Abstract
We previously reported that the chestnut blight fungus Cryphonectria parasitica expresses at least three G-protein α subunits and that Gα subunit CPG-1 is essential for regulated growth, pigmentation, sporulation, and virulence. We now report the cloning and characterization of a C. parasitica regulator of G-protein signaling (RGS) protein, CPRGS-1. The phylogenetic relationship of CPRGS-1 to orthologs from other fungi was inferred and found to be generally concordant with species relationships based on 18S ribosomal sequences and on morphology. However, Hemiascomycotine RGS branch lengths in particular were longer than for their 18S sequence counterparts, which correlates with functional diversification in the signaling pathway. Deletion of cprgs-1 resulted in reduced growth, sparse aerial mycelium, and loss of pigmentation, sporulation, and virulence. Disruption of cprgs-1 was also accompanied by a severe posttranscriptional reduction in accumulation of CPG-1 and Gβ subunit CPGB-1 and severely reduced expression of the hydrophobin-encoding gene cryparin. The changes in phenotype, cryparin expression, and CPGB-1 accumulation resulting from cprgs-1 gene deletion were also observed in a strain containing a mutationally activated copy of CPG-1 but not in strains containing constitutively activated mutant alleles of the other two identified Gα subunits, CPG-2 and CPG-3. Furthermore, cprgs-1 transcript levels were increased in the activated CPG-1 strain but were unaltered in activated CPG-2 and CPG-3 strains. The results strongly suggest that CPRGS-1 is involved in regulation of Gα subunit CPG-1-mediated signaling and establish a role for a RGS protein in the modulation of virulence, conidiation, and hydrophobin synthesis in a plant pathogenic fungus.
Heterotrimeric G proteins play an essential role in the ability of eukaryotic cells to respond to extracellular signals. Activation of the pathway occurs when extracellular ligands bind to transmembrane G-protein-coupled receptors. This allows the Gα subunit of the heterotrimeric G-protein complex to exchange GDP for GTP, which results in dissociation of the Gα and Gβγ subunits and activation of downstream effectors by both Gα and Gβγ (for a review, see reference 22). Gα and Gβ subunits have been identified in numerous filamentous fungi, and their importance in biological processes such as morphogenesis, asexual and sexual reproduction and pathogenesis has been firmly established (reviewed in 2 and 34).
An essential step in controlling the cellular response to G-protein signal transduction is termination of the signal, which is achieved through the intrinsic GTPase activity of the Gα subunit. The GTPase activity is relatively weak (21), and is greatly enhanced by interaction of the Gα subunit with regulator of G-protein signaling (RGS) proteins (reviewed in reference 47). Evidence for the existence of RGS proteins came initially from studies of the mating pheromone response in Saccharomyces cerevisiae, which is mediated via heterotrimeric G proteins. The RGS-encoding SST2 gene was shown to be indispensable for desensitization of the signaling pathway, allowing cells that fail to mate to resume normal growth and cell division (reviewed in reference 32). Sst2 was isolated in a complex with Gpa1 by immunoprecipitation, providing evidence for a direct interaction between Sst2 and Gpa1 (14). Subsequent studies in a range of eukaryotic organisms have led to the recognition of a gene family that shares an ∼130-amino-acid domain, termed the RGS domain. Many RGS proteins contain N- or C-terminal extensions with different functions, linking them to other signaling pathways. Several reviews have been published that focus on RGS proteins and their diverse roles in cellular functions (12, 26, 47).
Studies on the role of RGS proteins in regulation of G-protein signaling in filamentous fungi is currently limited to the model ascomycete Aspergillus nidulans (23, 33, 58) and the homobasidiomycete Schizophyllum commune (18). The FlbA RGS protein of A. nidulans, the second RGS protein to be identified as such (33), has been shown to negatively regulate signaling for vegetative growth and activate asexual sporulation, mediated through the Gα subuit FadA and the Gβ subunit SfaD (46, 58). Han et al. (23) recently described a second RGS protein in A. nidulans, RgsA, that appears to dampen stress responses and stimulate asexual sporulation through negative regulation of the Gα subunit GanB. The RGS protein encoded by the thn-1 gene of S. commune has been shown to be required for aerial-hyphae formation in monokaryons and fruiting body formation in dikaryons (18). Recent characterizations of constitutively activated Gα subunits in S. commune suggest the possibility that Thn-1 may regulate the activity of two Gα subunits, ScGP-A and ScGP-C (55). To further our understanding of the role of RGS proteins in fungal pathogenesis, we now report the cloning and characterization of the RGS-encoding gene, cprgs-1 from the chestnut blight pathogen Cryphonectria parasitica.
Previous studies on G-protein signaling in C. parasitica identified genes encoding three Gα subunits (cpg-1, cpg-2, and cpg-3), one Gβ subunit (cpgb-1), and a regulator of Gβ function (bdm-1) (19, 28, 29, 40). Gene deletion experiments revealed that Gα subunit CPG-1 is essential for growth, conidiation, pigmentation, and virulence, whereas CPG-2 is dispensable (19). A recently isolated third Gα subunit, CPG-3, awaits detailed characterization (40). The Gβ subunit CPGB-1 was found to be dispensable for growth, since Δcpgb-1 strains grew even faster than a wild-type strain. However, pigmentation, sporulation, and virulence were severely reduced, indicating a role for CPGB-1 in these processes (28). In a recent study, CPG-1 was further characterized by introducing mutations designed to abolish GTPase activity, resulting in constitutive activation. The resulting phenotypic changes resembled those caused by a cpg-1 deletion. Both mutant strains showed reduced growth and pigmentation, failed to produce conidia, and were completely avirulent, indicating that tight regulation of CPG-1-mediated signaling is required to control these processes. In contrast, opposing responses of Δcpg-1 and activated mutant strains were found to chronic heat, hyperosmolarity, and oxidative stress. Also, transcript levels of the hydrophobin-encoding gene cryparin were increased in the Δcpg-1 strain but nearly undetectable in mutants expressing activated CPG-1, suggesting a direct role for CPG-1 in regulating cryparin gene expression (49).
The characterization of cprgs-1 described here strongly suggests that CPG-1 is a target of CPRGS-1 regulation and establishes a role for an RGS protein in the modulation of virulence, conidiation, and hydrophobin synthesis in a plant pathogenic fungus. A posttranscriptional reduction in the accumulation of CPG-1 and the Gβ subunit CPGB-1 was also observed in the cprgs-1 deletion mutant, reinforcing our previous reports that mutations in fungal G-protein signaling components can have a dramatic effect on the accumulation of presumptive interacting components (29, 40, 49).
MATERIALS AND METHODS
Fungal strains, growth conditions, and transformation.
C. parasitica strains were maintained on potato dextrose agar (PDA; Difco) as described previously (24). Preparative cultures for protein or RNA isolation were grown for 7 days at room temperature under ambient light, with cellophane covering the growth medium to facilitate mycelial harvest. Preparation and transformation of C. parasitica spheroplasts was carried out essentially as described by Churchill et al. (7). Benomyl (0.5 μg/ml) or hygromycin (40 μg/ml) were included in the growth medium to provide for selection of transformants. Pathogenicity assays were performed as described by Choi and Nuss (6), with 10 replicates per strain.
Nucleic acid procedures.
To isolate the C. parasitica RGS homolog, a PCR was carried out by using a Platinum Taq DNA Polymerase High-Fidelity Supermix (Invitrogen, Carlsbad, Calif.) with degenerate primers RGS-F2 and RGS-R3 (see Table 1) and C. parasitica genomic DNA as a template. After a denaturing step of 3 min at 94°C, PCR amplification was carried out for 35 cycles of 30 s at 94°C, 30 s at 50°C, and 2 min at 72°C. The resulting 260-nucleotide PCR fragment was cloned into plasmid vector pPCR-Script by using the PCR-Script Amp cloning kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions and sequenced. This fragment was used subsequently to screen the cDNA library described in Dawe et al. (10), which yielded several cprgs-1 encoding cDNA clones, none of which contained the complete open reading frame. The Smart-RACE kit (Clontech/BD Biosciences, Bedford, Mass.) was used according to the manufacturer's specifications to isolate the 5′ portion of the cprgs-1 cDNA. General molecular biology procedures and screening of genomic and cDNA libraries were carried out by standard techniques (48).
