Summary
Notch signaling is critically involved in neural development, but the downstream effectors remain incompletely understood. In this study, we cultured neurospheres from Nestin-Cre-mediated conditional Rbp-j knockout (Rbp-j cKO) and control embryos and compared their miRNA expression profiles using microarray. Among differentially expressed miRNAs, miR-342-5p showed upregulated expression as Notch signaling was genetically or pharmaceutically interrupted. Consistently, the promoter of the miR-342-5p host gene, the Ena-vasodilator stimulated phosphoprotein-like (Evl), was negatively regulated by Notch signaling, probably through HES5. Transfection of miR-342-5p promoted the differentiation of neural stem cells (NSCs) into intermediate neural progenitors (INPs) in vitro and reduced the stemness of NSCs in vivo. Furthermore, miR-342-5p inhibited the differentiation of neural stem/intermediate progenitor cells into astrocytes, likely mediated by targeting GFAP directly. Our results indicated that miR-342-5p could function as a downstream effector of Notch signaling to regulate the differentiation of NSCs into INPs and astrocytes commitment.
Keywords: neural stem cells, intermediate neural progenitors, Notch, RBP-J, neuron, glia, miR-342-5p
Graphical Abstract
Highlights
-
•
miR-342-5p acts as a downstream effector of canonical Notch signaling
-
•
Notch signal inhibits miR-342-5p expression by regulating its host gene Evl
-
•
miR-342-5p promotes the transition of NSCs into INPs
-
•
Astrocyte commitment was suppressed by miR-342-5p targeting GFAP
In this article, Han and colleagues show that miR-342-5p acts as a downstream effector of Notch signaling in the mouse CNS. Notch signal inhibits miR-342-5p expression by regulating its host gene Evl. And with attenuated Notch signal in NSCs, miR-342-5p is upregulated to promote NSCs transition into INPs, and to inhibit astrocyte commitment by targeting GFAP.
Introduction
The mammalian CNS originates from neural stem cells (NSCs) which are multipotential to generate neurons, astrocytes and oligodendrocytes (Temple, 2001). NSCs present as neuroepithelial (NE) cells in early developmental stage and divide symmetrically to expand the neural epithelium (Franco and Muller, 2013, Gage and Temple, 2013, Kriegstein and Alvarez-Buylla, 2009, Merkle and Alvarez-Buylla, 2006, Noctor et al., 2004). Radial glial cells (RGCs) then replace NE cells and divide asymmetrically in the ventricular zone (VZ) of the developing brain to maintain a self-renewing stem cell pool and generate differentiating daughter cells. These daughter cells, defined as intermediate neural progenitors (INPs), migrate radially outward while dividing symmetrically to amplify themselves. Using a time-lapse imaging method, INPs have been shown to accumulate in the subventricular zone (SVZ) (Noctor et al., 2004), although other studies have also shown that INPs coexist with NSCs in the VZ of mouse telencephalon (Mizutani et al., 2007). NSCs and INPs could be distinguished by molecular markers as well as the type of progenies they generate, because INPs have limited differentiation potential compared with NSCs.
The development of NSCs and INPs is regulated by a series of extrinsic and intrinsic elements (Kohwi and Doe, 2013, Shi et al., 2010). Notch signaling mediates adjacent cell-cell communications. Upon activation by ligands, Notch receptors are processed by protease complexes containing γ-secretase to release the intracellular domain of Notch receptor (NICD). NICD translocates into nucleus and converts the transcription factor RBP-J from a repressor into an activator. The downstream targets regulated by NICD/RBP-J are mainly basic-helix-loop-helix (bHLH) transcription factors, such as the HES family proteins. The Notch pathway regulates multiple steps of NSC and INP development (Pierfelice et al., 2011, Yoon and Gaiano, 2005). Notch signaling promotes RGC identity and maintains their stemness (Gaiano and Fishell, 2002, Gaiano et al., 2000). Notch signaling also inhibits the differentiation of NSCs and INPs into neurons (Hitoshi et al., 2002), and directs several binary fate choices, such as NSCs differentiating into glia versus neurons and glial progenitors differentiating into astrocytes versus oligodendrocytes (Louvi and Artavanis-Tsakonas, 2006, Mizutani et al., 2007, Tanigaki et al., 2001, Zheng et al., 2009). It has been shown that Notch receptors are activated in both NSCs and INPs, but the downstream signals might be mediated in different ways, namely, through RBP-J-dependent signaling in NSCs and RBP-J-independent signaling in INPs (Gao et al., 2009, Mizutani et al., 2007). Knockdown of Rbp-j promotes the differentiation of NSCs into INPs, which preferentially differentiated into neurons versus astrocytes, and promotes migration of ventricular progenitor cells to the outside of the cortical plate (Mizutani et al., 2007). We have shown that neurospheres cultured from Rbp-j knockout embryonic forebrain contain more INPs in comparison with neurospheres derived from the control littermates that contain more NSCs, demonstrating that RBP-J-dependent Notch signaling inhibits differentiation of NSCs into INPs (Gao et al., 2009). The differentiation of these Rbp-j-deleted INPs also showed neurogenic preference against astrocytes. Moreover, Notch signaling is also reported to play a role in the apoptosis of progenitor cells and the maturation of neurons (Sestan et al., 1999, Redmond et al., 2000, Hoeck et al., 2010). However, downstream molecules through which Notch signaling regulates NSCs and neural development have been elusive.
MicroRNAs (miRNAs) are short non-coding RNA molecules that are abundant in the CNS and are crucial for neural development (Bian et al., 2013, Fineberg et al., 2009, Shi et al., 2010). Encouraged by other reports showing that some miRNAs are involved in the regulation of Notch signaling (Boucher et al., 2011, Hamidi et al., 2011, Roese-Koerner et al., 2016), in this study, we cultured neurospheres from E11.5 Rbp-j knockout and control embryos and compare their miRNA expression by using microarray hybridization. We found that among several differentially expressed miRNAs, miR-342-5p might be a downstream target of Notch signaling. Overexpression of miR-342-5p could promote the differentiation of NSCs into INPs, and inhibit an astrocyte fate, likely by direct targeting GFAP. Inhibition of miR-342-5p, on the contrary, could maintain the stemness of NSCs impaired by Notch blockade. Our results indicated that miR-342-5p could function as a downstream molecule of Notch signaling to regulate the differentiation of NSCs into INPs, and the commitment into astrocytes.
Results
Identification of miR-342-5p as a Downstream Molecule of Notch Signaling in Neural Stem/Intermediate Progenitor Cells
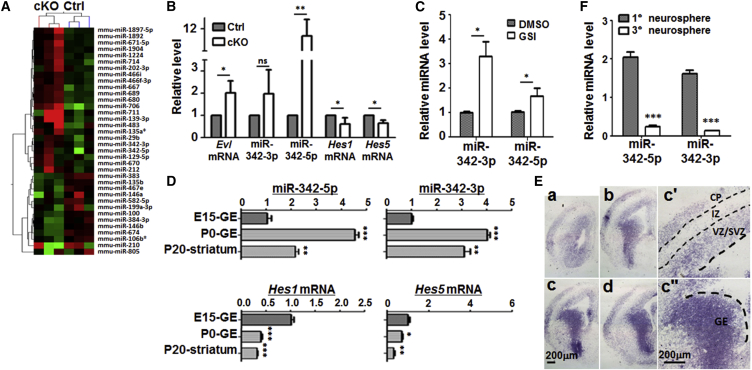
In order to find the downstream miRNAs of Notch signaling, we cultured neurospheres from ganglionic eminence (GE) of NesCre-Rbp-jf/f (Rbp-jcKO) and NesCre-Rbp-jf/+ (control) embryos on 11.5 dpc (days post coitus) (Gao et al., 2009). Total RNA was extracted from neurospheres, and their miRNA expression profiles were compared by using miRNA array hybridization. Cluster analysis indicated that a number of miRNAs were differentially expressed between RBP-J-deficient and control neurospheres (Figure 1A). Among these miRNAs, the levels of miR-342-5p and miR-342-3p increased apparently in RBP-J-deficient neurospheres. The genes of miR-342-5p and miR-342-3p are located in the third intron of Evl gene, which has been reported to play a role in neural development (Kwiatkowski et al., 2007, Vanderzalm and Garriga, 2007). We therefore tested the expressions of miR-342-5p, miR-342-3p, and Evl in Rbp-j-deleted neurospheres by real-time PCR analysis. The attenuated Notch signaling in Rbp-j-deleted neurospheres was validated by the decreased expression of Hes1 and Hes5, two bHLH downstream effectors of Notch signaling (Figure 1B). And the expression of miR-342-5p increased 11.67-fold compared with the control (Figure 1B). The expression of the host gene Evl also increased significantly. However, the expression of miR-342-3p showed a tendency to increase, but this was not statistically significant (Figure 1B). We therefore focused on miR-342-5p in the following analysis.
