Summary
Age-related skeletal degeneration in patients with osteoporosis is characterized by decreased bone mass and occurs concomitant with an increase in bone marrow adipocytes. Using microarray expression profiling with high temporal resolution, we identified gene regulatory events in early stages of osteogenic and adipogenic lineage commitment of human mesenchymal stromal cells (hMSCs). Data analysis revealed three distinct phases when cells adopt a committed expression phenotype: initiation of differentiation (0–3 hr, phase I), lineage acquisition (6–24 hr, phase II), and early lineage progression (48–96 hr, phase III). Upstream regulator analysis identified 34 transcription factors (TFs) in phase I with a role in hMSC differentiation. Interestingly, expression levels of identified TFs did not always change and indicate additional post-transcriptional regulatory mechanisms. Functional analysis revealed that forced expression of IRF2 enhances osteogenic differentiation. Thus, IRF2 and other early-responder TFs may control osteogenic cell fate of MSCs and should be considered in mechanistic models that clarify bone-anabolic changes during clinical progression of osteoporosis.
Keywords: human MSC differentiation, gene expression profiling, osteoblast, adipocyte, regenerative medicine, transcription factor, interferon, bone, bioinformatics
Graphical Abstract
Highlights
-
•
Determination of high temporal gene expression changes in differentiating hMSCs
-
•
Three transcriptional phases during early osteoblast and adipocyte differentiation
-
•
Transcription factors that change activity during early hMSC differentiation
-
•
IRF2 enhances osteogenic differentiation of hMSCs
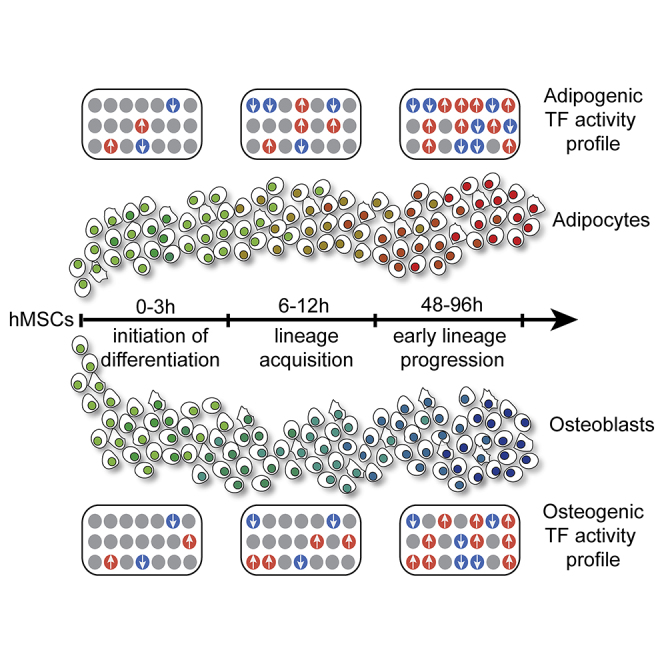
In this article, van Leeuwen and colleagues determine high temporal gene expression changes in human mesenchymal stromal cells upon induction of adipocyte and osteoblast differentiation. They identify three early differentiation phases within the first 4 days. Furthermore, novel transcription factors were identified that may control cell fate of hMSCs and should be considered in mechanistic models that clarify bone-anabolic changes.
Introduction
Mesenchymal stem/stromal cells (MSCs) are an excellent biological source for bone regenerative therapies, tissue engineering, and treatment of post-menopausal osteoporosis (Murphy et al., 2013, Steinert et al., 2012). Ex vivo expansion of autologous bone marrow stromal cells and systematic administration of hMSCs was proposed 20 years ago to treat patients with osteoporosis (Bruder et al., 1997). To date, treatment of osteoporosis using bone marrow-derived MSCs is not yet standard clinical practice. Donor variation among patients and unpredictable capacity for differentiation are among the current shortcomings of hMSCs for their application in regenerative cell therapies (Murphy et al., 2013).
In MSCs, transcription factors (TFs) such as RUNX2, SP7/Osterix, and SOX9 have been shown to play critical roles in differentiation of MSCs into osteoblasts or chondrocytes. Overexpression of RUNX2 in non-osteoblastic cells or in adipose-tissue-derived MSCs increases expression of osteoblastic markers and enhances osteoblast differentiation and mineralization (Ducy et al., 1997, Otto et al., 1997, Zhang et al., 2006). Moreover, homozygous RUNX2 mutant mice lack mature osteoblasts and mineralized bone, indicating that a single TF is important for bone development and osteoblast differentiation.
Besides differentiation into osteoblasts, MSCs can differentiate into other cell lineages such as adipocytes and chondrocytes (Pittenger et al., 1999). The identification of regulators of lineage commitment is therefore an essential step toward our understanding and control of human MSC differentiation into osteoblasts. The balance between osteoblast and adipocyte differentiation is of specific interest. Increased adipose tissue volume is observed in the bone marrow cavity in osteoporotic people where increased bone resorption is not sufficiently compensated by an increase in bone formation by osteoblasts (Justesen et al., 2001, Yeung et al., 2005). The hypothesis underlying the current study is that detailed analysis of gene expression changes upon induction of osteogenic and adipogenic differentiation of MSCs enables identification of TFs that change activity during early differentiation in both lineages. While previous gene expression studies identified important regulatory pathways and processes involved in MSC differentiation, the results are limited by differentiation into a single mesenchymal lineage, a later stage of differentiation, and/or a low temporal density at early time points (Hung et al., 2004, Kulterer et al., 2007, Ng et al., 2008, Piek et al., 2010). Because key lineage decisions are made during the early stages of mesenchymal differentiation, a high density of early time points of differentiation is critical for the identification of important regulators within the initial differentiation phases.
Here, we systematically investigated gene expression changes upon differentiation of human MSCs into adipocytes and osteoblasts with high temporal resolution. One of our key findings was the identification of three distinct sequential phases of differentiation in both lineages. Furthermore, we characterized genes and regulatory programs controlling the early stages of mesenchymal lineage commitment. These findings provide opportunities for designed engineering of hMSCs for applications in both personalized and regenerative medicine.