TABLE 1.
Oligonucleotides used in this studya
| Purpose and oligonucleotide | Sequence (5′-3′) |
|---|---|
| PCR-based cloning of cprgs-1 | |
| RGS-F2 | TGYGARYTNAAYATHGAYCA |
| RGS-R3 | TDRAAYTTNGGNACYGARTC |
| Mutated alleles of cpg-2 and cpg-3 | |
| C2-QLF | GTTGGAGGATTACGAAGCGAG |
| C2-QLR | CTCGCTTCGTAATCCTCCAAC |
| C3-QLF | GTCGGTGGGCTGCGATCAGAG |
| C3-QLR | CTCTGATCGCAGCCCACCGAC |
| C2-FH | TATAAGCTTCAAGATGGGTGCCTGC |
| C2-RS | TATGCATGCGTGATCACAAAATGCCTG |
| C3-FH | TATAAGCTTACAATGTGCTTCGGCAG |
| C3-RS | TATGCATGCTCAAACTCATAAGATCAG |
| S-tag constructs | |
| Stag-NF | TATATGCATAAAGAAACCGCTGCTGCT |
| Stag-NR | ATACTGCAGGCTGTCCATGTGCTGGCG |
| RGS-HF | ATAAAGCTTCACCTGCCGAAAATCG |
| RGS-NR | ATAGCGGCCGCGGTGAAAAGAGCGC |
| Real-time RT-PCR | |
| Cryp-probe | TCTACTTGCGCCACGTCCGGC |
| Cryp-F | CAGCGCATCAAGCTTCGA |
| Cryp-R | CGTGCAGCACTTGGCATCT |
| RGS-probe | ACGCCCAACGGCAAGGATCTCA |
| RGS-F | ACCGACCAAGCACTCCATTTA |
| RGS-R | CGCGAGTTCATGCCATTG |
Nucleotides in boldface introduce the desired mutation. Underlined nucleotides indicate restriction sites used for cloning.
A cprgs-1 cDNA fragment was used to screen a C. parasitica genomic library, and a 6.2-kb KpnI/XbaI genomic DNA fragment was subcloned into pBluescript, resulting in plasmid pRGSX. The insert was fully sequenced and shown to contain the complete cprgs-1 open reading frame, as well as 2.7 kb upstream of the start codon and 1.5 kb downstream of the stop codon (GenBank accession number AY673600). To generate a gene replacement construct, a 1,030-bp SphI/SmaI fragment within the cprgs-1 open reading frame was replaced with a gene cassette conferring hygromycin resistance (9), generating plasmid pRGSX-Hyg. The insert was released from the vector by digestion with KpnI/NotI, and the digestion mixture was used to transform wild-type strain EP155.
RNA was prepared from PDA-cellophane-grown mycelium that was ground to a powder in liquid nitrogen by using the RNaid Plus kit (Qbiogene, Carlsbad, Calif.) according to the manufacturer's instructions. Quantitative analysis of transcript accumulation was performed via reverse transcription and quantitative PCR as described previously (39) by using TaqMan reagents (Applied Biosystems, Foster City, Calif.) and a GeneAmp 5700 PCR apparatus. Transcripts were analyzed at least twice, in triplicate on each occasion, from at least two independent RNA preparations, with primers and probes specific for 18S rRNA, cprgs-1, cpg-1, cpgb-1, and cryparin (39, 40) (Table 1). Calculations to obtain transcript accumulation values in the mutants relative to those in EP155 were performed by using the comparative ΔCt method as described previously (39) using the 18S rRNA values to normalize for variations in template concentration.
The expression vectors used in the present study were based on pCPXHY1 (8). The C. parasitica GPD promoter and terminator region from pCPXHY1 were cloned into pGEM4Z (Promega, Madison, Wis.), and the original polylinker between the GPD promoter and terminator was replaced with a polylinker containing HindIII, HpaI, NotI, and SphI restriction sites. To make pCPX-Hy3, the HygB cassette from pCPXHY1 was cloned at the 5′ end of the GPD promoter. Plasmid pCPX-NBn2 was constructed by inserting the Neurospora crassa benomyl cassette (38) upstream of the GPD promoter.
To make a cprgs-1 overexpression construct, the cprgs-1 open reading frame was amplified by PCR with primers RGS-HF and RGS-NR (Table 1) using a full-length cDNA clone as a template. The PCR fragment was digested with HindIII and NotI and cloned into HindIII/NotI-digested pCPX-NBn2. The unique NsiI site at the start codon of the cprgs-1 open reading frame was used to introduce a S-tag (31), which was amplified by PCR from plasmid vector pET-36b (Novagen/EMD Biosciences, San Diego, Calif.) with the primers Stag-NF and Stag-NR (Table 1), resulting in plasmid pRSO. The S-tag was also introduced into the unique NsiI site in genomic clone pRGSX. The plasmids were sequenced to confirm the absence of mutations introduced by PCR.
PCRs to introduce mutations into cpg-2 and cpg-3 were performed as described previously (49) with the PCR primers listed in Table 1 and cDNA clones of cpg-2 and cpg-3 as templates. The PCR products were digested with HindIII and SphI. The mutated cpg-2 PCR fragment was cloned into pCPX-NBn2, resulting in plasmid pCPX-C2QL, and the cpg-3 PCR fragment was cloned into pCPX-Hy3, resulting in plasmid pCPX-C3QL. The coding regions were completely sequenced to ensure that only the desired mutations had been introduced. Plasmid pCPX-C2QL was transformed into the Δcpg-2 strain (19), and plasmid pCPX-C3QL was transformed into EP155.
DNA sequencing was performed by the CBR sequencing facility using the ABI-Prism BigDye Terminator Ready-Reaction Cycle Sequencing kit (Applied Biosystems) to prepare the sequencing reactions, which were analyzed on an ABI 3100 genetic analyzer (Applied Biosystems).
Protein extraction and analysis.
Protein was extracted from PDA-cellophane-grown mycelium as detailed in Parlsey et al. (40). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blotting, and immunodetection were performed as described in Sambrook and Russell (48). For detection of CPG-1, 20 μg of protein extract was loaded per well, whereas 30 μg of protein extract was loaded for detection of CPGB-1 or S-tagged CPRGS-1. Anti-CPG-1 antibodies (20) were used at a dilution of 1:2,500, and anti-CPGB-1 antibodies (29) were used at a dilution of 1:400. The S-tag HRP Lumiblot kit (Novagen/EMD Biosciences, San Diego, Calif.) was used according to the instructions provided with the kit.
Phylogenetic analysis.
