Abstract
Centromere-specific H3-like proteins (CenH3s) are conserved across the eukaryotic kingdom and are required for packaging centromere DNA into a specialized chromatin structure required for kinetochore assembly. Cse4 is the CenH3 protein of the budding yeast Saccharomyces cerevisiae. Like all CenH3 proteins, Cse4 consists of a conserved histone fold domain (HFD) and a divergent N terminus (NT). The Cse4 NT contains an essential domain designated END (for essential N-terminal domain); deletion of END is lethal. To investigate the role of the Cse4 NT in centromere targeting, a series of deletion alleles (cse4ΔNT) were analyzed. No part of the Cse4 NT was required to target mutant proteins to centromere DNA in the presence of functional Cse4. A Cse4 degron strain was used to examine targeting of a Cse4ΔNT protein in the absence of wild-type Cse4. The END was not required for centromere targeting under these conditions, confirming that the HFD confers specificity of Cse4 centromere targeting. Surprisingly, overexpression of the HFD bypassed the requirement for the END altogether, and viable S. cerevisiae strains in which the cells express only the Cse4 HFD and six adjacent N-terminal amino acids (Cse4Δ129) were constructed. Despite the complete absence of the NT, mitotic chromosome loss in the cse4Δ129 strain increased only 6-fold compared to a 15-fold increase in strains overexpressing wild-type Cse4. Thus, when overexpressed, the Cse4 HFD is sufficient for centromere function in S. cerevisiae, and no posttranslational modification or interaction of the NT with other kinetochore component(s) is essential for accurate chromosome segregation in budding yeast.
Each time a eukaryotic cell divides, it replicates its genome and accurately distributes a complete set of chromosomes to each daughter cell. To accomplish the complicated process of chromosome segregation, the cell assembles a spindle apparatus containing microtubules that interact with the centromere region of each chromosome. Interactions between the spindle fiber and centromere are mediated by the kinetochore, a DNA-protein complex common to all eukaryotic organisms (57). Budding yeast Saccharomyces cerevisiae chromosomes each have one well-defined “point” centromere that interacts with a single microtubule in vivo (34). Mammalian centromeres are highly complex, contain megabases of DNA, and interact with multiple spindle fibers (10). Despite large differences in size and complexity, the primary function of all kinetochores is to establish and maintain a stable connection between the chromosome and spindle apparatus. A structural feature common to all kinetochores is the packaging of the underlying centromeric DNA into nucleosomes containing a specialized, centromere-specific histone H3 variant called CenH3 (51).
The nucleosome is the fundamental repeating structural unit of eukaryotic chromosomal DNA. Each nucleosome consists of an octamer of the four core histones (H2A, H2B, H3, and H4), around which is wrapped ∼146 bp of DNA (30). Nucleosome assembly initiates with the deposition of an (H3-H4)2 tetramer onto DNA, each tetramer wrapping 120 bp of DNA (12, 15). Subsequent incorporation of two dimers (H2A-H2B) and additional DNA completes the nucleosome structure. In centromeric nucleosomes, the specialized CenH3 protein replaces the standard histone H3 in the nucleosome core (60). The CenH3 proteins are the most highly conserved components of eukaryotic kinetochores, now identified in many organisms, including mammals (CENP-A), flies (Cid), worms (HCP-3), Arabidopsis (HTR12), fission yeast (Cnp1), and S. cerevisiae (Cse4) (32).
CenH3 proteins have two major structural domains, a divergent N terminus and a conserved histone fold domain (HFD) which shares about 50% amino acid identity with the HFD of histone H3 (32). Several studies have shown that the specificity of CenH3s for centromere DNA is species dependent and conferred by the HFD. In these “domain swap” experiments, mutant CenH3 proteins in which all or part of the HFD, with or without the N terminus, was replaced by the homologous region of either H3 or a heterologous CenH3 were constructed. The chimeric proteins were expressed and analyzed for their ability to localize to the cognate centromere DNA. While the HFD was found to be necessary and sufficient for centromere targeting of CENP-A and Cid (16, 47, 59), the experimental systems utilized wild-type cells, and heterodimer formation with the endogenous CenH3 (i.e., “piggybacking”) was not ruled out. In addition, these studies did not determine whether the centromere-targeted HFDs actually functioned in the assembly of active kinetochores.
Compared to the HFD, the N termini of CenH3 proteins differ greatly in length and amino acid sequence (32), providing few clues to possible function. In Drosophila lineages, the N terminus of Cid has evolved adaptively, implying that it confers a function subjected to selective pressure (31), perhaps contributing a specificity determinant to CenH3 centromere DNA binding (33) or acting to stabilize Cid nucleosomes (59). The N terminus of CENP-A has been implicated in binding other mammalian kinetochore components (58). In S. cerevisiae, the Cse4 N terminus is 129 amino acid residues long and is required for cell viability (22). Genetic analysis has revealed that the function of the Cse4 N terminus is distinct from that of the HFD and that the region from amino acid residues 28 to 60, designated the END (for essential N-terminal domain), confers the essential function (9). When expressed in cells carrying a conditional lethal cse4 HFD mutation, Cse4 deletion derivatives lacking the END (Cse4ΔNT) localize to centromere DNA and functionally complement the HFD defect, suggesting that, as for Cid and CENP-A, the Cse4 N terminus is not required to confer centromere specificity (9). However, in this case, heterodimer formation between the END-less Cse4ΔNT proteins and the HFD mutant protein was shown to occur, begging the question of whether the Cse4 HFD would be sufficient for centromere localization (or function) in the absence of a functional heterodimerization partner.
