Abstract
The ABC transporter genes CDR1 and CDR2 can be upregulated in Candida albicans developing resistance to azoles or can be upregulated by exposing cells transiently to drugs such as fluphenazine. The cis-acting drug-responsive element (DRE) present in the promoters of both genes and necessary for their upregulation contains 5′-CGG-3′ triplets that are often recognized by transcriptional activators with Zn(2)-Cys(6) fingers. In order to isolate regulators of CDR1 and CDR2, the C. albicans genome was searched for genes encoding proteins with Zn(2)-Cys(6) fingers. Interestingly, three of these genes were tandemly arranged near the mating locus. Their involvement in CDR1 and CDR2 upregulation was addressed because a previous study demonstrated a link between mating locus homozygosity and azole resistance. The deletion of only one of these genes (orf19.3188) was sufficient to result in a loss of transient CDR1 and CDR2 upregulation by fluphenazine and was therefore named TAC1 (transcriptional activator of CDR genes). Tac1p has a nuclear localization, and a fusion of Tac1p with glutathione S-transferase could bind the cis-acting regulatory DRE in both the CDR1 and the CDR2 promoters. TAC1 is also relevant for azole resistance, since a TAC1 allele (TAC1-2) recovered from an azole-resistant strain could trigger constitutive upregulation of CDR1 and CDR2 in an azole-susceptible laboratory strain. Transcript profiling experiments performed with a TAC1 mutant and a revertant containing TAC1-2 revealed not only CDR1 and CDR2 as targets of TAC1 regulation but also other genes (RTA3, IFU5, and HSP12) that interestingly contained a DRE-like element in their promoters. In conclusion, TAC1 appears to be the first C. albicans transcription factor involved in the control of genes mediating antifungal resistance.
Several antifungal agents are available to treat infections due to Candida albicans. The success of antifungal treatments depends on the patient's condition, the type of antifungal agent given, and the biological response of the fungal pathogen. It has been demonstrated on many occasions that C. albicans can respond by developing specific resistance mechanisms upon exposure to antifungal drugs. Resistance to the class of azole antifungal agents used for the treatment of oropharyngeal candidiasis in human immunodeficiency virus-positive patients is a classical example.
In C. albicans, one of the well-documented mechanisms of resistance to azole antifungal agents is the upregulation of multidrug transporter genes (29, 33, 35, 39). The upregulation of multidrug transporter genes leads to the enhanced efflux of azoles and therefore results in decreased drug accumulation and reduced inhibition of their target encoded by the ERG11 gene. At least two families of multidrug transporters, the ABC (ATP-binding cassette) transporter family and the major facilitator superfamily (MFS), have been shown to be involved in resistance to azole antifungal agents. Cdr1p and Cdr2p (Candida drug resistance) from the family of ABC transporters and CaMdr1p (C. albicans multidrug resistance 1) from the family of MFS transporters are the principal mediators of resistance to azoles due to transport phenomena (33, 35). Each of the genes encoding these proteins can be upregulated in distinct clinical azole-resistant strains. The transcription of CaMDR1, the gene encoding CaMdr1p, is almost not detectable in azole-susceptible isolates but is measurable in some azole-resistant isolates. In contrast, the transcription of CDR1, the gene encoding Cdr1p, is detectable in azole-susceptible isolates but is increased to higher levels in some azole-resistant isolates. CDR1 is usually upregulated in these isolates together with CDR2, a gene with no detectable transcriptional activity in azole-susceptible isolates. Interestingly, CDR1 and CDR2 can also be induced transiently by treating cells with various drugs, such as estradiol and fluphenazine (8, 15), thus mimicking their expression in azole-resistant cells.
The upregulation of multidrug transporters in the development of antifungal resistance is well described in the baker's yeast Saccharomyces cerevisiae. Like C. albicans, S. cerevisiae possesses ABC transporters (for example, PDR5 [3, 4]) or MFS transporters (for example, FLR1 [1]) that are able to confer resistance to antifungal drugs. The regulation of ABC transporters and, more specifically, of PDR5, has been extensively documented. Two transcription factors, PDR1 and PDR3, encoding proteins with Zn(2)-Cys(6) finger motifs, have been described to regulate the transcription of PDR5 (7, 20). cis-Acting elements, so-called PDRE (pleiotropic drug response element), have been localized in the promoter of PDR5 and are the targets of PDR1 and PDR3 (21). PDRE have also been found in the promoters of several other genes in the S. cerevisiae genome and to be coregulated with PDR5 (9). The regulation of FLR1 is mediated by another transcriptional circuit that involves the leucine zipper transcription factor Yap1 (1).
In C. albicans, the molecular mechanisms governing the regulation of multidrug transporter genes have not been examined in great detail. No cis-acting element capable of regulating the expression of CaMDR1 has yet been determined. Up to now, elements permitting the basal activity of CDR1 and the upregulation of CDR1 and CDR2 have been determined. A 22-bp drug-responsive element (DRE) containing two 6-bp repetitive elements with 5′-CGG-3′ triplets is known to be required for the induction of both CDR1 and CDR2. Band shift assays revealed the presence of protein complexes binding to the DRE; however, their identity remains unknown (8). In the present study, we addressed the identification of trans-acting elements involved in the regulation of CDR1 and CDR2. We explored the C. albicans genome for genes encoding putative regulators with Zn(2)-Cys(6) finger motifs, since these factors can potentially bind 5′-CGG-3′ triplets that are present in the DRE of both the CDR1 and the CDR2 promoters (24). Among three genes tandemly arranged near the mating type locus, the deletion of one of them, now named TAC1, resulted in a loss of transient CDR1 and CDR2 upregulation after drug exposure.
MATERIALS AND METHODS
Strains, plasmids, and media.
The C. albicans and S. cerevisiae strains used in this study are listed in Table 1. The plasmids constructed in this study (see below for further details) are listed in Table 2. Yeast strains were grown either in complete medium (1% Bacto Peptone [Difco Laboratories, Basel, Switzerland], 0.5% yeast extract [Difco], 2% glucose [Fluka, Buchs, Switzerland]; YEPD) or in minimal medium (yeast nitrogen base [Difco], 2% glucose [Fluka]; YNB). For growth on solid media, 2% agar (Difco) was added to either of the media. C. albicans Gal− mutants were generated by the method described by Gorman et al. (14). Escherichia coli DH5α (16) was used as a host for plasmid constructions and propagation. DH5α was grown in Luria-Bertani broth or on Luria-Bertani agar plates supplemented with ampicillin (0.1 mg/ml) when required.
TABLE 1.
Strains used in this study
| Strain | Genotype | Parent strain | Reference or source |
|---|---|---|---|
| C. albicans | |||
| CAF2-1 | ura3Δ::imm434/URA3 | SC5314 | 11 |
| CAF4-2 | ura3Δ::imm434/ura3Δ::imm434 | CAF2-1 | 11 |
| CHY439 | ura3/ura3 MTLa/mtlα1::hisG mtlα2::hisG | CAI4 | 26 |
| CHY477 | ura3/ura3 mtla1::hisG/MTLα ade2::hisG-URA3-hisG/ade2::hisG | CAI4 | 26 |
| DSY294 | Azole-susceptible clinical strain (MTLa/MTLα) | 35 | |
| DSY296 | Azole-resistant clinical strain (MTLα/MTLα) | DSY294 | 35 |
| DSY296-3 | gal1 | DSY296 | This study |
| DSY2781 | GAL1/gal1 ura3/URA3 MTLa/mtlα1::hisG mtlα2::hisG/MTLα | DSY296-3, CHY439 | This study |
| DSY2875 | TAC1/tac1Δ::hisG-URA3-hisG | CAF4-2 | This study |
| DSY2883 | TAC1/tac1Δ::hisG | DSY2875 | This study |
| DSY2903 | tac1Δ::hisG/tac1Δ::hisG-URA3-hisG | DSY2883 | This study |
| DSY2906 | tac1Δ::hisG/tac1Δ::hisG | DSY2903 | This study |
| DSY2937 | tac1Δ::hisG/tac1Δ::hisG LEU2::TAC1/URA3 | DSY2906 | This study |
| DSY2925 | tac1Δ::hisG/tac1Δ::hisG LEU2::TAC1-1/URA3 | DSY2096 | This study |
| DSY2926 | tac1Δ::hisG/tac1Δ::hisG LEU2::TAC1-2/URA3 | DSY2096 | This study |
| DSY2884 | ZNC3/znc3Δ::hisG-URA3-hisG | CAF4-2 | This study |
| DSY2869 | ZNC1/znc1Δ::hisG-URA3-hisG | CAF4-2 | This study |
| DSY654 | cdr1Δ::hisG/cdr1Δ::hisG cdr2Δ::hisG/cdr2Δ::hisG-URA3-hisG | CAF4-2 | 33 |
| S. cerevisiae DSY669 | MATapdr1::TRP1 pdr3::HIS3 PDR5 ura3-52 leu2-Δ1 GAL2 | FY1679-28C | 7 |
TABLE 2.
Plasmids used in this study
| Plasmid | Parent plasmid | Descriptiona | Reference or source |
|---|---|---|---|
| pAU36 | See reference 38 | lacZ reporter system for C. albicans | 38 |
| pDS958 | pAU36 | CDR1-lacZ fusion cloned into pAU36 | This study |
| pDS959 | pAU36 | CDR2-lacZ fusion cloned into pAU36 | This study |
| pDS1093 | pGEX4-2T | Insertion of the N-ternimal end of TAC1 containing the DNA-binding domain into pGEX4-2T | This study |
| pDS178 | See reference 8 | pRC2312 with a pBluescript multiple-cloning site | 8 |
| pDS1097 | pDS178 | Insertion of TAC1 from SC5314 into pDS178 | This study |
| pDS1098 | pDS178 | Insertion of TAC1-1 from strain DSY294 into pDS178 | This study |
| pDS1099 | pDS178 | Insertion of TAC1-2 from strain DSY296 into pDS178 | This study |
| YIp353 | See reference 27 | lacZ reporter system in integrative S. cerevisiae plasmid | 27 |
| pDS1139 | YIp353 | CDR2-lacZ fusion cloned into YIp353 | This study |
| pDS1157 | pDS1139 | TAC1 cloned into pDS1139 | This study |
| pDS1161 | pDS1139 | Deletion of the DRE within the CDR2 promoter in pDS1139 | This study |
| pDS1180 | pDS1161 | TAC1 cloned into pDS1161 | This study |
| pDS1187 | pDS1097 | CDR2-lacZ fusion cloned into pDS1097 | This study |
| pDS1190 | pDS1097 | Deletion of the DRE within the CDR2 promoter in pDS1097 | This study |
| pDS1160 | CIpACT-C-ZZ | TAC1 fused to protein A in expression plasmid CIpACT-C-ZZ (5) | This study |
| pDS1202 | pDS1160 | TAC1 fused to GFP in pDS1160 | This study |
Detailed descriptions and constructions are given in Material and Methods.
