Abstract
Background
The optimisation of trypanosomosis control programs warrants a good knowledge of the main vector of animal and human trypanosomes in sub-Saharan Africa, the tsetse fly. An important aspect of the tsetse fly population is its trypanosome infection prevalence, as it determines the intensity of the transmission of the parasite by the vector. We therefore conducted a systematic review of published studies documenting trypanosome infection prevalence from field surveys or from laboratory experiments under controlled conditions. Publications were screened in the Web of Science, PubMed and Google Scholar databases. Using the four-stage (identification, screening, eligibility and inclusion) process in the PRISMA statement the initial screened total of 605 studies were reduced to 72 studies. The microscopic examination of dissected flies (dissection method) remains the most used method to detect trypanosomes and thus constituted the main focus of this analysis. Meta-regression was performed to identify factors responsible for high trypanosome prevalence in the vectors and a random effects meta-analysis was used to report the sensitivity of molecular and serological tests using the dissection method as gold standard.
Results
The overall pooled prevalence was 10.3% (95% confidence interval [CI] = 8.1%, 12.4%) and 31.0% (95% CI = 20.0%, 42.0%) for the field survey and laboratory experiment data respectively. The country and the year of publication were found to be significantly factors associated with the prevalence of trypanosome infection in tsetse flies. The alternative diagnostic tools applied to dissection positive samples were characterised by low sensitivity, and no information on the specificity was available at all.
Conclusion
Both temporal and spatial variation in trypanosome infection prevalence of field collected tsetse flies exists, but further investigation on real risk factors is needed how this variation can be explained. Improving the sensitivity and determining the specificity of these alternative diagnostic tools should be a priority and will allow to estimate the prevalence of trypanosome infection in tsetse flies in high-throughput.
Electronic supplementary material
The online version of this article (doi:10.1186/s12917-017-1012-9) contains supplementary material, which is available to authorized users.
Keywords: Meta-regression, Systematic review, Glossina, Trypanosome infection prevalence, Diagnostic methods
Background
Glossina species (commonly known as tsetse flies) are the major vectors of several Trypanosoma species, the causative agents of animal and human African trypanosomosis, also called Nagana and sleeping sickness, respectively [1–3]. Once established in the tsetse fly, trypanosomes undergo a developmental cycle within the tsetse fly with varying complexity depending on the species [4]. The infected tsetse fly then transmits the parasite to diverse host species during its blood meal. Tsetse flies infest an area of about 10 million km2 comprising 38 sub-Saharan African countries [5]. The disease constitutes a major veterinary and medical burden affecting the life of millions of people. Within affected regions, the density of the vector and the prevalence of trypanosome infections in the host is attributed to complex interactions between and among humans, domestic livestock, wildlife, tsetse flies, trypanosomes and various economic and ecological factors [6, 7].
The prevalence of trypanosome infections in the tsetse flies is often a neglected parameter probably due to the intensive labour required for its evaluation. Integrating this parameter in a monitoring program allows however a more precise evaluation of the risk of being infected in a particular region.
Dissection of flies remains the most common technique for detecting the presence of trypanosomes. Although molecular and serological techniques are assumed to detect far higher levels of genetic diversity with a higher sensitivity [8], the performance of such tests has been reported to be unsatisfactory [9–13]. For instance, PCR failed to detect trypanosomes in dissection positive flies or vice versa, and tsetse fly samples negative by PCR were positive by fluorescent fragment length barcoding tests even allowing the discovery of new genotypes [14, 15].
The aim of this systematic review was to (i) synthesize the limited information on the trypanosome prevalence in tsetse flies, and (ii) assess the sensitivity of various diagnostic methods for the detection of trypanosomes in the tsetse flies using the dissection method as gold standard.
Methods
Search strategy and inclusion of studies
Publications were screened in the Web of Science, PubMed and Google Scholar databases. The last search was done on July the 20th 2015. The following Boolean parameter combinations and Medical Subject Headings terms were used: “Trypanosomes” and “infection rate” and “tsetse fly or Glossina”. The retrieved articles were then first screened by title and abstract by two independent readers. Any discrepancies were discussed until consensus. Selected articles were retained for further full text analysis. The inclusion criteria for further data extraction and meta-analysis were the presence of the following data: (i) tsetse species, (ii) study type (laboratory or field), (iii) location (country) of study, (iv) trypanosome detection method, (v) type of tsetse sample examined, (vi) number and type of fly samples, and (vii) number of samples positive for trypanosomes. A flow chart describing the number of articles retrieved, screened and included or rejected is presented in Fig. 1.
Fig. 1.
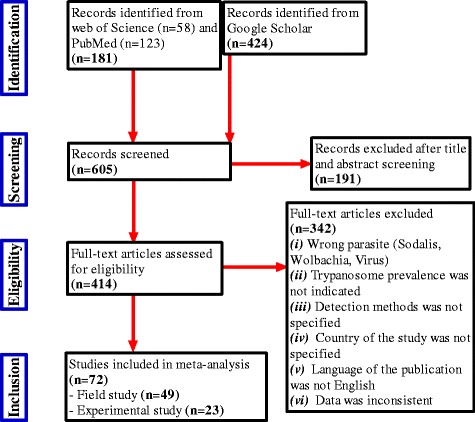
Flowchart detailing the number of studies excluded and included at each step for systematic review of the prevalence of trypanosome infection in tsetse flies
Meta-analysis
All meta-analyses were performed with STATA Version 12 (StataCorp LP, College Station, Texas). The core analysis focused on the prevalence of trypanosome infections in the tsetse flies as assessed by the dissection technique. Analyses are done separately for data obtained from field surveys and laboratory experiments. In laboratory experiments, blood meals and external conditions are standardized and all feeding flies ingest parasites. In the field surveys, the prevalence of trypanosomes in the host and the parasitaemia will be the determining factors explaining the prevalence in flies. First, pooled estimates were calculated based on the random effects model [16, 17] with study as random effect. Results were presented by forest plots. Next, factors that could be associated with the prevalence of infection were considered using logistic meta-regression analysis [18, 19] with country, tsetse species and tsetse organ as categorical and year of publication as continuous fixed effects factors. Odds ratios with their 95% confidence interval (CI) were used as summary statistic and testing was done at the 5% significance level.
A second part of the meta-analysis assessed the sensitivity of the alternative diagnostic tests using the dissection method as a gold standard. The sensitivity was evaluated using the random effects model.
Diagnostic tools to assess trypanosome infection in a tsetse fly
The most widely used method constitutes the dissection of tsetse flies and microscopic evaluation of the organs. It is cheap [20], but laborious, low in sensitivity and cannot differentiate mixed infections of trypanosome species and its different developmental stages in the fly [21]. The warm slide technique is occasionally used to assess the prevalence of trypanosomes in tsetse flies. Accordingly, tsetse flies are allowed to salivate (probe) on a warm slide, then, trypanosome examination is done by microscopy of the slide [22]. An alternative for parasite detection with higher sensitivity is the inoculation of dissolved organ contents of tsetse flies in rats or mice for xenodiagnosis. Its added merit is that field isolates from mammals or tsetse flies can be collected via rodent inoculation for further studies. Its disadvantage is that diagnosis is not immediate and that T. vivax and T. simiae do not infect rodents [23]. Tissue culture techniques (in vitro cultivation) can be another option using different culture media. Cultivation is widely done for species of the Trypanosoma brucei group. The culture method is vital as it can provide information on pathogen viability and susceptibility to drugs [23, 24]. Isoenzyme band pattern examination technique is also possible. In this technique, 10–20 enzymes extracts from the trypanosome cytoplasm common to nearly all trypanosome species are separated by native electrophoresis and visualized by native staining. It requires a minimum of 100 million trypanosomes to test positive [25, 26]. The dot-ELISA test is another option which is based on the preparation of suspensions of different organs of tsetse flies that are applied on nitrocellulose membranes. Trypanosome species-specific monoclonal antibodies are used to detect the presence of trypanosomes in the suspension. The test is highly specific as monoclonal antibodies are used and it is simple, rapid, and can detect mixed infections via testing of one sample multiple times using different monoclonal antibodies [21, 27, 28]. In the DNA probing technique, a denatured DNA sample (target) fixed on nitrocellulose is exposed to a radioactively labeled DNA-probe, which is a fragment of DNA of variable length. The probe - target complementary base pairing of the sequence in the probe is used to diagnose infection [29–31]. Conventional PCR has also been used [23, 32]. PCR works using either species-specific primers or generalist non-species-specific primers (e.g. ITS1) to differentiate trypanosomes. The advantage of ITS1 PCR is that only one test needs to be done to assess whether trypanosomes occur in the sample –regardless of the species, whereas in the species-specific PCR a sample must be tested repeatedly with each species-specific primer pair [33, 34]. Trypanosome detection by PCR is done using the entire tsetse body or different tsetse organs and recently also even anal and oral droppings are used [35, 36]. Another modern technique is the fluorescent fragment-length barcoding method (FFLB), which is a hybrid of PCR and sequencing. FFLB amplifies fragments with inter-species size variation by PCR using fluorescently tagged primers, then, the sizes of the fragments of the PCR product are determined accurately using an automated DNA sequencer. Therefore, it discriminates trypanosome species by size polymorphisms. However, FFLB is too advanced and expensive for routine use in Africa [14, 15, 33]. Real-time PCR has the inherent ability to detect and quantify the number of trypanosomes in a sample [37]. Finally, loop mediated isothermal amplification (LAMP) is a low-tech trypanosome detection technique. The target sequence is amplified by LAMP at a constant (isothermal) temperature of 60–65 °C using either two or three sets of primers and a polymerase with high strand displacement activity in addition to a replication activity. Typically, 4 different primers are used to identify 6 distinct regions on the target gene, which leads typically to good specificity. The added advantage of the LAMP technique is that it does not require experience nor instruments except a water bath or incubator and results are obtained quickly [38].
