Abstract
Yeast cells adapt to hyperosmotic shock by accumulating glycerol and altering expression of hundreds of genes. This transcriptional response of Saccharomyces cerevisiae to osmotic shock encompasses genes whose products are implicated in protection from oxidative damage. We addressed the question of whether osmotic shock caused oxidative stress. Osmotic shock did not result in the generation of detectable levels of reactive oxygen species (ROS). To preclude any generation of ROS, osmotic shock treatments were performed in anaerobic cultures. Global gene expression response profiles were compared by employing a novel two-dimensional cluster analysis. The transcriptional profiles following osmotic shock under anaerobic and aerobic conditions were qualitatively very similar. In particular, it appeared that expression of the oxidative stress genes was stimulated upon osmotic shock even if there was no apparent need for their function. Interestingly, cells adapted to osmotic shock much more rapidly under anaerobiosis, and the signaling as well as the transcriptional response was clearly attenuated under these conditions. This more rapid adaptation is due to an enhanced glycerol production capacity in anaerobic cells, which is caused by the need for glycerol production in redox balancing. Artificially enhanced glycerol production led to an attenuated response even under aerobic conditions. These observations demonstrate the crucial role of glycerol accumulation and turgor recovery in determining the period of osmotic shock-induced signaling and the profile of cellular adaptation to osmotic shock.
The ability to perceive and adapt to environmental changes is essential for all cells. Cellular adaptation to hyperosmotic shock in the yeast Saccharomyces cerevisiae requires accumulation of glycerol as compatible solute and is mediated by both transcriptional and metabolic alterations. Hyperosmotic shock evokes a broad transcriptional response, including that of several genes implicated in oxidative stress protection (8, 17, 25, 28, 39). In this work, we addressed the question of whether this is due to coregulation or due to osmotic shock resulting in oxidative stress. Such a link between osmotic shock and development of reactive oxygen species (ROS) could have specific regulatory and signaling roles, as suggested for liver cells (26, 27). Moreover, it has recently been reported that the yeast osmosensing high-osmolarity glycerol (HOG) pathway can also be stimulated by oxidative stress treatments (4). To study a possible link between osmotic and oxidative stress in yeast, we utilized the ability of S. cerevisiae to grow in the absence of oxygen, precluding the development of ROS.
Cells subjected to a hyperosmotic shock suffer loss of water and turgor pressure, transiently arrest proliferation, and recover by accumulating glycerol (reviewed in reference 18). Glycerol stimulates water uptake and restoration of turgor pressure, and the accumulation is controlled at three levels. Glycerol export is diminished due to closing of the turgor-responsive aquaglyceroporin Fps1 (33). In addition, glycolytic flux is stimulated by activation of phosphofructokinase (13). Finally, glycerol production capacity is increased by enhanced production of Gpd1 (glycerol-3-phosphate dehydrogenase) and Gpp2 (glycerol-3-phosphatase), the enzymes converting dihydroxyacetonephosphate to glycerol. Stimulated expression of the genes GPD1 and GPP2 is characteristic of the transcriptional response to osmotic shock (1, 22).
The transcriptional response to a hyperosmotic shock is largely mediated by the HOG signaling pathway (11, 18, 23). This branched mitogen-activated protein kinase (MAPK) pathway is controlled by different sensing systems converging on the MAPK kinase Pbs2, which activates the MAPK Hog1 by dual phosphorylation on threonine and tyrosine. One striking feature of the transcriptional response is its broad scope (8, 17, 25, 28, 39), encompassing genes lacking obvious roles, or even phenotypes (38), during osmotic adaptation. This general stress response (31), or environmental stress response (17), includes genes involved in the oxidative stress response, such as CTT1 (catalase T) and SOD2 (superoxide dismutase). The concurrent induction of the oxidative and osmotic stress responses suggests a fundamental coregulation. However, it could also be due to reduced intracellular water levels leading to generation of reactive oxygen species, for instance by affecting mitochondrial functions.
S. cerevisiae is a facultative anaerobe that can grow equally well aerobically and anaerobically in the presence of glucose (15, 16). While the glycolytic pathway is redox balanced (i.e., all NADH produced in the upper part is oxidized to NAD+ by conversion of pyruvate to ethanol), a net surplus of NADH is caused by conversion of a fraction of the glycolytic intermediates to biomass (30). Under anaerobic conditions, when respiration cannot account for NADH oxidation, reoxidation of NADH is achieved by reducing dihydroxyacetonephosphate to glycerol. This is a specific adaptation: expression of GPD2, which encodes an isoform of glycerol-3-phosphate dehydrogenase, is stimulated under anaerobic conditions, resulting in elevated glycerol production capacity (3, 5). The osmotic balance is maintained under these conditions by glycerol efflux across the plasma membrane via Fps1. Consistently, an fps1Δ mutant fails to grow under anaerobic conditions (33).
In this work, we have studied the osmotic stress response during anaerobiosis by using global gene expression analysis. We employed a novel two-dimensional visualization of hierarchical clusters to compare transcriptional expression data originating from aerobic and anaerobic cultures subjected to osmotic stress. This two-dimensional plotting of clustered data allows an easy comparison of two-variable queries, in this case osmotic stress and oxygen availability. It highlights the correlation between the data sets and facilitates interpretation. The comparison clearly showed that the osmotic stress response encompasses the presumed oxidative stress response genes even during anaerobic growth. However, anaerobic cells adapt faster and show a shorter, less pronounced response to osmotic shock. We show that this is due to the high intrinsic glycerol production, highlighting the role of glycerol accumulation and turgor recovery in the osmotic adaptation profile.
MATERIALS AND METHODS
Strains and plasmids.
The S. cerevisiae strains used in this work were W303-1A (MATaleu2-3/112 ura3-1 trp1-1 his3-11/15 ade2-1 can1-100 GAL SUC2 mal0) (35) and an isogenic prototrophic strain (9). The multicopy vector YEplac112, containing a 2.9ikb ClaI-SwaI fragment of GPD1 (3), was used to overexpress GPD1.
Cell culturing and sampling.
Cultivations were carried out under either aerobic or anaerobic conditions in a fermentor with a working volume of 750 ml (Belach Bioteknik AB). CBS medium (37) with 7.5 g of (NH4)2SO4 per liter as a nitrogen source and 20g of glucose per liter as a carbon and energy source was used for all experiments. Cultures with the auxotrophic W303-1A strain were supplied with 120 mg of each of the required amino acids per liter. The temperature was 30°C, the stirring rate 400 rpm, and the pH was kept constant at 5.0 by automatic addition of 1 M NaOH. The cultures were continuously flushed (35 liters/h) with either nitrogen gas for the anaerobic conditions or air for the aerobic conditions. The heat production rate (dQ/dt) was measured with a flowthrough microcalorimeter as described previously (36). Carbon evolution was continuously analyzed with a carbon dioxide and oxygen monitor (type 1308; Bruel and Kjaer). Growth of the cultures was monitored both by measuring the heat production rate and by performing on-line analysis of outgoing gas. Cells for the GPD1 overexpression experiment were cultivated in E-flasks. For osmotic shock treatment, 2.5 M NaCl was added to a final concentration of 0.5 M when the cells had reached mid-logarithmic phase (optical density of 0.8). For gene expression analysis, cells were harvested in ice-cold water, while cells for Western analysis were rapidly cooled in a dry-ice-ethanol bath.
