Abstract
Colorectal cancer (CRC) is one of the most prevalent malignancies in the world. CRC-associated morbidity and mortality is continuously increasing, in part due to a lack of early detection. The existing screening tools such as colonoscopy, are invasive and yet high cost, affecting the willingness of patients to participate in screening programs. In recent years, evidence is accumulating that the interaction of aberrant genetic and epigenetic modifications is the cornerstone for the CRC development and progression by alternating the function of tumor suppressor genes, DNA repair genes and oncogenes of colonic cells. Apart from the understanding of the underlying mechanism(s) of carcinogenesis, the aforementioned interaction has also allowed identification of clinical biomarkers, especially epigenetic, for the early detection and prognosis of cancer patients. One of the ways to detect these epigenetic biomarkers is the cell-free circulating DNA (circDNA), a blood-based cancer diagnostic test, mainly focusing in the molecular alterations found in tumor cells, such as DNA mutations and DNA methylation. In this brief review, we epitomize the current knowledge on the research in circDNA biomarkers - mainly focusing on DNA methylation - as potential blood-based tests for early detection of colorectal cancer and the challenges for validation and globally implementation of this emergent technology.
Keywords: Colorectal cancer early detection, Colorectal cancer screening, Circulating free DNA, Colorectal cancer blood-based biomarkers, DNA methylation blood biomarkers
Core tip: Colorectal cancer (CRC) is one of the most prevalent malignancies in the world. CRC-associated morbidity and mortality is continuously increasing, in part due to a lack of early detection. The main aim of this article is the brief description of the basic screening modalities and their efficacy for CRC detection, the process of colorectal carcinogenesis and how the molecular pathways of CRC (focusing on epigenetic modifications) influence the clinical application of new blood-based biomarkers such as circDNA. Then we will focus on the most recent findings concerning the studies on circDNA, mainly related to DNA methylation and the challenges for validation and globally implementation of this emergent technology.
INTRODUCTION
Colorectal cancer (CRC) is the most common malignancy presented in gastrointestinal (GI) tract and the third most frequent cancer globally, with an incidence approximately approaching 1.5 million cases per year[1,2]. Likewise, it is considered that over 600000 deaths occur each year by neoplasms of the large bowel making them, the third commonest cause of cancer-related deaths[2]. It is thought that the gradual adoption of Westernized lifestyle and dietary habits by the majority of the countries in association with aging of the population are responsible for the increase in morbidity and mortality rates from CRC. This is in accordance with the World Health Organization which estimates that a substantial increase in the number of newly diagnosed CRC cases and a 80% rise in deaths from CRC are expected by 2030[3]. It should also be pointed out that colorectal adenocarcinomas are distinctive for their relatively fast progression and late clinical presentation, characteristics that are fairly preventable if identified at an early stage. Nevertheless, the currently available screening tests for the early detection of CRC need improvement enough in order to increase their cost-effective status. Thus, it is conceivable that, there is a significant interest in using noninvasive blood biomarkers which could be of low cost and high sensitivity and specificity to help reduce the predicted surge in the incidence of CRC by identification and removal of a larger number of polyps that potentially could lead to CRC over time[3]. These biomarkers are designed to detect molecular indicators in the plasma or serum such as DNA, RNA or protein in order to expand the existing list of CRC screening modalities[4].
Εpigenetic phenomena contribute to colorectal neoplasia[5]. This term refers to the mechanisms that alter gene expression without changing their DNA sequence. Epigenetic phenomena may include DNA methylation, histone modification and chromatin regulation through non-coding RNAs (microRNAS, incRNAs, etc.)[6]. Since DNA methylation and DNA mutations detected in tumor cells, it is reasonable to assume that these alterations are reflected in circDNA released from neoplastic tissue into blood circulation. Testing for circDNA in the peripheral blood could serve as an important candidate biomarker for the detection of CRC at early stages. An existing paradigm of commercial blood test for CRC detection in the circDNA is the monitoring of methylation of the septin 9 gene (SEPT9) promoter region.
Therefore, the aim of this article is the brief description of the basic screening modalities and their efficacy for CRC detection, the process of colorectal carcinogenesis and how the molecular pathways of CRC (focusing on epigenetic modifications) influence the clinical application of new blood-based biomarkers such as circDNA. Then we will focus on the most recent findings concerning the studies on circDNA, mainly related to DNA methylation and the challenges for validation and globally implementation of this emergent technology.
EXISTING SCREENING MODALITIES TO CRC
Screening modalities
There are various strategies for screening nowadays; the most accepted being the colonoscopy, and the combination of sigmoidoscopy and fecal occult blood test (FOBT). The high sensitivity and specificity has established the colonoscopy the cornerstone for the early identification of colonic malignancies in the average-risk population[7,8]. There are some drawbacks that limit the desired wide acceptance. As an invasive examination, complications may be unavoidable, the most common being cardiovascular events during the procedure and the post-polypectomy bleeding and perforation[7]. Other disadvantages could be a significant miss rate of lesions even for large colonic abnormalities, its high cost and the low acceptance level by the population[9].
Compared to colonoscopy, sigmoidoscopy has quite few disadvantages due to low cost, less preparation time and no need for sedation[10]. The main problem is the ability to detect only the lesions of distal colon making the decision of performing colonoscopy a subject for controversy even to date.
The third and most frequently applied screening test is FOBT[11]. Although these tests are easier to perform than colonoscopy or sigmoidoscopy, they are associated with false positive and false negative results due to diet, other conditions like colitis and hemorrhoids and the effect of temperature on the samples[12]. Moreover, FOBT cannot be used as solo screening test, as a positive results lead to colonoscopy performance[13].
Thus, there is an emergent need for new screening tests such as blood-based test which could detect CRC earlier, increase patient participation with minimal risks, costs, and false positive and negative results.
MOLECULAR PATHWAYS IN CRC AND DNA METHYLATION
CRC molecular pathways
Colorectal cancer is a multifarious disease. The comprehension of the molecular pathways involved in its development, will help to optimize the screening procedure based on distinctive pathologic and molecular features of the malignancies. Three basic pathways of colorectal carcinogenesis have been recognized since 1990 that is, Chromosomal Instability (CIN), Microsatellite Instability (MSI) and CpG island methylator phenotype (CIMP) pathway[14].