In this study, we describe experiments designed to analyze the behavior of Cse4ΔNT proteins in the presence and absence of wild-type Cse4. Using a Cse4 degron strain, we found that the HFD of Cse4 is sufficient for centromere targeting in the absence of detectable wild-type protein. Unexpectedly, we also found that the essential function of the END is bypassed by overexpression of the Cse4 HFD and that under these conditions the Cse4 N terminus is entirely dispensable for cell viability. The overexpressed Cse4 HFD protein was more stable than wild-type Cse4, suggesting that the Cse4 N terminus influences Cse4 protein turnover in vivo. Most surprisingly, S. cerevisiae cells that overexpress the Cse4 HFD as the sole source of Cse4 protein exhibited only mild impairment of chromosome segregation. We conclude that in S. cerevisiae, the HFD of Cse4 is necessary and sufficient for both centromere targeting and propagation of active centromeres. We discuss our results in light of current models of kinetochore structure in S. cerevisiae and compare Cse4 function with what is known about CenH3 protein function in other eukaryotic organisms.
MATERIALS AND METHODS
S. cerevisiae strains and plasmids.
The media for S. cerevisiae growth were described previously (48). All minimal media containing galactose contained 2% galactose and 0.5% sucrose. S. cerevisiae strains used in this study are listed in Table 1. The Cse4 degron strain YC405 was constructed as described by Moqtaderi et al. (38) using a targeting plasmid derived from pZM168 by inserting DNA encoding residues 2 to 161 of Cse4. After linearization, integration of the plasmid results in disruption of the endogenous CSE4 gene and the simultaneous generation of the degron-tagged Cse4 allele. A haploid strain carrying the chromosomal cse4Δ::kanMX4 allele was obtained by transforming the heterozygous diploid strain 24898 (Invitrogen) with the URA3-CSE4-CEN-ARS plasmid pRB163 and obtaining a Kanr meiotic segregant. The trp1Δ63 allele (5) was introduced by a genetic cross. A MET15-marked chromosome fragment (CF) derived from chromosome III was obtained by the method of Shero et al. (49). The fragmentation vector pJS2 was modified by replacing the SmaI-SalI segment (SUP11-URA3) with a SmaI-SalI MET15 fragment produced by PCR, using pRS401 (5) as template. The resulting CF, originally formed in strain R332-1C was transferred to other strain backgrounds by a genetic cross. In all cases, the CF segregated 2:2.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotypea | Reference or source |
|---|---|---|
| KC100 | MATα ade2-101 his3-11,15 leu2-3 lys2-801 trp1Δ901 ura3-52 cse4::HIS3[pCL1] | 22 |
| ZMY60 | MATaade2 trp1-1 ura3-52 leu2::PET56 | 38 |
| YC405 | MATaade2 trp1-1 ura3-52 leu2::PET56 HA-tagged CSE4 dgrn::TRP1 | This study |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 5 |
| BY4734 | MATahis3Δ200 leu2Δ0 met15Δ0 ura3Δ0 trp1Δ63 | 5 |
| 24898 | MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 lys2Δ0/LYS2 ura3Δ0/ura3Δ0 cse4Δ::kanMX4/CSE4 | SGDPb |
| R332-5C | MATahis3Δ leu2Δ0 ura3Δ0 trp1Δ63 cse4Δ::kanMX4[pRB163] | This study |
| R332-4B | MATα his3Δ leu2Δ0 ura3Δ0 trp1Δ63 met15Δ0 lys2Δ0 | This study |
| R358 | MATa/MATα his3Δ/his3Δ leu2Δ0/leu2Δ0 met15Δ0/MET15 lys2Δ0/LYS2 ura3Δ0/ura3Δ0 trp1Δ63/trp1Δ63 cse4Δ::kanMX4/CSE4 | This study |
| R365 | MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 lys2Δ0/LYS2 ura3Δ0/ura3Δ0 trp1Δ63/trp1Δ63 gal-cse4Δ129::TRP1/CSE4 | This study |
| R365-5B | MATahis3Δ leu2Δ0 met15Δ0 lys2Δ0 ura3Δ0 trp1Δ63 gal-cse4Δ129::TRP1 | This study |
| R363-1D | MATahis3Δ leu2Δ0 met15Δ0 lys2Δ0 ura3Δ0 trp1Δ63 gal-CSE4::TRP1 | This study |
| R386-1C | MATα his3Δ leu2Δ0 ura3Δ0 trp1 Δ63 met15Δ0 lys2Δ0 [CFIII(D8B.d.R332-1C)MET15] | This study |
| R367-3A | MATahis3Δ leu2Δ0 met15Δ0 lys2Δ0 ura3Δ0 trp1Δ63 gal-cse4Δ129::TRP1 [CFIII(D8B.d.R332-1C)MET15] | This study |
| R368-2D | MATahis3Δ leu2Δ0 met15Δ0 lys2Δ0 ura3Δ0 trp1Δ63 gal-CSE4::TRP1 [CFIII(D8B.d.R332-1C)MET15] | This study |
| R373-2C | MATahis3Δ leu2Δ0 met15Δ0 lys2Δ0 ura3Δ0 trp1Δ63 | This study |
| 3677 | BY4741 mcm21Δ::kanMX4 | SGDP |
| 2613 | BY4741 mcm22Δ::kanMX4 | SGDP |
The starting plasmid for constructing Cse4 N-terminal deletions was pRB294, which contains the triple-hemagglutinin (HA)-tagged CSE4 allele (53) cloned into the CEN-ARS-TRP1 vector pRS314 (50). The CSE4 allele in this plasmid was altered to contain an NsiI site at codon 1. The CSE4 allele in pRB294 was put under control of the GAL1 promoter by exchanging the EcoRI-NsiI segment, containing the CSE4 upstream sequences, with an 814-bp PCR-generated EcoRI-NsiI fragment, containing the GAL1 upstream activation sequence (UAS) and initiator ATG, and fusing it to codon 1 of CSE4 (the NsiI site was designed into the downstream PCR primer). The resulting plasmid, pRB467, was used to construct various N-terminal deletions by replacing the NsiI-SphI segment with analogous fragments produced by PCR and carrying the desired deletions (the SphI site is within the Cse4 open reading frame, downstream of all deletion endpoints). For deletions beyond codon 50, DNA encoding the triple-HA tag was fused directly to the initiator ATG using the NsiI site, and the deleted CSE4 segments were inserted via the downstream NotI site of the tag-encoding DNA (53). All newly constructed alleles were sequenced and found to contain no additional mutations within the Cse4 open reading frame.