Figure 1.
miR-342-5p Was a Downstream miRNA of the Notch Pathway in NSCs
(A) Screening of Notch downstream miRNAs using microarray hybridization. Primary neurospheres were cultured with cells derived from the ganglion eminences (GE) of NesCre-Rbp-jf/f (cKO) and NesCre-Rbp-j+/f embryos (Ctrl) (E11.5). Differentially expressed miRNAs were compared by using microarray hybridization and clustered. RNA samples were derived from three different pairs of littermates.
(B) The expression of Evl mRNA, miR-342-3p, miR-342-5p, Hes1 mRNA, and Hes5 mRNA in Rbp-j cKO and control neurospheres was determined by qRT-PCR. Neurospheres were derived from four different pairs of littermates.
(C) Primary neurospheres were cultured by using cells derived from GE of wild-type embryos (E15.5). Cells were treated with 75 μM GSI for 12 hr, and the expression of miR-342-3p and miR-342-5p was examined by qRT-PCR. DMSO was used as a control. RNA samples were extracted from four pairs of GSI- and DMSO-treated neurospheres.
(D) The expression of miR-342-5p, miR-342-3p, Hes1 mRNA, and Hes5 mRNA in the GE and striatum during neural development in mice was determined by qRT-PCR. The brain tissues at the specific time point came from four mice).
(E) The expression of miR-342-5p in the telencephalon of E14.5 embryo was determined by in situ hybridization using a locked nucleic acid probe. The brain sections were derived from three different wild-type embryos. (a–d) Coronal sections of E14.5 telencephalon. (c′ and c″) The cortex and GE regions of the telencephalon section in (c) are shown in magnification, respectively. VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate.
(F) The expression of miR-342-5p and miR-342-3p in neurospheres of the first-generation (1°) and third-generation (3°) was determined by qRT-PCR. RNA samples were extracted from four pairs of passaged neurospheres.
Bars, means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, not significant.
In wild-type neurospheres treated with GSI, an inhibitor of Notch activation, the expression of miR-342-5p was upregulated compared with the control (Figure 1C), and the expression of miR-342-3p also increased in this model. These results indicated that the expression of miR-342-5p was increased in neural stem/intermediate progenitor cells (NS/PCs) after Notch signaling was either genetically or pharmaceutically blocked.
The upregulated expression of miR-342-5p and its host gene Evl could be a result of altered cell composition in neurospheres with Notch blockade (Mizutani et al., 2007, Gao et al., 2009). To clarify this, we treated neurospheres with GSI for different periods of time, and observed the expression of miR-342-5p and the NSC markers Nestin and Glast, and the INP marker Tα1 (Gao et al., 2009). The result showed that the expression of Nestin and Glast decreased 24 hr after GSI treatment, while the INP marker did not change until 96 hr after GSI treatment. However, the expression of miR-342-5p was upregulated as early as 6 hr after GSI treatment (Figure S1A), suggesting that the upregulation of miR-342-5p after Notch blockade was not a consequence of cell composition change. In addition, we examined the expression of Nestin, Glast, Tα1, and miR-342-5p in neurospheres derived from NSC-specific Notch activation mice (NICD transgene activated by Nestin-Cre, see below), and found that while the expression of Nestin, Glast, and Tα1 remained unchanged, the level of miR-342-5p was reduced in neurospheres of NICD transgenic mice (Figure S1B). Moreover, to access the relationship between miR-342-5p expression and Notch activation in vivo, we determined the expression of miR-342-5p and Hes1 and Hes5 in embryonic day (E15.0) GE and post natal striatum (P0, P20), which is derived from embryonic GE. The results showed that, compared with that of E15.0 GE, the expression of Hes1 and Hes5 decreased in P0 and P20 striatum, suggesting lowered Notch activation. Meanwhile, the expression of miR-342-5p and miR-342-3p in P0 and P20 striatum increased compared with that in E15.0 GE, consistent with a negative regulation of miR-342-5p expression by Notch signaling (Figure 1D). Next, we examined the expression of miR-342-5p by in situ hybridization on E14.5 brain sections. The results showed that from VZ to SVZ, the expression of miR342-5p gradually elevated either in the developing cortex or GE, indicating that its expression was upregulated along with NSC differentiation (Figure 1E). miR-342-5p also has a weak expression in the intermediate zone and a moderate expression in the cortical plate (Figure 1E). To validate the elevated expression of miR-342-5p in INPs compared with NSCs, we also examined the expression of miR-342-5p and miR-342-3p in neurospheres of the first-generation and third-generation spheres, because more NSCs were enriched in the 3° spheres as the passages through different generations had excluded the progenitor cells with less self-renewal ability. We found that their expression apparently decreased in the third-generation neurospheres (Figure 1F).
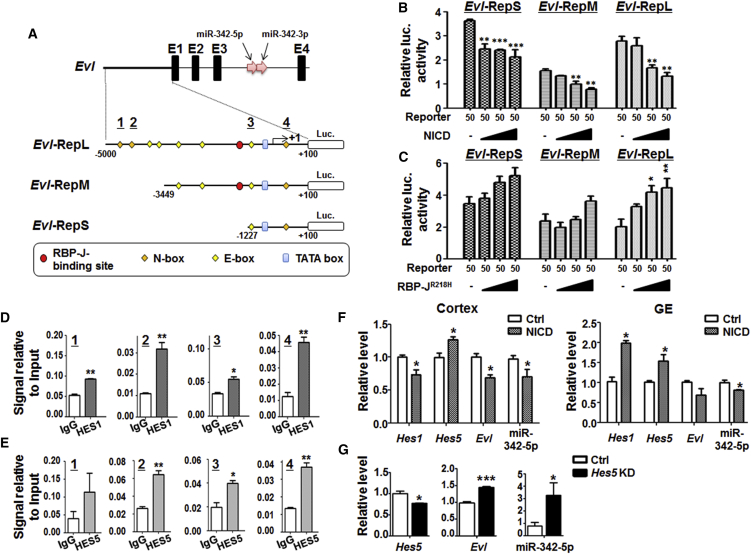
Notch Signaling Regulated the Promoter of Evl that Harbors miR-342
The Evl promoter region (Grady et al., 2008) harbors an RBP-J-binding site, three N boxes (−4,519 to −4,514, −3,942 to −3,937, −430 to −435) and five E boxes (Iso et al., 2003) (Figure 2A). We cloned different fragments of the Evl promoter region, and constructed reporter genes named Evl-RepS (−1,227 to +100), Evl-RepM (−3,449 to +100), and Evl-RepL (−5,000 to +100) encompassing different transcription recognition sites. NIH3T3 cells were transfected with different reporters and increasing amounts of pEFBOS-NICD, with a Renilla luciferase vector as an internal control. The result showed that, 48 hr after the transfection, overexpression of NICD resulted in reduced luciferase activity in cells transfected with each of the three reporters (Figure 2B). On the contrary, transfection of NIH3T3 cells with pCMV-RBP-JR218H (R218H), a dominant-negative form of RBP-J, led to increased luciferase expression (Figure 2C). Similar results were obtained when HEK293 or PC12 cells were employed in the reporter assays (data not shown). These results suggested that Notch signaling might negatively regulate the expression of Evl/miR-342-5p through the Evl promoter.