Chromosomal instability, also entitled “the suppressor pathway”, was first introduced in 1990 by Fearon et al[14] and is the most frequent etiology for gene alteration in colorectal neoplasia. Its main characteristic is the modification of whole chromosome or some of its regions, affecting important genes leading to carcinogenesis. These genomic defects provoke suppressor genes inactivation as Deleted in Colon Carcinoma (DCC), SMAD family member 2 (SMAD2), SMAD family member 4 (SMAD4), Adenomatosis polyposis coli (APC) and tumor protein p53 (TP53) and oncogene activation such as the human homolog of the Kirsten rat sarcoma-2 virus oncogene (KRAS)[15]. The accumulation of these modifications seems to play the most crucial role for cancer to develop and not the sequence of their presentation as once considered. The second model which involved in normal intestinal mucosa transformation to malignancy is the microsatellite instability. MSI is another type of genomic instability which refers to deletions or insertions of a few nucleotides in genes responsible for repair during DNA replication, the DNA mismatch repair (MMR) genes[16]. This aberrant genomic region mainly segregates in repetitive DNA nucleotide unit (microsatellites) throughout the genome resulting in the inactivation of MMR genes (i.e., MSH2, MLH1, MSH6, PMS1-2, MLH3, MSH3, ExoI). It is well-known that this route of carcinogenesis is involved in Lynch syndrome and for a notable proportion of sporadic CRC (15%-20%)[17]. The third model involved in the CRC development and progression, is CIMP which refers to the presence of simultaneous hypermethylation of multiple genes. It belongs to the epigenetic mechanisms leading to silence gene function after methylation at the 5’-CG-3’(CpG) dinucleotide in the promoter region of many genes (APC, MCC, MLH1, MGMT), resulting therefore, in inactivation of tumor suppressor genes[18]. CIMP is accountable for 15%-20% of sporadic CRC and according to the study of Jass[19], we are able to classify CRC according to the presence of MSI and CIMP as Figure 1 shows.
Figure 1.
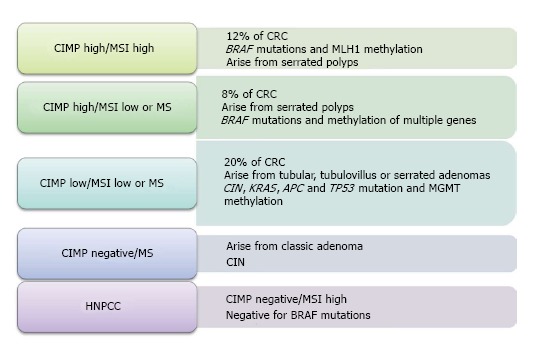
Molecular classification of colorectal cancer. The figure shows the different molecular profile and clinic-histopathological characteristics of each classification. CIMP: CpG island methylator phenotype; MSI: Microsatellite instability; MS: Microsatellite stability; CRC: Colorectal cancer; CIN: Chromosomal instability; HNPCC: Hereditary nonpolyposis colorectal cancer; MGMT: O-6-methylguanine-DNAmethyltransferase; BRAF: v-raf murine sarcoma viral oncogene homologe B1; MLH1: MutL homolog 1; KRAS: Kirsten rat sarcoma 2 viral oncogene homolog; APC: Adenomatosis polyposis coli; TP53: Tumor protein p53.
DNA methylation and its role in CRC carcinogenesis
DNA methylation mainly occurs in specific parts of the genome called, as we have seen previously, CpG islands. Considering the stability of DNA methylation compared to mutations, we may presume that methylation is a favorable area for biomarker exploration.
The concept that genome methylation may play a critical role in specific steps in the CRC carcinogenesis has been expressed in 1983 by Feinberg and Vogelstein[20] who showed that at early stages of CRC there is a DNA hypomethylation, mainly located at CpG islands. They also demonstrated that this loss of methylation was combined by hypermethylation and inactivation of tumor suppressor or DNA repair genes[21]. This epigenetic modification has recently been associated with the normal mucosa-aberrant crypt focus (ACF)-adenoma-carcinoma sequence, playing an important role in CRC development[22]. Consequently, DNA methylation appears to be one of the cornerstones of carcinogenesis because it occurs at the first steps of CRC process; involves CIMP pathway with MSI, as hypermethylation of MMR genes results in MSI sporadic CRC; through CIMP, it has been linked with CIN in colon malignancy (promoter methylation of GATA4, GATA5, p16 resulted in chromosomal loss or gain); and finally it is implicated in each of these paths through many abnormally methylated genes as recently studies have revealed[23-28] (Figure 2). The design of genetic and epigenetic biomarkers, especially those related to detection of aberrant methylated genes able to offer the maximum coverage of intestinal neoplasia, seems to be a reasonable approach. Accordingly, several studies have been performed the last decade, for the potential use of DNA markers in different biologic fluids as strategies for colorectal carcinoma early detection[29].
Figure 2.
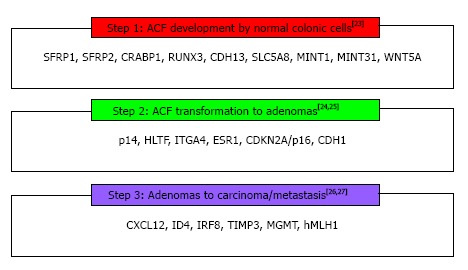
The figure exhibits the most frequent methylated genes/loci involved in step-by-step adenoma-carcinoma process in the context of colorectal cancer development. SFRP1: Secreted frizzled-related protein 1; CRABP1: Cellular retinoic acid binding protein 2; RUNX3: Runt-related transcription factor 3; CDH13: Cadherin 13; SLC5A8: Sodium solute symporter family 5 member 8; MINT1: Methylated in tumor locus 1; WNT5A: Wingless-type MMTV integration site family, member 5A; p14: Tumor protein 14; HLTF: Helicase-like transcription factor; ITGA4: Integrin, alpha 4; ESR1: Estrogen receptor 1; CDKN2A/p16: Cyclin-dependent kinase inhibitor 2A; CDH1: E-cadherin; CXCL12: Chemokine (C-X-C) ligand 12; ID4: Inhibitor of DNA binding 4; IRF8: Interferon regulatory factor 8; TIMP3: Tissue inhibitor of metalloproteinase 3; MGMT: O-6-methylguanine-DNAmethyltransferase; hMLH1: MutL homolog 1.
CELL-FREE CIRCULATING DNA AS A BLOOD-BASED BIOMARKER
Mandel and Metais[30] in 1948, were the pioneers who discovered the existence of cell-free nucleic acids (cfNAs) in blood, leading to ‘‘discrimination’’ of affected patients from healthy controls, even though the first reported presence of cfNAs was in 1869[31]. Since then, several studies have been made, especially related to cancer pathogenesis, showing that malignant-nucleic acids could be present in different ‘‘body fluids’’ (i.e., stool, blood, urine). Therefore, it was a matter of time the establishment of the potential advantages using cfNA as noninvasive neoplasia detection[32,33]. The first report referring to detection of ‘‘abnormal’’ cfNAs in CRC patients took place in 1992, when Vogelstein et al[34] discovered KRAS gene mutation in stools samples of CRC-affected individuals. Since then, a great number of studies have performed evaluating other than KRAS, genome modifications directly expressed by cfNAs. These modifications are characterized mainly by analysis of high mutation frequency genes (KRAS, TP53, and APC), MSI, Loss of Heterozygosity (LOH), DNA, and microRNA methylation changes. Except the latter one, all the others are key factors for colorectal carcinogenesis which could expressed by increased circDNA concentrations in blood of CRC patients compared with healthy controls, first mentioned by Leon et al[35] in 1977, followed by many other studies[35-37].