A TRP1 integration vector targeting the CSE4 locus was obtained by PCR amplifying 411 bp of CSE4 upstream DNA (coordinates −429 to −19 relative to the CSE4 initiator ATG) and inserting it between the PstI and SacI sites of pRS304 (50). Wild-type or mutant cse4 alleles under GAL1 control were then inserted as EcoRI-PstI fragments. Cleavage of the resulting plasmids by PstI produced linear DNA molecules with the structure CSE4 5′-flank-TRP1-GAL1 UAS-cse4ΔNT (or wild type)-CSE4 3′-flank.
Assays for Cse4 expression and function.
Functional complementation of the cse4Δ::kanMX4 null allele was tested using a plasmid shuffle assay (22). The tester strain was R332-5D, which contains wild-type CSE4 on the CEN-ARS-URA3 plasmid pRB163. This strain was transformed with CEN-ARS-TRP1 plasmids carrying the cse4ΔNT alleles to be tested, and transformants were selected on galactose-containing dropout medium. Trp+ colonies were picked, diluted, and plated on medium containing 5-fluoroorotic acid (FOA) to score loss of pRB163. Plates were photographed after 3 or 4 days.
For Western blots, cells were grown to an optical density at 600 nm of 1 to 8 in rich medium, a volume of culture containing the equivalent of 2 optical density units of cells was centrifuged, and the supernatant was removed. The resulting cell pellets were resuspended in 20 μl of Y-Per permeabilization buffer (Pierce) containing 0.1 M dithiothreitol and 1 mM phenylmethylsulfonyl fluoride. After incubation at room temperature for 20 min, 10 μl of 3× sodium dodecyl sulfate (SDS) sample buffer was added, and the samples were boiled for 2 min. Eight to 12 μl of sample were electrophoresed on SDS-12% polyacrylamide gels (25). After electrophoresis, proteins were transferred to Immobilon (Millipore) membranes, and the HA epitope was detected using anti-HA monoclonal antibody 12CA5 (Roche) and peroxidase-conjugated sheep anti-mouse immunoglobulin (Amersham Biosciences) by chemiluminescence (SuperSignal West Pico; Pierce). Primary and secondary antibodies were diluted 1:1,500 and 1:5,000, respectively.
Northern and Southern blots.
Total RNA was prepared, electrophoresed on formaldehyde-agarose gels, and transferred to nylon membranes as described previously (40). S. cerevisiae genomic DNA was prepared as described previously (48), and 0.5-μg samples were digested with restriction enzymes before electrophoresis on 0.7% agarose gels in standard Tris-borate-EDTA buffer. After electrophoresis, DNA was transferred to Zetaprobe nylon membranes (Bio-Rad) in 0.4 M NaOH using a Posi-Blot apparatus (Stratagene). Specific RNA or DNA on blots was detected using the AlkPhos Direct nonradioactive labeling system (Amersham Biosciences).
ChIP assay.
Chromatin immunoprecipitation (ChIP) analysis was performed as previously described (55). Briefly, approximately 109 cells were treated in situ with 1% formaldehyde for 45 min. Cross-linked chromatin was then prepared (450 μl) and sonicated to fragment the chromosomal DNA. Immunoprecipitation was performed with 400 μl of the chromatin solution using 20 μl of polyclonal mouse anti-HA antibody (Santa Cruz). The remaining 50 μl was used to prepare the total chromatin control samples. PCR was performed with 1/50 of the total chromatin or 1/10 of the immunoprecipitated chromatin using the primers described by Ortiz et al. (41). One-third of each PCR product was analyzed by agarose gel electrophoresis and stained with ethidium bromide.
CF loss assay.
Mitotic CF loss rates were determined by fluctuation analysis as described previously (2) with some modifications. When grown on medium containing lead (5), cells carrying the MET15-marked CF give rise to white Met+ colonies with brown Met− sectors due to CF loss during colony growth. Strains to be assayed were grown on methionine dropout medium to select for cells carrying the CF, then diluted, and plated on complete minimal medium. For each determination, five individual colonies were picked, resuspended in minimal medium lacking amino acids, sonicated briefly, diluted, and plated on lead indicator plates. After 5 to 7 days, the brown colonies (CF−) were counted, the total number of colonies were determined, and the loss rate was determined using the formulas of Lea and Coulson (26). Τo verify that the strain stocks used for the fluctuation analyses carried only a single copy of the CF, they were crossed to a wild-type parent and sporulated. The CF segregated 2:2 (CF+:CF−) in the resulting tetrads.