Yeast tranformation.
S. cerevisiae was transformed by a standard lithium acetate protocol as reported previously (13). For C. albicans transformation, cells from 0.2 ml of a stationary-phase culture were resuspended in 0.1 ml of a solution containing 200 mM lithium acetate (pH 7.5), 40% (wt/vol) polyethylene glycol 8000, 15 mg of dithiothreitol (DTT)/ml, and 250 μg of denatured salmon sperm DNA/ml. Transforming DNA (1 to 5 μg) was added to the yeast suspension, which was then incubated for 60 min at 43.5°C. Transformation mixtures were plated directly on selective plates.
Construction of promoter-lacZ fusions and measurement of β-galactosidase activities.
CDR1- and CDR2-lacZ fusions made for C. albicans were constructed by amplification of the CDR1 and CDR2 promoters with primers CDR1-KPN and CDR1-PST or primers CDR2-KPN and CDR2-PST (Table 3) and cloning into pAU36 (38) that had been digested with KpnI and PstI. The resulting plasmids, pDS958 and pDS959, were digested with SacI and SnaBI prior to transformation into C. albicans, thus resulting in their integration at the CDR1 and CDR2 loci, respectively. β-Galactosidase activities were measured as described by Uhl and Johnson (38).
TABLE 3.
Primers used in this study
| Primer | Sequencea |
|---|---|
| Znc2-5-BamB | GCAAGGATCCAAGAAGAAGTGGATAATTTTGATTAAC |
| Znc2-3-Xho | GCAACTCGAGAGTATATTCTGTTGGGAAAGGGGTGAG |
| Znc2-GST1 | AGCAAGGATCCATGGACACTTCACTGTCACTGGGA |
| Znc2-GST2 | GCTTTCTCGAGACTTGATTTGTTGTCATTTATGCCG |
| CDR1-F | CATGGTCAAGCCATTTTGTG |
| CDR1-R | ATCCATTCTGCTGGATTTGC |
| CDR2-F | CATGGTCAAGCCATTTTGTG |
| CDR2-R | ATCCATTCTGCTGGATTTGC |
| IFU5-F | AAACCCACCACAAGTTCCTG |
| IFU5-R | CTTGGGGCATTAGACCTTGA |
| RTA3-F | TACAGAATGGACTCCTACCT |
| RTA3-R | GCCGTACGATTTAATCGA |
| CDR1-PST | GCGCAAACTGCAGAATTTTTTTCTTTTTGACCTTTTAAAGAAA |
| CDR1-KPN | GCGCAAAGGTACCGGATCCTCGTTACTCAATAAGTATTAATTG |
| CDR2-PST | GCGCAAACTGCAGATGTTTTTATTGTATGTGTTAATTAGTGAA |
| CDR2-KPN | GCGCAAAGGTACCGGTTCCTCTAAATAAAAACTAGAAGGTTAT |
| CDR2-HIND | GCGCAAAGCTTTGTTGTGACTTGCAGTAGCAT |
| CDR2-PST-900 | GCAAACTGCAGGTTCCTCTAAATAAAAACTAGAAGG |
| CDR2-DEL5 | GTATTAATTTTTACGTATTTTCTTTGTGTTATTCAATTCTTGTTTTCAAAGCCT |
| CDR2-DEL3 | AGTATTCATAATAGAGGCTTTGAAAACAAA |
| CDR2-XHO | GCGCAAACTCGAGACATGAAAAATGAAATCAATTCAAACACAA |
| CDR2-ANTI | AATGTTTTTATTGTATGTGTTAATTAGTGAA |
| CDR2-LACZ | AACACATACAATAAAAACATATGAACATGACTGAAAAAATTCAAACTT |
| CDR2-SAC | GCGCAAAGAGCTCACATGAAAAATGAAATCAATTCAAACACAA |
| MAL2-BAM | GCGCAAAGGATCCCATACGCTTTGCAGGTGGTGTTGATCC |
| SALI-ZNC2C | GCGCAAAGTCGACATAATGGACACTTCACTGTCACTGGGA |
| SPHI-ZNC2C | GCGCAAAGCATGCAAATCCCCAAATTATTGTCAAAGAAAAA |
| GFP-5-SPHI | CGCGAAAGCATGCATTCTAAAGGTGAAGA |
| GFP-3-NHE | CGCGAAAGCTAGCTTATTTGTACAATTCATCCATACCATGGG |
Restriction sites are underlined.
The CDR2-lacZ fusion made for the expression of CDR2 promoter activity in S. cerevisiae was prepared by inserting the product of PCR amplification of the CDR2 promoter with primers CDR2-PST-900 and CDR2-HIND (Table 3) into the PstI and HindIII sites of YIp353, resulting in pDS1139. Deletion of the DRE (−221 to −220) within this promoter was carried out as described by De Micheli et al. (8) by sewing of PCR fragments obtained with primers CDR2-PST-900 and CDR2-DEL3, with primers CDR2-HIND and CDR2-DEL5 (Table 3), and with pDS246 (33) as a template. The final amplification product obtained with external primers CDR2-PST-900 and CDR2-HIND was inserted into YIp353, resulting in pDS1161. TAC1 alleles were obtained from pDS1097 (see below) and cloned into the BamHI and XhoI sites of pDS1139 and pDS1161, resulting in pDS1157 and pDS1180. These plasmids, together with pDS1139, were linearized with StuI to facilitate integration at the S. cerevisiae URA3 locus.
The DRE-containing CDR2-lacZ derivatives used for expression in C. albicans were prepared by sewing of PCR fragments amplified from the first 300 bp of the CDR2 promoter with primers CDR2-SACI and CDR2-anti, with primers CDR2-LACZ and MAL2-BAM, and with pDS246 (33) as a template. The final amplification product obtained with external primers CDR2-SACI and MAL2-BAM was subcloned into the SacI and BamHI sites of pDS1097, which contains TAC1, to yield pDS1187. The DRE-negative construct was prepared by PCR amplification with the same primers as those used to generate pDS1187 but with DRE-negative plasmid pDS1180 as a template. The final DRE-negative CDR2-lacZ fusion was created by PCR with primers CDR2-SACI and MAL2-BAM and by subcloning into pDS1097 to yield pDS1190. pDS1187 and pDS1190 were linearized with SalI before transformation into TAC1 mutant DSY2906, facilitating integration at the genomic LEU2 locus.
Construction of gene disruption cassettes.
For the disruption of ZNC1, ZNC2 (TAC1), and ZNC3, a region containing the entire open reading frame (ORF) or a portion of the ORF was amplified from genomic DNA with the cloning primers shown in Table 4. PCR fragments were cloned into pBluescript KS(+) to yield the cloning constructs shown in Table 4. Deletions within cloned regions were created by PCR with the deletion primers shown in Table 5 and with cloning constructs as templates. The 3.7-kb PstI-BglII fragment comprising the URA3-blaster cassette from pMB7 (11) was cloned into PCR fragments that had been digested with PstI and BglII to obtain deletion constructs (Table 5). For transformation in C. albicans, linear fragments were obtained by digestion of deletion constructs with ApaI and SacI.
TABLE 4.
Cloning constructs from plasmid Bluescript KS(+)
| Gene | Region amplified with respect to ATG | Cloning primer | Sequencea | Cloning construct (insert size, kb) |
|---|---|---|---|---|
| TAC1 | +1 to +2947 | CaZNC2-BamHI | GCGCAAAGGATCCTTAAATCCCCAAATTATTGTCAAAGAAAAA | pDS1048 (2.9) |
| CaZNC2-Xho | GCGCAAACTCGAGATGGACACTTCACTGTCACTGGGAACTCAC | |||
| ZNC1 | +1 to +2770 | CaZNC1-Xba2 | GCGCAAATCTAGATGCTGGATGTTAATGATACACTTAATCCT | pDS1047 (2.7) |
| CaZNC1-Xho | GCGCAAACTCGAGTTATATATTTTGCTCATTAAATCCTAAAGA | |||
| ZNC3 | +1 to +3034 | CaZNC3-Xho2 | GCGCAAACTCGAGTTAGTTATAAAATATATCAGGAAAGTTCAA | pDS1043 (3) |
| CaZNC3-Xba | GCGCAAATCTAGATGGATCCTGCTTATGATATACAATCGCA |
Restriction sites are underlined.
TABLE 5.
Deletion constructs from cloning vectors containing URA3-blaster from pMB7 (11)
| Gene | Region deleted with respect to ATG | Deletion primer | Sequencea | Disruption construct (deletion size) |
|---|---|---|---|---|
| TAC1 | +1154 to +1419 | Deletion performed by removal of an internal PstI-BglII fragment | pDS1052 (265 bp) | |
| ZNC1 | +550 to +1993 | CaZNC1-Pst | GCGCAAACTGCAGATCGTTATGGTTATATTCTATACTGCCATC | pDS1049 (1.4 kb) |
| CaZNC1-Bgl | CAATCAATCATTAGCTGGAGCTAGATCTATGTTG | |||
| ZNC3 | +531 to +2543 | CaZNC3-Bgl | GCGCAAAGATCTTGGAGAGGGCTAGATTTATCTGTCTTTGCT | pDS1065 (2 kb) |
| CaZNC3-Pst2 | GCGCAAACTGCAGCATCATCATCGACTTCTTTCCAACCACCTA |
Restriction sites are underlined.
For deletion of a region of TAC1, the 3.7-kb PstI-BglII fragment comprising the URA3-blaster cassette from pMB7 was ligated to pDS1048 that had been digested with BglII and PstI, resulting in an internal deletion of 265 bp within the TAC1 ORF and creating pDS1052. The linear fragment liberated after digestion of pDS1052 with ApaI and SacI was used to transform C. albicans.
Construction of revertant strains.