Thus, since circDNA in blood reflects significant genome alterations emerging during CRC carcinogenesis, it could be used systematically as a potential biomarker for early detection of colonic malignant tumors, especially after the recent advances in next generation sequencing (NGS) technology[38]. Here follows a discussion of methods to detect circDNA-based markers in blood and the studies focusing on description of these markers namely, aberrant DNA methylation and mutations, microsatellite alterations, DNA modifications in mitochondria, integrity and quantification of DNA.
Methods to detect methylation-related circDNA markers in blood
Methodologies suitable to investigate and detect in serum or plasma frequently methylated genomic regions, offering high detection rate is more than an appealing aim. These ideal markers should have low levels of background methylation so as to avoid the decreased specificity and increased false-positive results. Several molecular approaches are currently performed to identify circDNA in blood. Conventional methylation-specific PCR, though very sensitive method, presents high levels of false positive and negatives results and its qualitative method to interpreter the findings, limits its clinical utility[39]. On the other hand, methods based on quantitative methylation-specific PCR (i.e., MethyLight, SMART-methylation-specific PCR) offers the opportunity to select more easily the methylation thresholds and avoid false positive results identifying incomplete bisulfite conversion[40]. Other approaches include Methylation Array, DNA array, Surface-Enhanced Raman Scattering (SERS), and Restriction Fragment Length Polymorphism (RFLP). Another interesting and ‘‘more digital’’ methodology, called ‘‘Methyl-BEAMing’’, was demonstrated by Li et al[41] in order to digitally quantify cancer-derived vimentin DNA, confronting the issues with the small fraction of blood circDNA.
Given the great concerns arouse by previous approaches concerning the inability for reproducibility and high sensitivity, promising results revealed by the study of Leary et al[42]. The use of NGS approach showed encouraging results, distinguishing CRC patients at advanced stage from healthy controls. Thus, NGS could provide high sensitivity, covering large regions of genome for CRC early detection.
Cell-free circulating DNA-based markers
During the last two decades, circDNA has become a potential biomarker for diagnosis of malignant tumors, exhibiting their genetic and epigenetic modifications. It exists in the plasma or serum, being source of apoptotic, necrotic cancer cells or even living cells. It can appear as unbound DNA molecule; as histone part in nucleosome; or as portion of apoptotic cells. As already mentioned, there are several methods of assessing circDNA as a potential biomarker for detection of CRC at early stages. Herein, we would try to summarize the main characteristics of each method, noting presentative studies reflecting the potential clinical use of these circDNA-based modalities.
One of these methods is the quantification of circDNA levels in blood, studied thoroughly in CRC patients since the research by Leon et al[35] in 1977 who proved that concentration of circDNA was higher than that of healthy persons. Additional studies, such as the ones performed by Frattini, Schwarzenbach et al[43] respectively, verified the elevated circDNA levels in the plasma of CRC individuals compared with the non-cancerous controls[43]. Although, patients with malignancies may present greater levels of circDNA than normal persons, it should be emphasized that circDNA in plasma may also be observed in other clinical entities like trauma, inflammatory disorders even in healthy individuals[35].
In the recent years, it is well-established that the manner, with which the circDNA is released in bloodstream, reflects its size and morphology. The exact mechanism is yet to be clarified but it is believed that circDNA entered the blood by apoptosis and then it is fragmented by the action of nucleases or phagocytes, into small particles of 185 to 200 bp in length[44]. The measure of the ratio of long circDNA fragments to short ones mirrors circDNA integrity. A great number of studies have been performed, demonstrating inconsequent results (Table 1)[45-51] as concerns the sensitivity and specificity of circDNA integrity index for CRC early detection. Interestingly, a recent research by Hao et al[52] showed that the combination of DNA integrity index (ALU247/115 and ALU115 index) and carcinoembryonic antigen (CEA) detection may be efficient and reproducible method for early diagnosis of CRC. Therefore, larger clinical studies should be performed in order to limit the inconsistencies that circDNA integrity method exhibits.
Table 1.
DNA integrity index in colorectal cancer patients
Microsatellite alterations is another investigation field related to tumorigenesis of CRC and consist of MSI and LOH. Due to their presence in circDNA, it is assumed that they could be potential CRC biomarkers for early diagnosis of affected individuals. As we already have mentioned, MSI refers to deletions or insertions of a few nucleotides (1-6 bp in length) in genes responsible for repair during DNA replication, the MMR genes[16], while LOH analysis emphasizes the loss of chromosomal parts carrying tumor suppressor genes. These somatic alterations have been detected in blood, nearly in 35% of all CRC patients. The existence of MSI-related circDNA fragments is known since the ending of 20th century followed by many studies focusing on the presence of MSI and LOH in circDNA[53]. One of these by Hibi et al showed in 1999 that, although LOH and MSI found in 80% CRC patients when examined their microsatellite alterations, these shifts weren’t verified upon the corresponding serum-based circDNA. Therefore, the available data reveals the relatively low sensitivity and specificity in diagnosing CRC at early stages when microsatellite alterations are investigated.
Similar disappointing results have been arisen from the study of circulating mitochondrial DNA (mtDNA) as potential circDNA-based biomarker for premature diagnosis of colonic neoplasia. Mitochondria are the cornerstone in energy metabolism, aging, and apoptosis, playing a crucial role in shifting the cell from scheduled death to abnormal cell growth, thus having potential contribution to the carcinogenesis[54]. An important part of mtDNA is its D-loop region, a noncoding region which involves the expression and organization of the mitochondrial genome. It is hypothesized that this part of mtDNA is a hotspot of mutations leading to DNA instability, opinion that has been verified in several types of cancers such as, head and neck, colorectal, stomach, prostate, breast[55]. Despite the initial encouraging signs, mtDNA shows reduced detection rate of early stage CRC, as the study by Hibi et al[56] revealed, where the discovery of mtDNA modifications (somatic mutation in D-loop region) in tissues of early CRC patients haven’t been noted in their circDNA.