The closest homologs of the RGS sequence from C. parasitica (CPRGS-1) were identified by a BLASTP search of genomic sequences from five other ascomycotes and three basidiomycotes. The fungal species names are listed below, each followed by a possible synonym (when available), GenBank Taxonomy ID number, and GenBank accession number or the name of the RGS protein homolog, if known. The protein sequence of the CPRGS-1 homolog from Coprinopsis cinerea (syn. Coprinus cinereus) (no. 5346) was derived from the nucleotide sequence obtained by a tBlastN search of the database at the Center for Genome Research at the Broad Institute (Cambridge, Mass.). Protein sequences of RGS homologs from Emericella nidulans (syn. Aspergillus nidulans; no. 162425; FlbA) (33), Gibberella zeae (no. 5518; XP_386404), N. crassa (no. 5141; XP_329025), Saccharomyces cerevisiae (no. 4932; Sst2p) (13), Schizosaccharomyces pombe (no. 5334; Rgs1p) (53), S. commune (no. 5334; Thn1) (18), and Ustilago maydis (no. 5270; XP_399719) were obtained from the GenBank database. The sequences were aligned by using CLUSTALX ver. 1.81 (51) with the following alignment parameters: gap opening of 10 and gap extension of 0.20. From this alignment, a data set for phylogenetic analysis was constructed that excluded large insertions and deletions (indels). From C. parasitica, the following RGS residues shown in Fig. 1 were included in the phylogenetic data set: positions 215 to 399, 415 to 491, 579 to 617, 636 to 683, and 712 to 760. (The multiple sequence alignment in Fig. 1 for the three taxa is slightly different than the one obtained for nine taxa, which is not shown.) The total length of the nine sequences varied from 481 residues in Schizosaccharomyces sp. to 763 residues in Ustilago sp. The final data set—with large indel regions excluded—contained 389 characters. Approximately 2% of total sequence data still represented indels, and these were coded as completely ambiguous. Unweighted parsimony analysis of amino acid sequences was conducted with PAUP*4.0 software (D. L. Swofford; available from Sinauer Associates, Sunderland, Mass.). Analysis consisted of heuristic searches with TBR branch swapping and 100 random sequence addition replicates. Nonparametric bootstrap analysis (17) of 10,000 pseudoreplicate data sets was performed in a manner identical to that used for the parsimony analysis of the original data set. Bremer support values (3) were calculated by using TreeRot software (M. D. Sorenson, TreeRot, version 2c; Boston University, Boston, Mass.). Average distances across clades were manually calculated by using the table of pairwise difference values available with PAUP*4.0. Branch lengths were calculated by using MacClade 4 software (D. R. Maddison and W. P. Maddison; available from Sinauer Associates).
FIG. 1.
Alignment of RGS proteins from C. parasitica (CP-RGS-1) (GenBank accession number AY673600), A. nidulans (An-FlbA), and S. commune (Sc-Thn1). The N-terminal 200 amino acids of FlbA are not shown. Identical amino acids are indicated with black boxes and similar amino acids with gray boxes. Arrows F2 and R3 indicate the degenerate PCR primers used to isolate cprgs-1 (see Materials and Methods). The RGS domain is indicated by a “▸▸▸” underscore, and the two DEP domains are indicated by “░⃞” underscores.
RESULTS
Cloning and sequence analysis of cprgs-1.
RGS proteins play an essential role in regulating signal transduction through heterotrimeric G-protein pathways. A C. parasitica RGS homolog was isolated via a PCR-based approach with degenerate PCR primers that were derived from conserved amino acid sequences of several characterized fungal RGS-protein-encoding genes (18, 33). The resulting PCR fragment was sequenced and found to be homologous to FlbA and was subsequently used to screen cDNA and genomic libraries. Sequence analysis of a full-length 3.2-kb cDNA clone revealed several open reading frames. The longest open reading frame, spanning 1,533 nucleotides, encoded the RGS protein and was termed CPRGS-1. The predicted size of CPRGS-1 is 57 kDa, and the amino acid sequence contains the signature sequence of RGS proteins: the RGS domain, as well as two DEP domains. Figure 1 shows an alignment of the predicted amino acid sequences of RGS proteins from C. parasitica, A. nidulans (FlbA), and S. commune (Thn1). A 6.2-kb KpnI/XbaI fragment from the genomic library was subcloned (pRGSX) and sequenced, and found to contain the full coding region as well as the promoter and terminator. Comparison of the genomic and cDNA sequences showed the presence of two introns within the cprgs-1 coding region.
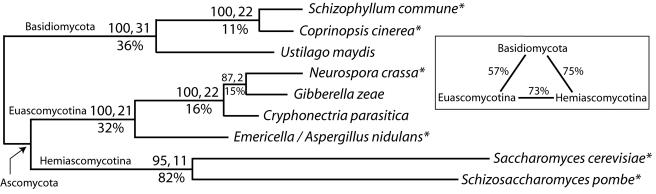
To place CPRGS-1 from C. parasitica within a phylogenetic context, the closest homologs within genomic sequence databases available from five other Ascomycota and three Basidiomycota were identified in silico. After alignment, relationships among these nine sequences were inferred by using the parsimony criterion (Fig. 2). Of the six dichotomous relationships recovered, five have bootstrap percentages and decay indices of at least 95% and 11, respectively, indicating that these RGS sequences manifest a prominent phylogenetic signal at this taxonomic level. However, pairwise divergences were large, particularly between the two Hemiascomycotine taxa (82%) and between them and the others (up to 75%). Greater confidence in the monophyly of Hemiascomycotina in particular will require greater taxon sampling to break up long branches and/or implementation of more highly parameterized models of sequence evolution than parsimony to minimize the risk of artifactual long-branch attraction (16). Regardless of topological uncertainties, including the position of the root for these nine taxa, the much greater branch lengths of both Hemiascomycotine taxa is very likely a real observation. This indicates that their rate of RGS sequence evolution is accelerated relative to that in Euascomycotina and Basidiomycota.
FIG. 2.
Phylogenetic analysis under the parsimony criterion of RGS protein sequences from nine taxa within Ascomycota and Basidiomycota. Taxa are referred to by their Linnaean names (see Materials and Methods for synonyms and gene names). Those genera followed by asterisks were also sampled in Bruns et al. (4). Number of most-parsimonious trees, 1; most-parsimonious tree length, 1006; number of parsimony-informative characters, 221; total number of included characters, 389; consistency index, 0.9324; retention index, 0.8265. Bootstrap percentages followed by Bremer support/decay indices are shown above the internal branches. The percent uncorrected average pairwise amino acid differences are shown below branches and in the enclosed box. Branch lengths are proportional to the number of amino acid changes and are optimized on a topology rooted between Ascomycota and Basidiomycota.
Phenotypic characterization of cprgs-1 deletion mutants.
To examine the effect of deletion of the cprgs-1 gene, a knockout construct was made by replacing a 1,030-bp SphI/SmaI fragment within the cprgs-1 open reading frame in pRGSX with a gene cassette that confers hygromycin resistance. The plasmid was digested with KpnI and NotI to release the insert from the vector and transformed into wild-type strain EP155. Of 50 independent transformants, 5 colonies showed an altered phenotype. PCR and genomic Southern blot analysis showed that three transformants contained one copy of the knockout construct, integrated at the cprgs-1 locus (results not shown). None of these three transformants had vector sequences integrated into the genome (not shown).
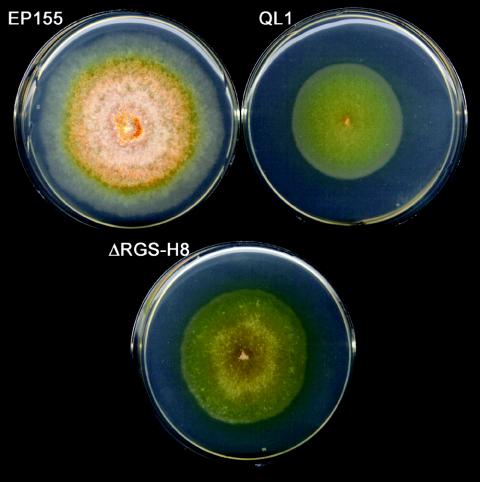
Since RGS proteins function to enhance the intrinsic GTPase activity of Gα, deletion of an RGS gene would result in a prolonged activity of its Gα subunit counterpart. Thus, the phenotypic effects of an RGS-null mutation are predicted to be similar to, but perhaps not as severe as, those caused by the presence of a constitutively active Gα allele. As shown in Fig. 3, the phenotype of a Δcprgs-1 strain mimicked that of a strain which contains a copy of a mutationally activated cpg-1 allele, QL1 (49). Both mutations resulted in colonies that showed a complete absence of sporulation and pigmentation and a reduction in radial growth in vitro on solid medium. The mutations also resulted in complete loss of virulence on dormant chestnut stems (not shown). The growth characteristics of the Δcprgs-1 and QL1 strains under conditions of stress were also remarkably similar. Radial growth was reduced under conditions of osmotic stress caused by the addition of KCl or sorbitol to the media and during growth at elevated temperature (34°C). In contrast, growth of both mutant strains on menadione-containing medium, which causes oxidative stress, resulted in faster radial growth, compared to standard conditions (Fig. 4 in reference 49 and results not shown).
