Abstract
Membrane fusion requires localized destabilization of two phospholipid bilayers, but unrestrained membrane destabilization could result in lysis. prm1 mutant yeast cells have a defect at the plasma membrane fusion stage of mating that typically results in the accumulation of prezygotes that have fingers of membrane-bound cytoplasm projecting from one cell of each pair into its mating partner in the direction of the osmotic gradient between the cells. However, some prm1 mating pairs fuse successfully whereas the two cells in other prm1 mating pairs simultaneously lyse. Lysis only occurs if both mating partners are prm1 mutants. Osmotic stabilization does not protect prm1 mating pairs from lysis, indicating that lysis is not caused by a cell wall defect. prm1 mating pairs without functional mitochondria still lyse, ruling out programmed cell death. No excess lysis was found after pheromone treatment of haploid prm1 cells, and lysis did not occur in mating pairs when prm1 was combined with the fus1 and fus2 mutations to block cell wall remodeling. Furthermore, short (<1 μm) cytoplasmic microfingers indicating the completion of cell wall remodeling appeared immediately before lysis. In combination, these results demonstrate that plasma membrane contact is a prerequisite for lysis. Cytoplasmic microfingers are unlikely to cause lysis since most prm1 mating pairs with microfingers do not lyse, and microfingers were also detected before fusion in some wild-type mating pairs. The lysis of prm1 mutant mating pairs suggests that the Prm1 protein stabilizes the membrane fusion event of yeast mating.
Phospholipid bilayer membranes form a barrier surrounding cells and intracellular organelles. Cellular membranes are inherently stable, but they are continually remodeled by budding and fusion events. During fusion, two membranes must be pulled together and reorganized in a controlled manner that avoids leakage or rupture.
The mechanism of membrane fusion is best understood for the fusion events that accompany infection by enveloped viruses. In the case of influenza virus, the hemagglutinin (HA) fusion protein in the viral envelope is activated to undergo a conformational shift by the pH change that accompanies virus internalization into endosomes. A fusion peptide that is initially buried within the hydrophobic core of an HA trimer is exposed and then inserted into the endosomal membrane. Assembly of an α-helical bundle then pulls the viral and cellular membranes together to drive fusion. Intracellular membrane fusion events during secretion and endocytosis have been proposed to occur via a similar mechanism. SNARE proteins extending from the cytoplasmic surfaces of vesicle and target membranes bind in trans to form an α-helical bundle that links the two membranes, ultimately leading to fusion.
In contrast to the extensive literature on viral and intracellular fusion, little is known about the mechanism of fusion between cells. In humans, cells fuse in a variety of contexts, including the fusion of sperm and egg to form a fertilized zygote, the fusion of trophoblasts to form the placenta, the fusion of myocytes to form muscle fibers, the fusion of fiber cells to form the lens of the eye, and the fusion of macrophage/monocyte-derived cells to form the large phagocytes involved in immune surveillance and bone resorption. No bona fide fusion proteins have been described for any of these cell fusion events, with the possible exception of trophoblast fusion, which can be blocked by antibodies against the fusion protein of an endogenous retrovirus (34).
We have chosen to study the cell fusion event of mating in Saccharomyces cerevisiae because this system offers the possibility of combining genetic, biochemical, and cell biological approaches. Yeast has two haploid mating types, MATa and MATα (reviewed in reference 31). Mating initiates when pheromones secreted by haploid yeast cells bind to receptors expressed on cells of the opposite mating type. A signaling pathway involving G proteins and mitogen-activated protein (MAP) kinases is activated in both haploid cells, resulting in arrest of the cell cycle before DNA synthesis, a shift in the cellular growth axis toward the pheromone source, and transcriptional induction of genes involved in the mating process. Haploid cells of opposite mating types bind to each other and then remodel their cell walls to allow their plasma membranes to contact each other and fuse. Later, the nuclei of the two parent cells fuse and a diploid daughter cell buds from the conjugation bridge connecting the two parents.
Cell fusion mutants have mating defects at a stage after haploid cells of the opposite mating type adhere to each other but before they fuse (46). Many of these mutants have a bilateral phenotype, meaning that cell fusion is more severely inhibited if the mutation is present in both cells of the mating pair (5, 45). Even in bilateral crosses, successful cell fusion generally occurs in a fraction of the mating pairs, suggesting that there is more than one pathway leading to cell fusion. In fus1, the first cell fusion mutant to be described, arrested prezygotes have cell walls separating the two plasma membranes, indicating a defect in cell wall remodeling rather than in membrane fusion (33, 45). The prezygotes that accumulate in the fus2, rvs161, spa2, pea2, bni1, and fig1 mutants also have intact cell walls (46).
In contrast to other cell fusion mutants, prm1 mating pairs arrest at a stage after cell wall remodeling but before fusion (18). The PRM1 gene encodes a pheromone-regulated membrane protein. This 115-kDa glycoprotein has five predicted transmembrane domains and is targeted to regions of contact between mating cells. No PRM1 homologues have been identified in mammalian cells, but there are homologous genes in Schizosaccharomyces pombe and Candida albicans. The prm1 mutant phenotype implicates Prm1 at the plasma membrane fusion stage of mating, but PRM1 is not essential for fusion because up to 40% of prm1 mating pairs are able to fuse.
We reexamined the prm1 mutant and found that 20 to 40% of prm1 mating pairs lyse instead of fusing or arresting as unfused prezygotes. Lysis only occurs after the plasma membranes of two prm1 mutants have come into contact and cannot be prevented by osmotic stabilization. Therefore, we propose that the primary function of Prm1 is to stabilize assembling fusion pores.
MATERIALS AND METHODS
The yeast strains used in this study were derived from strains produced by the yeast deletion consortium (http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html). Genes encoding freely soluble versions of the green fluorescent protein (GFP) and the Discoideum red fluorescent protein (DsRed) were inserted between a GPD1 promoter and a PGK1 terminator in the expression vector pEG311. pEG311 was constructed to provide uniform and high expression levels in Δura3 mutant strains. In addition to a URA3 selectable marker, the vector also contains sequences from the 5′ untranslated region and the 5′ end of the coding region of the SSO1 gene to direct chromosomal integration. The GFP-SSO2 expression plasmid pEG361 was constructed by an in-frame insertion of the SSO2 gene at the C terminus of the GFP-encoding gene of pEG311. In this work, MATa strains express GFP or GFP-SSO2 and MATα strains express DsRed.
A PRM1 CEN plasmid was constructed by gap repair. Sequences upstream (positions −975 to −320) and downstream (positions 156 to 510) of the PRM1 open reading frame were amplified by PCR and subcloned into pRS316 (42) to construct pEG383. The two inserts were in the same orientation, with the upstream sequences at the 5′ end. pEG383 was cut between the two inserts with XbaI and then transformed into a strain with a wild-type PRM1 gene. Plasmid pEG390 was rescued from this strain and sequenced to confirm recovery of the PRM1 gene. The PRM1 gene was then subcloned from pEG390 into p415ADH (36) to create pEG403, which has a LEU2 selectable marker. The prm1::HIS3 disruption plasmid pEG381 was created by subcloning sequences upstream (positions −536 to −217) and downstream (positions 156 to 511) of the PRM1 open reading frame into pRS303 (42) in an orientation such that cutting the plasmid between the two inserts with SpeI generated a linear fragment with the upstream and downstream sequences in a 5′-to-3′ orientation separated by the HIS3 gene and other plasmid sequences.