As it is stated previously, the blood-based circDNA in CRC patients is composed by important molecules which have implicated in tumorigenesis process. Since 1992 when Vogelstein et al[34] discovered KRAS gene mutation in stools samples of CRC-affected individuals, high mutation frequency genes, as KRAS, TP53, and APC have been used as potential markers in circDNA analysis for early diagnosis of colonic malignant lesions[57,58]. The results were discouraging due to low concentration of tumor circDNA (based on the somatic mutations analysis of KRAS/TP53/APC genes modification) in CRC patients compared with the wild-type circDNA in non-CRC individuals[58]. Moreover, it should be noted that even the use of NGS circDNA detecting method, hasn’t offered any improvement in detection of these aberrant tumor DNA mutations[59]. Finally, as aforementioned, genes such as APC, TP53, and KRAS are mutated in a great degree of CRC cases, spreading over different parts of genome, making mutational assessment difficult. Thus, it is reasonable to assume that very large genomic regions would need to be evaluated in order to obtain a respectable sensitivity and in combination of the unique presentation of modified genes in each patient, it is still difficult enough to use somatic mutations for CRC early detection.
Abnormal DNA methylation as a cell-free circulating DNA biomarker
As it has been already highlighted, the critical role of abnormal DNA methylation to specific steps in the CRC carcinogenesis has been expressed since 1983 from Feinberg and Vogelstein[20]. Since then and during the recent years, many studies have revealed that this epigenetic modification has been associated with the normal mucosa-ACF-adenoma-carcinoma sequence, playing an important role in CRC development, mainly, at early stages[22-27]. It is known that during DNA methylation, DNA methyl transferases (DNMTS) catalyze the addition of a methyl group (-CH3) to the fifth carbon position on cytosines within CpG dinucleotides. The latter, although spread over throughout the human genome, they are frequently discovered in the promoter regions of nearly 70% of genes, usually named as “CpG-islands”[60]. Furthermore, it is well-established by now that hypermethylation of tumor suppressor promoters genes could induce transcriptional gene silencing, resulting on aberrant cellular signaling and therefore potential initiation of tumorigenesis process[61]. Moreover, it is interesting that methylation could happen in CpG sites throughout the genomic body and not necessarily only in promoter regions leading though to transcriptional activation[62]. On the other hand, global hypomethylation which frequently presented prematurely during carcinogenesis, exhibits loss of DNA methylation throughout the genome, resulting on CIN and cell mutation[63]. Consequently, the significance of aberrant DNA methylation led to investigation and discovery of blood-based mainly, due to its noninvasiveness and cost-effectiveness, CRC detection biomarkers.
One of the most investigated genes is the SEPT9 gene involved in cellular proliferation control. The methylation of v2 promoter region of SEPT9 has been demonstrated in CRC biopsy lesions compared with normal tissues. According to Grützmann et al[64], its detection in plasma of CRC patients exhibited a sensitivity of 72% and specificity of 90%, something that was validated by the study of Warren et al[65]. Nevertheless, a recent prospective trial performed by Church et al[66] investigated the SEPT9 methylation in 7941 asymptomatic individuals during screening with available assay showing a CRC detection rate up to 48.2% and specificity up to 91.5%. Obviously, the need of further researches upon this commercially available test is indispensable not only to improve its detection rate but also to discover new assays for SEPT9 methylation detection. Furthermore, researchers understanding the usefulness of SEPT9 have assessed potential combinations with other methylation biomarkers. Tänzer et al[67] have shown that methylated DNA from advanced premalignant intestinal lesions could be discovered using the panel of aristaless-like homeobox 4 (ALX4), and SEPT9 markers. Similarly, Kostin et al[68] compared the methylation status of SEPT9, Helicase-like transcription factor (HLTF) and ALX4 genes in macroscopically findings compatible with colorectal cancer (n = 55) and morphologically intact areas of the large bowel (n = 71), showing that this panel of biomarkers characterized by a sensitivity nearly to 74%-88% and a specificity 90%-96% for CRC early identification. Finally, He et al[69] demonstrated high sensitivities (81%-84%) and specificities (87%-90%) for noninvasive blood-based testing for initial-phase CRC, using multiplex MethyLight PCR assay to detect concomitantly, aberrant methylation pattern of ALX4, SEPT9, or transmembrane protein with EGF-like, and two follistatin-like domains 2 (TMEFF2) genes.
Apart from the aforementioned SEPT9-combined panels, there are recently studies showing even greater CRC detection rates if combined analysis of several genes is used. Alhquist et al[70] presented high overall sensitivity (87%) for the CRC detection compared with SEPT9 (60%), using the combination of methylated genes such as bone morphogenetic protein (BMP3), N-myc downstream regulated family member 4 (NDRG4), vimentin, tissue factor pathway inhibitor-2 (TFPI2), mutant KRAS and β-actin. According to Carmona et al[71], there is a 78% sensitivity for CRC early diagnosis when combining angiotensin II receptor type 1 (AGTR1), wingless-type MMTV integration site family member 2 (WNT2), slit homolog 2 (Drosophila) (SLIT2) genes. Moreover, Cassinotti et al[72] exhibited the potential use of gene panel, consisting of D-type cyclin gene (CYCD2), hypermethylated in Cancer 1 (HIC1), PAX5, Ras association domain family 1, isoform A (RASSF1A), retinoblastoma tumor suppressor (RB1) and sheep red blood cells (SRBC) with sensitivity nearly 84% and specificity 68%. Comparable results revealed by others studies making these panels powerful tools for future large-scale trials[73-95] (Table 2).
Table 2.