Glycerol measurements.
For total glycerol, samples were taken and boiled at 100°C for 10 min. After sedimentation, the supernatant was kept for further analysis. For intracellular glycerol, cells from 1 ml of sample were resuspended in 1 ml of water and boiled at 100°C for 10 min, and the supernatant after sedimentation was kept as before. Determination of glycerol concentration was performed with a glycerin-glycerol kit (Roche) and a Biomek 2000 laboratory robot (Beckman Instruments).
RNA extraction and Northern blotting.
Total RNA extraction and Northern blotting were performed as described previously (12). Probes were generated by using α[32P]dCTP, the MegaPrime Kit (Amersham), and PCR fragments of the STL1 open reading frame. Primers used for amplification had the sequences (5′ to 3′) TAAGCAGAACCAGTCACTGG and GTAGATTGTTGCGAAGACCC. Probes were purified by using Nick columns (Amersham), and 1,000,000 cpm/ml was used for hybridization. The signal was measured with an FX molecular imager (Bio-Rad) and Kodak K-screens. Quantification was performed with Quantity One software (version 4.2.3; Bio-Rad).
Global gene expression analysis.
Global gene expression was measured by using GeneFilters (GF100; Invitrogen) as described previously (28), and the signal was measured in a phosphorimager as for Northern analysis. Image analysis and quantification were performed with the Pathways 4 software (Invitrogen). Each filter was manually inspected for flaws, and invalid spots were removed prior to export to Microsoft Excel, where further analysis was performed. The ratio between the aerobic and anaerobic series was calculated for each time point. The time course expression profile for each condition was calculated as the ratio of the value at each time point against the average of those at the two prestress time points. If one of these was missing, the calculation was based on the existing measurement only. The spot intensity data and the ratio data used for the analysis are available in Tables S1 and S2, respectively, in the supplemental material, according to the minimal information about a microarray experiment standards.
Filtering and clustering.
The expression profiles were filtered to select genes that displayed a reliable expression change following osmotic shock. Two selection criteria were applied. First, the expression level had to change at least twofold compared to that for both prestress samples at two consecutive time points. This criterion eliminated expression profiles with small relative change. Second, no more than three changes in the sign of the derivative were allowed after the addition of salt. This criterion eliminated dubious expression profiles.
The first criterion is by far the most stringent, eliminating 96% of the genes, while the second criterion eliminates only 35% of the genes if applied by itself. Applying the second criterion to the 229 genes that passed the first criterion eliminates only two more genes, <1%, showing that the criteria favor the same set of genes (P = 3.6 × 10−28 by the chi-square test). Five genes passed only one criterion under each condition but were eliminated by applying the criteria simultaneously, leaving a set of 222 genes. Relaxation of criterion 1 by lowering the threshold to an altered expression of 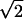 -fold instead of twofold results in a larger number of genes passing the filter while the two criteria still favor the same set of genes (P = 1.2 × 10−90 by the chi-square test).
-fold instead of twofold results in a larger number of genes passing the filter while the two criteria still favor the same set of genes (P = 1.2 × 10−90 by the chi-square test).
Genes passing both criteria were clustered by using Cluster and TreeView (http://rana.lbl.gov/EisenSoftware.htm) (14). The cluster analysis included three groups of genes: (i) those that displayed a significant expression change under both aerobic and anaerobic conditions, (ii) those with a significant expression change under aerobic conditions only, and (iii) those that showed a significant expression change under anaerobic conditions only. These groups were clustered separately for aerobic and anaerobic conditions, resulting in four distinct trees: tree A, aerobic expression profiles and significantly altered by osmotic shock; tree B, aerobic expression profiles but significantly altered by osmotic shock only under anaerobic conditions; tree C, anaerobic expression profiles and significantly altered by osmotic shock; and tree D, anaerobic expression profiles but significantly altered by osmotic shock only under aerobic conditions. These cluster trees were plotted against each other to produce Fig. 4.
FIG. 4.
Two-dimensional visualization of the correlation between aerobic and anaerobic expression. The upper (x) axis holds the cluster trees based on data obtained from the anaerobic culture. Clusters are labeled with roman numerals. The left (y) axis holds the cluster trees based on data obtained from the aerobic culture. These clusters are labeled with arabic numerals. Each gene occurs once on each axis, and ▪ marks the intersection between the positions on the two axes. The figure contains three different but connected plots that are separated by the dark grey lines. The upper left part contains the genes whose expression was significantly altered under both conditions (A versus C), the upper right part contains those whose expression was significantly altered under aerobic conditions only (A versus D), and the lower left contain those genes whose expression was significantly altered under anaerobic conditions only (B versus C). Cluster trees A and C are subdivided by general behavior, i.e., downregulation (group 1/I) or upregulation (group 2/II), by the lighter grey lines. These groups are further divided at a correlation of <0.75 by the thin black lines, grouping genes with closely related behavior. The grouping, rather than the physical distance, indicates similarity in behavior. Cluster trees B and D are subdivided by general behavior, i.e., downregulation (group 3/III), upregulation (group 5/V), and no or an ambiguous effect (groups 4/IV and 6). Figure 3 provides a numerical and simpler view. Further information can be found in Fig. S1 and S2 in the supplemental material. The color panels along the x and y axes show the induction profiles under anaerobic and aerobic conditions, respectively. Red indicates stimulated expression and green indicates diminished expression, compared to the average of the prestress time points. The time points are −30, −5, 10, 15, 20, 30, 40, and 60 min, starting from the periphery.
Protein extraction and Western blot analysis.