Results
Differentiation of hMSCs into Osteoblasts and Adipocytes
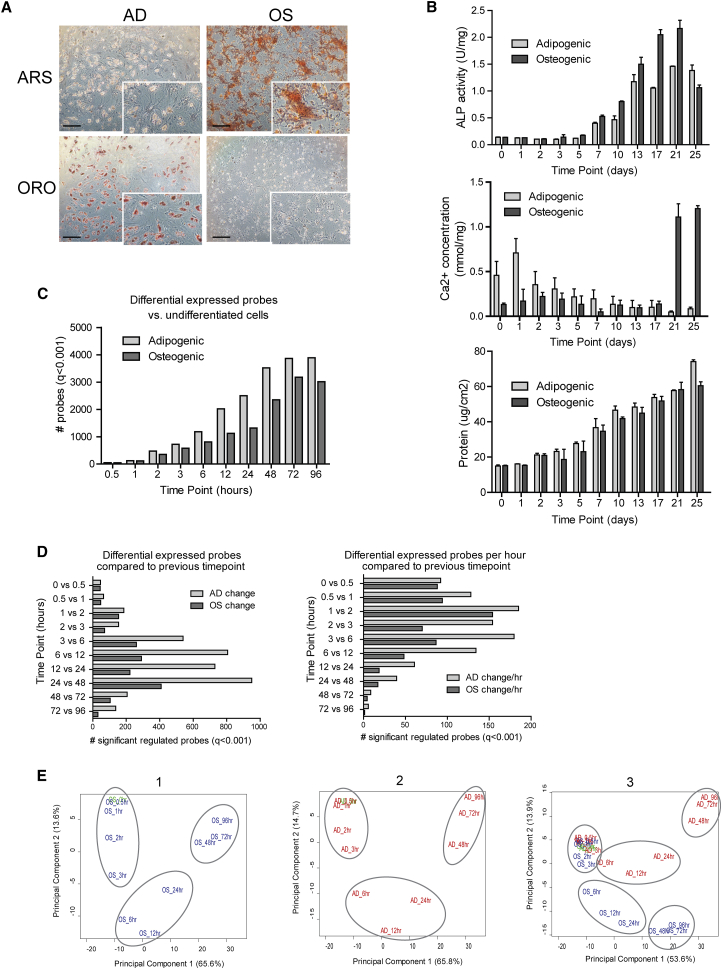
To analyze dynamic transcriptional networks in early differentiating human MSCs (hMSCs), we generated gene expression profiles with a high temporal density during the first 4 days of osteogenic and adipogenic differentiation. Histological staining for calcium and lipids in osteogenic and adipogenic differentiation cultures show that, respectively, the extracellular matrix (ECM) is mineralized and cells accumulate intracellular lipid vesicles as shown at day 25 (Figure 1A). Biochemical analyses of samples during differentiation show that total protein and alkaline phosphatase (ALP) activity transiently increases during osteogenic differentiation prior to a decrease of ALP upon mineralization (Figure 1B). ALP activity is similarly increased during adipogenic differentiation (Figure 1B). ECM mineralization is observed after 21 days of osteogenic differentiation as demonstrated by increased calcium deposition in the matrix (Figures 1A and 1B). Together, our observations establish that hMSCs differentiate into both lineages, consistent with their expected multi-lineage potential.
Figure 1.
Dynamic Transcriptional Changes upon Adipogenic and Osteogenic Differentiation of hMSCs
(A) Histochemical staining of calcium with Alizarin red (ARS) or adipocyte with oil red O (ORO) after 25 days of osteogenic (OS) or adipogenic (AD) differentiation. Scale bars, 200 μm.
(B) Biochemical analyses of osteogenic differentiating human mesenchymal stromal cells that were used to generated the gene expression profiles. Error bars indicate SD. Mean and SD of two independent experiments with two technical replicates.
(C) Number of significant differential probes (q < 0.001) relative to undifferentiated cells (t = 0) (based on three independent experiment with 10–20 technical replicated measurements in a single donor).
(D) Number of significant differential probes (q < 0.001) relative to the previous time point. The number of differential expressed probes was divided by the time differences between two time points (based on the same replicated samples as in C).
(E) Principle component analyses of the osteogenic (1), adipogenic (2), and all (3) gene expression profiling experiments using the 15,795 probes that were detected as expressed. Replicated (based on the same replicated samples as in C) probe intensities were averaged for each time point (blue, osteogenic; red, adipogenic). Gray circles indicate differentiation phases as described in the text.
Analysis of gene expression dynamics during hMSC differentiation reveals that transcript levels immediately change upon induction of differentiation (Figure 1C). To gain insight into the robustness of the gene expression changes, we selected genes that were specifically upregulated during osteogenic or adipogenic differentiation. We validated their change in expression in MSCs from an additional donor as well as during osteogenic differentiation of a committed human osteoblast cell line (NHOst). These analyses supported the observations obtained by the microarray gene expression analysis (Figure S1A). During induction of osteogenic differentiation, at least 44 gene probes are significantly different at 30 min and this increases further to 351 after 2 hr. The number of significantly modulated probes gradually increases and begins to level off after 2 days with a maximum number of 3,178 probes after 3 days of osteogenic differentiation. Comparable results were obtained during adipogenic differentiation where the number of differentially expressed probes was 46 after 30 min, 470 after 2 hr, and 3,863 by day 3. Remarkably, within the first 2 hr of differentiation, most of the genes that are modulated are upregulated (276 of 351 probes for osteogenic and 301 of 470 probes for adipogenic induction) (Figures S1B and S1C). Yet, the number of up- and downregulated genes is about the same in the two subsequent phases. These results suggest that transcription exceeds median mRNA degradation during the initial induction of differentiation. Gene expression at later stages of differentiation may be controlled by transcriptional repression combined with non-specific mRNA decay and/or constitutive transcription with enhanced mRNA destabilization.
Next, we calculated the number of significant differentially expressed probes at each time point compared with the preceding time point and divided the differences by the elapsed time (Figure 1D, right panel). Adipogenic differentiating cells showed a similar number of differentially expressed probes during the first 2 hr, but thereafter the number of gene expression changes per hour was almost two times higher than in the osteogenic differentiation cells and is in agreement with recent studies suggesting the default preference of bone marrow-derived MSCs for osteogenic differentiation (Meyer et al., 2016). In both conditions, the number of probes that changed per hour decreased drastically after 48 hr of differentiation, and only minor gene expression changes were evident between days 2 and 4 (Figure 1D).
More than 20% of genes change expression in the early phase (first 3 hr) of osteogenic differentiation (Figure 1C). We assessed whether this dramatic change is mostly due to uncoordinated gene activation and repression or proceeds in a more organized and sequential manner that reflects a well-defined single differentiation program in which the number of modulated mRNAs per time period increases during early lineage commitment and progression. Principal component analysis (PCA) was applied to examine whether expression produces a single dominant principal component (PC), and whether there is co-linearity between time and progression along that component. The PCA shows indeed a rather dominant first principal PC that encompasses two-thirds of the variation in expression (Figure 1E, 1 and 2). In this dimension, both lineages differentiate away from the undifferentiated MSCs. Gene expression (and its inherent variation) at a given biological time point generates time stamps. Since these time stamps occur in a precise order within the diagram, it appears that the process of differentiation of both lineages resembles an ordered program rather than an uncoordinated set of events.
By performing a single PCA on the data of both lineages, we addressed similarities and differences in the differentiation programs of the two lineages. We found that both lineages display a unique series of time stamps that occur in sequence on a single line (Figures 1E, 3, and S1D). The time stamp lines for the two lineages move in the direction of the main PC and diverge steadily over time, consistent with the early adoption of two distinct mesenchymal phenotypes. The osteogenic and adipogenic lineages already diverge within 2–3 hr upon induction of differentiation, and three phases can be discerned in each lineage (Figure 1E, 3).
Global gene expression analyses at high temporal resolution and unsupervised clustering of the microarray data defines three distinct sub-stages (phase 1, 0–3 hr; phase II, 6–24 hr; phase III, 48–96 hr) and dynamic transcriptional responses during differentiation of hMSCs into either osteogenic or adipogenic lineages (Figure 1E). Hence, mesenchymal differentiation into the cellular lineages that produce mature bone or fat tissue is a multi-stage process. Furthermore, phenotype commitment is initiated quite rapidly following induction.