Results
Inclusion of studies and data extraction
A total of 605 studies were initially screened of which 191 were excluded on the basis of their titles and abstracts. Of the remaining 414 which were fully evaluated, 72 were considered while 342 studies were excluded (Fig. 1). The 72 selected articles involved 23 countries for a total of 236,740 tsetse flies checked for trypanosome infection (Additional file 1). Of those 72 articles, 49 were reporting field studies with 202,182 tsetse flies analysed. The majority of the field studies (80%) used dissection (i.e. on 192,338 tsetse flies in total). The remaining 23 studies were laboratory experiments with 34,558 tsetse flies analysed of which 18 studies used dissection method. The studies included 12 different tsetse species. Samples analysed were saliva spit, anal droppings (diuresis fluid), midgut, proboscis, salivary glands and/or their DNA, DNA and pools of DNA from whole bodies. Methods used for the detection of trypanosomes were: dissection, microscopy of diuresis, probing on mice, warm slide probe, culture using media, isoenzyme analysis, DNA probing, Dot-ELISA, species specific PCR, ITS-1 PCR, real time PCR, species-specific LAMP and FFLB. Details are provided in Tables 1 and 2.
Table 1.
List of published articles included in the systematic review for field studies with dissection method used for diagnosis of trypanosome infection
| Variable | No studies | No species | No flies | References |
|---|---|---|---|---|
| Overall field studies | 49 | 12a | 202,182 | [8, 9, 14, 15, 27, 29, 30, 33–35, 44–80] |
| Country | ||||
| Angola | 1 | 1 | 62 | [54] |
| Burkina Faso | 1 | 2 | 435 | [44] |
| Cameroon | 3 | 5a | 6104 | [12, 77] |
| Democratic Republic of Congo | 1 | 1 | 254 | [76] |
| Equatorial Guinea | 1 | 1 | 62 | [35] |
| Ethiopia | 1 | 4 | 384 | [51] |
| Gambia | 2 | 4a | 3055 | [30, 61] |
| Ivory Coast | 3 | 5 | 3707 | [50, 52, 62] |
| Kenya | 7 | 4a | 41,959 | [13, 27, 29, 45, 71, 78, 79] |
| Liberia | 1 | 3 | 2224 | [72] |
| Nigeria | 4 | 3 | 27,502 | [56, 58, 73, 75] |
| Rwanda | 1 | 3 | 5496 | [65] |
| South Africa | 3 | 2a | 1323 | [53, 60, 68] |
| Southern Sudan | 1 | 1 | 117 | [66] |
| Tanzania | 10 | 4a | 43,923 | [8, 9, 14, 15, 33, 34, 46, 48, 59, 60] |
| Uganda | 3 | 4 | 16,350 | [55, 74, 80] |
| Zambia | 6 | 3a | 49,225 | [47, 49, 57, 64, 70, 72] |
| Detection method | ||||
| Culture media | 1 | 3 | 1112 | [62] |
| Dissection | 38 | 12a | 192,338 | [8, 9, 12, 13, 15, 27, 29, 30, 34, 44–52, 55, 56, 58, 59, 61–63, 65, 67–69, 71–75, 77–80] |
| Dot-ELISA | 1 | 2 | 494 | [27] |
| FFLB | 1 | a | 91 | [14] |
| ITS-1 PCR | 2 | 1a | 173 | [14, 59] |
| Sp. specific PCR | 11 | 4a | 7974 | [35, 53, 54, 57, 60, 64, 66, 70, 71, 76, 77] |
| Glossina sample type | ||||
| DNA DO and Pool NDO | 1 | a | 3638 | [71] |
| DNA WB | 4 | 3 | 1221 | [54, 57, 64, 66] |
| DNA WB & DO | 1 | 1 | 279 | [59] |
| Pool DNA WB | 2 | 1a | 312 | [35, 70] |
| Mid gut | 8 | 5a | 20,792 | [14, 33, 34, 48, 62, 63, 76, 77] |
| MP | 8 | 7a | 46,416 | [8, 9, 15, 29, 53, 56, 58, 68] |
| MS | 1 | 2 | 1221 | [50] |
| MPS | 19 | 10a | 73,793 | [12, 27, 30, 44, 46, 61, 64, 65, 68, 69, 71–75, 78–80] |
| Proboscis | 5 | 5a | 46,991 | [13, 47, 49, 60, 67] |
| SG | 1 | 1 | 7519 | [45] |
| Glossina species | ||||
| G. austeni | 1 | 1 | 40 | [68] |
| G. brevipalpis | 6 | 1 | 5870 | [9, 49, 55, 60, 65, 68] |
| G. fuscipes | 5 | 1 | 7071 | [49, 51, 66, 74, 80] |
| G. longipennis | 2 | 1 | 1305 | [27, 71] |
| G. medicorum | 1 | 1 | 10 | [52] |
| G. morsitans | 10 | 1 | 51,556 | [9, 30, 44, 49, 51, 56, 61, 65, 72, 75] |
| G. nigrofusca | 3 | 1 | 294 | [50, 62, 65] |
| G. pallicera | 2 | 1 | 76 | [62, 63] |
| G. pallidipes | 20 | 1 | 64,395 | [8, 9, 13, 15, 27, 29, 45, 47–49, 51, 55, 57, 59, 64, 65, 69, 71, 78, 79] |
| G. palpalis | 14 | 1 | 14,669 | [12, 30, 35, 50, 52, 54, 55, 58, 62, 63, 67, 73, 76, 77] |
| G. swynnertoni | 5 | 1 | 14,414 | [8, 9, 15, 46, 48] |
| G. tachinoides | 6 | 1 | 6367 | [44, 51, 52, 58, 73, 75] |
| Mixeda | 9 | 1 | 35,865 | [12, 14, 15, 30, 33, 34, 49, 53, 71] |
| Not determined | 1 | 1 | 250 | [70] |
a= mixed tsetse sp. examined besides the indicated number of tsetse sp., DNA DO and Pool NDO DNA of dissected organs and of a pool of negative tsetse organs, DNA WB DNA of whole fly body, DNA WB & DO DNA of whole fly body and of dissected organs, Pool DNA WB pooling DNA of whole fly body, MP Mid gut and proboscis, MS Mid gut and salivary gland, MPS Mid gut, proboscis and salivary gland, SG Salivary gland, SP Salivary gland and proboscis
Table 2.