Cells were resuspended in sodium dodecyl sulfate (SDS) loading buffer (100 mM Tris-HCl [pH 6.8], 200 mM dithiothreitol, 4% SDS, 20% glycerol, 0.2% bromophenol blue, 20 mM mercaptoethanol, 10 mM NaF, 0.1 mM Na-vanadate, protease inhibitor [complete EDTA-free protease inhibitor cocktail tablets; Roche]). Cell suspensions were first boiled at 100°C for 10 min and then centrifuged at 16,000 relative centrifugal force and 4°C for 10 min to obtain pure protein extracts. Twenty to 25 μg of proteins was loaded on an SDS-10% polyacrylamide gel and blotted on a polyvinylidene difluoride membrane (Hybond-P; Amersham). Membranes were blocked with 5% milk (Difco) in Tris-buffered saline-Tween 20 (TBS-T). An antibody recognizing dually phosphorylated Hog1 (phospho-p38 MAPK [Thr180/Tyr182]; Cell Signaling) was diluted 1:1,000 in 5% bovine serum albumin-TBS-T, and the membrane was incubated overnight at 4°C. An antibody recognizing Hog1 independently of phosphorylation status (Hog1 yC-20; Santa Cruz Biotechnology) was used as control. It was diluted 1:200 in 5% milk-TBS-T, and the membrane was incubated for 1 h at room temperature. Secondary antibodies (horseradish peroxidase-anti-rabbit immunoglobulin G [Cell Signaling] and horseradish peroxidase-donkey anti-goat immunoglobulin G [Santa Cruz Biotechnology]) were applied in TBS-T at 1:2,000 and 1:1,500 dilutions, respectively. The Lumi-Light Western blotting substrate (Roche) and an LAS-1000 camera (Fuji) were used for visualization. Quantification was performed with ImageGauge 3.46 (Fuji).
ROS detection and microscopy.
After reaching mid-logarithmic phase, cells were incubated with dihydroethidium (5 μg/ml) with or without a stress agent (NaCl, sorbitol, or tert-butyl hydroperoxide solution) for 25 min in the dark at 30°C and were subsequently sedimented and washed once with phosphate-buffered saline. Detection of ROS was performed with a DMRXA fluorescence microscope (Leica), utilizing a fluorescein isothiocyanate filter. Dead cells were observed by using methylene blue dying and the normal bright filter.
RESULTS
Osmotic shock causes no detectable production of ROS.
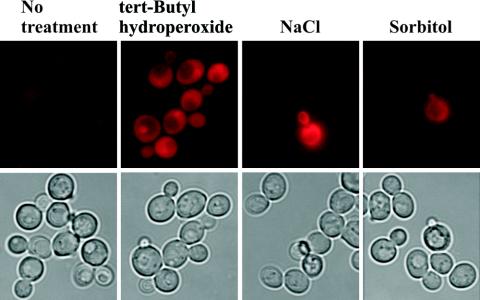
To determine if osmotic stress treatment resulted in development of ROS, cells were stained with dihydroethidium and inspected microscopically. While treatment with tert-butyl hydroperoxide, a ROS-generating compound, resulted in strong staining of all cells, addition of NaCl did not cause any staining above background in the majority of the cells (Fig. 1). However, a fraction of the cells showed strong staining following NaCl addition, and this fraction increased with increasing NaCl concentration (data not shown). The phase-contrast picture indicated that stained cells had not survived the treatment, as they showed an aberrant, granulated morphology typical of dead cells. Staining with methylene blue demonstrated that indeed such cells were dead (data not shown). Treatment with sorbitol instead of NaCl resulted in a similar picture, showing that osmotic shock does not result in strong development of ROS.
FIG. 1.
Osmotic shock does not result in detectable ROS production. The upper panels show ROS visualized with dihydroethidium in untreated samples or in samples treated with 1 mM tert-butyl hydroperoxide (positive control), 0.5 M NaCl, or 1 M sorbitol. The lower panels show the same cells through the bright filter.
Recovery from a hyperosmotic shock is faster during anaerobiosis.
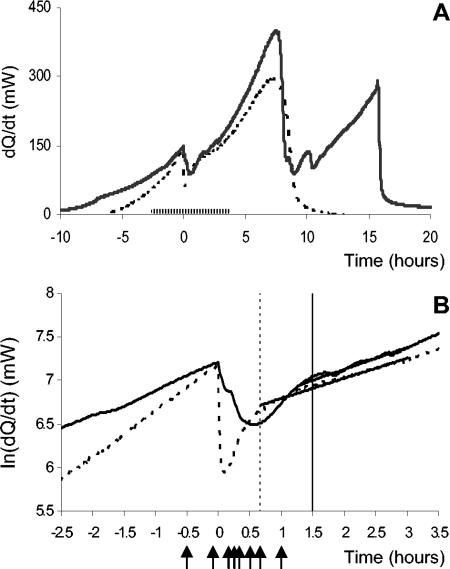
The osmotic shock responses in aerobic and anaerobic cultures were compared, with the latter precluding generation of ROS. The experiments were performed in controlled batch cultures with exponentially growing cells. Heat production was used as a measure of cellular metabolic activity, which reflects cell proliferation during logarithmic growth (6). Addition of NaCl to a final concentration of 0.5 M caused a transient drop in heat production (Fig. 2). This drop of metabolic activity was much more transient in the anaerobic culture, which, on average, recovered in around 40 min, compared to around 90 min for the aerobically grown cells. In contrast, the amplitudes of the drops were similar, on average around −50%, in the anaerobic and aerobic cultures. About half of this drop can be accounted for by dilution, as it occurs upon addition of water only but with no adaptation time. In the absence of salt stress, the heat generation profiles and generation times did not differ significantly between aerobic and anaerobic cultivation, in accordance with previous observations (6).
FIG. 2.
Yeast cells recover faster from osmotic shock under anaerobiosis. Heat production was used to monitor metabolic activity. Solid lines represent the aerobic culture, and dashed lines represent the anaerobic culture. The first peak in heat production represents fermentative growth, and the second, which is present only in the aerobic culture, represents ethanol respiration. Panel A shows the complete heat production profile of the two cultures, while B is a logarithmic display of the critical period indicated by a hatched bar on the x axis in panel A. Exponentially growing cells were subjected to an osmotic shock at time zero. The dashed and solid vertical lines indicate when the anaerobic and aerobic cultures, respectively, resumed exponential heat production, which occurred after approximately 40 min in the anaerobic culture and after 90 min in the aerobic culture. Arrows indicate sampling points for global expression analysis.
Time course analysis of global gene expression.
The differences in the global transcriptional response to hyperosmotic stress between aerobiosis and anaerobiosis were assessed with time course experiments. These were performed with gene filter arrays at 30 and 5 min before and at 10, 15, 20, 30, 40, and 60 min after the addition of NaCl, as indicated in Fig. 2B. Multiple Northern blot analyses of selected genes by using RNA from the same as well as additional cultures confirmed the results obtained with gene filters.
Comparison of our data with the two previously published microarray analyses that employed cells grown under anaerobic conditions revealed no significant overlap. This is likely due to different experimental conditions: while we employed batch cultures under glucose repression, ter Linde et al. (34) used glucose-limited respiratory cultures, and Kwast et al. (20) used galactose as a carbon source. Indeed, several genes classified as aerobic in these studies encode respiratory functions. Expression of those genes is repressed under our culture conditions.
