Abstract
In Candida albicans, drug resistance to clinically important antifungal drugs may be regulated through the action of transcription factors in a manner that may or may not be similar to regulation in Saccharomyces cerevisiae. A search of the C. albicans genome identified a single homolog of the S. cerevisiae transcription factor genes UPC2 (ScUPC2) and ECM22 (ScECM22) that have been associated with regulation of ergosterol biosynthesis. Sequence analysis of this C. albicans UPC2 (CaUPC2) gene identifies two domains, an anchoring transmembrane domain and a transcription factor region containing multiple nuclear localization signals and a fungal Zn(2)-Cys(6) binuclear cluster domain. Heterozygous deletion, homozygous deletion, and reconstructed strains of CaUPC2 as well as the parental strain were tested against several antifungal drugs, including ergosterol biosynthesis inhibitors. The CaUPC2 homozygous deletion strain showed marked hypersusceptibility to most drugs, compared to the parental and reconstructed strains. The deletion strains accumulate significantly less radiolabeled cholesterol, suggesting reduced ergosterol scavenging in those strains. When grown under azole drug pressure, the parental, heterozygous deletion and reconstructed strains of CaUPC2 upregulate the ERG2 and ERG11 ergosterol biosynthesis genes, while the homozygous deletion strain shows no such upregulation. Consistent with these results, CaUPC2 deletion strains show reduced ergosterol levels, which may explain the increased susceptibilities of the CaUPC2 deletion strains. Thus, it appears that CaUPC2 acts as a transcription factor involved in the regulation of ergosterol biosynthetic genes and as a regulator of sterol uptake across the plasma membrane.
Candida albicans is an opportunistic fungal pathogen that exists as a commensal in immunocompetent individuals. In persons with diminished immune function, such as the elderly or individuals with AIDS, C. albicans can cause oral candidiasis (16). In neutropenic populations, such as transplant and chemotherapy patients, C. albicans is the leading cause of bloodstream fungal infections (25). C. albicans infections are typically treated with azoles, which target the ergosterol biosynthetic pathway, or with polyenes that bind to ergosterol in the membrane. Resistance to the azole and polyene class drugs has been documented clinically (18). Azole resistance has been causally associated with mutations in the gene encoding the target of the azoles, ERG11, and upregulation of the efflux pump genes MDR1, CDR1 and CDR2 as well as ERG11 (28). It is not known whether the upregulation of these genes in C. albicans is due to alteration in their promoters, altered recruitment of transcriptional machinery due to changes in enhancers or repressors, increased mRNA stability, or an upregulation of relevant transcription factors.
Saccharomyces cerevisiae transcription factors that regulate the expression of genes homologous to the C. albicans drug resistance-associated genes have been identified (8, 15). The S. cerevisiae transcription factor gene ROX1 represses transcription of hypoxia-controlled genes, such as ERG11, in the presence of oxygen through recruitment of the Ssn6-Tup1 corepressor complex (10, 12). In the absence of ROX1 repression, ERG11 expression is upregulated and cells are up to 16-fold less susceptible to azole drugs (8).
ScUPC2 encodes a transcription factor originally characterized as a sterol regulatory gene, upregulating ERG2 and ERG3 in the ergosterol biosynthetic pathway in response to ergosterol starvation (27). ScUPC2 is a member of a family of fungal Zn(2)-Cys(6) binuclear cluster transcription factors that are highly conserved among fungal species but share no homology to the analogous mammalian sterol regulatory element binding proteins (SREBPs), which regulate cholesterol synthesis (27). ScUpc2p possesses a carboxy-terminal transmembrane domain that may act as a membrane anchor, keeping the protein in the cytoplasm. Under inducing conditions, the protein is internally cleaved, releasing an amino-terminal head group containing the transcription factor domain that translocates to the nucleus (27) (Fig. 1). Deletion of ScUPC2 alters susceptibility to lovastatin and amphotericin B (27). Deletion of ScUPC2 has been associated with lowered uptake of cholesterol across the plasma membrane (4)
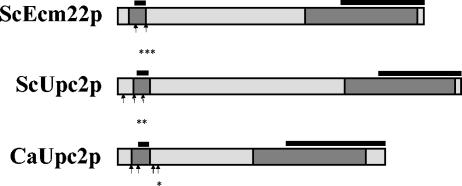
FIG. 1.
Alignment of C. albicans and S. cerevisiae proteins. Regions of significant homology are boxed in dark gray. The percent identity in the box near the amino terminus of CaUPC2 with both ScUPC2 and ScECM22 is 66%. The percent identity in the box near the carboxy terminus of CaUPC2 with both ScUPC2 and ScECM22 is 64%. The solid bars above the proteins near the amino termini designate Zn(2)-Cys(6) binuclear cluster transcription factor domains. The solid bars above the proteins near the carboxy termini designate transmembrane domains. Black arrowheads and asterisks identify nuclear localization signals (NLS). *, five NLS in this region and eight total NLS for CaUPC2. **, three NLS signals in this region and five total NLS for ScUPC2. ***, four NLS signals in this region and five total NLS for ScECM22.
ScECM22, a paralog of ScUPC2, was originally characterized in a screen for mutants hypersusceptible to the cell wall perturbing agent calcofluor white. It has been shown to regulate ERG2 and ERG3 in a manner similar to ScUPC2. ScECM22 is also a member of the fungal Zn(2)-Cys(6) binuclear cluster family and has similar transmembrane and nuclear localization domains (Fig. 1).
