Abstract
The soil-borne fungus Fusarium oxysporum causes vascular wilt of a wide variety of plant species. F. oxysporum produces three kinds of asexual spores, macroconidia, microconidia, and chlamydospores. Falcate macroconidia are formed generally from terminal phialides on conidiophores and rarely from intercalary phialides on hyphae. Ellipsoidal microconidia are formed from intercalary phialides on hyphae. Globose chlamydospores with thick walls are developed by the modification of hyphal and conidial cells. Here we describe FoSTUA of F. oxysporum, which differentially regulates the development of macroconidia, microconidia, and chlamydospores. FoSTUA encodes a basic helix-loop-helix protein with similarity to Aspergillus nidulans StuA, which has been identified as a transcriptional regulator controlling conidiation. Nuclear localization of FoStuA was verified by using strains expressing FoStuA-green fluorescent protein fusions. The FoSTUA-targeted mutants exhibited normal microconidium formation in cultures. However, the mutants lacked conidiophores and produced macroconidia at low frequencies only from intercalary phialides. Thus, FoSTUA appears to be necessary to induce conidiophore differentiation. In contrast, chlamydospore formation was dramatically promoted in the mutants. These data demonstrate that FoStuA is a positive regulator and a negative regulator for the development of macroconidia and chlamydospores, respectively, and is dispensable for microconidium formation in cultures. The disease-causing ability of F. oxysporum was not affected by mutations in FoSTUA. However, the mutants produced markedly fewer macroconidia and microconidia in infected plants than the wild type. These results suggest that FoSTUA also has an important role for microconidium formation specifically in infected plants.
Asexual sporulation is a common reproductive mode for fungi. Asexual spores of higher fungi are generally called conidia (16). In many plant-pathogenic fungi, asexual spores are the infectious propagules responsible for initiating infection as well as disease dissemination (18). Knowledge of the molecular mechanisms controlling asexual sporulation in plant-pathogenic fungi will contribute to the search for a target for disease control through reducing primary inocula and spread of disease. However, the mechanisms of asexual sporulation remain largely undefined in plant-pathogenic fungi.
The vascular wilt fungus Fusarium oxysporum is an economically important soil-borne pathogen with a worldwide distribution (7, 9). This species includes intraspecific variants, called formae speciales, which cause vascular wilt in ∼80 botanical species (7, 9).
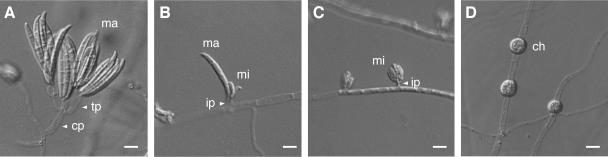
F. oxysporum is classified in the subdivision Deuteromycotina (Fungi Imperfecti) because it lacks sexual reproduction. Fusarium species that have sexual stages are classified in the subdivision Ascomycotina. F. oxysporum is unique in its asexual reproduction: it produces three kinds of asexual spores, macroconidia, microconidia, and chlamydospores (39, 40) (Fig. 1). Macroconidia are falcate and have three or four septa (Fig. 1A and B). Microconidia are ellipsoidal and have no septa or one septum (Fig. 1C). Globose chlamydospores have thick walls (Fig. 1D). Conidiogenesis of macroconidia and microconidia is phialidic (39, 40). Macroconidia are produced most often from terminal phialides that arise from conidiophores (Fig. 1A) and at low frequencies from intercalary phialides that arise directly from hyphae (Fig. 1B). Microconidia are produced from intercalary phialides in false heads (Fig. 1C). Chlamydospores are generally developed through the modification of hyphal and conidial cells through the condensation of their contents (Fig. 1D). These asexual spores play important roles in the disease cycle: macroconidia and microconidia are produced on the stem surfaces of infected plants and serve as secondary inocula to spread the fungus to neighboring host plants, and chlamydospores are endurance organs in soil and act as primary inocula when suitable host plants are planted in soil (17, 26, 39, 44, 45).
FIG. 1.
Conidiation of F. oxysporum. Strain Mel02010 was grown on SNA-paper at 25°C for 5 days. (A) Macroconidia (ma) are produced generally from terminal phialides (tp) on conidiophores (cp). (B) Macroconidia are also produced rarely from intercalary phialides (ip) on hyphae. mi, microconidia. (C) Microconidia are produced from intercalary phialides generally in false heads. (D) Chlamydospores (ch) are formed from hyphae. Bars, 10 μm.
A mutant screen of F. oxysporum f. sp. melonis, which causes vascular wilt of melon (31), was previously described; this screen was based on restriction enzyme-mediated integration mutagenesis (25, 42). A rensa mutant was isolated by screening conidiation mutants; “rensa” means catenation in Japanese (42). The affected gene, named REN1, which encodes a nuclear protein with similarity to MedA of Aspergillus nidulans and Acr1 of Magnaporthe grisea (42), was identified. MedA and Acr1 have been reported to act as developmental regulators of conidiation in these fungi (13, 15, 21, 30). Targeted mutation of REN1 results in the defective development of macroconidia and microconidia, and the mutants form rod-shaped, conidium-like cells directly from hyphae by acropetal division (42). However, the mutants exhibit normal growth and chlamydospore formation (42). Thus, REN1 is specifically required for the development of macroconidia and microconidia (42).
In A. nidulans, at least four regulatory genes (brlA, abaA, stuA, and medA) have been reported to control conidial development (1, 5, 12, 13, 15, 32-34). BrlA and AbaA comprise the core pathway required for the transition from vegetative hyphae to conidia, and brlA and abaA mutant strains are aconidial (1, 5, 12, 49). StuA and MedA are developmental modifiers required for correct cellular differentiation (13, 15, 19, 32, 33, 57); however, stuA and medA mutant strains produce conidia with normal viability (13, 15, 33).
The stuA gene encodes a basic helix-loop-helix (bHLH) transcriptional regulator (19). In A. nidulans, conidia are formed through the differentiation of a series of specialized cells: a conidiophore develops as an aerial hyphal branch that swells at its tip to form a globose vesicle, metulae bud from the vesicle, and then phialides bud from the metulae (2, 55). Conidia are formed through interstitial budding of the phialides in chains (2, 55). stuA mutants have extremely shortened conidiophores that lack metulae and phialides and produce conidia directly from conidiophore vesicles at low frequencies (15, 33). StuA has been reported to affect conidiation through the spatial and temporal regulation of brlA and abaA expression (19, 33). stuA mutants also show the stunted phenotype, resulting from shortened aerial hyphae, and are defective in sexual reproduction (15).
In other fungi, several proteins have been reported to contain segments highly similar to the bHLH region of StuA and have been termed APSES proteins (Asm-1, Phd1, StuA, Efg1, and Sok2) (6, 22, 53, 56). In Neurospora crassa, Asm-1 is involved in several processes, including spore germination, vegetative growth, and sexual sporulation (6). Phd1 and Sok2 of Saccharomyces cerevisiae and Efg1 of Candida albicans regulate pseudohyphal growth and morphology (22, 50, 53, 56).
Here we report the cloning and functional analysis of FoSTUA, the F. oxysporum homologue of stuA. FoStuA contains a segment highly similar to the bHLH motif of APSES proteins and localizes in nuclei. Targeted mutation of FoSTUA resulted in a lack of conidiophores, from which macroconidia are differentiated, and in increased chlamydospore formation. The mutants produced microconidia normally in cultures but not in infected plants. Thus, it appears that FoStuA differentially controls the developmental pathways for macroconidia, microconidia, and chlamydospores in F. oxysporum.
MATERIALS AND METHODS
Fungal strains, plasmids, and genomic library.
