Abstract
The Saccharomyces cerevisiae spore is protected from environmental damage by a multilaminar extracellular matrix, the spore wall, which is assembled de novo during spore formation. A set of mutants defective in spore wall assembly were identified in a screen for mutations causing sensitivity of spores to ether vapor. The spore wall defects in 10 of these mutants have been characterized in a variety of cytological and biochemical assays. Many of the individual mutants are defective in the assembly of specific layers within the spore wall, leading to arrests at discrete stages of assembly. The localization of several of these gene products has been determined and distinguishes between proteins that likely are involved directly in spore wall assembly and probable regulatory proteins. The results demonstrate that spore wall construction involves a series of dependent steps and provide the outline of a morphogenetic pathway for assembly of a complex extracellular structure.
As a response to nitrogen starvation in the presence of a poor carbon source, MATa/MATα diploid cells of the baker's yeast Saccharomyces cerevisiae exit the cell cycle, undergo meiosis, and form haploid spores (18). These spores are a quiescent, stress-resistant cell type that can survive until nutrients are reintroduced. Much of the spores' resistance to environmental damage is provided by a specialized extracellular coat, the spore wall (41).
The spore wall is a stratified extracellular matrix that is more complex than the normal vegetative cell wall (15, 31, 40). The vegetative wall consists primarily of an inner layer (closest to the plasma membrane) of β-glucans interspersed with a small amount of chitin and an outer layer of heavily mannosylated proteins (mannans) (15, 31). By contrast, the spore wall consists of four distinct layers. The first two strata, an innermost layer composed primarily of mannan and a second layer of β1-3-linked glucans, are similar in composition to the vegetative wall but are reversed in position with respect to the spore plasma membrane (16). The outer portion of the spore wall is comprised of two polymers that are unique to the spore and confer much of the spore's resistance to environmental damage (4, 32). Immediately outside of the β-glucan is a layer composed primarily of chitosan, a glucosamine polymer synthesized by the deacetylation of chitin (7, 25, 32). Outside of the chitosan is a layer that consists largely of cross-linked tyrosine molecules (4-6).
In addition to being more complex than the cell wall, the spore wall is also unique in that it is constructed without a preexisting matrix to act as a template. Spore morphogenesis begins with the formation of prospore membranes within the cytoplasm of the cell that envelop each of the haploid nuclei produced by the meiotic divisions (26). Closure of the prospore membrane results in each nucleus being surrounded by a double membrane (26). The unit membrane closest to the nucleus serves as the plasma membrane of the spore. The spore wall is constructed in the luminal space between these two membranes (23) until lysis of the unit membrane farthest from the nucleus, which occurs during the process of spore wall assembly. With the exception of mannoproteins, none of the spore wall polymers are present in the lumen in significant amounts during prospore membrane growth, indicating that they are synthesized only after capture of the nuclei (43). Thus, assembly of the spore wall provides a model system in which to study the construction of a multilaminar extracellular matrix de novo.
Several mutants that are defective in some aspect of spore wall construction have been described. Many of these mutants display terminal phenotypes with heterogeneous spore wall defects (12, 17, 39, 42, 45), which has made their specific roles in assembly difficult to define. Other mutants, however, produce clearly defined effects. The chitin synthase gene CHS3, for example, is required for the synthesis of chitin as a precursor to the chitosan layer (32). Mutation of CHS3 causes the loss of both the chitosan and dityrosine layers. Similarly, the DIT1 and DIT2 genes are required for the synthesis of the dityrosine monomers that serve as precursors for construction of the dityrosine layers (3). Mutation of either of these genes results in spores that lack the outermost spore wall layer.
Mutation of the GIP1 gene also causes arrest at a discrete point in spore wall development (43). Sporulating gip1 cells envelop nuclei within prospore membranes but never deposit any spore wall material, suggesting that some signal is required to initiate spore wall synthesis. Further, in wild-type cells, spore walls are formed by the ordered synthesis of the different layers: first mannan, then β-glucan, then chitosan, and finally dityrosine (43). Again, these results indicate that the assembly of spore walls occurs through a coordinated series of steps.
We report here the identification and characterization of mutants defective in various aspects of spore wall synthesis. A systematic characterization of the defects in these mutants reveals discrete blocks at various stages in spore wall assembly. The phenotypes of these mutants define specific stages of spore wall construction and provide the outline of a pathway of spore wall assembly.
MATERIALS AND METHODS
Yeast strains and media.
Unless otherwise noted, standard methods and media were used (37). The strains used are listed in Table 1. All strains used in this study are in the fast-sporulating SK-1 strain background. To construct the sps2 sps22 double mutant, one allele of each gene was disrupted in the diploid MCY387 (9). This double heterozygote was then sporulated, and appropriate haploid segregants were mated to generate MCY417. Strain AC6 was constructed by crossing spores from strain MYA-1844 to AN117-16D (29). The resulting strain was sporulated and dissected, and a haploid gis1 segregant was again crossed to AN117-16D. Two haploid segregants from this cross were then mated to generate AC6. Strains ADY14, AC34, AC35, AC30, and AC25 were constructed by PCR-mediated gene knockout of AMA1 or PCR-mediated tagging with the 13Xmyc epitope of SPO73, SPO77, OSW1, and OSW2, respectively, in strains AN117-4B and AN117-16D (29) and mating of the resulting haploids to generate homozygous diploids.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| K8409 | MATa/MATα his3/his3 HO/HO LEU2::pURA3-tetR-GFP/LEU2::pURA3-tetR-GFP lys2/lys2 REC8::HA3-URA3/REC8::HA3-URA3 trp1/trp1 URA3::tetO224/URA3::tetO224 | 35 |
| MYA-1915 | As K8409, plus ama1Δ::HIS3MX6/ama1Δ::HIS3MX6 | 35 |
| MYA-1872 | As K8409, plus spo73Δ::HIS3MX6/spo73Δ::HIS3MX6 | 35 |
| MYA-1970 | As K8409, plus spo75Δ::HIS3MX6/spo75Δ::HIS3MX6 | 35 |
| MYA-1998 | As K8409, plus spo77Δ::HIS3MX6/spo77Δ::HIS3MX6 | 35 |
| MYA-2057 | As K8409, plus ssp2Δ::HIS3MX6/ssp2Δ::HIS3MX6 | 35 |
| MYA-2058 | As K8409, plus osw1Δ::HIS3MX6/osw1Δ::HIS3MX6 | 35 |
| MYA-2061 | As K8409, plus mum3Δ::HIS3MX6/mum3Δ::HIS3MX6 | 35 |
| MYA-1844 | As K8409, plus gis1Δ::HIS3MX6/gis1Δ::HIS3MX6 | 35 |
| MYA-1983 | As K8409, plus osw2Δ::HIS3MX6/osw2Δ::HIS3MX6 | 35 |
| MCY417 | MATa/MATα ura3/ura3 leu2/leu2 trp1/trp1 his3/his lys2/lys2 ade2/ADE2 can1/CAN1 | This study |
| cyh2/CYH2 his4::HIS3/his4 sps2::URA3/sps2::URA3 ycl048w::LEU2/yc1048w::LEU2 | ||
| AN262 | MATa/MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3ΔSK/his3ΔSK lys2/lys2 arg4-NspI/ARG4 hoΔLYS2/hoΔLYS2 rme1::LEU2/RME1 chs3ΔHIS3MX6/chs3 ΔHIS3MX6 | 8 |
| AN264 | MATa/MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3ΔSK/his3ΔSK lys2/lys2 arg4-NspI/ARG4 hoΔLYS2/hoΔLYS2 rme1::LEU2/RME1 dit1 ΔHIS3MX6/dit1 ΔHIS3MX6 | 8 |
| ADY14 | MATa/MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3ΔSK/his3ΔSK lys2/lys2 arg4-NspI/ARG4 hoΔLYS2/hoΔLYS2 rme1::LEU2/RME1 ama1 ΔC.gTRP1/ama1ΔC.gTRP1 | This study |
| AC6 | MATa/MATα ura3/ura3 LEU2/leu2 trp1::hisG/trp1::hisG his3/his3 lys2/lys2 arg4-NspI/ARG4 hoΔLYS2/hoΔLYS2 gis1ΔHIS3MX6/gis1ΔHIS3MX6 | This study |
| AC25 | MATa/MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3ΔSK/his3ΔSK lys2/lys2 arg4/ARG4 hoΔLYS2/hoΔLYS2 rme1::LEU2/RME1 OSW2::13Xmyc(HIS3)/ OSW2::13Xmyc(HIS3) | This study |
| AC30 | MATa/MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3ΔSK/his3ΔSK lys2/lys2 arg4/ARG4 hoΔLYS2/hoΔLYS2 rme1::LEU2/RME1 OSW1::13Xmyc(HIS3)/ OSW1::13Xmyc(HIS3) | This study |
| AC34 | MATa/MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3ΔSK/his3ΔSK lys2/lys2 arg4/ARG4 hoΔLYS2/hoΔLYS2 rme1::LEU2/RME1 SPO73::13Xmyc(HIS3)/ SPO73::13Xmyc(HIS3) | This study |
| AC35 | MATa/MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3ΔSK/his3ΔSK lys2/lys2 arg4/ARG4 hoΔLYS2/hoΔLYS2 rme1::LEU2/RME1 SPO77::13Xmyc(HIS3) | This study |
| NY501 | MATa/MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3ΔSK/his3ΔSK lys2/lys2 arg4-NspI/ARG4 hoΔLYS2/hoΔLYS2 rme1::LEU2/RME1 dit1ΔHIS3MX6/dit1 ΔHIS3MX6 | 43 |
Screen for ether-sensitive mutants.