FIG. 3.
Phenotypes of wild-type strain EP155, a strain expressing constitutively active cpg-1 (QL1) and cprgs-1 deletion strain ΔRGS-H8, recorded 7 days after inoculation. Two other independent cprgs-1 deletion strains (H15 and H22) showed a phenotype identical to that of ΔRGS-H8 (not shown).
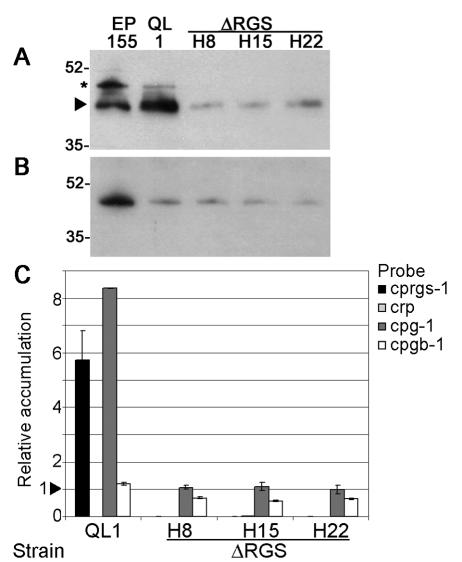
FIG. 4.
Real-time RT-PCR and Western blot analysis of gene and protein expression in a strain expressing activated CPG-1 (QL1) and the Δcprgs-1 strains ΔRGS-H8, -H15, and -H22. (A) Western blot analysis of CPG-1 protein accumulation. The arrow indicates the bands corresponding to CPG-1. The asterisk indicates a slower-migrating band that cross-reacts with α-CPG-1 antibody (40). Size markers are indicated in kilodaltons. (B) Western blot analysis of CPGB-1 protein accumulation. (C) Transcript accumulation of cprgs-1, cryparin (crp), cpg-1, and cpgb-1 in mutant strains relative to accumulation in the wild-type strain EP155. The arrowhead represents the reference value of 1.0 for EP155, calculated as described in Materials and Methods. Bars represent the standard error for each data set.
When cprgs-1 deletion mutant Δcprgs-1-H8 was transformed with the cprgs-1 genomic clone pRGSX, the wild-type phenotype was restored, indicating that the mutant phenotype was caused by specific deletion of RGS, and that the genomic clone pRGSX contains the sequences necessary for correct ectopic expression of the cprgs-1 gene (not shown). Insertion of an S-tag (31) in pRGSX, at the N terminus of the predicted open reading frame, also complemented the gene deletion mutation, indicating that the S-tag does not interfere with RGS function (results not shown).
Deletion of cprgs-1 affects accumulation of G-protein subunits and expression of cryparin.
We have previously shown that mutations in or deletions of G-protein subunits affect the stability of the predicted interacting subunits, resulting in reduced accumulation (29, 40, 49). This effect was posttranscriptional, since transcript levels of the corresponding genes remained unaltered. Since RGS proteins interact with Gα subunits, we determined the effect of cprgs-1-deletion on the protein and transcript accumulation of cpg-1 and cpgb-1, which encode Gα and Gβ subunits, respectively. Western blot analysis showed that the protein levels of both CPG-1 and CPGB-1 were dramatically reduced in the Δcprgs-1 strains (Fig. 4A and B). The level of reduction of CPGB-1 in Δcprgs-1 strains was comparable to that exhibited by the QL1 strain (Fig. 4B).
Transcript levels were analyzed by real-time reverse transcription-PCR (real-time RT-PCR) (Fig. 4C). The absence of cprgs-1 transcript in the Δcprgs-1 strain confirmed deletion of the cprgs-1 gene. Interestingly, the levels of cprgs-1 transcript were increased in the QL1 strain. The cpg-1 transcript levels, which were increased in the QL1 strain as shown before (49), were unchanged in the Δcprgs-1 strains. Transcript levels of cpgb-1 were slightly reduced (<2-fold) in the Δcprgs-1 strains but did not change in the QL1 strain compared to the wild-type strain EP155. The transcript levels of cryparin, which were previously shown by Northern blot analysis to be significantly reduced in activated CPG-1 strains (49), were also nearly undetectable in the Δcprgs-1 strains.
Overexpression of cprgs-1 results in hypersporulation.
Whereas deletion of an RGS-encoding gene is predicted to result in constitutive activation of its Gα partner, overexpression of rgs could result in reduced activity of Gα-mediated signaling. To study the effects of overexpression of cprgs-1, the cprgs-1 open reading frame containing a 5′ S-tag was cloned into expression vector pCPX-NBn2 (see Materials and Methods) and introduced into wild-type strain EP155. The phenotype of the colonies ranged from wild type-like to colonies that were reduced in radial growth and pigmentation but exhibited high levels of sporulation (Fig. 5). The N-terminal S-tag allowed detection of the tagged CPRGS-1 protein. Western blot analysis showed accumulation of a protein of ∼70 kDa in the transformants but not in wild-type strain EP155 (Fig. 6A).
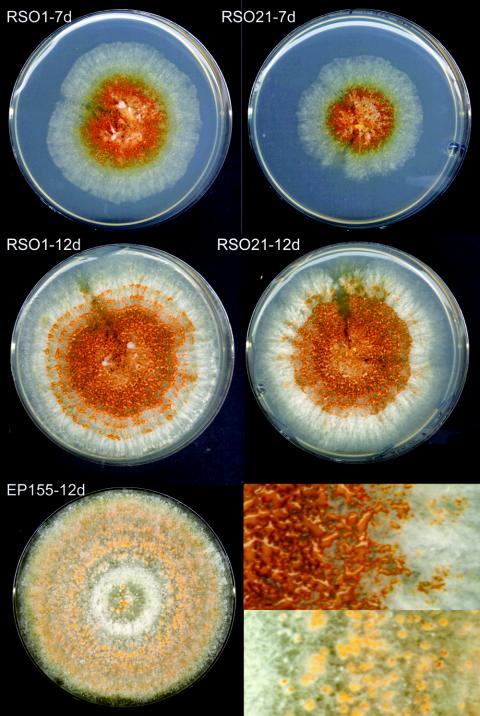
FIG. 5.
Phenotype of RGS-overexpressing strains 1 and 21, recorded 7 and 12 days after inoculation on PDA, compared to the phenotype of wild-type strain EP155. The inset shows a magnification of RSO21 (top) and EP155 (bottom) 12 days after inoculation. The RSO strain sporulates abundantly, as evidenced by the orange coalescent masses of spores, whereas the spores in EP155 are mostly contained within the pycnidia.
FIG. 6.