The fus1 prm1 and fus1 fus2 strains were constructed by crossing single mutants. The fus1 fus2 prm1 strains were constructed by transforming fus1 fus2 double mutants with pEG381. The fps1 phenotype was confirmed by measuring sensitivity to hypo-osmotic shock (44). The MATα fps1 strain failed this test. The fps prm1 strain was constructed by transforming an fps1 mutant with pEG381. Petite strains were derived by growth in ethidium bromide (15). Loss of mitochondrial function was confirmed by a failure to grow on yeast extract peptone glycerol medium and by the absence of 4′,6′-diamidino-2-phenylindole (DAPI)-stained mitochondrial DNA.
Cells expressing GFP were grown to log phase in yeast extract peptone dextrose (YPD) medium. Since DsRed takes 27 h to reach half-maximal fluorescent intensity at room temperature (1), the red fluorescent signal was maximized in the time-lapse microscopy experiments by diluting cells from a stationary phase culture into fresh YPD medium 3 to 6 h before mating. Substantially identical results were obtained by using DsRed cells in log phase, although the fluorescent signal intensity was reduced. Other DsRed variants, DsRed.T1 (3) and mDsRed (7), were not sufficiently bright or stable for this work.
In mating assays, equal amounts (0.1 optical density unit = 106 cells) of MATa and MATα cells were mixed and then collected on 2.5-cm-diameter cellulose ester filters (Millipore). The filters were placed cell side up on synthetic complete (SC) agar plates and incubated at 30°C. For the endpoint assays presented in Fig. 1A to 1F, 2 to 6, and 10, mated cells were collected from filters into ice-cold TAF buffer (20 mM Tris [pH 8.0], 20 mM NaN3, 20 mM NaF). The cells were concentrated by centrifugation for 5 s and then resuspended in 30 μl of TAF buffer.
FIG. 1.
prm1 mutant mating pairs. MATa prm1 GFP cells were mated with MATα prm1 DsRed cells for 2 h at 30°C. Three outcomes were observed: fusion (A to C), prezygote accumulation (D to F), and lysis (G to I). (A to C) The same fused zygote is shown through GFP filters (A), through DsRed filters (B), and merged (C). (D to F) Late prezygotes could have either GFP (D) or DsRed (E) fingers (arrows). Early prezygotes (F) have a flat interface between the cells (arrow). (G to I) The same mating pair is shown before (G) and after (H and I) lysis with merged GFP and DsRed fluorescence (G and H) or with DIC optics (I). Bar = 2 μm.
FIG. 2.
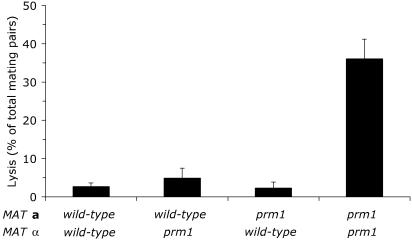
Lysis only occurs in bilateral prm1 mutant crosses. Wild-type, unilateral prm1, and bilateral prm1 mating mixtures were incubated for 3 h at 30°C. Lysis was scored by loss of cytoplasmic GFP fluorescence. For this experiment, four independent prm1::KanMX and PRM1 strains of each mating type were isolated from a genetic cross. Sixty to 80 mating pairs were analyzed for each combination. Data are presented as mean ± standard deviation.
FIG. 6.
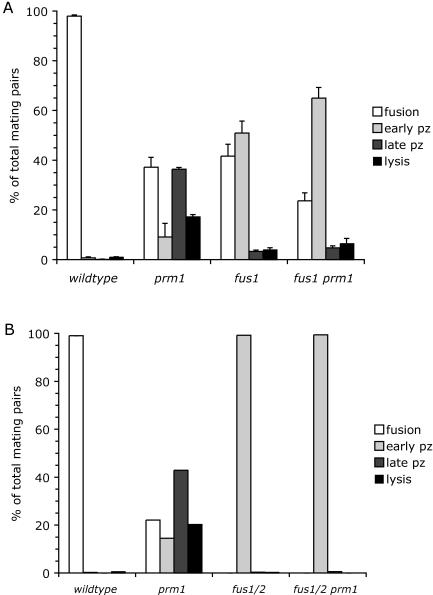
fus1 and fus2 are epistatic to prm1. (A) Bilateral matings were performed for 3 h between wild-type, prm1 and fus1 single-mutant, and prm1 fus1 double-mutant strains. (B) The same as panel A, except that a fus1 fus2 double mutant was used in place of the fus1 single mutant to completely block cell wall remodeling. Mating phenotypes were scored as described in the legend to Fig. 1. pz, prezygote.
FIG. 10.
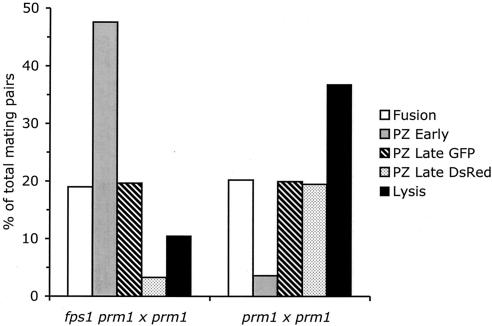
An osmotic gradient contributes to finger extension. MATa fps1 prm1 GFP (left) or MATa prm1 GFP (right) cells were mated with MATα prm1 DsRed cells for 45 min at 30°C on nutrient agar medium supplemented with 0.3 M sorbitol and then shifted to sorbitol-free medium for 1 h. Mating phenotypes were scored as described in the legend to Fig. 1 (n = >500). PZ, prezygote.
For time-lapse microscopy, mating mixtures were preincubated on filters over SC agar plates for 45 min. This step increases the efficiency of mating and delays the onset of hypoxic conditions that can result in photoconversion of GFP to a red fluorescent state (12). Cells were collected from the filters into 1 ml of SC medium and concentrated to 30 μl by centrifugation. A 1.6-μl aliquot was then pipetted onto a 1.5-mm-thick pad of SC medium with 3% agarose on a microscope slide. Application of an 18-mm2 coverslip caused the cell suspension to spread into an even layer. After the excess agar was trimmed away, the slides were sealed with VALAP (a 1:1:1 mixture of petrolatum [Vaseline], lanolin, and paraffin) and observed during the period from 1 to 2 h after mixing.