Gene Ontology Analysis Reveals General and Lineage-Specific Functional Processes during Differentiation
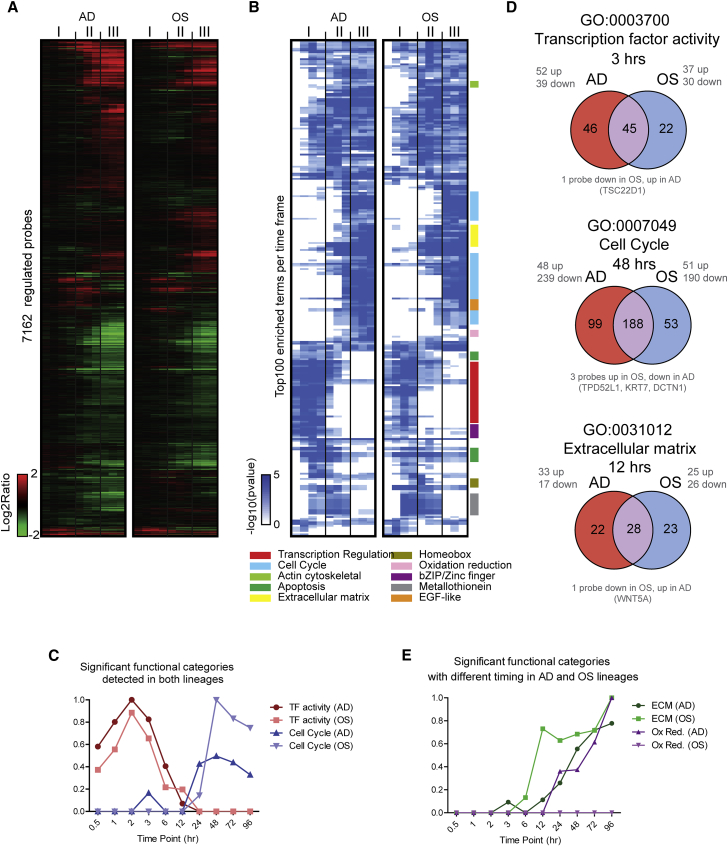
To understand mechanistic cellular changes upon induction of differentiation, we used the gene expression profiles to assess the functional categories that were significantly enriched at the different time points after induction of differentiation into both lineages. Differentially expressed probes at each individual time point (Figure 2A) were subjected to a gene ontology analyses, and the most significantly enriched functional categories were visualized in a hierarchical clustering diagram (Figure 2B and Table S1).
Figure 2.
Functional Annotation of Differential Expressed Genes
(A) Heatmap of differentially regulated probes compared with undifferentiated cells (t = 0 hr) Replicated (based on the same replicated samples as in Figure 1C) probe intensities were averaged for each time point.
(B) Analyses of enriched functional categories among the probes that were differentially expressed during adipogenic and osteogenic differentiating hMSCs. The cluster diagram depicts the −log10(p value) of the enrichment. Only significant (p < 0.05) instances are shown. Colors on the side of the cluster diagram depict the similarly associated functional categories.
(C) Significance of enrichment of the functional categories, TF activity and cell cycle, during differentiation.
(D) Venn diagram of the functional categories that are similarly enriched, TF activity and cell cycle, and extracellular matrix proteins, a functional category that is enriched earlier in osteoblasts. The numbers inside the Venn diagram are the number of significantly expressed probes at the time point indicated. The numbers of up- or downregulated probes are shown in gray type. The number of probes that are oppositely regulated in the two conditions are shown below the Venn diagrams.
(E) Significance of enrichment of functional categories, extracellular matrix proteins and oxidoreductase activity, that were enriched during osteoblast or adipocyte differentiating hMSCs, respectively.
The clustering of enriched functional categories illustrates that modulated genes in the first phase are highly different from those in the second and third phases. Importantly, the gene expression program of both lineages are enriched in similar gene ontology terms, consistent with classical models for cellular differentiation in which changes in cell proliferation accompany the acquisition of lineage-committed cellular phenotypes. These functional categories include transcriptional regulation (enriched in phase I), apoptosis (enriched in phases I and II), as well as cell-cycle/DNA replication and mitosis (enriched in phases II and III) (Figures 2B and 2C).
Because similar functional categories are enriched in both lineages, for each lineage we investigated which genes are differentially expressed. Expression of 45 probes linked to transcriptional control is modulated in both lineages within 3 hr of differentiation (phase I), and these genes may represent a class of common early-responder TFs in phase I (Figure 2D). We also observed 22 and 46 TF activity-linked probes that are specifically regulated within the initiation phase of osteogenic and adipogenic differentiation (phase I), respectively. Furthermore, 53 and 99 probes linked to cell-cycle mechanisms are differentially expressed in the osteogenic and adipogenic lineages at 48 hr, respectively, when cells progress from a lineage-acquisition phase (phase II) into the lineage-progression phase (phase III) (Figure 2D). The situation is clear but paradoxical: the two lineages are largely similar in terms of functional categories, yet distinctly; the differences between the two lineages set in early (second PC in Figure 1E and detailed differences of the differential expressed TFs in Figure 2D, upper panel).
We note that many probes (n = 188) associated with the cell cycle are differentially expressed in both lineages after 48 hr, and about 85% of these probes are downregulated (Figure 2D). This result indicates that induction of differentiation in both lineages coincides with downregulation of the expression program that mediates orderly progression through the mitotic cell cycle. More importantly, the modulated expression of 185 common probes may define a shared mesenchymal cell-cycle program of downregulated cell-cycle stimulatory factors (CCNA2, CCNB1, CCNB2, CCND1, CCNE1, CCNE2, and CCNF; and E2Fs: E2F2 and E2F7) and upregulated cell-cycle inhibitors (CDKN1C and CDKN2C) that is coordinately controlled in each lineage.
Besides functional categories that were enriched in both lineages (Figure S2A), we identified a number of gene ontology terms that are only enriched during adipogenic differentiation in phase III (Figures 2B and 2E) and not during osteoblast differentiation. The unique regulation of these gene categories (e.g., monocarboxylic acid binding [GO: 0033293] and oxidation reduction [GO: 0055114]) suggest changes in metabolic activity in the adipogenic lineage that do not occur during osteoblast differentiation.
Osteoblast differentiated MSCs are defined in part by the production of ECM proteins. Indeed, the number of genes enriched in phase II of osteogenic differentiation is related to cell adhesion (GO: 0007155), the ECM (GO: 0031012), and proteinaceous ECM (GO:0005578) (Figures 2E and S2A). Interestingly, genes with the same GO terms are also significantly enriched during adipogenic differentiation but at a later stage (phase III). Within 12 hr of differentiation, the expression data show that 23 of 51 and 22 of 50 modulated ECM genes are either osteoblast or adipocyte related, respectively (Figures 2D and 2E). Taken together, gene ontology analysis establishes that phenotype acquisition is initiated in phase I and continues into phases II and III.
Differential Expression of Transcription Regulators within 3 hr after Induction of Differentiation in Phase I
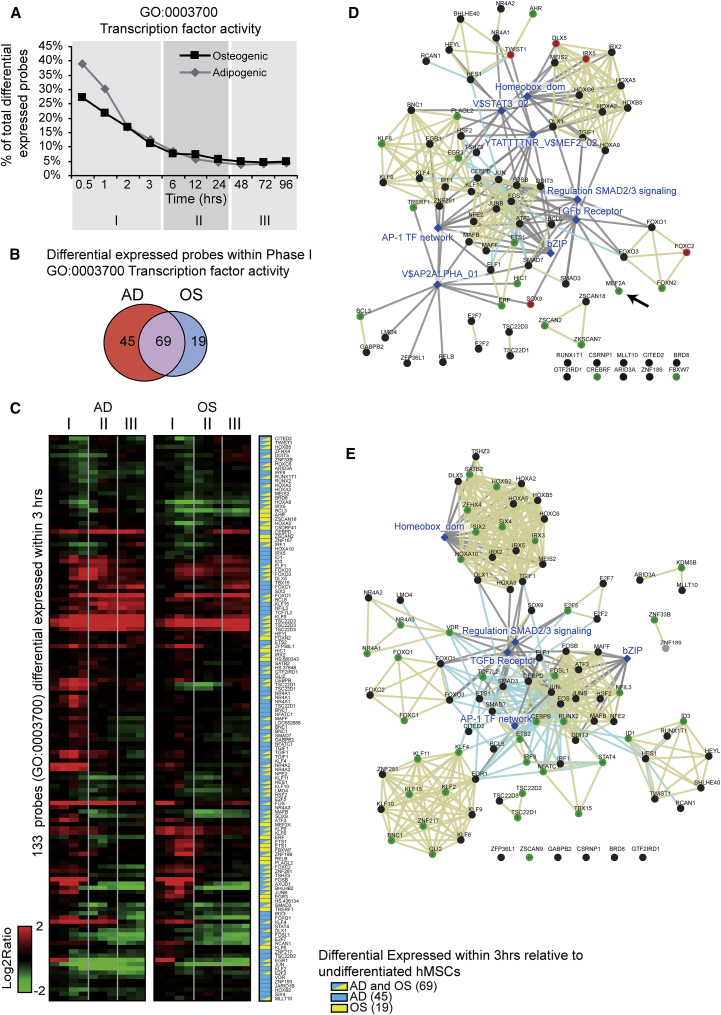
The main class of genes that is activated during phase I in both osteogenic and adipogenic differentiation is associated with regulation of transcription (Figure 3A). This interpretation is based on significant enrichment of functional categories such as homeobox TFs (IPR: 001356), basic-leucine zipper TFs (IPR: 004827), TF activity (GO: 0003700), DNA binding (GO: 0003677), as well as positive and negative regulation of transcription (GO: 0045941 and GO: 0016481) (Figure 2B and Table S1). Within 1 hr, more than 20% (osteoblast) and 30% (adipocyte) of the regulated genes are related to TF activity (GO: 0003700) and decrease below 10% in both lineages after 6 hr (Figure 3A). Hence, changes in mesenchymal phenotypes upon induction of differentiation appear to be mediated by rapid changes in the expression of TFs, which can function as inducers of secondary responses to sustain either osteogenic or adipogenic phenotype commitment.