Gene selection and filtering.
To characterize the differences between the transcriptional profiles following osmotic shock in anaerobic and aerobic cells, the analysis was limited to genes showing a significant osmotic upregulation or downregulation, as described in Materials and Methods. The result was a set of 222 genes showing stable and at least twofold up- or downregulation under at least one of the two conditions. These genes are listed in Table S3 in the supplemental material.
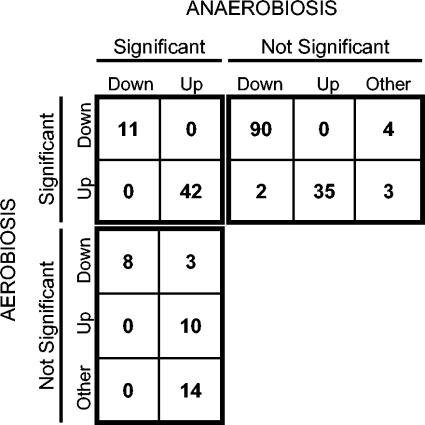
Of these 222 genes, 187 (84%) showed a significant osmotic response under aerobic conditions and 88 (40%) showed a significant osmotic response under anaerobic conditions, with an overlap of 53 (24%) that showed a significant response under both conditions (Fig. 3). This overlap is significant (P = 3.8 × 10−217 by the chi-square test). There was no single gene that showed significantly increased expression following osmotic shock under one condition and significantly decreased expression under the other condition, indicating that yeast cells respond to osmotic shock in basically similar ways under aerobic and anaerobic conditions (P = 1.3 × 10−11 by Fisher's exact test). Lowering the threshold for induction or repression to 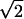 resulted in a larger data set but did not alter the general trends: 96% of the 166 genes that showed significantly altered expression changes under both conditions altered expression in the same direction (P = 1.2 × 10−33 by the chi-square test).
resulted in a larger data set but did not alter the general trends: 96% of the 166 genes that showed significantly altered expression changes under both conditions altered expression in the same direction (P = 1.2 × 10−33 by the chi-square test).
FIG. 3.
The correlation between the aerobic and anaerobic responses to osmotic stress is very high. The figure illustrates the behavior of the genes included in this study and is a numerical representation of the clustering results (see Fig. 4). Expression of 105 genes was significantly downregulated upon osmotic shock under aerobic conditions. Expression of 101 (96%) of those was clearly downregulated also under anaerobic conditions, but only 11 (10%) of those passed the filtering criteria (row 1). Likewise, expression of 82 genes was significantly upregulated by osmotic shock under aerobic conditions. Expression of 77 (94%) of those was clearly upregulated also under anaerobic conditions, but only 42 (51%) passed the filtering criteria (row 2). Moreover, all 19 genes whose expression was significantly downregulated upon osmotic shock under anaerobic conditions were also downregulated under aerobic conditions (column 1). In contrast, only 52 (75%) of the 69 genes whose expression was significantly upregulated under anaerobic conditions were also upregulated aerobically (column 2).
The transcriptional response to osmotic stress is quantitatively but not qualitatively different in aerobiosis and anaerobiosis.
To analyze the expression profiles in more detail, we employed hierarchical clustering (14). Clustering was performed independently for the aerobic and anaerobic expression profiles, resulting in four different trees: tree A, expression profiles from cells grown under aerobic conditions and significantly altered by osmotic shock; tree B, expression profiles from cells grown under aerobic conditions but significantly altered by osmotic shock only under anaerobic conditions; tree C, expression profiles from cells grown under anaerobic conditions and significantly altered by osmotic shock; and tree D, expression profiles from cells grown under anaerobic conditions but significantly altered by osmotic shock only under aerobic conditions. The clusters were then plotted against each other in a two-dimensional matrix to assess the degree of correlation between the data sets (Fig. 4; a numerical presentation is given in Fig. 3). Figure S1 in the supplemental material provides an extended presentation, including gene names.
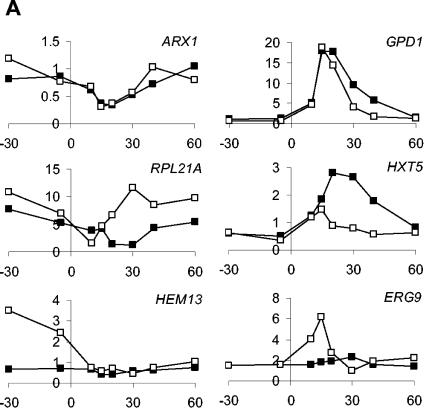
This presentation demonstrates the high degree of similarity in the response to osmotic shock under aerobic and anaerobic conditions. Three observations are of specific interest. First, as stated above, all genes displaying significantly altered expression changes under both aerobic and anaerobic conditions changed expression in the same direction under both conditions. Second, the vast majority (88%) of the osmoregulated genes showed similar expression profiles following osmotic shock under anaerobic and aerobic conditions, even though the expression change sometimes passed the significance criteria for only one condition. The main differences are quantitative, as expression changes were attenuated in cells growing under anaerobic conditions. Third, the genes that did not show similar behavior under aerobiosis and anaerobiosis were not randomly distributed. Indeed, they were concentrated in the group of genes significantly induced under anaerobic conditions (P = 1.2 × 10−5 by the chi-square test). This group, which contains several genes encoding enzymes in ergosterol biosynthesis, is discussed further below. Selected gene expression profiles are shown in Fig. 5A.
FIG. 5.
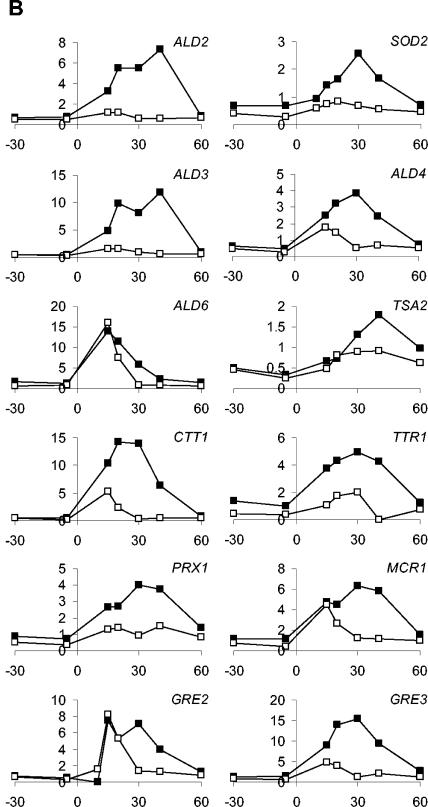
Expression profiles of selected genes. (A) Examples of expression profiles of genes discussed in this study. (B) Expression profiles of the 12 genes implicated in the oxidative stress response. The x axis displays time in minutes, and the y axis displays mRNA levels in arbitrary units. Closed squares represent aerobic expression patterns, and open squares represent anaerobic expression patterns.
