dyn1 mutant indicated a higher percentage of germ tubes of the dyn1 mutant that had not formed septa (19%), presumably due to nuclear migration defects. The percentage of septa close to the bud neck in the wild type and dyn1 mutant was found to correspond to the number of pseudohyphae that were formed.
FIG. 6.
Induction of hyphal growth in wild-type and mutant strains. Hypha formation on solid media was determined by plating the indicated strains on either (A) complete synthetic medium (CSM) containing 10% serum or (B) on Spider medium. All plates were incubated for 4 days at 37°C prior to photography. Note the abundant filamentation at the colony edges of both the wild type (SC5314) and heterozygous DYN1/dyn1 on both media; this filamentation is completely absent in the homozygous dyn1dyn1 mutant. Images representing magnifications of the colony edges were acquired via digital microscopy of the corresponding colonies.
FIG. 7.
In vivo time-lapse analyses of growth of wild-type and dyn1 mutant strains under hypha-inducing conditions. Representative frames of movies of wild-type (A) and Cadyn1 cells (B) are shown at the indicated time points. Cells were preincubated overnight in sterile water. Single cells were mounted on inducing solid CSM containing serum at 37°C. Time is in hours:minutes. Bars, 50 μm each. Movies are available at http://pinguin.biologie.uni-jena.de/phytopathologie/pathogenepilze/index.html.
TABLE 5.
Analysis of polarized morphogenesis
| Criterion | Wild-type SC5314 (%) | GC3 (dyn1/dyn1) (%) |
|---|---|---|
| Germ tube formation (after 3 h) of single cells/ pseudohyphae/germ tubes | 6/11/83 (n = 317) | 6/16/78 (n = 234) |
| Hyphal growth (after 6 h) of single cells/pseudohyphae/germ tubes or hyphae | 7/24/69 (n = 207) | 5/39/56 (n = 200) |
TABLE 6.
Position of septa in germ tubesa
| Distance (μm) | Wild-type SC5314 (%) | GC3 (dyn1/dyn1) (%) |
|---|---|---|
| No septum | 8 | 19 |
| 0-5 | 16 | 23 |
| 6-20 | 73 | 51 |
| >20 | 3 | 7 |
n > 170.
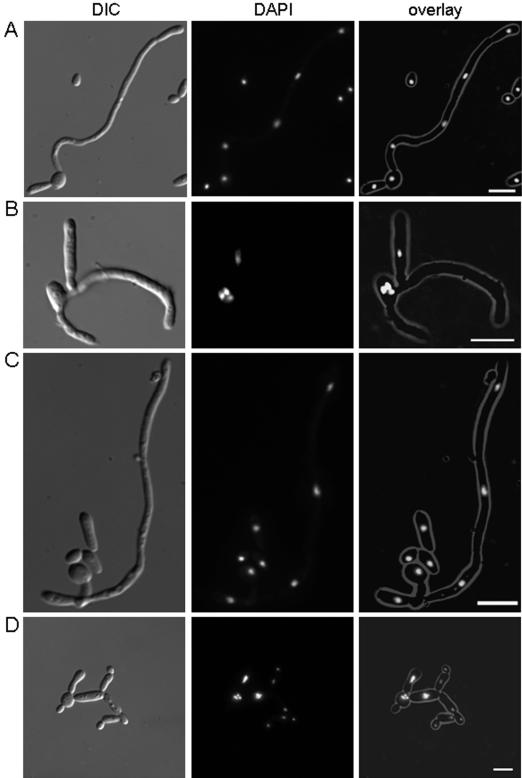
To demonstrate that the described phenotypes of the dyn1 mutant strain were due to the deletion of both alleles of DYN1, we used the heterozygous DYN1/dyn1::URA3 strain (GC1) and fused the remaining copy of DYN1 with the regulatable MAL2 promoter, again using a PCR-based approach. To determine nuclear positioning in germ tubes and hyphae of the wild type, the dyn1 mutant, and the strain expressing DYN1 from the inducible maltose promoter (CAT23), we used DAPI staining of fixed cells. Hyphal growth in the wild type resulted in nuclear migration that moved one nucleus into the hyphal tip and produced regular intervals of nuclear spacing within the hyphae. In the dyn1 mutant, few germ tubes were invaded by nuclei, so tips of germ tubes were found to be devoid of nuclei. The lack of sustained hyphal morphogenesis in the dyn1 mutant was therefore found to be accompanied by the failure of delivery of nuclei into the apical parts of germ tubes (Fig. 8). In the strain carrying the MAL2p-DYN1 construct, correct nuclear migration and positioning in germ tubes and hyphae was observed only during growth in maltose, whereas the mutant dyn1 phenotype occurred under repressible conditions during growth in glucose, indicating that Dyn1p is essential for nuclear migration and positioning in C. albicans hyphae. This also indicates that the defects in nuclear positioning and nuclear movement described in this report are, in fact, solely due to the deletion of DYN1 in C. albicans.