ScUPC2 and ScECM22 regulate expression of genes in the sterol biosynthetic pathway, binding to an 11-bp sterol response element (SRE) in the promoters of ERG2 and ERG3 in response to ergosterol starvation conditions, such as drug pressure (27).
Deletion of either gene in Saccharomyces is nonlethal. However, deletion of ScECM22 and ScUPC2 together has been reported to be viable (27) or nonviable (http://www.yeastgenome.org/), perhaps related to strain differences.
This study identifies and characterizes the Candida albicans UPC2 gene that is related to both ScUPC2 and ScECM22. A set of CaUPC2 deletion and reconstructed strains were used to analyze the effect of the gene on drug susceptibility, sterol accumulation, ergosterol levels, and ergosterol biosynthesis gene expression.
MATERIALS AND METHODS
Strains.
The Candida albicans strains used in this study are BWP17 (ura3Δ::λ434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG) and its derivatives S-7, D-6, EC-7, and EC-2 (see Table 1 for UPC2 genotype). Strains S-7 and D-6 were constructed through successive transformation of BWP17 with a PCR product containing 50-mer regions of homology to the UPC2 locus flanking either the URA3 or ARG4 marker genes as described previously (29). Strains EC-7 and EC-2 were constructed through transformation of the D-6 strain with a fragment containing the UPC2 locus and the HIS1 selection marker and subsequent integration at either allele.
TABLE 1.
C. albicans strains used in this study
| Strain | Identification no. | UPC2 genotype | Source or reference |
|---|---|---|---|
| BWP17 | TW14901 | UPC2/UPC2 | Wilson et al. (29) |
| S-7 | TW14902 | UPC2/upc2::URA3 | This study |
| D-6 | TW14903 | upc2::URA3/upc2::ARG4 | This study |
| EC-7 | TW14904 | upc2::URA3/upc2::UPC2-HIS1 | This study |
| EC-2 | TW14905 | upc2::ARG4/upc2::UPC2-HIS1 | This study |
Media.
Yeast extract-peptone-dextrose (YEPD) medium contains 20 g of dextrose, 20 g of Bacto peptone, and 10 g of Difco yeast extract per liter. It was supplemented with 20 mg of uridine/ml, 50 mg of arginine/ml, and 20 mg of histidine/ml for nonselective propagation of C. albicans strains. Selective propagation of C. albicans strains was done in YAD-CSM defined medium containing 1.77 g of yeast nitrogen base without amino acids or ammonium sulfate, 5 g of ammonium sulfate, 20 g of dextrose, and the appropriate CSM medium (Qbiogene, Carlsbad, Calif.) lacking the marker amino acids. The lipophilic dye medium consisted of YEPD medium supplemented with 7 mg of phloxine B/ml or 200 μg of Congo red/ml. Plates also contain 10 g of agar/liter.
Plasmids.
Plasmids pGEM-URA3 and pRS-ARG4ΔSpe1 (29) were used for construction of C. albicans strains S-7 and D-6. These plasmids contain a selectable marker, URA3 or ARG4, flanked by a polyclonal region. Oligonucleotides UPC2 5′ KO (5′ CTTTTTTCTTTCCCTTTTTCTTTAATTTATTACTACATCAAGTTTATTTATTTCCCAGTCACGACGTT 3′) and UPC2 3′ KO (5′ GCGAAACACCTTCTAAAAACCATACTTTGTCTTTCATTTCGGTTGTAAACTGTGGAATTGTGAGCGGATA 3′), with 50 bp of homology to the UPC2 locus linked to 18-mers with homology to the polyclonal region, were used to amplify marker genes from pGEM-URA3 or pRS-ARG4ΔSpe1 as described previously (29). Plasmid pTU-1 was constructed by ligation of a PCR fragment containing the entire UPC2 locus (797 bp upstream of the ATG to 291 bp downstream of the stop codon, amplified by PCR using oligonucleotides UPC2 upstream (5′ ATGGATGTTGGTATATCAGG 3′) and UPC2 downstream (5′ AACTGTATCTTTTTCCCACT 3′) into the pCR-Topo vector. pTU-1 was then digested with BamHI and ApaI, and the fragment containing the UPC2 locus was ligated to pGEM-HIS1 at the ApaI and BamHI sites. This plasmid, containing both the UPC2 locus and the HIS1 marker gene, was designated pHU-1.
C. albicans transformation.
Competent C. albicans cells were electroporated according to previously described methods (6, 26). One to five micrograms of a PCR product was used in each transformation. Electroporation was done at 1.6 kV, 25 μF, and 200 Ω in a gene pulser (Bio-Rad, Hercules, Calif.). After electroporation, 1 ml of 1 M sorbitol was added to the cells and they were plated immediately on selective medium. Cells were incubated at 30°C for 3 to 5 days. Single colonies were picked and passaged three times to ensure a homogenous population.
DNA and protein analysis.
Protein analysis was performed by using the PSORTII program (http://psort.nibb.ac.jp/form2.html). Identification of potential promoter SREs was accomplished by using the search pattern function available at http://genolist.pasteur.fr/CandidaDB/genome.cgi. The patterns searched were TCGTATA and TATACGA, the seven essential base pairs identified in S. cerevisiae (27).
Southern blot analysis.
Genomic DNA of each transformant was prepared by glass bead lysis as described previously (9). DNA was digested with BglII, separated in a 0.8% agarose gel, blotted, and hybridized with an end-labeled UPC2 5′ KO oligonucleotide as described previously (3, 13, 23).