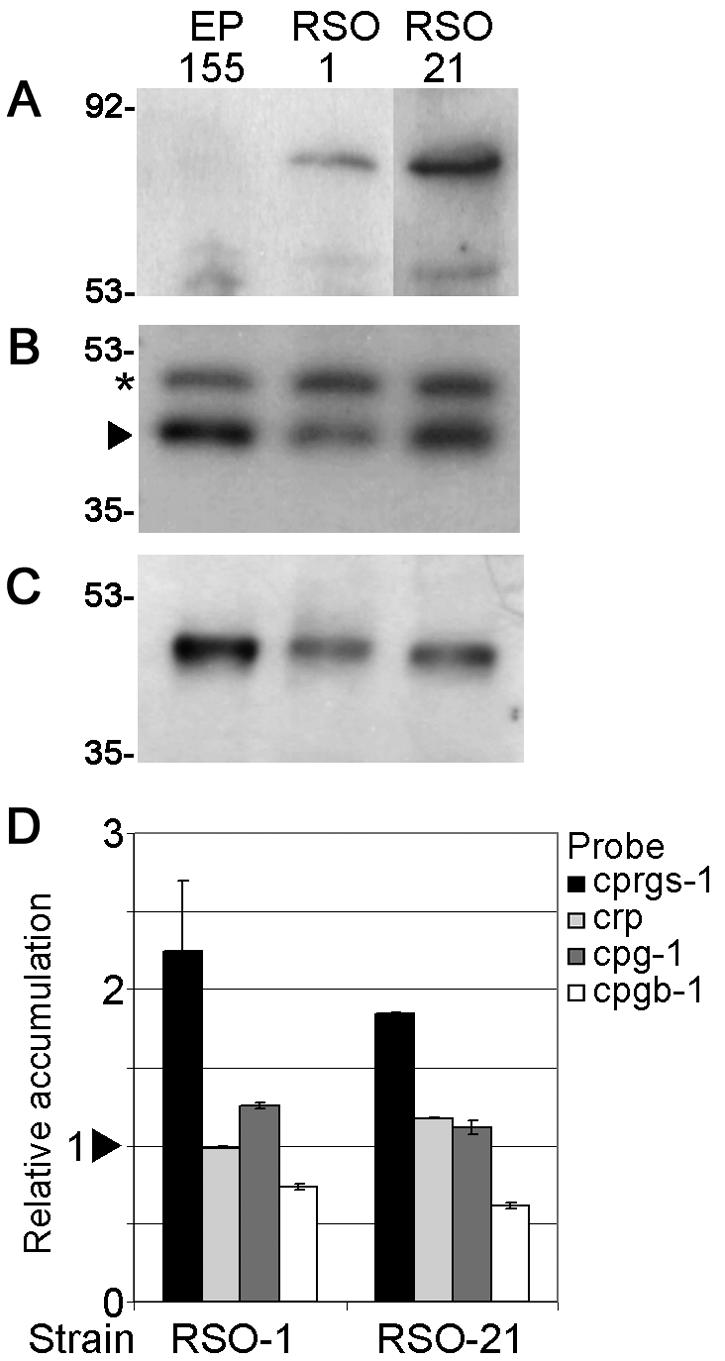
Western blot and real-time RT-PCR analysis of protein and gene expression in EP155 and strains overexpressing cprgs-1. (A) S-tagged CPRGS-1 accumulation in RSO strains. Wild-type strain EP155 was loaded as negative control. Size markers are indicated in kilodaltons. (B) Western blot analysis of CPG-1 protein accumulation. The arrow indicates the bands corresponding to CPG-1. The asterisk indicates a slower-migrating band that cross-reacts with α-CPG-1 antibody (40). (C) Western blot analysis of CPGB-1 protein accumulation. (D) Transcript accumulation of cprgs-1, cryparin (crp), cpg-1, and cpgb-1 in mutant strains, relative to accumulation in wild-type strain EP155. The arrowhead represents the reference value of 1.0 for EP155. Bars represent standard error for each data set.
The hypersporulating strains RSO1 and RSO21, termed RSO for “RGS S-tag overexpressing,” were chosen for further analysis, since Western blot analysis showed that these strains accumulated the highest levels of S-tagged RGS protein. These strains also showed a twofold increase in cprgs-1 transcript accumulation (Fig. 6D). The onset of sporulation in the hypersporulating strains did not change since the colonies started sporulating ca. 6 days after inoculation, similar to the wild-type strain EP155. No sporulation was observed when RSO strains were cultured in liquid medium, which suppresses sporulation in wild-type EP155. The CPG-1 levels were not changed in these cprgs-1-overexpressing strains (Fig. 6B), whereas the levels of CPGB-1 were slightly reduced (Fig. 6C). Analysis of transcript accumulation by real-time RT-PCR revealed that cryparin gene expression was not affected by overexpression of cprgs-1. The cpg-1 transcript levels were unaltered, whereas the transcript levels of cpgb-1 were reduced in the RSO strains (Fig. 6D).
Mutational activation of Gα subunits CPG-2 and CPG-3 causes phenotypic changes different from those caused by cprgs-1 deletion.
Although the experimental results described above suggest an interaction between CPRGS-1 and the Gα subunit cpg-1, we extended the study to include cpg-2 and the recently identified third Gα subunit cpg-3 (40). The conserved Gln-204 in one of the GTP-binding domains of CPG-1 is also conserved in CPG-2 (Q-207) and CPG-3 (Q-204) (30). To generate constitutively active alleles, point mutations were made in cDNA clones of cpg-2 and cpg-3 to change Gln-207 in CPG-2 and Gln-204 in CPG-3 to Leu. The mutated cDNAs were cloned into a pCPX expression vector (8; see also Materials and Methods) containing a selectable marker for either benomyl resistance (cpg-2) or hygromycin resistance (cpg-3) resulting in plasmids pCPX-C2QL and pCPX-C3QL, respectively. Plasmid pCPX-C2QL was transformed into the Δcpg-2 strain, and plasmid pCPX-C3QL was transformed into EP155.
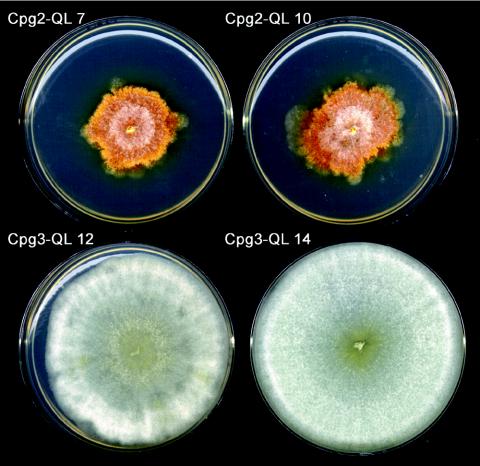
Mutational activation of CPG-2 resulted in colonies that grew slower than either the recipient Δcpg-2 strain or the wild-type strain EP155 and showed irregular colony margins but were not impaired in pigmentation and conidiation. Transformants expressing CPG-3-QL were suppressed in pigmentation and conidiation. Radial growth was not affected in the CPG-3-QL transformants (Fig. 7). Two independent transformants containing the CPG-2-QL allele and two independent transformants containing the CPG-3-QL allele were chosen for further analysis.
FIG. 7.
Phenotype of strains expressing CPG-2-QL and CPG-3-QL, recorded 7 days after inoculation.
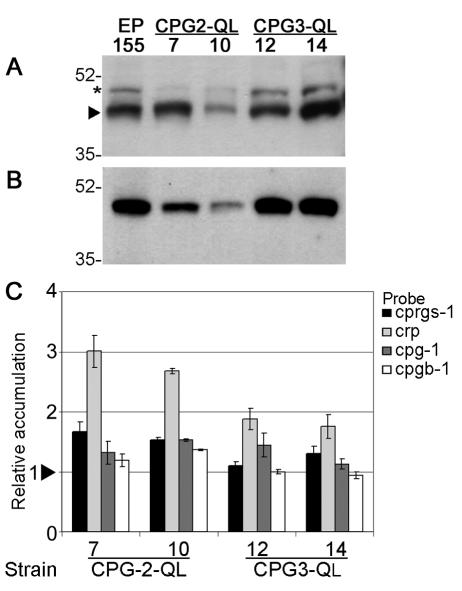
Western blot analysis of CPG-1 protein levels revealed that they were reduced in strain CPG-2-QL 10, but the levels in the other CPG-2-QL strain and both CPG-3-QL strains were not affected (Fig. 8A). The protein levels of CPGB-1 were reduced in the CPG-2-QL strains but remained unchanged in CPG-3-QL strains (Fig. 8B). The mutants were also tested for accumulation of mRNAs for the genes cprgs-1, cryparin, cpg-1, and cpgb-1 (Fig. 8C). cprgs-1 transcript levels did not change appreciably in the CPG-2-QL and CPG-3-QL strains, whereas the QL1 strain accumulated ∼6-fold more cprgs-1 transcript (Fig. 4C). In contrast to the QL1 strain, which expresses barely detectable levels of cryparin transcript, the levels of cryparin transcript were somewhat elevated in the activated CPG-2-QL and CPG-3-QL strains. The transcript levels of cpg-1 and cpgb-1 in the mutant strains were comparable to those in wild-type strain EP155.