Revertant strains of each homozygous mutant generated in this study were obtained by transformation of a C. albicans ura3 derivative with pRC2312-derived plasmid pDS178, containing the URA3 and LEU2 markers described previously (8). To generate revertants of TAC1 mutant DSY2906, ORFs flanked by 500 bp were amplified from genomic DNAs of strains SC5314, DSY294, and DSY296 with primers Znc2-5-BamB and Znc2-3-Xho (Table 3) and inserted into pDS178 that had been digested with BamHI and XhoI to yield pDS1097, pDS1098, and pDS1099, respectively. These plasmids were linearized with SalI and transformed into C. albicans, allowing integration at the genomic LEU2 locus.
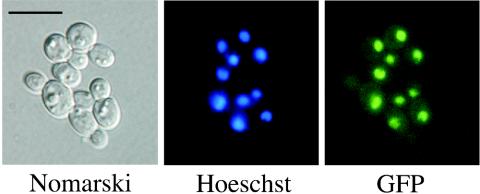
Construction and visualization of a Tac1p-GFP fusion.
Green fluorescent protein (GFP) was fused to Tac1p at the C-terminal end. The TAC1 ORF was amplified with primers SALI-ZNC2C and SPHI-ZNC2C and with pDS1097 as a template and was cloned into compatible SalI-SphI sites of CIpACT-C-ZZ (5) to yield pDS1160. GFP was amplified from yEGFP3 (6) with primers GFP-5-SPH and GFP-3-NHE and was cloned into compatible NheI-SphI sites of pDS1160, replacing protein A and yielding pDS1202. The final Tac1p-GFP fusion was under the control of the ACT1 promoter contained in CIpACT-C-ZZ. Plasmid pDS1202 was digested with StuI and transformed into DSY2906. Transformants were grown on selective medium to logarithmic phase, and GFP fluorescence was revealed by microscopy. Nuclear DNA was stained by the addition of Hoechst 33258 at a concentration of 2 μg/ml for 30 min at room temperature prior to microscopy. Fluorescence microscopy and phase-contrast microscopy were performed with a Zeiss Axioplan microscope equipped for epifluorescence microscopy with a 100-W mercury high-pressure bulb and Zeiss filter sets 9 (for GFP) and 15 (for Hoechst 33258). A DX30 digital camera with high resolution (Kappa Messtechnik GmbH, Gleichen, Germany) was used to record images.
Purification of Tac1p fused to GST.
A Tac1p-glutathione S-transferase (GST) fusion plasmid was constructed by introducing the first 129 amino acids of Tac1p in frame with GST into pGEX4-2T (Amersham Biosciences, Otelfingen, Switzerland). The fragment encoding the first 129 amino acids was first amplified with primers Znc2-GST1 and Znc2-GST2 (Table 3) and with genomic DNA as a template and then introduced into the BamHI-XhoI sites of pGEX4-2T to obtain pDS1093. Purification of the Tac1p-GST fusion from E. coli BL21 (E. coli B F− ompT hsdS [rB− mB−] gal dcm) transformed with pDS1093 was carried out by standard protocols provided by Amersham Biosciences.
Northern and Southern blotting.
Northern blotting was carried out as described previously (35). DNA probes used in this study were generated by PCR with the pairs of primers shown in Table 3. 32P-labeled DNA probes were generated by random priming (10). The TEF3 probe, used as an internal standard, originated from a 0.7-kb EcoRI-PstI fragment from pDC1 (18). Southern blotting was performed as described previously (35). Radioactive signals were revealed by exposure to Kodak BioMax MR film (Amersham Biosciences).
Band shift assays.
Total protein extraction from C. albicans was performed as described previously (8). The DNA-binding probe was obtained by annealing complementary oligonucleotides (7.5 μg) corresponding to probes 1 to 5 (see Fig. 4) in 10 mM Tris-HCl (pH 8.0)-100 mM NaCl-1 mM EDTA (100 μl) for 2 min at 85°C followed by stepwise 1-min incubations at 75, 65, 50, and 25°C. A 2-μl aliquot of the probe was labeled with 1.8 MBq of [γ-32P]dATP by using polynucleotide kinase (Roche, Basel, Switzerland) in a 10-μl reaction for 1 h at 37°C. The probe was purified by using a QiaQuick nucleotide removal kit (Qiagen Inc., Chatsworth, Calif.). The binding solution was obtained by adding sequentially 7 μl of binding buffer (20 mM HEPES [pH 7.9], 50 mM KCl, 0.5 mM EDTA, 0.1% NP-40, 1 mg of bovine serum albumin/ml, 5% glycerol), 1 μl of 0.2 M phenylmethylsulfonyl fluoride, 1 μl of 0.1 M DTT, 1 μl of poly(dI-dC) at 1 mg/ml, proteins, competitor if needed, and 2 to 3 μl of probe (at least 106 cpm). The total volume was adjusted to 20 μl with H2O. Binding was performed for 20 min at room temperature. Before samples were loaded onto the polyacrylamide gel, 3 μl of 40% glycerol was added to the samples and the gel was prerun for 30 min at 100 V. Samples were electrophoresed for 150 min at 200 V onto a 5% polyacrylamide gel (Protogel 37.5:1) in Tris-borate-EDTA buffer. The gel was dried on Whatman 3MM paper and exposed to Kodak BioMax MR film.
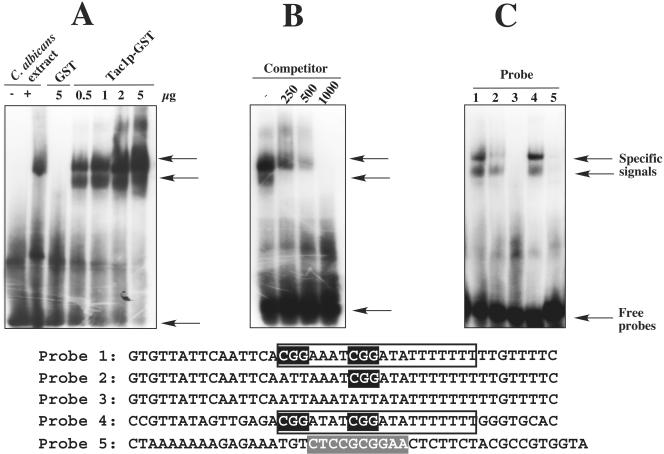
FIG. 4.
Tac1p binds to the CDR DRE. Labeled probes were separated as described in Materials and Methods. Arrows indicate the positions of specific complexes and of free labeled probes. (A) Binding saturation of the DRE by Tac1p-GST. Increasing amounts of Tac1p-GST were added to the reaction mixtures before loading. (B) Competition experiments with unlabeled DRE. A total of 1 μg of Tac1p-GST was added to the labeled probe along with increasing amounts of unlabeled probe. Probe 1 corresponding to the DRE region (DRE consensus sequence boxed with two CGG triplets highlighted) of the CDR2 promoter was used for the band shifts shown in panels A and B. (C) CGG triplet-dependent binding of Tac1p-GST. Probes 1, 2 (first CGG triplet changed), and 3 (first and second CGG triplets changed) correspond to the DRE region of the CDR2 promoter. Probes 4 and 5 correspond to the DRE region (DRE consensus sequence boxed with two CGG triplets highlighted) of the CDR1 promoter and to the PDRE (consensus sequence highlighted) of PDR5, respectively. Each probe was designed with two complementary oligonucleotides.
Immunoblotting.
C. albicans cell extracts for immunoblotting were prepared from cells grown to mid-log phase by an alkaline extraction procedure. Briefly, cells (optical density at 540 nm, 5) were resuspended in an Eppendorf tube with 500 μl of water and 150 μl of a solution containing 1.85 M NaOH and 7.5% β-mercaptoethanol. This mixture was incubated on ice for 10 min. Proteins were precipitated with 150 μl of a 50% trichloroacidic acid solution, and the suspension was left on ice for 10 min. Precipitated proteins were sedimented by centrifugation at 13,000 × g in a microcentrifuge for 15 min. The sediment was resuspended in 50 μl of loading buffer (40 mM Tris-HCl [pH 6.8], 8 M urea, 5% sodium dodecyl sulfate [SDS], 0.1 M EDTA, 1% β-mercaptoethanol, 0.1 mg of bromphenol blue/ml) and incubated at 37°C for 10 min. Nonsolubilized material was cleared by centrifugation for 10 min. Ten-microliter samples of solubilized yeast proteins were separated by SDS-10% polyacrylamide gel electrophoresis (PAGE) and transferred by Western blotting to a nitrocellulose membrane. Immunodetection of Cdr1p and Cdr2p was performed as described previously (8) with rabbit polyclonal anti-Cdr1p and anti-Cdr2p antibodies by chemiluminescence with an ECL kit according to the recommendations of the manufacturer (Amersham Biosciences).
Drug susceptibility testing.
Drug susceptibility testing was performed with microtiter plates and twofold serial dilutions of fluconazole (range, 128 to 0.06 μg/ml). Yeast cultures were grown overnight in YEPD and inoculated at a density of 104 cells/ml in a total volume of 200 μl containing a serial fluconazole dilution. Microtiter plates were incubated at 35°C for 24 h, and optical densities were read with a microtiter plate reader at a wavelength of 540 nm. The MIC was determined as the drug concentration required to decrease the optical density of the drug-free culture by at least 50%. Drug susceptibility testing was also performed by disk diffusion on YEPD agar plates onto which 105 cells had been dispersed by application of a filter disk containing 50 μg of fluconazole; the plates were incubated for 48 h at 30°C.
Microarray experiments.
C. albicans cDNA microarray slides from Eurogentec, Seraing, Belgium, were used in this study. Briefly, 6,039 ORFs from PCR amplicons of 300 bp were spotted in duplicate on each aldehyde-coated glass slide in 32 blocks. The microarray covers nearly 98% of the total number of C. albicans genes and includes 27 control genes in each block, including, for example, negative controls (intergenic regions), cross-hybridization controls (S. cerevisiae genes), and dynamic-range controls (serial dilution of the TEF3 gene from 1 to 1/32).
(i) RNA isolation and probe labeling.