Northern analysis.
Total RNA was prepared by using a glass bead phenol extraction as described previously (3, 13). Two 25-ml cultures were inoculated for each strain from overnight cultures to a starting optical density (OD) of 0.2 and grown for 4 to 8 h to an OD of between 0.8 and 1.0. One culture from each strain was grown in the presence of 5 μg of fluconazole/ml. RNA was blotted as described previously (13, 20) and probed with an end-labeled oligonucleotide, UPC2-AS (5′ GCTGGAAATGCAAATATACGTAAAATAAAGTCAGACTTGTATCTCTCCTT 3′), ERG2-AS (5′ GACCTTCAGTCCCAATTGCTGTACCGAAAAAAATCAAATATTCAGAAATT 3′), or ERG11-Coding (5′ CTAAGGGACAAAAAATAATTAATGCCATCAATGACAGTTTCAACAATAGCCATGATTGATAATTATTTGA 3′), with homology to the gene of interest. mRNA signals were quantified by using a Storm Phosphorimager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). All Northern signals were normalized to mRNA from the housekeeping gene ACT1 (5′ ATTTGAGTCATCTTTTCTCTGTTGGATTTTGGATTCATTGGAGCTTCGGT 3′), which allows for comparison of specific mRNA levels in reference to total mRNA.
MIC analysis.
Drug susceptibility was determined by using the standard NCCLS broth microdilution protocol that determines the MIC of drug needed to inhibit 80% of cell growth (14). Strains were grown in a 96-well plate containing a gradient of drug, typically eightfold above the MIC to eightfold below the MIC, by using serial twofold dilutions of drug. Etest strips (AB Biodisk, N.J.), plastic strips impregnated with gradients of drug, were used per the manufacturer's instructions. Each strip is placed onto an agar plate containing a lawn of cells, and growth inhibition is measured directly off the plate after 24 or 48 h.
Aerobic cholesterol accumulation.
An assay of total cholesterol accumulation with radiolabeled [14C]cholesterol (58.0 mCi/mmol) was performed as described previously (22) with the following modifications. Strains were grown in 5 ml of defined RPMI 1640 medium (Sigma-Aldrich, St. Louis, Mo.) supplemented with 50 mg of arginine/ml, 20 mg of histidine/ml, and 20 mg of uridine/ml in a 50-ml conical tube for 48 h. The media contained 14C-labeled cholesterol (5 nCi/ml). After 48 h of growth, the cells were pelleted, washed, and lyophilized overnight. Pellet weight was measured, each pellet was resuspended in 1 ml of scintillation fluid, and counts per minute were determined.
Ergosterol levels.
Ergosterol levels were measured by using a protocol described previously (2) with the following modifications. All strains were grown in YAD-CSM medium supplemented with 20 mg of uridine/ml, 50 mg of arginine/ml, and 20 mg of histidine/ml. Strains were grown for 48 h, and 100 ODs of cells at 600 nm [concentration of cells (OD/ml) × volume (ml) = 100 OD] were used in a heptane extraction of ergosterol. Ergosterol levels were determined on a spectrophotometer scanning from 230 to 310 nm.
RESULTS
C. albicans UPC2 homolog.
A search of the C. albicans genome database maintained at Stanford University was performed by using the ScUPC2 and ScECM22 sequences. Both ScUPC2, a gene associated with uptake of ergosterol across the plasma membrane and regulation of ERG2, and ScECM22, a gene associated with increased susceptibility to the chitin-disrupting drug calcofluor white, have homology (e−112 and 2e−87, respectively) to a single open reading frame (ORF) in C. albicans, ORF 19.391 (with allele ORF 19.8021) (http://www-sequence.stanford.edu/group/candida). The C. albicans ORF will be referred to subsequently as UPC2 (CaUPC2 for clarity in this publication).
The amino acid homology between these proteins is highly conserved within two regions, with 66% identity in the amino-terminal transcription factor domain and 64% identity in the carboxy-terminal transmembrane domain (Fig. 1). The amino head region of these proteins contains a highly conserved Zn(2)-Cys(6) DNA binding domain unique to fungi, with over 80 proteins described in this family (22, 27). These zinc finger domains are flanked by multiple nuclear localization signals in all three proteins. The carboxy-terminal region of these proteins contains a transmembrane domain proposed to act as a cytoplasmic anchor for the ScUPC2 protein, allowing the transcription factor domain to translocate to the nucleus only after cleavage (http://psort.nibb.ac.jp/form2.html) (22).
PCR-based disruption of C. albicans genes.
The first step in the analysis of the C. albicans UPC2 gene was a deletion of both alleles. BWP17, a strain with a triple auxotrophy (lacking Uri, Arg, and His), was used for the removal of two alleles and later complementation. The heterozygous deletion strain S-7 (UPC2/upc2::URA3) was generated through homologous recombination with a disruption amplicon containing the URA3 gene flanked by 50-mer regions of homology to the UPC2 gene. Colonies grew on plates lacking Uri and were confirmed by Southern blotting to have a UPC2 deletion (Fig. 2). The homozygous deletion strain D-6 (upc2::URA3/upc2::ARG4) was constructed through transformation of S-7 with an amplicon containing the ARG4 gene flanked by the same 50-mer regions of homology. Colonies grew on plates lacking Uri and Arg and were confirmed by Southern blotting to contain a deletion of the second allele. The genotypes of these strains are summarized in Table 1. To restore the gene, the entire UPC2 locus (797 bp upstream of the ATG to 291 bp downstream of the stop codon) was cloned into the pGEM-HIS1 vector upstream of the HIS1 gene at the BamHI and ApaI sites and confirmed by sequencing. A PCR amplification of the UPC2-HIS1 region was then used to transform the homozygous deletion strain D-6. Colonies grew on the medium lacking His, replacing the allele containing the ARG4 gene and the allele containing the URA3 gene at equal rates as measured by Southern blotting and growth on selective media. Figure 2 shows a Southern blot of genomic DNA from the strain constructs, illustrating the deletion of two UPC2 alleles and their reconstruction at the UPC2 locus.