Figure 3.
Genes Annotated as Transcription Factor Activity Are Immediately Enriched Upon Differentiation
(A) Percentage of probes annotated as TF activity during differentiation into osteoblasts and adipocytes of the total differential expressed probes.
(B) Venn diagram of all probes annotated as TF activity (GO: 0003700) that are differentially expressed within the first 3 hr during adipocyte and osteoblast differentiation.
(C) Hierarchical clustering of 133 probes of the gene expression study and annotated as TF activity (depicted in B). Red, upregulated; green, downregulated relative to undifferentiated cells; yellow, significant differentially expressed within 3 hr of osteoblast differentiation; blue, significant differentially expressed within 3 hr of adipocytes differentiation.
(D) GeneMania network analyses of the 77 (88 probes) TFs that were up- or downregulated during osteoblast differentiation. Gray and yellow edges illustrate consolidated pathways and protein domains with edges shared, respectively. Blue nodes are pathways; known osteoblast TFs are marked in red; and green nodes depict the TFs that are specifically regulated during osteogenesis. Arrow see text.
(E) GeneMania network analyses of the 95 (104 probes) TFs that were up- or downregulated during adipocyte differentiation. Edges are colored as in (D). Green nodes are TFs that are specifically regulated during adipogenesis.
Within the first phase of osteogenic and adipogenic differentiation, there are in total 133 probes (corresponding to 114 genes) related to transcriptional activity that are acutely regulated (Figures 3B and 3C). Most of the probe sets differ in expression in both lineages (i.e., 78%, 69 of 88 upon osteogenic induction; 61%, 69 of 114 upon adipogenic induction) (Figures 3B and 3C). A small number of the regulated transcription-related probes were specifically regulated in one of the two lineages and may represent lineage-specific regulators (i.e., 22%, 19 of 88 in osteogenic medium; 39%, 45 of 114 in adipogenic medium). Transient upregulation of early-responder TFs in phase I is occasionally followed by a return to basal levels of expression in phases II and III. This biphasic modulation indicates that induction of differentiation initiates a primary transcriptional program that may serve to induce a secondary program of phenotype-specific genes.
GeneMania network analyses illustrates that the 88 transcription-related probes (80 genes) regulated upon osteogenic differentiation are linked to established signaling pathways such as transforming growth factor β (TGF-β) receptor signaling, regulation of nuclear Smad2/3 signaling, and AP1 TF signaling (Figure 3D), perhaps indicating activation of a TGF-β-Smad2/3-AP1 network. Similar signaling pathways are evident among the 114 probes (97 genes) annotated for TFs that are controlled during adipogenic induction (Figure 3E). The TFs regulated in both lineages are associated with similar pathways, yet these pathways are linked to TFs with lineage-specific changes in expression. For example, expression of MEF2A and VDR is modulated only during osteogenesis and adipogenesis, respectively, and functionally related to TGF-β receptor signaling (Figure 3D, arrow). Other TFs regulated during osteogenic differentiation of MSCs include five TFs with well-known roles in skeletal morphogenesis (DLX5, FOXC2, IRX5, SOX9, and TWIST1) (Figure 3D). Furthermore, there are many homeobox-domain-containing TFs that are prominently regulated during either osteogenic (12 TFs) or adipogenic differentiation (19 TFs) (Figures 3D and 3E). Taken together, lineage-specific transient stimulation and suppression of early-responder TFs in phase I may represent a pre-commitment stage that drives activation of the target genes to establish the specialized mesenchymal phenotypes that support bone formation or fat metabolism.
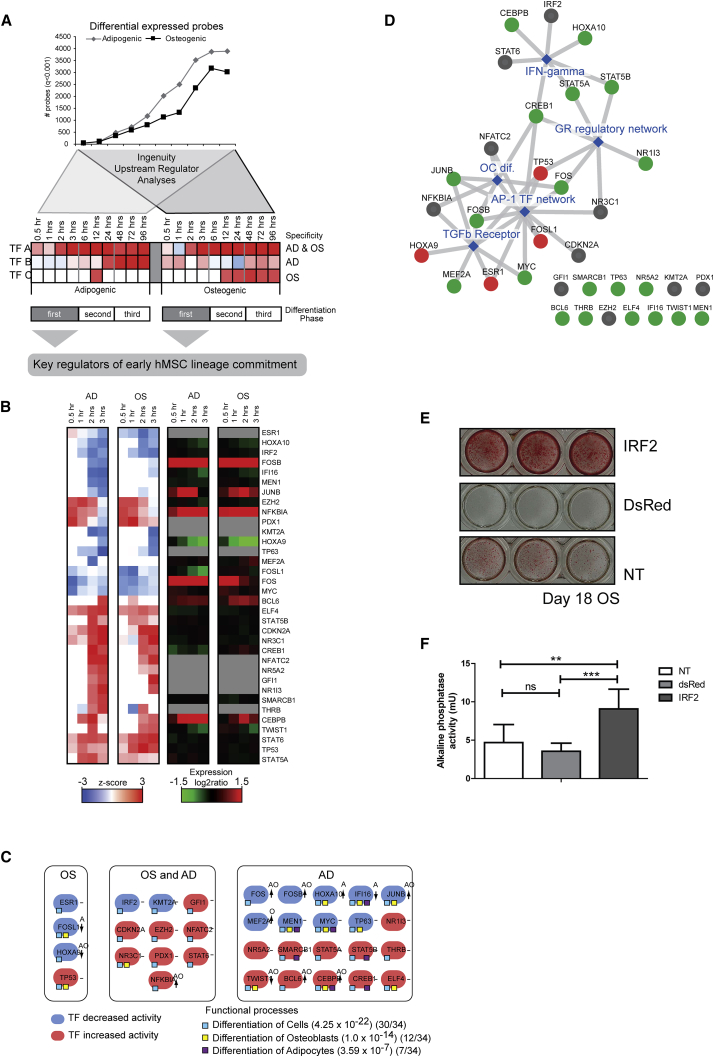
Analysis of Upstream Regulators Reveals a Coordinately Controlled TF-Gene Network
The regulated probes at each individual time point were selected and used for upstream regulator analyses (URA) in ingenuity pathway analyses (Figure 4A) to assess if the modulated genes correlate with TFs that change activity immediately upon differentiation. URA generated a pattern for 147 TFs of which the activities are modulated during osteogenic or adipogenic differentiation (Figure S3A). Two notable genes activated in both lineages are CDKNA2 and NR3C1. CDKN2A is a major regulator of cellular quiescence and senescence, and its activation blocks cell proliferation by ensuring that cell-cycle stimulating regulatory E2F factors remain sequestered by pRB/p105. NR3C1/GR encodes a key nuclear receptor that controls gene expression in response to glucocorticoids (e.g., dexamethasone), a known stimulant of both osteoblastogenesis and adipogenesis that is included as an inducer of differentiation in our cell-culture experiments. Interestingly, the change in TF activity of NR3C1 exemplifies that additional post-transcriptional regulations should occur since its expression level did not change.
