Abstract
The polysaccharide capsule is the main virulence factor of the basidiomycetous yeast Cryptococcus neoformans. Four genes (CAP10, CAP59, CAP60, and CAP64) essential for capsule formation have been previously identified, although their roles in the biosynthetic pathway remain unclear. A genetic and bioinformatics approach allowed the identification of six CAP64-homologous genes, named CAS3, CAS31, CAS32, CAS33, CAS34, and CAS35, in the C. neoformans genome. This gene family is apparently specific in a subclass of the basidiomycete fungi. Single as well as double deletions of these genes in all possible combinations demonstrated that none of the CAP64-homologous genes were essential for capsule formation, although the cas35Δ strains displayed a hypocapsular phenotype. The chemical structure of the glucuronomannan (GXM) produced by the CAS family deletants revealed that these genes determined the position and the linkage of the xylose and/or O-acetyl residues on the mannose backbone. Hence, these genes are all involved in assembly of the GXM structure in C. neoformans.
Cryptococcus neoformans is an encapsulated basidiomycetous yeast and is the most common cause of fungal meningoencephalitis, which is lethal if untreated (6). C. neoformans is a heterogeneous species with four serotypes that can be classified into three varieties. C. neoformans var. grubii (serotype A) is the most common, representing 90% of the clinical isolates around the world. C. neoformans var. neoformans corresponds to serotype D, and C. neoformans var. gatti includes both serotypes B and C (20, 33). The capsule of this yeast is the major virulence factor, along with the production of melanin (5). Acapsular strains are avirulent, and the capsule components modulate the host immune response (8, 49). A major component of C. neoformans capsule is glucuronoxylomannan (GXM), a large polymer of (1→3)-α-mannose with xylose, glucuronic acid, and O-acetyl branches. The serotypic specificity is determined by both the number and position of the O-acetyl and xylose residues (12). The minor components of the capsule include galactoxylomannan and mannoproteins (reviewed in reference 4). Cherniak and colleagues defined six elementary structures called structure reporter groups (SRGs), designated M1 to M6, which defined GXM's constitutive elements (12). The GXM structure of each strain is composed of a major SRG specific for each serotype and one or several minor SRGs. Eight chemotypes were thus defined according to the presence of one or the other minor SRG. In the same study, the authors demonstrated that the structure of GXM is very heterogeneous even within each serotype, although the main SRG is generally specific for some serotypes.
To date, only a few genes involved in capsule biosynthesis have been identified. In the 1990s, four genes necessary for capsule production were cloned by functional complementation of acapsular mutant strains (8-11). The functions of these genes, however, have not been clearly assigned, and since the homologs of these genes have been identified in several noncapsulated fungi, their roles might not be restricted to capsule production (4). More recently, a number of elements in the cyclic AMP signal transduction pathway have been shown to play an important role in environmental regulation of capsule size (4, 18). However, many environmental factors, such as pH, serum, carbon and nitrogen sources, iron deprivation, CO2 concentration, and osmolarity, influence the size and perhaps even the structure of the capsule (reviewed in reference 26). A number of other unidentified signaling pathways are also likely to be involved in the regulation of capsule production (4, 53). Even though most of the genes involved in the biosynthesis of capsule polysaccharide precursors have been identified (2, 37, 44, 52), only two genes potentially involved in the capsule polymerization have been studied to date. Genes involved in capsule secretion or precursor transport have yet to be identified, even though a role of Cap59p in capsule secretion has been invoked (22, 27, 44).
We previously isolated a group of mutant strains that synthesize capsule with a modified structure in comparison with the original strain. We used these mutants to isolate two genes (CAS1 and UXS1), which are necessary for O acetylation and xylosylation of the capsule, respectively (27, 37). Using isogenic sets of strains and purified polysaccharide, we demonstrated the influence of the capsule structure on the pathophysiology of cryptococcal infection in a murine model (31). In this study, we report the isolation of a new gene (CAS3) which is necessary to complete O acetylation of the capsule. We demonstrated that Cas3p belongs to a seven-member protein family that includes Cap64p, an essential component of capsule production (11). Furthermore, all of the genes from this family were systematically deleted, and mutants deficient in one or more of these genes were characterized. Study of the GXM structure of the deletion strains demonstrated that the proteins encoded by these genes determine the position and the linkage of the xylose and O-acetyl residues on the mannose backbone.
MATERIALS AND METHODS
Strains and culture media.
The C. neoformans strains used in this study are listed in Table 1. The strains were routinely cultured on yeast-peptone-dextrose medium at 30°C (42). Minimal medium (SD), induction medium, and 5-fluoroorotic acid medium were prepared as previously described (19, 27). Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.) was used for the propagation of all plasmids.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| H99 | MATα | 39 |
| JEC21 | MATα | 50 |
| JEC43 | MATαura5 | 50 |
| JEC155 | MATαura5 ade2 | 50 |
| JEC156 | MATaura5 ade2 | 50 |
| JEC33 | MATαlys2 | 50 |
| JEC32 | MATalys2 | 50 |
| TYCC77 | MATα cap64 Δ::ADE2 ura3 | 11 |
| NE159 | MATα cap64 Δ::ADE2 | This study |
| NE81 | MATα cas3-1 ura5 | 37 |
| NE84 | cas3 mutant | This study |
| NE68 | MATαcas3-2 ura5 | This study |
| NE87 | MATαcas3-4 ura5 | This study |
| NE113 | MATαcas3-5 ura5 | This study |
| NE53 | MATacas3Δ::ADE2 ura5 | This study |
| NE190 | MATαcas3Δ::ADE2 | This study |
| NE195 | MATαcas31Δ::URA5 ade2 | This study |
| NE197 | MATαcas32Δ::URA5 ade2 | This study |
| NE152 | MATαcas33Δ::URA5 ade2 | This study |
| NE156 | MATαcas34Δ::URA5 ade2 | This study |
| NE234 | MATαcas35Δ::ADE2 ura5 | This study |
| NE228 | MATαcas31Δ::URA5 | This study |
| NE230 | MATαcas32Δ::URA5 | This study |
| NE238 | MATαcas33Δ::URA5 | This study |
| NE232 | MATαcas34Δ::URA5 | This study |
| NE244 | MATαcas35Δ::ADE2 | This study |
| NE223 | MATαcas31Δ::URA5 cas3Δ::ADE2 | This study |
| NE224 | MATαcas32Δ::URA5 cas3Δ::ADE2 | This study |
| NE213 | MATαcas33Δ::URA5 cas3Δ::ADE2 | This study |
| NE218 | MATαcas34Δ::URA5 cas3Δ::ADE2 | This study |
| NE245 | MATαcas31Δ::URA5 cas3Δ::ADE2 | This study |
| NE253 | MATαcas32Δ::URA5 cas35Δ::ADE2 | This study |
| NE249 | MATαcas33Δ::URA5 cas35Δ::ADE2 | This study |
| NE257 | MATαcas34Δ::URA5 cas35Δ::ADE2 | This study |
MAbs.
The anticapsular monoclonal antibodies (MAbs) E1 (17); CRND-8 (kindly provided by T. Shinoda, Tokyo, Japan) (25); and 4H3, 2H1, and 5E4 (kindly provided by A. Casadevall, Albert Einstein College of Medicine, Bronx, N.Y.) (7) were used for immunoblotting and immunofluorescence experiments as previously described (16, 27). They were selected because they have different specificities (3).
Capsule measurements.
To determine the capsule thickness, cells were grown overnight at 30°C in induction medium. Yeast cells were suspended in India ink, the distance from the cell wall to the outer border of the capsule for 20 cells was measured under a magnification of ×100 by using a grid with a resolution of 0.10 μm, and the mean thickness was determined for each strain.