Oxidative stress genes.
We looked more closely at the genes whose expression was significantly upregulated by osmotic shock and that encoded functions known or presumed to be relevant for combating the toxicity of ROS or products thereof. The following genes were included in this analysis: ALD2, SOD2, ALD3, ALD4, ALD6, TSA2, CTT1, TTR1, PRX1, MCR1, GRE2, and GRE3 (Fig. 5B). This set followed the trend observed for the majority of upregulated genes; i.e., stimulated expression was on average twofold higher after osmotic shock under aerobic conditions. At the same time, it was apparent that all of the oxidative stress genes were also upregulated under anaerobic conditions. Statistical analysis showed that the induction levels of this set of genes did not differ significantly from those of the 70 other genes upregulated by osmotic shock (P = 0.35 by Student's t test).
Hog1-dependent genes and genes depending on different transcription factors.
Hog1 mediates osmotic upregulation of a large subset of genes, and it does so via different transcription factors, such as Hot1 and Msn2/Msn4 (18, 28). The present analysis included 28 (of 48), 6 (of 9), and 25 (of 46) of the genes previously listed as dependent on Hog1, Hot1, and Msn2/Msn4, respectively (28). Of the Sko1 transcription factor targets, only one gene was included (29). The induction levels of the Hog1-, Hot1-, and Msn2/Msn4-dependent subsets of genes did not show significant differences (by Student's t test) from each other or from those of the 43 upregulated genes not listed as targets of any of them. Therefore, differences in the induction levels upon osmotic shock under aerobic and anaerobic conditions are not biased towards any of the regulatory proteins considered in this analysis.
Genes encoding functions in ergosterol biosynthesis.
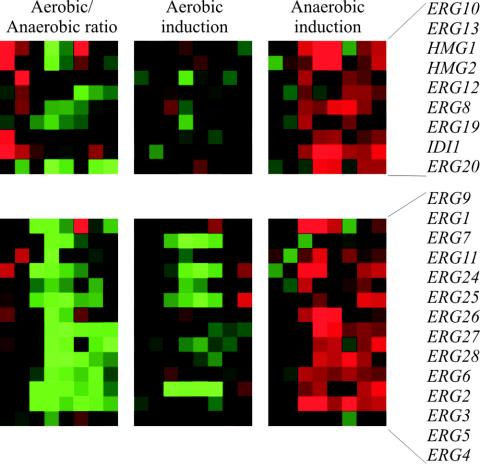
Among the 17 genes showing significantly enhanced expression following osmotic shock only under anaerobic conditions, we found ERG9, ERG10, ERG13, and ERG26, which encode enzymes in ergosterol biosynthesis. These constitute a significant overrepresentation (P = 1.8 × 10−60 by the chi-square test), and indeed, closer inspection revealed that expression of a majority of the genes encoding enzymes in ergosterol biosynthesis appeared to be upregulated following osmotic shock in the anaerobic but not in the aerobic culture (Fig. 6). Genes encoding enzymes in phospholipid and sphingolipid biosynthesis (10) did not show such behavior. Ergosterol is an essential yeast membrane sterol that requires molecular oxygen for production and hence is supplemented to the anaerobic culture. Expression of the ERG genes is controlled by oxygen levels (see reference 21 and references therein) and is repressed under aerobic conditions. However, the strict anaerobic conditions employed here did not result in significant upregulation of the ERG genes compared to aerobic conditions.
FIG. 6.
Expression of genes encoding enzymes of the ergosterol biosynthetic pathway. The genes are listed in the order in which their products function in ergosterol biosynthesis. Erg10 to Erg20 constitute the first part of the pathway, and Erg9 to Erg4 constitute the second part. The left panels display the ratio between aerobic conditions and anaerobic conditions. Red indicates higher expression under aerobic conditions, and green indicates higher expression under anaerobic conditions. The middle and right panels show the expression profiles under aerobic and anaerobic conditions, respectively. Red indicates stimulated expression and green diminished expression, compared to the average for the prestress time points. Note the presence of a clear trend of induction under anaerobic but not under aerobic conditions.
That expression of these genes was not stimulated by osmotic stress per se is clear from the lack of induction in the aerobic cultures. We tested whether this effect might have been a response caused by the minute amounts of oxygen possibly dissolved in the NaCl solution used to apply stress. Indeed, ERG gene upregulation could be provoked by addition of water only, even by water whose oxygen content was below the detection level of the gas analyzer (data not shown). Still, salt or water addition sufficed to briefly repress HEM13 (Fig. 5A), a known oxygen-repressed gene (2). We conclude that the expression of this set of genes is exquisitely sensitive to addition of even trace amounts of oxygen.
Faster glycerol accumulation accounts for more rapid osmotic adaptation under anaerobic conditions.
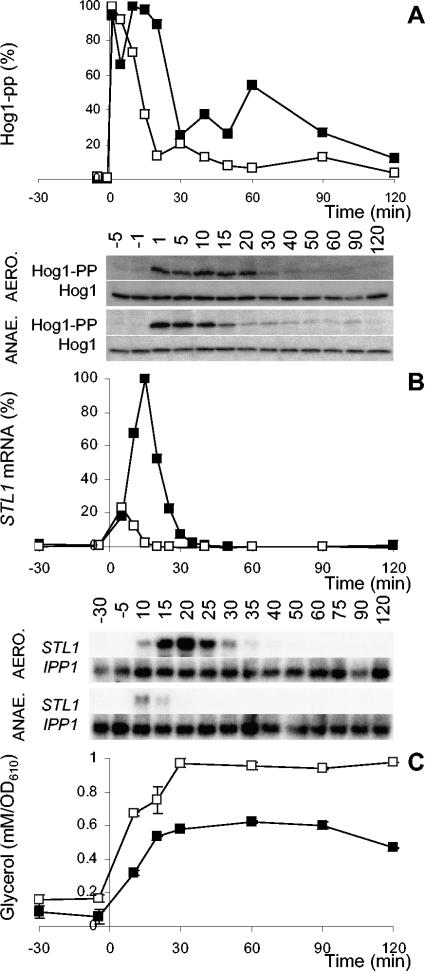
The profiles of heat production and gene expression suggested that yeast cells adapted faster to an osmotic shock under anaerobic conditions but did not provide an explanation for the phenomenon. In order to address this, parallel measurements of HOG pathway activity, target gene expression and glycerol accumulation were analyzed. Full activation, as measured by dual phosphorylation of Hog1, occurred within 1 min following osmotic shock in both the aerobic and anaerobic cultures (Fig. 7A). However, activation was more transient under anaerobic conditions. STL1 is the most strongly osmoinduced yeast gene, and its induction fully depends on the HOG pathway (28). Northern blot analysis confirmed gene filter data in that expression of STL1 was stimulated to a lower level and for a shorter period in the anaerobic cultures (Fig. 7B). Furthermore, the intracellular glycerol content increased more rapidly in the anaerobic culture (Fig. 7C).