The collection of knockout strains, obtained from the American Type Culture Collection arrayed in four 96-well microtiter dishes, were grown to saturation in yeast extract-peptone-dextrose (YPD) medium. The strains were then pinned onto YPD plates (one half of a microtiter dish per plate) and incubated at 30°C for 2 days, and each YPD plate was replica plated to two SPO (1% potassium acetate, 0.05% yeast extract, 0.05% glucose) plates. One SPO plate was incubated at 30°C, and one was incubated at 37°C. After 2 days of incubation, each SPO plate was replica plated to two YPD plates. One of these YPD plates was exposed to ether vapor for 5 min, and growth on both plates was examined after 24 h of incubation at 30°C. All candidates that showed ether-sensitive germination were repatched from the original YPD plates and retested twice to confirm the phenotype, and samples from the SPO plates were examined in the light microscope to determine whether visible spores were formed.
Quantitative ether and Zymolyase sensitivity assays.
For quantitative analysis of ether sensitivity, 100 μl of sporulated culture (approximately 107 asci) was washed once in distilled water (dH2O) and then resuspended in 667 μl of dH2O. Diethyl ether (333 μl) was added to this tube, and then at 2-min intervals 100 μl of cells was removed, rapidly diluted in YPD, and plated to determine the cell titer. To quantitate Zymolyase sensitivity, 100 μl of sporulated culture was washed once in dH2O and then resuspended in 990 μl of dH2O. Ten microliters of Zymolyase 100T (U.S. Biological, Swampscott, Mass.) at 10 mg/ml was added, and the cells were incubated at 37°C. At 10-min intervals, 100 μl of cells was removed, diluted in YPD, and plated to determine the titer of viable cells.
Plasmids.
The plasmid pHindIII-DIT1::lacZ, which carries a translational fusion of the start codon and ∼1 kb of the DIT1 promoter region to the lacZ gene, was provided by J. Segall (University of Toronto). The SPR3::lacZ reporter was pGK16 (43). For disruption of SPS2, plasmid p1884-3 was used (34). To disrupt SPS22, a 2.5-kb HindIII-XbaI fragment from a genomic DNA library plasmid (28) was subcloned into pBluescriptII KS(+) to create pMCB138. A SmaI-NheI LEU2 fragment from YdpL (2) was then used to replace an EcoRI (blunted)-SpeI segment of SPS22 to create pMCB142. This plasmid was digested with HindIII and XbaI before transformation.
β-Galactosidase assays.
Yeast strains were sporulated in liquid medium. Three aliquots were taken for each strain at each time point and assayed for β-galactosidase activity as described previously (41).
Quantitation of glucosamine and dityrosine.
Total glucosamine in spore walls was determined as described previously (46). Samples were hydrolyzed in 6 N HCl at 115°C for 14 h and dried in vacuo. The glucosamine content was determined with the Waters Pico-Tag amino acid analysis system according to the manufacturer's suggestions for amino sugars. To distinguish between chitin and chitosan, wall fractions were suspended in HNO2 prior to hydrolysis, since HNO2 destroys chitosan but leaves chitin intact. After 4 h at room temperature, insoluble material was removed by centrifugation, washed with water, hydrolyzed, and analyzed as described above.
For dityrosine analyses, strains were sporulated in liquid medium at a density of 107 cells/ml. After completion of sporulation, cells and medium were separated by centrifugation and analyzed separately. Cells (106) were hydrolyzed in 6 N HCl at 95°C for 3 h. Dried hydrolysates were dissolved in water and resolved by isocratic reverse-phase high-pressure liquid chromatography (HPLC), using an Agilent 1100 HPLC system with fluorescence detection and a Waters Nova-Pak C18 column as described previously (11). To determine the amount of dityrosine secreted to the medium, 100 μl of untreated medium was injected into the HPLC system, and the column (Waters Nova-Pak C18) was developed with a water-acetonitrile gradient (11). Concentrations of bisformyl dityrosine and its degradation product monoformyl dityrosine were determined by fluorescence detection. The sum of both substances was taken as the dityrosine concentration secreted by 106 cells.
Electron microscopy.
Cells were prepared for transmission electron microscopy (TEM) and scanning electron microscopy (SEM) essentially as described previously (8). For TEM analysis, all strains except MCY417 were visualized by using osmium and thiocarbohydrazide staining. Cells were sporulated in liquid medium and then fixed for 1 h in 3% glutaraldehyde in cacodylate buffer, washed once in cacodylate buffer (0.1 M sodium cacodylate [pH 7.4], 5 mM calcium chloride), resuspended in 1% osmium tetroxide and 1% potassium ferricyanide in cacodylate buffer, and incubated for 30 min at 23°C. The cells were then washed four times in dH2O, resuspended in 1% thiocarbohydrazide in water, and incubated for 5 min at 23°C. Cells were again washed in dH2O, incubated in 1% osmium tetroxide and 1% potassium ferricyanide in cacodylate buffer for an additional 5 min, and washed again in dH2O. The cells were then incubated in saturated uranyl acetate for 2 h and finally dehydrated through a graded series of acetone washes. The dehydrated samples were embedded in Epon 812 and sectioned, and images were collected on a JEOL 1200EX microscope at 80 kV. For strain MCY417, cells were prepared similarly except that a 1-h incubation in 2% potassium permanganate was used in place of the osmium tetroxide, potassium ferricyanide, and sodium thiocarbohydrazide incubations.
For SEM analysis of spores, cells were first grown to mid-log phase in YP-acetate and spheroplasted by using Zymolyase 100T, and spheroplasts were then sporulated in osmotically stabilized sporulation medium (1.4% potassium acetate, 0.7 M sorbitol). After sporulation, spheroplasts were washed once in 0.5% sodium dodecyl sulfate to remove the ascal membranes, rinsed once in dH2O, and then adhered to polylysine-coated glass coverslips. The cells on the coverslips were fixed in 3% glutaraldehyde in cacodylate buffer for 1 h at 23°C, stained for 1 h at 23°C in 1% osmium tetroxide and 1% potassium ferricyanide in cacodylate buffer, washed with dH2O, and then dehydrated by 10-min incubations in a graded acetone series. The coverslips were then critical point dried and sputter coated with 4-nm gold particles. Images were collected in a LEO1550 SEM at 2.5 kV using an in-lens detector.