Figure 2.
Notch Signaling Regulated the Evl Promoter
(A) Schematic illustration of the mouse Evl promoter, the reporter constructs, and the position of primers used in ChIP. Thick line, thin lines, and filled boxes represent 5′ upstream sequence, introns, and exons, respectively. The miR-342-5p gene and miR-342-3p gene are represented by two pink arrows. The putative RBP-J-binding site, N boxes, E boxes, and a TATA box are indicated by a red dot, orange quadrangles, yellow quadrangles, and a blue box, respectively. Different promoter fragments were cloned into the pGL3-basic vector to construct the reporters including Evl-RepL (−5,000 → +100 bp), Evl-RepM (−3449 → +100 bp), and Evl-RepS (−1,227 → +100 bp).
(B and C) Reporter assays. NIH3T3 cells were transfected with different reporters in combination with increasing amounts of pEFBOS-NICD (B) or pCMV-RBP-JR218H (C). A Renilla luciferase (luc) expression vector was co-transfected as an internal control. Cell lysates were prepared 48 hr after the transfection, and luciferase activity was determined. Reporter assays were done by four independent transfections, and three wells of 96-well plates were prepared as replicate samples in each group.
(D and E) ChIP assays. Crosslinked chromatins were prepared from cultured primary neurospheres (neurospheres derived from three different wild-type embryos), and immunoprecipitated with immunoglobulin G (IgG) and anti-HES1 (D) or anti-HES5 (E) antibody. After washing, the co-precipitated DNA was extracted and amplified by PCR primers targeting three different N boxes and an E box; numbers underlined in (A).
(F) Neurospheres cultured from E15.5 cortex and GE of NesCre-ROSA-Stopf/+-NIC (NICD) and ROSA-Stopf/+-NIC embryos (Ctrl) were harvested and total RNAs were extracted. The expressions of Hes1, Hes5, Evl, and miR-342-5p were determined by qRT-PCR. Neurospheres were derived from three different pairs of littermates.
(G) Neurospheres were transfected with siHes5 and control oligonucleotide (three independent transfections performed), and the expressions of Hes5, Evl, and miR-342-5p were analyzed by qRT-PCR.
Bars, means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
The repressing of the Evl promoter appeared not dependent on the RBP-J-binding site, suggesting that Notch signaling might regulate the Evl promoter through N- and E- boxes. Therefore we performed chromatin immunoprecipitation (ChIP) assays with the anti-HES1 and anti-HES5 antibodies using primary neurospheres cultured from E15.5 telencephalon. The result showed that, compared with the control (ChIP with immunoglobulin G), all three N boxes and the E box examined were enriched in the anti-HES1-precipitated chromatins (Figure 2D). It was the same for the anti-HES5-precipitated chromatins, except for the first N box (Figure 2E). These results suggested that Notch signaling might regulate the Evl promoter through HES1 and HES5.
To further verify the regulation of miR-342-5p and Evl expression by Notch signaling in NS/PCs, we took advantage of NesCre transgenic mice and ROSA-Stopf-NICD mice (Zhao et al., 2016) to obtain NS/PCs with activated Notch signaling by culturing neurospheres from the E15.5 cortex and GE. By qRT-PCR, we found that the expression of Hes5 increased, whereas that of Hes1 increased in the GE but decreased in the cortex (Figure 2F). In contrast to that of Hes5, the expression of Evl and miR-342-5p reduced (Figure 2F). These results showed that the activation of Notch signaling could inhibit the expression of miR-342-5p, which was consistent with the increased expression of miR-342-5p in our genetic or pharmaceutical Notch blockade models. We then transfected siHes5 or the scrambled control siRNA into cultured neurospheres, and found that the expressions of miR-342-5p and Evl were elevated with the knockdown of Hes5 (Figure 2G). These results implied that Notch signaling might inhibit the expression of Evl and miR-342-5p through the transcription repressor HES5 in vivo.
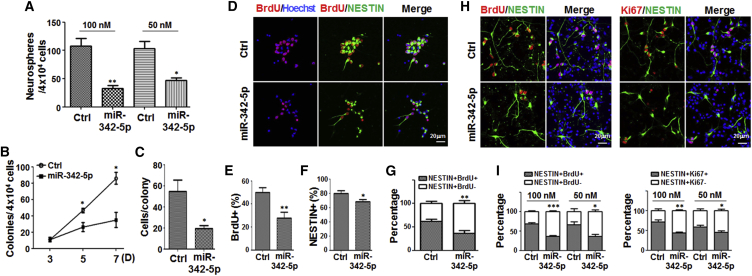
miR-342-5p Reduced the Formation of Neurospheres and NS/PC Colonies In Vitro
In order to reveal the role of miR-342-5p in NSCs, we transfected cultured NS/PCs with synthetic miR-342-5p mimics. Nearly 100% of the neurospheres could be transfected with Cy3-labeled scrambled control oligonucleotides by liposome-mediated transfection (Figure S2A). Therefore, we first cultured mouse primary NS/PCs under adherent conditions for 12 hr and transfected them with miR-342-5p mimics or control oligonucleotides. The cells were then detached and replated, and cultured under the neurosphere conditions for 7 days to observe the formation of neurospheres (Figure S2B). The result showed that upregulation of miR-342-5p led to reduced neurosphere formation (Figures 3A and S4A).
Figure 3.
Overexpression of miR-342-5p Repressed the Formation of Both Suspended Neurospheres and Adherent Colonies, and Repressed the Proliferation of NS/PCs
(A) Single-cell suspensions were prepared from the GE of E15.5 embryos and cultured on poly-L-lysine (PLL)-coated dishes in DF12 medium supplemented with N2, 20 ng/mL bFGF, and 20 ng/mL EGF. After 12 hr, NS/PCs were transfected with 50 or 100 nM miR-342-5p mimics or control oligonucleotides (Ctrl). Cells were detached 4 hr later and cultured under neurosphere conditions for 7 days. The cultures were photographed and the number of neurospheres was determined under a microscope. The transfections were done five times independently.
(B and C) NS/PCs derived from normal embryos were transfected with 100 nM miR-342-5p mimics or control as in (A) (five independent transfections performed). Cells were then replated at clonal density on PLL-coated dishes for 7 days. The number of colonies (B) and cell number per colony (C) were determined.
(D–I) NS/PCs were transfected with miR-342-5p mimics and the control, and were cultured adherently at clonal density for 7 days (D–G) (five independent transfections performed) or at normal density for 48 hr (H and I) (five independent transfections performed). BrdU was added into the medium 18 hr before the end of the experiment. Cells were fixed and stained with anti-BrdU and anti-NESTIN antibodies by immunofluorescence staining (D) or with anti-BrdU, anti-NESTIN, and anti-Ki67 antibodies (H). Nuclei were counterstained with a Hoechst stain. The percentages of BrdU+ (E), NESTIN+ (F), and NESTIN+ BrdU+ (G) cells within one colony were determined (50 colonies counted in each group). The percentages of NESTIN+ BrdU+ cells and NESTIN+ Ki67+ cells within one field were determined (I) (ten fields counted in each group).