RESULTS
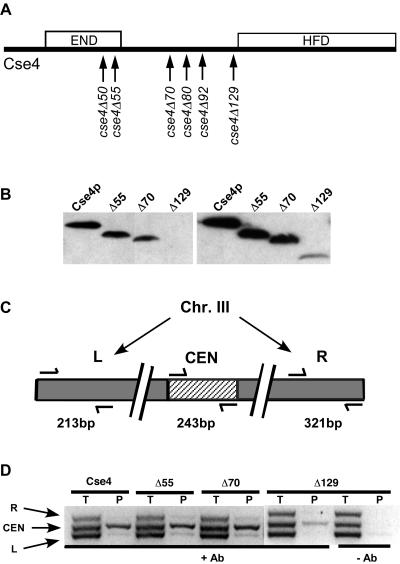
The Cse4 N terminus is not required for CEN localization. In previous studies, we constructed and analyzed cse4 alleles with mutations altering the Cse4 N terminus and found that deletion of the Cse4 END region (amino acids 28 to 60) is lethal, although the proteins with the END region deleted localize to centromere DNA in the presence of wild-type Cse4, in some cases by forming heterodimers with the wild-type protein (9). To further characterize the function(s) of the Cse4 N terminus, we constructed a series of deletion alleles encoding Cse4 proteins with increasing N-terminal deletions (cse4ΔNT) (Fig. 1A). The Cse4ΔNT proteins were analyzed in cells that also express wild-type Cse4. Western blot analysis showed that with the exception of the Cse4Δ129 protein, the truncated proteins were present at levels similar to that of wild-type Cse4 (Fig. 1B and data not shown). ChIP assays were performed to determine whether the Cse4Δ55, Cse4Δ70, and Cse4Δ129 proteins localized to CEN DNA. In these assays, DNA in the Cse4 immunoprecipitates was amplified by PCR using primer pairs designed to amplify CEN3 and two other regions located on the arms of chromosome III (Fig. 1C). The wild-type Cse4 and all three Cse4ΔNT mutant proteins were specifically localized to CEN3 as indicated by enrichment of CEN3 DNA in the immunoprecipitates relative to DNA derived from the L and R control regions (Fig. 1D). Both CEN and non-CEN sequences are detected in the input DNA samples (Fig. 1D). The reduced CEN3 signal from the cse4Δ129 strain reflects the much lower concentration of Cse4Δ129 protein in the cell relative to those of other Cse4 proteins. We conclude that the END is not required to target Cse4 to centromere DNA if wild-type Cse4 is present.
FIG. 1.
CEN localization of Cse4ΔNT proteins expressed in the presence of wild-type Cse4. (A) Schematic diagram showing the endpoints of cse4ΔNT deletions. The cse4ΔNT alleles were named for the number of the last amino acid deleted. The Cse4Δ129 protein contains, in addition to the HFD, the five N-terminal residues that enter the nucleosome core between the DNA gyres and the residue connecting them to the N-helix of the HFD (30). The locations of the END (residues 28 to 60) and HFD (residues 136 to 229) are indicated by the boxes. (B) Western blot showing Cse4ΔNT proteins expressed from native CSE4 promoters. The left and right panels show different exposures of the same blot (2 and 10 min, respectively). Δ55, Cse4Δ55; Δ70, Cse4Δ70; Δ129, Cse4Δ129. (C) PCR primer sets used in the ChIP assays and their relative positions on chromosome III (Chr. III). (D) ChIP assays showing CEN localization of Cse4ΔNT proteins in the presence of endogenous wild-type Cse4 and in the presence (+) and absence (−) of antibody (Ab). Total (T) and immunoprecipitated (P) DNA was used for multiplex PCR with the primer sets shown in panel C.
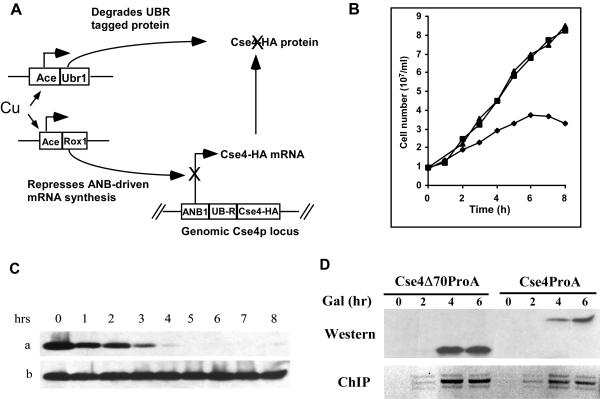
To learn about targeting of the cse4 mutant proteins in the absence of wild-type Cse4, we utilized the S. cerevisiae degron system of Moqtaderi et al. (38). The Cse4 degron construct allowed us to trigger degradation of the wild-type Cse4 prior to inducing expression of the cse4 mutant proteins to be studied. In the presence of copper, the existing Cse4, tagged by HA, is degraded via the ubiquitin pathway with simultaneous inhibition of transcription of the degron allele (Fig. 2A). Genes encoding the mutant proteins, tagged with ProA, were placed under the control of the glucose-repressible GAL1 promoter and introduced into the degron strain on low-copy-number plasmids. After growth on medium containing glucose and copper to eliminate preexisting Cse4, expression of the ProA-tagged Cse4 proteins was induced by switching the cells to media containing galactose and copper.
FIG. 2.