Abnormally methylated genes as potential circDNA blood-based colorectal cancer detection biomarkers
| Potential biomarkers | CRC sensitivity (%) | CRC specificity (%) | Ref. |
| ALX4 | 40-83 | 70-82 | [67,76] |
| TFPI2 | 76-89 | - | [77-80] |
| SDC2 | 92 | - | [81] |
| RUNX3 | 65 | 100 | [82,83] |
| NEUROG1 | 52-64 | 91 | [84] |
| MGMT | 39 | 96 | [74] |
| RARβ2 | 24 | 100 | [74] |
| NGFR | 51 | 84 | [85] |
| 9-Sep | 48-90 | 86-93 | [64-67,70,86] |
| TMEFF2 | 65 | 69 | [85] |
| Vimentin | 59 | 93 | [41] |
| RASSF2A | 58 | 100 | [74] |
| Wif-1 | 74 | 98 | [74] |
| APC | 6 | 100 | [87] |
| hMLH1 | 43 | 98 | [87] |
| HTLF | 21-34 | 98-100 | [87,88] |
| SFRP2 | 67 | 94 | [89] |
| CDKN2A/P16 | 71 | 100 | [83] |
| Panel: SEPT9, HLTF and ALX4 | 74-88 | 90-96 | [68] |
| Panel: SEPT9 and ALX4 | - | - | [67] |
| Panel: MGMT, RASSF2A, Wif-1 gene | 86.5 | - | [74] |
| Panel: BMP3, NDRG4, vimentin, TFPI2, mutant KRAS and β-actin | 87 | - | [69] |
| Panel: AGTR1, WNT2, SLIT2 | 78 | - | [71] |
| Panel: CINP1, FBN1, INA, SNCA, MAL and SPG20 | 90-99 | - | [73] |
| Panel: CYCD2, HIC1, PAX5, RASSF1A, RB1 and SRBC | 84 | 68 | [72] |
| Panel: THBD and C9orf50 | 71 | 80 | [75] |
| RASSF1A, E-cadherin | - | - | [72,90] |
| CAHM | - | - | [91] |
| FRP2, TPEF/HPP1 | - | - | [83,84,92] |
ALX4: Aristaless-like homeobox 4; TFPI2: Tissue factor pathway inhibitor 2; SDC2: Syndecan 2; RUNX3: Runt-related transcription factor 3; NEUROG1: Neurogenin 1; MGMT: O-6-methylguanine-DNAmethyltransferase; RARβ2: Retinoic acid receptor β2; NGFR: Nerve growth factor receptor; SEPT9: Septin 9; TMEFF2: Transmembrane protein with EGF-like, and two follistatin-like domains 2; RASSF2A: Ras association domain family 2 (isoform A); Wif-1: Wnt inhibitory factor-1; APC: Adenomatosis polyposis coli; hMLH1: MutL homolog 1; HTLF: Helicase-like transcription factor; SFRP2: Secreted frizzled-related protein 2; CDKN2A/P16: Cyclin-dependent kinase inhibitor 2A; BMP3: Bone morphogenetic protein; NDRG4: N-myc downstream regulated family member 4; KRAS: Kirsten rat sarcoma 2 viral oncogene homolog; AGTR1: Tissue fac angiotensin II receptor type 1; WNT2: Wingless-type MMTV integration site family member 2; SLIT2: Slit homolog 2 (Drosophila); CINP1 FBN1: Fibrillin 1; SNCA: α-synuclein gene; SPG20: Spastic paraplegia-20; CYCD2: D-type cyclin gene; HIC1: Hypermethylated in cancer 1; RASSF1A: Ras association domain family 1 (isoform A); RB1: Retinoblastoma tumor suppressor; SRBC: Sheep red blood cells; THBD: Thrombomodulin; C9orf50: Chromosome 9 open reading frame 50; FRP2: Frizzled related protein 2; TPEF/HPP1: Transmembrane protein containing epidermal growth factor, follistatin domain/hyperplastic polyposis 1.
In parallel, several blood-based methylated genes as potential biomarkers have been studied either alone or within panels as previously demonstrated, and a summary of them exhibited in Table 2, concerning their detection rate[41,64-67,70,72,74-92]. Some of them (SEPT9, ALX4, SDC2, RUNX3, TMEFF2, NEUROG1) present high sensitivity and specificity for CRC detection during initial stages when analyzing methylation status of circDNA[67,69]. Although the evaluation some of these aberrant methylated genes may demonstrate better diagnostic results than the SEPT9 analysis, their cost effectiveness, further technical improvement and low testing uptake issues impede their use within large-scale clinical trials[70,93]. Thus, SEPT9 as the most common blood-based methylation analysis biomarker holds promising example of sending on real life the laboratory methylation studies upon circDNA, for early CRC diagnosis of average-risk individuals.
circDNA pre-analysis considerations
As we stated before, analysis of SEPT9 gene methylation could be subject of further technical advance[93]. Therefore, it is reasonable to presume that several factors could play crucial role such as blood sampling and circDNA processing. It is well-established that blood-based circDNA could be extracted from both plasma and serum with the latter one exhibiting higher concentration of DNA[94,95]. However there are studies suggesting that this high amount of DNA in serum reflects the in vitro lysis of leucocytes when the procedures of coagulation and/or fibrinolysis take place[95]. Another theory highlights the significant effect that chemicals differences between serum and plasma have during DNA extraction[96]. Other factors that researches should take into account are: The interval time of blood drawn and centrifugation; the sample storage modality; the anticoagulant used; temperature; and the plasma-based DNA isolation protocol[96]. All these parameters exhibit enormous significance as concerns the efficiency and quality of circDNA analysis, illustrating the reliability that newer methods of circDNA analysis, should have.
CONCLUSION AND FUTURE ASPIRATIONS
Colorectal cancer is one of the deadliest malignancies to date even though various techniques are available to prevent and detect its emergence. Although these preventive modalities (sigmoidoscopy, colonoscopy, FOBT, FIT) exhibit high CRC detection sensitivity and specificity, the acceptance rate among population remains low. In parallel, the rapid progression of molecular biology has revealed new translational research fields related to discovery of potential CRC biomarkers in body “liquid fluids”. These markers evaluate the fragments of DNA, RNA or proteins in the blood or feces demonstrating an increasingly cost-effective and sensitive way to detect premalignant modification of genome in individuals on average risk for CRC development. Thus, with this review we tried to highlight those circDNA blood-based biomarkers that offer an easy, cost-effective and with minimal invasiveness diagnosis of colonic neoplasia (Table 2). We believe that this research demonstrate in depth the need for further studies to be done which should be large randomized and will try to evaluate or elucidate the clinical value of all these new proposed screening tests which could be combined the older ones as a critical strategy to improve quality of the existing life expectancy as well as to advance the latter one.