Microscopy was performed with an Axioplan 2 motorized microscope (Zeiss) outfitted with a mercury arc lamp, band pass filters (Chroma), differential interference contrast (DIC) optics, and an Orca ER digital camera (Hamamatsu). Single images were collected with a 100× objective. For time-lapse microscopy, the 63× objective lens (Plan Apochromat) and microscope stage were heated to 30°C. Automated data collection and analysis were performed with Openlab software (Improvision). Binning (2×) was used to reduce exposure times and minimize photobleaching. Sets of GFP, DsRed, and DIC images were collected sequentially at 2- to 10-s intervals.
RESULTS
Mating phenotype of the prm1 mutant.
MATa prm1 cells expressing GFP as a soluble cytoplasmic protein were mated with MATα prm1 cells expressing DsRed as a soluble cytoplasmic protein. When cell fusion occurs, the two fluorescent proteins diffuse in opposite directions across the fusion pore connecting the two mating cells, producing a zygote that contains both GFP and DsRed (Fig. 1A to C). The rates, but not the extents, of fusion are similar in prm1 and wild-type mating pairs. In both cases, fusion is essentially complete after 2.5 h at 30°C. Consistent with a previous report, more than half of the mating pairs arrested as unfused prezygotes (18). In some prezygotes, a projection of green (GFP) or red (DsRed) cytoplasm extended from one cell into its mating partner (Fig. 1D and E). These cytoplasmic projections always remained connected to the cell of origin, so we refer to them as fingers rather than bubbles. GFP fingers and DsRed fingers occurred with equal frequencies, but an individual prm1 mating pair could have only one finger. Since fingers can only form if the cell walls separating the two mating partners have been degraded, we classify prezygotes with fingers as late prezygotes. In contrast, the early prezygotes of fus1 mating pairs have intact cell walls separating the two partner cells, and there is a flat GFP-DsRed interface between the cells. Some prm1 prezygotes also have a flat interface between the two cells, suggesting the presence of an intact cell wall (Fig. 1F). Although these are classified as early prm1 prezygotes, we cannot exclude the possibility that they have completed cell wall remodeling because they might have fingers that are too small to detect. Alternatively, a flat GFP-DsRed interface could occur after cell wall remodeling if both mating partners attempted to form a cytoplasmic finger at the same place with the same force.
Remarkably, a significant number of prm1 mating pairs lysed. After lysis, cytoplasmic proteins including GFP and/or DsRed diffuse into the surrounding medium, leaving behind a ghost that can be recognized in DIC optics by the high-contrast profile of vacuoles (Fig. 1G to I). Since lysis was not described in the initial report on the PRM1 gene (18), we considered the possibility that a secondary mutation in our strains contributed to the phenotype. To address this issue, we confirmed that expression of PRM1 from a CEN plasmid rescued both mating phenotypes: lysis and prezygote accumulation. In addition, we found that the two prm1 phenotypes were linked in a genetic cross and confirmed that the original prm1 mutant strains also lyse when mated. Lysis was only observed when both partners in the mating pair carried a prm1 deletion (Fig. 2). In agreement with previous results, it was also necessary for both mating partners to be prm1 mutants in order to find prezygote accumulation.
Osmotic stabilizers do not protect prm1 mutant mating pairs.
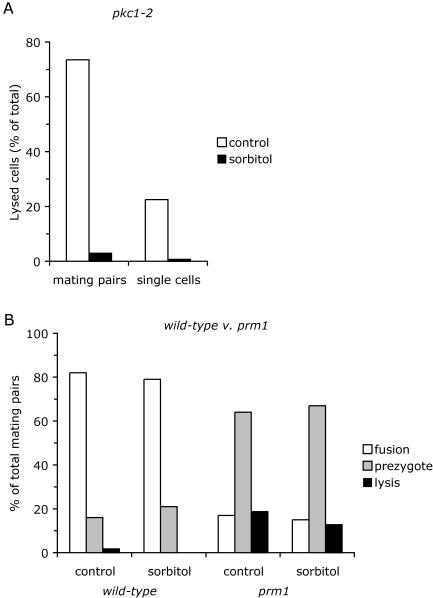
We considered the possibility that PRM1 regulates cell wall remodeling. The cell wall provides structural support to the plasma membrane, thereby protecting yeast cells from osmotic lysis. The growth defect of many cell wall remodeling mutants can be remediated by adding an osmotic stabilizer such as 1 M sorbitol to the growth medium. For example, osmotic support is required for growth of the pkc1-2 mutant at elevated temperatures (25). When two pkc1-2 mutants were mated at 35°C, 73% of the mating pairs lysed (Fig. 3A). Adding 1 M sorbitol to the growth medium reduced lysis to 3%. Cells in the mating reaction that did not bind to a partner of the opposite mating type lysed less frequently than mating cells, but single cells were also protected by sorbitol. The extensive lysis of mating pkc1-2 cells emphasizes the importance of proper cell wall regulation during mating.
FIG. 3.
Sorbitol does not protect prm1 mutant mating pairs from lysis. (A) Sorbitol protects the pkc1-2 mutant from lysis. pkc1-2 mutant mating mixtures were collected on filters and incubated for 2 h at 35.5°C on standard YPD plates or on YPD plates supplemented with 1 M sorbitol. Lysis was scored by the absence of GFP and DsRed fluorescence. (B) Wild-type and prm1 mutant mating mixtures were collected on filters and incubated for 2 h at 30°C on YPD or YPD sorbitol plates. Mating phenotypes were scored as described in the legend to Fig. 1.
To test the prm1 mutant for cell wall remodeling defects, prm1 mutant and wild-type control mating reactions were set up in the presence or absence of sorbitol (Fig. 3B). Sorbitol slightly reduced lysis in both strains. Since the extent of protection provided by sorbitol was independent of the prm1 mutation, the lysis phenotype of prm1 mating pairs is not due to a cell wall remodeling defect.
Lysis of prm1 mating pairs is not a consequence of programmed cell death.
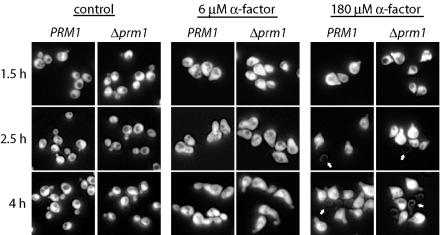
Yeast cells lyse in response to extended treatment with large doses of mating pheromone (24, 27, 40). This lysis has been proposed to represent a form of programmed cell death to enhance the fitness of a population of yeast cells by removing cells that fail to mate (43). Since PRM1 expression is induced by α-factor and many prm1 mating pairs fail to mate, we asked whether α-factor-induced lysis would be enhanced in the prm1 mutant. The results show that wild-type and prm1 mutant MATa haploids responded identically to α-factor (Fig. 4). At a moderate dose (6 μM), both strains exhibited a typical morphogenic response by extending mating projections (shmoos) at 90 min. At later time points, they adapted to the pheromone and resumed budding. Thus, lysis in the prm1 mutant is not a consequence of the morphogenic response to α-factor. At a higher α-factor concentration (180 μM), 12% of the prm1 cells lysed after 2.5 h but a similar percentage (14%) of wild-type cells also lysed. Thus, the increased lysis of prm1 mating pairs compared to wild-type mating pairs can be distinguished from α-factor-induced lysis, which is PRM1 independent.