Bars, means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
When the transfected adherent NS/PCs were replated onto poly-L-lysine-coated plates at a very low clonal density (Figure S2C), individual NS/PCs could form spatially isolated colonies (Molofsky et al., 2003). The transfection of miR-342-5p reduced the total number of colonies (86 ± 7.21 colonies in control versus 35 ± 9.17 colonies in the miR-342-5p group) (Figure 3B). The average cell number in each colony also decreased dramatically after transfection of miR-342-5p (54.57 ± 10.84 versus 19.33 ± 2.96) (Figure 3C). These results suggested that upregulation of miR-342-5p could inhibit the formation of NS/PC colonies in both suspension and under the adherent condition.
miR-342-5p Inhibited the Proliferation of NS/PCs
The reduced neurospheres and colony formation might be caused by enhanced apoptosis and decreased proliferation of NS/PCs, we then examined their apoptosis and proliferation. We performed TUNEL staining to detect apoptosis in NS/PCs transfected with miR-342-5p. The result showed that under the clonal-density adherent conditions, NS/PCs transfected with miR-342-5p mimics exhibited a slight increase in the percentage of TUNEL-positive cells compared with the control (Figures S4B and S4C). Moreover, immunofluorescence staining of activated caspase-3 (aCASP3) in cells cultured under the normal-density adherent conditions also indicated that overexpression of miR-342-5p increased aCASP3-positive cells (Figures S4D and S4E). These results suggested that miR-342-5p might induce apoptosis in NS/PCs. However, we also found that NESTIN-positive cells hardly showed aCASP3-positive signals (Figure S4D). Therefore, we analyzed the proliferation and differentiation phenotypes within the NESTIN-positive NS/PCs to avoid the influence of cells that underwent subsequent apoptosis.
We then examined the proliferation of NS/PCs in the clonal-density adherent culture model using bromodeoxyuridine (BrdU) incorporation. NS/PCs were transfected with miR-342-5p mimics and the control as above, and cultured under the clonal-density adherent conditions for 7 days (Figure S2D). BrdU was added into the medium 18 hr before the harvest of the cells. Immunofluorescence staining with anti-BrdU antibody showed that transfection of the miR-342-5p mimics reduced the incorporation of BrdU within each colony (49.91% ± 4.15% in the control group versus 27.7% ± 5.26% in the miR-342-5p-transfected group) (Figures 3D and 3E). The percentage of NESTIN-positive cells slightly reduced (Figures 3D and 3F), however, the percentage of BrdU-positive cells in NESTIN-positive fraction decreased dramatically in the miR-342-5p-transfected group (Figure 3G) (63.46% ± 6.01% in the control group versus 37.77% ± 4.60% in the miR-342-5p-transfected group). Similar result was obtained when transfected NS/PCs were cultured under the normal-density adherent condition (Figures 3H, 3I, and S2E). These results suggested that overexpression of miR-342-5p inhibited NS/PC proliferation.
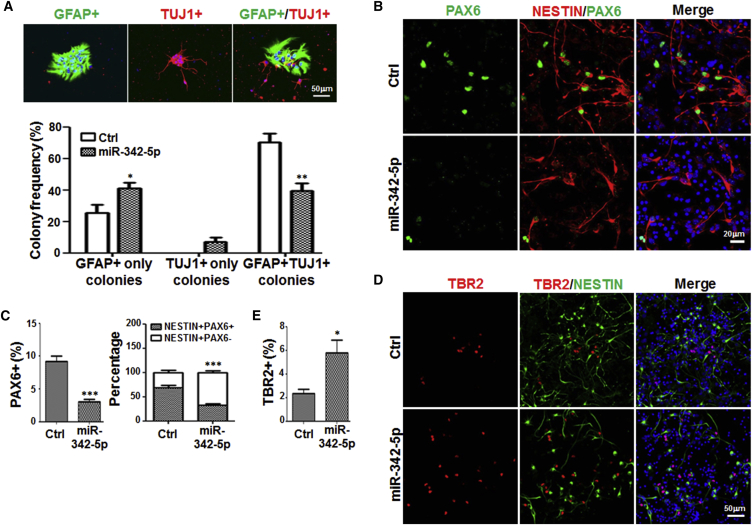
miR-342-5p Promoted the Differentiation of NSCs into INPs In Vitro
RBP-J-dependent Notch signaling has been implicated in the differentiation of NSCs into INPs (Mizutani et al., 2007). To determine NSCs and INPs in cultured NS/PCs, we employed the clonal-density adherent culture system which allows distinguishing NSC colonies from INP colonies retrospectively after they were induced to differentiate (Figure S2F). Colonies differentiating into a single type of progeny, either neurons or astrocytes, were INP colonies, while colonies giving both neurons and astrocytes were NSC colonies. The results showed that, compared with the control, NS/PCs transfected with miR-342-5p contained fewer bipotential colonies (GFAP+ TUJ1+) (70.23% ± 5.57% in control versus 39.32% ± 4.99% in miR-342-5p transfected cells). Meanwhile, mono-potential colonies, either astrocytes (GFAP+) or neurons (TUJ1+), increased remarkably after transfection with miR-342-5p (Figure 4A). Furthermore, cortex NS/PCs were cultured and transfected with miR-342-5p mimics or the control, followed by the normal-density adherent culture in differentiation medium for 2 days. Immunofluorescence staining with anti-PAX6 and TBR2, which were markers for cortical NSCs and INPs, respectively (Englund et al., 2005), showed that transfection of miR-342-5p mimics led to reduced numbers of both total PAX6-positive cells and NESTIN-positive PAX6-positive cells (Figures 4B and 4C). In contrast, TBR2-positive cells increased in the miR-342-5p-transfected NS/PCs (Figures 4D and 4E). These results suggested that overexpression of miR-342-5p enhanced the differentiation of NSCs into INPs.
Figure 4.
miR-342-5p Promoted the Differentiation of NS/PCs into INPs In Vitro
(A) NS/PCs were transfected with miR-342-5p mimics or control (five independent transfections performed), and cultured under the adherent condition to proliferate at clonal density for 7 days. The newly formed colonies were further induced to differentiate for 48 hr with low mitogens (DF12 containing 1/2 × N2, 5 ng/mL bFGF and 5 ng/mL EGF) supplemented with 5% FBS. Cells were then fixed and stained with anti-GFAP and anti-TUJ1. The frequencies of GFAP+, TUJ1+, and GFAP+ TUJ1+ colonies were compared (50 colonies counted in each group).
(B and C) Adherently cultured NS/PCs were stained with anti-PAX6 and anti-NESTIN (B), and percentages of PAX6+ cells in total cells or NESTIN+ cells were determined (C) (eight fields counted in each group).
(D and E) NS/PCs were stained with anti-TBR2 and anti-NESTIN (D), and percentage of TBR2+ cells were determined (E) (eight fields counted in each group).