FIG. 8.
Analysis of nuclear positioning in wild-type and dyn1 mutant strains. Cells were incubated for 10 h under hypha-inducing conditions (CSM in the presence of 0.5 g of proline/liter). DIC images and images showing the DAPI fluorescence of stained nuclei were merged into an overlay. Characteristic nuclear positioning in the wild type (A) shows elongated hyphae with regular nuclear spacing, septal intervals, and one nucleus in the apical compartment, whereas inducing conditions resulted in germ tube formation in the dyn1 mutant (B) in which nuclei were either trapped in the germ cell or did not migrate into the hyphal tip. (C) Hyphal induction using C. albicans strain CAT23 (MAL2p-DYN1:HIS1/dyn1::URA3) in maltose-containing medium shows wild-type-like nuclear positioning, whereas hyphal induction on CSM containing glucose (D) reveals the dyn1 mutant phenotype.
DISCUSSION
Cytoplasmic dynein is a minus-end-directed motor protein that is involved in transport processes of various organelles. In fungi, dynein is required for nuclear migration and nuclear positioning (30). Functional analyses of dynein have been performed with a variety of unicellular and filamentous fungal organisms, including S. cerevisiae, U. maydis, A. nidulans, N. crassa, and A. gossypii (2, 19, 23, 29). In the dimorphic plant pathogen U. maydis, the dynein heavy chain is encoded by two genes, dyn1 and dyn2 (23). However, in U. maydis nuclear migration differs from that in ascomycetous yeasts. During yeast-like growth, premitotic movement of the nucleus into the bud occurs, followed by mitosis and migration of one nucleus back into the mother cell. Upon loss of dynein function this nuclear migration is eliminated, resulting in mitotic division in the mother cell. This defect, in contrast to similar defects in S. cerevisiae and as we describe here for C. albicans, is lethal in U. maydis (23). C. albicans is one of the most important human fungal pathogens, and its ability to change from yeast to hyphal stages has been shown to contribute significantly to the virulence of this organism (15). We were, therefore, interested in elucidating the role of C. albicans dynein in nuclear migration and correct positioning as well as its relation to polarized hyphal growth. An influence on morphogenesis was revealed for the dynein/dynactin complex in N. crassa. Neurospora ropy mutants show distorted hyphal morphologies resulting in spiralized hyphal growth. This indicated a role of dynein in the positioning of the Spitzenkörper, which acts as a vesicle supply center that directs the trajectory of hyphal growth (21).
Functional analysis of CaDYN1: morphological defects.
Rapid and efficient strain constructions were achieved by PCR-based gene targeting using 100 bp of target homology regions. C. albicans dyn1 mutant strains showed prolonged cell cycle times and a more elongated yeast cell shape than those of the wild type. The latter phenotype was accompanied by the generation of cell aggregates in the dyn1 strain that did not occur in the wild type; this activity was monitored by in vivo time-lapse microscopy. Under hypha-inducing conditions, vigorous filamentation occurred in the wild type, resulting in the formation of branched mycelia. C. albicans dyn1 strains failed to support hyphal growth both on solid and in liquid media. Germ tube formation was initiated in the dynein mutant with wild-type frequencies, but hyphal growth after application of inducing extracellular stimuli came to a halt and swollen cells were generated that failed to produce elongated hyphal filaments. The germ tubes appeared to be in the hyphal state, because septa were frequently placed within the germ tubes of the dyn1 mutant. Severe growth defects were also observed in dynein mutants of A. nidulans and A. gossypii. These were based either on a lack of nuclear migration into the hyphal tip in A. nidulans or on the trapping of nuclei in apical parts of hyphae in A. gossypii (2, 29).