List of published articles included in the systematic review for laboratory experimental studies with dissection method used for diagnosis of trypanosome infection
| Variable | No studies | No species |
No flies |
References |
|---|---|---|---|---|
| Overall experimental studies | 23 | 7a | 34,558 | [21, 22, 24, 28, 31, 36, 37, 81–97] |
| Country | ||||
| Belgium | 3 | 2 | 1559 | [24, 85, 88] |
| Burkina Faso | 2 | 1a | 1443 | [36, 87] |
| BFZ | 1 | 2a | 1092 | [95] |
| France | 1 | 1 | 594 | [97] |
| Ghana | 2 | 2 | 540 | [28, 86] |
| Kenya | 5 | 7 | 19,592 | [21, 31, 81–83] |
| Tanzania | 1 | 1 | 3274 | [22] |
| Uganda | 2 | 4 | 2011 | [89, 90] |
| United Kingdom | 4 | 2 | 721 | [37, 91, 94, 96] |
| Zambia | 1 | 1 | 1796 | [92] |
| Zimbabwe | 1 | 1 | 1936 | [93] |
| Detection method | ||||
| Dissection | 17 | 7a | 22,478 | [21, 24, 36, 81–83, 85, 87–95, 97] |
| DNA probe | 1 | 1 | 15 | [31] |
| Dot-ELISA | 3 | 2 | 1240 | [21, 28, 86] |
| Microscopy of diuresis | 1 | a | 266 | [95] |
| Sp. Specific PCR | 2 | 3a | 1246 | [21, 95] |
| probing on mice | 1 | 1 | 300 | [96] |
| Real Time PCR | 1 | 1 | 150 | [37] |
| Warm slide probe | 5 | 1a | 8863 | [22, 82, 93–95] |
| Glossina sample type | ||||
| DF | 1 | a | 532 | [95] |
| Mid gut | 5 | 2 | 2205 | [28, 31, 37, 92, 94] |
| MP | 3 | 2 | 1573 | [85, 88, 95] |
| MS | 2 | 2 | 1754 | [22, 93] |
| MPS | 5 | 2a | 4263 | [21, 24, 86, 87, 91] |
| proboscis | 3 | 6 | 12,315 | [81–83] |
| SS | 5 | 1a | 9011 | [22, 84, 93–95] |
| SG | 2 | 4 | 2411 | [90, 97] |
| SP | 1 | 1 | 194 | [89] |
| N.A | 1 | 1 | 300 | [96] |
| Glossina species | ||||
| G. austeni | 1 | 1 | 1062 | [83] |
| G. brevipalpis | 2 | 1 | 1256 | [83, 90] |
| G. fuscipes | 2 | 1 | 1570 | [83, 90] |
| G. morsitans | 19 | 1 | 22,607 | [22, 24, 28, 31, 37, 81–83, 85, 86, 89–97] |
| G. pallidipes | 4 | 1 | 2619 | [21, 86, 90, 94] |
| G. palpalis | 4 | 1 | 2184 | [36, 83, 88, 95] |
| G. tachinoides | 1 | 1 | 1009 | [83] |
| Mixeda | 2251 | [87, 95] | ||
a= mixed tsetse sp. examined besides the indicated number of tsetse sp., DF Diuresis fluid, MP Mid gut and proboscis, MS Mid gut and salivary gland, MPS Mid gut, proboscis and salivary gland, SS Saliva spit, SG Salivary gland, SP Salivary gland and proboscis, N.A. Not available
One third of the studies compared dissection positive results with at least one alternative serological or molecular technique: species-specific PCR (n = 15), DNA probe (n = 4), fluorescent fragment length barcoding (n = 3), ITS-1 PCR (n = 2), and dot-ELISA (n = 1).
Meta-analysis of dissection based field studies
The overall trypanosome prevalence of flies in the field studies (n = 39) was 10.3% (95% CI = 8.1, 12.4) (Fig. 2). Significant between-study heterogeneity was observed (P < 0.001). Different factors were thus further analysed. Results are presented in Table 3. The prevalence of trypanosomes decreases with publication year (P = 0.035). Trypanosome prevalence differs significantly between countries (P = 0.004). The prevalence ranged from 4.1% in Rwanda to 40.5% in Burkina Faso. The type of tsetse fly sample (body part) did not have a significant effect on the prevalence (P = 0.2155). The prevalence of trypanosomes ranged from 6.5% in midguts to 30.8% in the pooled midgut/salivary glands samples. Tsetse fly species or group (morsitans, fusca, palpalis) were not significant factors (P = 0.1466). The highest trypanosome prevalence was observed in G. negrofusca (26%, n = 3, fusca group) and the lowest was observed in G. longipennis (0.2%, n = 2, fusca group). The two variables that were significant in the univariate analysis (Table 3) remained significant with minor changes in the estimates of the odds ratios in the multivariate meta-regression analysis (results not shown).
Fig. 2.
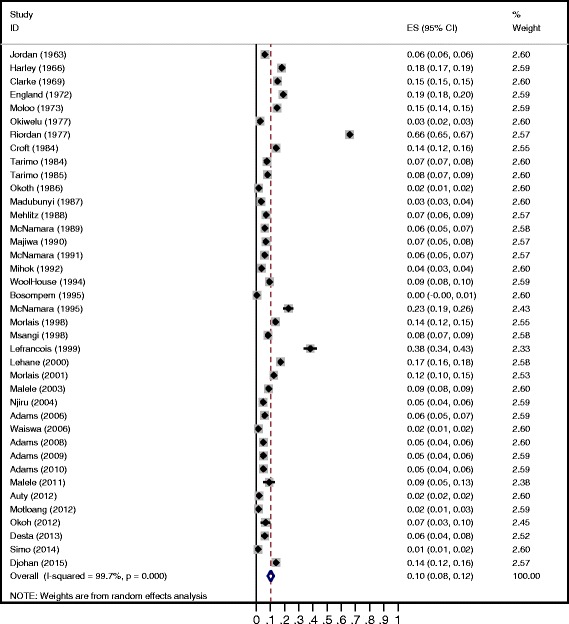
Forest plot of the prevalence of trypanosome infection in tsetse flies by the dissection method for field studies
Table 3.
Univariate meta-regression analysis of factors for the prevalence of trypanosome infection based on the field studies with dissection method used for diagnosis of trypanosome infection
| Variables | Prevalence | Odds ratio | |||||
|---|---|---|---|---|---|---|---|
| Estimate | 95% CI | Estimate | 95% CI | p-value | |||
| Year of publication | - | - | 0.998 | 0.996 | 0.999 | 0.035 | |
| Country | |||||||
| Burkina Faso | 40.5 | 28.1 | 53.0 | Ref. | |||
| Ivory Coast | 18.6 | 13.7 | 23.6 | 0.82 | 0.69 | 0.97 | 0.021 |
| Nigeria | 18.0 | 5.0 | 30.9 | 0.80 | 0.67 | 0.95 | 0.010 |
| Cameroon | 10.9 | 2.9 | 18.8 | 0.74 | 0.62 | 0.89 | 0.002 |
| Uganda | 10.0 | 3.0 | 17.0 | 0.74 | 0.62 | 0.88 | 0.001 |
| Zambia | 8.6 | 3.1 | 14.1 | 0.73 | 0.61 | 0.86 | <0.001 |
| Liberia | 8.2 | 4.4 | 12.1 | 0.74 | 0.61 | 0.89 | 0.002 |
| Kenya | 7.4 | 3.5 | 11.4 | 0.72 | 0.61 | 0.85 | <0.001 |
| Tanzania | 7.0 | 5.2 | 8.9 | 0.71 | 0.61 | 0.84 | <0.001 |
| South Africa | 6.8 | 0.2 | 20.5 | 0.71 | 0.58 | 0.88 | 0.002 |
| Gambia | 6.0 | 5.2 | 6.8 | 0.71 | 0.59 | 0.85 | <0.001 |
| Ethiopia | 4.9 | 0.6 | 9.2 | 0.70 | 0.58 | 0.84 | <0.001 |
| Rwanda | 4.1 | 3.0 | 5.1 | 0.70 | 0.58 | 0.84 | <0.001 |
| Glossina sample type | |||||||
| MS | 30.8 | 7.6 | 53.9 | Ref. | |||
| Salivary gland | 19.3 | 18.4 | 20.2 | 0.91 | 0.69 | 1.19 | 0.472 |
| MPS | 11.2 | 8.5 | 14.0 | 0.84 | 0.72 | 0.98 | 0.023 |
| Proboscis | 10.6 | 4.3 | 16.9 | 0.83 | 0.70 | 0.98 | 0.029 |
| Mid gut | 6.5 | 4.6 | 8.3 | 0.83 | 0.70 | 0.98 | 0.027 |
| Mid gut and proboscis | 6.5 | 5.0 | 8.0 | 0.80 | 0.68 | 0.94 | 0.007 |
| Glossina species | |||||||
| G. nigrofusca | 25.9 | 1.9 | 49.9 | Ref. | |||
| G. pallicera | 23.5 | 12.4 | 34.7 | 1.04 | 0.81 | 1.34 | 0.751 |
| G. medicorum | 20.0 | 4.8 | 44.8 | 0.96 | 0.66 | 1.39 | 0.811 |
| G. tachinoides | 17.7 | 8.5 | 26.9 | 0.93 | 0.78 | 1.12 | 0.454 |
| G. morsitans | 15.8 | 9.0 | 22.6 | 0.92 | 0.77 | 1.08 | 0.304 |
| G. austeni | 15.0 | 3.9 | 2.16 | 0.91 | 0.68 | 1.22 | 0.520 |
| G. palpalis | 10.5 | 6.4 | 14.6 | 0.87 | 0.74 | 1.03 | 0.107 |
| G. mixed species | 9.6 | 4.3 | 14.8 | 0.86 | 0.72 | 1.03 | 0.102 |
| G. swynnertoni | 9.2 | 3.0 | 15.4 | 0.86 | 0.71 | 1.03 | 0.101 |
| G. pallidipes | 8.3 | 5.8 | 10.8 | 0.85 | 0.72 | 1.00 | 0.052 |
| G. brevipalpis | 5.8 | 2.3 | 9.3 | 0.83 | 0.69 | 1.00 | 0.046 |
| G. fuscipes | 1.1 | 0.1 | 2.1 | 0.79 | 0.65 | 0.96 | 0.017 |
| G. longipennis | 0.2 | 0.1 | 0.5 | 0.78 | 0.63 | 0.98 | 0.031 |
MS Mid gut and salivary gland, MPS Mid gut, proboscis and salivary gland
Meta-analysis of dissection based laboratory experiments
The overall trypanosome prevalence of flies in the laboratory experiments (n = 18) was 31.0% (95% CI = 20.0, 42.0) (Fig. 3). Significant between-study heterogeneity was also observed in these studies (P < 0.001). However, the trypanosome prevalence did not differ significantly between countries (P = 0.0916) nor as a function of the publication year (P = 0.184) (Table 4). No significant (P = 0.9545) differences in trypanosome prevalence among the seven tsetse species were observed. The sample type (body part) was significantly (P = 0.0122) associated with trypanosome prevalence. The highest trypanosome prevalence was observed in the proboscis.