Each C. albicans strain was pregrown overnight in 5 ml of YEPD with constant agitation at 30°C. For RNA extraction, cultures were diluted to a density of 0.75 × 107 cells/ml in 15 ml of fresh YEPD and were grown at 30°C with agitation for 2 h to reach a density of 1.5 × 107 cells/ml. At this point, cultures of DSY294, DSY296, DSY2925, and DSY2926 were centrifuged for 5 min at 5,500 × g and 4°C for subsequent RNA isolation. Strains CAF2-1 and DSY2906 were exposed for 20 min to 10 μg of fluphenazine (Sigma-Aldrich, Buchs, Switzerland)/ml at 30°C with agitation. CAF2-1 also was cultivated in parallel under the same conditions but without drugs. After drug exposure, cultures were centrifuged at 4°C and 5,500 × g for RNA isolation. Total RNA was extracted by using glass beads as described previously (32). A least 200 μg of each RNA was purified as described by the RNeasy kit supplier (Qiagen). The concentration of purified RNA was measured spectrophotometrically at A260 and A280 and adjusted to 2.5 mg/ml. RNA was stored at −80°C until use.
For RNA labeling, 25 μg of total RNA was mixed with 8 μl of 5× First-Strand buffer (Invitrogen, Basel, Switzerland), 1 μl of 0.1 μM C. albicans-specific Primer Mix Plus [including T20VN and oligo(dT)18-21] (Eurogentec), 3 μl of 6.67 mM deoxynucleoside triphosphates (Roche), 1 μl of 1 mM dCTP (Roche), 1.5 μl of 1 mM cyanine 3 (Cy3)-dCTP or 1.5 μl of 1 mM cyanine 5(Cy5)-dCTP (Amersham Biosciences), 4 μl of 100 mM DTT (Invitrogen), 1 μl of RNasin (20 to 40 U/μl; Promega, Wallisellen, Switzerland), and diethyl pyrocarbonate-treated water to a volume of 40 μl. The mixture was heated at 65°C for 5 min and cooled at 42°C for 5 min. The reverse transcription reaction was performed for 1 h at 42°C with 1 μl of RNasin and 200 U of SuperScript II reverse transcriptase (Invitrogen). The reaction was stopped, and RNA was degraded by the addition of 5 μl of EDTA (50 mM, pH 8.0) and 2 μl of NaOH (10 N) and incubation at 65°C for 20 min. The reaction was neutralized with 4 μl of acetic acid (5 M). The labeled probes (cDNA) were purified according to the QiaQuick PCR purification kit protocol (Qiagen). The elution step was performed twice with prewarmed H2O (42°C), and the samples were centrifuged at 13,000 × g for 1 min. The purified labeled probes were concentrated to a final volume of 5 μl by using a Microcon-30 filter (Amicon, Wallisellen, Switzerland).
(ii) Hybridization of microarrays.
Cy5- and Cy3-labeled cDNAs synthesized from RNAs of cells for which transcription profiles were to be compared were pooled with 5 μl of heat-denatured salmon sperm DNA. Four different transcription profile comparisons were performed: (i) Cy3-labeled cDNA from CAF2-1 not exposed to fluphenazine with Cy5-labeled cDNA from the same strain exposed to fluphenazine (Flu experiment, as labeled in the data file array_TAC1_Sanglard.xls); (ii) Cy3-labeled cDNA from CAF2-1 with Cy5-labeled cDNA from DSY2906, both exposed to fluphenazine (TAC1 flu experiment); (iii) Cy3-labeled cDNA from DSY2925 with Cy5-labeled cDNA from DSY2926 (TAC1 allele experiment); and (iv) Cy3-labeled cDNA from DSY294 with Cy5-labeled cDNA from DSY296 (CDR experiment). Each DNA pool was boiled at 95°C for 2 min, chilled on ice, and mixed with 40 μl of hybridization buffer (DIG easy hyb; Roche). The mixture was applied to C. albicans microarray slides, covered with Lifterslips (25 by 44 mm; Erie Scientific Co., Portsmouth, N.H.) for all hybridization steps, and loaded into the hybridization chamber (Corning Inc., Corning, N.Y.), which was kept wet by loading 10 μl of H2O in each existing well. After overnight hybridization at 42°C, coverslips were removed from slides by dipping in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS. Slides were washed for 5 min at room temperature with occasional agitation and rinsed with 0.2× SSC for 5 min at room temperature with occasional agitation. Slides were spin dried (500 × g, 5 min) and stored protected from light until scanning.
(iii) Quantitative analysis of microarrays.
Hybridized microarray slides were scanned with a ScanArray 4000 scanner (Perkin-Elmer, Schwerzenbach, Switzerland) at a 10-μm resolution. The following wavelengths were used for photoexcitation: 532 nm for Cy3-labeled cDNA and 635 nm for Cy5-labeled cDNA. The intensity of each laser light was determined by equalizing the Cy3 and the Cy5 fluorescence signals of the dynamic-range controls (TEF3) in three different blocks for each slide. The resulting 16-bit TIFF files of each signal were quantified and converted to a text file by using ImaGene software, version 4.0 (BioDiscovery Inc., El Segundo, Calif.).
(iv) Normalization control.
Data from each hybridization experiment were normalized by using a LOWESS (locally weighted scatter plot smooth) analysis for print-tip variability and for slide-to-slide variability with publicly available software (from Lund University; http://www.braju.com/R/com.braju.sma/) run in the R-project environment (http://cran.r-project.org/). In order to determine a significant threshold, LOWESS normalization was performed only on the TEF3 spots, TEF3 being a housekeeping gene. A significant threshold was designated as being above the mean of the TEF3 ratios and twice the standard deviation. A twofold differential expression limit was therefore chosen for the comparison of pairwise experiments in this study. Normalized data can be downloaded from http://www.hospvd.ch/imul/ under the file name Microarray Data (array_TAC1_sanglard.xls).
RESULTS
Genome information as a tool for the discovery of CDR1 and CDR2 regulators.
The cloning of transcriptional activators of CDR1 and CDR2 can be approached by several strategies. Since multidrug transporters of the ABC family share common functions in C. albicans and S. cerevisiae, it could be assumed that their regulation could be controlled by transcriptional factors with some degree of similarity. Attempts to clone C. albicans transcriptional activators of ABC transporters by use of S. cerevisiae resulted in the isolation of FCR1 and FCR3 (37, 40) and of three additional genes (SHY1, SHY2, and SHY3 [suppressor of hypersusceptibility]) encoding proteins with Zn(2)-Cys(6) binuclear cluster domains. However, the deletion of these genes in C. albicans had no effect on CDR1 and CDR2 regulation (D. Sanglard, unpublished data).
Since the nucleotide sequence of the C. albicans genome is available, an alternative strategy consisting of a systematic search of ORFs encoding proteins with Zn(2)-Cys(6) binuclear cluster domains was undertaken. This strategy was based on the fact that transcription factors containing these motifs often recognized in cis-acting regulatory regions 5′-CGG-3′ triplets (22) that are present in the DRE of CDR1 and CDR2 (8). Using the data available from the most recent assembly of the C.albicans genome (http://www-sequence.stanford.edu/group/candida/search.html), we identified a total of 72 ORFs encoding proteins with Zn(2)-Cys(6) DNA-binding domains; of these gene products, 62 had these domains at their N-terminal ends. A cluster analysis of these ORFs revealed a group of related genes, i.e., orf19.3187 (ZNC1 [zinc-Cys 1]), orf19.3188 (ZNC2), and orf19.3190 (ZNC3) (see the supplemental material). Interestingly, these genes are clustered physically within a 13-kb region present in the nucleotide sequences of contigs 19 to 10070 and contigs 19 to 20070. This region, the so-called zinc cluster region (Fig. 1A), is located on chromosome 5 of C. albicans, as deduced from data available at http://candida.bri.nrc.ca/candida/. Moreover, the zinc cluster region is situated approximately 14 kb upstream of the mating locus (either MTLa or MTLα). Interestingly, homozygosity at the mating locus has been linked to the development of azole resistance in specific clinical strains (31). We therefore hypothesized that the genes contained in the zinc cluster region could be involved in this phenomenon. To test this hypothesis, individual mutants lacking each of the genes in this cluster were constructed and subjected to an analysis of CDR1 and CDR2 transcriptional regulation. The construction of homozygous mutants could be accomplished only with ZNC2. Only heterozygous mutants were obtained with ZNC1 and ZNC3. Attempts to obtain homozygous mutants with these genes failed even after the screening of approximately 100 transformants from the disruption of the second allele of each gene.
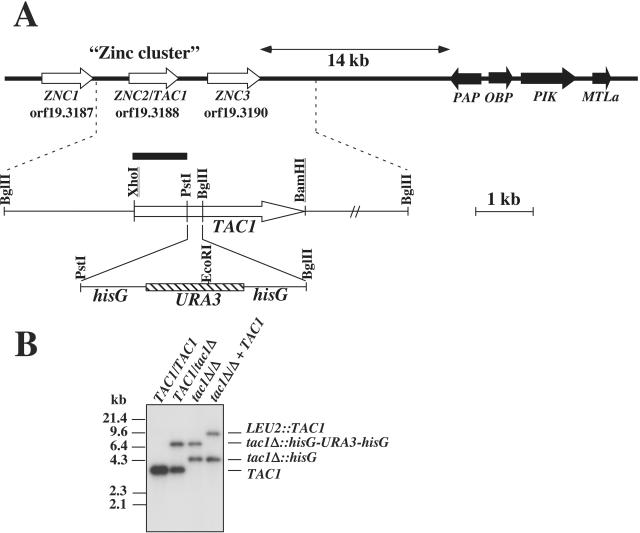
FIG. 1.
(A) Restriction map of zinc cluster genes. The physical map shows the zinc cluster and the neighboring MTL locus (MTLa). White arrows show the positions of ZNC ORFs, and black arrows show the positions of MTLa locus genes. A second map with a higher scale for ZNC2 (TAC1) focuses on the location of the TAC1 deletion constructed with the URA3-blaster cassette. The entire TAC1 ORF was used for cloning and construction of disruption cassettes (see Material and Methods for details). Underlined restriction sites were created by PCR cloning. (B) Southern analysis of the TAC1 disruption. Genomic DNA was digested with BglII. The identity of each band of the expected size is shown at the right side of the Southern blot. The positions of molecular size standards are shown at the left side. The following strains correspond to the indicated genotypes: TAC1/TAC1, CAF2-1; TAC1/tac1Δ, DSY2875; tac1Δ/Δ, DSY2903; and tac1Δ/Δ + TAC1, DSY2937.
ZNC2 (TAC1) disruption affects CDR1 and CDR2 upregulation.