Strain Mel02010 (JCM9288) of F. oxysporum f. sp. melonis (38) was used in this study. Mel02010 and its transformants were maintained routinely on potato dextrose agar (PDA) (Difco).
The integrative transformation vectors pSH75 (28) and pII99 (37) were used for the transformation of F. oxysporum. These vectors carry hph (23) and nptII (8), respectively, fused to the A. nidulans trpC promoter and terminator (36).
A genomic cosmid library of Mel02010 constructed in cosmid vector pMLF2 (4) was described previously (42). Screening of the library by colony hybridization was conducted by the standard method (46).
Fungal transformation.
Protoplast preparation and transformation of F. oxysporum were performed as previously described (24, 25). Transformants carrying hph or nptII were selected on regeneration medium containing hygromycin B (Wako Pure Chemicals) at 60 μg/ml or Geneticin (Gibco) at 180 μg/ml, respectively (24, 25).
Test for conidiation.
To induce conidiation in F. oxysporum strains, two media were used: synthetic low-nutrient agar medium (SNA), containing (in weight/volume) 0.1% KH2PO4, 0.1% KNO3, 0.05% MgSO4 · 7H2O, 0.05% KCl, 0.02% glucose, 0.02% sucrose, and 2% agar (41), and liquid carboxymethyl cellulose medium (CMC), containing (in weight/volume) 1.5% carboxymethyl cellulose, 0.1% yeast extract, 0.1% NH4NO3, 0.1% KH2PO4, and 0.05% MgSO4 · 7H2O (14). Sterilized filter paper (1 by 2 cm) was placed on the SNA surface before the inoculation of fungal strains (41).
Strains were grown on PDA at 25°C for 5 days. Agar blocks (3 mm in diameter) carrying mycelia were prepared from the resulting colonies and inoculated onto SNA 5 mm away from the filter paper pieces. After incubation at 25°C for 5 days under continuous black blue light (Toshiba FL15BLB), conidiation was observed with a light microscope (Olympus BX50). The number of chlamydospores formed in a 5-mm square between the agar block and the filter paper was counted.
A PDA block carrying mycelium was inoculated into 50 ml of CMC in a 100-ml Erlenmeyer flask and incubated at 25°C for 4 days on an orbital shaker (100 rpm) under continuous fluorescent light (14). The numbers of macroconidia and microconidia were counted with a microscope. Conidia were fixed in 0.4% p-formaldehyde and stained with Fluostain I (25 μg/ml; Dojin) and Hoechst 33258 (250 μg/ml; Wako) to visualize cell walls and nuclei, respectively, as previously described (42). Stained cells were observed with a BX50 fluorescence microscope and a U-MWU filter (Olympus).
For the time course study of conidial development, a PDA block carrying mycelium was inoculated into 25 ml of liquid complete medium (CM) (47) in a 50-ml Erlenmeyer flask and incubated at 25°C for 18 h on an orbital shaker (100 rpm). The resulting mycelium was collected by centrifugation at 1,600 × g for 10 min, inoculated into 50 ml of CMC in a 100-ml Erlenmeyer flask, and incubated as described above. During incubation, the numbers of macroconidia and microconidia were counted with a microscope at 12-h intervals from 0 to 132 h. At 0, 12, 24, 36, 48, 60, 72, 96, and 120 h after inoculation into CMC, fungal tissues were collected from five flasks by centrifugation and subjected to RNA isolation as described below.
Test for vegetative growth and pathogenicity.
To test for the vegetative growth of F. oxysporum strains, the strains were grown on three agar media: PDA, complete medium agar (CMA) (47), and minimal medium agar (MMA) (47). PDA blocks (3 mm in diameter) carrying mycelia were inoculated onto PDA, MMA, and CMA. After incubation at 25°C for 5 days, colony growth and morphology were observed.
Pathogenicity was tested by a root dip method with susceptible melon (Cucumis melo L.) cultivar Amus and conidial suspensions (∼107 conidia/ml in sterilized water) as previously described (25). Disease symptoms were assessed 3 weeks after inoculation.
Nucleic acid analysis.
Isolation of total DNA from F. oxysporum and DNA gel blot hybridization were performed as previously described (25, 38). Isolation of total RNA from F. oxysporum, preparation of poly(A)+ RNA, and RNA gel blot hybridization were performed as previously described (42, 43).
For analysis of nucleotide sequences, DNA was cloned in pBluescript KS(+) (Stratagene) or pGEM-T Easy (Promega). DNA sequences were determined by using a BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) and an automated DNA sequencer (model 373A; Applied Biosystems). DNA sequences were analyzed with BLAST (3). Nucleotide and amino acid sequences were aligned with the CLUSTAL W program (54).
Isolation of FoSTUA.
The FoSTUA fragment was amplified from total DNA of strain Mel02010 by PCR with the primers stuA-1f (AATGGCATGATCAACGGCAC) and stuA-1r (AGATGCATCGGGCCAATCTT) and with Taq DNA polymerase (Takara). These primers were designed on the basis of the conserved bHLH-encoding regions of A. nidulans stuA (19, 33) and N. crassa asm-1 (6). The PCR products were cloned in vector pGEM-T Easy. Several clones were sequenced, and one clone was found to contain a PCR product with the expected size of ∼130 bp and to encode a peptide with strong similarity to corresponding regions of APSES proteins. This PCR product was used as a probe for screening of a genomic cosmid library of Mel02010, and a positive clone, named pcSTUAh-1, was isolated (Fig. 2A). A 6.0-kb region containing the stuA homologue was sequenced, and a putative open reading frame (ORF) for FoSTUA was identified.
FIG.2.
F. oxysporum FoSTUA gene. (A) Map of the FoSTUA locus. The bars pointing to the right indicate FoSTUA with introns (white segments). The 0.8-kb BglII-SalI fragment of FoSTUA was cloned in pBluescript KS(+) to make pS1BS, which was used as a probe for RNA gel blot analysis. Arrowheads above FoSTUA denote the orientations and locations of the oligonucleotide primers (Sf1 and Sr1) used in RT-PCR experiments. B, BglII; H, HindIII; S, SalI. (B) Similarity of FoStuA to other proteins. The amino acid sequence of FoStuA (Fo) was aligned with those of Asm-1 of N. crassa (Nc; Genbank accession no. CAD70882), StuA of A. nidulans (An; accession no. AAA33325), and StuA of P. marneffei (Pm; accession no. AAM27919). Amino acids that are conserved between FoStuA and any of the others are indicated as white letters on a black background. The most conserved bHLH motifs among the four sequences are underlined (amino acids 84 to 184 of FoStuA).
FoSTUA cDNA was isolated by reverse transcription (RT)-PCR by using an RNA PCR kit, version 2.1 (Takara). The cDNA was amplified from total RNA (1 μg) of strain Mel02010 with the primers Sf1 (ATGAACCAAGGCCATCCCCA) and Sr1 (CTAACCGAAAGATTGTTGTC) (Fig. 2A) according to the manufacturer's instructions; Sf1 and Sr1 contain the FoSTUA initiation and termination codons (italic type), respectively. The RT-PCR products were cloned in vector pGEM-T Easy to determine the sequences.
Construction of FoSTUA-GFP gene fusion vectors.
The green fluorescent protein (GFP) expression vectors pYTGFP-N and pYTGFP-C were used to make FoSTUA-GFP gene fusion vectors. These vectors contain the GFP (enhanced GFP [EGFP]) ORF fused to the A. nidulans trpC promoter and terminator (24, 27, 36). Vectors pYTGFP-N and pYTGFP-C were made for N-terminal and C-terminal fusions of GFP, respectively, to target proteins (24). As a control, we used plasmid pYTGFPc, which carries only the GFP ORF under the control of the trpC promoter and terminator (24).