Nuclear migration in the C. albicans wild type.
We used GFP-tagged histone H4 and α-tubulin to monitor nuclear migration in C. albicans. The Hhf1p-GFP label proved to be strong enough for in vivo time-lapse microscopy, whereas Tub1p-GFP was too weak and did not sustain cell viability after prolonged UV exposure to capture fluorescent images. Our Hhf1p-GFP in vivo time-lapse recordings were efficiently run over several hours, enabling us to follow several cell cycles. Compared to the nuclear migration phases that were discerned in S. cerevisiae, in C. albicans several key differences were evident: (i) nuclear movement to the bud site, as shown for S. cerevisiae (10, 32), was not that pronounced in C. albicans; (ii) oscillatory movements of elongated spindles upon insertion into the bud neck could not be observed in C. albicans; (iii) as a consequence of the low frequency of nuclear migration to the bud neck, we observed spindle elongation in a manner not coordinated with the mother bud axis, which then required realignment of elongated spindles with the mother-bud axis in the C. albicans wild type. This realignment of the spindle ensured that mitoses resulted in correct nuclear distribution between mother and daughter cells, as there were very few instances in the wild type in which mitosis was completed in a mother cell. Such a realignment of elongated spindles is a very infrequent event in S. cerevisiae (10). In baker's yeast, the nucleus is moved close to the bud neck early in G1/S phase via a Bim1p- and Kar9p-dependent search-and-capture mechanism that already ensures correct positioning of the spindle pole body with respect to the mother-daughter axis (11, 12, 17, 22, 30). Within the Candida genome sequence, open reading frames that encode potential homologs of S. cerevisiae Bim1p (orf19.00676.prot) and Kar9p (YPL269w) (orf19.05011.prot) proteins were identified which allow further analysis of a similar mechanism in C. albicans.
Nuclear migration in the C. albicans dyn1 mutant.
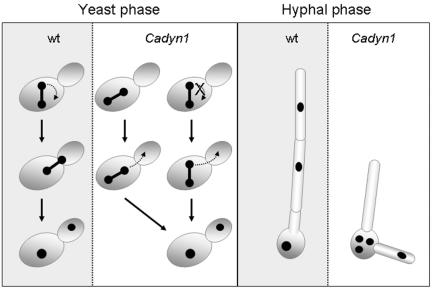
The dyn1 mutant exhibited a failure to move the nucleus to the bud site. On the other hand, only those nuclei that obtained a position close to the bud neck exhibited spindle elongation in the mother-bud axis, which then also resulted in correct nuclear distribution between mother and daughter cells during mitosis. Two striking defects of dyn1 mutant yeast cells were the lack of any spindle realignment and the subsequent completion of mitosis in the mother cell in those cases were the spindle was not realigned (Fig. 9). As in S. cerevisiae, those C. albicans cells in which mitosis occurred in the mother cell still managed to distribute their nuclei evenly between mother and daughter cells. This is contrasted by U. maydis mutants deficient in dynein, in which mitosis aberrantly occurs in the mother cell but postmitotic nuclear migration is missing, resulting in lethality of the cells (23). Postmitotic nuclear migration in Cadyn1 yeast cells largely prevents the generation of binucleate cells. As in S. cerevisiae, this nuclear migration may be triggered by a Kip3p homolog in C. albicans (see below). Under hypha-inducing conditions, however, nuclear migration defects became more dramatic in C. albicans dyn1 cells. Mitotic divisions frequently took place in the dyn1 mother cells even though germ tubes were formed. Therefore, the C. albicans dyn1 defect resembles the nudA1 phenotype in A. nidulans, which refers to clusters of nuclei in the germ cell (29). In C. albicans as in A. nidulans, these clusters are located in the germ cells, whereas in A. gossypii dynein mutant nuclei accumulate at the hyphal tips (2). Mitosis that took place in dyn1 germ cells is in contrast to that of the C. albicans wild type, in which serum-induced cells predominantly undergo their first mitotic divisions in the germ tubes (24). Lack of long-range nuclear migration in dyn1 hyphae finally results in the breakdown of polarized hyphal growth in dyn1 cells (Fig. 9). Other microtubule-based motor proteins, for example, Kip2p and Kip3p, for which homologs exist in Candida, may be responsible for postmitotic nuclear