Preparation and analysis of nucleic acid.
Purification of genomic DNA was carried out as previously described (21). The RNeasy Mini and Oligotex kits (Qiagen S.A., Courtaboeuf, France) were used to purify total RNA and mRNA, respectively, from C. neoformans cells. Southern blotting and colony hybridization were carried out by using standard protocols (40). Probe labeling, hybridization, washing, and detection of hybridized bands were performed with the digoxigenin nonradioactive nucleic acid labeling and detection system (Roche Diagnostic, Meylan, France) according to the manufacturer's instructions. DNA was sequenced by ESGS (Evry, France) with synthetic primers. The University of Wisconsin Genetics Computer Group programs were used for analysis of nucleic acid sequences (15). Restriction endonuclease digestions and ligations were carried out by standard methods, as recommended by the suppliers.
Cloning of CAS3 from C. neoformans var. neoformans.
Strain NE81 was transformed by electroporation (19) with a genomic library constructed in plasmid pCnTEL-1, kindly provided by B. Wickes (San Antonio, Tex.). By using a procedure described previously (27, 37), one plasmid able to complement the mutation of strain NE81 was isolated. Following subcloning, a 2.7-kb EcoRI-EcoRI DNA fragment was sequenced.
3′ and 5′ RACE analysis.
The C and N termini of Cas3p, Cas31p, Cas32p, Cas33p, and Cas34p were determined using the Smart RACE (rapid amplification of cDNA ends) system kit from Clontech (Palo Alto, Calif.) as previously described (27). The primers CAS3XF and CAS3XR used for the 5′- and 3′-end amplifications are listed at http://www.pasteur.fr/recherche/unites/Mymol/. The cDNA fragments were then cloned into the pGEM-T Easy vector (Promega, Madison, Wis.) and sequenced.
Chromosome separation by pulsed-field gel electrophoresis.
C. neoformans chromosome plugs for pulsed-field gel electrophoresis were prepared as previously described (51), and the chromosomes were separated on a 0.9% agarose gel (1× Tris-acetate-EDTA) by using a CHEF DR II apparatus (Bio-Rad) with the following parameters: 12°C, 16-h pulse from 100 to 200 s followed by 52-h pulse from 200 to 450 s, 2 V cm−1.
Disruption of the genes.
(i) CAS31. Two 1-kb DNA fragments specific for the CAS31 upstream and downstream regions were amplified by PCR with the HFPCR kit from Clontech and the primer pairs CAS31-5′5-CAS31-5′3 and CAS31-3′5-CAS31-3′3, respectively. Each amplified fragment was then cloned into the pGEM-t Easy cloning vector (Promega), resulting in the plasmids pNE195 and pNE170, respectively. The SpeI-SmaI fragment of pNE195 was then cloned into the SpeI-SmaI sites of the pNE170 plasmid to construct plasmid pNE196. Finally, plasmid pNE203 was obtained by inserting the XhoI-BamHI fragment of the pCIP3 plasmid (8), which contains the URA3 gene at the SmaI site of pNE196, after filling in of the cohesive ends.
(ii) CAS32.
By using the same strategy as for CAS31, two 1-kb DNA fragments specific for the CAS32 upstream and downstream regions were amplified by PCR with the HFPCR kit from Clontech and the primer pairs CAS325′5-CAS32-5′3 and CAS32-3′5-CAS32-3′3, respectively. Each amplified fragment was then cloned in the pGEM-t Easy cloning vector (Promega) to construct plasmids pNE169 and pNE168, respectively. The SpeI-BamHI fragment of pNE169 was then cloned into the SpeI-BamHI sites of the pNE168 plasmid to construct pNE176. Finally, plasmid pNE190 was obtained by inserting the XhoI-BamHI fragment of the pCIP3 plasmid (8), which contains the URA3 gene at the XhoI-BamHI sites of pNE176.
(iii) CAS3, CAS33, CAS34, and CAS35.
The disruption cassettes for CAS3, CAS33, CAS34, and CAS35 were constructed by PCR fusion by using a strategy similar to that described by Kuwayama and colleagues for Clostridium difficile (32). Briefly, upstream and downstream gene fragments and the selection marker genes (ADE2 or URA5) were PCR amplified from plasmids pRCD28 (46) and pCIP3 (8), respectively, by using the HFPCR kit from Clontech. The primers used for these amplifications are listed at http://www.pasteur.fr/recherche/unites/Mymol/ (see Fig. 3 for their positions). CAS3X-5′3 and CAS3X-3′5 contain sequences recognized by the M13R and M13F primers, respectively. Similarly, the MKRCAS3Xf and MKRCAS3XR primers were designed to anneal to both M13R and M13F, as well as CAS3X-5′3, CAS3X-3′5, respectively. Consequently, the CAS3X-3′5 and CAS3X-5′3 sequences contain the reverse complements of MKRCAS3Xf and MKRCAS3Xr, respectively. In addition, 5 ng of each of the three amplified and gel-purified fragments was utilized as the templates for PCR fusion with the primers CAS3X-5′5 and CAS3X-3′3 according to the following program: 95°C for 5 min, followed by 35 cycles of 94°C for 30 s and 68°C 5 min. The final PCR-amplified fragment represented the cas33Δ::URA5, cas34Δ::URA5, cas35Δ::ADE2, and cas3Δ::ADE2 alleles.
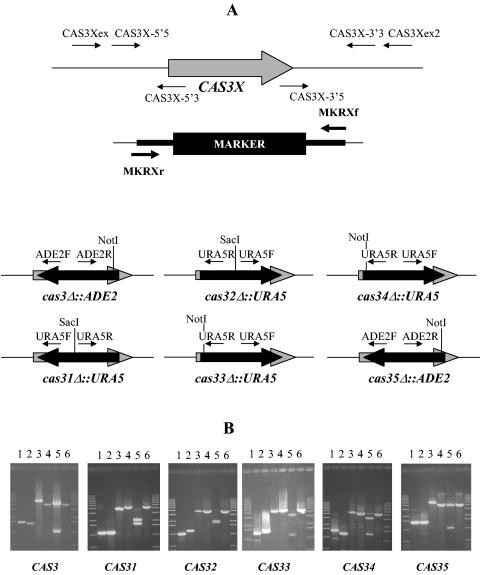
FIG. 3.
Gene disruption. (A) Positions of the different primers used to construct the disruption cassettes and for screening the transformants (X indicates 1, 2, 3, 4, or 5). (B) PCR analysis of the gene disruptants. For CAS3 and CAS35, the primers used were (lane 1) CAS3Xex-ADE2F, (lane 2) CAS3Xex2-ADE2R, and (lanes 3 to 6) CAS3Xex-CAS3Xex2. For CAS31, the primers used were (lane 1) CAS31ex-URA5F, (lane 2) CAS31ex2-URA5R, and (lanes 3 to 6) CAS31ex-CAS31ex2. For CAS32 to CAS34, the primers used were (lane 1) CAS3Xex-URA5R, (lane 2) CAS3Xex2-URA5F, and (lane 3 to 6) CAS3Xex-CAS3Xex2. Genomic DNA of strain JEC21 (MATα) (lanes 4 and 6) or of the corresponding cas3XΔ mutant (lanes 1, 2, 3, and 5) was used as the substrate. Lanes 5 and 6 the PCR amplification product were gel purified and digested with the adapted restriction enzyme (NotI or SacI).
The NotI-linearized plasmids pNE190 and pNE203 for CAS32 and CAS31, respectively, or the PCR-amplified fragments for the four other genes were used to transform strains JEC155 and JEC156 by biolistic DNA delivery. Transformants were selected on minimal medium with 1 M sorbitol but lacking uracil or adenine, depending on the markers used (48).