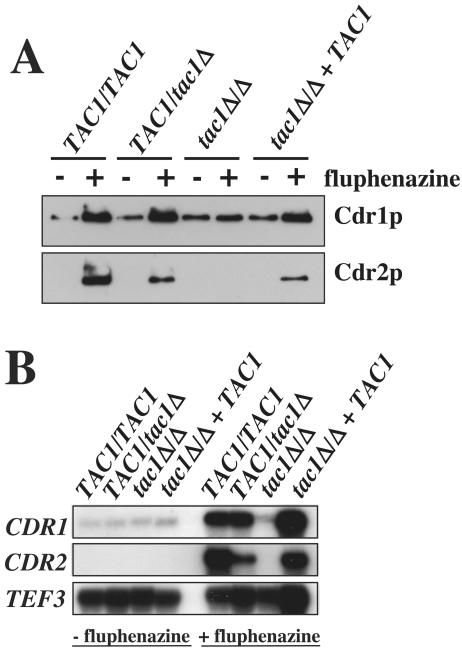
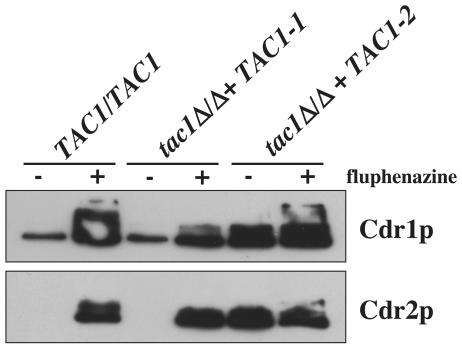
Southern analysis of the ZNC2 (now renamed TAC1 [transcriptional activator of CDR genes]; see below) disruption revealed the expected restriction fragments in the analyzed mutants (Fig. 1B). These C. albicans strains, i.e., the heterozygous TAC1/tac1Δ mutant DSY2875, the homozygous tac1Δ/Δ mutant DSY2903, and the revertant (tac1Δ/Δ TAC1) DSY2937, were next subjected to an analysis of CDR1 and CDR2 expression under conditions of fluphenazine exposure or no fluphenazine exposure, which are known to affect the transporters. While the levels of Cdr1p and Cdr2p were elevated by fluphenazine exposure in wild-type strain CAF2-1, these levels remained unchanged in the tac1Δ/Δ strain (Fig. 2A). Intermediate increases in the levels of Cdr1p and Cdr2p were observed in the TAC1/tac1Δ strain and the revertant DSY2937. Northern analysis of the same strains with labeled CDR1 and CDR2 probes (Fig. 2B) demonstrated that TAC1 controls the upregulation of CDR1 and CDR2 at the transcriptional level: while CDR1 and CDR2 were upregulated in CAF2-1 and the TAC1/tac1Δ strain, upregulation was absent in the tac1Δ/Δ strain after exposure to fluphenazine but was restored in the revertant DSY2937. Furthermore, relative β-galactosidase activities of CDR1- and CDR2-lacZ fusions introduced in the ura3 derivative of the tac1Δ/Δ strain DSY2903 were not increased after fluphenazine exposure (data not shown). Taken together, these data strongly suggest that the gene previously named ZNC2 is a positive regulator of both CDR1 and CDR2 and therefore can be renamed TAC1 (transcriptional activator of CDR genes).
FIG. 2.
TAC1 functions as a transcriptional activator of CDR1 and CDR2. (A) Immunodetection of Cdr1p and Cdr2p in TAC1 mutant and revertant strains. Protein extracts of each strain were separated by SDS-10% PAGE and immunoblotted with rabbit polyclonal anti-Cdr1p and anti-Cdr2p antibodies as described previously (8). C. albicans strains were grown in YEPD to mid-log phase and exposed (+) or not exposed (−) to fluphenazine (10 μg/ml) for 20 min. See the legend to Fig. 1 for strain and genotype designations. (B) Northern analysis of TAC1 mutant and revertant strains with CDR1 and CDR2 probes.
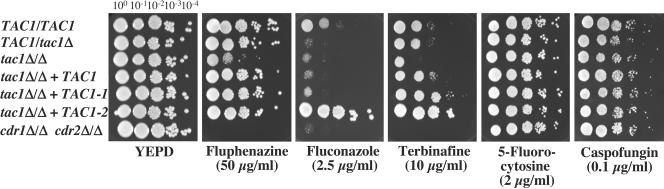
As expected from the role of TAC1 in CDR1 and CDR2 upregulation, the disruption of TAC1 alleles in C. albicans affected susceptibility to antifungal drugs and to other metabolic inhibitors: the tac1Δ/Δ strain was more susceptible to fluphenazine, fluconazole, or terbinafine than the wild-type strain, and this phenotype was reversed in strain DSY2937, into which the TAC1 allele from SC5314 was reintroduced (Fig. 3). The reversal of the drug susceptibility phenotype was best observed in the revertant DSY2937 on plates containing fluphenazine and terbinafine. However, the susceptibility of the tac1Δ/Δ mutant was not as pronounced as that of the cdr1Δ/Δ cdr2Δ/Δ mutant DSY654 in medium containing fluphenazine, fluconazole, or terbinafine: serially diluted cells of the cdr1Δ/Δ cdr2Δ/Δ mutant did not grow in the presence of these drugs, while residual growth was still observed for the tac1Δ/Δ mutant at low dilutions. These results can be explained by the contribution of the still measurable CDR1 expression in the tac1Δ/Δ mutant to the observed differences in drug susceptibility. On the other hand, when compounds such as flucytosine or caspofungin were tested with the tac1Δ/Δ mutant, no difference in susceptibility between the mutant and the wild type could be observed (Fig. 3). This effect is consistent with studies reporting that these two antifungal agents are not considered to be substrates of Cdr1p and Cdr2p (2, 34).
FIG. 3.
Drug susceptibility testing of C. albicans TAC1 mutant and revertant strains. Spotting assays were performed with serial dilutions of overnight cultures on YEPD containing the indicated drugs. Plates were incubated for 48 h at 35°C. The following strains correspond to the indicated genotypes: tac1Δ/Δ + TAC1-1, DSY2925; tac1Δ/Δ + TAC1-2, DSY2926; and cdr1Δ/Δ cdr2Δ/Δ, DSY654. See the legend to Fig. 1 for other strain and genotype designations.
TAC1 binds to the DRE of CDR1 and CDR2 and is localized in the nucleus.
De Micheli et al. showed previously that CDR1 and CDR2 contain a cis-acting element, the so-called DRE, that is important for their upregulation after drug exposure or for their constitutive high level of expression in azole-resistant isolates (8). Since TAC1 regulates CDR1 and CDR2, it is possible that the protein encoded by this gene binds to the DRE. As a first approach, TAC1 was expressed in both S. cerevisiae and C. albicans, and its activity was tested with the help of species-specific CDR2-lacZ chimeric constructs, one of which was devoid of the DRE. The S. cerevisiae integrative YIp353-derived plasmids contained the CDR2 promoter with or without the DRE (pDS1157 or pDS1180, respectively) and TAC1 from strain SC5314. The C. albicans integrative plasmids contained the CDR2 promoter with or without the DRE (pDS1187 or pDS1190, respectively) fused to Streptococcus thermophilus lacZ (38). The CDR2-lacZ fusions were subcloned into pDS1097, which contained TAC1 from strain SC5314. β-Galactosidase activities measured in S. cerevisiae revealed that the presence of TAC1 in pDS1157 could elevate reporter activities by approximately 40-fold (Table 6). However, the absence of the DRE in the CDR2 promoter subcloned in pDS1180 decreased reporter activities to almost background levels (Table 6). In C. albicans, β-galactosidase activities derived from the DRE-containing CDR2-lacZ fusion were near background levels under normal growth conditions; however, as expected, they were increased by fluphenazine treatment by approximately 50-fold. The absence of the DRE in plasmid pDS1190 abolished the increase in reporter activities (Table 6). Taken together, the data summarized in Table 6 demonstrate that TAC1 needs the DRE to activate the transcription of CDR2 not only in S. cerevisiae but also in C. albicans.
TABLE 6.
CDR2-lacZ reporter activities mediated by TAC1 in S. cerevisiae and C. albicans
| Type of promoter-lacZ fusion (plasmid) | Mean ± SD β-galactosidase activity (Miller units)a
|
|
|---|---|---|
| Without fluphenazine | With fluphenazine | |
| S. cerevisiae | ||
| CDR2-lacZ | 2.4 ± 0.4 | ND |
| TAC1-CDR2-lacZ (pDS1157)b | 96 ± 10 | ND |
| TAC1-CDR2 (lacking DRE)-lacZ (pDS1180)b | 0.7 ± 0.3 | ND |
| YIp353 (parent plasmid) | <0.1 | ND |
| C. albicans | ||
| TAC1-CDR2-lacZ (pDS1187)c | 0.3 ± 0.05 | 14 ± 4 |
| TAC1-CDR2 (lacking DRE)-lacZ (pDS1190)c | <0.1 | <0.1 |
| pDS178 (parent plasmid) | <0.1 | <0.1 |
Values are from three measurements. ND, not determined. Fluphenazine treatment was performed with 20 μg/ml for 60 min.
DSY669 was the recipient of pDS1157 and pDS1180.
DSY2906 was the recipient of pDS1187 and pDS1190.
To show further that Tac1p is able to bind to the DRE, a Tac1p chimeric protein was constructed by fusion of the first 129 amino acids of Tac1p with GST. The first 129 amino acids of Tac1p contained the Zn(2)-Cys(6) binuclear cluster domain expected to interact with DNA. This fusion construct enabled the production of a soluble protein in E. coli and its recovery by affinity purification. After purification of the fusion protein, band shift assays were performed with Tac1p-GST and a labeled 43-bp DNA fragment based on the CDR2 promoter sequence and containing the DRE. As shown in Fig. 4A and B, specific Tac1p-GST complexes with the labeled probe were visible. The formation of these complexes was dependent on the amount Tac1p-GST added to the reaction (Fig. 4A) and could be inhibited by increasing the addition of cold competitor from a 250- to a 1,000-fold excess (Fig. 4B). The binding of Tac1p-GST and a 43-bp fragment corresponding to the DRE-containing portion of the CDR1 promoter was also observed (Fig. 4C, probe 4), indicating that the DRE consensus sequence (boxed in the sequences given at the bottom of Fig. 4) rather than nucleotide sequences upstream or downstream of the DRE consensus sequence is responsible for binding. The formation of two specific complexes in these band shifts could be due to the formation of protein multimers, either from the presence of GST or from the binding of several protein moieties to the DRE.