Light microscopy.
The mannan, β-glucan, and chitosan layers of the spore wall were visualized using fluorescein isothiocyanate-concanavalin A (FITC-ConA), anti β-glucan antibodies and Calcofluor White, respectively, as described previously (43). Briefly, sporulated cells were fixed in formaldehyde and then spheroplasted with Zymolyase. The spheroplasts were incubated for 5 min in SHA buffer (1 M sorbitol, 0.1 M HEPES, pH 7.5, 5 mM sodium azide) containing 0.1% Triton X-100 and for 10 min in SHA buffer containing 0.2 mg of Calcofluor White (Sigma Aldrich) per ml. The cells were then adhered to polylysine-coated slides, treated with ice-cold methanol for 6 min, and rinsed in acetone for 30 s. The slides were then blocked for 1 h in 5% bovine serum albumin in phosphate-buffered saline (PBS) and incubated overnight with a 1:300 dilution of an anti-β-1,3-glucan monoclonal antibody (Biosupplies, Parkville, Australia). Wells were then rinsed with PBS, incubated for 1 h with a 1:400 dilution of Alexa 546 goat anti-mouse antibody (Molecular Probes) and 0.1 mg of FITC-ConA (Sigma Aldrich) per ml, washed again in PBS, and finally mounted under a coverslip for examination in the microscope. To localize the 13Xmyc-tagged proteins, essentially the same protocol was used except that the primary antibody used was an undiluted tissue culture supernatant containing the anti-myc monoclonal antibody 9E10, the secondary antibody was Alexa 488 goat anti-mouse (Molecular Probes), and no Calcofluor White or FITC-ConA was included.
RESULTS
Ether screen of the Rabitsch mutant collection.
Rabitsch et al. (35) constructed a set of ∼300 strains, each with a different meiotically induced gene deleted, and screened microscopically for mutations causing defects in chromosome segregation or spore formation. Approximately 10% of the strains displayed a phenotype in one of these two assays (35). Their screen, however, would not identify genes required for proper spore wall construction that still produced visible spores. In order to identify such mutants, we reanalyzed this set of strains for sporulation defects by using an alternative assay, sensitivity to ether vapor (36). All of the mutants that displayed poor germination on the ether-treated plates were examined directly for sporulation with the light microscope. This analysis identified 38 mutants that displayed an apparent sensitivity to ether. Included in this set were 28 meiosis and sporulation mutants identified in the original screen by Rabitsch et al. (35). In addition, 10 new mutants that produced visible spores that were sensitive to ether were identified (Table 2).
TABLE 2.
Mutants identified in the ether sensitivity screen
| Group (reference) | Open reading frame | Gene name |
|---|---|---|
| Mutants also identified in | YBR045c | GIP1 |
| previous cytological | YBR233w | PBP2 |
| screen (35) | YCR010c | ADY2 |
| YCR086w | CSM1 | |
| YDL149w | APG9 | |
| YDL239c | ADY3 | |
| YDR065W | ||
| YDR104c | SPO71 | |
| YER046w | SPO73 | |
| YFR021w | AUT10 | |
| YGL170c | SPO74 | |
| YGL183c | MND1 | |
| YGR225w | AMA1 | |
| YHL024w | RIM4 | |
| YHR184w | SSP1 | |
| YHR185c | ADY1 | |
| YIL073c | SPO22 | |
| YIR025w | MND2 | |
| YLL005c | SPO75 | |
| YLR341w | SPO77 | |
| YML066c | SMA2 | |
| YMR048w | CSM3 | |
| YOL091c | SPO21 | |
| YOR177c | MPC54 | |
| YOR242c | SSP2 | |
| YPL018w | CTF19 | |
| YPL027w | SMA1 | |
| YPL121c | MEI5 | |
| Mutants that form ether- | YCL048w | SPS22 |
| sensitive spores | YDR096w | GIS1 |
| YDR260c | SWM1 | |
| YDR326c | ||
| YLR054c | OSW2 | |
| YLR238w | FAR10 | |
| YOR255w | OSW1 | |
| YOR279c | RFM1 | |
| YOR298w | MUM3 | |
| YPL077c |
These new mutants likely carry mutations that cause defects in assembly of the spore wall that lead to sensitivity to ether vapor but are not so severe as to block the appearance of refractile spores. One of the mutants, the swm1 mutant, has been previously described to have a spore wall phenotype, though in the published study swm1 mutants did not make visible spores (45). This may be due to the different strain backgrounds used in this work (SK-1) and the previous study (W303). Four of the mutants (the gis1, mum3, yor255c, and ylr054c mutants) were chosen for further examination. YOR255c and YLR054c have been designated OSW1 and OSW2 (for outer spore wall), respectively. In addition, the mutant gene of one of the other ether-sensitive mutants, ycl048w (now designated SPS22), is homologous to a second sporulation-induced gene, SPS2 (34). The double mutant strain was constructed, and this proved to have a more severe sporulation defect, lacking visible spores. The sps2 sps22 double mutant was also analyzed further as described below.
Finally, the sporulation defect in the previously described “no spore and ascus formation” class (35), the members of which have normal meiotic divisions but produce no visible spores, was examined. These strains were transformed with a plasmid carrying a green fluorescent protein (GFP) marker for the prospore membrane (27), sporulated, and analyzed both for progression through meiosis by DAPI (4′,6′-diamidino-2-phenylindole) staining and for prospore membrane development. Of these previously reported mutants there were six, the ama1, gip1, spo73, spo75, spo77, and ssp2 mutants, that made no visible spores but had normal prospore membranes, suggesting that these mutants are required after prospore membrane formation for proper spore wall assembly. Consistent with this, two of these mutants, the gip1 and ssp2 mutants, have been previously described to have spore wall defects (38, 43).
In sum, the screen identified 38 mutants with reduced germination after the spores were exposed to ether. Of these, 16 were candidates for having roles in spore wall morphogenesis, including 13 newly identified as having spore wall phenotypes.
Characterization of spore wall defects.
To more precisely define the roles of these gene products in spore wall assembly, the spore wall mutants were examined in several ways. All of the mutants identified as having normal prospore membranes but not producing visible spores (the ama1, spo73, spo75, spo77, ssp2, and sps2 sps22 mutants), as well as mutants that made strongly ether-sensitive spores (the gis1, osw1, osw2, and mum3 mutants), were examined. For comparison, mutations in chs3, which removes both the outer chitosan and dityrosine layers, and in dit1, which removes only the dityrosine layer, were examined. To directly visualize the spore walls, each mutant was sporulated and its terminal phenotype was examined in the transmission electron microscope. TEM micrographs of each of the mutants are shown in Fig. 1. Because differentiation of the spore wall layers in the TEM can be problematic, the presence or absence of individual layers was also examined by using fluorescence assays for mannan, β-glucan, and chitosan. The results of these assays are given in Table 3.
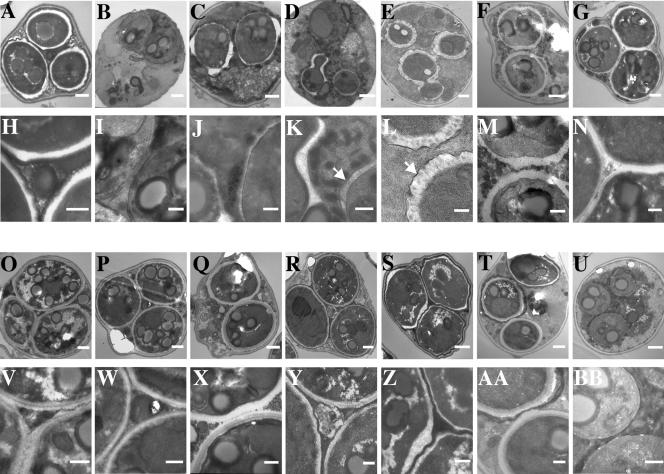
FIG. 1.