FoSTUA cDNA was amplified from poly(A)+ RNA of strain Mel02010 by RT-PCR with the primers FoSTUA1N-f (AAAGGATCCTGATGAACCAAGGCCATCCCC) and FoSTUAN-r (CCCCTGCAGCTAACCGAAAGATTGTTGTCT); FoSTUAN-f has a BamHI site (underlined) with the initiation codon (italic type), and FoSTUAN-r has a PstI site (underlined) with the termination codon (italic type). The amplified DNA was digested with BamHI and PstI and cloned into the BamHI-PstI site of pYTGFP-N to make pGFP-FoSTUA, resulting in an N-terminal fusion of GFP to FoStuA.
FoSTUA cDNA was also prepared from poly(A)+ RNA of strain Mel02010 by RT-PCR with the primers FoSTUAC-f (ACATCTAGATGAACCAAGGCCATCCCCAGC) and FoSTUAC-r (CTCCTGCAGTGCTCTTGGTGCAGGTTCTGC); FoSTUAC-f contains an XbaI site (underlined) with the initiation codon (italic type), and FoSTUAC-r contains a PstI site (underlined) fused to the last codon of FoSTUA. The amplified DNA was digested with XbaI and PstI and cloned into the XbaI-PstI site of pYTGFP-C to make pFoSTUA-GFP, resulting in a C-terminal fusion of GFP to FoStuA.
All of the PCR products cloned in the vectors were sequenced to confirm the fact that no nucleotide substitution had occurred during amplification.
Nucleotide sequence accession number.
The FoSTUA sequence has been deposited in the DDBJ/EMBL/GenBank databases under accession number AB180746.
RESULTS
Isolation of FoSTUA.
To isolate the APSES gene homologue from F. oxysporum, part of the homologue was amplified from DNA of strain Mel02010 by PCR. The PCR product was used as a probe to screen a cosmid genomic library of Mel02010. A positive clone, pcSTUAh-1, was isolated and partially restriction mapped (Fig. 2A). Sequencing of a 6.0-kb region in pcFoSTUA-1 revealed a putative ORF encoding a protein with similarity to APSES proteins.
cDNA was prepared from total RNA of strain Mel02010 by RT-PCR with primers Sf1 and Sr1 (Fig. 2A). RT-PCR amplification produced a 1.7-kb fragment of DNA. Comparison of the genomic sequence with that of the RT-PCR product indicated that the gene has two introns (52 and 65 bp) and three exons (340, 133, and 1177 bp) and potentially encodes a 550-amino-acid protein.
A BLAST database search revealed that the deduced amino acid sequence has significant similarity to those of APSES proteins, such as N. crassa Asm-1 (6), Penicillium marneffei StuA (11), and A. nidulans StuA (33)—32.1, 28.1, and 27.1% identical, respectively (Fig. 2B). Thus, the gene containing the ORF of interest was named FoSTUA. The similarity was most evident in amino acids 84 to 184 of F. oxysporum, which correspond to the bHLH region (6, 11, 19, 33) (Fig. 2B).
Expression of FoSTUA.
Fusarium species are known to produce conidia when they are grown in CMC (14). Ohara et al. previously observed that F. oxysporum also produces macroconidia and microconidia in CMC but not in CM or liquid minimal medium (MM) (42).
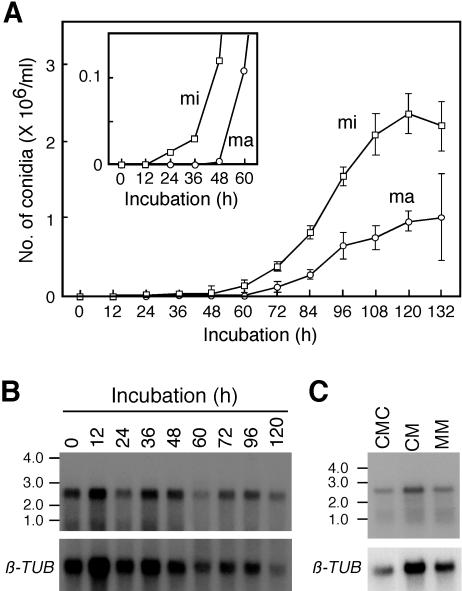
The timing of conidial development and FoSTUA expression was observed in CMC. Strain Mel02010 was grown in CM at 25°C for 18 h. The resulting mycelia were inoculated into CMC and incubated at 25°C on an orbital shaker. Microconidia and macroconidia first appeared at 24 and 48 h, respectively, after inoculation into CMC, and their numbers increased up to 120 h (Fig. 3A). Poly(A)+ RNA was prepared from fungal tissues, and the gel blot was probed with pS1BS containing the FoSTUA fragment (Fig. 2A). The probe hybridized to 2.7-kb bands in RNAs from inoculated mycelia which had been grown in CM and from fungal tissues grown in CMC with similar signal intensities (Fig. 3B). This result suggested that FoSTUA is constitutively expressed during conidiation as well as during vegetative growth.
FIG. 3.
Time course of conidial development and FoSTUA expression. (A) Time course of conidial development. Mycelia of strain Mel02010 grown in CM were inoculated into CMC and incubated at 25°C for 132 h. The numbers of macroconidia (ma) and microconidia (mi) were counted at 12-h intervals with a microscope; data represent the means and standard deviations of five replications. The inset shows the time course of conidial development during the initial 60 h of incubation on an expanded scale. (B) Time course of FoSTUA expression during conidiation. Fungal tissues of strain Mel02010 grown for the indicated periods were collected, and poly(A)+ RNA was prepared from the tissues. Poly(A)+ RNA (∼5 μg/lane) was electrophoresed in a 1.5% agarose gel containing 2.2 M formaldehyde. The blot was probed with pS1BS containing the FoSTUA fragment (Fig. 2A). Sizes (in kilobases) of marker RNA fragments (Novagen) are indicated on the left. The blot was also probed with pFOTUB1 containing the β-tubulin gene fragment (β-TUB) (42). (C) FoSTUA expression during conidiation and vegetative growth. Strain Mel02010 was grown in CMC, CM, and MM at 25°C for 4 days, and poly(A)+ RNA was prepared from the cultures. The RNA gel blot was probed with pS1BS and pFOTUB1.
To verify the expression of FoSTUA during vegetative growth, strain Mel02010 was grown in CM and MM, in which conidiation is not induced, at 25°C for 4 days, and poly(A)+ RNA was prepared from the cultures. The RNA gel blot was probed with pS1BS (Fig. 2A). The probe hybridized to 2.7-kb bands in RNAs from fungal tissues grown in CMC, CM, and MM (Fig. 3C). Thus, FoSTUA is expressed by F. oxysporum not only during conidiation but also during vegetative growth.
Intracellular localization of GFP-tagged FoStuA.
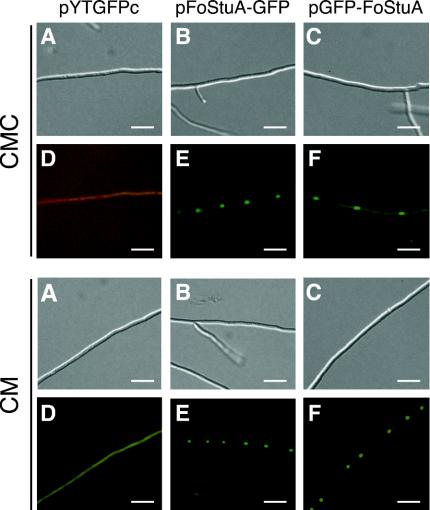
FoStuA contains a bHLH structure, and its amino acid sequence is significantly similar to those of APSES proteins (Fig. 2B), which have been considered to be transcriptional regulators (6, 11, 19, 22, 53, 56). To test the intracellular localization of FoStuA, we made strains expressing FoStuA-GFP and GFP-FoStuA fusions and observed the intracellular distribution of the GFP fluorescence in the strains by fluorescence microscopy. We constructed FoSTUA-GFP and GFP-FoSTUA gene fusions under the control of the A. nidulans trpC promoter as pFoStuA-GFP and pGFP-FoStuA, respectively. These constructs were introduced into strain Mel02010 by cotransformation with plasmid pSH75, conferring hygromycin B resistance. As a control, Mel02010 was transformed with plasmid pYTGFPc, which carries the GFP gene under the control of the trpC promoter.