Polysaccharide analysis.
Each strain was grown for 3 days in induction medium, and GXM was prepared as previously reported (37). Isolated GXMs were sonicated and lyophilized three times before being dissolved in 0.5 ml of 99.96% D2O (Sigma, St. Louis, Mo.) for nuclear magnetic resonance (NMR) analysis. NMR spectra were acquired with a Bruker Avance 600 NMR spectrometer, using a 5-mm (1H, 13C) inverse-detection dual-frequency probe, operating at 600.13 and 150.913 MHz, respectively. The temperature was maintained at 70°C during the NMR measurements. 1H NMR spectra were acquired with a spectral width of 3,000 Hz, a time domain of 32 K, 128 acquisitions, and a repetition time of 6.5 s. Two-dimensional NMR spectra were acquired by using standard Bruker pulse sequences (XWINNMR 3.5). Double-quantum-filtered (1H, 1H) COSY, TOCSY, and NOESY spectra were obtained in the phase-sensitive mode. The spectral width was 3,000 Hz, and 256 increments of 16 acquisitions were acquired, each containing 2,000 data points. The acquisition time was 340 ms, and the relaxation delay was 1.5 s. Squared sine-bell functions shifted by π/2 were applied for data processing in t1 and t2. The mixing time for the TOCSY spectra was either 40 or 150 ms. (1H, 13C) single-bond shift correlation (HSQC) spectra were obtained in the 1H detection mode. Acquisition parameters were as follows: spectral widths in t1 and in t2, 12,000 and 3,000 Hz, respectively; optimization for 1JC,H coupling constants, 140 Hz; acquisition of 256 increments in t1, each consisting of 32 to 64 acquisitions of 2,000 data points; acquisition time, 170 ms; relaxation delay, 1s. A sine-bell function shifted by π/2 was applied in t2 and a Gaussian-Lorentzian function was applied in t1. Cross-peak volumes were determined for comparison of relative intensity ratios of carbohydrate residues. (1H, 13C) multiple-bond heteronuclear correlation (HMBC) spectra were acquired similarly to the HSQC spectra. The spectral width in t1 was 15,000 Hz. Experiments were optimized for nJC,H long-range coupling constants of approximately 2 Hz to determine the linkage between carbohydrate moieties. The number of acquisitions per increment was 64.
Nucleotide sequence accession numbers.
The sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers AY421722, AY421723, AY421724, AY421725, AY421726, AY428736, and AY063511.
RESULTS
Cloning of CAS3.
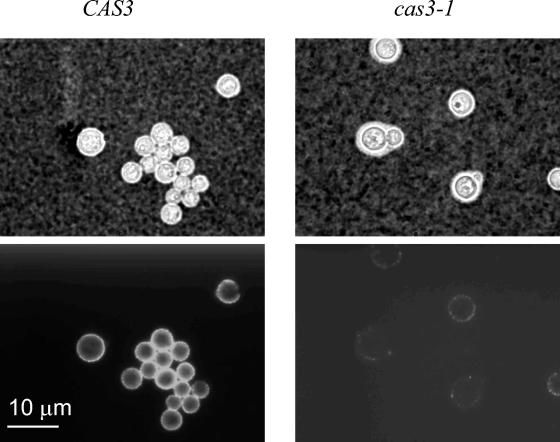
As reported previously, the phenotype of cas3 mutant strains differed from that of cas1Δ or uxs1Δ strains (27, 37). As shown in Fig. 1, binding of the anti-GXM-specific MAb CRND-8 to cells of strain NE81 was significantly weaker than that to cells of the original strain JEC43. This phenotype was not complemented by any of the previously identified CAS genes, suggesting that the gene mutated in strain NE81 was not one of the previously identified CAS genes. We then used functional complementation to clone the CAS3 gene as previously described for the CAS1 and UXS1 genes (27, 37). Following subcloning and complementation analysis, the sequence of a 2,663-bp EcoRI-EcoRI DNA fragment capable of complementing the mutation in NE81 was identified. The complete cDNA sequence of the CAS3 gene was obtained after 3′ and 5′ RACE amplifications. The CAS3 gene encodes a protein of 57 kDa with 34% similarity to the capsule protein Cap64p (11). We sequenced the 5′ ends of five independent cDNA clones and determined that the transcription start site was located between 76 and 80 nucleotides upstream of the AUG site. Sequencing of the 3′ end showed that the polyadenylation site was located 154 bp downstream of the stop codon. Alignment of the cDNA and genomic sequences revealed the presence of 12 introns having an average length of 55.25 nucleotides, with the 12th intron being located downstream of the stop codon. BLAST searches in the general databases did not reveal any other homologous sequence except for the CAP64 homologs in the Homobasidiomycete fungi Pleurotus ostreatus (GenBank accession no. AJ318523) and Phanerochaete chrysosporium (http://www.jgi.doe.gov/programs/whiterot.htm). However, no homolog was present in the genome of the Heterobasidiomycete Ustilago maydis or any other eukaryotic or prokaryotic genome.
FIG. 1.
Comparison of the cas3-1 (NE81) and CAS3 (JEC43) strains by using India ink negative staining (top panels) and immunofluorescence labeling with the MAb CRND-8 (bottom panels). Cells were grown to stationary phase in capsule-inducing medium at 30°C (see Materials and Methods), washed with sterile water, and stained with India ink or labeled with the MAb.
Identification and cloning of CAS3 homologs in the C. neoformans genome.
A search for homologous sequences in the genome of C. neoformans strain JEC21 at the Institute for Genomic Research website (http://www.tigr.org/tdb/e2k1/cna1/) revealed the presence of five other putative proteins sharing homologies with Cas3p and Cap64p. Specific primers were chosen, and 5′ and 3′ RACE amplification allowed the elucidation of the entire cDNA sequence. The proteins encoded by these sequences were named Cas31p, Cas32p, Cas33p, Cas34p, and Cas35p. Cas31p corresponds to the Cap64p homolog that has been shown to copurify with the α-1,3-mannosyl transferase protein Cmt1p (44). Using a specific probe, we located CAS3 on one of the largest chromosomes in serotype D (JEC20 and JEC21) and serotype A (H99) genomes (data not shown). This result is consistent with the last assembly of the strain JEC21 genome, where CAS3 is located in chromosome scaffold 2. In this assembly, CAS31, CAS32, and CAS35 are located in chromosome scaffold 1, CAP64 and CAS34 are located in chromosome scaffold 3, and CAS33 is located in scaffold 11. However, the genes located on the same chromosome are separated by large regions, and a simple explanation for the organization of these genes is not readily apparent.