Fig. 3.
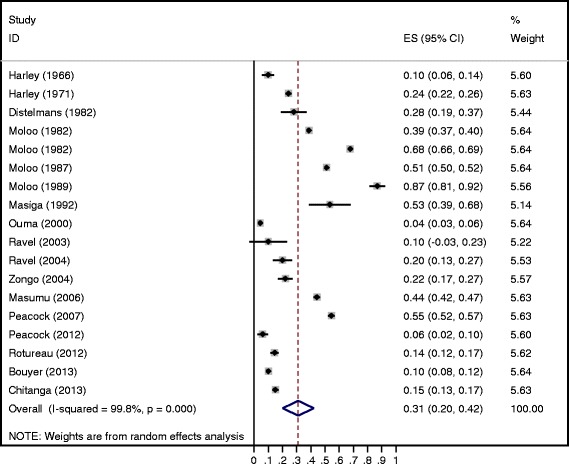
Forest plot of the prevalence of trypanosome infection in tsetse flies by the dissection method for laboratory experiments
Table 4.
Univariate meta-regression analysis of factors for the prevalence of trypanosome infection based on the laboratory experimental studies with dissection method used for diagnosis of trypanosome infection
| Variables | Prevalence (%) | Odds ratio | |||||
|---|---|---|---|---|---|---|---|
| Estimate | 95% CI | Estimate | 95% CI | p-value | |||
| Year of publication | - | - | 0.996 | 0.99 | 1.00 | 0.184 | |
| Country | |||||||
| Zimbabwe | 54.5 | 52.2 | 56.8 | Ref | |||
| Kenya | 50.9 | 31.7 | 70.0 | 0.96 | 0.62 | 1.51 | 0.866 |
| Zambia | 44.3 | 42.0 | 46.6 | 0.90 | 0.49 | 1.65 | 0.726 |
| Uganda | 21.4 | 14.9 | 27.9 | 0.71 | 0.45 | 1.15 | 0.155 |
| Belgium | 20.8 | 13.9 | 27.8 | 0.72 | 0.44 | 1.18 | 0.181 |
| United Kingdom | 20.5 | 0.0 | 41.3 | 0.71 | 0.43 | 1.17 | 0.171 |
| Burkina Faso and Zimbabwe | 20.1 | 9.1 | 31.0 | 0.71 | 0.42 | 1.22 | 0.192 |
| France | 14.5 | 11.6 | 17.3 | 0.67 | 0.37 | 1.22 | 0.181 |
| Burkina Faso | 10.0 | 8.4 | 11.5 | 0.64 | 0.38 | 1.08 | 0.093 |
| Glossina sample type | |||||||
| Proboscis | 56.1 | 43.3 | 68.8 | Ref | |||
| Mid gut and salivary gland | 32.7 | 0.0 | 76.3 | 0.80 | 0.59 | 1.09 | 0.145 |
| Salivary gland | 22.3 | 16.8 | 27.8 | 0.71 | 0.58 | 0.89 | 0.004 |
| Mid gut and proboscis | 20.2 | 13.5 | 26.9 | 0.70 | 0.56 | 0.89 | 0.005 |
| Mid gut | 19.0 | 0.0 | 51.4 | 0.69 | 0.53 | 0.89 | 0.007 |
| MPS | 17.6 | 10.1 | 25.1 | 0.71 | 0.56 | 0.89 | 0.006 |
| Salivary gland and proboscis | 9.8 | 5.6 | 14.0 | 0.63 | 0.42 | 0.95 | 0.028 |
| Glossina species | |||||||
| G. brevipalpis | 51.9 | 0.0 | 100.0 | Ref | |||
| G. tachinoides | 40.2 | 37.2 | 43.3 | 0.89 | 0.48 | 1.66 | 0.698 |
| G. morsitans | 37.9 | 23.6 | 52.2 | 0.87 | 0.59 | 1.28 | 0.455 |
| G. fuscipes | 31.7 | 11.2 | 52.2 | 0.82 | 0.49 | 1.36 | 0.415 |
| G. austeni | 31.5 | 28.7 | 34.2 | 0.81 | 0.44 | 1.52 | 0.499 |
| G. palpalis | 31.0 | 23.4 | 38.7 | 0.796 | 0.52 | 1.22 | 0.278 |
| G. pallidipes | 11.9 | 0.0 | 24.0 | 0.67 | 0.42 | 1.07 | 0.088 |
| Mixed | 10.0 | 8.4 | 11.5 | 0.66 | 0.35 | 1.22 | 0.174 |
MPS Mid gut, proboscis and salivary gland
Sensitivity of advanced detection methods
The results of the meta-analysis of the 25 studies using alternative diagnostic tests on dissection positive samples are shown in Fig. 4. With the exception of the dot-ELISA (sensitivity of 91%, which was represented only by one study), the remaining methods had similar levels of sensitivity ranging from 43% for DNA probe to 62% for fluorescent fragment length barcoding.
Fig. 4.
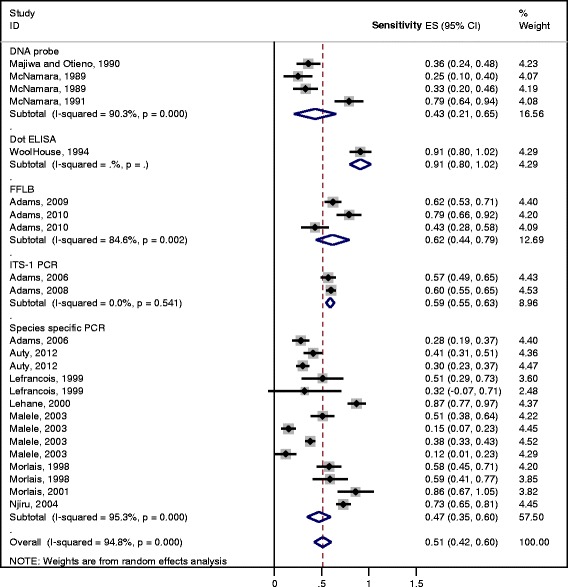
Forest plot for the sensitivity of molecular and serological detection methods of trypanosome infection in the tsetse flies using the dissection method as gold standard
Discussion
The scientific literature on trypanosome detection methods and prevalence in tsetse flies published in English since more than a half century covering natural and experimental infections of tsetse flies was reviewed in this paper.