FIG. 7.
Anaerobic cultures display more transient Hog1p phosphorylation and target gene activation as well as faster glycerol accumulation. Osmotic stress activates the HOG pathway rapidly during both aerobiosis and anaerobiosis, but the signal is more transient in the anaerobically growing cells, indicative of a faster adaptation (A). Accordingly, induction of the HOG pathway-dependent STL1 gene is strongly reduced during anaerobic conditions, in both amplitude and duration (B). Furthermore, the accumulation of intracellular glycerol occurs faster during anaerobiosis (C). Closed squares represent the aerobic culture, and open squares represent the anaerobic culture. OD610, optical density at 610 nm.
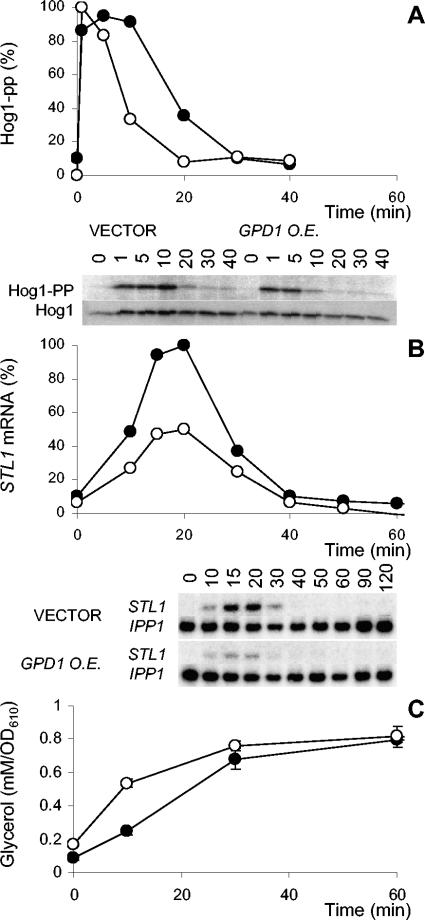
In order to determine whether the quicker adaptation in anaerobic cultures was due to the intrinsically higher glycerol production capacity or any other property of anaerobic growth, we tested the effect of GPD1 overexpression on the osmotic adaptation profile in an aerobic culture. The GPD1-overexpressing cells, like the anaerobic cultures, showed a more transient Hog1 phosphorylation (Fig. 8A), reached only a lower level of STL1 mRNA (Fig. 8B), and accumulated glycerol faster (Fig. 8C). Hence, a higher intrinsic capacity to produce glycerol prepares yeast cells for, and allows a faster adaptation to, osmotic shock.
FIG. 8.
GPD1 overexpression mimics anaerobic conditions. Cells overexpressing (O.E.) GPD1, i.e., with an increased intrinsic glycerol production capacity, show a more transient HOG pathway activation (A). Furthermore, these cells show a reduced STL1 induction (B) and a faster accumulation of intracellular glycerol (C). Closed circles represent the wild type, and open circles represent the GPD1-overexpressing strain. OD610, optical density at 610 nm.
DISCUSSION
Osmotic shock, like other acute stress conditions, leads to a broad transcriptional response in which many genes displaying stimulated expression do not appear to have a significant role in adaptation. In this work, we addressed the question of whether osmotic shock causes oxidative stress or whether the oxidative component of the transcriptional response constitutes an integral part of the overall response. Our data show that when oxygen is excluded from the culture, the osmotic stress response does not differ qualitatively from that under aerobic conditions and, in particular, encompasses the oxidative stress response genes. Furthermore, the analysis showed that cells grown anaerobically are preconditioned to osmotic shock, since the enhanced intrinsic glycerol production capacity allows cells to adapt and resume growth faster, resulting in an attenuated osmotic response.
The high intrinsic glycerol production capacity during anaerobiosis preconditions cells to hyperosmotic shock.
Cells growing under anaerobiosis adapt faster to osmotic shock, as illustrated by the much more transient drop in heat production and as further confirmed by the profiles of HOG signaling and gene expression changes. Hog1 phosphorylation, a measure for HOG pathway activity, was more short-lived in anaerobic than in aerobic cultures. As a consequence, HOG-dependent, as well as HOG-independent, gene expression changes generally reached lower amplitudes and/or were more transient. Faster adaptation is specifically well reflected in expression changes of the genes encoding functions in translation, which make up by far the largest class of downregulated genes. This downregulation reflects the proliferation arrest following stress treatment and is consequently more transient the faster the cell adapts and resumes proliferation (17, 28).
The faster adaptation is likely due to the higher capacity to produce glycerol in anaerobic cells, which essentially preconditions cells to osmotic stress. Under anaerobic conditions, yeast cells produce glycerol for redox regulation and expression of GPD2 is stimulated, leading to a higher capacity to produce glycerol (3), which is then accumulated faster following osmotic shock. Overexpression of GPD1 caused a similar faster glycerol accumulation and shorter periods of HOG pathway and HOG-dependent gene expression activation. Hence, faster adaptation of anaerobic cells is due to enhanced glycerol production and not to any other property of anaerobic growth.
These observations confirm the well-established central role of glycerol accumulation and hence turgor control in osmotic adaptation. This is in contrast to the facts that so many genes are upregulated following osmotic shock and that very few of these upregulated genes seem to have a functional role in adaptation to osmotic shock (38). GPD1 is one of them.
At the same time, the data demonstrate the less well established role of glycerol accumulation in determining the profile of osmotic adaptation. The shorter period of Hog1 phosphorylation in anaerobic cells and cells overexpressing GPD1 illustrates the important role of glycerol accumulation in determining the profile of HOG pathway activity. We have previously provided evidence, both experimentally and by mathematical modeling (19), that feedback control of the HOG pathway depends on successful osmotic adaptation and turgor resumption rather than activation of protein phosphatases. The data presented here add further evidence in support of this. We have previously shown that the inability to rapidly accumulate glycerol causes a longer period of Hog1 phosphorylation (19), while we show here that faster glycerol accumulation results in shorter periods of HOG pathway activity. Hence, the period of Hog1 phosphorylation is not determined by intrinsic feedback mechanisms alone but mainly by the ability of the cell to adapt to osmotic shock.