Bars, means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
miR-342-5p Promoted Precocious Differentiation of NSCs In Vivo
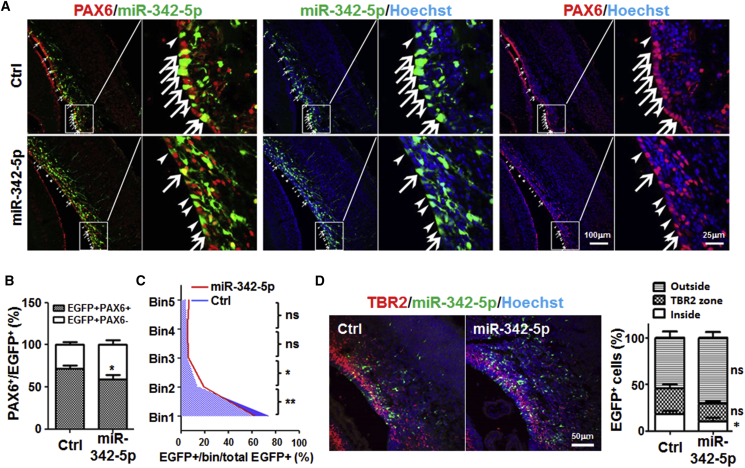
In order to verify the role of miR-342-5p in vivo, we performed in utero embryonic electroporation of E15.5 mice embryo with the miR-342-5p expression vector (pcDNA6.2-GW/EmGFP-miR-342-5p), which could also express EGFP. The embryos were collected on E18.5, cryosectioned, and stained with anti-PAX6. The result showed that in the control group (electroporated with pcDNA6.2-GW/EGFP-negative control), 71.94% ± 3.329% of EGFP-positive cells were PAX6-positive (Figure 5A, arrows). But in mice electroporated with the miR-342-5p-expressing plasmid, the number of EGFP- and PAX6-double-positive cells was reduced significantly (58.98% ± 5.295%) (Figures 5A and 5B), suggesting a decrease of NSCs. Moreover, in the miR-342-5p group, it appeared that more EGFP-positive cells migrated out of the germinal zone (bin 1, p = 0.0052) toward the basal side (pial surface) of the cortex (bins 2–5, p = 0.0333 in bin 2), consistent with precocious differentiation of NSCs (Figures 5A and 5C). Moreover, the samples were stained with anti-TBR2 antibody by using immunofluorescence. The result showed that although the percentage of TBR2-positive cells within the total EGFP-positive cells had no change between the miR-342-5p and the control groups, more miR-342-5p-transfected cells migrated out of the VZ zone which was outlined within TBR2-positive cell bands (Figure 5D). No differences in proliferation or apoptosis were detected between the two groups (data not shown). In summary, our in vivo electroporation data suggested that overexpression of miR-342-5p could promote precocious differentiation of NSCs.
Figure 5.
miR-342-5p Promoted Precocious Differentiation of NSCs In Vivo
(A) The forebrain areas of E15.5 mouse embryos were electroporated in utero with pcDNA6.2-GW/EmGFP-miR-342-5p (miR-342-5p) that could express miR-342-5p and EGFP, or with pcDNA6.2-GW/EGFP-negative control (Ctrl) that expressed only EGFP as a control. At E18.5, the mice were perfused and brains were fixed, cryosectioned, and stained with anti-PAX6 using immunofluorescence. Nuclei were counterstained with a Hoechst stain. Arrows and arrowheads represent EGFP- and PAX6-double-positive cells and EGFP-single-positive cells, respectively. Five pairs of littermates successfully electroporated with miR-342-5p and Ctrl separately were obtained.
(B) The percentage of PAX6-positive cells in EGFP-positive cells was compared (12 fields counted in each group).
(C) The EGFP-positive cells in each bin of five bins of the transfected cortical region were counted, and the percentage of EGFP-positive cells of each bin in total EGFP-positive cells was determined. Asterisks represent statistical analysis of the difference between the miR-342-5p-transfected and the control groups in the bin 1 and bin 2 areas (ten fields counted in each group).
(D) Samples in (A) were stained with anti-TBR2. The EGFP-positive cells were compared in the SVZ (TBR2+ zone), VZ (inside SVZ), and cortex (outside the SVZ) regions (ten fields counted in each group).
Bars, means ± SD. ∗p < 0.05, ∗∗p < 0.01, ns, not significant.
miR-342-5p Targeted GFAP and Regulated Astrocyte Differentiation
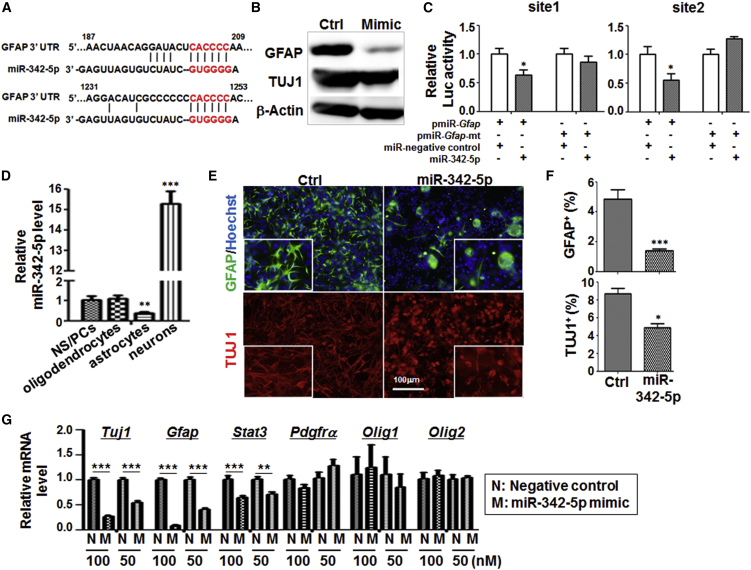
Bioinformatic searching of miR-342-5p-targeted genes suggested that miR-342-5p might regulate GFAP expression through its 3′ UTR, which harbors two miR-342-5p recognition sites (Figure 6A). Indeed, western blotting of NS/PCs transfected with miR-342-5p mimics indicated that GFAP expression was reduced dramatically (Figure 6B). We then cloned the two fragments of 3′ UTR of mouse Gfap cDNA containing the two miR-342-5p recognition sites, respectively, and constructed reporter plasmids with the wild-type or mutant miR-342-5p seed sequences using the pMIR-REPORT luciferase vector. Luciferase assays showed that miR-342-5p could repress luciferase expression by targeting either pmiR-GFAP-site1 or pmiR-GFAP-site2, and the mutation of site 1 or site 2 abolished the repression by miR-342-5p (Figure 6C). This result suggested that miR-342-5p could directly target GFAP and inhibit its expression, through binding site 1 and site 2 in the 3′ UTR of Gfap mRNA.
Figure 6.
miR-342-5p Targeted GFAP and Regulated Astrocyte Differentiation
(A) The sequence of the 3′ UTR of Gfap was aligned with the seed sequence of miR-342-5p. The recognized sequences (nucleotides 202–207 and 1,246–1,251) are marked in red.
(B) Adherently cultured NS/PCs were transfected with miR-342-5p mimics or the control (five independent transfections performed), and 4 hr later 5% FBS medium was applied to induce the differentiation for another 48 hr. Cells were collected and cell lysates were analyzed by western blot using anti-GFAP, anti-TUJ1, and anti-β-actin.
(C) Reporter assays. Cos7 cells were transfected with different reporter constructs, pcDNA6.2-GW/EmGFP-miR-342-5p and pcDNA6.2-GW/EmGFP-miR-negative control, and phRL-TK-expressing Renilla luciferase as an internal control. Cells were lysed 48 hr after the transfection, and luciferase activities in the cell lysates were determined. Reporter assays were done by four independent transfections, and three wells of a 96-well-plate were prepared as replicate samples in each group.
(D) Primary neurospheres (NS/PCs), oligodendrocytes, astrocytes, and neurons were isolated from rats (each type of cells derived from three different rat samples). Total RNA was extracted from cells and the expression of miR-342-5p was determined by qRT-PCR, with U6 RNA as an endogenous reference.
(E and F) NS/PCs were transfected with miR-342-5p or the control (five independent transfections performed), and cells were induced to differentiate by 5% FBS for 48 hr, and then fixed and stained with anti-GFAP and anti-TUJ1. Nuclei were counterstained with a Hoechst stain. The percentages of GFAP+ cells and TUJ1+ cells were compared (eight fields counted in each group).
(G) After differentiation, cells were collected, and the expressions of genes related with progeny cells were analyzed by qRT-PCR. Transfections of miR-342-5p mimic and the negative control were performed with 50 and 100 nM dosages (three independent transfections performed).
Bars, means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, not significant.