Footnotes
Conflict-of-interest statement: No potential conflicts of interest. No financial support. All the authors declare that they have no conflict of interest.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Greece
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
Peer-review started: August 1, 2016
First decision: December 1, 2016
Article in press: March 14, 2017
P- Reviewer: Doll D, Hansen TF, Ju SQ, Kadiyska TK S- Editor: Gong ZM L- Editor: A E- Editor: Lu YJ
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, Forman D, Mathers C and Parkin DM. Lyon, France: International Agency for Research on Cancer; 2010. GLOBOCAN 2008 v2.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10 [Internet] Available from: http://globocan.iarc.fr/ [Google Scholar]
- 3.World Cancer Report. Cancer syte by syte-colorectal cancer. In: Boyle P, Levin B, editors. World cancer report 2008. Lyon: International Agency for Research on Cancer; 2008. pp. 374–379. [Google Scholar]
- 4.Tänzer M, Liebl M, Quante M. Molecular biomarkers in esophageal, gastric, and colorectal adenocarcinoma. Pharmacol Ther. 2013;140:133–147. doi: 10.1016/j.pharmthera.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Migliore L, Migheli F, Spisni R, Coppedè F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011;2011:792362. doi: 10.1155/2011/792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodenhiser D, Mann M. Epigenetics and human disease: translating basic biology into clinical applications. CMAJ. 2006;174:341–348. doi: 10.1503/cmaj.050774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segnan N, Patnick J, von Karsa L, editors . Luxembourg: European Commission, Publications Office of the European Union; 2010. European guidelines for quality assurance in colorectal cancer screening and diagnosis. European guidelines for quality assurance in colorectal cancer screening and diagnosis - First edition. [DOI] [PubMed] [Google Scholar]
- 8.Brenner H, Chang-Claude J, Jansen L, Knebel P, Stock C, Hoffmeister M. Reduced risk of colorectal cancer up to 10 years after screening, surveillance, or diagnostic colonoscopy. Gastroenterology. 2014;146:709–717. doi: 10.1053/j.gastro.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Corley DA, Jensen CD, Marks AR. Can we improve adenoma detection rates? A systematic review of intervention studies. Gastrointest Endosc. 2011;74:656–665. doi: 10.1016/j.gie.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Schoen RE, Pinsky PF, Weissfeld JL, Yokochi LA, Church T, Laiyemo AO, Bresalier R, Andriole GL, Buys SS, Crawford ED, et al. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N Engl J Med. 2012;366:2345–2357. doi: 10.1056/NEJMoa1114635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaukat A, Mongin SJ, Geisser MS, Lederle FA, Bond JH, Mandel JS, Church TR. Long-term mortality after screening for colorectal cancer. N Engl J Med. 2013;369:1106–1114. doi: 10.1056/NEJMoa1300720. [DOI] [PubMed] [Google Scholar]
- 12.Kaminski MF, Regula J, Kraszewska E, Polkowski M, Wojciechowska U, Didkowska J, Zwierko M, Rupinski M, Nowacki MP, Butruk E. Quality indicators for colonoscopy and the risk of interval cancer. N Engl J Med. 2010;362:1795–1803. doi: 10.1056/NEJMoa0907667. [DOI] [PubMed] [Google Scholar]
- 13.Binefa G, Rodríguez-Moranta F, Teule A, Medina-Hayas M. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol. 2014;20:6786–6808. doi: 10.3748/wjg.v20.i22.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 15.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boland CR, Sinicrope FA, Brenner DE, Carethers JM. Colorectal cancer prevention and treatment. Gastroenterology. 2000;118:S115–S128. doi: 10.1016/s0016-5085(00)70010-2. [DOI] [PubMed] [Google Scholar]
- 17.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong JJ, Hawkins NJ, Ward RL. Colorectal cancer: a model for epigenetic tumorigenesis. Gut. 2007;56:140–148. doi: 10.1136/gut.2005.088799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 20.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 21.Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 22.Yamashita K, Dai T, Dai Y, Yamamoto F, Perucho M. Genetics supersedes epigenetics in colon cancer phenotype. Cancer Cell. 2003;4:121–131. doi: 10.1016/s1535-6108(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 23.Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143:1442–1460.e1. doi: 10.1053/j.gastro.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo L, Chen WD, Pretlow TP. CpG island methylation in aberrant crypt foci and cancers from the same patients. Int J Cancer. 2005;115:747–751. doi: 10.1002/ijc.20936. [DOI] [PubMed] [Google Scholar]
- 25.Subramaniam MM, Chan JY, Soong R, Ito K, Yeoh KG, Wong R, Guenther T, Will O, Chen CL, Kumarasinghe MP, et al. RUNX3 inactivation in colorectal polyps arising through different pathways of colonic carcinogenesis. Am J Gastroenterol. 2009;104:426–436. doi: 10.1038/ajg.2008.141. [DOI] [PubMed] [Google Scholar]
- 26.Wendt MK, Johanesen PA, Kang-Decker N, Binion DG, Shah V, Dwinell MB. Silencing of epithelial CXCL12 expression by DNA hypermethylation promotes colonic carcinoma metastasis. Oncogene. 2006;25:4986–4997. doi: 10.1038/sj.onc.1209505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umetani N, Takeuchi H, Fujimoto A, Shinozaki M, Bilchik AJ, Hoon DS. Epigenetic inactivation of ID4 in colorectal carcinomas correlates with poor differentiation and unfavorable prognosis. Clin Cancer Res. 2004;10:7475–7483. doi: 10.1158/1078-0432.CCR-04-0689. [DOI] [PubMed] [Google Scholar]
- 28.Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Notterman DA, Alon U, Sierk AJ, Levine AJ. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61:3124–3130. [PubMed] [Google Scholar]
- 30.Mandel P, Metais P. Les acides nucléiques du plasma sanguine chezl’homme. C R Seances Soc Biol Fil. 1948;142:241–243. [PubMed] [Google Scholar]
- 31.Asworth TR. A case of cancer in which cells similar to those in tumors were seen in the blood after death. Aust Med J. 1869;14:146–149. [Google Scholar]
- 32.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–2714. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 33.Müller HM, Oberwalder M, Fiegl H, Morandell M, Goebel G, Zitt M, Mühlthaler M, Ofner D, Margreiter R, Widschwendter M. Methylation changes in faecal DNA: a marker for colorectal cancer screening? Lancet. 2004;363:1283–1285. doi: 10.1016/S0140-6736(04)16002-9. [DOI] [PubMed] [Google Scholar]
- 34.Sidransky D, Tokino T, Hamilton SR, Kinzler KW, Levin B, Frost P, Vogelstein B. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102–105. doi: 10.1126/science.1566048. [DOI] [PubMed] [Google Scholar]