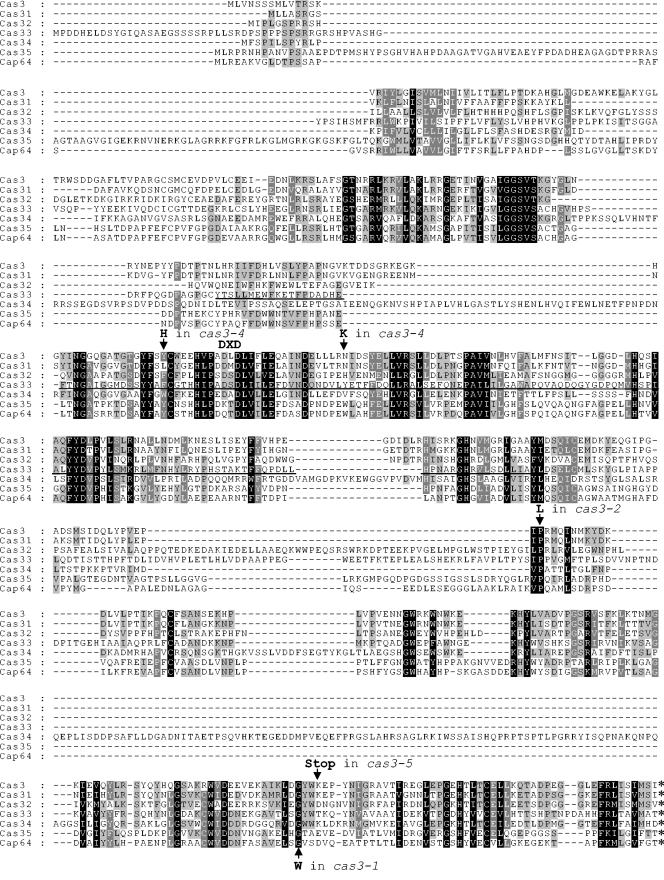
The proteins encoded by these genes are comparable in size (506, 481, 551, 603, 674, and 628 amino acids for Cas3p, Cas31p, Cas32p, Cas33p, Cas34p, and Cas35p, respectively) and have a putative N-terminal hydrophobic domain similar to that of Cap64p. Table 2 illustrates the degree of homology between the different sequences, and the sequence alignment is presented in Fig. 2. A search for sequences homologous to Cas31p, Cas32p, Cas33p Cas34p, and Cas35p did not reveal any homologs in addition to those previously identified with Cas3p. However, analysis of the Cas33p sequence showed the presence of a domain between amino acids 191 and 353 (COG2755.1, TesA) that is conserved in different esterases and lysophospholipases (Fig. 2). Interestingly, this domain was present in different esterases or transferases involved in polysaccharide synthesis or degradation in microorganisms, such as the acetyl xylan esterase in the fungus Neocallimastix patriciarum (13), the acylneuraminate cytidylyltransferase in Streptococcus agalactiae (24), and the rhamnogalacturonan acetylesterase in Xanthomonas campestris (14).
TABLE 2.
Similarities between the different members of the Cas3p/Cap64p family
| Protein | % Similarity to:
|
|||||
|---|---|---|---|---|---|---|
| Cas3p | Cas31p | Cas32p | Cas33p | Cas34p | Cas35p | |
| Cas3p | ||||||
| Cas31p | 77 | |||||
| Cas32p | 44 | 44 | ||||
| Cas33p | 36 | 36 | 35 | |||
| Cas34p | 34 | 35 | 29 | 26 | ||
| Cas35p | 31 | 30 | 31 | 36 | 23 | |
| Cas64p | 34 | 35 | 34 | 42 | 25 | 62 |
FIG. 2.
Regions conserved between different members of the Cas3/Cap64 family. Sequences were aligned by using ClustalW 1.83 (47) and edited with Genedoc (38). The TesA conserved domain identified in the Cas33p sequence is underlined. The positions and natures of sequence alterations in the different cas3 alleles are indicated by arrows.
CAS3 sequence modifications.
In our initial screen for cas mutant strains, we isolated five strains displaying the same alteration of anticapsular MAb binding pattern, defined as the Cas3 phenotype, including strain NE81, which was used to clone CAS3 (37). Transformation of the remaining four mutant strains with an autonomously replicating plasmid containing the wild-type CAS3 gene restored binding of the antibodies to wild-type levels in each strain (data not shown). The CAS3 gene was amplified from each mutant, and two independent PCR products were sequenced. With the exception of strain NE84, mutations were identified within the CAS3 gene, suggesting the importance of the corresponding residues for CAS3 function. As shown in Fig. 2, the cas3-1 allele from strain NE81 encodes a tryptophan residue in place of the conserved glycine residue at position 457. The cas3-2 allele from strain NE68 revealed a mutation corresponding to position 356, where a conserved proline residue was replaced with a leucine residue. The cas3-4 allele from strain NE87 contained two mutations, encoding Y195H and N222K, located within a highly conserved region encompassed by the putative TesA domain. Finally, the protein encoded by the cas3-5 allele in strain NE113 was truncated due to the presence of an ochre mutation at position 460. In strain NE84, a mutation in the CAS3 gene was not found. However, a mutation might be in a regulatory region of CAS3 outside the amplified fragment.
The comparison of the CAS gene sequences revealed the presence of a partially conserved DXD motif essential for the activity of different glycosyltransferases. We used site-directed mutagenesis to replace the first two amino acids of this motif with two alanine residues but these substitutions had no detectable effect on the function of Cas3p (data not shown). In contrast, tagging the C-terminal region of Cas3p with His6 completely abolished the Cas3p function (data not shown). Thus, the C-terminal region is essential for the function of Cas3p.
Gene disruption.
We disrupted every gene of the CAS3 family with either the ADE2 or URA5 selection marker. Correct integration of the cassette was determined by PCR with a primer that annealed to a region outside the disruption cassette (CAS3Xex) and a primer that annealed to sequences within the marker (URA5F, URAR, or ADE2a) (Fig. 3). We screened 20 clones resulting from each transformation and identified two putative homologous integrants each for CAS3, CAS33, and CAS34 and four each for CAS31, CAS32, and CAS35. A second pair of primers was used to determine the correct integration of the cassette (Fig. 3). PCR amplifications and restrictions of the CAS3Xex-CAS3Xex2 region were used to verify the disruption of the wild-type gene in each putative deletion strain. In addition, Southern analysis was used to confirm replacement of the genes with the corresponding maker (data not shown). To construct isogenic sets of strains, genetic crosses were performed. Strain NE53 (MATa cas3Δ::ADE2 ura5) was crossed with strain JEC33 (MATα lys2), and the prototrophic progeny strains of this cross were selected on SD medium. Strains containing disrupted or wild-type genes were identified by PCR, and one strain with each possible genotype (MATa cas3Δ::ADE2, MATα cas3Δ::ADE2, MATa CAS3, and MATα CAS3) was then stored for further studies. Similar procedures were used for CAS31, CAS32, CAS33, CAS34, and CAS35 to isolate different sets of strains. Finally, a large set of double mutants were selected by using the different auxotrophic markers. For example, strain NE53 (MATa cas3Δ::ADE2 ura5) was crossed with strains NE195 (MATα cas31Δ::URA5 ade2), NE197 (MATα cas32Δ::URA5 ade2), NE152 (MATα cas33Δ::URA5 ade2), and NE156 (MATα cas34Δ::URA5 ade2). Prototrophic progeny strains were then selected on SD medium, and their genotypes were confirmed by using PCR. Strains of each possible genotype (MATa cas3Δ::ADE2 cas3XΔ::URA5 or MATα cas3Δ::ADE2 cas3XΔ::URA5) were stored in duplicate for further studies (Table 1).
Phenotypic analysis.
(i) Growth rate, colony morphology and sensitivity to drugs. The growth rates of all of the strains inoculated on yeast-peptone-dextrose and incubated at both 30 and 37°C were compared, and no significant differences were found (data not shown). In addition, the colony morphologies of mutants were indistinguishable from that of the wild type, except that the cas35Δ and cap64Δ strains produced drier colonies (data not shown). We tested the mutant and wild-type strains for their sensitivities to caspofungin and amphotericin B and found no significant differences between the mutants and the parental strains (data not shown).
(ii) Capsule sizes.