CEN targeting of Cse4Δ70 protein in the absence of wild-type Cse4. (A) In S. cerevisiae strain YC405, the CSE4 locus has been modified so that the wild-type gene product is tagged with HA and is a substrate for rapid, Ubr1p-dependent degradation. Addition of copper to the medium induces expression of both Ubr1p and Rox1p. Ubr1p targets HA-Cse4 for degradation via the ubiquitin pathway, while Rox1p represses the ANB1 promoter and inhibits HA-tagged CSE4 (Cse4-HA) transcription. (B) Growth of strain YC405 (diamonds) and its parent ZMY60 (squares) after the addition of 0.5 mM CuSO4. As a control, YC405 was also grown in the absence of copper (triangles). The CSE4 locus is unmodified in ZMY60. (C) Western blot analysis of HA-Cse4 in YC405 cells grown for 8 h in medium containing 0.5 mM CuSO4 (a) or not containing CuSO4 (b). (D) YC405 cells carrying plasmid-borne ProA-tagged cseΔ70 or ProA-tagged wild-type CSE4 under GAL1 transcriptional control were shifted to medium containing galactose (Gal) and copper after a 5-h incubation in medium containing glucose and 0.5 mM CuSO4 to deplete preexisting Cse4. Aliquots of both cultures were taken at the indicated times after galactose shift and analyzed by Western blotting and ChIP.
After 5 to 6 h in copper-containing medium, the HA-tagged Cse4 (HA-Cse4)-depleted cells stopped dividing (Fig. 2B) and arrested predominantly as large-budded cells (>74% [data not shown]). Western blot analysis showed that HA-Cse4 was not detected 5 h after degron induction (Fig. 2C). Cells carrying ProA-tagged Cse4 (ProA-Cse4) or ProA-Cse4Δ70 plasmids were grown on medium containing glucose and copper for 5 h and then switched to medium containing galactose and copper to induce expression. The ProA-tagged Cse4 proteins were detected 4 h after galactose induction (Fig. 2D, top gel). ChIP assays demonstrated that both ProA-Cse4 and ProA-Cse4Δ70 proteins successfully targeted the CEN DNA under these conditions (Fig. 2D, bottom gel), demonstrating that the first 70 amino acids of Cse4, including the END, are not required for CEN localization of the mutant protein even in the absence of functional wild-type Cse4.
Cse4 overexpression bypasses the requirement for END function.
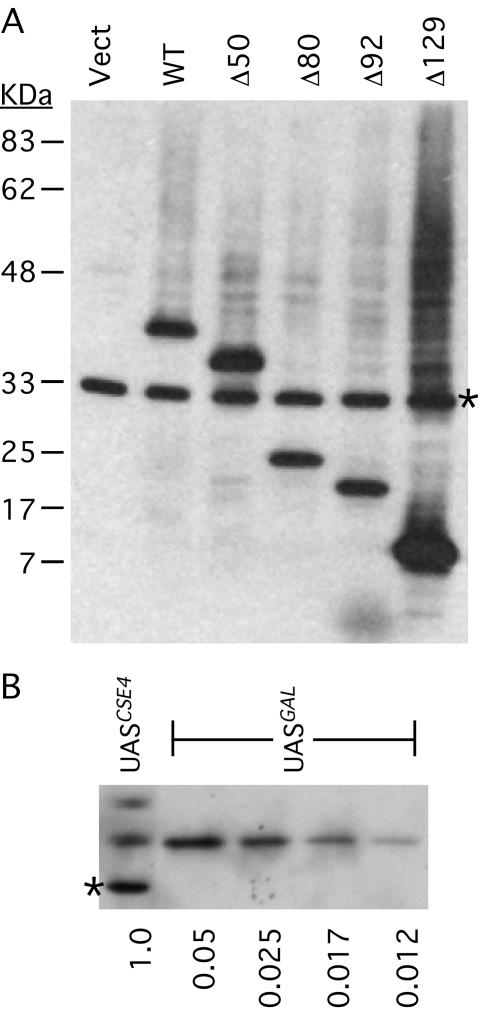
When characterizing the Cse4 degron strains, we noticed that after galactose induction of the Cse4Δ70 protein, the copper-grown cells appeared to reenter the cell cycle. Thus, not only was the Cse4Δ70 protein (Cse4Δ70p) able to target centromere DNA in the absence of wild-type Cse4, but the END-deleted protein also appeared to support chromosome segregation. We considered the possibility that the requirement for END function was bypassed due to the higher level of expression driven by the GAL1 promoter. To test this hypothesis, we placed several cse4ΔNT alleles under GAL1 control and tested them for the ability to complement a cse4 null allele (cse4Δ::kanMX4) using a plasmid shuffle assay (Fig. 3). Four alleles were tested in addition to cse4Δ70 (cse4Δ50, cse4Δ80, cse4Δ92, and cse4Δ129). All of the mutants provided sufficient Cse4 function to rescue the cse4Δ null allele when expression was induced by galactose (medium containing FOA and galactose), but not under repressing conditions imposed by glucose (medium containing FOA and glucose). Western blot analyses of the HA-tagged Cse4ΔNT proteins showed that all were expressed at high levels, with the level of Cse4Δ129p reproducibly higher than that of the others (Fig. 4). Direct comparison to wild-type Cse4 expression levels revealed that the GAL1 promoter induces a 40-fold increase in the steady-state level of Cse4. The Cse4Δ129 protein consists of only the Cse4 HFD and the six adjacent N-terminal amino acids that are ordered in the nucleosome crystal structure (30). Thus, the Cse4 HFD appears to be entirely sufficient for cell viability when it is overexpressed.
FIG. 3.
Complementation of cse4Δ::kanMX4 by GAL1-driven cse4ΔNT alleles. Growth on media containing FOA and either galactose or glucose (FOA/Gal and FOA/Glu, respectively) indicates complementation of the lethal cse4Δ null allele (see Materials and Methods). −Trp/Gal, without tryptophan but with galactose; WT, wild type.