Transformants were grown in CMC and CM, and their mycelia were observed with a fluorescence microscope. Of 10 pYTGFPc transformants, 7 expressed GFP. In these transformants, GFP fluorescence did not localize in any cell components throughout mycelia grown in CMC and CM (Fig. 4). Of 10 pFoStuA-GFP transformants, 6 emitted GFP fluorescence; of 13 pGFP-FoStuA transformants, 6 emitted GFP fluorescence. In all of the transformants expressing FoStuA-GFP or GFP-FoStuA fusions, GFP fluorescence was targeted to nuclei in mycelial cells grown in CMC and CM (Fig. 4), suggesting that FoStuA localizes in nuclei. These results were consistent with the prediction that FoStuA acts as a transcriptional regulator.
FIG. 4.
Intracellular localization of GFP-tagged FoStuA. Transformants with pYTGFPc (A and D), pFoStuA-GFP (B and E), and pGFP-FoStuA (C and F) were grown in CMC and CM at 25°C for 4 days. The resulting mycelia were observed with a fluorescence microscope and a U-MWIG filter (Olympus) for GFP fluorescence. Bars, 20 μm. (A to C) Differential interference contrast images. (D to F) GFP fluorescence images.
Targeted deletion of FoSTUA.
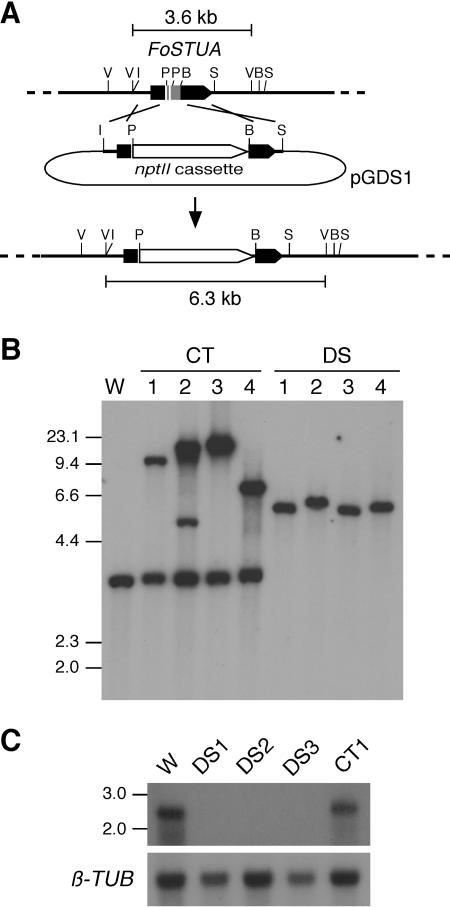
To determine the function of FoSTUA in developmental processes in F. oxysporum, homologous recombination was used to replace FoSTUA with plasmid pGDS1, which contains the nptII cassette flanked by 5′ and 3′ sequences from FoSTUA (Fig. 5A). Strain Mel02010 was transformed with pGDS1, and 36 transformants were isolated. Transformants were grown on SNA-paper. All of the transformants produced macroconidia, microconidia, and chlamydospores, which were similar in morphology to those of the wild type (Fig. 1). However, 29 transformants were distinguishable from the wild type in the quantities of macroconidia and chlamydospores produced: the numbers of macroconidia and chlamydospores were markedly decreased and increased, respectively, in these transformants. Microscopic observation showed that these transformants completely lacked conidiophores and formed macroconidia only from intercalary phialides that arose directly from hyphae.
FIG. 5.
Transformation-mediated targeting of FoSTUA. (A) Structure of the FoSTUA locus before and after homologous integration of the FoSTUA-targeting vector pGDS1. To make pGDS1, a 0.9-kb EcoRI-PvuII fragment and a 0.8-kb BglII-SalI fragment containing the 5′ and 3′ regions of FoSTUA, respectively, were cloned into transformation vector pII99. B, BglII; I, EcoRI; V, EcoRV; P, PvuII; S, SalI. (B) DNA gel blot analysis of pGDS1 transformants. Total DNA (∼2 μg/lane) from the wild-type strain and the pGDS1 transformants was digested with EcoRV and fractionated in a 0.8% agarose gel. The blot was probed with the 0.8-kb BglII-SalI fragment from pGDS1. Sizes (in kilobases) of marker DNA fragments (HindIII-digested λ DNA) are indicated on the left. W, wild-type strain Mel02010; CT1 to CT4, transformants showing normal conidiation; DS1 to DS4, transformants lacking conidiophores. (C) RNA gel blot analysis of pGDS1 transformants. The wild-type strain and the pGDS1 transformants were grown in CMC at 25°C for 4 days, and poly(A)+ RNA was prepared from the cultures. Poly(A)+ RNA (∼5 μg/lane) was electrophoresed in a 1.5% agarose gel containing 2.2 M formaldehyde. The blot was probed with pS1BS containing the FoSTUA fragment (Fig. 2A). Sizes (in kilobases) of marker RNA fragments are indicated on the left. The blot was also probed with pFOTUB1 containing the β-tubulin gene fragment (β-TUB) (42).
The mode of integration of pGDS1 in four transformants (DS1 to DS4) lacking conidiophores was analyzed by DNA gel blot analysis. As controls, four transformants (CT1 to CT4) showing wild-type conidiation were also used. Total DNAs of strain Mel02010 and transformants were digested with EcoRV, and the blot was probed with the 0.8-kb BglII-SalI fragment integrated in pGDS1 (Fig. 5A). The probe hybridized to ∼3.6-kb EcoRV fragments in Mel02010 and the CT transformants (Fig. 5B). However, all of the DS transformants lacked 3.6-kb bands (Fig. 5B). DS1, DS3, and DS4 had ∼6.3-kb bands resulting from homologous recombination, and DS2 had an ∼6.4-kb band (Fig. 5B). The homologous integration of pGDS1 in DS2 was probably accompanied by a deletion of the flanking region.
The expression of FoSTUA in the FoSTUA deletion (ΔFoSTUA) strains was determined by RNA gel blot analysis. Poly(A)+ RNAs were prepared from fungal tissues of strains Mel02010, DS1, DS2, DS3, and CT1 grown in CMC. The RNA gel blot was probed with pS1BS (Fig. 2A). The probe hybridized to 2.7-kb bands in Mel02010 and CT1 but not to any bands in DS1, DS2, and DS3 (Fig. 5C). DNA and RNA gel blot analyses clearly showed that targeted mutation of FoSTUA caused a deficiency in conidiophore development and promoted chlamydospore formation in DS transformants.
Macroconidia, microconidia, and chlamydospores of ΔFoSTUA mutants.