Figure 4.
Ingenuity Upstream Regulator Analyses Identifies General and Lineage-Specific TF Activity Changes
(A) Procedure of the upstream regulator function in Ingenuity (www.ingenuity.com).
(B) Cluster diagram of all TFs that a Z-score of>2 or <−2 in either lineages within the first 3 hr of osteogenic and adipogenic differentiation. Next to the activity patterns is the relative expression during differentiation of the TFs. Expression patterns of genes that are gray were not expressed or not present on the array, and therefore no expression data were available.
(C) All TFs that were changing activity (blue, Z-score of<−2; or red, Z-score of>2) within the first 3 hr of differentiation of osteoblasts (OS), adipocytes (AD), or both (OS and AD).
(D) GeneMania network analyses of the 34 TFs identified. The connected edges in gray illustrate the consolidated pathways (blue); red, the osteoblast-specific TFs; green, the adipocyte-specific TFs; gray, the TFs that we identified in both lineages.
(E) Alizarin red staining of hMSC transduced with pLenti6.3-DsRED, pLenti6.3-IRF2, and non-transduced (NT) and cultured for 21 days in osteogenic differentiating conditions.
(F) ALP levels of transduced cells after 7 days osteogenic differentiation. n = 3 independent experiments with three replicates per experiment, mean ± SEM; statistical significance using a one-way ANOVA; ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant.
URA also identified two other proteins (TP53 and EZH2) that have direct gene regulatory functions specifically during hMSC differentiation. TP53 encodes the tumor suppressor protein p53 that is required for normal osteoblast differentiation, while EZH2 is a histone methyltransferase that is known to suppress CDKN2A and control mesenchymal differentiation during skeletal development (Dudakovic et al., 2015). Beyond CDKN2A and NR3C1, there are eight other gene regulators (ELF4, FOSB, MYC, NFKBIA, SMARCB1/BAF45, STAT5A, NR1I3, and THRB/NR1A2) that are identified by URA in phase I of adipogenic differentiation (Figure S3A) and have direct connections with adipogenic differentiation based on well-established pathways. Collectively, our high-density temporal analysis combined with URA validates the known osteogenic and/or growth inhibitory functions of four gene regulatory proteins (i.e., CDKNA2, NR3C1, TP53, and EZH2) during differentiation of hMSCs.
Many TFs have lineage-independent activation/inhibition patterns. For example, 71% (phase I), 60% (phase II), and 59% (phase III) of TFs modulated during osteogenesis are activated or inhibited during the same phases of adipogenesis (Figure S3B). These results show that initiation of either osteogenic or adipogenic differentiation is mediated by shared regulatory pathways, and a select number of critical cell-fate-determining proteins may coordinate activation of mesenchymal lineage-specific gene expression programs.
Interestingly, URA predicts that many TFs change their activity without a change in their expression level. During osteoblast differentiation, only 14%, 18%, and 23% of the TFs identified by URA exhibit a change in gene expression in each of the three different phases, while 30%, 29%, and 33% of these TFs change during adipocyte differentiation (Figure S3A). Thus, the mRNA-independent activation and inhibition of TFs predicted by URA suggest phase transitions during differentiation that may occur via a post-transcriptional mechanism including translation, protein modification, and/or subcellular translocation.
Analyses of Early Activated and Inhibited TFs
We subsequently focused our studies on TFs that change activity within the first phase (0–3 hr) when cells are postulated to make important lineage decisions (Figures 2 and 4B). Functional annotation illustrated that nearly all identified TFs are involved in cellular differentiation (30 of 34) (Figure 4C). More specifically, 7 and 11 of the 34 TFs have been linked to differentiation of adipocytes and osteoblasts, respectively (Figure 4C). Consolidated pathway analyses illustrated that the identified TFs are involved in well-known osteoblast and adipocytes signaling pathways such as GR, AP1, TGF-β, and interferon γ (IFN-γ) signaling (Figure 4D) (Augello and De Bari, 2010). Interestingly, the downstream analyses of four TFs that changed activity only during osteogenic differentiation (i.e., TP53, FOSl1, ESR1, and HOXA9), suggest that they may regulate 36.6% of the genes that were differential expressed within this first phase (Table 1). This number increased to 52% and 58% of the differential expressed genes in phases II and III. Importantly, this analysis demonstrates that activation or inhibition of only four TFs within the first 3 hr is linked to the regulation of more than 50% of the genes at later stages. Taken together, our analyses validate known regulators of osteoblast differentiation and identify candidates that may regulate many downstream genes involved in osteoblast differentiation.
Table 1.
Number of Genes Regulated Downstream of the Activated/Inhibited TFs from Phase I
| Number of TFs Act/Inh in Phase I | Regulated Genes in Osteogenic Phase I | Regulated Genes in Osteogenic Phase II | Regulated Genes in Osteogenic Phase III | |
|---|---|---|---|---|
| OS only | 4 | 112 | 260 | 438 |
| OS and AD | 10 | 151 | 272 | 441 |
| All OS | 14 | 192 | 408 | 693 |
| Osteoblast specific | 36.6% | 52.3% | 57.5% | |
| Number of TFs Act/Inh in Phase I | Regulated Genes in Adipogenic Phase I | Regulated Genes in Adipogenic Phase II | Regulated Genes in Adipogenic Phase III | |
| AD only | 20 | 170 | 492 | 647 |
| OS and AD | 10 | 181 | 431 | 533 |
| All AD | 30 | 235 | 706 | 964 |
| Adipocyte specific | 31.8% | 55.9% | 66.6% | |
One of the TFs we identified is interferon-regulating factor 2 (IRF2). This conclusion was based on the direction of regulation of five IRF2 target genes: IRF1, IL6, CDKN1B, SOCS1, and PTGS2. To assess the robustness of this conclusion, we studied the expression of these five target genes in MSCs obtained from four different donors. As shown in Figure S4, the change in expression was identical in all donors at 3 or 6 hr (Figure S4A and S4B) after the start of osteogenic differentiation. Forced expression of IRF2 in MSCs resulted in increased ALP activity and ECM mineralization (Figures 4E and 4F). Thus, IRF2 is functionally rate limiting for osteogenic lineage commitment and progression of hMSC differentiation and mineralization.
Discussion
The present study provides key insights into mechanistic events that direct osteoblast and adipocyte differentiation, which occur before time windows examined by previous studies (Hung et al., 2004, Kulterer et al., 2007, Ng et al., 2008, Piek et al., 2010). The high-density temporal dynamic gene expression profiles we generated permit a mechanistic description of functional processes that change during the first hours and days of osteoblast and adipocytes differentiation. Among the main findings of this study is the definition of at least three distinct early differentiation phases, each with their own mRNA dynamics, and the identification of candidates for early regulation of hMSC differentiation. We used isolated MSCs, which represent a heterogeneous cell population. However, by using dexamethasone as inducer of both osteogenic and adipogenic differentiation, we synchronized the clock of the cells (So et al., 2009), which is coupled to the cell cycle (Feillet et al., 2015), and thereby limited cell-cycle heterogeneity and explained the identification of cell-cycle-related genes.