- 35.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 36.Chen K, Zhang H, Zhang LN, Ju SQ, Qi J, Huang DF, Li F, Wei Q, Zhang J. Value of circulating cell-free DNA in diagnosis of hepatocelluar carcinoma. World J Gastroenterol. 2013;19:3143–3149. doi: 10.3748/wjg.v19.i20.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi J, Qian C, Shi W, Wu X, Jing R, Zhang L, Wang Z, Ju S. Alu-based cell-free DNA: a potential complementary biomarker for diagnosis of colorectal cancer. Clin Biochem. 2013;46:64–69. doi: 10.1016/j.clinbiochem.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 38.Kaiser J. Medicine. Keeping tabs on tumor DNA. Science. 2010;327:1074. doi: 10.1126/science.327.5969.1074. [DOI] [PubMed] [Google Scholar]
- 39.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kristensen LS, Mikeska T, Krypuy M, Dobrovic A. Sensitive Melting Analysis after Real Time- Methylation Specific PCR (SMART-MSP): high-throughput and probe-free quantitative DNA methylation detection. Nucleic Acids Res. 2008;36:e42. doi: 10.1093/nar/gkn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Chen WD, Papadopoulos N, Goodman SN, Bjerregaard NC, Laurberg S, Levin B, Juhl H, Arber N, Moinova H, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27:858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, O’Shaughnessy J, Kinzler KW, Parmigiani G, Vogelstein B, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarzenbach H, Stoehlmacher J, Pantel K, Goekkurt E. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Ann N Y Acad Sci. 2008;1137:190–196. doi: 10.1196/annals.1448.025. [DOI] [PubMed] [Google Scholar]
- 44.Rykova EY, Morozkin ES, Ponomaryova AA, Loseva EM, Zaporozhchenko IA, Cherdyntseva NV, Vlassov VV, Laktionov PP. Cell-free and cell-bound circulating nucleic acid complexes: mechanisms of generation, concentration and content. Expert Opin Biol Ther. 2012;12 Suppl 1:S141–S153. doi: 10.1517/14712598.2012.673577. [DOI] [PubMed] [Google Scholar]
- 45.Umetani N, Kim J, Hiramatsu S, Reber HA, Hines OJ, Bilchik AJ, Hoon DS. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: direct quantitative PCR for ALU repeats. Clin Chem. 2006;52:1062–1069. doi: 10.1373/clinchem.2006.068577. [DOI] [PubMed] [Google Scholar]
- 46.da Silva Filho BF, Gurgel AP, Neto MÁ, de Azevedo DA, de Freitas AC, Silva Neto Jda C, Silva LA. Circulating cell-free DNA in serum as a biomarker of colorectal cancer. J Clin Pathol. 2013;66:775–778. doi: 10.1136/jclinpath-2013-201521. [DOI] [PubMed] [Google Scholar]
- 47.Leszinski G, Lehner J, Gezer U, Holdenrieder S. Increased DNA integrity in colorectal cancer. In Vivo. 2014;28:299–303. [PubMed] [Google Scholar]
- 48.Mouliere F, Robert B, Arnau Peyrotte E, Del Rio M, Ychou M, Molina F, Gongora C, Thierry AR. High fragmentation characterizes tumour-derived circulating DNA. PLoS One. 2011;6:e23418. doi: 10.1371/journal.pone.0023418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mead R, Duku M, Bhandari P, Cree IA. Circulating tumour markers can define patients with normal colons, benign polyps, and cancers. Br J Cancer. 2011;105:239–245. doi: 10.1038/bjc.2011.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mouliere F, El Messaoudi S, Pang D, Dritschilo A, Thierry AR. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol Oncol. 2014;8:927–941. doi: 10.1016/j.molonc.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yörüker EE, Özgür E, Keskin M, Dalay N, Holdenrieder S, Gezer U. Assessment of circulating serum DNA integrity in colorectal cancer patients. Anticancer Res. 2015;35:2435–2440. [PubMed] [Google Scholar]
- 52.Hao TB, Shi W, Shen XJ, Qi J, Wu XH, Wu Y, Tang YY, Ju SQ. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br J Cancer. 2014;111:1482–1489. doi: 10.1038/bjc.2014.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lazarev I, Leibovitch L, Czeiger D, Sion-Vardi N, Geffen DB, Douvdevani A, Ariad S. Cell-free DNA blood levels in colorectal cancer patients do not correlate with mismatch repair-proficiency. In Vivo. 2014;28:349–354. [PubMed] [Google Scholar]
- 54.Cavalli LR, Liang BC. Mutagenesis, tumorigenicity, and apoptosis: are the mitochondria involved? Mutat Res. 1998;398:19–26. doi: 10.1016/s0027-5107(97)00223-6. [DOI] [PubMed] [Google Scholar]
- 55.Zhu W, Qin W, Bradley P, Wessel A, Puckett CL, Sauter ER. Mitochondrial DNA mutations in breast cancer tissue and in matched nipple aspirate fluid. Carcinogenesis. 2005;26:145–152. doi: 10.1093/carcin/bgh282. [DOI] [PubMed] [Google Scholar]
- 56.Hibi K, Nakayama H, Yamazaki T, Takase T, Taguchi M, Kasai Y, Ito K, Akiyama S, Nakao A. Detection of mitochondrial DNA alterations in primary tumors and corresponding serum of colorectal cancer patients. Int J Cancer. 2001;94:429–431. doi: 10.1002/ijc.1480. [DOI] [PubMed] [Google Scholar]
- 57.Lecomte T, Berger A, Zinzindohoué F, Micard S, Landi B, Blons H, Beaune P, Cugnenc PH, Laurent-Puig P. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer. 2002;100:542–548. doi: 10.1002/ijc.10526. [DOI] [PubMed] [Google Scholar]
- 58.Diehl F, Li M, Dressman D, He Y, Shen D, Szabo S, Diaz LA, Goodman SN, David KA, Juhl H, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci USA. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin H, Liang Y, Yan Z, Liu B, Su Q. Mutation spectrum in human colorectal cancers and potential functional relevance. BMC Med Genet. 2013;14:32. doi: 10.1186/1471-2350-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 62.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 63.Peeters M, Douillard JY, Van Cutsem E, Siena S, Zhang K, Williams R, Wiezorek J. Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: assessment as prognostic and predictive biomarkers of response to panitumumab. J Clin Oncol. 2013;31:759–765. doi: 10.1200/JCO.2012.45.1492. [DOI] [PubMed] [Google Scholar]
- 64.Grützmann R, Molnar B, Pilarsky C, Habermann JK, Schlag PM, Saeger HD, Miehlke S, Stolz T, Model F, Roblick UJ, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One. 2008;3:e3759. doi: 10.1371/journal.pone.0003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warren JD, Xiong W, Bunker AM, Vaughn CP, Furtado LV, Roberts WL, Fang JC, Samowitz WS, Heichman KA. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011;9:133. doi: 10.1186/1741-7015-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rösch T, Osborn N, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–325. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tänzer M, Balluff B, Distler J, Hale K, Leodolter A, Röcken C, Molnar B, Schmid R, Lofton-Day C, Schuster T, et al. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One. 2010;5:e9061. doi: 10.1371/journal.pone.0009061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kostin PA, Zakharzhevskaia NB, Generozov EV, Govorun VM, Chernyshov SV, Shchelygin IuA. [Hypermethylation of the CDH1, SEPT9, HLTF and ALX4 genes and their diagnostic significance in colorectal cancer] Vopr Onkol. 2010;56:162–168. [PubMed] [Google Scholar]