To further analyze the role of FoSTUA in the conidiation of F. oxysporum, strain Mel02010, ΔFoSTUA mutants (DS1 to DS3), and an ectopic transformant (CT1) were grown in CMC, and their macroconidia and microconidia were qualitatively and quantitatively observed. The ΔFoSTUA mutants produced macroconidia and microconidia which were not different in morphology from those of Mel02010 and CT1 (Fig. 6A). Conidia were stained with Fluostain I and Hoechst 33258 to visualize cell walls and nuclei, respectively. Macroconidia and microconidia from Mel02010 and the mutants contained a single nucleus in each cell (Fig. 6A), suggesting that mutation of FoSTUA does not affect the coupling of cell division and nuclear division during conidiation.
FIG. 6.
Conidiation of ΔFoSTUA mutants. (A) Macroconidia and microconidia of wild-type and ΔFoSTUA mutant strains. Conidia were stained with Fluostain I and Hoechst 33258 to visualize cell walls and nuclei, respectively, and were observed with differential interference contrast (DIC) and fluorescence microscopes. Bars, 10 μm. (B and C) Numbers of macroconidia and microconidia produced by wild-type and ΔFoSTUA mutant strains. Strains were grown in CMC at 25°C for 4 days. The resulting macroconidia and microconidia were counted; data represent the means and standard deviations of three replications. Mel02010, wild type; DS1 to DS3, ΔFoSTUA mutants; CT1, ectopic transformant.
The formation of macroconidia and the formation of microconidia were quantitatively compared between wild-type and mutant strains. The numbers of macroconidia were clearly reduced in the ΔFoSTUA mutants (Fig. 6B). Observation of fungal tissues grown in CMC showed that strains Mel02010 and CT1 produced many conidiophores. The mutants, however, produced no conidiophore-like structures (data not shown), suggesting that the mutants produced macroconidia only from intercalary phialides on hyphae. In contrast, the numbers of microconidia did not differ significantly between wild-type and mutant strains (Fig. 6C). These results indicate that FoSTUA is essential for the development of conidiophores, from which macroconidia arise, and is dispensable for microconidium formation in cultures.
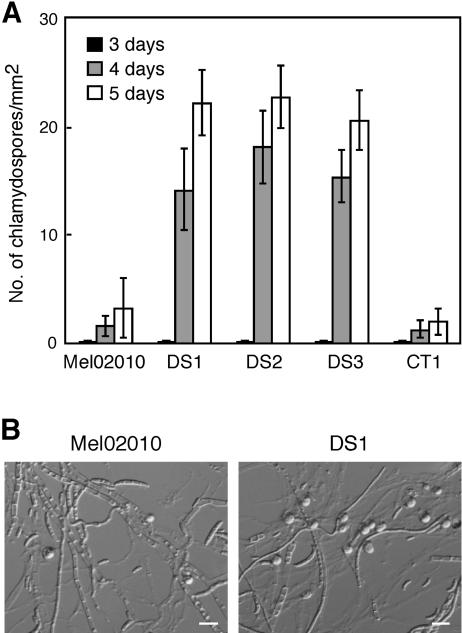
To observe chlamydospore formation, strain Mel02010, three mutants (DS1 to DS3), and strain CT1 were grown on SNA-paper. Wild-type and mutant strains produced chlamydospores from 3 days after inoculation, and the numbers increased by 4 and 5 days (Fig. 7A). Chlamydospores of the mutants were similar in morphology to those of the wild type (Fig. 7B). However, the mutants produced many more chlamydospores than the wild type—6.7 to 7.5 times more than Mel02010 at 5 days of incubation (Fig. 7). These results indicate that mutation of FoSTUA leads to marked promotion of chlamydospore formation. It is likely that FoStuA has contrary functions in the developmental pathways of macroconidia and chlamydospores.
FIG. 7.
Chlamydospore formation by ΔFoSTUA mutants. (A) Numbers of chlamydospores produced by wild-type and ΔFoSTUA mutant strains. Strains were grown on SNA-paper at 25°C, and the numbers of chlamydospores produced were counted at 3, 4, and 5 days of incubation. Data represent the means and standard deviations of three replications. Mel02010, wild type; DS1 to DS3, ΔFoSTUA mutants; CT1, ectopic transformant. (B) Microscopic photographs of strains Mel02010 and DS1 grown on SNA-paper. Photographs were taken at 5 days of incubation. Globose organs are chlamydospores. Bars, 30 μm.
Vegetative growth and pathogenicity of ΔFoSTUA mutants.
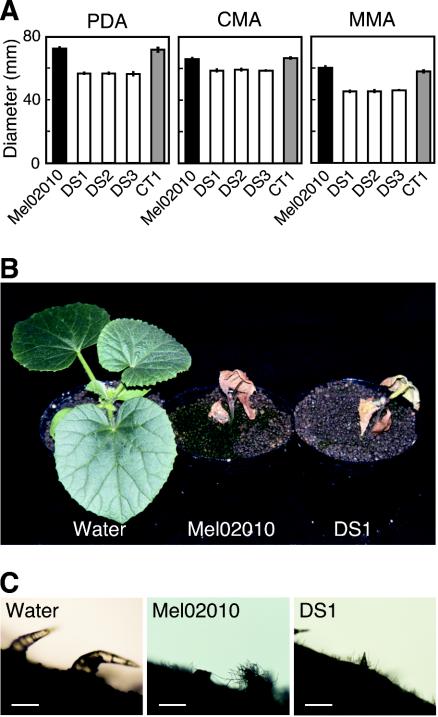
Vegetative growth of ΔFoSTUA mutants (DS1 to DS3) was evaluated by measuring the diameters of colonies grown on three agar media, PDA, CMA, and MMA, at 25°C for 5 days. On all of the media, the colonies of the mutants were slightly smaller than those of strains Mel02010 and CT1 (Fig. 8A). Although the wild-type strain produced abundant aerial hyphae on all media, the mutants produced fewer aerial hyphae, especially on MMA (data not shown). These results indicate that FoSTUA is involved in but is not essential for vegetative growth.
FIG. 8.
Vegetative growth and pathogenicity of ΔFoSTUA mutants. (A) Colony growth of wild-type and ΔFoSTUA mutant strains. Strains were grown on PDA, CMA, and MMA at 25°C for 5 days. Colony diameters were measured; data represent the means and standard deviations of three replications. Mel02010, wild type; DS1 to DS3, ΔFoSTUA mutants; CT1, ectopic transformant. (B and C) Pathogenicity of and colonization by wild-type and ΔFoSTUA mutant strains. Seedlings of cultivar Amus with a single true leaf were inoculated with conidial suspensions (∼107 conidia/ml) of wild-type strain Mel02010 and ΔFoSTUA mutant DS1 by a root dip method. Control plants were immersed in water. Photographs of plants were taken 3 weeks after inoculation (B). Stem surfaces immediately above crowns were observed with a microscope (C). Bars, 100 μm.
ΔFoSTUA mutants (DS1 to DS3) were tested for pathogenicity for melon plants. Fifteen seedlings of susceptible cultivar Amus with a single true leaf were inoculated with a conidial suspension of each strain by the root dip method (25). The mutants caused typical wilt symptoms on all seedlings, as did the wild type. Figure 8B shows disease symptoms caused by strains Mel02010 and DS1 as examples. The timing of symptom appearance and the severity of symptoms were almost the same on plants inoculated with wild-type and mutant strains, indicating that the ΔFoSTUA mutants retained the abilities to infect host plant tissues and to cause disease symptoms under the conditions tested.
F. oxysporum invades from roots of host plants, colonizes roots and stems, and produces conspicuous masses of macroconidia and microconidia on stem surfaces at the late stage of symptom development (26, 44, 45). We observed mycelia of wild-type and mutant strains growing on stem surfaces 3 weeks after inoculation. The mutants colonized stems and produced mycelia on the surfaces, as did the wild type (Fig. 8C), suggesting that FoSTUA is dispensable for colonization of host plants.