FIG. 2.
Southern blot analysis of CaUPC2 strains. DNA from UPC2 deletion strains and the parental was digested with BglII, run on an agarose gel, blotted onto a nitrocellulose membrane, and then probed with the radiolabeled UPC2 5′ KO oligonucleotide (see Materials and Methods). The blot shows the replacement of a UPC2 allele with URA3 in strain S-7 and then the replacement of the remaining allele with ARG4 in strain D-6, confirming the disruption of both UPC2 alleles. Strains EC-7 and EC-2 are single-allele reconstructions replacing either the URA3 or ARG4 locus with a UPC2-HIS1 fragment. The additional lower band in the EC-7 lane is the result of the plasmid constructs.
Increased susceptibility in UPC2 deletion strains.
Because the ScECM22 gene was initially identified in a screen for increased susceptibility to calcofluor white (1), and because the ScUPC2 deletion has been associated with altered sterol accumulation and regulation of sterol biosynthesis genes (27), it was of interest to test the C. albicans UPC2 deletions against a panel of antifungal drugs (Table 2). For the azole drugs ketoconazole, itraconazole, and fluconazole, which target the product of the ERG11 gene, there was a marked increase in the susceptibility of the UPC2 deletion strains (8-, 8-, and 11-fold, respectively). The UPC2 deletion strains were also tested against drugs that act on ergosterol biosynthesis at sites upstream and downstream of ERG11, including terbinafine, which targets the product of the ERG1 gene (squalene epoxidase), fenpropimorph, which targets the product of the ERG2 gene (C-8 sterol isomerase), and lovastatin, which targets the product of the HMG1 gene (HMG-CoA reductase) and is used clinically as an anticholesterol drug. For each of these drugs, an increase in susceptibility was observed (800-, 25-, and 16-fold, respectively). Drugs that act to disrupt the synthesis or deposition of chitin in the cell wall, such as nikkomycin Z and calcofluor white, were also tested with the UPC2 deletions strains. The strains showed an increased susceptibility to the drugs (21- and 32-fold). As shown in Table 2, the reconstructed strains (EC-7 and EC-2) consistently returned drug susceptibility to that of the single allele or wild-type (WT) level.
TABLE 2.
Susceptibility testing of UPC2 disruption strainsa
| Drug | Drug target | MIC (μg/ml)
|
Fold changeb | ||||
|---|---|---|---|---|---|---|---|
| BWP17 | S-7 | D-6 | EC-7 | EC-2 | |||
| Fluconazole | Erg11p | 2 | 1 | 0.19 | 1.5 | 2 | 11 |
| Itraconazole | Erg11p | 0.016 | 0.008 | 0.002 | 0.032 | 0.016 | 8 |
| Ketoconazole | Erg11p | 0.016 | 0.008 | 0.002 | 0.016 | 0.012 | 8 |
| Terbinafine | Erg1p | 100 | 1 | 0.125 | 100 | 12.5 | 800 |
| Fenpropimorph | Erg2p | 6.3 | 6.3 | 0.25 | 6.25 | 12.5 | 25 |
| Lovastatin | Hmg1p | 16 | 16 | 1 | 16 | 16 | 16 |
| Nikkomycin Z | Chitin synthetase | 125 | 3.9 | 3.9 | 125 | 125 | 32 |
| Calcofluor white | Chitin deposition | 64 | 15.6 | 3.1 | 64 | 128 | 21 |
| Amphotericin B | Ergosterol | 0.19 | 0.19 | 0.19 | 0.25 | 0.19 | 1 |
| Cycloheximide | LSU rRNA | 250 | 250 | 250 | 250 | 250 | 1 |
| DTT | Reducing agent | 0.5 | 1 | 1 | 1 | 1 | 1 |
| SDS | Plasma membrane | 0.025 | 0.025 | 0.013 | 0.025 | 0.025 | 2 |
All strains were grown in RPMI 1640 medium supplemented with arginine, histidine, and uridine for 48 h. Data reflect the MIC80 cutoff. MICs for itraconazole and amphotericin B were determined by Etest. Other MICs were determined by the standard NCCLS microdilution protocol.
Fold change = WT/D6 (homozygous deletion strain).
Altered drug susceptibility for CaUPC2 deletion strains to all drugs was not observed. Amphotericin B, which binds directly to ergosterol in the plasma membrane, resulting in cell lysis, showed no change in susceptibility between BWP17 and the UPC2 deletion strains. Cycloheximide, a potent inhibitor of protein synthesis, also showed no difference in its effect on parental and UPC2 deletion strains of C. albicans. For several compounds (e.g., sodium dodecyl sulfate [SDS] and dithiothreitol [DTT]) known to have a general effect on cell wall structure, there was no change in susceptibility between BWP17 and the UPC2 deletion strains. Similarly, no change in growth was observed for cells grown at high salt concentrations (2.5 M NaCl) or under osmotic stress (2 M sorbitol) (data not shown).