- 69.He Q, Chen HY, Bai EQ, Luo YX, Fu RJ, He YS, Jiang J, Wang HQ. Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet Cytogenet. 2010;202:1–10. doi: 10.1016/j.cancergencyto.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 70.Ahlquist DA, Taylor WR, Mahoney DW, Zou H, Domanico M, Thibodeau SN, Boardman LA, Berger BM, Lidgard GP. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol. 2012;10:272–277.e1. doi: 10.1016/j.cgh.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carmona FJ, Azuara D, Berenguer-Llergo A, Fernández AF, Biondo S, de Oca J, Rodriguez-Moranta F, Salazar R, Villanueva A, Fraga MF, et al. DNA methylation biomarkers for noninvasive diagnosis of colorectal cancer. Cancer Prev Res (Phila) 2013;6:656–665. doi: 10.1158/1940-6207.CAPR-12-0501. [DOI] [PubMed] [Google Scholar]
- 72.Cassinotti E, Melson J, Liggett T, Melnikov A, Yi Q, Replogle C, Mobarhan S, Boni L, Segato S, Levenson V. DNA methylation patterns in blood of patients with colorectal cancer and adenomatous colorectal polyps. Int J Cancer. 2012;131:1153–1157. doi: 10.1002/ijc.26484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lind GE, Danielsen SA, Ahlquist T, Merok MA, Andresen K, Skotheim RI, Hektoen M, Rognum TO, Meling GI, Hoff G, et al. Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas. Mol Cancer. 2011;10:85. doi: 10.1186/1476-4598-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, Park J, Kim DH. Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res. 2009;15:6185–6191. doi: 10.1158/1078-0432.CCR-09-0111. [DOI] [PubMed] [Google Scholar]
- 75.Lange CP, Campan M, Hinoue T, Schmitz RF, van der Meulen-de Jong AE, Slingerland H, Kok PJ, van Dijk CM, Weisenberger DJ, Shen H, et al. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PLoS One. 2012;7:e50266. doi: 10.1371/journal.pone.0050266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ebert MP, Model F, Mooney S, Hale K, Lograsso J, Tonnes-Priddy L, Hoffmann J, Csepregi A, Röcken C, Molnar B, et al. Aristaless-like homeobox-4 gene methylation is a potential marker for colorectal adenocarcinomas. Gastroenterology. 2006;131:1418–1430. doi: 10.1053/j.gastro.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 77.Glöckner SC, Dhir M, Yi JM, McGarvey KE, Van Neste L, Louwagie J, Chan TA, Kleeberger W, de Bruïne AP, Smits KM, et al. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 2009;69:4691–4699. doi: 10.1158/0008-5472.CAN-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hibi K, Goto T, Kitamura YH, Yokomizo K, Sakuraba K, Shirahata A, Mizukami H, Saito M, Ishibashi K, Kigawa G, et al. Methylation of TFPI2 gene is frequently detected in advanced well-differentiated colorectal cancer. Anticancer Res. 2010;30:1205–1207. [PubMed] [Google Scholar]
- 79.Hibi K, Goto T, Shirahata A, Saito M, Kigawa G, Nemoto H, Sanada Y. Detection of TFPI2 methylation in the serum of colorectal cancer patients. Cancer Lett. 2011;311:96–100. doi: 10.1016/j.canlet.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 80.Takada H, Wakabayashi N, Dohi O, Yasui K, Sakakura C, Mitsufuji S, Taniwaki M, Yoshikawa T. Tissue factor pathway inhibitor 2 (TFPI2) is frequently silenced by aberrant promoter hypermethylation in gastric cancer. Cancer Genet Cytogenet. 2010;197:16–24. doi: 10.1016/j.cancergencyto.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 81.Oh T, Kim N, Moon Y, Kim MS, Hoehn BD, Park CH, Kim TS, Kim NK, Chung HC, An S. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn. 2013;15:498–507. doi: 10.1016/j.jmoldx.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 82.Nishio M, Sakakura C, Nagata T, Komiyama S, Miyashita A, Hamada T, Kuryu Y, Ikoma H, Kubota T, Kimura A, et al. RUNX3 promoter methylation in colorectal cancer: its relationship with microsatellite instability and its suitability as a novel serum tumor marker. Anticancer Res. 2010;30:2673–2682. [PubMed] [Google Scholar]
- 83.Tan SH, Ida H, Lau QC, Goh BC, Chieng WS, Loh M, Ito Y. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol Rep. 2007;18:1225–1230. [PubMed] [Google Scholar]
- 84.Herbst A, Rahmig K, Stieber P, Philipp A, Jung A, Ofner A, Crispin A, Neumann J, Lamerz R, Kolligs FT. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol. 2011;106:1110–1118. doi: 10.1038/ajg.2011.6. [DOI] [PubMed] [Google Scholar]
- 85.Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, Song X, Lesche R, Liebenberg V, Ebert M, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414–423. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- 86.deVos T, Tetzner R, Model F, Weiss G, Schuster M, Distler J, Steiger KV, Grützmann R, Pilarsky C, Habermann JK, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009;55:1337–1346. doi: 10.1373/clinchem.2008.115808. [DOI] [PubMed] [Google Scholar]
- 87.Leung WK, To KF, Man EP, Chan MW, Bai AH, Hui AJ, Chan FK, Sung JJ. Quantitative detection of promoter hypermethylation in multiple genes in the serum of patients with colorectal cancer. Am J Gastroenterol. 2005;100:2274–2279. doi: 10.1111/j.1572-0241.2005.50412.x. [DOI] [PubMed] [Google Scholar]
- 88.Wallner M, Herbst A, Behrens A, Crispin A, Stieber P, Göke B, Lamerz R, Kolligs FT. Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clin Cancer Res. 2006;12:7347–7352. doi: 10.1158/1078-0432.CCR-06-1264. [DOI] [PubMed] [Google Scholar]
- 89.Tang D, Liu J, Wang DR, Yu HF, Li YK, Zhang JQ. Diagnostic and prognostic value of the methylation status of secreted frizzled-related protein 2 in colorectal cancer. Clin Invest Med. 2011;34:E88–E95. doi: 10.25011/cim.v34i1.15105. [DOI] [PubMed] [Google Scholar]
- 90.Pack SC, Kim HR, Lim SW, Kim HY, Ko JY, Lee KS, Hwang D, Park SI, Kang H, Park SW, et al. Usefulness of plasma epigenetic changes of five major genes involved in the pathogenesis of colorectal cancer. Int J Colorectal Dis. 2013;28:139–147. doi: 10.1007/s00384-012-1566-8. [DOI] [PubMed] [Google Scholar]
- 91.Pedersen SK, Mitchell SM, Graham LD, McEvoy A, Thomas ML, Baker RT, Ross JP, Xu ZZ, Ho T, LaPointe LC, et al. CAHM, a long non-coding RNA gene hypermethylated in colorectal neoplasia. Epigenetics. 2014;9:1071–1082. doi: 10.4161/epi.29046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suzuki H, Yamamoto E, Maruyama R, Niinuma T, Kai M. Biological significance of the CpG island methylator phenotype. Biochem Biophys Res Commun. 2014;455:35–42. doi: 10.1016/j.bbrc.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 93.Potter NT, Hurban P, White MN, Whitlock KD, Lofton-Day CE, Tetzner R, Koenig T, Quigley NB, Weiss G. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60:1183–1191. doi: 10.1373/clinchem.2013.221044. [DOI] [PubMed] [Google Scholar]
- 94.Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Warton K, Lin V, Navin T, Armstrong NJ, Kaplan W, Ying K, Gloss B, Mangs H, Nair SS, Hacker NF, et al. Methylation-capture and Next-Generation Sequencing of free circulating DNA from human plasma. BMC Genomics. 2014;15:476. doi: 10.1186/1471-2164-15-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jung M, Klotzek S, Lewandowski M, Fleischhacker M, Jung K. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin Chem. 2003;49:1028–1029. doi: 10.1373/49.6.1028. [DOI] [PubMed] [Google Scholar]