FIG. 8.
Real-time RT-PCR and Western blot analysis of gene and protein expression in strains expressing constitutively activated CPG-2 (Cpg-2-QL-7 and -10) or constitutively activated CPG-3 (Cpg-3-QL-12 and -14). (A) Western blot analysis of CPG-1 protein accumulation in wild-type strain EP155 and strains expressing constitutively activated CPG-2 or constitutively activated CPG-3. The arrow indicates the bands corresponding to CPG-1. The asterisk indicates a slower-migrating band that cross-reacts with α-CPG-1 antibody (40). (B) Western blot analysis of CPGB-1 protein accumulation. (C) Transcript accumulation of cprgs-1, cryparin (crp), cpg-1, and cpgb-1 in strains expressing CPG-2-QL or CPG-3-QL. Values are relative to accumulation in wild-type strain EP155. The arrowhead represents the reference value of 1.0 for EP155, calculated as described in Materials and Methods. Bars represent standard error for each data set.
DISCUSSION
Heterotrimeric G proteins play crucial roles in the regulation of fungal developmental processes, as well as pathogenicity (2). Tight regulation of G-protein-mediated signaling is essential, since both null and activating mutations of the same Gα subunit result in debilitating effects on fungal growth, reproduction, and pathogenicity (15, 19, 27, 35, 45, 49, 56, 58-60). RGS proteins provide an essential function as negative regulators, since they enhance the intrinsic GTPase activity of the Gα subunit, which is a critical step in the deactivation of the cellular response. A large number of mammalian RGS proteins have been isolated and shown to play roles in various signaling pathways (26). Molecular and functional characterization of fungal RGS homologs has been limited to FlbA (33, 58) and the recently identified RgsA (23) in A. nidulans and Sst2 in S. cerevisiae (reviewed in reference 32). The characterization of CPRGS-1 described here establishes a role for a RGS protein in the modulation of virulence, conidiation, and hydrophobin gene transcription in a plant pathogenic fungus.
C. parasitica CPRGS-1 shows an organization similar to several RGS proteins from other lower eukaryotes, such as FlbA (A. nidulans), Thn1 (S. commune), Sst2 (S. cerevisiae), and RGS1 (S. pombe). All have the RGS domain, which is involved in binding the Gα subunit at the C terminus, as well as additional N-terminal DEP domains. The DEP domain was initially identified in the proteins Dishevelled (Drosophila melanogaster), Egl-10 (Caenorhabditis elegans), and human Pleckstrin (43) and was later identified in numerous other signaling proteins (NCBI Conserved Domain Database). In a number of cases, DEP domains have been shown to be necessary for membrane localization (1, 25, 36, 41). Furthermore, studies with a S. cerevisiae strain that constitutively expressed the N-terminal DEP domain of Sst2 showed that the DEP domain is involved in regulating the expression of stress-responsive genes through interaction with other proteins (5). Further studies are necessary to determine the function of the N-terminal DEP domains of CPRGS-1.
Phylogenetic analysis of RGS sequences from nine taxa, all within Ascomycota and Basidiomycota, has yielded a fairly robust result (Fig. 2). Significantly, the relationships are congruent for the six genera cosampled in a previous 18S ribosomal study (see Fig. 4B in reference 4) and with morphology generally (50), indicating that the RGS sequences are likely to be orthologous and that both RGS and 18S gene trees are tracking the species relationships. In the 18S study (4), monophyly of Hemiascomycotina was sensitive to the method of analysis and inclusion of outgroups, reinforcing our cautionary statement (see Results) about the large distances among RGS sequences for these same taxa.
It is particularly interesting that 18S sequences from S. cerevisiae and S. pombe are not long branched, in contrast to their RGS counterparts (4). Although requiring data from additional genes, we could hypothesize that this difference means that the taxa themselves are not undergoing accelerated evolution; rather, acceleration is restricted to a subset of genes. A study by Bölker (2) provides suggestive evidence that the Gα subunits Gpa1p from S. cerevisiae and S. pombe are also long branched. Since these Gα subunits interact with the RGS proteins Sst2 and Rgs1, respectively, we could further hypothesize that multiple sequences within the G-protein signaling pathway have undergone accelerated evolution, perhaps in a coevolutionary sense (see, for example, reference 42). A clearer picture of the evolution of RGS and other molecules within the G-protein signaling pathway will require functional studies from additional species within Hemiascomycotina (and closely related groups), followed by their placement within a phylogenetic context.
It is noteworthy that, despite 69% sequence identity between CPRGS-1 and FlbA from A. nidulans, the respective knockout phenotypes were very different. This follows a trend which has also been observed for Gα subunits, where mutations or deletions of homologous Gα subunits in closely related fungi often cause vastly different effects, as discussed by Dawe and Nuss (11). Deletion of cprgs-1 resulted in reduced growth, aerial mycelium production, and absence of conidiation, whereas flbA deletion resulted in a so-called fluffy phenotype, characterized by abundant aerial mycelium production, which autolyses as the colony matures and reduced conidiation (33). Inactivation of the RGS-encoding gene thn1 in S. commune resulted in phenotypic changes that were similar to deletion of cprgs-1 in C. parasitica. Thn1 deletion mutants lacked aerial mycelium, and dikaryons that were homozygous for the thn1 mutation did not produce fruiting bodies (44).
Overexpression of both cprgs-1 and flbA (33) caused hypersporulation in C. parasitica and A. nidulans, respectively. However, whereas the flbA-overexpressing strain produced conidia in liquid culture, which represses sporulation of a wild-type strain, the cprgs-1 overexpressing mutants RSO1 and RSO21 did not sporulate during growth in liquid culture. The hypersporulating phenotype of the cprgs-1 overexpressing mutants is probably not caused through inactivation of CPG-1, since Δcpg-1 strains do not conidiate, but rather through interaction with another protein target.
A function for FlbA and Thn1 as RGS proteins modulating Gα subunit-mediated signaling was indicated by the observation that null mutations in these genes resembled the phenotypes caused by constitutive activation of Gα subunit FadA in A. nidulans (58) and of Gα subunits ScGP-A and ScGP-C in S. commune (55). Deletion of cprgs-1 resulted in a phenotype that was remarkably similar to that obtained by introduction of a mutationally activated cpg-1 allele, QL1 (49). Furthermore, the response of Δcprgs-1 to chronic heat, hyperosmolarity, and oxidative stress was similar to that of CPG-1-QL strains (reference 49 and results not shown), and neither CPG-1-QL nor Δcprgs-1 was able to infect chestnut stems (results not shown), suggesting that CPRGS-1 mainly interacts with CPG-1. In contrast, constitutive activation of Gα subunit CPG-2 did not cause loss of pigmentation or conidiation. In comparison, constitutive activation of Gα subunit CPG-3 gave rise to colonies that were in some respects similar to CPG-1-QL mutants but were not impaired in growth and produced abundant aerial mycelia which collapsed in the center of the colony. Interestingly, the phenotype observed for CPG-3-QL mutants was similar to that of the Gβ subunit cpgb-1 deletion mutant (28), although CPGB-1 levels in the CPG-3-QL strains were unaltered. This observation suggests that CPG-3 interacts with CPGB-1 to regulate pigmentation and conidiation and warrants further investigation.