FIG. 4.
α-Factor pheromone treatment of wild-type and prm1 mutant MATa cells. GFP-expressing cells were treated with moderate (6 μM) or uncommonly high (180 μM) doses of α-factor for the indicated times and then imaged with a combination of bright-field and fluorescence optics. Live cells have bright GFP fluorescence, whereas lysed cells (arrows) do not.
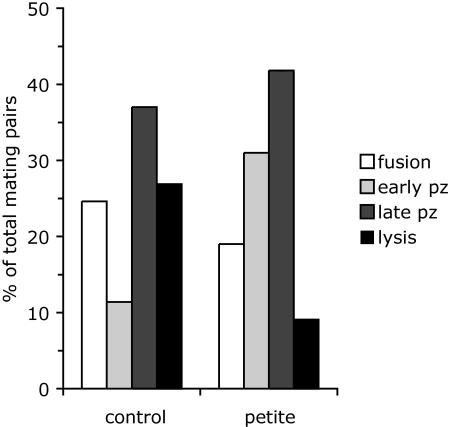
Programmed cell death typically involves the release of cytochrome c from mitochondria, and death in response to α-factor is completely dependent upon mitochondrial function (40). To determine whether lysis of prm1 mating pairs is a form of programmed cell death, we grew prm1 haploid strains in medium containing ethidium bromide to create petite mutants lacking mitochondrial function. Petite prm1 mating pairs lysed, but they did so at a lower frequency than prm1 mating pairs with functional mitochondria (Fig. 5). The reduced lysis of petite prm1 mating pairs is in part due to a kinetic delay in these slow-growing strains. When the standard 3-h incubation was extended to 4.5 h, the percentage of lysed mating pairs increased from 9 to 20%, with a corresponding decrease in the percentage of early prezygotes. Therefore, lysis of prm1 mating pairs is independent of programmed cell death and other mitochondrial functions, including respiration.
FIG. 5.
Lysis does not require functional mitochondria. Haploid prm1 mutants were grown in medium containing 30 μM ethidium bromide to poison mitochondrial DNA replication. The resulting petite mutants were mated for 3 h at 30°C. Mating phenotypes were scored as described in the legend to Fig. 1. pz, prezygote.
Lysis of prm1 mutant mating pairs requires plasma membrane contact.
To determine whether cell wall remodeling must occur before lysis of prm1 mutant mating pairs, we examined the mating of fus1 prm1 double-mutant strains (Fig. 6A). In the fus1 single mutant, mating is arrested prior to cell wall remodeling, leading to an accumulation of early prezygotes (33, 45). Early prezygote accumulation was also the predominant phenotype observed in the fus1 prm1 double mutant, consistent with the expectation that fus1 acts upstream of prm1 in the mating pathway. Importantly, the fus1 prm1 double mutant had significantly less lysis and fewer late prezygotes compared with the prm1 single mutant, suggesting that cell wall remodeling is required for lysis, as well as for cytoplasmic-finger formation.
Cell fusion is inhibited by only 60% in fus1 mating pairs, and a similar incomplete mating defect was found in fus2 mating pairs (45). Mating is almost completely blocked when the fus1 and fus2 mutations are combined, suggesting that fus1 and fus2 act on parallel pathways leading to cell wall remodeling (45). fus1 fus2 prm1 triple-mutant strains were mated to definitively determine whether lysis can occur prior to cell wall remodeling (Fig. 6B). Essentially no lysis was detected, indicating that plasma membrane contact is a prerequisite for lysis.
prm1 mating pairs lyse before or during fusion.
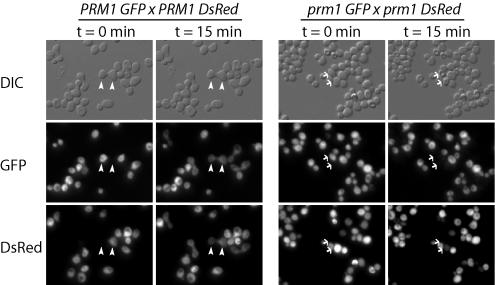
Since some prm1 mutant mating pairs can fuse, we asked whether lysis occurs before, during, or after fusion by monitoring the interaction between MATa GFP cells and MATα DsRed cells over time. We first developed conditions for time-lapse microscopy that permit efficient mating of wild-type partners (Fig. 7). When a fusion pore opens, the fluorescent proteins diffuse through the pore until the cytoplasmic concentration in the two cells equilibrates. GFP typically diffuses to equilibrium within 2 min, but actual diffusion rates vary depending upon the size and opening rate of the fusion pore. DsRed diffuses more slowly than GFP because the fluorescent form of DsRed is a tetramer of GFP-sized subunits (1). In contrast to the fusion that occurs in all wild-type mating pairs, most prm1 mating pairs lyse (Fig. 7). Indeed, lysis of prm1 mating pairs occurs more frequently relative to mating in time-lapse recordings, possibly because of the lack of gas exchange after cells are sealed under a glass coverslip. Although no obvious differences were noted between the rates of GFP diffusion through prm1 mutant and wild-type fusion pores, a quantitative comparison of pore permeances is in progress and will be reported elsewhere. Thirty-nine prm1 lysis events and 22 prm1 fusion events were analyzed, but lysis never occurred after GFP had diffused to equilibrium. Thus, lysis must occur before or during fusion.
FIG. 7.
Specific lysis of prm1 mating pairs under time-lapse conditions. For time-lapse microscopy, MATa GFP and MATα DsRed cells were applied to a nutrient agar pad and then sealed under a coverslip (see Materials and Methods). Fusion was detected by following the transfer of GFP and DsRed into the mating partner (arrowheads). Wild-type cells fuse efficiently under these conditions. In contrast, prm1 mating pairs lyse, resulting in loss of GFP and DsRed fluorescence (arrows).
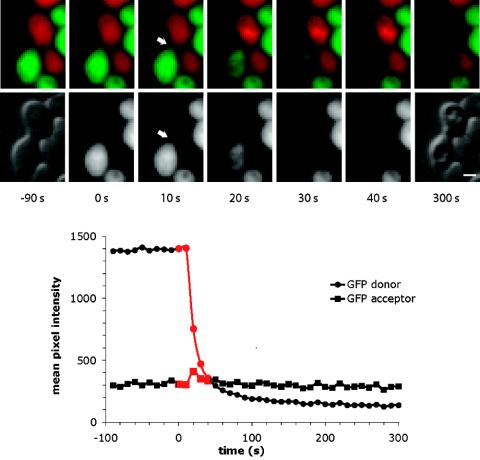
To determine whether lysis and fusion can occur simultaneously, the time-lapse series documenting lysis were examined for evidence of GFP transfer between cells. One example is shown in Fig. 8 and in Movie 1 of the supplemental material. Boundaries were drawn around the two cells of the mating pair, and the mean intensity of GFP fluorescence in each cell was measured as a function of time. When the MATa cell lysed, its GFP intensity declined precipitously. At this time, a small amount of GFP was transiently detected in the MATα cell, suggesting simultaneous opening of a fusion pore. Since these experiments were performed with a wide-field microscope, we cannot exclude the possibility that GFP diffused to a location above or below the mating partner. GFP transfer was detected in 4 out of the 39 mating pairs. DsRed was not detected in the corresponding MATa cells of these mating pairs but did transfer before the lysis of other mating pairs. In normal fusion events, DsRed transfer is often detected later than GFP transfer because pores must open wider before they can allow passage of the larger DsRed probe. Thus, lysis and fusion pore opening may occur at the same time.