FIG. 4.
Expression of Cse4ΔNT proteins driven by the GAL1 UAS control region. S. cerevisiae strains carrying GAL1-driven alleles were grown in medium containing galactose and prepared for SDS-polyacrylamide gel electrophoresis as described in Materials and Methods. (A) Western blot of extracts prepared from cells carrying the indicated HA-tagged cse4ΔNT alleles on low-copy-number plasmid vectors. Vect, empty vector; WT, wild-type. The band marked with an asterisk is a cross-reacting background protein. (B) Western blot comparing expression of endogenous CSE4 (UASCSE4) with GAL1-driven CSE4 (UASGAL) in strains grown on galactose. The relative amount of extract loaded is indicated below the gel.
A protein stability determinant in the Cse4 N terminus.
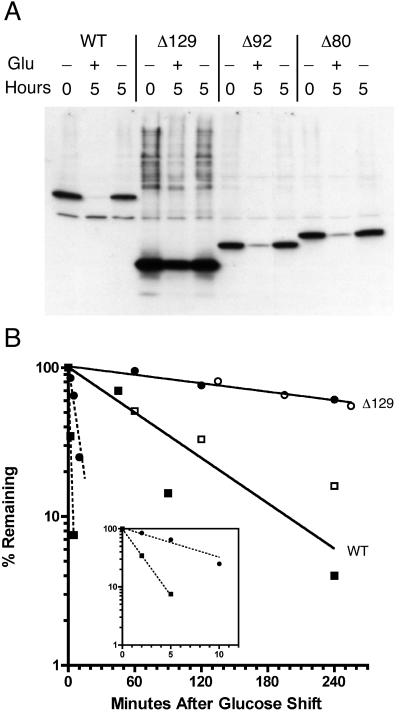
Steady-state levels of Cse4Δ129 were consistently higher than those of the wild-type Cse4 protein and other Cse4 proteins in which portions of the N terminus had been deleted (Fig. 4). Since the promoter was the same in all cases, it seemed likely that the elevated level of Cse4Δ129 was due to increased stability of the protein or mRNA (or both). When cells were shifted to glucose, which represses transcription from the GAL1 promoter, the wild-type protein was lost and had a half-life of 59 min, while Cse4Δ129 had a half-life of greater than 5 h (Fig. 5B). This difference accounts for the roughly fivefold difference in protein levels. Northern blots of total RNA revealed that the level of cse4Δ129 RNA was higher than the level of RNA of the wild-type gene driven by the same GAL1 promoter; however, both RNA species were found to be unstable, having half-lives of 1.3 and 6 min for wild-type and cse4Δ129 RNA, respectively (Fig. 5B, insert). This difference in RNA half-lives is not sufficient to account for the difference observed in protein half-lives.
FIG. 5.
A stability determinant in the Cse4 N terminus. (A) A wild-type (WT) CSE4 strain carrying the indicated HA-tagged cse4ΔNT alleles on low-copy-number plasmids were grown in galactose-containing medium and shifted to fresh medium containing either glucose (Glu) (repressing) (+) or galactose (inducing) (−). After 5 h, cell extracts were prepared, and the proteins were analyzed by Western blotting. (B) Cultures were treated as in panel A, except samples were taken at various time points and processed for Western blotting or Northern blot analysis. Protein or RNA bands on the films were quantitated by densitometry. Protein decay curves (solid lines) and RNA decay curves (broken lines) are shown. Both lines were fit by least-squares analysis using Prism 4 software (GraphPad). The data from two independent experiments (white and black symbols) are shown. (Insert) RNA decay curves on an expanded x axis. Squares, wild type (WT); circles, cse4Δ129.
The Cse4 HFD alone (Cse4Δ129 protein) assembles functional CEN chromatin.
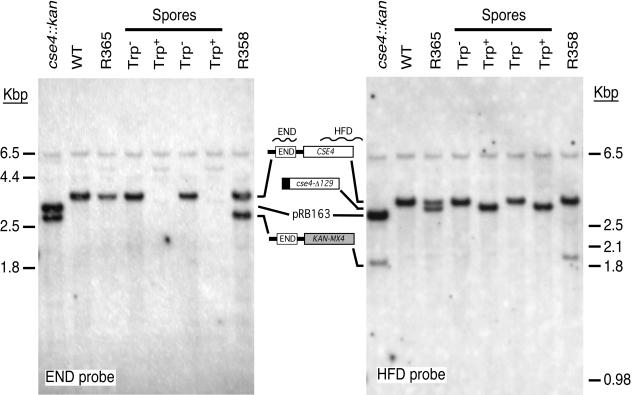
The cse4Δ::kanMX4 allele in the cse4Δ complementation tester strain R332-5D does not remove the entire CSE4 open reading frame. (The cse4Δ::kanMX4 allele was generated by the Saccharomyces Genome Deletion Project [45]. Due to an error in the original chromosome XI database sequence, the in-frame methionine at CSE4 codon 93 was annotated as the initiator ATG.) CSE4 codons 1 to 92, which include the sequence encoding the Cse4 END, are still present in the R332-5D cells, raising the formal possibility that END-containing polypeptides expressed from the cse4Δ::kanMX4 locus might contribute an essential END function in trans. This seemed unlikely, because similarly truncated Cse4ΔNT polypeptides are not detected in vivo (unpublished results) and are unable to complement conditional cse4 END alleles (9). Nonetheless, to rule out possible interallelic complementation of a cse4ΔNT allele by cse4Δ::kanMX4, an S. cerevisiae strain that expresses Cse4Δ129 and that completely lacks DNA sequences encoding the wild-type Cse4 N terminus was constructed.