Further evidence for an interaction between CPRGS-1 and CPG-1 was obtained by real-time RT-PCR analysis of cryparin gene expression. Deletion of cprgs-1 or constitutive activation of cpg-1 both resulted in a severe reduction in cryparin transcript levels. In contrast, expression of cryparin was slightly increased in the strains containing cpg-2-QL and cpg-3-QL alleles. Interestingly, S. commune thn1 deletion mutants were shown to be deficient in expression of Sc3 (54), a hydrophobin-encoding gene that is abundantly expressed during formation of aerial hyphae (37). Further evidence is needed to determine whether a common regulatory pathway for hydrophobin gene expression, regulated through RGS proteins and Gα subunits, exists in filamentous fungi.
As we have noted in earlier studies, mutations in G-protein subunits can have dramatic effects on posttranscriptional accumulation of their presumptive G-protein partners (28, 40, 49). This observation was recently confirmed in N. crassa (57), indicating the existence of a regulatory pathway common to other fungi and possibly other organisms. In the present study we have shown that deletion of the gene encoding CPRGS-1, which is predicted to interact with Gα subunit CPG-1, also results in severe posttranscriptional reduction of CPG-1 levels and CPGB-1 levels. These observations suggest that G-protein subunit stability and degradation provides an additional level of control on G-protein-mediated signaling and reinforces the fact that the effects of a posttranscriptional control mechanism must be considered when results of mutations in signaling components are interpreted.
In addition, we observed an effect of CPG-1 on the transcription of cprgs-1. cprgs-1 transcript levels increased dramatically in response to constitutive activation of CPG-1, suggesting the existence of a control mechanism on the transcriptional level to regulate activity of the CPG-1-mediated signaling pathway. This effect was not observed in CPG-2-QL or CPG-3-QL strains, providing additional support for a specific interaction between CPRGS-1 and CPG-1.
Although CPG-1 protein levels were severely reduced in Δcprgs-1 strains, the changes observed in cprgs-1 deletion strains are likely to be mediated through the CPG-1 signaling pathway. CPG-1-QL has a strong, dominant effect, since transformation of a cpg-1-QL expressing plasmid into a wild-type, Δcpgb-1, or Δcpg-1 strain resulted in the same phenotype, regardless of the genetic background of the recipient strain (49). Although interaction of CPRGS-1 with CPG-2 or CPG-3 cannot be ruled out, it is likely that C. parasitica expresses several RGS-encoding genes, each with specificity to a particular G α subunit. For example, S. cerevisiae has two RGS encoding genes, Sst2 and Rgs2, which regulate activity of Gα subunits Gpa1 and Gpa2, respectively. These two RGS proteins share little homology outside their RGS domains and have nonoverlapping specificity for their respective Gα subunits (52). It will be of interest to identify and characterize additional RGS proteins in C. parasitica and determine the specificities of each RGS protein for the various Gα subunits and their corresponding regulatory pathways.
Acknowledgments
This study was supported by Public Health Service grant GM55981 to D.L.N.
REFERENCES
- 1.Axelrod, J. D., J. R. Miller, J. M. Shulman, R. T. Moon, and N. Perrimon. 1998. Differential recruitment of Dishevelled provides signaling specificity in the planar cell polarity and Wingless signaling pathways. Genes Dev. 12:2610-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bölker, M. 1998. Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet. Biol. 25:143-156. [DOI] [PubMed] [Google Scholar]
- 3.Bremer, K. 1988. The limits of amino acid sequence data in angiosperm phylogenetic reconstruction. Evolution 42:795-803. [DOI] [PubMed] [Google Scholar]
- 4.Bruns, T. D., R. Vilgalys, S. M. Barns, D. Gonzalez, D. S. Hibbett, D. J. Lane, L. Simon, S. Stickel, T. M. Szaro, W. G. Weisburg, and M. L. Sogin. 1992. Evolutionary relationships within the fungi: analyses of nuclear small subunit rRNA sequences. Mol. Phylogenet. Evol. 1:231-241. [DOI] [PubMed] [Google Scholar]
- 5.Burchett, S. A., P. Flanary, C. Aston, L. Jiang, K. H. Young, P. Uetz, S. Fields, and H. G. Dohlman. 2002. Regulation of stress response signaling by the N-terminal dishevelled/EGL-10/pleckstrin domain of Sst2, a regulator of G protein signaling in Saccharomyces cerevisiae. J. Biol. Chem. 277:22156-22167. [DOI] [PubMed] [Google Scholar]
- 6.Choi, G. H., and D. L. Nuss. 1992. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257:800-803. [DOI] [PubMed] [Google Scholar]
- 7.Churchill, A. C. L., L. M. Ciufetti, D. R. Hansen, H. D. Van Etten, and N. K. Van Alfen. 1990. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17:25-31. [Google Scholar]
- 8.Craven, M. G., D. M. Pawlyk, G. H. Choi, and D. L. Nuss. 1993. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67:6513-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen, D., S. A. Leong, L. J. Wilson, and D. J. Henner. 1987. Transformation of Aspergillus nidulans with the hygromycin-resistance gene, hph. Gene 57:21-26. [DOI] [PubMed] [Google Scholar]
- 10.Dawe, A. L., V. C. McMains, M. Panglao, S. Kasahara, B. Chen, and D. L. Nuss. 2003. An ordered collection of expressed sequences from Cryphonectria parasitica and evidence of genomic microsynteny with Neurospora crassa and Magnaporthe grisea. Microbiology 149:2373-2384. [DOI] [PubMed] [Google Scholar]
- 11.Dawe, A. L., and D. L. Nuss. 2001. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annu. Rev. Genet. 35:1-29. [DOI] [PubMed] [Google Scholar]
- 12.De Vries, L., B. Zheng, T. Fischer, E. Elenko, and M. G. Farquhar. 2000. The regulator of G protein signaling family. Annu. Rev. Pharmacol. Toxicol. 40:235-271. [DOI] [PubMed] [Google Scholar]
- 13.Dietzel, C., and J. Kurjan. 1987. Pheromonal regulation and sequence of the Saccharomyces cerevisiae SST2 gene: a model for desensitization to pheromone. Mol. Cell. Biol. 7:4169-4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dohlman, H. G., J. Song, D. Ma, W. E. Courchesne, and J. Thorner. 1996. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit). Mol. Cell. Biol. 16:5194-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang, E. G., and R. A. Dean. 2000. Site-directed mutagenesis of the magB gene affects growth and development in Magnaporthe grisea. Mol. Plant-Microbe Interact. 13:1214-1227. [DOI] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1978. Cases in which parsimony and compatibility methods will be positively misleading. Syst. Zool. 27:401-410. [Google Scholar]
- 17.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 18.Fowler, T. J., and M. F. Mitton. 2000. Scooter, a new active transposon in Schizophyllum commune, has disrupted two genes regulating signal transduction. Genetics 156:1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, S., and D. L. Nuss. 1996. Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc. Natl. Acad. Sci. USA 93:14122-14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, S., and D. L. Nuss. 1998. Mutagenesis of putative acylation sites alters function, localization, and accumulation of a Giα subunit of the chestnut blight fungus Cryphonectria parasitica. Mol. Plant-Microbe Interact. 11:1130-1135. [DOI] [PubMed] [Google Scholar]
- 21.Gilman, A. G. 1987. G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 56:615-649. [DOI] [PubMed] [Google Scholar]
- 22.Hamm, H. E. 1998. The many faces of G protein signaling. J. Biol. Chem. 273:669-672. [DOI] [PubMed] [Google Scholar]