We then examined the role of miR-342-5p in the differentiation of astrocytes, in which Gfap is specifically expressed. qRT-PCR indicated that, compared with NS/PCs, the expression of miR-342-5p was downregulated in astrocytes but strongly upregulated in neurons (Figure 6D). We then transfected miR-342-5p mimics into adherent NS/PCs, and induced the differentiation of the transfected NS/PCs by culturing them in the presence of serum for 48 hr (Figure S2G). Differentiated cells were fixed and stained with different antibodies. The result showed that GFAP-positive astrocytes reduced significantly in miR-342-5p-transfected cells (4.8% in the control versus 1.7% in miR-342-5p-transfected cells) (Figures 6E and 6F). The morphology of astrocytes also apparently changed with retracted processes and round cell bodies (Figure 6E). The number of TUJ1-positive neurons also reduced but to a less extent (Figure 6F, lower). These results suggested that miR-342-5p repressed the differentiation of astrocytes, likely through directly targeting GFAP.
Further analyses on mRNA expression of NS/PC differentiation was performed by qRT-PCR (Figure 6G). The results showed that the expression of astrocyte-related genes Gfap and Stat3 was decreased after overexpression of miR-342-5p in a dose-dependent manner. This was the same with the neuronal marker Tuj1, but the oligodendrocyte-related genes Pdgfrα, Olig1, and Olig2, showed no obvious changes. These results were in accordance with the immunocytofluorescence results (Figures 6E and 6F), and altogether suggested that miR-342-5p repressed the commitment of NS/PCs into astrocytes.
Inhibition of miR-342-5p Could Rescue Part of the Phenotypes of Notch Blockade in NS/PCs
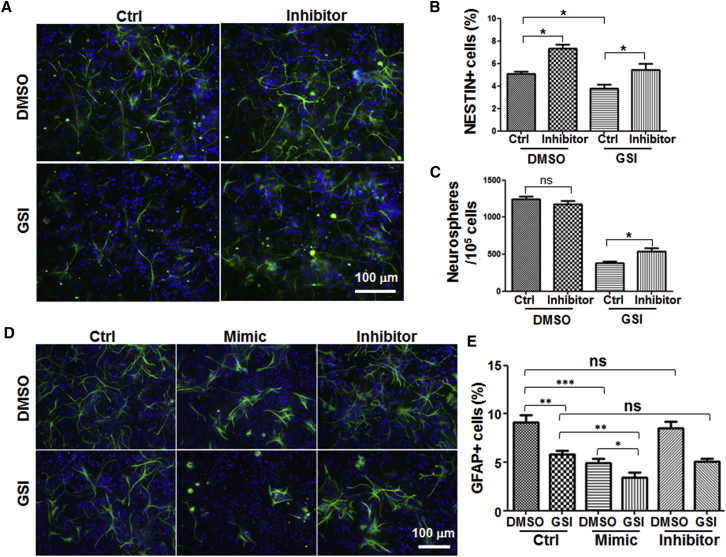
To further validate the functions of miR-342-5p by loss of function analyses, we transfected a miR-342-5p inhibitor into GSI-treated NS/PCs and analyzed the proliferation and differentiation of NS/PCs. The transfected NS/PCs cultured with DMSO showed increased NESTIN+ NSCs (Figures 7A and 7B), and when the Notch signal was inhibited by GSI, NS/PCs transfected with miR-342-5p inhibitor showed increased NESTIN+ NSCs and sphere formation compared with those of the control (Figures 7A–7C). These results indicated that miR-342-5p acts as a downstream effector to promote NSC differentiation upon Notch blockade.
Figure 7.
Inhibition of miR-342-5p Could Rescue Part of the Phenotypes Caused by Notch Blockade
(A and B) Adherent NS/PCs were treated with GSI or DMSO, and miR-342-5p inhibitor or control were transfected into these cells (four independent transfections performed). After 48 hr, NS/PCs were fixed and stained with anti-NESTIN antibody (A). The percentage of NESTIN+ cells within cells of one field was calculated (B) (eight fields counted in each group).
(C) NS/PCs treated as those in (A) were resuspended to form neurospheres. After 7 days, neurospheres were counted and compared. Four independent transfections were performed, and three wells of a 12-well-plate were counted as replicate samples in each group.
(D and E) NS/PCs treated as those in (A) were induced to differentiate for another 48 hr. Cells were then collected and stained with anti-GFAP antibody (D). The percentage of GFAP+ cells within cells of one field were analyzed (E) (eight fields counted in each group).
Bars, means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, not significant.
We then accessed the relationship between Notch signaling and miR-342-5p in astrocyte differentiation by blocking Notch signaling with GSI and transfecting miR-342-5p mimics or inhibitors. As shown in Figures 7D and 7E, GSI treatment reduced GFAP-positive astrocytes. When cells were transfected with miR-342-5p mimics and treated with GSI simultaneously, GFAP-positive cells were reduced further, suggesting that GSI and miR-342-5p repressed astrocyte differentiation additively. The miR-342-5p inhibitor did not promote astrocyte differentiation or rescue the GSI-induced astrocyte repression, suggesting that the miR-342-5p was necessary but not sufficient for astrocyte differentiation. It was likely that there were additional downstream targets of Notch signaling affecting astrocyte differentiation besides miR-342-5p, therefore inhibition of miR-342-5p alone failed in rescuing astrocyte differentiation.
Discussion
Notch signaling has been reported to play multiple roles during the CNS development. In progenitor cells, Notch signaling inhibits the differentiation of NSCs into INPs (Gao et al., 2009, Mizutani et al., 2007, Pierfelice et al., 2011). There are at least three types of neural progenitor cells in embryonic neocortex, including NSCs in the VZ, INPs in the VZ, and INPs in the SVZ. While a majority of INPs are in the SVZ, a fraction of INPs coexists with NSCs in the VZ. The Notch pathway is differentially used by these different types of progenitor cells. NSCs in the VZ, which have self-renewal and multipotent differentiation characteristics, exhibit RBP-J dependence. INPs in the VZ, in contrast, display attenuated RBP-J activity and are predominantly neurogenic. In addition, INPs in the SVZ signal to adjacent cells by expressing Notch ligands. A major unanswered question about the role of Notch signaling in NSCs-INPs is the downstream molecules controlling NSC differentiation. In this study, we have identified miR-342-5p as one of the downstream miRNAs of Notch signaling during NSC differentiation based on the following findings.
The expression of miR-342-5p is inversely correlated with Notch signaling in the tissues and cells examined. The in situ hybridization results of miR-342-5p showed that its expression gradually elevated from the VZ to the SVZ, which was complementary to the pattern of attenuated RBP-J-dependent Notch signaling from the VZ to the SVZ (Mizutani et al., 2007). In addition, modulations of Notch signaling led to changes in the expression of miR-342-5p. Blocking Notch signaling genetically or pharmaceutically in NS/PCs induced increased miR342-5p expression, whereas the activation of Notch signaling by NSC-specific NICD overexpression led to decreased miR342-5p expression. Since INPs increased in Notch blockade spheres (Gao et al., 2009), and miR-342-5p expression was upregulated in INPs compared with NSCs, it was difficult to judge whether the increase of miR-342-5p expression in Rbp-j KO spheres was only the result of INP enrichment or that miR-342-5p was directly regulated by Notch signaling, which then regulated the differentiation of NS/PCs in turn. To clarify this, we tested the expression profiles of miR-342-5p and INP marker Tα1 in neurospheres treated with GSI for different periods of time, and found that miR-342-5p was upregulated prior to the increase of Tα1, indicating that the increase of miR-342-5p expression after Notch blockade was not only a consequence of INP enrichment. Therefore, Notch signaling might directly regulate miR-342-5p expression. Although the intronic miRNA expression often occurs independent of host gene transcription, previous research has shown that the methylation of the Evl promoter region silenced the expression of both Evl and miR-342 together (Grady et al., 2008), indicating that the expression of miR-342 might be regulated by the same promoter of its host gene. Therefore, we investigated the regulation of the Evl promoter by Notch signaling. By using reporter assays, we have demonstrated that Notch signaling could directly regulate the transactivation of the promoter of Evl that harbors miR-342-5p and miR-342-3p genes. Because NICD repressed a truncated reporter in which the putative RBP-J-binding site was deleted, we propose that Notch signaling likely modulates the Evl promoter through both RBP-J and HES proteins. Indeed, ChIP assays have shown that HES1 and HES5 could bind to multiple N boxes and at least one E box in the Evl promoter. However, further investigations on NSC-specifically NICD-overexpressed mice showed that Hes5 expression was elevated, whereas Hes1 expression showed different changes in the cortex and GE, and miR-342-5p and Evl expressions were decreased on Notch activation. In addition, the knockdown of Hes5 by siRNA transfection into NS/PCs resulted in upregulated miR-342-5p and Evl expressions. Therefore, we thought that HES5 might directly regulate miR-342-5p expression through the Evl promoter instead of HES1 in vivo.