Since Zn(2)-Cys(6) fingers are thought to bind to cis-acting elements via CGG triplets, transversions were performed on those of the DRE and the resulting probes were incubated with Tac1p-GST. As shown in Fig. 4C, the removal of one CGG triplet (probe 2) decreased the binding of Tac1p-GST compared to the results obtained with probe 1, while the removal of both CGG triplets (probe 3) completely abolished the binding of the fusion protein. However, CGG triplets must be present in the context of the DRE, since Tac1p-GST could not bind to the PDRE of PDR5 (probe 5), which also contains CGG triplets recognized by Zn(2)-Cys(6) binuclear cluster domains. That fact that Tac1p-GST can bind to the DRE strongly suggests that Tac1p is involved in the regulation of CDR1 and CDR2. The binding of this factor is probably a necessary step for the activation process resulting in the upregulation of both genes. Due to its DNA-binding activity, Tac1p is expected to be localized in the nucleus. To address this notion, GFP was fused to the C-terminal end of Tac1p, and the resulting fusion protein was observed by fluorescence microscopy. The fusion protein was transcriptionally functional, since it was able to restore CDR2 upregulation when present in the tac1Δ/Δ mutant (data not shown). Figure 5 shows that the fusion protein was localized in the nuclei of growing cells, since staining of nuclear DNA colocalized with GFP in the majority of the observed cells. The nuclear localization of Tac1p agrees with its function as a transcription factor.
FIG. 5.
Nuclear localization of Tac1p. DSY2906 transformed with pDS1202 was grown in liquid selective medium to a cell density of 107 cells/ml with constant agitation. Culture aliquots were sampled for nuclear staining as described in Materials and Methods and visualized by microscopy. Digital images were further processed with the computer program Adobe Photoshop 7.0 (Adobe Systems Incorporated, Mountain View, Calif.). Bar, 10 μm.
A TAC1 allele from an azole-resistant clinical isolate can confer azole resistance to a susceptible isolate.
Azole resistance can be coupled to the upregulation of CDR1 and CDR2. Several investigators demonstrated this feature in several azole-resistant isolates (28, 35, 39). One possible hypothesis to explain the constitutive upregulation of these transporter genes in C. albicans is that it exists in several TAC1 alleles and some of them are in a hyperactive state. To test this hypothesis, TAC1 alleles from an azole-susceptible strain (DSY294) and its azole-resistant parent (DSY296, a strain in which CDR1 and CDR2 are upregulated) were recovered and introduced into a ura3 derivative of the tac1Δ/Δ mutant, as described for the revertant strain DSY2937. The transformants (DSY2925 [tac1Δ/Δ TAC1-1], from DSY294, and DSY2926 [tac1Δ/Δ TAC1-2], from DSY296) then were exposed to fluphenazine to induce the expression of CDR1 and CDR2, as described above. Western blot analysis of the transformants with anti-Cdr1p and anti-Cdr2p antibodies revealed that, while the levels of these proteins were elevated by fluphenazine treatment in the revertant strain DSY2925 containing the TAC1-1 allele from DSY294, these levels were already elevated in the transformant DSY2926 containing a TAC1-2 allele from the azole-resistant strain DSY296 (Fig. 6). Our current data reveal that only TAC1-2 alleles have been recovered so far from the azole-resistant strain DSY296.
FIG. 6.
The TAC1-2 allele functions as a constitutive transcriptional activator of CDR1 and CDR2. Protein extracts of each strain were separated by SDS-10% PAGE and immunoblotted with rabbit polyclonal anti-Cdr1p and anti-Cdr2p antibodies as described previously (8). C. albicans strains were grown in YEPD to mid-log phase and exposed (+) or not exposed (−) to fluphenazine (10 μg/ml) for 20 min. See the legends to Fig. 1 and 3 for strain and genotype designations.
Drug susceptibility testing of these strains was performed by using a microtiter plate format with YEPD, increasing concentrations of fluconazole, and incubation for 24 h. The fluconazole MICs for strain CAF2-1, the tac1Δ/Δ mutant, and strain DSY2925 were between 0.5 and 1.0 μg/ml, but the MIC for strain DSY2926 was 16- to 32-fold higher (16 μg/ml), indicating that TAC1-2 mediates resistance to this specific agent. Furthermore, the presence of the TAC1-2 allele decreased susceptibility to other compounds, as shown for strain DSY2926 (Fig. 3, serial dilutions of tac1Δ/Δ TAC1-2). Azole resistance in strain DSY2926 could be explained by the increased expression of CDR1 and CDR2 and corresponding proteins—a consequence of the presence of TAC1-2. Experiments are in progress to demonstrate this hypothesis through the expression of the TAC1-2 allele in a cdr1Δ/Δ cdr2Δ/Δ mutant. Our present results demonstrate a direct relationship among the presence of TAC1-2 from an azole-resistant isolate, high cellular levels of Cdr1p and Cdr2p (Fig. 6), and the development of azole resistance.
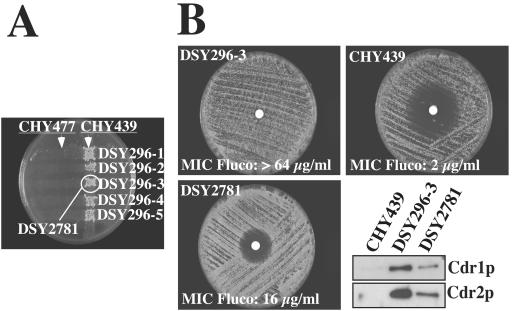
Codominance of azole resistance in C. albicans.
It was previously reported that homozygosity at the mating locus was strongly linked to the development of resistance to azoles (31). We partially confirmed this observation, as we determined, using the method described by Rustad et al. (31), that from among our collection of clinical isolates and among 13 azole-resistant matched isolates upregulating CDR1 and CDR2, 8 (2 MTLa/MTLa and 6 MTLα/MTLα) were homozygous at the mating locus (data not shown). In particular, the azole-resistant strain DSY296, from which only TAC1-2 alleles were recovered, was homozygous for MTLα, while the matched azole-susceptible strain DSY294 was still MTLa/MTLα. Since TAC1 is located upstream of the mating locus and since TAC1-2 is involved in CDR1 and CDR2 upregulation, it is possible that homozygosity at the mating locus can favor the development of azole resistance by rendering the MTL-associated TAC1-2 locus homozygous. In other words, gain-of-function TAC1-2 alleles could be codominant and need to be homozygous to confer sufficient CDR1 and CDR2 upregulation. To address this possibility, we crossed the Gal− derivative of DSY296 containing the TAC1-2 allele (DSY296-3 [MTLα/MTLα Gal−]) with the test strain CHY439 (MTLa/MTLa Gal+ Ade− Ura−). One of the resulting fusion products (DSY2781 [Gal+ Ura+ Ade+]) (Fig. 7A) had an intermediate phenotype with respect to azole susceptibility, as shown by fluconazole disk diffusion assays and MIC determinations. The fluconazole MICs for strains DSY296-3 and CHY439 were ≥64 and 2 μg/ml, respectively, while that for strain DSY2781 was 16 μg/ml (Fig. 7B). Accordingly, Cdr1p and Cdr2p levels were intermediate between DSY296-3 and CHY439 (Fig. 7B). Since TAC1-2 from DSY296-3 is responsible for CDR1 and CDR2 upregulation (Fig. 6), the reduced Cdr1p and Cdr2p levels in the fusion product DSY2781 are consistent with a codominance phenotype of the TAC1-2 allele. Moreover, the codominance of the gain-of-function TAC1-2 allele could explain the need for homozygosity at the MTL-associated TAC1 locus in strain DSY296 for high-level expression of the CDR1 and CDR2 genes.
FIG. 7.
Codominance of azole resistance in C. albicans. (A) Strain DSY296, which is homozygous at the MTLα locus, was mutagenized to obtain gal1 strains (numbered 1 to 5). These strains were crossed with mating test strains CHY477 (MTLα) and CHY439 (MTLa) (26) and replica plated on YNB containing galactose. (B) Fluconazole (Fluco) susceptibility of strain CHY439, strain DSY296-3, and fusion product DSY2781 tested by diffusion disk assays and by corresponding Western blotting with anti-Cdr1p and anti-Cdr2p antibodies.
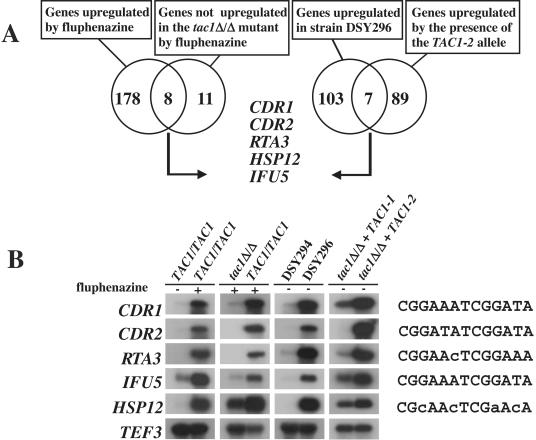
Gene expression profiling experiments reveal potential targets of TAC1.
CDR1 and CDR2 are not the only DRE-containing genes in the C. albicans genome; thus, it is possible that TAC1 could regulate these additional genes. TAC1 might also regulate genes through other indirect processes. In order to determine additional TAC1 gene targets, a gene expression profiling approach was used with microarrays of C. albicans. Several experiments were designed for this purpose. The numbers of genes upregulated in the presence of the hyperactive TAC1 allele (in a comparison of C. albicans strains DSY2925 and DSY2926 containing TAC1-1 and TAC1-2 alleles in a tac1Δ/Δ mutant, respectively) and of genes upregulated in the azole-resistant strain DSY296, from which the TAC1-2 allele originated (in a comparison of strains DSY294 and DSY296), were determined. We found 103 genes that were upregulated in the azole-resistant strain DSY296 compared to the azole-susceptible strain DSY294, 89 that were upregulated due to the presence of the TAC1-2 allele compared to the presence of the TAC1-1 allele in the tac1Δ/Δ mutant, and finally 7 genes that were upregulated in both experiments (Fig. 8A). We reasoned that genes upregulated in both experiments could be considered to be potential targets of TAC1. As expected, genes upregulated under both conditions included CDR1 and CDR2 but also RTA3 (probable transmembrane protein similar to S. cerevisiae YOR049c), HSP12 (heat shock protein), IPF14285 (unknown function), CDC23 (probable member of the anaphase-promoting complex), and IPF8179 (protein with high similarity to S. cerevisiae YJL068p, a putative esterase).