- 23.Han, K. H., J. A. Seo, and J. H. Yu. 2004. Regulators of G-protein signaling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (Gα) signaling. Mol. Microbiol. 53:529-540. [DOI] [PubMed] [Google Scholar]
- 24.Hillman, B. I., R. Shapira, and D. L. Nuss. 1990. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80:950-956. [Google Scholar]
- 25.Hoffman, G. A., T. R. Garrison, and H. G. Dohlman. 2000. Endoproteolytic processing of Sst2, a multidomain regulator of G protein signaling in yeast. J. Biol. Chem. 275:37533-37541. [DOI] [PubMed] [Google Scholar]
- 26.Hollinger, S., and J. R. Hepler. 2002. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 54:527-559. [DOI] [PubMed] [Google Scholar]
- 27.Ivey, F. D., P. N. Hodge, G. E. Turner, and K. A. Borkovich. 1996. The Gαi homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 7:1283-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasahara, S., and D. L. Nuss. 1997. Targeted disruption of a fungal G-protein β subunit gene results in increased vegetative growth but reduced virulence. Mol. Plant-Microbe Interact. 10:984-993. [DOI] [PubMed] [Google Scholar]
- 29.Kasahara, S., P. Wang, and D. L. Nuss. 2000. Identification of bdm-1, a gene involved in G protein β-subunit function and α-subunit accumulation. Proc. Natl. Acad. Sci. USA 97:412-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaziro, Y., H. Itoh, T. Kozasa, M. Nakafuku, and T. Satoh. 1991. Structure and function of signal-transducing GTP-binding proteins. Annu. Rev. Biochem. 60:349-400. [DOI] [PubMed] [Google Scholar]
- 31.Kim, J. S., and R. T. Raines. 1993. Ribonuclease S-peptide as a carrier in fusion proteins. Protein Sci. 2:348-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurjan, J. 1993. The pheromone response pathway in Saccharomyces cerevisiae. Annu. Rev. Genet. 27:147-179. [DOI] [PubMed] [Google Scholar]
- 33.Lee, B. N., and T. H. Adams. 1994. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol. Microbiol. 14:323-334. [DOI] [PubMed] [Google Scholar]
- 34.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, S., and R. A. Dean. 1997. G protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant-Microbe Interact. 10:1075-1086. [DOI] [PubMed] [Google Scholar]
- 36.Martemyanov, K. A., P. V. Lishko, N. Calero, G. Keresztes, M. Sokolov, K. J. Strissel, I. B. Leskov, J. A. Hopp, A. V. Kolesnikov, C. K. Chen, J. Lem, S. Heller, M. E. Burns, and V. Y. Arshavsky. 2003. The DEP domain determines subcellular targeting of the GTPase activating protein RGS9 in vivo. J. Neurosci. 23:10175-10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulder, G. H., and J. G. H. Wessels. 1986. Molecular-Cloning of RNAs differentially expressed in monokaryons and dikaryons of Schizophyllum commune in relation to fruiting. Exp. Mycol. 10:214-227. [Google Scholar]
- 38.Orbach, M. J., E. B. Porro, and C. Yanofsky. 1986. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol. Cell. Biol. 6:2452-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parsley, T. B., B. Chen, L. M. Geletka, and D. L. Nuss. 2002. Differential modulation of cellular signaling pathways by mild and severe hypovirus strains. Eukaryot. Cell 1:401-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parsley, T. B., G. C. Segers, D. L. Nuss, and A. L. Dawe. 2003. Analysis of altered G-protein subunit accumulation in Cryphonectria parasitica reveals a third Gα homologue. Curr. Genet. 43:24-33. [DOI] [PubMed] [Google Scholar]
- 41.Patikoglou, G. A., and M. R. Koelle. 2002. An N-terminal region of Caenorhabditis elegans RGS proteins EGL-10 and EAT-16 directs inhibition of Gαo versus Gαq signaling. J. Biol. Chem. 277:47004-47013. [DOI] [PubMed] [Google Scholar]
- 42.Peixoto, A. A., J. M. Hennessy, I. Townson, G. Hasan, M. Rosbash, R. Costa, and C. P. Kyriacou. 1998. Molecular coevolution within a Drosophila clock gene. Proc. Natl. Acad. Sci. USA 95:4475-4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponting, C. P., and P. Bork. 1996. Pleckstrin's repeat performance: a novel domain in G-protein signaling? Trends Biochem. Sci. 21:245-246. [PubMed] [Google Scholar]
- 44.Raper, J. R., and P. G. Miles. 1958. The genetics of Schizophyllum commune. Genetics 43:530-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Regenfelder, E., T. Spellig, A. Hartmann, S. Lauenstein, M. Bölker, and R. Kahmann. 1997. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 16:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosen, S., J. H. Yu, and T. H. Adams. 1999. The Aspergillus nidulans sfaD gene encodes a G protein β subunit that is required for normal growth and repression of sporulation. EMBO J. 18:5592-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ross, E. M., and T. M. Wilkie. 2000. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69:795-827. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Segers, G. C., and D. L. Nuss. 2003. Constitutively activated Gα negatively regulates virulence, reproduction and hydrophobin gene expression in the chestnut blight fungus Cryphonectria parasitica. Fungal Genet. Biol. 38:198-208. [DOI] [PubMed] [Google Scholar]
- 50.Taylor, J. W., B. H. Bowman, M. L. Berbee, and T. J. White. 1993. Fungal model organisms: phylogenetics of Saccharomyces, Aspergillus, and Neurospora. Syst. Biol. 42:440-457. [Google Scholar]
- 51.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Versele, M., J. H. de Winde, and J. M. Thevelein. 1999. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 18:5577-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson, P., K. Davis, M. Didmon, P. Broad, and J. Davey. 1999. An RGS protein regulates the pheromone response in the fission yeast Schizosaccharomyces pombe. Mol. Microbiol. 33:623-634. [DOI] [PubMed] [Google Scholar]
- 54.Wessels, J. G., O. M. de Vries, S. A. Asgeirsdottir, and J. Springer. 1991. The thn mutation of Schizophyllum commune, which suppresses formation of aerial hyphae, affects expression of the Sc3 hydrophobin gene. J. Gen. Microbiol. 137(Pt. 10):2439-2445. [DOI] [PubMed] [Google Scholar]
- 55.Yamagishi, K., T. Kimura, M. Suzuki, and H. Shinmoto. 2002. Suppression of fruit-body formation by constitutively active G-protein α-subunits ScGP-A and ScGP-C in the homobasidiomycete Schizophyllum commune. Microbiology 148:2797-2809. [DOI] [PubMed] [Google Scholar]
- 56.Yang, Q., and K. A. Borkovich. 1999. Mutational activation of a Gαi causes uncontrolled proliferation of aerial hyphae and increased sensitivity to heat and oxidative stress in Neurospora crassa. Genetics 151:107-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, Q., S. I. Poole, and K. A. Borkovich. 2002. A G-protein β subunit required for sexual and vegetative development and maintenance of normal Gα protein levels in Neurospora crassa. Eukaryotic Cell 1:378-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu, J. H., J. Wieser, and T. H. Adams. 1996. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO J. 15:5184-5190. [PMC free article] [PubMed] [Google Scholar]
- 59.Zuber, S., M. J. Hynes, and A. Andrianopoulos. 2003. The G-protein α-subunit GasC plays a major role in germination in the dimorphic fungus Penicillium marneffei. Genetics 164:487-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuber, S., M. J. Hynes, and A. Andrianopoulos. 2002. G-protein signaling mediates asexual development at 25°C but has no effect on yeast-like growth at 37°C in the dimorphic fungus Penicillium marneffei. Eukaryot. Cell 1:440-447. [DOI] [PMC free article] [PubMed] [Google Scholar]