Functional analysis has shown that miR-342-5p promotes the differentiation of NSCs into INPs. Upregulation of miR-342-5p in cultured NS/PCs in vitro leads to reduced neurosphere formation and colony formation in the floating culture system and adherent culture system, respectively. Clonal analysis indicated that NSC clones were reduced while INP clones were increased. By using in utero electroporation in the embryo neocortex, we also found that upregulation of miR-342-5p reduced PAX6-positive NSCs and increased migrating neural cells that might undergo differentiating. These results suggest that enhanced expression of miR-342-5p could promote the differentiation of NSCs into INPs, and these phenotypes are consistent with that in Notch signal-blocked models (Gao et al., 2009, Hitoshi et al., 2002, Mizutani et al., 2007). Moreover, downregulation of miR-342-5p by its inhibitor transfection could partially rescue the decreased self-renewal and number of NSC neurospheres caused by the interrupted Notch signal. In summary, these results indicated that miR-342-5p acts as downstream effector to promote NSC differentiation into INPs on Notch blockade.
In addition to restricting NSCs differentiating into INPs, Notch signaling regulates differentiation of NS/PCs into astrocytes against neurons (Gaiano and Fishell, 2002). The expression of miR-342-5p is highest in neurons and lowest in astrocytes, suggesting a role of miR-342-5p in neuron and/or astrocyte differentiation. Indeed, our data have shown that miR-342-5p could inhibit the differentiation of NS/PCs into astrocytes. Furthermore, transfection of miR-342-5p in NS/PCs resulted in obvious morphological changes of differentiated astrocytes (and neurons to a lesser extent). On the other hand, neuronal differentiation was not enhanced at the expense of astrocyte commitment. Since miR-342-5p overexpression caused dramatically reduced numbers of astrocytes, which was supportive for neurons, the reduced number of neurons might be a successive defect of astrocyte deficiency. Further analyses should be done to clarify the role of miR-342-5p in neuronal differentiation in the future.
Although a number of target molecules of miR-342-5p have been predicted, with some of them verified by experiments (Wei et al., 2013), our results have identified GFAP as a target regulated by miR-342-5p. Since GFAP expression is fundamental for maintaining the identities of both RGCs and astrocytes, we could infer that as RBP-J-dependent Notch signaling is becoming attenuated in NSCs with their development, miR-342-5p increases and targets GFAP, which might result in NSCs losing their identity as radial glia and their potential change into astrocytes. In addition, we have found that the phosphorylated protein level of STAT3 was decreased after miR-342-5p overexpression in NS/PCs (Figure S5). STAT3 signaling has been shown to inhibit NSC transition to progenitors and promote astrocyte differentiation (Hong and Song, 2015). Therefore, the downregulation of STAT3 signaling might be one of the mechanisms of miR-342-5p functions on NS/PCs. Further analyses need to be done to investigate the direct or indirect regulation of STAT3 signaling by miR-342-5p during neurogenesis.
Besides the function on NS/PC differentiation, miR-342-5p overexpression inhibits proliferation and promotes apoptosis of NS/PCs. Further molecular analysis revealed that the total protein level of AKT was attenuated after miR-342-5p overexpression in NS/PCs, although the phosphorylated protein level of AKT was approximately stable (Figure S4F). These results indicate that the biological function of miR-342-5p on NS/PC proliferation and apoptosis might partially depend on targeting AKT, reminiscent of the same targeting mechanism of miR-342-5p in inflammatory macrophages during atherosclerosis (Wei et al., 2013). However, the accurate regulation relationship between miR-342-5p and AKT during neurogenesis, and their function on NS/PC proliferation and apoptosis, need further investigations.
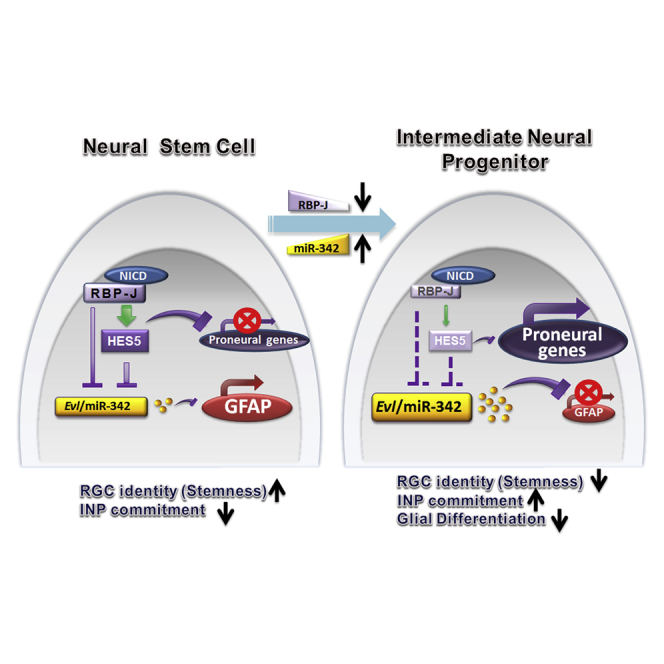
In summary, our results have shown that Notch signaling could inhibit the expression of miR-342-5p. In NSCs where Notch signaling is activated, the level of miR-342-5p is low and NSCs maintain the potential to differentiate into GFAP-positive astrocytes. When some NSCs gradually differentiate into INPs, RBP-J expression decreases and miR-342-5p expression increases in these cells (Figure S6). These cells might lose the ability to differentiate into GFAP-positive astrocytes, and become neurogenic INPs. Therefore, miR-342-5p could perform as a downstream molecule of Notch signaling to regulate the proliferation and differentiation of NSCs.
Experimental Procedures
All animal experiments were approved by the Animal Experiment Administration Committee of Fourth Military Medical University. All animal manipulations were carried out in accordance with the National Institute of Health guide for the care and use of Laboratory animals (NIH Publications, eighth edition), and all efforts were made to minimize the number of animals and their suffering.
Mice
Normal C57BL/6 mice, Rbp-j-floxed (Rbp-jf) mice, ROSA26-Stop-floxed-NICD mice (ROSA-Stopf-NICD), and Nestin-Cre (NesCre) mice were as described (Gao et al., 2009, Han et al., 2002, Zhao et al., 2016). NesCre mice were mated with Rbp-jf mice to obtain NesCre-Rbp-jf/f mice as cKO mice and NesCre-Rbp-jf/+ mice as controls. To achieve Notch activation in a wider range of NSCs, we utilized another NesCre mice stain (Jackson Laboratory 002859) to cross with ROSA-Stopf-NIC mice to obtain NesCre-ROSA-Stopf/+-NIC mice as conditional overexpressed mice and ROSA-Stopf/+-NIC mice as controls. All mice were maintained in a specific-pathogen-free facility. All animal experiments were approved by the Animal Experiment Administration Committee of the Fourth Military Medical University.