Transmission electron micrographs of spore wall mutants. Wild type (A, H); ama1 (B, I); spo73 (C, J); spo77 (D, K); sps2 sps22 (E, L); ssp2 (F, M); chs3 (G, N); osw1 (O, V); mum3 (P, W); gis1 (Q, X); dit1 (R, Y); osw2 (S, Z); spo75 (T, U, AA, BB). Panels H to N and V to BB are higher-magnification images of the cells in panels A to G and O to U, respectively. Bars: panels A to G and O to U, 500 nm; panels H to N and V to BB, 200 nm. The arrows in panels K and L indicate the outer membrane derived from the prospore membrane.
TABLE 3.
Fluorescence assays of spore wall mutants
| Mutant | Mannana | β-Glucanb | Chitosanc | Protein notes |
|---|---|---|---|---|
| Wild type | Yes | Yes | Yes | |
| ama1 | Yes | No | No | Cdc20/Fizzy family member |
| spo73 | Yes | No | No | Dysferlin motif |
| spo77 | Yes | Weak | Weak | |
| sps2 sps22 | Yes | NDd | Yes | Predicted secreted, GPI-anchored protein |
| ssp2 | Yes | Yes | No | |
| chs3 | Yes | Yes | No | Chitin synthase |
| osw1 | Yes | Yes | Weak | |
| mum3 | Yes | Yes | Weak | Homology to acyltransferases |
| gis1 | Yes | Yes | Yes | Cys2-His2 Zn2+ finger |
| dit1 | Yes | Yes | Yes | N-Formyltransferase |
| osw2 | Yes | Yes | Yes | Distantly related to ketopantoate reductase |
| spo75 | Yes | Yes, variable | Yes, variable | Eleven predicted transmembrane domains |
Determined by staining with FITC-ConA.
Determined by staining with anti-β-1,3-glucan antibodies.
Determined by staining with Calcofluor White.
ND, not determined.
Finally, direct biochemical measurements of the specific spore wall components glucosamine and dityrosine were made (Table 4). Total glucosamine in the spore wall fraction was measured in the wild type and in each of the mutant strains, and the amounts were normalized as a percentage of the wild-type amount. Also, for each strain, the fraction of glucosamine present in the form of chitosan, as opposed to chitin, was determined. In all of the mutants with significant amounts of glucosamine, the glucosamine was found predominantly as chitosan, indicating that none of the mutations affect the activity of the chitin deacetylases. For dityrosine, the total amount present in both the cells and culture medium and the amount present only in the cellular fraction were quantitated and normalized to those of the wild type as 100%. Analysis of the culture medium is based on the finding that in most spore wall mutants, unincorporated dityrosine does not remain in the spores but is secreted to the medium (11). Thus, this assay allows us to distinguish between mutants such as the dit1 mutant that fail to synthesize dityrosine and mutants such as the chs3 mutant that synthesize dityrosine but fail to incorporate it into the spore wall. The results for each mutant are summarized below.
TABLE 4.
Biochemical assays of spore wall mutants
| Mutant | % Glucosamine content of spore wallsa | % Glucosamine in chitosanb | % Total dityrosinec | % Cellular dityrosined |
|---|---|---|---|---|
| Wild type | 100 | 92 | 100 | 100 |
| ama1 | 0 | NDe | 8 | 1 |
| spo73 | 0 | ND | 5 | 0.5 |
| spo77 | 15 | ND | 140 | 12 |
| ssp2 | 5 | ND | 84 | 7 |
| chs3 | 44 | 3 | ||
| osw1 | 35 | 89 | 18 | 3 |
| mum3 | 15 | 83 | 37 | 5 |
| gis1 | 31 | 91 | Trace | Trace |
| dit1 | 85 | 90 | ||
| osw2 | 115 | 90 | 95 | 77 |
| spo75 | 15 | ND | 70 | 17 |
Relative amount of glucosamine present in the spore wall fraction.
Fraction of spore wall glucosamine present as chitosan.
Relative amount of dityrosine present in cells plus the culture medium.
Relative amount of dityrosine present in the cellular fraction.
ND, not determined.
(i) ama1.
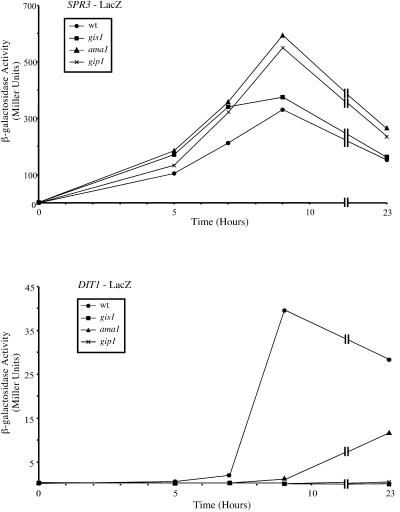
ama1 mutants underwent arrest very early in spore wall formation. In TEM, the cells displayed little or no expansion of the prospore membrane lumen, suggesting that spore wall precursors were not being delivered to this compartment (Fig. 1B). Consistent with this, both β-glucan and chitosan were absent as determined by the fluorescence assays. In the biochemical analyses, glucosamine, which is indicative of chitosan synthesis, was absent and dityrosine was greatly reduced. In this regard, the ama1 mutant resembles the previously described gip1 mutant, which undergoes arrest at the onset of spore wall synthesis (43). In addition to these phenotypes, gip1 mutants fail to induce expression of the DIT1 gene and have defects in localization of the septin proteins during prospore membrane growth (43). By contrast, ama1 mutants, despite the lack of dityrosine, showed delayed but significant expression of a DIT1::lacZ fusion and displayed normal septin localization (Fig. 2) (A. M. Neiman, unpublished observations). Further, complementary overexpression studies failed to reveal any genetic interactions between GIP1 and AMA1 (H. Tachikawa and A. M. Neiman, unpublished observations). Thus, AMA1 is required for the initiation of spore wall assembly, perhaps on a pathway independent from that for GIP1.
FIG. 2.
DIT1::lacZ expression in ama1 and gis1 mutants. Strains AN120 (wild type [wt]), ADY14 (ama1), AC6 (gis1), and NY501 (gip1) were transformed with plasmids carrying an SPR3::lacZ or a DIT1::lacZ reporter gene. The strains were sporulated in liquid and, at intervals, assayed for β-galactosidase activity. Upper panel, time course of the induction of the middle sporulation reporter SPR3::lacZ. Lower panel, time course of the induction of DIT1::lacZ.
(ii) spo73.
From both the fluorescence and biochemical data, spo73 mutants appear to be very similar to ama1 mutants, lacking β-glucan, chitosan, and dityrosine. However, the TEM images revealed deposition of a thin rim of spore wall material around the prospores, indicating that spo73 mutants progress farther than ama1 mutants. The absence of β-glucan staining in the fluorescence assay suggests that the spore wall material seen in spo73 cells is mannan. Interestingly, the outer membrane derived from the prospore membrane is absent in spo73 cells (Fig. 1J).
(iii) spo77.
The TEM analysis indicates that spo77 mutants progress farther than spo73 mutants, displaying a greater deposition of spore wall material. In the fluorescence assays, spo77 cells stained weakly with probes for both β-glucan and chitosan, indicating deposition of some amount of both polymers. Consistent with this, a measurable amount of glucosamine was present in the spo77 spore walls. While spo77 cells synthesized as much dityrosine as wild-type cells, only a small amount was retained in the spore wall. Further, despite the presence of these spore wall precursors, organized strata of these polymers were not seen in TEM. Additionally, a small fraction of the spo77 spores display an abnormal, blebbed morphology (Fig. 1D), and, unlike in spo73 cells, the outer membrane derived from the prospore membrane is intact in spo77 cells (Fig. 1K).
(iv) sps2 sps22.
The sps2 sps22 double mutant strain produced aberrant, blebbed spores that were surrounded by an extensive, very abnormal spore wall. The spore walls in these cells have a mottled appearance, suggesting that the different layers are intermingled rather than stratified. The fluorescence data also indicate that both β-glucan and chitosan are present in the mutant. As in spo77 cells, the outer membrane derived from the prospore membrane is present.