As expected, the prevalence of trypanosome infection in tsetse flies is higher in laboratory experiments than in field collections of tsetse flies with an overall prevalence of 31% and 10% respectively. In laboratory experiments, blood meals and external conditions are standardized and all feeding flies ingest parasites. In field collected tsetse flies, the prevalence of trypanosome infected hosts and their parasitaemia will be the determining factors explaining the prevalence in flies.
From our meta-analysis, it appears that differences in prevalence exist in field collected tsetse flies according to year and country. The factor “country” should be interpreted with care as many factors can explain the differences between countries, e.g., the ecological context and national vector control measures. However, original studies didn’t investigate the real ecological, entomological, parasitological, tsetse-host contact and intervention factors to identify the actual risk factors responsible for the infection of tsetse flies. If our review allowed pinpointing spatial differences, it could not clearly bring a precise explanation of the variations in infection prevalence.
The negative relationship between the year of publication and the prevalence of infection is a paradox. Encroachment, i.e., the degrading effect of human activities on the environment, has taken place in most regions of sub-Saharan Africa. Encroachment causes a decrease of the tsetse fly population size. However, as counterbalancing effect, the prevalence of the trypanosome infection of the flies has been observed to increase allowing for persistent transmission even when the tsetse fly vectors are scarce [39, 40]. However, the opposite effect is observed. This might be due to the higher intensity of drug treatment of the livestock by the farmers. Indeed, most farmers in endemic areas treat their herds regularly using trypanocidal drugs [41]. It is known that prolonged and persistent use of trypanocidal drugs in the field decreases/disrupts the transmission of trypanosomes by the tsetse flies [42, 43] thus reducing the risk of tsetse infection.
Our last objective was to assess the sensitivity of various diagnostic methods for the detection of trypanosomes in the tsetse flies using the dissection method as gold standard. The sensitivity of molecular/serological tests that were performed on positive samples (i.e. by dissection) was only around 50%. The alternative diagnostic tools applied to the dissection positive samples were thus characterised by low sensitivity, and no information on specificity is available at all. The currently available molecular and serological techniques are developed and optimized for trypanosome detection in the host; their detection performance in the insect (tsetse fly) is a different story. This study revealed that the tests apparently work suboptimal for tsetse fly samples. Sample processing conditions and specimens used are not standardized or externally controlled for detection of trypanosomes in tsetse flies. Comparing several tests on the same specimen panel would allow more accurate comparisons of the sensitivity and specificity.
Conclusions
Dissection remains the gold standard for the determination of the infection status of tsetse flies. Alternative molecular and serological techniques have currently too low sensitivity and their specificity is unknown, which warrants further investigation before they can be employed on a routine basis. Both temporal and spatial variation in trypanosome infection prevalence of field collected tsetse flies exists, but it needs to be investigated further how this variation can be explained by thorough real risk factor investigation for tsetse fly infection.
Acknowledgments
This work was financially supported by the VLIR-UOS South Initiative. We would like to thank Dr. Olana Merera, Dr. Megersa Bedasa, and Dr. Biyansa Adugna for their critical comments and editing the retrieved data.
Funding
This work was financially supported partly by the VLIR-UOS South Initiative.
Availability of data and materials
Additional file 1. Supplementary excel file is included.
Authors’ contributions
RD and LD conceived the idea. RD and WG designed the study and retrieved study articles. RD and GEA analysed the data. RD and LD wrote the manuscript. VD, MB, TVL and LD reviewed the manuscript. All authors read, commented and approved the final submitted version.
Competing interests
The authors declare that they have no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- FFLB
Fluorescent fragment-length barcoding
- LAMP
Loop mediated isothermal amplification
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
Additional file
Raw data for meta-analysis. (XLSX 14 kb)
References
- 1.Bhalla N. Pan African group takes lead against the tsetse fly. Lancet. 2002;359(9307):686. doi: 10.1016/S0140-6736(02)07843-1. [DOI] [PubMed] [Google Scholar]
- 2.Murray M, Gray AR. The current situation on animal trypanosomiasis in Africa. Prev Vet Med. 1984;2(1):23–30. doi: 10.1016/0167-5877(84)90045-X. [DOI] [Google Scholar]
- 3.Brun R, Blum J, Chappuis F, Burri C. Human African trypanosomiasis. Lancet. 2010;375(9709):148–159. doi: 10.1016/S0140-6736(09)60829-1. [DOI] [PubMed] [Google Scholar]
- 4.Vickerman K, Tetley L, Hendry KA, Turner CMR. Biology of African trypanosomes in the tsetse fly. Biol Cell. 1988;64(2):109–119. doi: 10.1016/0248-4900(88)90070-6. [DOI] [PubMed] [Google Scholar]
- 5.Kristjanson PM, Swallow BM, Rowlands GJ, Kruska RL, De Leeuw PN. Measuring the costs of African animal trypanosomosis, the potential benefits of control and returns to research. Agric Syst. 1999;59(1):79–98. doi: 10.1016/S0308-521X(98)00086-9. [DOI] [Google Scholar]
- 6.Duguma R, Tasew S, Olani A, Damena D, Alemu D, Mulatu T, et al. Spatial distribution of Glossina sp. and Trypanosoma sp. in south-western Ethiopia. Parasit Vectors. 2015;8:430. doi: 10.1186/s13071-015-1041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steverding D. The history of African trypanosomiasis. Parasit Vectors. 2008;1:3. doi: 10.1186/1756-3305-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams ER, Hamilton PB, Gibson WC. African trypanosomes: celebrating diversity. Trends Parasitol. 2010;26(7):324–328. doi: 10.1016/j.pt.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Malele I, Craske L, Knight C, Ferris V, Njiru ZK, Hamilton P, et al. The use of specific and generic primers to identify trypanosome infections of wild tsetse flies in Tanzania by PCR. Infect Genet Evol. 2003;3(4):271–279. doi: 10.1016/S1567-1348(03)00090-X. [DOI] [PubMed] [Google Scholar]
- 10.Solano P, Argiro L, Reifenberg JM, Yao Y, Duvallet G. Field application of the polymerase chain reaction (PCR) to the detection and characterization of trypanosomes in Glossina longipalpis (Diptera: Glossinidae) in Côte d'Ivoire. Mol Ecol. 1995;4(6):781–786. doi: 10.1111/j.1365-294X.1995.tb00280.x. [DOI] [Google Scholar]
- 11.Woolhouse MEJ, McNamara JJ, Hargrove JW, Bealby KA. Distribution and abundance of trypanosome (subgenus Nannomonas) infections of the tsetse fly Glossina pallidipes in southern Africa. Mol Ecol. 1996;5(1):11–18. doi: 10.1111/j.1365-294X.1996.tb00287.x. [DOI] [PubMed] [Google Scholar]