We also observed conidiation of wild-type and mutant strains in infected plants. Stems (∼1.0 cm) immediately above crowns were cut out from diseased plants and soaked in sterilized water for suspending conidia. The numbers of macroconidia and microconidia in the suspensions were counted under a microscope. Although the wild-type strain produced large number of macroconidia in infected plants, the mutants produced very few macroconidia (Table 1). Although the mutants showed normal microconidium formation in CMC (Fig. 6C), they produced 10- to 20-fold fewer microconidia than the wild type in infected plants (Table 1). This result suggests that FoSTUA may have more critical roles in conidiation in host plants than in cultures.
TABLE 1.
In planta conidiation of ΔFoSTUA mutantsa
| Inoculumb | Mean ± SD no. (104/cm of stem) of:
|
|
|---|---|---|
| Macroconidia | Microconidia | |
| Water | None | None |
| Mel02010 | 62.6 ± 21.7 | 5.0 ± 1.2 |
| DS1 | 0.4 ± 0.6 | 0.2 ± 0.5 |
| DS2 | 0.2 ± 0.5 | 0.2 ± 0.5 |
| DS3 | 0.4 ± 0.6 | 0.4 ± 0.6 |
| CT1 | 50.6 ± 17.8 | 4.0 ± 0.7 |
Seedlings of cultivar Amus were inoculated with a conidial suspension (∼107 conidia/ml) of each strain by a root dip method. Control plants were immersed in water. Stem segments (∼1.0 cm) immediately above crowns were cut out from inoculated plants 3 weeks after inoculation. The segments were soaked in sterilized water (1.0 ml) and vigorously shaken to suspend conidia. The numbers of macroconidia and microconidia in the suspensions were counted with a microscope; data are from five replications.
Mel02010, wild type; DS1 to DS3, ΔFoSTUA mutants; CT1, ectopic transformant.
DISCUSSION
FoSTUA is essential for conidiophore development.
In this study, we isolated FoSTUA, which differentially regulates the development of three kinds of asexual spores, macroconidia, microconidia, and chlamydospores, in F. oxysporum. FoStuA belongs to a group of APSES proteins which contain highly conserved bHLH motifs of ∼100 amino acids (6, 19, 22, 33, 53, 56). The APSES proteins have been identified to regulate developmental processes in fungi, such as the formation of metulae and phialides in A. nidulans and P. marneffei (11, 19, 33), sexual reproduction in N. crassa (6), pseudohypal growth in S. cerevisiae (22, 50, 56), and dimorphic switching and chlamydospore formation in C. albicans (51, 53).
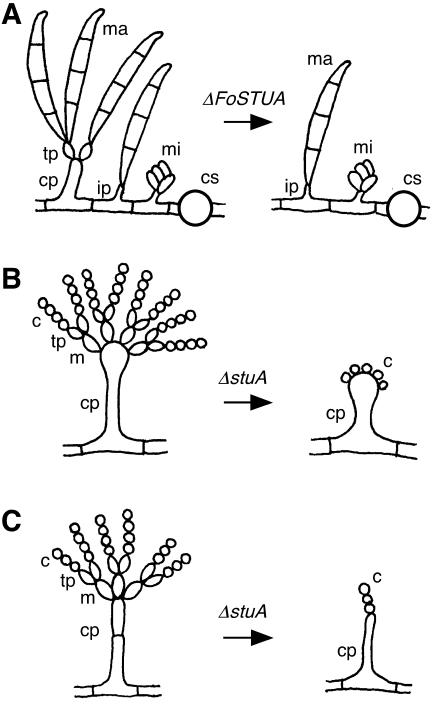
Targeted mutation of FoSTUA in F. oxysporum causes a lack of production of conidiophores, from which terminal phialides differentiate, followed by macroconidia that arise from phialides by basipetal division (Fig. 9A). ΔFoSTUA mutants, however, retain the abilities to differentiate intercalary phialides and to form macroconidia from intercalary phialides at low frequencies (Fig. 9A). Thus, the mutants produce macroconidia at lower frequencies than the wild type. In contrast, the mutants normally produce microconidia, because microconidia are generated from intercalary phialides. These results indicate that FoSTUA is required specifically for the initiation of conidiophore differentiation from hyphae in macroconidium development.
FIG. 9.
Schematic representation of conidiogenesis of ΔFoSTUA, ΔstuA, and ΔstuA mutants of F. oxysporum (A), A. nidulans (B), and P. marneffei (C), respectively. ma, macroconidium; mi, microconidium; tp, terminal phialide; ip, intercalary phialide; cp, conidiophore; cs, chlamydospore; m, metula; c, conidium.
Of the fungal APSES proteins, the StuA proteins of A. nidulans and P. marneffei have been identified to be required for asexual spore development (11, 19, 33, 57). The modes of conidiogenesis in these two fungi are similar and consist of the sequential development of conidiophores, metulae, phialides, and conidia (Fig. 9B and C). In addition to stuA, the abaA homologue of P. marneffei has been isolated and characterized as a conidiation regulator (10). ΔstuA mutants of these fungi can form conidiophores but not metulae and phialides (11, 19, 33, 57) (Fig. 9B and C). The mutants, however, produce conidia directly from conidiophores (11, 19, 33, 57) (Fig. 9B and C). Thus, the StuA proteins of these fungi are essential for the differentiation of two types of sterigmata (metulae and phialides). In contrast, FoStuA of F. oxysporum is required primarily for the development of conidiophores but not phialides. These data suggest that FoStuA of F. oxysporum has a role in conidiation that is different from the roles of the StuA proteins of A. nidulans and P. marneffei.
Ohara et al. isolated REN1, which encodes a putative transcriptional regulator, from a restriction enzyme-mediated integration transformant showing abnormal conidiation (42). Ren1 shows significant similarity to A. nidulans MedA (13). Mutation of medA results in aberrant conidiophores with branching chains of metulae, delayed differentiation of conidia, and frequent reinitiation of secondary conidiophores (13, 15, 21). Conidiation of the medA mutant is not completely blocked but is delayed (13, 15, 21). However, mutation of REN1 in F. oxysporum leads to a lack of normal conidiophores and phialides and to the production of chains of rod-shaped, conidium-like cells directly from hyphae (42). The conidium-like cells also branch, unlike cells and conidia during normal conidiogenesis. Thus, Ren1 appears to be a core component of the conidiogenesis pathway in F. oxysporum, while its homologue MedA acts only as a modifier of this pathway in A. nidulans (13, 15, 21). Our data concerning the functions of FoStuA and Ren1 suggest that the molecular machinery underlying conidiation in F. oxysporum differs significantly from that in A. nidulans.
Recently, the genome sequence of F. graminearum became available (http://www-genome.wi.mit.edu/annotation/fungi/fusarium/). On the website, we searched for the F. graminearum homologues of four genes (brlA, abaA, stuA, and medA) of A. nidulans that control conidiogenesis and found two putative genes that encode proteins significantly similar to StuA and MedA. These genes have exon and intron organizations similar to those of FoSTUA and REN1 and encode proteins strongly similar to FoStuA and Ren1 (data not shown). However, no brlA or abaA homologues were found in the F. graminearum genome, although BrlA and AbaA are core components of the conidiogenesis pathway in A. nidulans (1, 5, 12, 49). These results suggest that conidiation regulators and their functions differ between Fusarium species and A. nidulans. Together with the fact that FoStuA and Ren1 localize in nuclei, we propose that Ren1 and FoStuA are transcriptional regulators in the core pathway for conidiation in F. oxysporum. Identification of the target genes of FoStuA and Ren1 is needed to assess the precise functions of these regulators in F. oxysporum conidiation. Analysis of the N. crassa genome sequence showed that N. crassa also possesses no brlA or abaA homologues (20).