(v) ssp2.
In ssp2 cells, both the mannan and β-glucan layers were visible in TEM, but the outer chitosan and dityrosine layers were not. The absence of these outer layers was also indicated by the fluorescence and biochemical data. ssp2 mutants stained brightly with the anti-β-glucan antibodies but not with Calcofluor White. Glucosamine levels were low, suggesting that the lack of staining is due to a defect in chitosan synthesis. Consistent with the absence of chitosan, ssp2 cells synthesized dityrosine but released it into the culture medium. Thus, ssp2 mutants appear to arrest spore wall assembly after synthesis of the β-glucan layer. The β-glucan layer of ssp2 spores must not be entirely wild type, however, because these spores are not refractile in the light microscope (38). The outer membrane derived from the prospore membrane was absent in these cells and in all of the mutants described below.
(vi) chs3.
The chs3 mutant displayed normal-looking mannan and β-glucan layers. As expected from previous studies (32), the chitosan and dityrosine layers were completely absent. These spores were visible in the light microscope, indicating that refractility does not require the outer spore wall layers. Surprisingly, total synthesis of dityrosine was also slightly reduced in the chs3 mutant.
(vii) osw1.
In TEM, osw1 mutants appeared very similar to chs3 mutants in that the chitosan and dityrosine layers were absent from the spores. However, unlike chs3 mutants, osw1 mutants still synthesized chitosan as evidenced by weak staining with Calcofluor White and the presence of significant amounts of glucosamine. Thus, the defect in osw1 mutants may be in the organization, rather than the synthesis, of chitosan.
(viii) mum3.
mum3 mutants appeared similar in most respects to osw1 mutants except that glucosamine levels were lower.
(ix) gis1.
gis1 mutants displayed a slightly heterogeneous phenotype in TEM, with some cells lacking both chitosan and dityrosine and some lacking only the dityrosine layer. In fluorescence assays, gis1 mutants stained well with Calcofluor White, and significant amounts of glucosamine were present in the spore wall, so the absence of a chitosan layer presumably reflects an assembly defect. Additionally, dityrosine was almost undetectable in the gis1 cells, as was also seen in dit1 mutants. Because GIS1 has been previously shown to encode a transcription factor (14, 33), we tested whether GIS1 was required for transcription of the DIT1 gene. Consistent with the absence of dityrosine, DIT1::lacZ was not expressed in the gis1 mutant, although the induction of the middle sporulation reporter SPR3::lacZ was unaffected (Fig. 2).
(x) dit1.
As described previously (3), dit1 mutants had normal mannan, β-glucan, and chitosan layers but completely lacked dityrosine.
(xi) osw2.
Surprisingly, osw2 mutants appeared nearly indistinguishable from the wild type in all assays used.
(xii) spo75.
Unlike the other mutants examined, which displayed fairly homogeneous cytological phenotypes, spo75 mutants displayed a very heterogeneous phenotype. The phenotypes of spores in spo75 cells ranged from those that had little or no spore wall material to ones that had a wild-type appearance (Fig. 1T and U). Prospores with different levels of spore wall assembly were visible within the same ascus. This heterogeneity was also reflected in the fluorescence and biochemical data.
Comparison of ether and Zymolyase sensitivities.
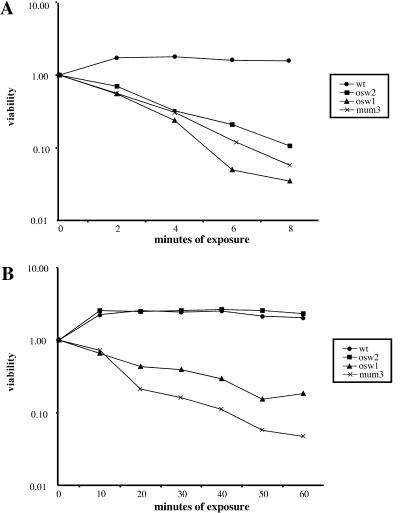
All of the mutants described were isolated on the basis of sensitivity to ether vapor in a patch assay. To examine this phenotype more closely, the ether sensitivities of several mutants that made visible spores were quantitated. As a comparison, the sensitivity of the spores to a different treatment, digestion by the glucanase Zymolyase, was also measured. As shown in Fig. 3, all of the mutants tested were at least 10-fold more sensitive to ether than were wild-type spores. Similarly, almost all of the mutants showed greatly increased sensitivity to Zymolyase. The sole exception to this pattern was osw2 cells, which were sensitive to ether but as resistant to Zymolyase digestion as were wild-type cells. These results suggest that the spore walls of osw2 mutants can provide resistance to large molecules such as Zymolyase but not to small molecules like ether.
FIG. 3.
Quantitation of ether and Zymolyase sensitivities in outer spore wall mutants. (A) Ether treatment. Spores of each strain were exposed to ether for the indicated times and then plated on rich medium to determine the titer of viable cells. For each strain viability is expressed as a fraction of the viable cells present at time zero. (B) Zymolyase treatment. Spores were exposed to Zymolyase, and titers were determined as for panel A.
Effects of the mutants on interspore bridges.
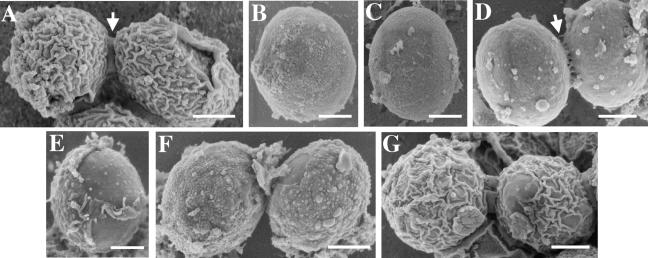
In a recent study, we demonstrated that the outer spore wall layers of adjacent spores in an ascus are connected by bridges that are composed partly of chitosan (8). In the absence of bridges, individual spores disperse when released from the ascus (8). To examine the effects of the new spore wall mutants on bridge formation, the mutants that produced visible spores were examined by SEM (Fig. 4). As expected from the TEM results, osw1 and mum3 mutants displayed a surface texture similar to that of chs3 mutants, suggesting that little or no chitosan layer overlying the β-glucan is formed in these mutants. Also similar to chs3 mutants, osw1 mutants lacked bridges and the spores were dispersed. By contrast, spores in the mum3 mutant were connected by vestigial bridges (Fig. 4D). The fact that mum3 mutants retain some bridge material even though they display less residual wall glucosamine than osw1 spores (Table 4) suggests that osw1 may play a more direct role in bridge construction.
FIG. 4.
Scanning electron micrographs of spore wall mutants. Wild type (A); chs3 (B); osw1 (C); mum3 (D); gis1 (E); dit1 (F); osw2 (G). Bars, 500 nm. The arrows in panels A and D show positions of the interspore bridges.
As in TEM, the gis1 mutant showed a heterogeneous phenotype in SEM, including individual spores that seemed to have only partial chitosan coats (Fig. 4E). In this respect, the gis1 phenotype is more severe than that of dit1 mutants, and thus the failure to express DIT1 cannot fully account for the gis1 phenotypes. Also consistent with the TEM analyses, osw2 mutants appeared similar to the wild type in the SEM images.
Localization of gene products.
Our results identify several new genes involved in spore wall assembly. To gain more insight into the function of these gene products, we sought to identify their subcellular localization. Ssp2p has been previously reported to localize to the spore wall (38), and Gis1p has been previously localized to the nucleus (13). Tagging of Sps2 and Sps22 was not attempted because these proteins have predicted amino-terminal signal sequences and carboxy-terminal GPI (glycophosphatidylinositol) anchor signals, both of which are cleaved during protein processing. However, their Schizosaccharomyces pombe ortholog has been reported to localize to the spore wall (44). For Ama1p, a C-terminal GFP fusion showed a nuclear localization (Neiman, unpublished observations). However, the fusion was nonfunctional, so the significance of this localization is unclear. For SPO73 and SPO77, GFP fusions were functional but no localizations were detectable. Therefore, carboxy-terminal fusions to a 13Xmyc epitope were constructed for the remaining genes, and the proteins were localized by immunofluorescence. Two of the 13Xmyc fusions, those with MUM3 and SPO75, were undetectable in immunofluorescence; the localizations of the other proteins are shown in Fig. 5.