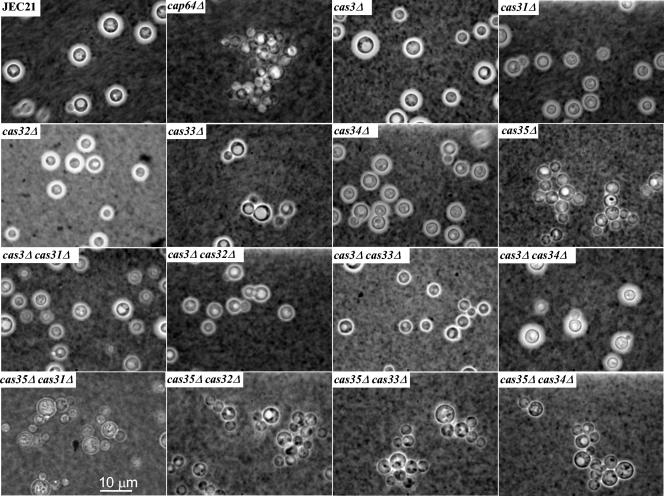
The capsule size of each strain was analyzed by using India ink negative staining and microscopic examination. As shown in Fig. 4, only mutations in CAP64 and CAS35 seemed to affect the size of capsule. Rigorous measurements of capsule sizes confirmed these results (1.5 ± 0.48 μm for JEC21, 1.7 ± 0.49 μm for the cas3Δ strains, 1.80 ± 0.43 μm for the cas31Δ strains, 1.25 ± 0.50 μm for the cas32Δ strains, 1.8 ± 0.4 μm for the cas32Δ strains, 2.0 ± 0.47 μm for the cas33Δ strains, 1.9 ± 0.55 μm for the cas34Δ strains, and 0.71 ± 0.40 μm for the cas35Δ strains). Strains with the CAS35 gene disrupted were thus hypocapsular, and an analysis of 32 progeny strains obtained from a cross between NE234 (MATα cas35Δ::ADE2 ura5) and JEC32 (MATa lys2) showed linkage between the hypocapsular phenotype and the cas35 deletion. Thus, all of the progeny strains containing the disruption cassette were hypocapsular, while all of the encapsulated ones contained the wild-type allele. The hypocapsular phenotype was, as expected, associated with a strong virulence defect of the cas35Δ strains in a mouse model (Y. C. Chan and K. J. Kwon-Chung, unpublished data). Double mutants deficient in CAS35 and any other genes displayed no additive defects in capsule production.
FIG. 4.
Analysis of capsule size. Cells were grown to stationary phase in capsule-inducing medium (see Materials and Methods), washed with sterile water, and stained with India ink.
(iii) Antibody analysis of capsule structure.
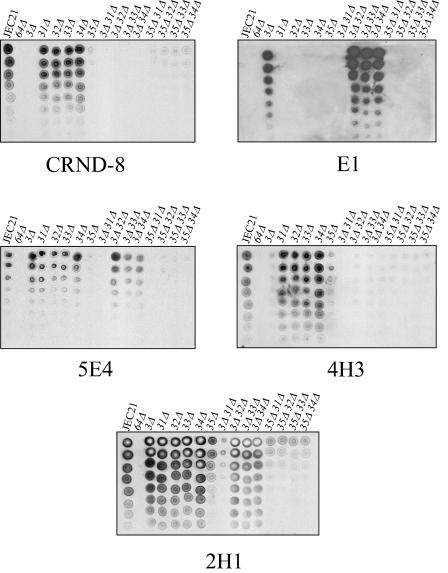
We previously performed a dot blot assay with different GXM-specific monoclonal antibodies to discriminate between modifications in capsule structure (27, 37). We used the same assay to analyze the different mutant strains obtained in this study, and the results are presented in Fig. 5. Mating type does not appear to influence capsule structure, as MATa strains were identical to MATα strains in the dot blot assay (data not shown). The phenotypes of the cas3Δ strains constructed for this report were the same as those of Cas3 mutant strains (37). Moreover, backcrosses with the cas3Δ mutants and the original strain demonstrated that this phenotype was genetically linked with the CAS3 disruption. We could not detect any difference in capsule structure in the other single mutant capsular strains. Although lighter signals were obtained with all of the MAbs and the cas35Δ strains, the decreased binding could have been due to the smaller size of the capsule in these mutants. As expected, the acapsular cap64Δ strain did not react with any of the antibodies tested. With the exception of the cas3Δ cas31Δ double mutant strains, a synthetic phenotype associated with a combination of gene deletions was not observed. Strains lacking CAS3 and CAS31 were not recognized by any of the antibodies used in this study which correspond to a phenotype previously observed with the cas1Δ strains (27).
FIG. 5.
Antibody reactivities of different GXM-specific MAbs with the mutant strains. Serial twofold dilutions of cell suspensions were spotted (starting with 3 × 104 cells on the first spot at the top of the lane) on a nitrocellulose membrane and probed with a given concentration of each MAb (37).
(iv) Analysis of polysaccharide structure.
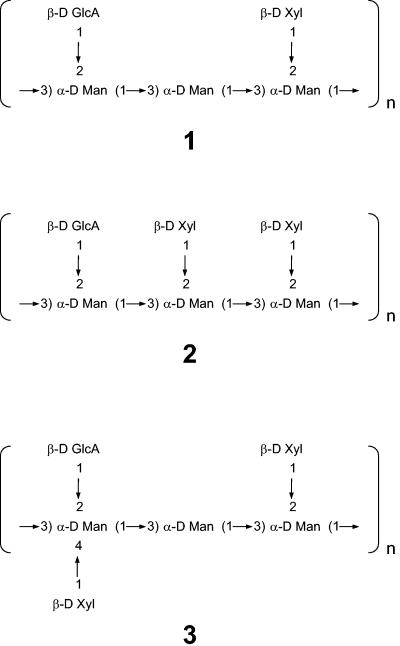
We purified GXM from all of the single capsular mutant strains, as well as the double mutant deficient in CAS31 and CAS3. The chemical structures of their polysaccharide were analyzed by NMR. Due to strong resonance overlap in the two-dimensional correlation spectra for minor residues in particular, not all chemical shifts could be determined unequivocally. Connectivity was determined for the smallest detectable repeating moiety. The smallest repeating moieties (Fig. 6) were determined by the assessment of connectivity between carbohydrate residues, using (1H, 13C) HMBC spectra and the chemical shifts in mutant strains compared to that of the parental strain as well as to those of previous work (1, 27, 37, 41). The structure of the GXM isolated from the parental strain was identical to that of the chemotype Chem1, which is dominant in serotype D isolates (12, 43) (moiety 1 in Fig. 6).
FIG. 6.
Smallest repeating carbohydrate moieties identified in GXMs isolated from the mutated strains. Additional variability was due to different 6-O acetylations of the mannosyl residues.
The GXM isolated from cas3Δ strains contained partly 6-O-acetylated mannosyl residues. The acetylation ratio relative to the amount of mannosyl residues was only half of that in the parental strain (Table 3). The 1H, 13C HSQC and HMBC spectra showed that the chemical shifts of H-5/C-5 and H-6/C-6 of the mannosyl residue in moiety 1 have shifted from 4.08/73.6 ppm and 4.30 to 4.35/66.5 ppm in the wild type to 3.91/75.2 ppm and 3.78 to 3.84/62.8 ppm in the cas3Δ strain, indicating that this residue is not 6-O acetylated in the mutated strain (27). In contrast to the previously reported 6-O-deacetylated GXM from the cas1Δ mutant (27), a second minor carbohydrate moiety was found in cas3Δ strain GXM. The most obvious difference in this minor repeating carbohydrate moiety was the presence of an additional xylosyl residue. The HMBC spectra indicated that the xylosyl residue was 1→2 connected to a mannosyl residue, which was 1→3 connected to another similar mannosyl residue (1→2 xylose substituted) and 3→1 connected to a mannosyl residue with a 1→2 glycuronic acid substitution (moiety 2 in Fig. 6). This structure has been previously defined as the SRG M2 (12). Based on the (1H, 13C) HSQC spectrum and integration of the H-1/C-1 cross-peaks of the xylosyl residues in the two moieties, the amount of the second moiety 2 relative to the major moiety 1 (also present in the wild type) was estimated to be 10 to 15% (Table 3). The structure and chemical shifts of the additional repeating carbohydrate moiety 2 were similar to those reported for 6-O-deacetylated GXM of serotype A (41). However, due to extensive resonance overlap between the two repeating sequences, it was not possible to determine whether the two sequences were present in parallel or whether the actual smallest repeating sequence contained more than three mannosyl residues, incorporating both repeating sequences. When CAS3 was reintroduced into the cas3Δ strain, the O-acetylation level was fully restored and the additional minor repeating carbohydrate disappeared in the GXM structure (data not shown).