Two-dimensional comparison of hierarchical clusters.
We utilized a novel two-dimensional visualization method to discern differences and similarities between different sets of expression data. We propose that this method can be extended to an increased number of dimensions and thus conditions. One of the main limitations of hierarchical clusters is their one-dimensional nature. Clustering (14) results in a sorted data set based on similarity, but with no regard for complex patterns with multiple components. Still, it is an excellent tool for single-variable queries, such as which genes are stimulated in a similar time-dependent pattern in response to one alteration in growth conditions. It is, however, not an optimal tool for the analysis of the differences between two or more data sets, as it looks for only one pattern. In order to circumvent this limitation and profit from the virtues of the method, we used hierarchical clustering for the two data sets independently and subsequently plotted the results in a two-dimensional matrix in order to visualize the correlation between the data sets. The two-dimensional matrix provides a strong visualization aid, and, supported with mathematical tools, it should prove useful for the analysis of pattern correlation between any number of experiments by extending the number of dimensions.
The osmotic stress responses under aerobic and anaerobic conditions do not differ qualitatively.
As illustrated in Fig. 4, there is an almost perfect overlap between the genes showing up- or downregulated expression following osmotic shock under aerobic or anaerobic conditions. As discussed above, the differences are largely quantitative; i.e., they affect the period and amplitude of expression changes rather than the expression profile. In particular, the expression of a set of genes whose products are known or expected to function in adaptation to oxidative stress was upregulated even under anaerobic conditions, where generation of ROS is excluded. Although formation of non-oxygen-based radicals cannot be ruled out, the reducing environment makes the generation of radicals highly unlikely. In fact, as mentioned in the introduction, anaerobiosis leads to accumulation of surplus reducing power that the cells can eliminate only by producing glycerol. As no evidence for generation of ROS upon osmotic shock even under aerobic conditions could be found, it appears that the oxidative stress genes are coregulated with the general stress response genes, even when there is no obvious need for that.
The genes included in this list of oxidative stress genes are controlled by different factors. For instance, expression of CTT1 and ALD3 is known to be upregulated by means of the transcription factors Msn2 and Msn4, which are responsible for a general stress response (7, 32). Hence, expression of these genes is probably stimulated by all conditions that prevent yeast cells from optimal proliferation. Expression of many of the other genes in this list is at least partly controlled by Msn2 and Msn4 as well as by other, stress-specific regulators. GRE2, on the other hand, is an example of a gene that does not seem to be controlled by Msn2 and Msn4 but rather seems to be controlled by stress-specific mechanisms only, i.e., Hog1 and Sko1 under osmotic stress and Yap1 under oxidative stress (29). Hence, the apparent general stress-responsive behavior of these genes is not due to one single control mechanism.
Our observations essentially exclude the possibility that generation of ROS, as a consequence of osmotic shock, plays a significant role in sensing and signaling of osmotic stress. Such a scenario has been proposed for liver cells (26, 27). On the other hand, links between the osmotic and oxidative stress responses do exist. The HOG pathway can be activated not only by osmotic shock but also by treatments causing oxidative stress, although activation occurs much more slowly under these conditions, which could point to an indirect effect (4). In contrast, the analogous Sty1 pathway in fission yeast can readily be activated by osmotic shock as well as oxidative stress. It apparently lacks an obvious Sln1-like osmosensor, while putative oxidative stress sensors have been described (reviewed in reference 18). Another link between the osmotic and oxidative stress responses concerns the Hog1-dependent upregulation of a small set of genes encoding (presumed) functions in redox metabolism. It appears that the Sko1 transcription factor is largely dedicated to controlling this set of genes (29). Finally, the hog1Δ mutant, as well as mutants deficient in glycerol production, are sensitive to oxidative stress (4, 24). The mechanisms behind these observations remain to be established. In any case, while a close link between osmotic and ROS signaling may occur in some systems, this does not seem to be the case for S. cerevisiae, which is consistent with its life style as a facultative anaerobe.
Activation of components of the oxidative stress response in an environment devoid of such stress appears to be wasteful and implies a lack of optimization. In fact, most genes whose expression is stimulated in response to osmotic stress cause no phenotype when deleted (38) and are thus not essential parts of the adaptation process, at least not under laboratory conditions. This crude regulation limits the needs for signaling systems and could be rationalized as more economical than maintenance of elaborate sensing and signaling systems. A broad transcriptional response may simply be more cost-efficient than a larger number of regulatory components, as it allows the cell the flexibility to respond to a wide range of circumstances via few signaling mechanisms.
It appears that the selective pressure has not been on the stringency of transcriptional regulation. It may be argued that yeast, as a unicellular organism, manages to navigate a changeable environment with few signaling pathways, as it does not need to react with the same stringency and sensitivity as cells in multicellular organisms. Presumably, the selective pressure on stringency is higher in multicellular organisms, as the cost for individual cell failures is higher. This can be illustrated by the development of cancer, where defects in communication and cell cycle regulation in one cell result in the death of the entire organism containing billions of cells. On the other hand, every organism needs to balance the use of its resources between maintenance and growth, in reality making optimization of singular properties unrealistic even in higher organisms. Thus, it appears that stringency is sacrificed for flexibility, in a balance that we would expect to be the rule even in higher organisms, such as humans.
Supplementary Material
Acknowledgments
We thank the members of our group, as well as Jonas Warringer and Morgan Andreasson, for critical reading of the manuscript.