An integrating plasmid containing cse4Δ129 was used to transform a diploid S. cerevisiae strain heterozygous for cse4Δ::kanMX4. Transformants were screened for loss of kanamycin resistance to obtain an S. cerevisiae strain (R365) in which the integrating cse4Δ129 plasmid, marked with TRP1, had replaced cse4Δ::kanMX4. Tetrad analysis revealed that the glucose sensitivity phenotype segregated 2:2 and was linked to the TRP1 marker (no recombinants among 12 tetrads). Southern hybridization confirmed the structure of the integrated cse4Δ129::TRP1 allele and the loss of END-coding sequences in the genomes of Trp+ segregants (Fig. 6). No remarkable phenotypic defects were observed for cse4Δ129::TRP1 strains (Table 2). Although the cse4Δ129::TRP1 cells grew significantly slower than the isogenic wild-type control strain, the 2.3-h doubling time was not significantly different from the 2.2-h doubling time of a similar strain expressing wild-type CSE4 under GAL1 control (Table 2). It appears that overexpression of Cse4 with or without the END is deleterious to cell growth.
FIG. 6.
Life without END. The cse4Δ::kanMX4 strain R332-5D (cse4::kan) carrying wild-type CSE4 on plasmid pRB163 was crossed with CSE4 wild-type strain R332-4C (WT) and cured of plasmid to obtain the heterozygous diploid strain R358. R358 was then transformed with PstI-linearized plasmid pRB588 to replace the cse4Δ::kanMX4 allele with GAL1-driven cse4Δ129::TRP1. A Kans transformant (R365) was picked and sporulated. Dissected tetrads each yielded 2 Trp+ spores and 2 Trp− spores, and all 22 Trp+ spores were also glucose sensitive. DNA from the indicated strains was cleaved with EcoRI, separated on an agarose gel, transferred to a nylon membrane, and hybridized with a probe homologous to DNA encoding the Cse4 END. The blot was stripped and rehybridized with a probe homologous to DNA encoding the Cse4 HFD. The HFD probe extends beyond the CSE4 coding region and thereby detects the cse4Δ::kan genomic EcoRI fragment.
TABLE 2.
Cse4 overexpression phenotypes
| Genotype | Doubling time (h) | CF loss rate (no. of events/division) (104) |
|---|---|---|
| CSE4 | 1.6 | 3.7 ± 0.5 |
| gal-CSE4 | 2.2 | 55 ± 17 |
| gal-cse4Δ129 | 2.3 | 21 ± 4 |
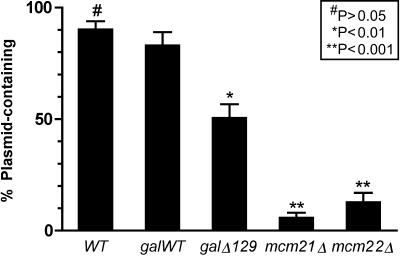
Centromere function in the cse4Δ129 cells was assessed by measuring chromosome and minichromosome loss rates. For the minichromosome loss assay, a 10.6-kb plasmid (pMW29) containing CEN4 was introduced into cells by transformation, and plasmid retention was measured after 8 to 10 generations of nonselective growth. In the experiment shown in Fig. 7, approximately 90% of the CSE4 wild-type control cell population retained the minichromosome. By contrast, the plasmid was retained in only 6 and 13% of cells in mcm21Δ and mcm22Δ strains, respectively. Mutations in the MCM (minichromosome maintenance) genes are known to compromise CEN plasmid stability (44). An intermediate Mcm phenotype was observed for the cse4Δ129 strain in which plasmid was retained in 51% of the cells (galΔ129). The corresponding strain containing a GAL1-driven full-length CSE4 allele (galWT) retained plasmid in 83% of cells, not significantly different from that observed with the CSE4 wild-type strain. Thus, the Mcm-like phenotype observed for cse4Δ129 can be attributed to the END deletion.
FIG. 7.
Minichromosome mitotic stability assay. The 10.6-kb plasmid minichromosome pMW29, which contains CEN4 and the selectable marker URA3, was introduced into the test strains by transformation. Transformant colonies were grown for 8 to 10 generations in nonselective medium, and the fraction of plasmid-containing cells was determined by plating identical aliquots on selective and nonselective medium. Five colonies were scored for each strain, except for the cse4Δ129 strain for which 10 colonies were scored. The means are plotted, with error bars showing the standard errors of the means. Statistical significance of the differences between the mean of galWT and all others was assessed by Bonferroni's multiple comparison test, computed using Prism 4 statistics software (GraphPad). WT, wild-type.
Centromere function in the chromosomal context was assayed by measuring mitotic loss of a nonessential chromosome fragment (CF) derived from chromosome III (49). This marker chromosome was lost from the wild-type strain at a rate of 3.2 × 10−4 per division, in agreement with previous results (2). The mitotic loss rate was elevated approximately sixfold in the cse4Δ129::TRP1 background; however, this increase was not as great as the 15-fold increase observed with the control strain overexpressing wild-type Cse4 (Table 2). Thus, in contrast to the minichromosome loss phenotype, the cse4Δ129::TRP1 allele does not compromise chromosomal CEN function more than overexpression of full-length Cse4. The 6- to 15-fold increase in CF loss rates is comparable to that observed for nonlethal cse4 HFD mutations (22).