FoSTUA acts as a repressor of chlamydospore formation.
FoSTUA is expressed in CMC and also in CM and MM, in which conidiation is not induced. GFP-tagged FoStuA is localized in nuclei in mycelial cells grown in CMC and CM. These features of FoSTUA and its product seemed to be responsible for the phenotypes of the ΔFoSTUA mutants. The colony growth of the mutants was slightly slower on solid media, and the mutants produced fewer aerial hyphae. This growth phenotype resembles that of the ΔstuA mutant of A. nidulans (15). In A. nidulans, stuA is expressed in both conidiating and vegetative hyphae (32). However, in P. marneffei, stuA is not expressed in vegetative hyphae, and deletion of stuA causes no abnormalities in growth or morphology of vegetative hyphae (11).
F. oxysporum has the ability to produce thick-walled cells, termed chlamydospores, in hyphae and conidia under certain environmental conditions (17, 39, 48, 52). Chlamydospores of Fusarium species provide the principal means of long-time survival during unfavorable periods in soil and play an important role as the primary inocula infecting plants (17, 39, 48, 52). Ohara et al. observed that a mutation of REN1 has no effect on chlamydospore formation (42). Thus, we concluded that the developmental pathway of chlamydospores is genetically independent of that of macroconidia or microconidia. However, mutation of FoSTUA leads to the marked promotion of chlamydospore formation. FoStuA appears to be involved not only in conidiophore development but also in chlamydospore formation, although it controls these two developmental processes in opposite manners.
Among fungi from which APSES proteins have been characterized, C. albicans also produces chlamydospores. Chlamydospores of F. oxysporum and C. albicans are formed under similar conditions: chlamydospore formation is induced by aging and by unfavorable conditions of growth, such as low temperatures and carbon starvation (29, 35, 39, 48, 51, 52). In C. albicans, deletion of EGF1, encoding an APSES protein, results in a complete deficiency in chlamydospore formation (51). Thus, FoStuA and Egf1 have opposite functions in chlamydospore formation. Egf1 of C. albicans has a dual role in hyphal development and chlamydospore formation, in which Egf1 acts as an activator. In contrast, FoStuA functions as a repressor of chlamydospore formation but also as an activator of the development of conidiophores and macroconidia. The molecular mechanisms for chlamydospore formation in these fungi are unexplored; identification of target genes of FoStuA may be useful for understanding the molecular mechanisms of the chlamydospore formation that is an important trait of F. oxysporum.
FoSTUA is dispensable for disease-causing ability but important for in planta conidiation.
ΔFoSTUA mutants of F. oxysporum caused wilt symptoms in susceptible melon plants, as does the wild type, under the conditions tested. The mutants also grew invasively in stems at the late stage of infection. These results strongly suggest that FoSTUA is not involved in invading root tissues, colonizing roots and stems, and causing symptoms.
After symptom development, F. oxysporum pathogens produce conspicuous masses of macroconidia and microconidia on the surfaces of infected plants (39, 48, 52). Macroconidia are formed predominantly on plant surfaces, although microconidia are generally predominant in artificial culture media. Macroconidia and microconidia serve to spread the fungus within the plant as well as outside the plant (39, 48, 52). In the final stages of wilt disease development, chlamydospores arise on macroconidia, microconidia, and mycelia (39, 48, 52). Conidiation in infected plants is an essential step for the disease cycle. Macroconidium formation by ΔFoSTUA mutants in infected plants was very poor. Although the mutants showed normal microconidium formation in cultures, they produced markedly fewer microconidia in infected plants than the wild type. These results suggest that FoSTUA has an important role in microconidium formation, specifically in infected plants, and that FoSTUA is essential for the life cycle of F. oxysporum.
Acknowledgments
We are grateful to Sally A. Leong and Yoshitaka Takano for providing pMLF2 and the GFP gene, respectively. We thank Takayuki Aoki, Hirofumi Yoshioka, Kazuhito Kawakita, and Noriyuki Doke for valuable suggestions and the Radioisotope Research Center, Nagoya University, for technical assistance.
This work was supported by research grants from the Japanese Society for Promotion of Sciences (grants 13460022, 14656017, 15208005, and 16658019).
REFERENCES
- 1.Adams, T. H., M. T. Boylan, and W. E. Timberlake. 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54:353-362. [DOI] [PubMed] [Google Scholar]
- 2.Adams, T. H., J. K. Wieser, and J. H. Yu. 1998. Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62:35-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäfer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An, Z., M. L. Farman, A. Budde, S. Taura, and S. A. Leong. 1996. New cosmid vectors for library construction, chromosome walking and restriction mapping in filamentous fungi. Gene 176:93-96. [DOI] [PubMed] [Google Scholar]
- 5.Andrianopoulos, A., and W. E. Timberlake. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14:2503-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aramayo, R., Y. Peleg, R. Addison, and R. Metzenberg. 1996. asm-1+, a Neurospora crassa gene related to transcriptional regulators of fungal development. Genetics 144:991-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armstrong, G. M., and J. K. Armstrong. 1981. Formae speciales and races of Fusarium oxysporum causing wilt diseases, p. 391-399. In P. E. Nelson, T. A. Toussoun, and R. J. Cook (ed.), Fusarium: diseases, biology, and taxonomy. The Pennsylvania State University Press, University Park.
- 8.Beck, E., G. Ludwig, E. A. Auerswald, B. Reiss, and H. Schaller. 1982. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon. Tn5. Gene 19:327-336. [DOI] [PubMed] [Google Scholar]
- 9.Beckman, C. H. 1987. The nature of wilt diseases of plants. The American Phytopathological Society Press, St. Paul, Minn.