migration in the dyn1 mutant during yeast-like growth but fail to achieve a nuclear distribution that can support hyphal development in C. albicans. This indicates a dependency of polarized hyphal growth on the faithful delivery of a nucleus in the case of C. albicans (or nuclei in the case of true filamentous ascomycetes, such as A. nidulans) to the tip compartment, even if establishment of polarized hyphal growth in ascomycetes may be solely a function carried out by the actin cytoskeleton (9). Efficient nuclear migration in S. cerevisiae is carried out by two partially redundant pathways centering on dynein and Kar9p (16). This is in line with the observation that deletion of KAR9 in S. cerevisiae is synthetically lethal with deletions in DYN1. Analyses of the C. albicans set of motor proteins with respect to their contribution to nuclear migration, particularly during the hyphal growth stage, need to be performed in the future.
FIG. 9.
Summary of nuclear migration defects in dyn1 compared to migration of the wild type (wt). During the yeast phase, spindle realignment with the mother-bud neck occurs frequently in the wild type but is absent in dyn1. In the dyn1 mutant, spindle elongation may occur in the mother-bud axis, resulting in correct nuclear migration, or else leads to mitosis in the mother cell. The generation of binucleate cells in dyn1 is prevented (with high frequency) by postmitotic nuclear migration delivering one nucleus into the daughter cell (see also Table 4). Hyphal growth in the wild type results in nuclear migration and in evenly distributed nuclei along the hyphal segments. In dyn1 cells induced for hyphal formation failure in nuclear migration leads to anucleate hyphal tips that cease growth and thus establish a filamentation defect.
Acknowledgments
We thank Judith Berman for discussions of the manuscript and for communicating results prior to publication. Sequence data for C. albicans were obtained from the Stanford Genome Technology Center website at http://www-sequence.stanford.edu/group/candida.
This research was supported by the Deutsche Forschungsgemeinschaft, the Friedrich-Schiller University, and the Hans-Knöll Institute. Sequencing of Candida albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund.
REFERENCES
- 1.Adames, N. R., and J. A. Cooper. 2000. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149:863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberti-Segui, C., F. Dietrich, R. Altmann-Johl, D. Hoepfner, and P. Philippsen. 2001. Cytoplasmic dynein is required to oppose the force that moves nuclei towards the hyphal tip in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 114:975-986. [DOI] [PubMed] [Google Scholar]
- 3.Bloom, K. 2001. Nuclear migration: cortical anchors for cytoplasmic dynein. Curr. Biol. 11:R326-R329. [DOI] [PubMed] [Google Scholar]
- 4.Brachat, A., J. V. Kilmartin, A. Wach, and P. Philippsen. 1998. Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol. Biol. Cell. 9:977-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai, A., and T. J. Mitchison. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83-117. [DOI] [PubMed] [Google Scholar]
- 5a.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garnjobst, L., and E. L. Tatum. 1967. A survey of new morphological mutants in Neurospora crassa. Genetics 57:579-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gola, S., R. Martin, A. Walther, A. Dünkler, and J. Wendland. 2003. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20:1339-1347. [DOI] [PubMed] [Google Scholar]
- 8.Gundersen, G. G., and A. Bretscher. 2003. Cell biology. Microtubule asymmetry. Science 300:2040-2041. [DOI] [PubMed] [Google Scholar]
- 9.Heath, I. B., G. Gupta, and S. Bai. 2000. Plasma membrane-adjacent actin filaments, but not microtubules, are essential for both polarization and hyphal tip morphogenesis in Saprolegnia ferax and Neurospora crassa. Fungal Genet. Biol. 30:45-62. [DOI] [PubMed] [Google Scholar]