- 12.Morlais I, Grebaut P, Bodo JM, Djoha S, Cuny G, Herder S. Detection and identification of trypanosomes by polymerase chain reaction in wild tsetse flies in Cameroon. Acta Trop. 1998;70(1):109–117. doi: 10.1016/S0001-706X(98)00014-X. [DOI] [PubMed] [Google Scholar]
- 13.Lehane MJ, Msangi AR, Whitaker CJ, Lehane SM. Grouping of trypanosome species in mixed infections in Glossina pallidipes. Parasitology. 2000;120(06):583–592. doi: 10.1017/S0031182099005983. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton PB, Adams ER, Malele II, Gibson WC. A novel high throughput technique for species identification reveals a new species of tsetse-transmitted trypanosome related to Trypanosoma brucei. Infect Genet Evol. 2008;8:26–33. doi: 10.1016/j.meegid.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Adams ER, Hamilton PB, Rodrigues AC, Malele II, Delespaux V, Teixeira MMG, Gibson W. New Trypanosoma (Duttonella) vivax genotypes from tsetse flies in East Africa. Parasitology. 2009;137(04):641–650. doi: 10.1017/S0031182009991508. [DOI] [PubMed] [Google Scholar]
- 16.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 17.Kontopantelis E, Reeves D. Performance of statistical methods for meta-analysis when true study effects are non-normally distributed: a simulation study. Stat Methods Med Res. 2012;21(4):409–426. doi: 10.1177/0962280210392008. [DOI] [PubMed] [Google Scholar]
- 18.Harbord RM, Higgins JP. Meta-regression in Stata. Stata J. 2008;8(4):493–519. [Google Scholar]
- 19.Thompson SG, Higgins JP. How should meta-regression analyses be undertaken and interpreted? Stat Med. 2002;21(11):1559–1573. doi: 10.1002/sim.1187. [DOI] [PubMed] [Google Scholar]
- 20.Lloyd LI, Johnson WB. The trypanosome infections of tsetse-flies in northern Nigeria and a new method of estimation. Bull Entomol Res. 1924;14:265–288. [Google Scholar]
- 21.Ouma JO, Masake RA, Masiga DK, Moloo SK, Njuguna JT, Ndung’u JM. Comparative sensitivity of dot-ELISA, PCR and dissection method for the detection of trypanosome infections in tsetse flies (Diptera: Glossinidae) Acta Trop. 2000;75:315–321. doi: 10.1016/S0001-706X(00)00065-6. [DOI] [PubMed] [Google Scholar]
- 22.Burtt E. Salivation by Glossina morsitans on to glass slides; a technique for isolating infected flies. Ann Trop Med Parasitol. 1946;40:141–144. doi: 10.1080/00034983.1946.11685271. [DOI] [PubMed] [Google Scholar]
- 23.Gibson W, Stevens J, Truc P. Identification of trypanosomes: from morphology to molecular biology. In: Dumas M, Bouteille B, Buguet A, editors. Progress in human African Trypanosomiasis, sleeping sickness. Paris: Springer; 1999. pp. 7–29. [Google Scholar]
- 24.Zongo I, Mbahin N, Van den Abbeele J, De Deken R, Van den Bossche P. Comparison of the infection rate of tsetse, Glossina morsitans morsitans, fed in vitro or in vivo. Med Vet Entomol. 2004;18(1):64–66. doi: 10.1111/j.1365-2915.2004.0474.x. [DOI] [PubMed] [Google Scholar]
- 25.Gashumba JK, Baker RD, Godfrey DG. Trypanosoma congolense: the distribution of enzymic variants in East and West Africa. Parasitology. 1988;96(03):475–486. doi: 10.1017/S0031182000080112. [DOI] [PubMed] [Google Scholar]
- 26.Godfrey DG, Baker RD, Rickman LR, Mehlitz D. The distribution, relationships and identification of enzymic variants within the subgenus Trypanozoon. Adv Parasitol. 1990;29:1–74. doi: 10.1016/S0065-308X(08)60104-9. [DOI] [PubMed] [Google Scholar]
- 27.Bosompem KM, Masake RA, Assoku RK, Opiyo EA, Nantulya VM. Field evaluation of a dot-ELISA for the detection and differentiation of trypanosome species in infected tsetse flies (Glossina spp.) Parasitology. 1995;112:205–211. doi: 10.1017/S0031182000084778. [DOI] [PubMed] [Google Scholar]
- 28.Bosompem KM, Assoku RKG, Nantulya VM. Hydrogenperoxide destaining: a new method for removing non-specific stains in nitrocellulose membrane-based dot-ELISA for detecting trypanosomes in tsetse flies (Glossina spp.) J Immunol Methods. 1995;187:23–31. doi: 10.1016/0022-1759(95)00163-5. [DOI] [PubMed] [Google Scholar]
- 29.Majiwa PAO, Otieno LH. Recombinant DNA probes reveal simultaneous infection of tsetse flies with different trypanosome species. Mol Biochem Parasitol. 1990;40:245–254. doi: 10.1016/0166-6851(90)90046-O. [DOI] [PubMed] [Google Scholar]
- 30.McNamara J, Dukes P, Snow WF, Gibson WC. Use of DNA probes to identify Trypanosoma congolense and T. simiae in tsetse from the Gambia. Acta Trop. 1989;46:55–61. doi: 10.1016/0001-706X(89)90016-8. [DOI] [PubMed] [Google Scholar]
- 31.Kukla BA, Majiwa PAO, Young CJ, Moloo SK, Ole-Moiyoi OK. Use of species-specific DNA probes for the detection and identification of trypanosome infections in tsetse flies. Parasitology. 1987;95:1–26. doi: 10.1017/S0031182000057498. [DOI] [PubMed] [Google Scholar]
- 32.Deborggraeve S, Büscher P. Molecular diagnostics for sleeping sickness: what is the benefit for the patient? Lancet Infect Dis. 2010;10(6):433–439. doi: 10.1016/S1473-3099(10)70077-3. [DOI] [PubMed] [Google Scholar]
- 33.Adams E, Hamilton PB, Malele I, Gibson WC. The identification, diversity and prevalence of trypanosomes in field caught tsetse in Tanzania using ITS-1 primers and fluorescent fragment length barcoding. Infect Genet Evol. 2008;8:439–444. doi: 10.1016/j.meegid.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 34.Adams ER, Malele II, Msangi AR, Gibson WC. Trypanosome identification in wild tsetse populations in Tanzania using generic primers to amplify the ribosomal RNA ITS-1 region. Acta Trop. 2006;100:103–109. doi: 10.1016/j.actatropica.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Ferreira F, Cano J, Furtado A, Ndong-Mabale N, Ndong-Asumu P, Benito A, et al. An alternative approach to detect Trypanosoma in Glossina (Diptera, Glossinidae) without dissection. J Infect Dev Ctries. 2008;2(1):63–67. doi: 10.3855/jidc.324. [DOI] [PubMed] [Google Scholar]
- 36.Ravel S, Grébaut P, Cuisance D, Cuny G. Monitoring the developmental status of Trypanosoma brucei gambiense in the tsetse fly by means of PCR analysis of anal and saliva drops. Acta Trop. 2003;88(2):161–165. doi: 10.1016/S0001-706X(03)00191-8. [DOI] [PubMed] [Google Scholar]
- 37.Ahmed HA, MacLeod ET, Welburn SC, Picozzi K. Development of real time PCR to study experimental mixed infections of T. congolense Savannah and T. B. brucei In Glossina morsitans morsitans. PLoS One. 2015;10(3):e0117147. doi: 10.1371/journal.pone.0117147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuboki N, Inoue N, Sakurai T, Di Cello F, Grab DJ, Suzuki H, et al. Loop-mediated isothermal amplification for detection of African trypanosomes. J Clin Microbiol. 2003;41(12):5517–5524. doi: 10.1128/JCM.41.12.5517-5524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mweempwa C, Marcotty T, De Pus C, Penzhorn BL, Dicko AH, Bouyer J, De Deken R. Impact of habitat fragmentation on tsetse populations and trypanosomosis risk in eastern Zambia. Parasit Vectors. 2015;8:406. doi: 10.1186/s13071-015-1018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ducheyne E, Mweempwa C, De Pus C, Vernieuwe H, De Deken R, Hendrickx G, Van den Bossche P. The impact of habitat fragmentation on tsetse abundance on the plateau of eastern Zambia. Prev Vet Med. 2009;91(1):11–18. doi: 10.1016/j.prevetmed.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grace D, Randolph T, Affognon H, Dramane D, Diall O, Clausen PH. Characterisation and validation of farmers’ knowledge and practice of cattle trypanosomosis management in the cotton zone of West Africa. Acta Trop. 2009;111(2):137–143. doi: 10.1016/j.actatropica.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Lutumba P, Robays J, Miaka mia Bilenge C, Betu Ku Mesu VK, Molisho D, Declercq J, et al. Trypanosomiasis control, Democratic Republic of Congo, 1993-2003. Emerg Infect Dis. 2005;11(9):1382–1388. doi: 10.3201/eid1109.041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hargrove JW, Ouifki R, Kajunguri D, Vale GA, Torr SJ. Modeling the control of trypanosomiasis using trypanocides or insecticide-treated livestock. PLoS Negl Trop Dis. 2012;6(5):e1615. doi: 10.1371/journal.pntd.0001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lefrancois T, Solano P, Bauer B, Kabore I, Toure' SM, Cuny G, Duvallet G. Polymerase chain reaction characterization of trypanosomes in Glossina morsitans sub morsitans and G. tachinoides collected on the game ranch of Nazinga, Burkina Faso. Acta Trop. 1999;72:65–77. doi: 10.1016/S0001-706X(98)00080-1. [DOI] [PubMed] [Google Scholar]