FIG. 5.
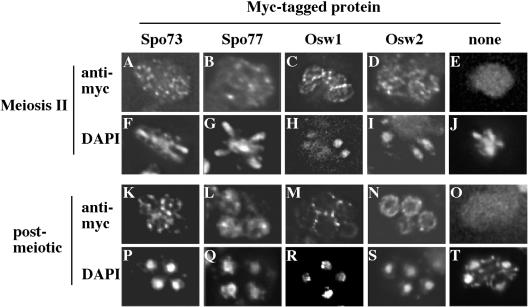
Localization of Spo73p, Spo77p, Osw1p, and Osw2p. Fluorescence micrographs of sporulating cells expressing Spo73-myc (A, F, K, P); Spo77-myc (B, G, L, Q); Osw1-myc (C, H, M, R); Osw2-myc (D, I, N, S); or no myc-tagged protein (E, J, O, T) are shown. A to E and K to O, anti-myc staining. F to J and P to T, corresponding DAPI staining of cells in panels A to E and K to O, respectively.
In meiosis II cells, the Spo73-myc protein displayed a punctate localization throughout the cytosol. In postmeiotic cells, these Spo73-myc puncta were organized around the periphery of the spore. By contrast, Spo77-myc protein localized diffusely throughout the cell at all stages of sporulation. The Osw1-myc and Osw2-myc proteins displayed distinct localizations. During meiosis II, both proteins were localized around the prospore membrane, with Osw1-myc found in a more uniform distribution than Osw2-myc. However, in postmeiotic cells, Osw2-myc was found within the spore cytoplasm, whereas Osw1-myc appears to be localized at the spore wall, predominantly at sites of spore-spore contact. The apparent enrichment of Osw1-myc at sites of spore wall contact is particularly interesting in light of the absence of interspore bridges in the osw1 mutant.
DISCUSSION
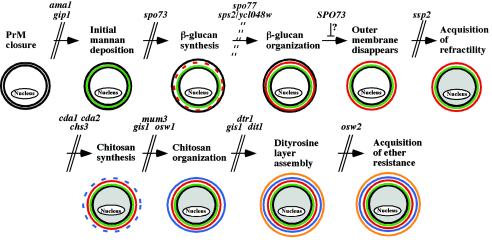
We present the identification of mutants defective in spore wall synthesis from a collection of strains with sporulation-induced genes deleted. A systematic cytological and biochemical analysis of 10 mutants, including 9 newly demonstrated to have spore wall defects, was performed. Given that several previously described spore wall mutants display heterogeneous defects in spore wall assembly (12, 17, 39, 42, 45), it was something of a surprise to find that most of the mutants analyzed here displayed homogeneous defects suggesting that they were arrested at specific stages in assembly of the spore wall. A previous study has established that the spore wall layers are deposited in the following temporal sequence: mannan, β-glucan, chitosan, and dityrosine (43). With that general sequence, the mutant phenotypes described here define an overall morphogenetic pathway of spore wall assembly (Fig. 6). The steps of assembly thus defined are as follows.
FIG. 6.
Pathway of spore wall assembly. All of the mutants indicated were characterized in this work, except for the gip1 (43), cda1 cda2 (7, 25), and dtr1 (11) mutants. Parallel lines indicate a block at a specific step in assembly caused by the indicated mutation. Dashed lines indicate that the mutants are defective in that step but that the mutations do not cause an arrest in the pathway. The diagrams depict the appearance of the spores at each stage in assembly. Black lines, membrane; green, mannan; red, β-glucan; blue, chitosan; orange, dityrosine.
Initiation of spore wall deposition.
The action of AMA1 is required to initiate spore wall assembly. Mutants with deletion of the gene have normal prospore membrane synthesis but deposit little or no spore wall material. The previously described GIP1 gene is also required at this stage. Although both of these mutations block the onset of spore wall formation, their phenotypes differ in some details, such as the expression of the DIT1 gene. It is possible, therefore, that they represent two different regulatory mechanisms. For example, GIP1 has been suggested to link the onset of spore wall synthesis to closure of the prospore membrane (43). By contrast, the homology of AMA1 to known cell cycle regulators suggests that it may play a role in linking spore wall synthesis to meiotic exit.
Deposition of mannoproteins.
Mannoproteins are present from the earliest stages of prospore membrane formation (43). However, additional deposition occurs before the onset of β-1,3-glucan synthesis. This step is inferred from the phenotype of spo73 mutants, which are seen to deposit some spore wall material but are without any β-glucan, chitosan, or dityrosine, as shown by both the fluorescence and biochemical analyses.
Organization of the β-glucan layer.
Once glucan synthesis is triggered, SPO77 and a redundant pair of genes, SPS2 and SPS22, are required to organize the glucans into a coherent layer. Presumably as a result of improper β-glucan organization, spores in both of these mutants (occasionally in spo77 and often in sps2 sps22) display a blebbed phenotype in which the spores are not round but have bud-like appendages. We have not seen this phenotype in any of the other mutants. Deposition of spore wall material is more extensive in the sps2 sps22 cells, but the spore wall layers in these cells appear mixed rather than stratified as in the wild type. In both the blebbing of the spores and the mottled appearance of the spore wall, sps2 sps22 cells closely resemble the phenotypes of spores in S. pombe strains carrying a mutation of the SPS2/SPS22 ortholog meu10 (44). This family of proteins thus seems to play a conserved role in spore wall assembly, possibly in creating a discrete layer of β-1,3-glucan, which is found in the spore walls of both yeasts (22, 24, 40). Unlike most of the other mutations we characterized, spo77 and sps2 sps22 do not appear to arrest the pathway of spore wall assembly. Despite the absence of an organized β-glucan layer, these cells proceed to synthesize chitosan and dityrosine but cannot arrange them into coherent layers.
Disappearance of the outer membrane.
At some point during or after the synthesis of the β-glucan layer, but before the initiation of chitosan synthesis, the outer membrane derived from the prospore membrane lyses. This membrane disappears abnormally early in the spo73 mutant, suggesting the possibility that SPO73 is important in maintaining the integrity of the outer membrane. The Spo73 protein contains a partial copy of a dysferlin motif. Although of unknown function, this motif is found in multiple copies in the dysferlin protein. Mutations in the dysferlin gene cause degenerative muscle disease (21). The dysferlin protein has been shown to be involved in the repair of membrane damage, protecting muscle cells from inappropriate lysis (1, 20), an interesting parallel to the spo73 phenotype.
Acquisition of refractility.
After the organization of the β-1,3-glucan polymers into a discrete layer but before the assembly of the chitosan layer, the spores become visible by phase-contrast microscopy. Deletion of SSP2 blocks spore wall assembly after synthesis of the β-glucan layer but before chitosan synthesis. Mutation of the chitin synthase gene CHS3, which catalyzes the formation of the chitosan polymer, is also blocked at this step (32). However, ssp2 and chs3 mutants differ in that chs3 mutants produce visible spores and ssp2 mutants do not. Thus, these results define an SSP2-dependent step at the end of β-glucan synthesis: the acquisition of refractility in the light microscope.
Synthesis of chitosan.
Chitosan is synthesized by the combined actions of Chs3p and Cda1/2p (7, 25, 32).
Organization of chitosan.
Two of the mutants, the mum3 and osw1 mutants, appear to be defective in the assembly of the chitosan layer. In the fluorescence and biochemical analyses these mutants incorporate a reduced amount of glucosamine into their spore walls. However, the EM analyses show that neither mutant produces an organized chitosan layer. In addition, these mutants, particularly the osw1 mutant, display defects in interspore bridge formation, providing support for the idea that bridges are connections between the chitosan layers of adjacent spores (8).