FIG. 8.
Time course of fusion and lysis in a prm1 mutant mating pair. The upper micrographs show merged GFP and DsRed images. After lysis, GFP and DsRed leak out of the dead mating pair. The lower micrographs show GFP-only images for times of 0 to 40 s and DIC images from before (−90 s) and after (300 s) lysis. The arrow in each 10-s image marks a small GFP finger. In the 20-s image, a small amount of GFP can be seen throughout the MATα cell. Vacuoles in the lysed mating pair appear highly refractile in the DIC image at 300 s. Bar = 2 μm. The graph in the lower panel shows the GFP intensity in the two mating cells as a function of time. For this measurement, the boundary of the MATα cell was set to exclude the site of the invading GFP finger in order to demonstrate transient influx of GFP into the entire acceptor cell at 20 s.
Cytoplasmic microfingers are exchanged before lysis and fusion.
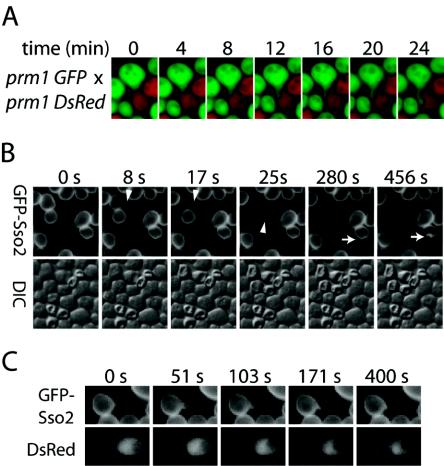
In the mating pair shown in Fig. 8, a cytoplasmic finger extended from the MATa cell into the MATα cell immediately before lysis. Indeed, cytoplasmic fingers containing either GFP or DsRed could be detected before (and during) most of the lytic events between prm1 mutants. When a GFP-Sso2 fusion protein on the plasma membrane of the MATa prm1 cell was used as a more sensitive marker for finger extension, fingers could be detected before essentially all lytic events (17 of 18).
Formation of a cytoplasmic finger clearly indicates that the cell wall between the two cells is degraded before lysis, but finger extension could also cause lysis by exerting mechanical force on the mating partner. Cytoplasmic fingers can be arbitrarily classified as microfingers and long fingers. Newly formed microfingers are less than 1 μm in length. If they continue to grow, they become long fingers, which can reach 4 μm in length over a period of 90 min (Fig. 9A; Movie 2 of the supplemental material). These long fingers can retract as well as extend. On every occasion (seven of seven) when lysis occurred after a finger grew longer than 1 μm, the invaded cell lysed before the cell that extended the finger, consistent with the possibility that invasion by a long finger causes lysis. Lysis of the invaded cell creates a void that may promote subsequent lysis of the cell extending the finger, but the cell extending the finger survived on two occasions. The survivors reoriented their growth axis and either attempted to mate with another partner or budded off a haploid daughter cell. Although these results are consistent with the possibility that long fingers cause lysis of the invaded cell, most (12 of 17) prm1 lytic events involve microfingers that formed less than 30 s prior to lysis.
FIG. 9.
Cytoplasmic fingers in prm1 mutant and wild-type mating pairs. (A) Growth of a 4-μm-long cytoplasmic finger between MATα prm1 GFP and MATα prm1 DsRed cells. (B) Microfingers are shown before lysis in a cross of MATa prm1 GFP-SSO2 and MATα prm1 mating pairs (arrowheads). Microfingers can also be detected in mating pairs that do not lyse (arrows). (C) A microfinger before fusion of a wild-type mating pair.
If local tension exerted by microfingers caused lysis, one would expect that the two cells would lyse at different times, as observed in mating pairs with long fingers. Instead, time-lapse DIC and GFP-Sso2 images revealed that when the two cells within a mating pair with a transient microfinger lyse, they always (17 out of 17 times) do so within the same 15-s interval (Fig. 9B). Furthermore, most cytoplasmic fingers (103 of 120) form in mating pairs that do not lyse. Thus, microfinger extension does not necessarily lead to lysis.
Movies of wild-type mating pairs were reexamined to look for cytoplasmic microfingers. Microfingers were detected before 5 of 53 fusion events (Fig. 9C; Movie 3 of the supplemental material). This observation suggests that microfingers can form whenever there is a delay between the completion of cell wall remodeling and the initiation of fusion. Furthermore, microfingers do not form purely as a consequence of the prm1 mutation.
Osmotic pressure regulates finger extension.
Cytoplasmic-finger extension could be driven by an osmotic gradient between mating cells. To examine this possibility, we asked whether an fps1 deletion would influence the direction of finger extension. FPS1 encodes a glycerol efflux channel in the plasma membrane (44). Yeast cells accumulate glycerol when grown in media of high osmolarity. After a hypoosmotic shift, the Fps1 channel opens to facilitate a return to normal turgor. fps1 mutants fail to release glycerol and therefore have higher cytoplasmic osmolarity. fps1 mutants are also known to have a partial cell wall remodeling defect during mating (39). To explore the relationship between osmotic pressure and finger extension, MATa fps1 prm1 GFP cells were mated with MATα prm1 DsRed cells and then subjected to a mild hypoosmotic shock (Fig. 10). The ratio of GFP fingers to DsRed fingers was 6:1, whereas a 1:1 ratio was observed in a control prm1 × prm1 mating. Thus, fingers preferentially extend from the cell with higher osmotic pressure.
DISCUSSION
PRM1 was originally proposed to act at the plasma membrane fusion stage of yeast mating based upon an accumulation of unfused mating pairs that had completed cell wall remodeling (18). A PRM1-independent fusion pathway was also postulated since some prm1 mutant mating pairs are able to fuse. The fact that many prm1 mating pairs lyse instead of either arresting as prezygotes or fusing suggests a different model: Prm1 stabilizes fusion pore assembly. In the absence of Prm1, the failure to assemble fusion proteins into a pore-forming complex usually results in a block prior to membrane fusion. However, if the fusion process proceeds beyond a committed step, the result will be either lysis or fusion, depending upon whether membrane merger has proceeded beyond a critical intermediate stage before membrane destabilization leads to lysis.