- 45.England EC, Baldry DAT. The hosts and trypanosome infection rates of Glossina pallidipes in the Lambwe and Roo valleys. Bull World Health Organ. 1972;47(6):785–788. [PMC free article] [PubMed] [Google Scholar]
- 46.Moloo SK, Steiger RF, Brun R. Trypanosome infection rates in Glossina swynnertoni and G. pallidipes in Ikoma, Musoma district, Tanzania. Parasitology. 1973;66(02):259–267. doi: 10.1017/S0031182000045194. [DOI] [PubMed] [Google Scholar]
- 47.Woolhouse MEJ, Bealby K, McNamara JJ, Silutongwe J. Trypanosome infections of the tsetse fly Glossina pallidipes in the Luangwa Valley Zambia. Int J Parasitol. 1994;24(7):987–993. doi: 10.1016/0020-7519(94)90164-3. [DOI] [PubMed] [Google Scholar]
- 48.Auty H, Anderson NE, Picozzi K, Lembo T, Mubanga J, Hoare R, et al. Trypanosome diversity in wildlife species from the Serengeti and Luangwa valley ecosystems. PLoS Negl Trop Dis. 2012;6(10) doi: 10.1371/journal.pntd.0001828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke JE. Trypanosome infection rates in the mouthparts of Zambian tsetse flies. Ann Trop Med Parasitol. 1969;63(1):15–34. doi: 10.1080/00034983.1969.11686596. [DOI] [PubMed] [Google Scholar]
- 50.Croft SL, Kuzoe FA, Ryan L, Molyneux DH. Trypanosome infection rates of Glossina spp.(Diptera: Glossinidae) in transitional forest-savannah near Bouafle, Ivory Coast. Tropenmed Parasitol. 1984;35(4):247–250. [PubMed] [Google Scholar]
- 51.Desta M, Menkir S, Kebede M. The study on tsetse fly (Glossina species) and their role in the trypanosome infection rate in Birbir valley, Baro Akobo River system, western Ethiopia. J Vet Med Anim Health. 2013;5(7):186–194. [Google Scholar]
- 52.Djohan V, Kaba D, Rayaissé JB, Dayo GK, Coulibaly B, Salou E, et al. Detection and identification of pathogenic trypanosome species in tsetse flies along the Comoé River in Côte d’Ivoire. Parasite. 2015;22:18. doi: 10.1051/parasite/2015018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gillingwater K, Mamabolo MV, Majiwa PAO. Prevalence of mixed Trypanosoma congolense infections in livestock and tsetse in KwaZulu-Natal, South Africa. J S Afr Vet Assoc. 2010;81(4):219–223. doi: 10.4102/jsava.v81i4.151. [DOI] [PubMed] [Google Scholar]
- 54.Gomes J, Leão C, Ferreira F, Afonso M, Santos C, Josenando T, et al. Molecular identification of T. brucei s.L. in tsetse flies after long-term capture in field traps. J Infect Dev Ctries. 2009;3(9):735–738. doi: 10.3855/jidc.389. [DOI] [PubMed] [Google Scholar]
- 55.Harley JMB. Further studies on age and trypanosome infection rates in Glossina pallidipes Aust., G. palpalis fuscipes Newst. And G. brevipalpis Newst. In Uganda. Bull Entomol Res. 1967;57(03):459–477. doi: 10.1017/S0007485300050203. [DOI] [PubMed] [Google Scholar]
- 56.Jordan AM. Trypanosome infection rates in Glossina morsitans submorsitans Newst. In Northern Nigeria. Bull Entomol Res. 1963;55:219–231. doi: 10.1017/S0007485300049415. [DOI] [Google Scholar]
- 57.Konnai S, Mekata H, Odbileg R, Simuunza M, Chembensof M, Witola WH, et al. Detection of Trypanosoma brucei in field-captured tsetse flies and identification of host species fed on by the infected flies. Vector Borne Zoonotic Dis. 2008;8(4):565–574. doi: 10.1089/vbz.2007.0223. [DOI] [PubMed] [Google Scholar]
- 58.Madubunyi LC. Trypanosome infections in Glossina spp. inhabiting peridomestic agro-ecosystems in Nsukka area, Anambra state, Nigeria. Ann Trop Med Parasitol. 1987;81(3):319–329. doi: 10.1080/00034983.1987.11812126. [DOI] [PubMed] [Google Scholar]
- 59.Malele II, Magwisha HB, Nyingilili HS, Mamiro KA, Rukambile EJ, Daffa JW, et al. Multiple Trypanosoma infections are common amongst Glossina species in the new farming areas of Rufiji district, Tanzania. Parasites Vectors. 2011;4:217. doi: 10.1186/1756-3305-4-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mamabolo MV, Ntantiso L, Latif A, Majiwa PAO. Natural infection of cattle and tsetse flies in South Africa with two genotypic groups of Trypanosoma congolense. Parasitology. 2009;136(04):425–431. doi: 10.1017/S0031182009005587. [DOI] [PubMed] [Google Scholar]
- 61.McNamara JJ, Snow WF. Improved identification of Nannomonas infections in tsetse flies from the Gambia. Acta Trop. 1991;48:127–136. doi: 10.1016/0001-706X(90)90052-2. [DOI] [PubMed] [Google Scholar]
- 62.McNamara JJ, Laveissiere C, Masiga DK. Multiple trypanosome infections in wild tsetse in cote d’Ivoire detected by PCR analysis. Acta Trop. 1995;59:85–92. doi: 10.1016/0001-706X(94)00087-H. [DOI] [PubMed] [Google Scholar]
- 63.Mehlitz D, Tietjen U. Trypanosome infection rates in tsetse mid guts using a short-term in vitro culture technique. Acta Trop. 1988;45(2):183–184. [PubMed] [Google Scholar]
- 64.Mekata H, Konnai S, Simuunza M, Chembensofu M, Kano R, Witola WH, et al. Prevalence and source of trypanosome infections in field-captured vector flies (Glossina pallidipes) in south eastern Zambia. J Vet Med Sci. 2008;70(9):923–928. doi: 10.1292/jvms.70.923. [DOI] [PubMed] [Google Scholar]
- 65.Mihok S, Otieno LH, Tarimo CS. Trypanosome infection rates in tsetse flies (Diptera: Glossinidae) and cattle during tsetse control operations in the Kagera River region of Rwanda. Bull Entomol Res. 1992;82(03):361–367. doi: 10.1017/S0007485300041158. [DOI] [Google Scholar]
- 66.Mohammed YO, Mohamed-Ahmed MM, Lubna TK, Rayah IE. Detection of Trypanosoma brucei gambiense and T. b. rhodesiense in Glossina fuscipes fuscipes (Diptera: Glossinidae) and Stomoxys flies using the polymerase chain reaction (PCR) technique in southern Sudan. Afr J Biotechnol. 2010;9(38):6408–6412. [Google Scholar]
- 67.Morlais I, Ravel S, Grébaut P, Dumas V, Cuny G. New molecular marker for Trypanosoma (Duttonella) vivax identification. Acta Trop. 2001;80(3):207–213. doi: 10.1016/S0001-706X(01)00160-7. [DOI] [PubMed] [Google Scholar]
- 68.Motloang M, Masumu J, Mans B, Van den Bossche P, Latif A. Vector competence of Glossina austeni and Glossina brevipalpis for Trypanosoma congolense in KwaZulu-Natal, South Africa. Onderstepoort J Vet Res. 2012;79(1):1–6. doi: 10.4102/ojvr.v79i1.353. [DOI] [PubMed] [Google Scholar]
- 69.Msangi AR, Whitaker CJ, Lehane MJ. Factors influencing the prevalence of trypanosome infection of Glossina pallidipes on the Ruvu flood plain of eastern Tanzania. Acta Trop. 1998;70(2):143–155. doi: 10.1016/S0001-706X(98)00013-8. [DOI] [PubMed] [Google Scholar]