Synthesis of the dityrosine layer.
Previous studies have identified Dit1p and Dit2p as spore-specific proteins required for the production of the dityrosine monomers and the Dtr1 protein as a transporter necessary to export the monomers from the spore cytoplasm to the spore wall (3, 4, 11). We identified one additional mutant, the gis1 mutant, which is defective in dityrosine production. GIS1 encodes a zinc finger family transcription factor (3, 4), so its role in synthesis of the dityrosine layer is likely indirect. GIS1 is required for transcription of the DIT1 gene (Fig. 2). However, the gis1 phenotype is more extreme than that of dit1, with some cells lacking both the chitosan and dityrosine layers, suggesting that GIS1 may regulate additional genes that are important for spore wall assembly. While assembly of the dityrosine into a layer requires the prior formation of a chitosan layer, synthesis and export of the dityrosine molecules require only that spore wall formation proceed past the SPO73-dependent step.
Acquisition of ether resistance.
Mutants with deletions of OSW2 appear like the wild type in every biochemical and cytological assay. Spores formed in this mutant are as resistant to Zymolyase digestion as wild-type spores are. However, they are extremely sensitive to ether vapor. OSW2 thus seems to be required for a very late step in spore wall synthesis, perhaps in completion of the dityrosine layer, which is required for the acquisition of ether resistance.
One implication of this pathway is that spore wall assembly consists of a series of dependent steps. This is true in the sense that the β-glucan layer must be properly assembled for the chitosan layer to form, and, in turn, the chitosan layer must be complete for the dityrosine layer to assemble. However, this is also true in the sense that if some early stages of spore wall assembly are incomplete, then the synthesis of the precursors for later stages of assembly is reduced. For example, in the ssp2 mutant, β-glucan assembly was not properly completed and no chitosan staining or glucosamine production was seen, suggesting that activation of Chs3p cannot proceed in this mutant. Similarly, in the early ama1 and spo73 mutants, no production of any of the outer spore wall polymers was seen. Genes such as these, in which mutations lead to discrete arrest points, are likely required either for the completion of a specific step or for signaling the completion of one step and triggering the start of the next. In either case, there must be some monitoring system to ensure proper order of assembly. The genes identified here may be components of this regulatory system.
As noted earlier, several previously described mutants that are defective in spore wall assembly display heterogeneous defects in spore wall construction. In particular, the protein kinase genes MPK1, CAK1, SPS1 and SMK1 are reported to have such heterogeneous phenotypes (12, 17, 39, 42). One of the mutants we have characterized, the spo75 mutant, also displays a heterogeneous phenotype. In spo75 cells, the spore wall phenotype in any individual spore ranged from the complete absence of wall components to wild-type spore wall formation. Differences in spore wall development between different spores in the same ascus were readily apparent (Fig. 1). One possibility is that mutants with heterogeneous phenotypes are involved in regulatory pathways coordinating the stages of spore wall assembly. In these mutants, because the order of deposition is uncontrolled, each individual spore reaches a different level of spore wall maturation. That several of the genes with heterogeneous phenotypes encode protein kinases is consistent with a regulatory role. Also, the observation that a graded series of hypomorphic smk1 alleles display discrete, rather than heterogeneous, spore wall defects suggests that SMK1 might be involved in signaling at multiple steps in spore wall assembly (46). SPO75 encodes a protein with multiple transmembrane domains and so could function as a transmembrane sensor in one or more of these hypothetical pathways.
For the remaining mutants described here, their specific roles in spore wall assembly remain unclear but they may be grouped broadly into two potential classes: those likely involved in regulation of assembly and those which may participate directly in the assembly process. In the first group are Ama1p, the transcription factor Gis1p, and the apparently cytoplasmic proteins Spo73p and Osw2p. The predicted GPI-anchored proteins Sps2p and Sps22p are more likely to be directly involved in wall assembly, as are the spore wall-localized proteins, Osw1p and Ssp2p. Mum3p has homology to a family of acyltransferases (30), suggesting that it has an enzymatic activity that plays a role in assembly of the outer spore wall.
The localization of Osw1p and Ssp2p to the spore wall is also of interest, because neither of these proteins contains predicted signal sequences or transmembrane domains to indicate that they traffic to the spore wall via the secretory pathway. An alternative possibility is that these proteins may reach the spore wall by being retained in the ascal cytoplasm at the time of prospore membrane closure. They could then gain access to the spore wall when the outer membrane is broken down. It has been reported that specific mRNAs are enriched in the ascal cytoplasm (19). Possibly, these could represent transcripts of proteins destined to reach the spore wall by this route.
For two of the mutants we characterized, our results differ slightly from previous reports. In particular, mutations in AMA1 have been reported to cause an arrest in meiosis I, although the same study reported that ama1 spo13 double mutants displayed a spore wall phenotype consistent with that described here (10). A second study noted a spore formation phenotype but no meiotic defect for ama1 (35). We found that ama1 mutants can show a delay in meiosis I but that this phenotype is inconsistent between experiments, ranging from several hours to no delay at all (A. Diamond and A. M. Neiman, unpublished observations). This variability, which was observed with both the previously published strains, may account for the discrepancies between the different studies. In all of our experiments, ama1 mutants displayed the terminal phenotype described here. Finally, a previous characterization of the ssp2 mutant reported phenotypes very similar to those described here, except that the ssp2 mutant was reported to stain with Calcofluor White, indicating some synthesis of chitosan (38). By contrast, we find no Calcofluor White staining and no glucosamine present in ssp2 cells. The reasons for this difference are unclear.
In sum, our initial studies of a set of spore wall-defective mutants have defined multiple steps in the assembly of this structure. These results provide the outline of a morphogenetic pathway of spore wall assembly. Further studies to determine the molecular mechanism by which each of the gene products described here functions to promote assembly as well as the determination of where previously identified spore wall-defective mutants act in this pathway will provide a more complete understanding of the de novo formation of a complex extracellular matrix.
Acknowledgments
We thank Greg Rudomen and James Quinn for assistance with the electron microscopy and Jacqueline Segall (University of Toronto) for plasmids.
This work was supported by grant GM62184 to A.M.N. and the Austrian Fonds zur Förderung der Wissenschaftlichen Forschung, project P14735-B09, to P.B. M.N.C. was supported by grant HN5-042 from the Oklahoma Center for the Advancement of Science and Technology (OCAST).