FIG. 8.
Microarray analysis reveals a subset of TAC1-dependent genes. (A) Venn diagram analysis of genes upregulated by fluphenazine in strain CAF2-1 and of genes no more upregulated under the same conditions in a TAC1 mutant strain (left side) and of genes commonly upregulated in strain DSY2926 expressing the TAC1-2 allele and in azole-resistant strain DSY296 (right side). DSY296 upregulates CDR1 and CDR2 and is the strain from which TAC1-2 originates. Genes common to these experiments are listed. A threshold of twofold was used to determine genes that were significantly upregulated. (B) Northern analysis of TAC1-regulated genes. DRE-like elements are shown for each gene investigated by Northern analysis. Lowercase letters show nucleotides different from those in the DRE of CDR1 and CDR2. See the legends to Fig. 1 and 3 for strain and genotype designations. −, no fluphenazine; +, fluphenazine.
We further extended our analysis by determining genes upregulated by fluphenazine treatment (in a comparison of strain CAF2-1 exposed to fluphenazine and strain CAF2-1 not exposed to fluphenazine) and next by identifying genes not upregulated by fluphenazine in a tac1Δ/Δ mutant strain compared to a wild-type strain (in a comparison of strain CAF2-1 exposed to fluphenazine and the tac1Δ/Δ mutant strain exposed to fluphenazine). We found 178 genes that were specifically upregulated by the addition of fluphenazine in the culture of the wild-type strain, 11 genes that were no more upregulated in the presence of fluphenazine in the tac1Δ/Δ mutant strain, and 8 genes that were upregulated by the addition of fluphenazine but that were also TAC1 dependent (Fig. 8A). The combination of these conditions revealed CDR1, CDR2, HSP12, and RTA3 but also IPF885 [protein with weak similarity to Xog1p, an exo-β-(1,3)-glucanase involved in cell wall metabolism], IPF1514 (unknown function), IPF17283 (unknown function), GRP2 (probable reductase similar to S. cerevisiae YOL151w), and GPX1 (probable glutathione peroxidase similar to the S. cerevisiae HYR1 gene product). Only four genes were common to these two microarray experiments, i.e., CDR1, CDR2, and RTA3, which all contain a consensus DRE in their promoters, and HSP12 (Fig. 8A).
In order to confirm the microarray results, Northern blotting was performed with total RNAs extracted from strains grown under the conditions tested in the microarray analysis. As shown in Fig. 8B, mRNA signals obtained for CDR1, CDR2, RTA3, and HSP12 confirmed the microaray results. These genes were upregulated in strain DSY2926 containing the hyperactive TAC1-2 allele, in strain CAF2-1 exposed to fluphenazine, and in the azole-resistant strain DSY296. It was reported previously that IFU5 (LPG20) was a DRE-containing gene (8). This gene failed to appear in the microarray analysis because it was flagged in two sets of experiments. However, IFU5 was coordinately upregulated with CDR1, CDR2, RTA3, and HSP12, as shown by Northern analysis (Fig. 8B). Taken together, our results indicate that the CDR1, CDR2, RTA3, IFU5, and HSP12 genes constitute a group of TAC1-dependent genes, consistent with the presence of a DRE-like region in these genes and with the ability of Tac1p to bind to the DRE.
DISCUSSION
Isolation of transcriptional activators of CDR1 and CDR2 by mining in the C. albicans genome.
In this article, we report the isolation of a transcription factor involved in the regulation of CDR1 and CDR2 by inspection of genome data. We focused on Zn(2)-Cys(6) transcription factors because these factors are able to target 5′-CGG-3′ motifs that are present in the DRE of CDR1 and CDR2 promoters. These transcription factors have in common a Zn(2)-Cys(6) DNA-binding domain which targets sequences with direct, inverted, or everted palindromic repeats containing a 5′-CGG-3′ motif with various numbers of nucleotides in the intervening spacer region. Several examples can be cited from studies performed with S. cerevisiae: Hap1p recognizes the direct repeat 5′-CGGNNNTANCGG-3′; the heterodimer Pip2p/Oaf1p binds to inverted repeats of the consensus sequence 5′-CGGN15-18-CCG-3′; Pdr8p and Yrr1p, which are factors involved in multidrug resistance, target DNA-binding motifs containing the 5′-CGG-3′ triplet in dyad symmetry; and Pdr1p and Pdr3p regulate their targets via the everted sequence motif 5′-TCCG/aC/tGG/cA/g-3′ (where lowercase letters indicate variable bases in a consensus motif) (9, 17, 23). More recently, it was shown that War1p binds the cis-acting element 5′-CGG-N23-CCG-3′, which is essential for PDR12 activation and resistance to weak acid stress (22).
Using the most recent assembly of the C. albicans genome, we were able to enumerate at least 72 ORFs with a Zn(2)-Cys(6) DNA-binding motif. Some ORFs might have been missed in this analysis; therefore, this number may be revised in future studies. Since no systematic collection of deletion mutants exists for C. albicans, potential candidates were selected based on their closest similarity to C. albicans genes that could interact with 5′-CGG-3′ triplets. Of particular interest was a cluster of three such genes (ZNC1 to ZNC3) located in a tandem repeat (the zinc cluster) (Fig. 1A) but showing a low level of similarity to each other. The inactivation of ZNC2 (TAC1) was sufficient to perturb the regulation of CDR1 and CDR2. Several pieces of experimental evidence were provided here to establish TAC1 as a positive activator of CDR1 and CDR2 through the cis-acting regulatory DRE: (i) a mutant lacking TAC1 was not capable of upregulating CDR1 and CDR2 after drug exposure, and this phenotype was restored in a revertant containing a single TAC1 copy (Fig. 2); (ii) a Tac1p-GST fusion containing the N-terminal DNA-binding domain could bind a 43-bp sequence containing a DRE previously identified as a cis-acting regulatory element of CDR1 and CDR2 (Fig. 4); (iii) a gain-of-function TAC1 allele from an azole-resistant isolate (TAC1-2) resulted in constitutive high-level expression of CDR1 and CDR2 in the laboratory strain CAF4-2 (Fig. 6); and (iv) the reporter activities of a CDR2-lacZ chimeric promoter were dependent on the presence of TAC1 and of the DRE either in S. cerevisiae or in C. albicans (Table 6).
Tac1p is a 981-amino-acid protein with typical features of zinc finger proteins of the Zn(2)-Cys(6) family. It contains, besides the DNA-binding domain (positions 33 to 80), an activation domain rich in acidic residues at the C terminus (from position 880) and a region (approximately from positions 400 to 510), called the middle homology region, that may be involved in the regulation of transcriptional activity (36). A BLAST search of the available GenBank data indicated the highest score (although the overall similarity was below 20%) for the S. cerevisiae protein Hal9p, which is a transcription factor that increases salt tolerance through the increased expression of ENA1 (Na+/Li+ extrusion pump) (25). The closest partners of Tac1p in C. albicans were those of the zinc cluster, especially the ZNC1 gene product (30% similarity); however, similarity to the other 71 members was generally below 20%. TAC1 is composed of two ORFs in the most recent assembly of the C. albicans genome (orf19.3188 and orf19.3189), since a nucleotide change introduces a stop codon. Our own verification of the TAC1 sequence resulted in a noninterrupted ORF. In conclusion, Tac1p is a protein with no known equivalent deposited in available protein databases, a result which could be expected from the motif of its DNA target, the DRE, which also has no equivalent among DNA-binding motifs in eukaryotes.
The removal of TAC1 abolished the upregulation of CDR2; thus, this factor represents a major regulator of this gene, even though the possibility that other, not-yet-identified factors could contribute to CDR2 upregulation cannot be excluded. The situation is slightly different for CDR1, which shows basal expression affected little by the absence of TAC1. Therefore, additional regulators for this gene can be expected. In a previous study, De Micheli et al. showed that the CDR1 promoter contained a basal expression element (8). Moreover, Gaur et al. (12) reported that the CDR1 promoter contained negative regulatory elements. Karnani et al. (19) described cis-acting elements that are different from the above-mentioned DRE and that are responsible for the upregulation of CDR1 by steroids. The protein factors responsible for these effects are still unknown.
Functional aspects and relevance of TAC1 for antifungal drug resistance.
The pathway leading to the activation of CDR1 and CDR2 through Tac1p is not yet established. Here we described two different possibilities for the upregulation of CDR1 and CDR2: either drug exposure or the expression of a mutant TAC1-2 allele. Zinc finger regulators can be activated by mechanisms such as overexpression, nuclear-cytoplasmic shuttling, DNA binding, phosphorylation, and unmasking of the activation domain. Interestingly, it was shown earlier that the intensity of the band shift was increased in cells exposed to CDR1 and CDR2 inducers. Even though the amounts of Tac1p in C. albicans still cannot be precisely quantified, these results suggest that activation could be due to the enhanced expression of TAC1 after drug induction (8). On the other hand, the fact that protein complexes probably containing Tac1p can bind to the DRE under normal growth conditions, as described by De Micheli et al. (8), suggests that Tac1p binds constitutively to the DRE. We showed here that Tac1p has a constitutive nuclear localization (Fig. 5); therefore, its activation is due not to migration into the nucleus but rather to a posttranslational modification(s). This feature was described recently for War1p. This zinc finger transcription factor has a nuclear localization and binds to its target in a constitutive manner. War1p can activate its target gene (the ABC transporter gene PDR12) through exposure to weak acid in a manner similar to that used here for Tac1p—exposure to fluphenazine. War1p is phosphorylated under these conditions, probably resulting in the activation of PDR12 (22). Such an activation mechanism could be envisaged for Tac1p, and experiments are currently under way to address this hypothesis.
The TAC1-2 allele isolated here from an azole-resistant strain does not require activation for the high-level expression of CDR1 and CDR2 (Fig. 6). Since the activation pathway for Tac1p is still not understood, it is difficult to provide explanations for this phenotype. However, since hyperactive alleles of transcription factors were described for PDR1 and PDR3 in S. cerevisiae and since these alleles contained gain-of function mutations, it is likely that TAC1-2 contains one or several mutations enabling the constitutive upregulation of CDR1 and CDR2. Preliminary data obtained from TAC1-1 and TAC1-2 sequencing revealed several substitutions for both alleles, but their individual contributions to CDR1 and CDR2 upregulation remain to be determined (Sanglard, unpublished).