A thorough analysis of the lysis phenotype established that lysis does not occur until the plasma membranes of two prm1 mutant cells come into contact. prm1 mutant strains do not lyse during normal growth because PRM1 is not normally expressed in nonmating cells. prm1 mutant cells also do not lyse when treated with moderate doses of α-factor, which activates a variety of responses normally associated with mating. fus1 and fus2 mutations were used to inhibit the cell wall remodeling step that must occur before membranes contact. Compared with the prm1 single mutant, lysis was strongly reduced in the prm1 fus1 double mutant and completely eliminated in the prm1 fus1 fus2 triple mutant, indicating that a cell wall barrier between plasma membranes prevents lysis. Further evidence that plasma membrane contact precedes lysis was provided by movies of prm1 mutant mating pairs. Essentially all prm1 lysis events were preceded by the extension of a cytoplasmic finger. Since cytoplasmic fingers cannot form until plasma membrane contact has been achieved, the two plasma membranes must come into contact before lysis can occur. A requirement for contact before fusion also suggests that the lytic lesion forms at sites of plasma membrane contact.
In yeast, lysis typically indicates a cell wall defect. During mating, the normal cell wall remodeling processes that accompany bud growth are modified to permit cell fusion without exposing either cell to the external medium. First, the individual cell walls of the two mating partners are combined into a unified barrier surrounding the mating pair. The vestigial cell wall separating the two plasma membranes is then degraded. Cell wall remodeling during mating must be carefully regulated to ensure against osmotic lysis (39). One aspect of this regulation is PKC1-dependent activation of the cell wall integrity MAP kinase pathway (6, 13). Consistent with previous results obtained with the mpk1 MAP kinase mutant (13), we found that the pkc1-2 mutant is especially prone to osmotic lysis during mating. We therefore considered an alternative explanation for the prm1 mutant phenotype: PRM1 is a negative regulator of cell wall remodeling. In this scenario, lysis could occur if unregulated glucanase activity resulted in a lesion in the cell wall, and cytoplasmic fingers could form if cell wall remodeling was completed before activation of the plasma membrane fusion machinery. The critical test to determine whether PRM1 regulates cell wall remodeling was to mate prm1 mutants in the presence of sorbitol, which protects cell wall remodeling mutants from lysis. prm1 mutant mating pairs still lyse in medium supplemented with 1 M sorbitol, indicating that the prm1-encoded phenotype does not result from unregulated cell wall remodeling.
Yeast cells lyse by a mechanism resembling apoptosis in response to oxidative stress, aging, and high doses of α-factor (24, 27, 40). Altruistic suicide of yeast cells that fail to mate has been suggested to be a communal behavior to eliminate weak individuals from the population (40, 43). Indeed, mating yeast cells were reported to lyse by programmed cell death when assembly of stable mating pairs was inhibited by chloroquine at a dose that has no effect on the growth rate of haploid cells (40). These results suggest the possibility that prm1 mating pairs also lyse by programmed cell death. However, we found that prm1 mating pairs lacking functional mitochondria still lysed, even though mitochondria are absolutely required for programmed cell death of pheromone-treated cells (40). Although much remains to be learned about the mechanism and physiology of programmed cell death in unicellular organisms, the hypothesis that a block in mating induces cell death is contradicted by the observation that lysis does not occur in fus1 fus2 mating pairs.
The most obvious mating phenotype of the prm1 mutant is an accumulation of mating pairs with long cytoplasmic fingers. These fingers can continue growing for up to 2 h and can reach a length of 5 μm. At an earlier stage of mating, we observed extension of cytoplasmic fingers less than 1 μm in length. These microfingers were primarily observed in prm1 mutant prezygotes that neither fused nor lysed, suggesting that most microfingers are intermediates in the growth of larger cytoplasmic fingers. However, microfingers were also detected before essentially all prm1 lysis events and before 10% of the fusion events between wild-type mating pairs.
The force for extending cytoplasmic fingers could come from osmotic pressure differences between the two cells or from an actin-dependent process analogous to the mechanism for extension of lamellipodia in mammalian cells (38). We tested the actin-based model by observing finger extension in prm1 mating pairs in which one cell expressed either Abp1-GFP to mark actin patches or Abp140-GFP to mark actin cables (11, 47). Fingers were identified as invaginations into the DsRed-labeled cytoplasm of the mating partner. We found nascent microfingers that excluded actin filaments labeled with either fluorescent actin-binding protein. Thus, actin does not appear to be required for the initiation of microfinger extension. At later times, both Abp-GFPs entered all fingers.
The osmotic-pressure model of finger extension was tested in prm1 mating pairs in which one partner had higher turgor pressure because of an fps1 mutation blocking glycerol export. This experiment was complicated by the accumulation of early prezygotes in the fps1 mating pairs because of activation of a cell wall remodeling checkpoint (39) and the observation that a large (1 M) hypoosmotic shift, which might have produced a steeper osmotic gradient, prevented the formation of late prezygotes in the fps1 mating pairs. Nevertheless, 78% of the fingers formed after a mild hypoosmotic shift extended from the fps1 cell. Therefore, an osmotic gradient can clearly contribute to finger extension, but it is possible that osmotic pressure reinforces fingers initiated by other mechanisms. Osmotic pressure from both mating partners could contribute to fusion by promoting close apposition of the two membranes.
What is the mechanism of lysis? Since cytoplasmic microfingers take time to form, the appearance of microfingers before virtually all prm1 lysis events, and their absence in 90% of wild-type fusion events, suggests a kinetic delay after the completion of cell wall remodeling. Such a delay could facilitate lysis of membranes that are stressed and destabilized by processes and proteins (other than Prm1) that prepare membranes for fusion. The fusion protein for mating is a likely contributor to lysis because many viral fusion proteins contain amphipathic fusion peptides with intrinsic membrane destabilization activity (23). Multiple fusion proteins act in concert during the formation of both viral and intracellular fusion pores (17, 19, 30). HA fusion proteins have been proposed to form a fence surrounding the nascent fusion pore (9). PRM1 might stabilize an analogous complex during yeast mating.
Once two plasma membranes have come into contact, lysis could be mediated by stress on the membranes that is not on the main pathway to fusion. What that stress might be is difficult to imagine. It is not osmotic stress because lysis cannot be prevented with sorbitol. It is not related to programmed cell death or oxidative respiration because lysis occurs in the absence of functional mitochondria. It is not related to finger extension because less than 20% of the mating pairs with microfingers lysed. It must act quickly because most mating pairs lyse within 30 s after the first appearance of a microfinger. Finally, the plasma membranes in prm1 mating pairs can be highly elastic since we have observed a finger that expanded to fill >50% of the cytoplasmic volume of the mating partner in 12 s without lysing.