Ergosterol levels are lower in UPC2 deletion strains.
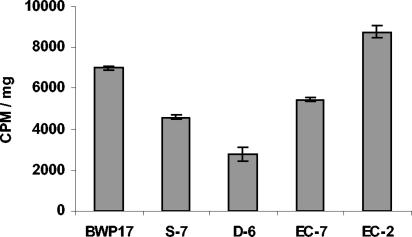
In S. cerevisiae, the UPC2 gene has been associated with regulation of sterol uptake across the plasma membrane. To investigate whether the C. albicans gene was behaving in a similar manner, accumulation of [14C]cholesterol was measured (Fig. 3). Each strain was incubated in radioactive cholesterol for 48 h under aerobic conditions, and total accumulation of [14C]cholesterol was measured. The WT strain BWP17 and one reconstructed strain, EC-2, exhibited comparable cholesterol accumulation. The heterozygous deletion, S-7, and the reconstructed strain EC-7 had similar intermediate levels of cholesterol accumulation. The homozygous deletion strain, D-6, exhibited the lowest level of [14C]cholesterol accumulation.
FIG. 3.
Analysis of [41C]cholesterol uptake in CaUPC2 deletion strains. All strains were grown aerobically in the presence of 14C-labeled cholesterol for 48 h, washed, and lyophilized. Pellets were then counted and normalized to weight. Values on the y axis are counts per minute (CPM)/mg.
C. albicans UPC2 deletion strains also demonstrate an increased susceptibility to ergosterol biosynthetic pathway inhibitors as well as to drugs acting at the cell wall (Table 2). To investigate whether this susceptibility correlates with lower levels of ergosterol in the plasma membrane, an ergosterol extraction where ergosterol was measured as an absorption spectra between 230 nm and 310 nm was performed (Fig. 4). The parental BWP17 strain had the highest level of ergosterol, the heterozygous deletion had an intermediate level of ergosterol, and the homozygous deletion strain had the lowest level of ergosterol. The reconstructed strains, EC-2 (Fig. 4) and EC-7 (data not shown), had ergosterol levels comparable to that of the heterozygous disruption strain, S-7.
FIG. 4.
Analysis of ergosterol in CaUPC2 deletion strains. Wild-type and UPC2 deletion strains were grown in YAD-CSM medium supplemented with 20 mg of uridine/ml, 50 mg of arginine/ml, and 20 mg of histidine/ml for 48 h, followed by a heptane extraction and spectrophotometric scan to detect ergosterol levels. Extracts were scanned from 230 to 310 nm, and the data were plotted. Strains are represented as BWP17 (open diamonds), UPC2 heterozygous deletion strain S-7 (open squares), UPC2 homozygous deletion strain D-6 (open triangles), and reconstructed strain EC-2 (open circles). The EC-7 strain had levels equivalent to S-7 and EC-2 but was omitted for clarity (data not shown). Abs., absorption.
To determine if the UPC2 gene deletion in C. albicans alters accumulation of ergosterol specifically or lipophilic molecules in general, strains were grown on medium containing two lipophilic dyes, Congo red and phloxine B (Fig. 5) (5). On each agar plate, the level of dye accumulation was highest for BWP17. Dye accumulation was severely reduced in the deletion strains, especially in the homozygous deletion strain. Dye accumulation was restored to heterozygous deletion levels in the reconstructed strain EC-7, while the reconstructed strain EC-2 restored dye uptake to wild-type levels. Dye accumulation was determined visually by colony color.
FIG. 5.
Lipophilic dye accumulation in CaUPC2 deletion strains. All strains were grown on YEPD plates containing the lipophilic dyes Congo red (A) or phloxine B (B) for 72 h at 30°C and photographed.
Ergosterol biosynthesis genes are unable to respond to drug pressure in UPC2 deletion strains.
The ScUPC2 and ScECM22 genes have been shown to regulate transcription of the ERG2 and ERG3 genes in the ergosterol biosynthetic pathway in response to cellular ergosterol levels (27). In order to investigate the effect of a CaUPC2 deletion on the expression of genes in the ergosterol biosynthetic pathway, strains were grown under drug pressure and the transcription of the ERG2, ERG11, and UPC2 genes was measured (Fig. 6A). Parental and UPC2 deletion strains were grown in culture without drug and in the presence of 5 μg of fluconazole/ml. All strains showed similar baseline levels of expression for the ERG2 and ERG11 genes in the absence of drug. All strains except the UPC2 homozygous deletion strain, D-6, showed an increase in the expression of ERG2 and ERG11 when grown in the presence of drug (up to 16-fold compared to strain D-6) (Fig. 6B). As expected, the UPC2 deletion strain, D-6, did not show any expression of the UPC2 gene when grown with or without drug. All other strains showed background levels of expression of UPC2 in the absence of fluconazole and an increase in expression when exposed to 5 μg of fluconazole/ml, similar to the ERG2 and ERG11 genes (Fig. 6A). As UPC2 expression levels are minimal in the absence of drug, quantification of induction is difficult for UPC2 (data not shown).
FIG. 6.