This work was supported by the Swedish Research Council (Vetenskapsrådet) (a research position to S.H.), the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas) (a research grant to S.H. for a Ph.D. position for M.K.), the National Research School for Genomics and Bioinformatics (Ph.D. positions for B.N. and M.J.), and the Swedish Energy Agency (a Ph.D. position for H.V.).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Albertyn, J., S. Hohmann, J. M. Thevelein, and B. A. Prior. 1994. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14:4135-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amillet, J. M., N. Buisson, and R. Labbe-Bois. 1996. Characterization of an upstream activation sequence and two Rox1p-responsive sites controlling the induction of the yeast HEM13 gene by oxygen and heme deficiency. J. Biol. Chem. 271:24425-24432. [DOI] [PubMed] [Google Scholar]
- 3.Ansell, R., K. Granath, S. Hohmann, J. M. Thevelein, and L. Adler. 1997. The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J. 16:2179-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilsland, E., C. Molin, S. Swaminathan, A. Ramne, and P. Sunnerhagen. 2004. Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol. Microbiol. 53:1743-1746. [DOI] [PubMed] [Google Scholar]
- 5.Björkqvist, S., R. Ansell, L. Adler, and G. Lidén. 1996. Glycerol-3-phosphate dehydrogenase mutants of Saccharomyces cerevisiae grown under aerobic and anaerobic conditions. Appl. Environ. Microbiol. 63:128-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomberg, A., C. Larsson, and L. Gustafsson. 1988. Microcalorimetric monitoring of growth of Saccharomyces cerevisiae: osmotolerance in relation to physiological state. J. Bacteriol. 170:4562-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boy-Marcotte, E., M. Perrot, F. Bussereau, H. Boucherie, and M. Jacquet. 1998. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costenoble, R., H. Valadi, L. Gustafsson, C. Niklasson, and C. J. Franzen. 2000. Microaerobic glycerol formation in Saccharomyces cerevisiae. Yeast 16:1483-1495. [DOI] [PubMed] [Google Scholar]
- 10.Daum, G., N. D. Lees, M. Bard, and R. Dickson. 1998. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast 14:1471-1510. [DOI] [PubMed] [Google Scholar]
- 11.de Nadal, E., P. M. Alepuz, and F. Posas. 2002. Dealing with osmostress through MAP kinase activation. EMBO Rep. 3:735-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Winde, J. H., M. Crauwels, S. Hohmann, J. M. Thevelein, and J. Winderickx. 1996. Differential requirement of the yeast sugar kinases for sugar sensing in establishing the catabolite-repressed state. Eur. J. Biochem. 241:633-643. [DOI] [PubMed] [Google Scholar]
- 13.Dihazi, H., R. Kessler, and K. Eschrich. 2004. HOG-pathway induced phosphorylation and activation of 6-phosphofructo-2-kinase are essential for glycerol accumulation and yeast cell proliferation under hyperosmotic stress. J. Biol. Chem. 279:23961-23968. [DOI] [PubMed] [Google Scholar]
- 14.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gancedo, C., and R. Serrano. 1989. Energy-yielding metabolism, p. 205-257. In A. H. Rose and J. S. Harrison (ed.), The yeasts, 2nd ed., vol. 3. Academic Press, New York, N.Y.
- 16.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klipp, E., B. Nordlander, R. Krüger, P. Gennemark, and S. Hohmann. Submitted for publication. [DOI] [PubMed]
- 20.Kwast, K. E., L. C. Lai, N. Menda, D. T. James III, S. Aref, and P. V. Burke. 2002. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184:250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lees, D. N., and M. Bard. 2004. Sterol biochemistry and regulation in the yeast Saccharomyces cerevisiae, p. 213-240. In G. Daum (ed.), Lipid metabolism and membrane biogenesis. Springer-Verlag, Heidelberg, Germany.
- 22.Norbeck, J., A. K. Påhlman, N. Akhtar, A. Blomberg, and L. Adler. 1996. Purification and characterization of two isoenzymes of dl-glycerol-3-phosphatase from Saccharomyces cerevisiae. Identification of the corresponding GPP1 and GPP2 genes and evidence for osmotic regulation of Gpp2p expression by the osmosensing mitogen-activated protein kinase signal transduction pathway. J. Biol. Chem. 271:13875-13881. [DOI] [PubMed] [Google Scholar]
- 23.O'Rourke, S. M., I. Herskowitz, and E. K. O'Shea. 2002. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 18:405-412. [DOI] [PubMed] [Google Scholar]
- 24.Påhlman, A. K., K. Granath, R. Ansell, S. Hohmann, and L. Adler. 2001. The yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J. Biol. Chem. 276:3555-3563. [DOI] [PubMed] [Google Scholar]
- 25.Posas, F., J. R. Chambers, J. A. Heyman, J. P. Hoeffler, E. de Nadal, and J. Arino. 2000. The transcriptional response of yeast to saline stress. J. Biol. Chem. 275:17249-17255. [DOI] [PubMed] [Google Scholar]
- 26.Reinehr, R., S. Becker, A. Hongen, and D. Haussinger. 2004. The Src family kinase Yes triggers hyperosmotic activation of the epidermal growth factor receptor and CD95. J. Biol. Chem. 279:23977-23987. [DOI] [PubMed] [Google Scholar]
- 27.Reinehr, R., F. Schliess, and D. Haussinger. 2003. Hyperosmolarity and CD95L trigger CD95/EGF receptor association and tyrosine phosphorylation of CD95 as prerequisites for CD95 membrane trafficking and DISC formation. FASEB J. 17:731-733. [DOI] [PubMed] [Google Scholar]
- 28.Rep, M., M. Krantz, J. M. Thevelein, and S. Hohmann. 2000. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275:8290-8300. [DOI] [PubMed] [Google Scholar]
- 29.Rep, M., M. Proft, F. Remize, M. Tamas, R. Serrano, J. M. Thevelein, and S. Hohmann. 2001. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol. Microbiol. 40:1067-1083. [DOI] [PubMed] [Google Scholar]
- 30.Rigoulet, M., H. Aguilaniu, N. Averet, O. Bunoust, N. Camougrand, X. Grandier-Vazeille, C. Larsson, I. L. Pahlman, S. Manon, and L. Gustafsson. 2004. Organization and regulation of the cytosolic NADH metabolism in the yeast Saccharomyces cerevisiae. Mol. Cell. Biochem. 256-257:73-81. [DOI] [PubMed] [Google Scholar]
- 31.Ruis, H., and C. Schüller. 1995. Stress signaling in yeast. Bioessays 17:959-965. [DOI] [PubMed] [Google Scholar]
- 32.Schüller, G., J. L. Brewster, M. R. Alexander, M. C. Gustin, and H. Ruis. 1994. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 13:4382-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamás, M. J., K. Luyten, F. C. W. Sutherland, A. Hernandez, J. Albertyn, H. Valadi, H. Li, B. A. Prior, S. G. Kilian, J. Ramos, L. Gustafsson, J. M. Thevelein, and S. Hohmann. 1999. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 31:1087-1104. [DOI] [PubMed] [Google Scholar]
- 34.ter Linde, J. J., H. Liang, R. W. Davis, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 1999. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J. Bacteriol. 181:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas, B. J., and R. J. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 36.Valadi, H., C. Larsson, and L. Gustafsson. 1998. Improved ethanol production by glycerol-3-phosphate dehydrogenase mutants of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 50:434-439. [DOI] [PubMed] [Google Scholar]
- 37.Verduyn, C. 1991. Physiology of yeasts in relation to biomass yields. Antonie Leeuwenhoek 60:325-353. [DOI] [PubMed] [Google Scholar]
- 38.Warringer, J., E. Ericson, L. Fernandez, O. Nerman, and A. Blomberg. 2003. High-resolution yeast phenomics resolves different physiological features in the saline response. Proc. Natl. Acad. Sci. USA 100:15724-15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yale, J., and H. J. Bohnert. 2001. Transcript expression in Saccharomyces cerevisiae at high salinity. J. Biol. Chem. 276:15996-16007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.