Although we have investigated in detail the differentiation of bone marrow-derived MSCs, further in-depth studies are needed to establish whether MSCs from adipose tissue or different anatomical locations undergo a similar cascade of transcriptional events. Although adipose-derived MSCs could function as a source of hMSCs for regenerative medicine, previous studies illustrated that MSCs isolated from different adipose depots respond very differently with respect to their clonogenic potential, doubling time, and differentiation (Russo et al., 2014). Therefore, the gene signature identified here may aid in the selection of cells from adipose tissue or other anatomical sites with increased regenerative capacity for the treatment of skeletal disorders.
Gene expression analyses revealed an increased number of genes differentially regulated in osteoblasts and adipocytes. Yet, the enriched biological processes and TFs that change activity are mostly identical between the lineages within the first 4 days and therefore suggest that the timing of expression modulation and the specific types of genes within each functional category differ between the two lineages.
Based on the number of genes regulated and the unsupervised cluster analyses, we discriminated three different phases within the first 4 days of differentiation. The third and last phase (early lineage progression, 48–96 hr) is characterized by a stabilization of the number of differentially expressed genes and suggests that the differentiating cells have reached a stable phenotype. The second phase (lineage acquisition, 6–24 hr) represents a transition phase in which the two differentiating lineages begin to deviate, as reflected by many transcriptional changes that are a direct result of the changes in the first phase. Downstream analyses of the four TFs that were identified during the first phase of osteogenic differentiation showed that these are capable of regulating more than 50% of the regulated genes in the second and third phases.
Within the third phase of osteogenic and adipogenic differentiation, most of the genes associated with the cell cycle were downregulated. This illustrates the transition from proliferation to differentiation and reflects the inverse correlation between these processes as has been described in various other differentiating cells. The SWI/SNF chromatin remodeling complex is important for this regulation (Ruijtenberg and van den Heuvel, 2016). Moreover, since RUNX2-dependent skeletal gene expression requires SWI/SNF, it agrees with the sequential process from proliferation to differentiation of hMSCs (Young et al., 2005). Nevertheless, we need to investigate whether small molecules that inhibit proliferation are sufficient for the induction of differentiation of hMSCs.
In the regulatory model derived from our data, the first phase (0–3 hr) represents the initiation stage of the differentiation program of hMSCs. This phase is characterized by expression changes of many transcription-related genes that regulate lineage commitment and set the stage for further differentiation toward a stable phenotype. We identified many TFs with homeobox and bZip domains that were regulated in both lineages. Homeobox TFs have been extensively studied and are generally important for their regulatory effects during development, as well as MSC differentiation (Stains and Civitelli, 2003). Strikingly, the homeobox TF DLX5 is upregulated within the first hours of differentiation. Dlx5 is epigenetically unlocked during DMSO-induced osteogenic differentiation (Thaler et al., 2012), activates the osteoblast TF Runx2 (Lee et al., 2005), and is required for mesenchymal cell proliferation and differentiation (Samee et al., 2008). Interestingly, seven homeobox TFs (e.g., HOXA10, HOXB2, IRX3, SATB2, SIX2, SIX4, and ZFHX4) are only regulated upon adipocyte differentiation. Apart from HOXA10, these TFs have not yet been described to be involved in adipocyte differentiation and hold the potential of early regulators for lineage specificity and commitment.
Consolidated pathway analyses of the immediately early-regulated TFs identify Smad2/3-TGF-β and AP1 signaling. TGF-β and AP1 signaling are also enriched in the URA of the regulated genes in the first phase. Because most of the TFs within these pathways are regulated in both differentiating osteoblasts and adipocytes, we hypothesize that the initiation of hMSC differentiation is similarly activated in both lineages and that changes in the combination of these signaling pathways are necessary to exit the immature multi-potent cell stage (loss of stemness) to allow acquisition of a specialized mesenchymal phenotype. Indeed, the dominant PC of expression changes may correspond to this loss of stemness. The TGF-β family member Activin A inhibits differentiation and bone formation of committed osteoblasts (Eijken et al., 2007) by altering the ECM composition (Alves et al., 2013). Because most TFs associated with TGF-β signaling (e.g., ESR1, FOSB, HOXA9, JUNB, MEF2A, and MYC) appear to be inhibited in our URA and because the Activin-antagonist follistatin enhances osteoblast differentiation (Eijken et al., 2007), we hypothesize that inhibition of TGF-β signaling is essential for early initiation of osteogenic differentiation of hMSCs.
Our studies also identified IRF2 as a regulator of osteoblastogenesis in hMSCs. Irf2 is an antagonist of Irf1 and inhibits the transcriptional activation by IFN-α and -β (Zhang et al., 2015), and has a separate function in cell proliferation (Vaughan et al., 1995). In addition, we found that IFN-γ-pathway-associated TFs (e.g., CEBPβ, CREB1, HOXA10, STAT5A, STAT5B, and STAT6) change activity within the first phase. Consistent with these findings, IFNs do not affect induction of osteogenic differentiation in hMSCs, but they inhibit mineralization when administered after lineage commitment (2 days after initiating osteogenic differentiation) (Woeckel et al., 2012) or to pre-committed immortalized human fetal osteoblasts. Taken together, these findings indicate that regulation of IFN signaling is important for the osteogenic differentiation of hMSCs.
In conclusion, our data show that a stable osteoblast or adipocyte phenotype is established within the first 2 days upon induction of lineage commitment in hMSC. Three distinct early phases with characteristic cellular responses and differentially expressed TFs are evident during both adipogenic and osteogenic differentiation. We observed that adipogenic differentiation of MSCs derived from young healthy individuals required a higher number of genes to change in expression than osteogenic differentiation. This observation together with the known shift in balance between adipocyte and osteoblast differentiation with aging (Li et al., 2016) motivates further studies to investigate the extent of transcriptional changes as a function of age or gender. Interestingly, changes in TF activity that occur within the first 3 hr may control regulation of subsequent later phases of mesenchymal differentiation. Upstream regulator analyses identified TFs in both canonical and less explored signaling pathways. The latter finding opens up possibilities for studies on small molecules that target early regulators to efficiently induce osteoblast and adipocytes differentiation, as part of a bone anabolic strategy for osteoporosis.
Experimental Procedures
Cell Culture
Bone marrow-derived hMSCs and NHOst from healthy individuals were obtained from Lonza and differentiated as previously described (Bruedigam et al., 2011). The donor characteristics of the different hMSC batches were as follows: donor 1, 19 years, male, ethnicity unknown (microarray study); donor 2, 22 years, male, white; donor 3, 20 years, male, black; donor 4, 33 years, male, black. Briefly, hMSC and NHOst at passage 2 were expanded in Mesenchymal Stem Cell Growth Media or Osteoblast Basal Growth Media (Lonza, Switzerland), respectively, and and 5 × 103 vital cells/cm2 (hMSC) or 1 × 104 vital cells/cm2 (NHOst) were seeded in 12-well cell-culture plates in αMEM (Life Technologies, including 50 μg/mL ascorbic acid) supplemented with 10% heat-inactivated fetal calf serum (Invitrogen), 20 mM HEPES (Sigma), 1.8 mM CaCl2 (Sigma), and adjusted to pH 7.5. Two days after seeding, cells were differentiated into adipocytes using αMEM supplemented with 100 nM dexamethasone (Sigma), 60 μM 3-isobutyl-1-methylxanthine (Sigma) and 500 μM indomethacin (Sigma), or differentiated into osteoblasts using αMEM supplemented with 100 nM dexamethasone (Sigma) and 10 mM β-glycerophosphate (Sigma) and cultured at 37°C and 5% CO2 in a humidified atmosphere. Differentiation media were replaced every 3–4 days and not during the first 96 hr of the experiment.