- 10.Hoepfner, D., A. Brachat, and P. Philippsen. 2000. Time-lapse video microscopy analysis reveals astral microtubule detachment in the yeast spindle pole mutant cnm67. Mol. Biol. Cell. 11:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korinek, W. S., M. J. Copeland, A. Chaudhuri, and J. Chant. 2000. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science 287:2257-2259. [DOI] [PubMed] [Google Scholar]
- 12.Lee, L., J. S. Tirnauer, J. Li, S. C. Schuyler, J. Y. Liu, and D. Pellman. 2000. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science 287:2260-2262. [DOI] [PubMed] [Google Scholar]
- 13.Li, Y. Y., E. Yeh, T. Hays, and K. Bloom. 1993. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. USA 90:10096-10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 15.Lo, H. J., J. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 16.Miller, R. K., K. K. Heller, L. Frisen, D. L. Wallack, D. Loayza, A. E. Gammie, and M. D. Rose. 1998. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell 9:2051-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, R. K., D. Matheos, and M. D. Rose. 1999. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J. Cell Biol. 144:963-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris, N. R. 2003. Nuclear positioning: the means is at the ends. Curr. Opin. Cell Biol. 15:54-59. [DOI] [PubMed] [Google Scholar]
- 19.Plamann, M., P. F. Minke, J. H. Tinsley, and K. S. Bruno. 1994. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J. Cell Biol. 127:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reck-Peterson, S. L., and R. D. Vale. 2004. Molecular dissection of the roles of nucleotide binding and hydrolysis in dynein's AAA domains in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:1491-1495. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Riquelme, M., G. Gierz, and S. Bartnicki-Garcia. 2000. Dynein and dynactin deficiencies affect the formation and function of the Spitzenkorper and distort hyphal morphogenesis of Neurospora crassa. Microbiology 146:1743-1752. [DOI] [PubMed] [Google Scholar]
- 22.Snyder, M., S. Gehrung, and B. D. Page. 1991. Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. J. Cell Biol. 114:515-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Straube, A., W. Enard, A. Berner, R. Wedlich-Soldner, R. Kahmann, and G. Steinberg. 2001. A split motor domain in a cytoplasmic dynein. EMBO J. 20:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sudbery, P. E. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41:19-31. [DOI] [PubMed] [Google Scholar]
- 25.Tinsley, J. H., P. F. Minke, K. S. Bruno, and M. Plamann. 1996. p150Glued, the largest subunit of the dynactin complex, is nonessential in Neurospora but required for nuclear distribution. Mol. Biol. Cell 7:731-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vierula, P. J., and J. M. Mais. 1997. A gene required for nuclear migration in Neurospora crassa codes for a protein with cysteine-rich, LIM/RING-like domains. Mol. Microbiol. 24:331-340. [DOI] [PubMed] [Google Scholar]
- 27.Walther, A., and J. Wendland. 2003a. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr. Genet. 42:339-343. [DOI] [PubMed] [Google Scholar]
- 28.Walther, A., and J. Wendland. 2004. Polarized hyphal growth in Candida albicans requires the WASP homolog Wal1p. Eukaryot. Cell. 3:471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiang, X., S. M. Beckwith, and N. R. Morris. 1994. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 91:2100-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang, X., and R. Fischer. 2004. Nuclear migration and positioning in filamentous fungi. Fungal Genet. Biol. 41:411-419. [DOI] [PubMed] [Google Scholar]
- 31.Xiang, X., and N. R. Morris. 1999. Hyphal tip growth and nuclear migration. Curr. Opin. Microbiol. 2:636-640. [DOI] [PubMed] [Google Scholar]
- 32.Xiang, X., and M. Plamann. 2003. Cytoskeleton and motor proteins in filamentous fungi. Curr. Opin. Microbiol. 6:628-633. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto, A., and Y. Hiraoka. 2003. Cytoplasmic dynein in fungi: insights from nuclear migration. J. Cell Sci. 116:4501-4512. [DOI] [PubMed] [Google Scholar]
- 34.Yeh, E., R. V. Skibbens, J. W. Cheng, E. D. Salmon, and K. Bloom. 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130:687-700. [DOI] [PMC free article] [PubMed] [Google Scholar]