(A) Northern blot analysis of UPC2 strains. BWP17 and UPC2 deletions strains were grown in minimal medium in the absence or presence of 5 μg of fluconazole/ml for 6 h to an OD of 0.8 to 1.0. Total RNA was harvested, run on an agarose gel, blotted onto a nitrocellulose membrane, and then probed with radiolabeled oligonucleotides specific for each gene (see Materials and Methods). A probe of the ACT1 housekeeping gene served as the loading control for each blot; all strains had equivalent levels of ACT1 mRNA (data not shown). (B) Quantification of Northern blot results. mRNA levels for BWP17 and UPC2 deletion strains were measured by using a Storm Phosphorimager. After normalizing each band to its ACT1 loading control, a comparison of the relative intensities between strains grown with and without drug showed a 6- to 16-fold increase in the expression of ERG2 (light bars) and ERG11 (dark bars) in the presence of 5 μg of fluconazole/ml. UPC2 expression changes could not be measured, as signal in the no-drug lanes was not significantly above background.
Research on S. cerevisiae has identified an 11-bp sterol response element (5′ CTCGTATAAGC 3′) upstream of many ergosterol biosynthesis genes that acts as a binding site for ScUPC2 and ScECM22 (27). A core of 7 of the 11 bp (TCGTATA) appears to be essential for binding in S. cerevisiae. A search of promoters of C. albicans ergosterol biosynthetic genes revealed sequences identical to the S. cerevisiae SRE core upstream of many genes, including ERG1, ERG2, and ERG11 (Table 3). This SRE core was not found in the promoters of genes not associated with ergosterol biosynthesis, such as the housekeeping genes ACT1 or ENO1.
TABLE 3.
Genes with SREs in their promotersa
| Gene | Putative SRE | Coordinates |
|---|---|---|
| POT14 | TCGTCGTATAGAA | −267 to −255 |
| ERG13 | TTCTATACGACGA | −427 to −415 |
| TCGTCGTATAGAT | −322 to −310 | |
| HMG1 | CTGTCGTATAAAT | −1449 to −1437 |
| IDII | TATTATACGATAT | −11 to +1 |
| ERG1 | TCATCGTATATTT | −376 to −364 |
| ERG11 | ATGTCGTATATTC | −235 to −223 |
| ATCTATACGACGA | −489 to −477 | |
| ERG24 | CGTTATACGACCG | −142 to −130 |
| ERG25 | AGTTATACGACCA | −432 to −420 |
| ERG251b | AGCTATACGATGG | −502 to −490 |
| ATCTATACGATCA | −122 to −110 | |
| ERG26 | CTCTATACGACAA | −168 to −156 |
| ERG27 | GTTTATACGATCG | −154 to −142 |
| ERG6 | TTCTCGTATAGAA | −558 to −546 |
| ERG2 | TTGTCGTATAAAA | −220 to −208 |
| ERG5 | AATTATACGACCA | −580 to −568 |
This table shows the location of putative SREs in the promoters of ergosterol biosynthesis genes. The critical bases (in S. cerevisiae) TCGTATA are shown in bold face type. Some sequences are on the complementary strand. Two thousand base pairs upstream of the ATG start codon were searched for SREs. Genes are given in the order they occur in the ergosterol biosynthetic pathway. Coordinates are relative to the ATG start codon.
ERG251 appears to be an allele or second copy of the ERG25 gene.
DISCUSSION
The effect of a UPC2 deletion in Candida albicans has been investigated. Deletion strains exhibited a variety of phenotypes, including increased drug susceptibility; altered gene expression for ergosterol biosynthesis genes; an altered ergosterol profile; and altered accumulation of lipophilic molecules, including cholesterol.
CaUPC2 is closely related to the paralogs ScUPC2 and ScECM22 with protein homology BLAST scores of e−112 and 2e−87, respectively. There is a single copy of the diploid CaUPC2 gene (Fig. 1) that corresponds to both ScUPC2 and ScECM22 genes; this correspondence may be due to the genome duplication hypothesized to have occurred in S. cerevisiae (11) and later genetic drift. These C. albicans and S. cerevisiae genes are all members of a highly conserved fungus-specific family of transcription factors with a Zn(2)-Cys(6) binuclear cluster domain. This family, with over 80 members (many of which being transcriptional regulators), appears to be responsible for regulation of ergosterol synthesis in yeasts but bears no sequence homology to the analogous family of transcriptional regulators in humans, the SREBPs (21). Mammalian SREBPs are also highly conserved and have been shown to anchor in cytoplasmic membranes. Subsequent proteolytic cleavage allows a DNA-binding domain to enter the nucleus and regulate sterol biosynthesis genes (17, 19, 21). Analysis of the ScUPC2 and ScECM22 genes suggests a membrane anchorage and regulation of sterol biosynthesis genes, specifically ERG2 and ERG3 (27). ScUPC2 binds to a specific sequence (5′ CTCGTATAAGC 3′) that has been identified upstream of many genes in the ergosterol biosynthetic pathway (27). The protein homology between the CaUPC2 gene and the ScUPC2 gene suggests that CaUPC2 may play a similar regulatory role in C. albicans (Fig. 1).