The ability of PRM1 to protect against lysis when expressed in only one cell of a mating pair indicates that the function of Prm1 extends beyond merely protecting the cell in which it is expressed. If Prm1 promotes lateral interactions between fusion proteins as proposed above, Prm1 expressed in one cell could stabilize a fusion protein complex that spans both plasma membranes. In contrast, mechanisms involving homotypic interactions between Prm1 proteins expressed on different cells can be ruled out. An alternative model of membrane fusion proposes that fusion pores are formed by an interaction between preexisting channels in each membrane, analogous to the assembly of a gap junction (2, 26). Although there would have to be some mechanism to prevent channel opening before the channels in opposing membranes formed a tight seal, it is difficult to imagine how a Prm1 protein located in one membrane could regulate the channel in another membrane. Similarly, Prm1 is unlikely to function in an intercellular signal transduction pathway preventing lysis because it is difficult to imagine how the unidirectional signaling likely to occur in a wild-type × prm1 mutant mating pair could protect both cells from lysis. PRM1 is not the only yeast cell fusion gene that must be mutated in both mating partners to significantly inhibit fusion. However, there is a reasonable explanation for the bilateral mating phenotype of cell fusion mutants in which mating is blocked prior to cell wall remodeling: glucanases secreted by one cell are sufficient to degrade both cell walls at the site of cell-cell contact.
One unresolved issue is how far membrane fusion can proceed in a mating pair that lyses. Transfer of a small amount of GFP from the MATa cell into the MATα cell at the time of lysis supports the possibility that lysis and fusion occurred simultaneously. However, it is not certain that GFP actually entered the cytoplasm of the MATα cell. Instead of passing through a transient fusion pore, GFP released from a lysing MATa cell could diffuse into an expanded periplasmic space surrounding the shriveled remnant of its mating partner. Arguing against this possibility, >70% of GFP fluorescence is rapidly quenched in the growth medium, which has an initial pH of 5.2. Nevertheless, even if a GFP-permeable pore (>2.5 nm) does not form, a smaller ion-permeable pore might connect the cytoplasm of the two cells before or during lysis. In this case, prm1 mutant mating pairs would lyse as a consequence of a defect in fusion pore expansion rather than in the initial opening of the pore.
Are lysis and fusion associated in other systems? In a classic experiment, acidification activated the HA fusion protein for both membrane fusion and hemolysis (28, 29). However, hemolysis was only observed with virus that had been sonicated or frozen, indicating that hemoglobin escaped through pre-existing holes in the viral envelope rather that through the fusion pore itself (29). In reconstituted systems using purified components, membrane-destabilizing agents such as polyethylene glycol, Ca2+ ions, and the amphipathic fusion peptides of viral fusion proteins can induce fusion, leakage, or rupture of phospholipids vesicles (32, 37). In these in vitro systems, the relative frequencies of the three outcomes depend upon experimental conditions, including the amount of destabilizing agent, and the composition, size, and concentration of vesicles. In a more physiologically relevant system, influenza virus at low pH induced leakage pores in liposomes that were permeable to 10-kDa dextrans (4, 41). More recently, ion-permeable leakage pores were observed during the initial stages of HA-mediated fusion between cellular membranes (16). However, these leakage pores resealed as the fusion pore enlarged.
Evidence of leakage during fusion challenges the widely accepted stalk-pore model for the mechanism of membrane fusion, in which leakage is avoided by sequential fusion of the proximal and distal leaflets of the two membranes (10). An alternative fusion mechanism whereby stalk formation between two membranes nucleates holes in one or both bilayers is supported by a course-grained molecular dynamic simulation of fusion (35). Leakage across cellular membranes during fusion would disrupt transmembrane voltage, ion, and pH gradients, which could have detrimental effects on regulated exocytosis and other repetitive intracellular fusion events. Our findings obtained with the prm1 mutant suggest that additional attention should be paid to mechanisms for maintaining membrane integrity during exocytosis.
Medically, the most important example of a linkage between membrane fusion and lysis comes from human immunodeficiency virus (HIV)-infected T cells. Infection of T cells by HIV involves a membrane fusion event between the viral envelope and the host cell plasma membrane that is mediated by the gp120 viral fusion protein. In some circumstances, HIV-infected T cells expressing gp120 fuse with their neighbors to form syncytia. A more common fate for HIV-expressing T cells is single-cell lysis, which requires simultaneous expression of a fusion-competent form of gp120 and its receptor, CD4 (8, 14, 20-22).
The contact-dependent lysis phenotype of prm1 mutant mating pairs suggests that the Prm1 protein stabilizes nascent fusion pores. Stabilization is likely to require a physical interaction between Prm1 and core components of the fusion machinery because the sequence of Prm1 does not reveal any domains with potential catalytic activity. If this model is correct, a search for Prm1-binding proteins could reveal the elusive fusion protein for yeast mating.
Supplementary Material
Acknowledgments
This work was supported by laboratory startup funds from the Johns Hopkins Bloomberg School of Public Health.
Thanks to David Levin, Susan Michaelis, Janice Evans, Leonid Chernomordik, and an anonymous reader for critical comments; to David Levin, Peter Walter, David Drubin, and Lisa Pon for strains; and to Laurence Pelletier for assistance with the first time-lapse movie of prm1 mating. Preliminary work on this project was performed in Peter Novick's laboratory at Yale University.
Footnotes
Supplemental material for this article can be found at http://ec.asm.org.
REFERENCES
- 1.Baird, G. S., D. A. Zacharias, and R. Y. Tsien. 2000. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 97:11984-11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer, M. J., C. Reese, S. Buhler, C. Peters, and A. Mayer. 2003. Vacuole membrane fusion: V0 functions after trans-SNARE pairing and is coupled to the Ca2+-releasing channel. J. Cell Biol. 162:211-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20:83-87. [DOI] [PubMed] [Google Scholar]
- 4.Bonnafous, P., and T. Stegmann. 2000. Membrane perturbation and fusion pore formation in influenza hemagglutinin-mediated membrane fusion. A new model for fusion. J. Biol. Chem. 275:6160-6166. [DOI] [PubMed] [Google Scholar]
- 5.Brizzio, V., A. E. Gammie, and M. D. Rose. 1998. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J. Cell Biol. 141:567-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buehrer, B. M., and B. Errede. 1997. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6517-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, J., I. W. Park, A. Cooper, and J. Sodroski. 1996. Molecular determinants of acute single-cell lysis by human immunodeficiency virus type 1. J. Virol. 70:1340-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chernomordik, L. V., V. A. Frolov, E. Leikina, P. Bronk, and J. Zimmerberg. 1998. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J. Cell Biol. 140:1369-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chernomordik, L. V., and M. M. Kozlov. 2003. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 72:175-207. [DOI] [PubMed] [Google Scholar]