DNA, Protein, Alkaline Phosphatase Activity, and Mineralization Assays
DNA, protein, ALP activity, and calcium measurements were performed as previously described (Bruedigam et al., 2011).
RNA Isolation
For each time point, nine individual wells with MSCs were induced to differentiate into the osteogenic as well as the adipogenic lineage. To obtain enough RNA for the gene expression profiling analyses, we pooled three individual cultures in TRIzol (Life Technologies), resulting in a total of three experimental samples per time point (11 time points) per lineage (osteogenic or adipogenic) to be used for gene expression profiling analyses (66 samples in total). RNA was isolated as previously described (Bruedigam et al., 2011). The quality of isolated RNA was assessed on a 2100 Bioanalyzer (Agilent Technologies).
Illumina Gene Chip-Based Gene Expression Profiling
Illumina HumanHT-12 v3 BeadChip (Illumina) human whole-genome expression arrays were used. RNA from three biological replicates for each condition and time point were analyzed. Total RNA (100 ng) of each sample was amplified using an Illumina TotalPrep RNA Amplification Kit (Ambion). cRNA (750 ng) was hybridized using the standard Illumina protocol and scanned on an iScan.
Microarray Analysis
Raw data were background subtracted using Illumina GenomeStudio (V2010.1, Illumina), and further processed using the Bioconductor R2.10.1 lumi-package (Du et al., 2008). Data were transformed using variance stabilization and quantile normalized. Probes that were present at least once in the experiments (detection p value <0.01) were considered to be expressed (15,795 probes) and further analyzed. Differential expressed probes, q < 0.001, were calculated using the Bioconductor package limma (Smyth, 2004).
Enriched Functional Categories, Ingenuity Pathway Analysis, and GeneMania Network Generation
For each time point, we selected the significant regulated probes (q value <0.001 versus undifferentiated hMSCs), and enriched functional categories were calculated using DAVID v6.7 (http://david.abcc.ncifcrf.gov/ (Huang et al., 2009)). The 100 most significant functional categories (based on p value) per phase (0–3 hr, 6–24 hr, 48–96 hr) and lineage (adipogenic, osteogenic) were selected. The enrichment p values for each time point and treatment were added to the functional categories (in total 285, see Table S1). The p values were −log10 transformed and visualized in a cluster tree using JavaTreeView. Consolidated pathway analyses were performed with GeneMania (www.genemania.org). The URA for each time point was performed using ingenuity pathway analysis (www.ingenuity.com). TFs with a Z score below −2 or above 2 were considered inhibited or activated. For the downstream analyses depicted in Table 1, we counted the number of genes in phases II and III that can be regulated by the TFs in the first phase. The cluster analyses of the Z scores were visualized using JavaTreeView.
Generation of Overexpression Constructs
Overexpression constructs were generated using Gateway cloning (Invitrogen). Constructs containing the gene of interest (pCMV-SPORT6-IRF2: 3920890) were ordered from Open Biosystems. PCR products without a stop codon were generated from these constructs (primers: IRF2-for, caccatgccggtggaaaggatgc; IRF2-rev, acagctcttgacgcgggcctg) and ligated into pENTR-topo (Invitrogen) using the supplier's protocol. The generated Entry clone was sequenced and the insert was transferred into pLenti6.3/v5-DEST (Invitrogen) destination vector using an LR-gateway reaction according to the supplier's protocol. Virus production was carried out as previously described using Virapower lentiviral packaging constructs (Drabek et al., 2011). Subsequently, virus-containing media were concentrated using ultracentrifugation at 22,800 rpm in a SW32ti rotor (Beckman Coulter) at 4°C.
Author Contributions
Conceptualization, J.P. and J.L.; Methodology, J.P.; Investigation, J.P., T.S., and J.T.; Formal Analysis, J.P.; Writing – Original Draft, J.P. and J.L.; Writing – Review & Editing, J.P., H.W., A.W., and J.L.; Funding Acquisition, J.L.
Acknowledgments
The authors thank Marijke Schreuders-Koedam and Bianca Boers-Sijmons for technical assistance. This work was supported by the Dutch Institute for Regenerative medicine (NIRM: grant no. FES0908), Erasmus MC Stem Cell and Regenerative Medicine Institute, and Erasmus Medical Center (EMC-MM-01-39-02) as well as NIH grant R01 AR049069 (A.J.v.W.). We also acknowledge the generous philanthropic support of William and Karen Eby, as well as the charitable foundation in their names. H.W. is supported by Synpol, EU-FP7 (KBBE.2012.3.4–02 #311815); Corbel, EU-H2020 (INFRADEV-4-2014-2015 #654248); Epipredict, EU-H2020; MSCA-ITN-2014-ETN, Marie Skłodowska-Curie Innovative Training Networks (ITN-ETN) (#642691), by the Netherlands Organization for Scientific Research (NWO) in the integrated program of WOTRO (W01.65.324.00/project 4) Science for Global Development, and by BBSRC China (BB/J020060/1).
Published: March 23, 2017
Footnotes
Supplemental Information includes four figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.02.018.
Accession Numbers
The gene expression data analyzed here are publicly available and can be retrieved from the Gene Expression Omnibus (GEO) at the NCBI under accession number GEO: GSE80614.