Deletion of the CaUPC2 gene results in increased susceptibility to a variety of antifungal drugs, including many in clinical use (Table 2). This increased susceptibility is seen with drugs targeting the ergosterol biosynthesis pathway (azoles, terbinafine, and fenpropimorph) and may reflect the inability of the homozygous deletion strain to upregulate expression of ergosterol biosynthesis genes, such as ERG2 and ERG11, in the presence of ergosterol starvation conditions (for instance, those associated with drug exposure) (Fig. 6) (23, 24). Increased susceptibility is also noted for drugs that act to disrupt the cell wall, such as calcofluor white, a chitin deposition inhibitor (1), and nikkomycin Z, an antifungal drug that competitively inhibits chitin synthetase in the fungal cell (7). This increased susceptibility is unlikely to be related to the weakened cell wall these drugs produce; it is more likely to be related to a weakened plasma membrane, which may lyse in the presence of an unstable cell wall. Compounds that stress the cell wall, such as SDS and DTT, showed no difference in susceptibilities for the UPC2 deletion strains; this finding may be due to a threshold effect where moderate plasma membrane differences are undetectable when strains are challenged with compounds that have severe effects on cell walls. Similarly, no alteration in susceptibilities was observed with other stress conditions, such as high salt concentrations, osmotic stress, and heat shock (data not shown). CaUPC2 deletion strains did not show altered hyphal development compared to the BWP17 parental strain (data not shown).
The MICs of both amphotericin B, which targets ergosterol in the plasma membrane, and cycloheximide, which interferes with the large subunit rRNA, were unchanged in the UPC2 deletion strains. When exposed to amphotericin B, a potent cellular inhibitor, altered ergosterol levels associated with UPC2 deletion may be below the threshold of detection (there may be enough ergosterol in the membrane of all UPC2 strains to facilitate cell death). Cycloheximide, a potent inhibitor of protein synthesis, may be unaffected by the potential plasma membrane changes related to UPC2 deletion and retain activity against the basic cell process in all strains equally.
The accumulation of radiolabeled [14C]cholesterol by CaUPC2 deletion strains was reduced compared to the parental strain (Fig. 3). Additionally, the accumulation of other lipophilic molecules, such as the dyes phloxine B and Congo red, was reduced in the CaUPC2 deletion strains (Fig. 5). These data suggest that CaUPC2 is involved in the uptake and/or retention of lipophilic molecules. This may be related to membrane changes associated with lowered ergosterol levels. Such changes may alter the accumulation of lipophilic molecules, such as sterols, perhaps through an affected pump. The upc2-1 strain of S. cerevisiae shows increased uptake of radiolabeled cholesterol under aerobic conditions, related to a point mutation in ScUPC2 (4). The amino acid change in S. cerevisiae that allows aerobic sterol uptake is present in C. albicans, and its modification might result in increased sterol uptake. We are currently testing for a similar aerobic sterol exclusion phenotype in C. albicans.
Ergosterol levels in the UPC2 deletion strains were lower than in the parental strain (Fig. 4). C. albicans strains with lower endogenous levels of ergosterol, such as the UPC2 deletion strain, may be more susceptible to antifungal drugs due to a smaller ergosterol reservoir. C. albicans strains grown in the presence of static antifungal drugs, such as fluconazole, grow for several hours before the available ergosterol is depleted and growth is inhibited (23). In strains with a lower starting ergosterol reservoir, drug inhibition of cell growth would happen more rapidly.
The two reconstructed strains, EC-7 and EC-2, have a UPC2-HIS1 replacement of either the upc2::URA3 or upc2::ARG4 allele and therefore have a single copy of UPC2 driven by an endogenous promoter. The dissimilarity seen between the two reconstructed strains in lipophilic molecule accumulation (Fig. 3 and 5) may be the result of different promoter activities of the two endogenous alleles.
ScUPC2 has been shown to act as a regulator of the ERG2 and ERG3 genes in the Saccharomyces ergosterol biosynthetic pathway, upregulating expression in the presence of lovastatin. CaUPC2 appears to have a similar effect, upregulating the expression of ERG2 and ERG11 in the presence of ergosterol starvation conditions, in this case fluconazole drug pressure (Fig. 6). The homozygous deletion strain of CaUPC2 D-6 has lost the ability to upregulate the expression of these genes under drug pressure. No difference was observed in expression levels of ERG2 and ERG11 in the absence of ergosterol starvation (no drug pressure), indicating that CaUPC2 has no effect on the basal transcription of these genes. Thus, CaUPC2 does appear to be involved in upregulation in response to sterol stress as determined by fluconazole pressure (Fig. 6).
A decrease in lipophilic molecule accumulation (Fig. 3 and 5) may limit the ability of the cell to scavenge extracellular ergosterol and would exacerbate the need for sterol biosynthesis in the homozygous deletion strain D-6. The need for sterol synthesis results in the upregulation of the ERG2 and ERG11 genes under ergosterol starvation conditions. Thus, the homozygous deletion strain may be less able to produce sufficient endogenous ergosterol (Fig. 6) while concurrently lacking the ability to import exogenous sterols (Fig. 3 and 5).
The identification of potential CaUPC2 SRE sites in the promoters of the ERG2 and ERG11 genes (Table 3) suggests that the CaUPC2 regulation of those genes (Fig. 6) may be facilitated through these SREs, as previously shown with Saccharomyces (27). In addition, the presence of SREs in the promoters of 4 out of the 9 early ergosterol biosynthesis genes (acetate to squalene) and 9 of the 12 late ergosterol biosynthesis genes (squalene to ergosterol) suggests that CaUPC2 may be involved in regulation of this pathway at many stages (Table 3).