TABLE 3.
Concentration ratios of carbohydrate residues as determined from the NMR spectra
| Strain | Concn ratio (mean ± SD)
|
||
|---|---|---|---|
| O-acetyl/Mana | (Xyl + GlcA)/Manb | Xyl 1/2/3c | |
| JEC21 | 2.31 ± 0.2 | 1.85 ± 0.1 | 10:0:0d |
| cas3Δ | 1.23 ± 0.1 | 1.83 ± 0.1 | 10:1:0 |
| cas31Δ | 2.12 ± 0.1 | 1.97 ± 0.1 | 10:1.5:0 |
| cas32Δ | 2.07 ± 0.1 | 2.06 ± 0.1 | 10:2:0 |
| cas33Δ | 2.33 ± 0.2 | 1.87 ± 0.1 | 10:1:0 |
| cas34Δ | 2.01 ± 0.2 | 2.26 ± 0.1 | 10:3:0 |
| cas35Δ | 1.86 ± 0.1 | 2.33 ± 0.2 | 10:3:1 |
| cas3Δcas31Δ | 0.00 | 2.44 ± 0.1 | 10:4:0 |
The acetyl/mannosyl ratio was determined from the ID 1H NMR spectra by integration of the acetyl resonances (2.1 to 2.3 ppm) and the H-1 mannosyl resonances (5.1 to 5.4 ppm).
The (xylosyl + GlcA)/manosyl ratio was determined from the 1D H NMR spectra by integration of the Xyl H-2, H-3, and H-5a resonances and the GlcA, H-2 and H-3 resonances (3.1 to 3.55 ppm) relative to the H-1 mannosyl resonances (5.1 to 5.4 ppm).
The ratio of the xylosyl residues from structures 1, 2, and 3 was determined by integration of the (1H, 13C) HSQC cross peaks for H-1/C-1 of the respective xylosyl residues.
Resonances from minor unassigned residues were present (<5%).
The GXMs of all other cas mutants were 6-O acetylated, similar to what has been reported for the GXMs from wild-type strains (27). The acetyl/mannosyl ratios varied between 1.8 and 2.3 (Table 3). GXM 1, which is the dominant GXM in serotype D isolates, was also dominant in all other mutant strains. As in the cas3Δ mutant, the repeating sequence 2 was identified in all other mutant strains. Table 3 shows the ratio of moiety to moiety 2. Structure 2 was confirmed by the (1H, 13C) HMBC spectra. In addition, the mannosyl residue, to which the additional xylosyl residue is 1→2 linked, showed changes in the H-1/C-1 and H-2/C-2 chemical shifts, similar to those for cas3Δ. Interestingly, the mannosyl residue, which is not xylosylated in moiety 1 but is xylosylated in the minor moiety 2, was 6-O acetylated in moiety 1 but not in moiety 2, confirming previous findings that mannosyl residues that are 2-xylosylated are not 6-O acetylated (27). Most likely, this has contributed to the minor changes in the acetylation ratio that correlated with the amount of the repeating moiety 2 in the respective GXM. Only minor differences in the GXM composition were found between the cas31Δ, cas32Δ, cas33Δ, and cas34Δ mutants, which concern the ratio of the repeating moieties 1 and 2 (Table 3).
GXMs isolated from the cas35Δ strain showed the most deviated form of polysaccharides. The cas35Δ mutant contains a third repeating moiety, shown in Fig. 6 (moiety 3), in addition to moieties 1 and 2. As in the other strains, it was not possible to determine whether the smallest repeating carbohydrate sequences were part of a larger repeating sequence or if they were separate entities present in parallel. This additional moiety varied from the GXM of the wild type by an additional 1→4 substitution of a xylose residue to the 1→2 GlcA substituted mannosyl residue. Although resonance overlapping made assignment difficult, the most likely explanation is a repeating sequence 3, which is identical to one that was found in a Cap70 mutant (1). However, it cannot be fully excluded that the Man[1→2GlcA, 1→ 4Xyl] residue is 1→3 linked to another 1→2 substituted mannosyl residue (found in serotype B and C isolates [1, 44]), in contrast to the unsubstituted mannosyl residue in structure 3. The chemical shifts of H-5/C-5 and H-6/C-6 indicate that ManGlcA,Xyl is 6-O deacetylated in moiety 3.
The cas3Δ cas31Δ double mutant was the only one that exhibited a synthetic phenotype in the dot blot assay. Chemical analysis of the purified GXM from this mutant strain confirmed these results: the cas3Δ cas31Δ GXM was completely de-O acetylated, and a cumulative effect on the ratio of xylosyl residues associated with structure 2 was observed (Table 3).
DISCUSSION
In C. neoformans, four CAP genes have been demonstrated to be necessary for capsule production (8-11). The CAP59 and CAP60 genes, which have been identified by functional complementation of acapsular mutants, encode homologous proteins and share a significant degree of homology with the recently identified α-1,3-mannosyltransferase Cmt1p (8, 9, 44). Moreover, the recently released complete genome sequences of a serotype D and a serotype A strains of C. neoformans have allowed us to identify the fourth member of this protein family (26). Likewise, four proteins homologous to Cap10p (among them, Cap1p, encoded by a gene located in the mating type locus [34]) could also be identified in these genomes (26). In the present study, we demonstrated the existence of six proteins that are homologous to Cap64p. The existence of other Cap protein families orthologous to these proteins could be found in the proteomes of noncapsulated organisms, suggesting that the activities of the CAP gene products are not likely to be restricted to capsule biosynthesis (4). The Cas3p/Cap64p family, however, appears to be restricted to Homobasidiomycetous fungi. Among all the available genome data, Pleurotus ostreatus and Phanerochaete chrysosporium were the only organisms from which we were able to identify the orthologous genes. U. maydis, a Heterobasidiomycete, did not contain any ortholog of this family in its genome.
As for the other Cap proteins, the role of Cap64p in the capsule biosynthesis remains unclear, although several nonexclusive hypotheses have been made as to their function (capsule secretion, capsule biosynthesis regulation, or direct involvement in capsule biosynthesis) (4, 36). All of the mutant strains presented here except the cap64 mutant produce capsule, and we demonstrated that Cas3p, Cas31p, Cas32p, Cas33p, Cas34p, and Cas35p are involved in xylose branching and/or O acetylation of the capsule. These results rule out the possibility of secretory function and seem to support the notion that this Cap protein family is directly involved in capsule synthesis. The presence of an esterase motif in the conserved domain of the Cas33p sequence also supports this hypothesis. We cannot, however, eliminate the possibility that these genes regulate the transcription or the activities of some genes directly involved in capsule biosynthesis.