- 10.Borneman, A. R., M. J. Hynes, and A. Andrianopoulos. 2000. The abaA homologue of Penicillium marneffei participates in two developmental programmes: conidiation and dimorphic growth. Mol. Microbiol. 38:1034-1047. [DOI] [PubMed] [Google Scholar]
- 11.Borneman, A. R., M. J. Hynes, and A. Andrianopoulos. 2002. A basic helix-loop-helix protein with similarity to the fungal morphological regulators, Phd1p, Efg1p and StuA, controls conidiation but not dimorphic growth in Penicillium marneffei. Mol. Microbiol. 44:621-631. [DOI] [PubMed] [Google Scholar]
- 12.Boylan, M. T., P. M. Mirabito, C. E. Willet, C. R. Zimmermann, and W. E. Timberlake. 1987. Isolation and physical characterization of three essential conidiation genes from Aspergillus nidulans. Mol. Cell. Biol. 7:3113-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busty, T. M., K. Y. Miller, and B. L. Miller. 1996. Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of the bristle and stunted genes. Genetics 143:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cappellini, R. A., and J. L. Peterson. 1965. Macroconidium formation in submerged cultures by a nonsporulating strain of Gibberella zeae. Mycologia 57:962-966. [Google Scholar]
- 15.Clutterbuck, A. J. 1969. A mutational analysis of conidial development in Aspergillus nidulans. Genetics 63:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole, G. T. 1986. Models of cell differentiation in conidial fungi. Microbiol. Rev. 50:95-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couteaudier, Y., and C. Alabouvette. 1990. Survival and inoculum potential of conidia and chlamydospores of Fusarium oxysporum f. sp. lini in soil. Can. J. Microbiol. 36:551-556. [DOI] [PubMed] [Google Scholar]
- 18.Dahlberg, K. R., and J. L. Van Etten. 1982. Physiology and biochemistry of fungal sporulation. Annu. Rev. Phytopathol. 20:218-301. [Google Scholar]
- 19.Dutton, J. R., S. Johns, and B. L. Miller. 1997. StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J. 16:5710-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L.-J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. C. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 21.Gems, D. H., and A. J. Clutterbuck. 1994. Enhancers of conidiation mutants in Aspergillus nidulans. Genetics 137:79-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gimeno, C. J., and G. R. Fink. 1994. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Cell. Biol. 14:2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gritz, L., and J. Davies. 1983. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25:179-188. [DOI] [PubMed] [Google Scholar]
- 24.Inoue, I., F. Namiki, and T. Tsuge. 2002. Plant colonization by the vascular wilt fungus Fusarium oxysporum requires FOW1, a gene encoding a mitochondrial protein. Plant Cell 14:1869-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue, I., T. Ohara, F. Namiki, and T. Tsuge. 2001. Isolation of pathogenicity mutants of Fusarium oxysporum f. sp. melonis by insertional mutagenesis. J. Gen. Plant Pathol. 67:191-199. [Google Scholar]
- 26.Katan, T., E. Shlevin, and J. Katan. 1997. Sporulation of Fusarium oxysporum f. sp. lycopersici on stem surface of tomato plants and aerial dissemination of inoculum. Phytopathology 87:712-719. [DOI] [PubMed] [Google Scholar]
- 27.Kimura, A., Y. Takano, I. Furusawa, and T. Okuno. 2001. Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13:1945-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimura, N., and T. Tsuge. 1993. Gene cluster involved in melanin biosynthesis of the filamentous fungus Alternaria alternata. J. Bacteriol. 175:4427-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kono, Y., H. Yamamoto, M. Takeuchi, and H. Komada. 1995. Alteration in superoxide dismutase and catalase in Fusarium oxysporum during starvation-induced differentiation. Biochim. Biophys. Acta 1268:35-40. [DOI] [PubMed] [Google Scholar]
- 30.Lau, G., and J. E. Hamer. 1998. Acropetal: a genetic locus required for conidiophore architecture and pathogenicity in the rice blast fungus. Fungal Genet. Biol. 24:228-239. [DOI] [PubMed] [Google Scholar]
- 31.Leach, J. G., and T. M. Currence. 1938. Fusarium wilt of muskmelon in Minnesota. Minn. Agric. Exp. Stn. Bull. 129:32. [Google Scholar]
- 32.Miller, K. Y., T. M. Toennis, T. H. Adams, and B. L. Miller. 1991. Isolation and transcriptional characterization of a morphological modifier: the Aspergillus nidulans stunted (stuA) gene. Mol. Gen. Genet. 227:285-292. [DOI] [PubMed] [Google Scholar]
- 33.Miller, K. Y., J. Wu, and B. L. Miller. 1992. StuA is required for correct cell pattern formation in Aspergillus. Genes Dev. 6:1770-1782. [DOI] [PubMed] [Google Scholar]
- 34.Mirabito, P. M., T. H. Adams, and W. E. Timberlake. 1989. Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57:859-868. [DOI] [PubMed] [Google Scholar]
- 35.Montazeri, H., and H. G. Hedrick. 1984. Factors affecting spore formation in a Candida albicans strain. Appl. Environ. Microbiol. 47:1341-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullaney, E. J., J. E. Hamer, K. A. Roberti, M. M. Yelton, and W. E. Timberlake. 1985. Primary structure of the trpC gene from Aspergillus nidulans. Mol. Gen. Genet. 199:37-45. [DOI] [PubMed] [Google Scholar]
- 37.Namiki, F., M. Matsunaga, M. Okuda, I. Inoue, K. Nishi, Y. Fujita, and T. Tsuge. 2001. Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol. Plant-Microbe Interact. 14:580-584. [DOI] [PubMed] [Google Scholar]
- 38.Namiki, F., T. Shiomi, T. Kayamura, and T. Tsuge. 1994. Characterization of the formae speciales of Fusarium oxysporum causing wilts of cucurbits by DNA fingerprinting with the nuclear repetitive DNA sequences. Appl. Environ. Microbiol. 60:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson, P. E. 1981. Life cycle and epidemiology of Fusarium oxysporum, p. 51-80. In M. E. Mace, A. A. Bell, and C. H. Beckman (ed.), Fusarium wilt diseases of plants. Academic Press, Inc., New York, N.Y.
- 40.Nelson, P. E., T. A. Toussoun, and W. F. O. Marasas. 1983. Fusarium species: an illustrated manual for identification. The Pennsylvania State University Press, University Park.
- 41.Nirenberg, H. I. 1990. Recent advances in the taxonomy of Fusarium. Stud. Mycol. 32:91-101. [Google Scholar]
- 42.Ohara, T., I. Inoue, F. Namiki, H. Kunoh, and T. Tsuge. 2004. REN1 is required for development of microconidia and macroconidia, but not of chlamydospores, in the plant pathogenic fungus Fusarium oxysporum. Genetics 166:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okuda, M., K. Ikeda, F. Namiki, K. Nishi, and T. Tsuge. 1998. Tfo1: an Ac-like transposon from the plant pathogenic fungus Fusarium oxysporum. Mol. Gen. Genet. 258:599-607. [DOI] [PubMed] [Google Scholar]
- 44.Rekah, Y., D. Shtienberg, and J. Katan. 2000. Disease development following infection of tomato and basil foliage by airborne conidia of the soilborne pathogens Fusarium oxysporum f. sp. radicis-lycopersici and F. oxysporum f. sp. basilici. Phytopathology 90:1322-1329. [DOI] [PubMed] [Google Scholar]
- 45.Rowe, R. C., J. D. Farely, and D. C. Coplin. 1977. Airborne spore dispersal and recolonization of steamed soil by Fusarium oxysporum in greenhouses. Phytopathology 67:1513-1517. [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Sanderson, K. E., and A. M. Srb. 1965. Heterokaryosis and parasexuality in the fungus Ascochyta imperfecta. Am. J. Bot. 52:72-81. [PubMed] [Google Scholar]
- 48.Schippers, B., and W. H. van Eck. 1981. Formation and survival of chlamydospores in Fusarium, p. 250-260. In P. E. Nelson, T. A. Toussoun, and R. J. Cook (ed.), Fusarium: diseases, biology, and taxonomy. The Pennsylvania State University Press, University Park.
- 49.Sewall, T. C., C. W. Mims, and W. E. Timberlake. 1990. abaA controls phialide differentiation in Aspergillus nidulans. Plant Cell 2:731-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shenhar, G., and Y. Kassir. 2001. A positive regulator of mitosis, Sok2, functions as a negative regulator of meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:1603-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sonneborn, A., D. P. Bockmuhl, and J. F. Ernst. 1999. Chlamydospore formation in Candida albicans requires the Efg1p morphogenetic regulator. Infect. Immun. 67:5514-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevenson, I. L., and S. A. W. Becker. 1972. The fine structure and development of chlamydospores of Fusarium oxysporum. Can. J. Microbiol. 18:997-1002. [DOI] [PubMed] [Google Scholar]
- 53.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a covered class of bHLH proteins regulating morphogeneic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, J. D., D. C. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Timberlake, W. E. 1990. Molecular genetics of Aspergillus development. Annu. Rev. Genet. 24:5-36. [DOI] [PubMed] [Google Scholar]
- 56.Ward, M. P., C. J. Gimeno, G. R. Fink, and S. Garrett. 1995. SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol. Cell. Biol. 15:6854-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, J., and B. L. Miller. 1997. Aspergillus asexual reproduction and sexual reproduction are directly affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol. Cell. Biol. 17:6191-6201. [DOI] [PMC free article] [PubMed] [Google Scholar]