Author Contributions
H.H., M.H.Z., and G.J. designed and supervised the study. F.G. and Y.F.Z. cultured the primary NSCs and did the transfections. Z.P.Z. and L.A.F. performed analysis on transfected NSCs. X.L.C. produced all the mice used in this article. Y.Z.Z., C.J.G., and Y.Y.H. conducted the qRT-PCR analysis. X.C.Y. and Q.C.Y. constructed all the plasmids used in reporter assays and electroporation, F.G. and M.H.Z. performed the electroporation and in situ hybridization experiments. S.X.W., Y.Z.W., and X.H.Z. gave suggestions on the research and the manuscript. H.H., M.H.Z., and F.G. wrote the manuscript. All authors reviewed and approved the final manuscript.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation (31471044, 31071291, 31101041, 31130019, and 91339115) and the Ministry of Science and Technology of China (2015AA020918). The study was performed in the Graduates Innovation Center of the Fourth Military Medical University.
Published: March 23, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.02.017.
Contributor Information
Gong Ju, Email: jugong@fmmu.edu.cn.
Min-Hua Zheng, Email: minhua_zheng@126.com.
Hua Han, Email: huahan@fmmu.edu.cn.
Supplemental Information
References
- Bian S., Xu T.L., Sun T. Tuning the cell fate of neurons and glia by microRNAs. Curr. Opin. Neurobiol. 2013;23:928–934. doi: 10.1016/j.conb.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher J.M., Peterson S.M., Urs S., Zhang C., Liaw L. The miR-143/145 cluster is a novel transcriptional target of Jagged-1/Notch signaling in vascular smooth muscle cells. J. Biol. Chem. 2011;286:28312–28321. doi: 10.1074/jbc.M111.221945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C., Fink A., Lau C., Pham D., Daza R.A., Bulfone A., Kowalczyk T., Hevner R.F. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineberg S.K., Kosik K.S., Davidson B.L. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Franco S.J., Muller U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77:19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage F.H., Temple S. Neural stem cells: generating and regenerating the brain. Neuron. 2013;80:588–601. doi: 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- Gaiano N., Fishell G. The role of notch in promoting glial and neural stem cell fates. Annu. Rev. Neurosci. 2002;25:471–490. doi: 10.1146/annurev.neuro.25.030702.130823. [DOI] [PubMed] [Google Scholar]
- Gaiano N., Nye J.S., Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Gao F., Zhang Q., Zheng M.H., Liu H.L., Hu Y.Y., Zhang P., Zhang Z.P., Qin H.Y., Feng L., Wang L. Transcription factor RBP-J-mediated signaling represses the differentiation of neural stem cells into intermediate neural progenitors. Mol. Cell Neurosci. 2009;40:442–450. doi: 10.1016/j.mcn.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Grady W.M., Parkin R.K., Mitchell P.S., Lee J.H., Kim Y.H., Tsuchiya K.D., Washington M.K., Paraskeva C., Willson J.K., Kaz A.M. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene Evl in colorectal cancer. Oncogene. 2008;27:3880–3888. doi: 10.1038/onc.2008.10. [DOI] [PubMed] [Google Scholar]
- Hamidi H., Gustafason D., Pellegrini M., Gasson J. Identification of novel targets of CSL-dependent Notch signaling in hematopoiesis. PLoS One. 2011;6:e20022. doi: 10.1371/journal.pone.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H., Tanigaki K., Yamamoto N., Kuroda K., Yoshimoto M., Nakahata T., Ikuta K., Honjo T. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- Hitoshi S., Alexson T., Tropepe V., Donoviel D., Elia A.J., Nye J.S., Conlon R.A., Mak T.W., Bernstein A., van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16:846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeck J.D., Jandke A., Blake S.M., Nye E., Spencer-Dene B., Brandner S., Behrens A. Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat. Neurosci. 2010;13:1365–1372. doi: 10.1038/nn.2644. [DOI] [PubMed] [Google Scholar]
- Hong S., Song M.R. Signal transducer and activator of transcription-3 maintains the stemness of radial glia at mid-neurogenesis. J. Neurosci. 2015;35:1011–1023. doi: 10.1523/JNEUROSCI.2119-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T., Kedes L., Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Kohwi M., Doe C.Q. Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 2013;14:823–838. doi: 10.1038/nrn3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A., Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski A.V., Rubinson D.A., Dent E.W., Edward van Veen J., Leslie J.D., Zhang J., Mebane L.M., Philippar U., Pinheiro E.M., Burds A.A. Ena/VASP is required for neuritogenesis in the developing cortex. Neuron. 2007;56:441–455. doi: 10.1016/j.neuron.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Merkle F.T., Alvarez-Buylla A. Neural stem cells in mammalian development. Curr. Opin. Cell Biol. 2006;18:704–709. doi: 10.1016/j.ceb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Mizutani K., Yoon K., Dang L., Tokunaga A., Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Molofsky A.V., Pardal R., Iwashita T., Park I.K., Clarke M.F., Morrison S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor S.C., Martinez-Cerdeno V., Ivic L., Kriegstein A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Pierfelice T., Alberi L., Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Redmond L., Oh S.R., Hicks C., Weinmaster G., Ghosh A. Nuclear Notch1 signaling and the regulation of dendritic development. Nat. Neurosci. 2000;3:30–40. doi: 10.1038/71104. [DOI] [PubMed] [Google Scholar]
- Roese-Koerner B., Stappert L., Berger T., Braun N.C., Veltel M., Jungverdorben J., Evert B.O., Peitz M., Borghese L., Brustle O. Reciprocal regulation between bifunctional miR-9/9( ∗) and its transcriptional modulator Notch in human neural stem cell self-renewal and differentiation. Stem Cell Rep. 2016;7:207–219. doi: 10.1016/j.stemcr.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestan N., Artavanis-Tsakonas S., Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- Shi Y., Zhao X., Hsieh J., Wichterle H., Impey S., Banerjee S., Neveu P., Kosik K.S. MicroRNA regulation of neural stem cells and neurogenesis. J. Neurosci. 2010;30:14931–14936. doi: 10.1523/JNEUROSCI.4280-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigaki K., Nogaki F., Takahashi J., Tashiro K., Kurooka H., Honjo T. Notch1 and Notch3 instructively restrict bFGF-responsive multipotent neural progenitor cells to an astroglial fate. Neuron. 2001;29:45–55. doi: 10.1016/s0896-6273(01)00179-9. [DOI] [PubMed] [Google Scholar]
- Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- Vanderzalm P., Garriga G. Losing their minds: Mena/VASP/EVL triple knockout mice. Dev. Cell. 2007;13:757–758. doi: 10.1016/j.devcel.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Wei Y., Nazari-Jahantigh M., Chan L., Zhu M., Heyll K., Corbalan-Campos J., Hartmann P., Thiemann A., Weber C., Schober A. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation. 2013;127:1609–1619. doi: 10.1161/CIRCULATIONAHA.112.000736. [DOI] [PubMed] [Google Scholar]
- Yoon K., Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat. Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- Zhao J., Huang F., He F., Gao C., Liang S., Ma P., Dong G., Han H., Qin H. Forced activation of Notch in macrophages represses tumor growth by upregulating miR-125a and disabling tumor-associated macrophages. Cancer Res. 2016;76:1403–1415. doi: 10.1158/0008-5472.CAN-15-2019. [DOI] [PubMed] [Google Scholar]
- Zheng M.H., Shi M., Pei Z., Gao F., Han H., Ding Y.Q. The transcription factor RBP-J is essential for retinal cell differentiation and lamination. Mol. Brain. 2009;2:38. doi: 10.1186/1756-6606-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.