Families of genes encoding partially or completely redundant functions are common in polysaccharide synthesis pathways. Thus, the mannosyltransferase activities are encoded by different families of genes in Saccharomyces cerevisiae (35, 45). Consequently, single gene mutations are often either silent or barely express a phenotype, as proteins encoded by other paralogous genes tend to compensate for the lack of the activity of the mutated gene. Multiple mutations in different members of the gene family are therefore necessary to produce a phenotype. In this study, we constructed a set of single and double mutant strains to identify the ones which might be involved in similar functions. We hypothesized that a double mutation in genes encoding similar functions would have a synthetic phenotype, whereas a double mutation in genes acting at different steps of the biosynthesis would have a simple additive effect. Even though we observed a synthetic phenotype for only one double mutant strain (the cas3Δ cas31Δ), the phenotypes of the single mutants and the sequence similarities of the different proteins helped us to draw several hypotheses on their functions.
The GXM from the cas31Δ strain has the same O acetylation as that of the parental strain, the GXM from the cas3Δ strain has 30% of the O-acetyl residues, and the cas3Δ cas31Δ double mutant is completely de-O acetylated. These genes also appear to be the only ones of this family that are involved in O acetylation of the capsule. However, we cannot rule out that other combinations of double or triple mutations could reveal importance of the other genes in O acetylation. The Cas31p sequence is also most similar to that of Cas3p, suggesting that these two proteins have the same function but are differentially regulated. Indeed, Northern blot analysis demonstrated significantly weaker expression of CAS31 compared to CAS3 in rich medium (A. Alspaugh, personal communication).
Cas35p is most similar to Cap64p in its amino acid sequence, and while Cap64p is essential for capsule biosynthesis, cas35Δ mutant strains are hypocapsular. Moreover, the GXM of cas35Δ strains contained a unique repeating carbohydrate moiety (structure 3). Two hypotheses can be drawn to explain these results. First, Cas35p influences the substitution of the mannosyl backbone by directing an additional xylose residue to the ManGlcA residue. Second, this triad exists in the parental strain but at a very low concentration. It is detectable only due to a down-regulation of the M1 synthesis. Cas35p would be then necessary for the full expression of the major triad (M1) in serotype D. In the mutant strain, the synthesis of M1 would be strongly repressed, and the relative concentration of the cas35Δ-specific triad would be artificially increased. The identification of the same triad in another hypocapsulated mutant, Cap70 (1), supports the latter hypothesis, even though we cannot rule out that this strain has a mutation within CAS35.
The three remaining single mutant strains have very similar capsular phenotypes. A mutation in each gene does not modify the size or the O acetylation of the capsule directly. They do not have any synthetic phenotype associated with a mutation in CAS3 or CAS35. Finally, all three mutations increased the proportion of the minor SRG (M2) marginally without increasing that of the cas35Δ-specific triad. The only difference between M1 and M2 is the presence a β-1,2-xylose residue on the second mannose residue of the triad (Fig. 6). It remains to be demonstrated whether these proteins promote the addition of a β-1,2-xylose in position 3 of the triad or inhibit the β-1,2-xylosylation of the second mannose.
Sommer et al. have demonstrated recently that the protein Cmt1p, which has an α-1,3-mannosyltransferase activity, copurified with a Cap64p-paralogous protein (44). Here, we identified this protein as being Cas31p. This result strongly suggests the existence of protein complex involved in capsule biosynthesis. Such protein complexes have been observed in S. cerevisiae for the synthesis of the mannan moiety of the glycoproteins (28, 30). Thus, two multiprotein complexes (M-Pol I and M-Pol II) are necessary for the synthesis of the mannan backbone of the N-linked glycan (29), whereas Pmt proteins initiate the assembly of O-mannosyl glycans from heteromeric and homomeric complexes (23). In this study, we demonstrated that Cas31p is involved in capsule O acetylation and xylosylation, suggesting that this potential protein complex is multifunctional.
As the proteins from this family, except for Cap64p, are involved in the position and linkage of xylose and O-acetyl residues on the mannose backbone, they represent good candidates for the source of C. neoformans serospecificity. We indeed identified the seven members of this protein family in the serotype A and serotype B genomes (http://www.bcgsc.bc.ca/cgi-bin/crypto_data/blast_wm276.pl). Whether these homologous genes have analogous functions in different serotypes or are regulated differently remains to be determined. The molecular and biochemical research on capsule formation thus far reported is beginning to uncover the complexity of the GXM biosynthetic pathways. Comprehensive studies on genetics, cell biology, enzymology, and glycochemistry are required for an understanding of the prospective function of each component.
Acknowledgments
We are grateful to T. Shinoda (Tokyo, Japan) for the generous gift of MAb CRND-8; to B. Wickes (San Antonio, Tex.) for the C. neoformans genomic library; to A. Casadevall (Bronx, N.Y.) for MAbs 4H3, 5E4, and 2H1; and to J. Heitman (Durham, N.C.) for plasmid pRCD28. We are grateful to the C. neoformans genome sequencing projects at the Duke Center for Genome Technology (http://cgt.duke.edu/), at the Institute for Genomic Research (http://www.tigr.org/tdb/e2k1/cna1/), at the Stanford Genome Technology Center at StanfordUniversity (http://www-sequence.stanford.edu/group/C.neoformans/index.html), and at the Genome Sequence Centre at the British Columbia Cancer Research Centre (http://www.bcgsc.bc.ca/). We also thank the C. neoformans cDNA sequencing project at the University of Oklahoma (http://www.genome.ou.edu/cneo.html). U.H. is grateful to T. C. Sorrell and the Centre for Infectious Diseases, Westmead Hospital (Australia), for support of the NMR work.
The C. neoformans cDNA sequencing project at the University of Oklahoma is funded by NIH/NIAID grant AI47079. Studies of capsule synthesis are supported by Ensemble contre le SIDA (grant AO13-3) and the Pasteur Institute (to G.J.). The NMR work was funded by the National Health and Medical Research Council of Australia (grants 990738 and 980116 to U.H.).
REFERENCES
- 1.Bacon, B. E., R. Cherniak, K. J. Kwon-Chung, and E. S. Jacobson. 1996. Structure of the O-deacetylated glucuronoxylomannan from Cryptococcus neoformans Cap70 as determined by 2D NMR spectroscopy. Carbohydr. Res. 283:95-110. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Peled, M., C. L. Griffith, and T. L. Doering. 2001. Functional cloning and characterization of a UDP-glucuronic acid decarboxylase: the pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc. Natl. Acad. Sci. USA 98:12003-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belay, T., R. Cherniak, T. R. Kozel, and A. Casadevall. 1997. Reactivity patterns and epitope specificities of anti-Cryptococcus neoformans monoclonal antibodies by enzyme-linked immunosorbent assay and dot enzyme assay. Infect. Immun. 65:718-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bose, I., A. J. Reese, J. J. Ory, G. Janbon, and T. L. Doering. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchanan, K. L., and J. W. Murphy. 1998. What makes Cryptococcus neoformans a pathogen? Emerg. Infect. Dis. 4:71-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. American Society for Microbiology, Washington, D.C.