- 70.Mwandiringana E, Gori E, Nyengerai T, Chidzwondo F. Polymerase chain reaction (PCR) detection of mixed trypanosome infection and blood meal origin in field captured tsetse flies from Zambia. Afr J Biotechnol. 2012;11(79):14490–14497. [Google Scholar]
- 71.Njiru ZK, Makumi JN, Okoth S, Ndungu JM, Gibson WC. Identification of trypanosomes in Glossina pallidipes and G. longipennis in Kenya. Infect Genet Evol. 2004;4:29–35. doi: 10.1016/j.meegid.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 72.Okiwelu SN. Host preference and trypanosome infection rates of Glossina morsitans morsitans Westwood in the Republic of Zambia. Ann Trop Med Parasitol. 1977;71(1):101–107. doi: 10.1080/00034983.1977.11687166. [DOI] [PubMed] [Google Scholar]
- 73.Okoh KE, Anavhe A, Ayakpat HN, Onotu CS, Anchau R, Ajakaiye JJ. Trypanosomes infection in field-captured tsetse flies of the subgenus: Nemorhina in southern Guinea savannah zone of Nigeria. Curr Res J Biol Sci. 2012;4(6):713–716. [Google Scholar]
- 74.Okoth JO, Kapaata R. Trypanosome infection rates in Glossina fuscipes fuscipes Newst. In the Busoga sleeping sickness focus, Uganda. Ann Trop Med Parasitol. 1986;80(4):459–461. doi: 10.1080/00034983.1986.11812048. [DOI] [PubMed] [Google Scholar]
- 75.Riordan K. Long term variations in trypanosome infection rates in highly infected tsetse flies on a cattle route in south-western Nigeria. Ann Trop Med Parasitol. 1977;71(1):11–20. doi: 10.1080/00034983.1977.11687156. [DOI] [PubMed] [Google Scholar]
- 76.Simo G, Silatsa B, Njiokou F, Lutumba P, Mansinsa P, Madinga J, et al. Identification of different trypanosome species in the mid-guts of tsetse flies of the Malanga (Kimpese) sleeping sickness focus of the Democratic Republic of Congo. Parasit Vectors. 2012;5:201. doi: 10.1186/1756-3305-5-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simo G, Fogue PS, Melachio TTT, Njiokou F, Kuiate JR, Asonganyi T. Population genetics of forest type of Trypanosoma congolense circulating in Glossina palpalis palpalis of Fontem in the south-West region of Cameroon. Parasit Vectors. 2014;7(1):385. doi: 10.1186/1756-3305-7-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tarimo SRA, Snow WF, Butler L. Trypanosome infections in wild tsetse, Glossina pallidipes Austen on the Kenya coast. Int J Trop Insect Sci. 1984;5(05):415–418. doi: 10.1017/S1742758400008754. [DOI] [Google Scholar]
- 79.Tarimo SA, Snow FW, Butler L, Dransfield R. The probability of tsetse acquiring trypanosome infection from single blood meal in different localities in Kenya. Acta Trop. 1985;42(3):199–207. [PubMed] [Google Scholar]
- 80.Waiswa C, Rannaleet AH. Entrepreneurship initiatives in the control of sleeping sickness: experiences of the stamp out sleeping sickness (SOS) initiative in Uganda. J Small Business Entrepreneurship. 2010;23(4):555–564. doi: 10.1080/08276331.2010.10593501. [DOI] [Google Scholar]
- 81.Moloo SK, Gray MA. New observations on cyclical development of Trypanosoma vivax in Glossina. Acta Trop. 1989;46(3):167–172. doi: 10.1016/0001-706X(89)90033-8. [DOI] [PubMed] [Google Scholar]
- 82.Moloo SK. Studies on the infection rates of a West African stock of Trypanosoma vivax in Glossina morsitans morsitans and G. M. centralis. Ann Trop Med Parasitol. 1982;76(3):355–359. doi: 10.1080/00034983.1982.11687552. [DOI] [PubMed] [Google Scholar]
- 83.Moloo SK, Kutuza SB, Desai J. Comparative study on the infection rates of different Glossina species for East and West African Trypanosoma vivax stocks. Parasitology. 1987;95(03):537–542. doi: 10.1017/S0031182000057966. [DOI] [PubMed] [Google Scholar]
- 84.Moloo SK, Kutuza SB. Comparative study on the infection rates of different laboratory strains of Glossina species by Trypanosoma congolense. Med Vet Entomol. 1988;2(3):253–257. doi: 10.1111/j.1365-2915.1988.tb00192.x. [DOI] [PubMed] [Google Scholar]
- 85.Chitanga S, Namangala B, De Deken R, Marcotty T. Shifting from wild to domestic hosts: the effect on the transmission of Trypanosoma congolense to tsetse flies. Acta Trop. 2013;125(1):32–36. doi: 10.1016/j.actatropica.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 86.Bosompem KM, Moloo SK, Assoku RKG, Nantulya VM. Detection and differentiation between trypanosome species in experimentally-infected tsetse flies (Glossina spp.) using dot-ELISA. Acta Trop. 1995;60:81–96. doi: 10.1016/0001-706X(95)00111-Q. [DOI] [PubMed] [Google Scholar]
- 87.Bouyer J, Koné N, Bengaly Z. Dynamics of tsetse natural infection rates in the Mouhoun river, Burkina Faso, in relation with environmental factors. Front Cell Infect Microbiol. 2013;3:47. doi: 10.3389/fcimb.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Distelmans W, D’Haeseleer F, Kaufman L, Rousseeuw P. The susceptibility of Glossina palpalis palpalis at different ages to infection with Trypanosoma congolense. Ann Soc Belg Med Trop. 1982;62(1):41–47. [PubMed] [Google Scholar]
- 89.Harley JMB. Further studies on age and trypanosome infection rates in Glossina pallidipes Aust., G. palpalis fuscipes Newst. And G. brevipalpis Newst. In Uganda. B Entomol Res. 1966;57(03):459–477. doi: 10.1017/S0007485300050203. [DOI] [PubMed] [Google Scholar]
- 90.Harley JM. Comparison of the susceptibility of infection with Trypanosoma rhodesiense of Glossina pallidipes, G. morsitans, G. fuscipes and G. brevipalpis. Ann Trop Med Parasitol. 1971;65(2):185–189. doi: 10.1080/00034983.1971.11686744. [DOI] [PubMed] [Google Scholar]
- 91.Masiga DK, Smyth AJ, Hayes PJ, Bromidge TJ, Gibson WC. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol. 1992;22:909–918. doi: 10.1016/0020-7519(92)90047-O. [DOI] [PubMed] [Google Scholar]
- 92.Masumu J, Marcotty T, Ndeledje N, Kubi C, Geerts S, Vercruysse J, et al. Comparison of the transmissibility of Trypanosoma congolense strains, isolated in a trypanosomiasis endemic area of eastern Zambia, by Glossina morsitans morsitans. Parasitology. 2006;133(03):331–334. doi: 10.1017/S0031182006000369. [DOI] [PubMed] [Google Scholar]
- 93.Peacock L, Ferris V, Bailey M, Gibson W. Dynamics of infection and competition between two strains of Trypanosoma brucei brucei in the tsetse fly observed using fluorescent markers. Kinetoplastid Biol Dis. 2007;6(1):4. doi: 10.1186/1475-9292-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peacock L, Cook S, Ferris V, Bailey M, Gibson W. The life cycle of Trypanosoma (Nannomonas) congolense in the tsetse fly. Parasit Vectors. 2012;5(1):109. doi: 10.1186/1756-3305-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ravel S, Grébaut P, Mariani C, Jamonneau V, Cuisance D, Gooding RH, Cuny G. Monitoring the susceptibility of Glossina palpalis gambiensis and G. morsitans morsitans to experimental infection with savannah-type Trypanosoma congolense, using the polymerase chain reaction. Ann Trop Med Parasit. 2004;98(1):29–36. doi: 10.1179/000349804225003028. [DOI] [PubMed] [Google Scholar]
- 96.Roberts LW. Probing by Glossina morsitans morsitans and transmission of Trypanosoma (Nannomonas) congolense. Am J Trop Med Hyg. 1981;30(5):948–951. doi: 10.4269/ajtmh.1981.30.948. [DOI] [PubMed] [Google Scholar]
- 97.Rotureau B, Subota I, Buisson J, Bastin P. A new asymmetric division contributes to the continuous production of infective trypanosomes in the tsetse fly. Development. 2012;139(10):1842–1850. doi: 10.1242/dev.072611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Additional file 1. Supplementary excel file is included.