In this study, we showed that the gain-of-function TAC1-2 allele could be codominant and showed that homozygosity at the MTL-associated locus could be needed for the high-level expression of CDR1 and CDR2 (Fig. 7). The results of an analysis of MTL locus homozygosity in azole-resistant isolates from our collection are in good agreement with the results obtained by Rustad et al. (31), where a link between mating locus homozygosity and the development of azole resistance was observed in different sets of clinical isolates. However, Pujol et al. (30) reported contradictory results showing that mating locus homozygosity was not correlated with azole resistance in different sets of clinical isolates. The observations made by these authors led us to conclude that other gain-of-function TAC1 alleles with a dominant phenotype can exist, thus making it possible that some azole-resistant isolates upregulating CDR1 and CDR2 are still heterozygous at the mating locus. To examine this possibility, current work in our laboratory is under way to characterize other TAC1 alleles from several azole-resistant isolates as being homozygous or heterozygous at the MTL locus. These TAC1 alleles will be also helpful for mapping of the mutations responsible for the constitutive high-level expression of CDR1 and CDR2.
TAC1 and gene targets.
We showed here that in addition to CDR1 and CDR2, Tac1p has other putative target genes, as inferred from a microarray analysis. In a previous study, De Micheli identified by inspection of genome data two genes (IFU5/LPG20) and RTA3/YFLO10C) containing a DRE in their promoters (8). These genes were upregulated by exposure to estradiol, one of the agents that is able to upregulate CDR1 and CDR2. Interestingly, through the use of a genome-wide approach with four microarray experiments (Fig. 8), these putative additional targets, i.e., RTA3 and IFU5, were reconfirmed as TAC1 targets. HSP12 also belongs to this group of coregulated genes. This gene contains a DRE-like region with four mismatches compared to the consensus sequence for CDR1, CDR2, IFU5, and RTA3. It is difficult to predict whether this DRE-like region is a target of Tac1p, since the minimal requirements of a fully functional DRE are still not known. Therefore, it will be necessary to further verify the possible occupancy of Tac1p on the promoters of these genes by coimmunoprecipitation experiments. Moreover, it is still possible that additional TAC1 targets will be identified in the future, since the microarrays used in this study do not include the entire collection of ORFs contained in the C. albicans genome.
In conclusion, we demonstrated here that TAC1 is a major factor needed for the regulation of CDR1 and CDR2, which are the main ABC transporter genes responsible for antifungal drug resistance in C. albicans. Other genes are under the control of TAC1, but their role in the development of antifungal drug resistance is still not known (but is under investigation). More important is that the identification of TAC1 provides new perspectives for the characterization of additional elements linked to the regulatory circuit controlling antifungal drug resistance.
ADDENDUM IN PROOF
Recent experiments have shown that the expression of the TAC1-2 allele in the background of a mutant lacking TAC1, CDR1, and CDR2 did not result in fluconazole resistance, thus demonstrating that TAC1-2 mediates azole resistance through the presence of both CDR1 and CDR2.
Supplementary Material
Acknowledgments
Sequence data for C. albicans were obtained from the Stanford Genome Technology Center website at http://www-sequence.stanford.edu/group/candida.
Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund. This research was supported by grants from the EC (QLK2-CT-2001-02377) and from the Swiss National Research Foundation (3200B0-100747/1).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alarco, A. M., I. Balan, D. Talibi, N. Mainville, and M. Raymond. 1997. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 272:19304-19313. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann, S. P., T. F. Patterson, and J. L. Lopez-Ribot. 2002. In vitro activity of caspofungin (MK-0991) against Candida albicans clinical isolates displaying different mechanisms of azole resistance. J. Clin. Microbiol. 40:2228-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balzi, E., and A. Goffeau. 1994. Genetics and biochemistry of yeast multidrug resistance. Biochim. Biophys. Acta 1187:152-162. [DOI] [PubMed] [Google Scholar]
- 4.Bissinger, P. H., and K. Kuchler. 1994. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J. Biol. Chem. 269:4180-4186. [PubMed] [Google Scholar]
- 5.Blackwell, C., C. L. Russell, S. Argimon, A. J. Brown, and J. D. Brown. 2003. Protein A-tagging for purification of native macromolecular complexes from Candida albicans. Yeast 20:1235-1241. [DOI] [PubMed] [Google Scholar]
- 6.Cormack, B. P., G. Bertram, M. Egerton, N. A. Gow, S. Falkow, and A. J. Brown. 1997. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology 143:303-311. [DOI] [PubMed] [Google Scholar]
- 7.Delaveau, T., A. Delahodde, E. Carvajal, J. Subik, and C. Jacq. 1994. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol. Gen. Genet. 244:501-511. [DOI] [PubMed] [Google Scholar]
- 8.De Micheli, M., J. Bille, C. Schueller, and D. Sanglard. 2002. A common drug-responsive element mediates the upregulation of the Candida albicans ABC transporters CDR1 and CDR2, two genes involved in antifungal drug resistance. Mol. Microbiol. 43:1197-1214. [DOI] [PubMed] [Google Scholar]
- 9.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 10.Feinberg, A., and B. Vogelstein. 1984. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137:266-267. [DOI] [PubMed] [Google Scholar]
- 11.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaur, N. A., N. Puri, N. Karnani, G. Mukhopadhyay, S. K. Goswami, and R. Prasad. 2004. Identification of a negative regulatory element which regulates basal transcription of a multidrug resistance gene CDR1 of Candida albicans. FEMS Yeast Res. 4:389-399. [DOI] [PubMed] [Google Scholar]
- 13.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorman, J. A., J. W. Gorman, and Y. Koltin. 1992. Direct selection of galactokinase-negative mutants of Candida albicans using 2-deoxy-galactose. Curr. Genet. 21:203-206. [DOI] [PubMed] [Google Scholar]
- 15.Gupta, V., A. Kohli, S. Krishnamurthy, N. Puri, S. A. Aalamgeer, S. Panwar, and R. Prasad. 1998. Identification of polymorphic mutant alleles of CaMDR1, a major facilitator of Candida albicans which confers multidrug resistance, and its in vitro transcriptional activation. Curr. Genet. 34:192-199. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning. A practical approach. IRL Press, Oxford, England.
- 17.Hikkel, I., A. Lucau-Danila, T. Delaveau, P. Marc, F. Devaux, and C. Jacq. 2003. A general strategy to uncover transcription factor properties identifies a new regulator of drug resistance in yeast. J. Biol. Chem. 278:11427-11432. [DOI] [PubMed] [Google Scholar]
- 18.Hube, B., M. Monod, D. A. Schofield, A. J. Brown, and N. A. Gow. 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 14:87-99. [DOI] [PubMed] [Google Scholar]
- 19.Karnani, N., N. A. Gaur, S. Jha, N. Puri, S. Krishnamurthy, S. K. Goswami, G. Mukhopadhyay, and R. Prasad. 2004. SRE1 and SRE2 are two specific steroid-responsive modules of Candida drug resistance gene 1 (CDR1) promoter. Yeast 21:219-239. [DOI] [PubMed] [Google Scholar]
- 20.Katzmann, D. J., P. E. Burnett, J. Golin, Y. Mahe, and W. S. Moye-Rowley. 1994. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol. Cell. Biol. 14:4653-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katzmann, D. J., T. C. Hallstrom, Y. Mahe, and W. S. Moye-Rowley. 1996. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J. Biol. Chem. 271:23049-23054. [DOI] [PubMed] [Google Scholar]
- 22.Kren, A., Y. M. Mamnun, B. E. Bauer, C. Schuller, H. Wolfger, K. Hatzixanthis, M. Mollapour, C. Gregori, P. Piper, and K. Kuchler. 2003. War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol. Cell. Biol. 23:1775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Crom, S., F. Devaux, P. Marc, X. Zhang, W. S. Moye-Rowley, and C. Jacq. 2002. New insights into the pleiotropic drug resistance network from genome-wide characterization of the YRR1 transcription factor regulation system. Mol. Cell. Biol. 22:2642-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mamane, Y., K. Hellauer, M. H. Rochon, and B. Turcotte. 1998. A linker region of the yeast zinc cluster protein leu3p specifies binding to everted repeat DNA. J. Biol. Chem. 273:18556-18561. [DOI] [PubMed] [Google Scholar]
- 25.Mendizabal, I., G. Rios, J. M. Mulet, R. Serrano, and I. F. de Larrinoa. 1998. Yeast putative transcription factors involved in salt tolerance. FEBS Lett. 425:323-328. [DOI] [PubMed] [Google Scholar]
- 26.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293-302. [DOI] [PubMed] [Google Scholar]
- 27.Myers, A. M., A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299-310. [DOI] [PubMed] [Google Scholar]
- 28.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perea, S., J. L. Lopez-Ribot, B. L. Wickes, W. R. Kirkpatrick, O. P. Dib, S. P. Bachmann, S. M. Keller, M. Martinez, and T. F. Patterson. 2002. Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 46:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pujol, C., S. A. Messer, M. Pfaller, and D. R. Soll. 2003. Drug resistance is not directly affected by mating type locus zygosity in Candida albicans. Antimicrob. Agents Chemother. 47:1207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rustad, T. R., D. A. Stevens, M. A. Pfaller, and T. C. White. 2002. Homozygosity at the Candida albicans MTL locus associated with azole resistance. Microbiology 148:1061-1072. [DOI] [PubMed] [Google Scholar]
- 32.Sanglard, D., F. Ischer, D. Calabrese, P. A. Majcherczyk, and J. Bille. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC-transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 34.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40:2300-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schjerling, P., and S. Holmberg. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24:4599-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talibi, D., and M. Raymond. 1999. Isolation of a putative Candida albicans transcriptional regulator involved in pleiotropic drug resistance by functional complementation of a pdr1 pdr3 mutation in Saccharomyces cerevisiae. J. Bacteriol. 181:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhl, M. A., and A. D. Johnson. 2001. Development of Streptococcus thermophilus lacZ as a reporter gene for Candida albicans. Microbiology 147:1189-1195. [DOI] [PubMed] [Google Scholar]
- 39.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, X., D. Talibi, S. Weber, G. Poisson, and M. Raymond. 2001. Functional isolation of the Candida albicans FCR3 gene encoding a bZip transcription factor homologous to Saccharomyces cerevisiae Yap3p. Yeast 18:1217-1225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.