Supplemental Information
References
- Alves R.D.A.M., Eijken M., Bezstarosti K., Demmers J.A.A., van Leeuwen J.P.T.M. Activin A suppresses osteoblast mineralization capacity by altering extracellular matrix (ECM) composition and impairing matrix vesicle (MV) production. Mol. Cell. Proteomics. 2013;12:2890–2900. doi: 10.1074/mcp.M112.024927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augello A., De Bari C. The regulation of differentiation in mesenchymal stem cells. Hum. Gene Ther. 2010;21:1226–1238. doi: 10.1089/hum.2010.173. [DOI] [PubMed] [Google Scholar]
- Bruder S.P., Jaiswal N., Haynesworth S.E. Growth kinetics, self-renewal, and the osteogenic potential of purified human mesenchymal stem cells during extensive subcultivation and following cryopreservation. J. Cell. Biochem. 1997;64:278–294. doi: 10.1002/(sici)1097-4644(199702)64:2<278::aid-jcb11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Bruedigam C., Driel M.v, Koedam M., Peppel J.v, van der Eerden B.C.J., Eijken M., van Leeuwen J.P.T.M. Basic techniques in human mesenchymal stem cell cultures: differentiation into osteogenic and adipogenic lineages, genetic perturbations, and phenotypic analyses. Curr. Protoc. Stem Cell Biol. 2011;Chapter 1 doi: 10.1002/9780470151808.sc01h03s17. Unit1H.3. [DOI] [PubMed] [Google Scholar]
- Drabek K., van de Peppel J., Eijken M., van Leeuwen J.P.T.M. GPM6B regulates osteoblast function and induction of mineralization by controlling cytoskeleton and matrix vesicle release. J. Bone Miner. Res. 2011;26:2045–2051. doi: 10.1002/jbmr.435. [DOI] [PubMed] [Google Scholar]
- Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Ducy P., Zhang R., Geoffroy V., Ridall A.L., Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Dudakovic A., Camilleri E.T., Xu F., Riester S.M., McGee-Lawrence M.E., Bradley E.W., Paradise C.R., Lewallen E.A., Thaler R., Deyle D.R. Epigenetic control of skeletal development by the histone methyltransferase Ezh2. J. Biol. Chem. 2015;290:27604–27617. doi: 10.1074/jbc.M115.672345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijken M., Swagemakers S., Koedam M., Steenbergen C., Derkx P., Uitterlinden A.G., van der Spek P.J., Visser J.A., de Jong F.H., Pols H.A.P. The activin A-follistatin system: potent regulator of human extracellular matrix mineralization. FASEB J. 2007;21:2949–2960. doi: 10.1096/fj.07-8080com. [DOI] [PubMed] [Google Scholar]
- Feillet C., van der Horst G.T.J., Levi F., Rand D.A., Delaunay F. Coupling between the circadian clock and cell cycle oscillators: implication for healthy cells and malignant growth. Front. Neurol. 2015;6:96. doi: 10.3389/fneur.2015.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hung S.-C., Chang C.-F., Ma H.-L., Chen T.-H., Low-Tone Ho L. Gene expression profiles of early adipogenesis in human mesenchymal stem cells. Gene. 2004;340:141–150. doi: 10.1016/j.gene.2004.06.028. [DOI] [PubMed] [Google Scholar]
- Justesen J., Stenderup K., Ebbesen E.N., Mosekilde L., Steiniche T., Kassem M. Adipocyte tissue volume in bone marrow is increased with aging and in patients with osteoporosis. Biogerontology. 2001;2:165–171. doi: 10.1023/a:1011513223894. [DOI] [PubMed] [Google Scholar]
- Kulterer B., Friedl G., Jandrositz A., Sanchez-Cabo F., Prokesch A., Paar C., Scheideler M., Windhager R., Preisegger K.-H., Trajanoski Z. Gene expression profiling of human mesenchymal stem cells derived from bone marrow during expansion and osteoblast differentiation. BMC Genomics. 2007;8:70. doi: 10.1186/1471-2164-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-H., Kim Y.-J., Yoon W.-J., Kim J.-I., Kim B.-G., Hwang Y.-S., Wozney J.M., Chi X.-Z., Bae S.-C., Choi K.-Y. Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. J. Biol. Chem. 2005;280:35579–35587. doi: 10.1074/jbc.M502267200. [DOI] [PubMed] [Google Scholar]
- Li J., Liu X., Zuo B., Zhang L. The role of bone marrow microenvironment in governing the balance between osteoblastogenesis and adipogenesis. Aging Dis. 2016;7:514. doi: 10.14336/AD.2015.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M.B., Benkusky N.A., Sen B., Rubin J., Pike J.W. Epigenetic plasticity drives adipogenic and osteogenic differentiation of marrow-derived mesenchymal stem cells. J. Biol. Chem. 2016;291:17829–17847. doi: 10.1074/jbc.M116.736538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M.B., Moncivais K., Caplan A.I. Mesenchymal stem cells: environmentally responsive therapeutics for regenerative medicine. Exp. Mol. Med. 2013;45:e54. doi: 10.1038/emm.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng F., Boucher S., Koh S., Sastry K.S.R., Chase L., Lakshmipathy U., Choong C., Yang Z., Vemuri M.C., Rao M.S. PDGF, TGF-beta, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood. 2008;112:295–307. doi: 10.1182/blood-2007-07-103697. [DOI] [PubMed] [Google Scholar]
- Otto F., Thornell A.P., Crompton T., Denzel A., Gilmour K.C., Rosewell I.R., Stamp G.W., Beddington R.S., Mundlos S., Olsen B.R. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Piek E., Sleumer L.S., van Someren E.P., Heuver L., de Haan J.R., de Grijs I., Gilissen C., Hendriks J.M., van Ravestein-van Os R.I., Bauerschmidt S. Osteo-transcriptomics of human mesenchymal stem cells: accelerated gene expression and osteoblast differentiation induced by vitamin D reveals c-MYC as an enhancer of BMP2-induced osteogenesis. Bone. 2010;46:613–627. doi: 10.1016/j.bone.2009.10.024. [DOI] [PubMed] [Google Scholar]
- Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., Moorman M.A., Simonetti D.W., Craig S., Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Ruijtenberg S., van den Heuvel S. Coordinating cell proliferation and differentiation: antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle. 2016;15:196–212. doi: 10.1080/15384101.2015.1120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo V., Yu C., Belliveau P., Hamilton A., Flynn L.E. Comparison of human adipose-derived stem cells isolated from subcutaneous, omental, and intrathoracic adipose tissue depots for regenerative applications. Stem Cells Transl. Med. 2014;3:206–217. doi: 10.5966/sctm.2013-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samee N., Geoffroy V., Marty C., Schiltz C., Vieux-Rochas M., Levi G., de Vernejoul M.-C. Dlx5, a positive regulator of osteoblastogenesis, is essential for osteoblast-osteoclast coupling. Am. J. Pathol. 2008;173:773–780. doi: 10.2353/ajpath.2008.080243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- So A.Y.-L., Bernal T.U., Pillsbury M.L., Yamamoto K.R., Feldman B.J. Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proc. Natl. Acad. Sci. USA. 2009;106:17582–17587. doi: 10.1073/pnas.0909733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stains J.P., Civitelli R. Genomic approaches to identifying transcriptional regulators of osteoblast differentiation. Genome Biol. 2003;4:222. doi: 10.1186/gb-2003-4-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert A.F., Rackwitz L., Gilbert F., Nöth U., Tuan R.S. Concise review: the clinical application of mesenchymal stem cells for musculoskeletal regeneration: current status and perspectives. Stem Cells Transl. Med. 2012;1:237–247. doi: 10.5966/sctm.2011-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler R., Spitzer S., Karlic H., Klaushofer K., Varga F. DMSO is a strong inducer of DNA hydroxymethylation in pre-osteoblastic MC3T3-E1 cells. Epigenetics. 2012;7:635–651. doi: 10.4161/epi.20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan P.S., Aziz F., van Wijnen A.J., Wu S., Harada H., Taniguchi T., Soprano K.J., Stein J.L., Stein G.S. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- Woeckel V.J., Eijken M., van de Peppel J., Chiba H., van der Eerden B.C.J., van Leeuwen J.P.T.M. IFNβ impairs extracellular matrix formation leading to inhibition of mineralization by effects in the early stage of human osteoblast differentiation. J. Cell. Physiol. 2012;227:2668–2676. doi: 10.1002/jcp.23009. [DOI] [PubMed] [Google Scholar]
- Yeung D.K.W., Griffith J.F., Antonio G.E., Lee F.K.H., Woo J., Leung P.C. Osteoporosis is associated with increased marrow fat content and decreased marrow fat unsaturation: a proton MR spectroscopy study. J. Magn. Reson. Imaging. 2005;22:279–285. doi: 10.1002/jmri.20367. [DOI] [PubMed] [Google Scholar]
- Young D.W., Pratap J., Javed A., Weiner B., Ohkawa Y., van Wijnen A., Montecino M., Stein G.S., Stein J.L., Imbalzano A.N. SWI/SNF chromatin remodeling complex is obligatory for BMP2-induced, Runx2-dependent skeletal gene expression that controls osteoblast differentiation. J. Cell. Biochem. 2005;94:720–730. doi: 10.1002/jcb.20332. [DOI] [PubMed] [Google Scholar]
- Zhang X., Yang M., Lin L., Chen P., Ma K.T., Zhou C.Y., Ao Y.F. Runx2 overexpression enhances osteoblastic differentiation and mineralization in adipose–derived stem cells in vitro and in vivo. Calcif. Tissue Int. 2006;79:169–178. doi: 10.1007/s00223-006-0083-6. [DOI] [PubMed] [Google Scholar]
- Zhang X.-J., Jiang D.-S., Li H. The interferon regulatory factors as novel potential targets in the treatment of cardiovascular diseases. Br. J. Pharmacol. 2015;172:5457–5476. doi: 10.1111/bph.12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.