- 7.Casadevall, A., and M. D. Scharff. 1991. The mouse antibody response to infection with Cryptococcus neoformans: VH and VL usage in polysaccharide binding antibodies. J. Exp. Med. 174:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14:4912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. C., and K. J. Kwon-Chung. 1998. Isolation of the third capsule-associated gene CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 66:2230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. C., and K. J. Kwon-Chung. 1999. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 181:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 64:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherniak, R., H. Valafar, L. C. Morris, and F. Valafar. 1998. Cryptococcus neoformans chemotyping by quantitative analysis 1H nuclear magnetic resonance spectra of glucuronoxylomannans with a computer-simulated artificial neural network. Clin. Diagn. Lab. Immunol. 5:146-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalrymple, B. P., D. H. Cybinski, I. Layton, C. S. McSweeney, G. P. Xue, Y. J. Swadling, and J. B. Lowry. 1997. Three Neocallimastix patriciarum esterases associated with the degradation of complex polysaccharides are members of a new family of hydrolases. Microbiology 143:2605-2614. [DOI] [PubMed] [Google Scholar]
- 14.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 15.Devereux, J. P., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acid Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dromer, F., E. Guého, O. Ronin, and B. Dupont. 1993. Serotyping of Cryptococcus neoformans by using a monoclonal antibody specific for capsular polysaccharide. J. Clin. Microbiol. 31:359-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dromer, F., J. Salamero, A. Contrepois, C. Carbon, and P. Yeni. 1987. Production, characterization, and antibody specificity of a mouse antibody reactive with Cryptococcus neoformans capsular polysaccharide. Infect. Immun. 55:742-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Souza, C. A., J. H. Alspaugh, C. Yue, T. Harashima, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edman, J. C., and K. J. Kwon-Chung. 1990. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol. Cell. Biol. 10:4538-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzot, S. P., I. F. Salkin, and A. Casadevall. 1999. Cryptococcus neoformans var. grubii: separate status for Cryptococcus neoformans serotype A isolates. J. Clin. Microbiol. 37:838-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Hermoso, D., G. Janbon, and F. Dromer. 1999. Epidemiological evidence for dormant Cryptococcus neoformans infection. J. Clin. Microbiol. 37:3204-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.García-Rivera, J., Y. C. Chang, K. J. Kwon-Chung, and A. Casadevall. 2004. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girrbach, V., and S. Strahl. 2003. Members of the evolutionary conserved PMT family of protein O-mannosyltransferases from distinct protein complexes among themselves. J. Biol. Chem. 278:12554-12562. [DOI] [PubMed] [Google Scholar]
- 24.Haft, R. F., M. R. Wessels, M. F. Mebane, N. Conaty, and C. E. Rubens. 1996. Characterization of cpsF and its product CMP-N-acetylneuraminic acid synthetase, a group B streptococcal enzyme that can function in K1 capsular polysaccharide biosynthesis in Escherichia coli. Mol. Microbiol. 19:555-563. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda, R., S. Nishimura, A. Nishikawa, and T. Shinoda. 1996. Production of agglutinating monoclonal antibody against antigen 8 specific for Cryptococcus neoformans serotype D. Clin. Diagn. Lab. Immunol. 3:89-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janbon, G. 2004. Cryptococcus neoformans capsule biosynthesis and regulation. FEMS Yeast Res. 8:765-771. [DOI] [PubMed] [Google Scholar]
- 27.Janbon, G., U. Himmelreich, F. Moyrand, L. Improvisi, and F. Dromer. 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42:453-469. [DOI] [PubMed] [Google Scholar]
- 28.Jungmann, J., and S. Munro. 1998. Multiprotein complexes in the cis Golgi of Saccharomyces cerevisiae with a-1,6-mannosyltransferase activity. EMBO J. 17:423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jungmann, J., J. C. Rayner, and S. Munro. 1999. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J. Biol. Chem. 274:6579-6585. [DOI] [PubMed] [Google Scholar]
- 30.Knauer, R., and L. Lehle. 1999. The oligosaccharyltransferase complex from yeast. Biochim. Biophys. Acta 1426:259-273. [DOI] [PubMed] [Google Scholar]
- 31.Kozel, T. R., S. M. Levitz, F. Dromer, M. A. Gates, P. Thorkildson, and G. Janbon. 2003. Antigenic and biological characteristics of mutant strains of Cryptococcus neoformans lacking capsular O-acetylation or xylosyl side chains. Infect. Immun. 71:2868-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuwayama, H., S. Obara, T. Morio, M. Katoh, H. Urushibara, and Y. Tanaka. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwon-Chung, K. J., and J. E. Bennett. 1984. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 120:123-130. [DOI] [PubMed] [Google Scholar]
- 34.Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester, F. S. Dietrich, and J. Heitman. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1:704-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lussier, M., A. M. Sdicu, and H. Bussey. 1999. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1426:323-334. [DOI] [PubMed] [Google Scholar]
- 36.McFadden, D. C., and A. Casadevall. 2001. Capsule and melanin synthesis in Cryptococcus neoformans. Med. Mycol. 39:19-30. [PubMed] [Google Scholar]
- 37.Moyrand, F., B. Klaproth, U. Himmelreich, F. Dromer, and G. Janbon. 2002. Isolation and characterization of capsule structure mutant strains of Cryptococcus neoformans. Mol. Microbiol. 45:837-849. [DOI] [PubMed] [Google Scholar]
- 38.Nicholas, K. B., H. B. J. Nicholas, and D. W. I. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. Embnew. News 4:14. [Google Scholar]
- 39.Perfect, J. R., S. D. R. Lang, and D. T. Durack. 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 101:177-194. [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sheng, S., and R. Cherniak. 1997. Structure of the 13C-enriched O-deacetylated glucuronoxylomannan of Cryptococcus neoformans serotype A determined by NMR spectroscopy. Carbohydr. Res. 301:33-40. [DOI] [PubMed] [Google Scholar]
- 42.Sherman, F. 1992. Getting started with yeast, p. 3-21. In C. Guthrie and G. R. Fink (ed.), Guide to yeast genetics and molecular biology, vol. 194. Academic Press, San Diego, Calif.
- 43.Skelton, M. A., R. Cherniak, L. Poppe, and H. van Halbeek. 1991. Structure of the de-O-acetylated glucuronoxylomannan from Cryptococcus neoformans serotype, as determined by 2D NMR spectroscopy. Magn. Res. Chem. 29:786-793. [Google Scholar]
- 44.Sommer, U., H. Liu, and T. L. Doering. 2003. An α-1,3-mannosyltransferase of Cryptococcus neoformans. J. Biol. Chem. 278:47724-47730. [DOI] [PubMed] [Google Scholar]
- 45.Strahl-Bolsinger, S., M. Gentzsch, and W. Tanner. 1999. Protein O-mannosylation. Biochim. Biophys. Acta 1426:297-307. [DOI] [PubMed] [Google Scholar]
- 46.Sudarshan, S., R. C. Davidson, J. Heitman, and J. A. Alspaugh. 1999. Molecular analysis of the Cryptococcus neoformans ADE2 gene, a selectable marker for transformation and gene disruption. Fungal Genet. Biol. 27:36-48. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vecchiarelli, A. 2000. Immunoregulation by capsular components of Cryptococcus neoformans. Med. Mycol. 38:407-417. [DOI] [PubMed] [Google Scholar]
- 50.Wickes, B. L., and J. C. Edman. 1995. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol. Microbiol. 16:1099-1109. [DOI] [PubMed] [Google Scholar]
- 51.Wickes, B. L., T. D. E. Moore, and K. J. Kwon-Chung. 1994. Comparison of the electrophoretic karyotypes and chromosomal location of ten genes in the two varieties of Cryptococcus neoformans. Microbiology 140:543-550. [DOI] [PubMed] [Google Scholar]
- 52.Wills, E. A., I. S. Roberts, M. Del Poeta, J. Rivera, A. Casadevall, G. M. Cox, and J. R. Perfect. 2001. Identification and characterization of the Cryptococcus neoformans phosphoisomannose isomerase-encoding gene, MAN1, and its impact on pathogenicity. Mol. Microbiol. 40:610-620. [DOI] [PubMed] [Google Scholar]
- 53.Zaragoza, O., B. C. Fries, and A. Casadevall. 2003. Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO2. Infect. Immun. 71:6155-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]