These data show that the CaUPC2 gene is an important regulator of sterol biosynthesis, with activities similar to both ScUPC2 and ScECM22. CaUPC2 is a member of the fungal equivalent of the mammalian SREBP, sharing functional similarity but no sequence similarity. This finding provides an opportunity for drug development as well as new insights into sterol biosynthesis regulation. The increased drug susceptibilities of CaUPC2 deletions strains coupled with the decreased sterol uptake data suggest that further investigation of this gene may reveal novel drug targets. Future research will investigate other genes regulated by CaUPC2, the mechanism of CaUPC2 regulation, and the cellular changes associated with CaUPC2 deletion and overexpression.
Acknowledgments
We thank Aaron Mitchell (Columbia University, New York, N.Y.) for providing us with the BWP17 strain and pGEM and pRS plasmids. We thank members of the White laboratory for their support and valuable comments on the manuscript.
This research was funded by NIH NIDCR grants R01 DE11367 and R01 DE14161 and the M.J. Murdock Charitable Trust. T.C.W. was the recipient of a New Investigator Award in Molecular Pathogenic Mycology from the Burroughs Wellcome Fund. B.G.O. was supported by NIH Pathobiology training grant T32 AI 07509.
REFERENCES
- 1.Akache, B., K. Wu, and B. Turcotte. 2001. Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res. 29:2181-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthington Skaggs, B. A., H. Jradi, T. Desai, and C. J. Morrison. 1999. Quantitation of ergosterol content: novel method for determination of fluconazole susceptibility of Candida albicans. J. Clin. Microbiol. 37:3332-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Crowley, J. H., F. W. Leak, Jr., K. V. Shianna, S. Tove, and L. W. Parks. 1998. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol. 180:4177-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cserhati, T., and G. Oros. 2000. Relationship between the lipophilicity and specific hydrophobic surface area of non-homologous series of synthetic dyes. Croat. Chemica Acta 73:293-303. [Google Scholar]
- 6.De Backer, M. D., D. Maes, S. Vandoninck, M. Logghe, R. Contreras, and W. Luyten. 1999. Transformation of Candida albicans by electroporation. Yeast 15:1609-1618. [DOI] [PubMed] [Google Scholar]
- 7.Ernst, E. J. 2001. Investigational antifungal agents. Pharmacotherapy 21:165S-174S. [DOI] [PubMed] [Google Scholar]
- 8.Henry, K. W., J. T. Nickels, and T. D. Edlind. 2002. ROX1 and ERG regulation in Saccharomyces cerevisiae: implications for antifungal susceptibility. Eukaryot. Cell 1:1041-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 10.Kwast, K. E., L. C. Lai, N. Menda, D. T. James III, S. Aref, and P. V. Burke. 2002. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184:250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lafontaine, I., G. Fischer, E. Talla, and B. Dujon. 2004. Gene relics in the genome of the yeast Saccharomyces cerevisiae. Gene 335:1-17. [DOI] [PubMed] [Google Scholar]
- 12.Lowry, C. V., and R. S. Zitomer. 1988. ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4651-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 14.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard. NCCLS document M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 15.Nguyen, D. T., A. M. Alarco, and M. Raymond. 2001. Multiple Yap1p-binding sites mediate induction of the yeast major facilitator FLR1 gene in response to drugs, oxidants, and alkylating agents. J. Biol. Chem. 276:1138-1145. [DOI] [PubMed] [Google Scholar]
- 16.Odds, F. C. 1988. Candida and candidosis: a review and bibliography. Baillière Tindall, London, United Kingdom.
- 17.Rawson, R. B. 2003. The SREBP pathway—insights from Insigs and insects. Nat. Rev. Mol. Cell Biol. 4:631-640. [DOI] [PubMed] [Google Scholar]
- 18.Rex, J. H., M. G. Rinaldi, and M. A. Pfaller. 1995. Resistance of Candida species to fluconazole. Antimicrob. Agents Chemother. 39:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai, J., and R. B. Rawson. 2001. The sterol regulatory element-binding protein pathway: control of lipid homeostasis through regulated intracellular transport. Curr. Opin. Lipidol. 12:261-266. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Sato, R., J. Yang, X. Wang, M. J. Evans, Y. K. Ho, J. L. Goldstein, and M. S. Brown. 1994. Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1). J. Biol. Chem. 269:17267-17273. [PubMed] [Google Scholar]
- 22.Shianna, K. V., W. D. Dotson, S. Tove, and L. W. Parks. 2001. Identification of a UPC2 homolog in Saccharomyces cerevisiae and its involvement in aerobic sterol uptake. J. Bacteriol. 183:830-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song, J. L., J. B. Harry, R. T. Eastman, B. G. Oliver, and T. C. White. 2004. The Candida albicans lanosterol 14-α-demethylase (ERG11) gene promoter is maximally induced after prolonged growth with antifungal drugs. Antimicrob. Agents Chemother. 48:1136-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song, J. L., J. B. Harry, R. G. Eastman, B. G. Oliver, and T. C. White. 2004. Promoter analysis of Candida albicans lanosterol 14-alpha-demethylase (ERG11) gene promoter is maximally induced after prolonged growth with antifungal drugs. Antimicrob. Agents Chemother. 48:1136-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swerdloff, J. N., S. G. Filler, and J. E. Edwards, Jr. 1993. Severe candidal infections in neutropenic patients. Clin. Infect. Dis. 2:S457-S467. [DOI] [PubMed] [Google Scholar]
- 26.Thompson, J. R., E. Register, J. Curotto, M. Kurtz, and R. Kelly. 1998. An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast 14:565-571. [DOI] [PubMed] [Google Scholar]
- 27.Vik, A., and J. Rine. 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6395-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]