- 11.Doyle, T., and D. Botstein. 1996. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc. Natl. Acad. Sci. USA 93:3886-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elowitz, M. B., M. G. Surette, P. E. Wolf, J. Stock, and S. Leibler. 1997. Photoactivation turns green fluorescent protein red. Curr. Biol. 7:809-812. [DOI] [PubMed] [Google Scholar]
- 13.Errede, B., R. M. Cade, B. M. Yashar, Y. Kamada, D. E. Levin, K. Irie, and K. Matsumoto. 1995. Dynamics and organization of MAP kinase signal pathways. Mol. Reprod. Dev. 42:477-485. [DOI] [PubMed] [Google Scholar]
- 14.Etemad-Moghadam, B., D. Rhone, T. Steenbeke, Y. Sun, J. Manola, R. Gelman, J. W. Fanton, P. Racz, K. Tenner-Racz, M. K. Axthelm, N. L. Letvin, and J. Sodroski. 2001. Membrane-fusing capacity of the human immunodeficiency virus envelope proteins determines the efficiency of CD4+ T-cell depletion in macaques infected by a simian-human immunodeficiency virus. J. Virol. 75:5646-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox, T. D., L. S. Folley, J. J. Mulero, T. W. McMullin, P. E. Thorsness, L. O. Hedin, and M. C. Constanzo. 1991. Analysis and manipulation of yeast mitochondrial genes, p. 149-168. In C. Guthrie and G. R. Fink (ed.), Guide to yeast genetics and molecular biology, vol. 194. Academic Press, San Diego, Calif. [DOI] [PubMed]
- 16.Frolov, V. A., A. Y. Dunina-Barkovskaya, A. V. Samsonov, and J. Zimmerberg. 2003. Membrane permeability changes at early stages of influenza hemagglutinin-mediated fusion. Biophys. J. 85:1725-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grote, E., M. Baba, Y. Ohsumi, and P. J. Novick. 2000. Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion. J. Cell Biol. 151:453-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heiman, M. G., and P. Walter. 2000. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 151:719-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua, Y., and R. H. Scheller. 2001. Three SNARE complexes cooperate to mediate membrane fusion. Proc. Natl. Acad. Sci. USA 98:8065-8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalski, M., L. Bergeron, T. Dorfman, W. Haseltine, and J. Sodroski. 1991. Attenuation of human immunodeficiency virus type 1 cytopathic effect by a mutation affecting the transmembrane envelope glycoprotein. J. Virol. 65:281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaBonte, J. A., N. Madani, and J. Sodroski. 2003. Cytolysis by CCR5-using human immunodeficiency virus type 1 envelope glycoproteins is dependent on membrane fusion and can be inhibited by high levels of CD4 expression. J. Virol. 77:6645-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaBonte, J. A., T. Patel, W. Hofmann, and J. Sodroski. 2000. Importance of membrane fusion mediated by human immunodeficiency virus envelope glycoproteins for lysis of primary CD4-positive T cells. J. Virol. 74:10690-10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau, W. L., D. S. Ege, J. D. Lear, D. A. Hammer, and W. F. DeGrado. 2004. Oligomerization of fusogenic peptides promotes membrane fusion by enhancing membrane destabilization. Biophys. J. 86:272-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laun, P., A. Pichova, F. Madeo, J. Fuchs, A. Ellinger, S. Kohlwein, I. Dawes, K. U. Frohlich, and M. Breitenbach. 2001. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol. Microbiol. 39:1166-1173. [PubMed] [Google Scholar]
- 25.Levin, D. E., and E. Bartlett-Heubusch. 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol. 116:1221-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindau, M., and W. Almers. 1995. Structure and function of fusion pores in exocytosis and ectoplasmic membrane fusion. Curr. Opin. Cell Biol. 7:509-517. [DOI] [PubMed] [Google Scholar]
- 27.Madeo, F., E. Herker, C. Maldener, S. Wissing, S. Lachelt, M. Herlan, M. Fehr, K. Lauber, S. J. Sigrist, S. Wesselborg, and K. U. Frohlich. 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9:911-917. [DOI] [PubMed] [Google Scholar]
- 28.Maeda, T., K. Kawasaki, and S. Ohnishi. 1981. Interaction of influenza virus hemagglutinin with target membrane lipids is a key step in virus-induced hemolysis and fusion at pH 5.2. Proc. Natl. Acad. Sci. USA 78:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeda, T., and S. Ohnishi. 1980. Activation of influenza virus by acidic media causes hemolysis and fusion of erythrocytes. FEBS Lett. 122:283-287. [DOI] [PubMed] [Google Scholar]
- 30.Markovic, I., E. Leikina, M. Zhukovsky, J. Zimmerberg, and L. V. Chernomordik. 2001. Synchronized activation and refolding of influenza hemagglutinin in multimeric fusion machines. J. Cell Biol. 155:833-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsh, L., and M. D. Rose. 1997. The pathway of cell and nuclear fusion during mating in S. cerevisiae, p. 827-888. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Massenburg, D., and B. R. Lentz. 1993. Poly(ethylene glycol)-induced fusion and rupture of dipalmitoylphosphatidylcholine large, unilamellar extruded vesicles. Biochemistry 32:9172-9180. [DOI] [PubMed] [Google Scholar]
- 33.McCaffrey, G., F. J. Clay, K. Kelsay, and G. F. Sprague, Jr. 1987. Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol 7:2680-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mi, S., X. Lee, X. Li, G. M. Veldman, H. Finnerty, L. Racie, E. LaVallie, X. Y. Tang, P. Edouard, S. Howes, J. C. Keith, Jr., and J. M. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785-789. [DOI] [PubMed] [Google Scholar]
- 35.Muller, M., K. Katsov, and M. Schick. 2003. A new mechanism of model membrane fusion determined from Monte Carlo simulation. Biophys. J. 85:1611-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 37.Nir, S., and J. L. Nieva. 2000. Interactions of peptides with liposomes: pore formation and fusion. Prog. Lipid Res. 39:181-206. [DOI] [PubMed] [Google Scholar]
- 38.Pantaloni, D., C. Le Clainche, and M. F. Carlier. 2001. Mechanism of actin-based motility. Science 292:1502-1506. [DOI] [PubMed] [Google Scholar]
- 39.Philips, J., and I. Herskowitz. 1997. Osmotic balance regulates cell fusion during mating in Saccharomyces cerevisiae. J. Cell Biol. 138:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Severin, F. F., and A. A. Hyman. 2002. Pheromone induces programmed cell death in S. cerevisiae. Curr. Biol. 12:R233-R235. [DOI] [PubMed] [Google Scholar]
- 41.Shangguan, T., D. Alford, and J. Bentz. 1996. Influenza-virus-liposome lipid mixing is leaky and largely insensitive to the material properties of the target membrane. Biochemistry 35:4956-4965. [DOI] [PubMed] [Google Scholar]
- 42.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skulachev, V. P. 2002. Programmed death in yeast as adaptation? FEBS Lett. 528:23-26. [DOI] [PubMed] [Google Scholar]
- 44.Tamas, M. J., K. Luyten, F. C. Sutherland, A. Hernandez, J. Albertyn, H. Valadi, H. Li, B. A. Prior, S. G. Kilian, J. Ramos, L. Gustafsson, J. M. Thevelein, and S. Hohmann. 1999. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 31:1087-1104. [DOI] [PubMed] [Google Scholar]
- 45.Trueheart, J., J. D. Boeke, and G. R. Fink. 1987. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell. Biol. 7:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White, J. M., and M. D. Rose. 2001. Yeast mating: getting close to membrane merger. Curr. Biol. 11:R16-R20. [DOI] [PubMed] [Google Scholar]
- 47.Yang, H. C., and L. A. Pon. 2002. Actin cable dynamics in budding yeast. Proc. Natl. Acad. Sci. USA 99:751-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.