REFERENCES
- 1.Bansal, D., K. Miyake, S. S. Vogel, S. Groh, C. C. Chen, R. Williamson, P. L. McNeil, and K. P. Campbell. 2003. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature 423:168-172. [DOI] [PubMed] [Google Scholar]
- 2.Berben, G., J. Dumont, V. Gilliquet, P. A. Bolle, and F. Hilger. 1991. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast 7:475-477. [DOI] [PubMed] [Google Scholar]
- 3.Briza, P., M. Breitenbach, A. Ellinger, and J. Segall. 1990. Isolation of two developmentally regulated genes involved in spore wall maturation in Saccharomyces cerevisiae. Genes Dev. 4:1775-1789. [DOI] [PubMed] [Google Scholar]
- 4.Briza, P., M. Eckerstorfer, and M. Breitenbach. 1994. The sporulation-specific enzymes encoded by the DIT1 and DIT2 genes catalyze a two-step reaction leading to a soluble ll-dityrosine-containing precursor of the yeast spore wall. Proc. Natl. Acad. Sci. USA 91:4524-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briza, P., A. Ellinger, G. Winkler, and M. Breitenbach. 1990. Characterization of a dl-dityrosine-containing macromolecule from yeast ascospore walls. J. Biol. Chem. 265:15118-15123. [PubMed] [Google Scholar]
- 6.Briza, P., A. Ellinger, G. Winkler, and M. Breitenbach. 1988. Chemical composition of the yeast ascospore wall. The second outer layer consists of chitosan. J. Biol. Chem. 263:11569-11574. [PubMed] [Google Scholar]
- 7.Christodoulidou, A., V. Bouriotis, and G. Thireos. 1996. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 271:31420-31425. [DOI] [PubMed] [Google Scholar]
- 8.Coluccio, A., and A. M. Neiman. 2004. Interspore bridges: a new feature of the Saccharomyces cerevisiae spore wall. Microbiology 150:3189-3196. [DOI] [PubMed] [Google Scholar]
- 9.Conrad, M. N., A. M. Dominguez, and M. E. Dresser. 1997. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276:1252-1255. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, K. F., M. J. Mallory, D. B. Egeland, M. Jarnik, and R. Strich. 2000. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc. Natl. Acad. Sci. USA 97:14548-14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felder, T., E. Bogengruber, S. Tenreiro, A. Ellinger, I. Sa-Correia, and P. Briza. 2002. Dtrlp, a multidrug resistance transporter of the major facilitator superfamily, plays an essential role in spore wall maturation in Saccharomyces cerevisiae. Eukaryot. Cell 1:799-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friesen, H., R. Lunz, S. Doyle, and J. Segall. 1994. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 8:2162-2175. [DOI] [PubMed] [Google Scholar]
- 13.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 14.Jang, Y. K., L. Wang, and G. B. Sancer. 1999. RPH1 and GIS1 are damage-responsive repressors of PHR1. Mol. Cell. Biol. 19:7630-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klis, F. M., P. Mol, K. Hellingwerf, and S. Brul. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26:239-256. [DOI] [PubMed] [Google Scholar]
- 16.Kreger-Van Rij, N. J. 1978. Electron microscopy of germinating ascospores of Saccharomyces cerevisiae. Arch. Microbiol. 117:73-77. [DOI] [PubMed] [Google Scholar]
- 17.Krisak, L., R. Strich, R. S. Winters, J. P. Hall, M. J. Mallory, D. Kreitzer, R. S. Tuan, and E. Winter. 1994. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 8:2151-2161. [DOI] [PubMed] [Google Scholar]
- 18.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular biology of the yeast Saccharomyces, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Kurtz, S., and S. Lindquist. 1986. Subcellular differentiation in sporulating yeast cells. Cell 45:771-779. [DOI] [PubMed] [Google Scholar]
- 20.Lennon, N. J., A. Kho, B. J. Bacskai, S. L. Perlmutter, B. T. Hyman, and R. H. Brown, Jr. 2003. Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J. Biol. Chem. 278:50466-50473. [DOI] [PubMed] [Google Scholar]
- 21.Liu, J., M. Aoki, I. Illa, C. Wu, M. Fardeau, C. Angelini, C. Serrano, J. A. Urtizberea, F. Hentati, M. B. Hamida, S. Bohlega, E. J. Culper, A. A. Amato, K. Bossie, J. Oeltjen, K. Bejaoui, D. McKenna-Yasek, B. A. Hosler, E. Schurr, K. Arahata, P. J. de Jong, and R. H. Brown, Jr. 1998. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 20:31-36. [DOI] [PubMed] [Google Scholar]
- 22.Liu, J., X. Tang, H. Wang, and M. Balasubramian. 2000. Bgs2p, a 1,3-beta glucan synthase subunit, is essential for maturation of ascospore wall in Schizosaccharomyces pombe. FEBS Lett. 478:105-108. [DOI] [PubMed] [Google Scholar]
- 23.Lynn, R. R., and P. T. Magee. 1970. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J. Cell Biol. 44:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, V., J. C. Ribas, E. Carrero, A. Duran, and Y. Sanchez. 2000. bgs2+, a sporulation-specific glucan synthase homologue is required for proper ascospore wall maturation in fission yeast. Mol. Microbiol. 38:308-321. [DOI] [PubMed] [Google Scholar]
- 25.Mishra, C., C. E. Semino, K. J. McCreath, H. de la Vega, B. J. Jones, C. A. Specht, and P. W. Robbins. 1997. Cloning and expression of two chitin deacetylase genes of Saccharomyces cerevisiae. Yeast 13:327-336. [DOI] [PubMed] [Google Scholar]
- 26.Moens, P. B. 1971. Fine structure of ascospore development in the yeast Saccharomyces cerevisiae. Can. J. Microbiol. 17:507-510. [DOI] [PubMed] [Google Scholar]
- 27.Nakanishi, H., P. de los Santos, and A. M. Neiman. 2004. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell 15:1802-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nehlin, J. O., M. Carlberg, and H. Ronne. 1989. Yeast galactose permease is related to yeast and mammalian glucose transporters. Gene 85:313-319. [DOI] [PubMed] [Google Scholar]
- 29.Neiman, A. M., L. Katz, and P. J. Brennwald. 2000. Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics 155:1643-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neuwald, A. F. 1997. Barth syndrome may be due to an acyltransferase deficiency. Curr. Biol. 7:R465-R466. [DOI] [PubMed] [Google Scholar]
- 31.Orlean, P. 1997. Biogenesis of yeast wall and surface components, p. 229-262. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cell cycle and biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Pammer, M., P. Briza, A. Ellinger, T. Schuster, R. Stucka, H. Feldmann, and M. Breitenbach. 1992. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast 8:1089-1099. [DOI] [PubMed] [Google Scholar]
- 33.Pedruzzi, I., N. Burckert, P. Egger, and C. De Virgilio. 2000. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 19:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Percival-Smith, A., and J. Segall. 1986. Characterization and mutational analysis of a cluster of three genes expressed preferentially during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 6:2443-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabitsch, K. P., A. Toth, M. Galova, A. Schleiffer, G. Schaffner, E. Aigner, C. Rupp, A. M. Penkner, A. C. Moreno-Borchart, M. Primig, R. E. Esposito, F. Klein, M. Knop, and K. Nasmyth. 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11:1001-1009. [DOI] [PubMed] [Google Scholar]
- 36.Rockmill, B., E. J. Lambie, and G. S. Roeder. 1991. Spore enrichment. Methods Enzymol. 194:146-149. [DOI] [PubMed] [Google Scholar]
- 37.Rose, M. D., and G. R. Fink. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sarkar, P. K., M. A. Florczyk, K. A. McDonough, and D. K. Nag. 2002. SSP2, a sporulation-specific gene necessary for outer spore wall assembly in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics 267:348-358. [DOI] [PubMed] [Google Scholar]
- 39.Schaber, M., A. Lindgren, K. Schindler, D. Bungard, P. Kaldis, and E. Winter. 2002. CAK1 promotes meiosis and spore formation in Saccharomyces cerevisiae in a CDC28-independent fashion. Mol. Cell. Biol. 22:57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smits, G. J., H. van den Ende, and F. M. Klis. 2001. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147:781-794. [DOI] [PubMed] [Google Scholar]
- 41.Stern, M., R. Jensen, and I. Herskowitz. 1984. Five SWI genes are required for expression of the HO gene in yeast. J. Mol. Biol. 178:853-868. [DOI] [PubMed] [Google Scholar]
- 42.Straight, P. D., T. H. Giddings, Jr., and M. Winey. 2000. Mps1p regulates meiotic spindle pole body duplication in addition to having novel roles during sporulation. Mol. Biol. Cell 11:3525-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tachikawa, H., A. Bloecher, K. Tatchell, and A. M. Neiman. 2001. A Gip1p-Glc7p phosphatase complex regulates septin organization and spore wall formation. J. Cell Biol. 155:797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tougan, T., Y. Chiba, Y. Kakihara, A. Hirata, and H. Nojima. 2002. Meu10 is required for spore wall maturation in Schizosaccharomyces pombe. Genes Cells 7:217-231. [DOI] [PubMed] [Google Scholar]
- 45.Ufano, S., P. San-Segundo, F. del Rey, and C. R. Vazquez de Aldana. 1999. SWM1, a developmentally regulated gene, is required for spore wall assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2118-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, M., P. Briza, M. Pierce, and E. Winter. 1999. Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics 151:1327-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]