DISCUSSION
Several studies have indicated that the specificity of CenH3 proteins for centromere DNA is conferred by the HFD and that the N terminus is dispensable for centromere targeting under conditions where endogenous wild-type CenH3 protein is also present (9, 47, 59). Our results confirm and expand this conclusion in two important ways. First, we show that the HFD of S. cerevisiae Cse4 is sufficient for localizing the protein to centromere DNA even in the absence of detectable wild-type protein, definitively ruling out a role for the Cse4 N terminus in the centromere targeting process. These results can be extended to include the highly conserved HFD sequences found in the CenH3 proteins identified in other eukaryotes. On the basis of protein sequence similarities, we predict that the different CenH3 HFDs will perform similar if not identical functions in vivo regardless of the origin of the CenH3 protein. Second, we demonstrate that the overexpressed, centromere-targeted Cse4 HFD alone is sufficient for the assembly of functional centromeric chromatin and kinetochores capable of surprisingly accurate mitotic chromosome segregation. Construction and characterization of the S. cerevisiae cse4Δ129 strain, which relies on overexpression of the Cse4 HFD for cell viability, offers an important opportunity to study the fundamental features of the S. cerevisiae kinetochore, i.e., those structures and processes that depend on the Cse4 HFD and are completely independent of the Cse4 N terminus.
The HFD of Cse4 (residues 136 to 229) is homologous to the four α-helices and three loop regions of the histone H3 HFD (22). The Cse4Δ129 protein, which potentially represents the minimal functional region of Cse4, also contains six amino acids (residues 130 to 135) on the amino-terminal side of the N helix. In histone H3, these six residues are ordered within the nucleosome crystal structure, with five of them contacting DNA (30). By analogy, we consider this short “tail” to be part of the Cse4 HFD, although mutational analysis has shown that substitution of this region by other amino acids does not significantly impair the function of Cse4 (9).
Current models of the S. cerevisiae point centromere depict a surprisingly complex structure consisting of reasonably well-defined inner and outer kinetochore complexes connected by less well-defined complexes of the central kinetochore (7, 35). The inner kinetochore consists of the CBF3 complex (Ndc10p, Cep3p, Ctf13p, and Skp1p), Cbf1p, and the Cse4 nucleosome, all of which bind directly to CEN DNA (3, 27, 37, 41). The DASH complex (also known as the Dam1 complex), composed of 10 proteins including Dam1p, Spc19p, and Spc34p, directly binds microtubules and forms the outer kinetochore (6, 29). Connections between the inner and outer kinetochores are provided by the Ndc80 (Ndc80p, Spc25p, Spc24p, and Nut2p), COMA (Ctf19p, Okp1p, Mcm21p, and Ame1p), and MIND (Mtw1p, Nsl1p, Nnf1p, and Dsn1p) complexes (11, 19, 61). Several additional kinetochore proteins are found associated with the COMA and MIND complexes, including Cse4 (11). Cse4 is also found in a complex with Mif2p, a kinetochore-associated protein thought to bind the AT-rich CDEII element of CEN DNA (36, 61). With Cse4 embedded in the CEN chromatin, the extended Cse4 N terminus, protruding away from the nucleosome core, would be accessible for forming specific interactions with one or more proteins of the central kinetochore, thereby connecting the central kinetochore to the CEN DNA. The Cse4 N terminus interacts with Ctf19p (9), possibly providing such a connection; however, the Cse4Δ129 overexpression studies suggest that, despite the attractiveness of this model, no protein-protein interaction mediated by the Cse4 N terminus is essential for chromosome segregation or cell cycle progression.
In S. cerevisiae, like other eukaryotes, the N termini of core histones H3 and H4 are the targets of multiple posttranslational modifications that regulate a variety of chromatin-based processes, including nucleosome assembly and remodeling, gene transcription and silencing, and DNA condensation (54). The N terminus of CENP-A is phosphorylated by aurora kinases A and B at serine 7, and inhibition of this phosphorylation disrupts chromosome alignment at metaphase (24, 62). Although the Cse4 N terminus might require phosphorylation or some other posttranslational modification to function properly at wild-type Cse4 expression levels, the present results show that the entire Cse4 N terminus, including potentially modified serine-rich regions, is dispensable for viability when the cell expresses high levels of Cse4. Thus, no posttranslational modification of the Cse4 N terminus is essential for centromere function in S. cerevisiae.
What then is the function of the Cse4 N terminus? Mutational analysis clearly shows that the Cse4 N terminus contains a domain (END) that is essential when Cse4 is expressed at wild-type (i.e., relatively low) levels (9, 22). The observation that the essential END function is bypassed by high levels of Cse4 protein suggests that in wild-type cells the presence of the END acts by some means to increase the effective concentration of Cse4 in the cell. Given that relatively few Cse4 nucleosomes (possibly as few as one per chromosome) exist within a sea of conventional nucleosomes, several steps in the Cse4 nucleosome assembly pathway are expected to be strongly concentration dependent, including transport into the nucleus, retention in the nucleus, interactions with core histones (notably H4), and specific recognition of centromere DNA. On the other hand, functions executed by Cse4 after it is stably incorporated into CEN chromatin, such as binding to other kinetochore proteins, should not be affected by changes in the concentration of Cse4 that is not bound to DNA. Therefore, we believe that the essential function of the Cse4 END is executed prior to the incorporation of Cse4 into CEN chromatin, and we propose that the Cse4 END can be viewed as a cis-acting assembly factor for the Cse4 HFD. As such, the END function is not required when the cellular concentration of the Cse4 HFD is artificially high, in this case due to expression driven by the strong GAL1 promoter.
Although overexpression of wild-type Cse4 or Cse4Δ129 rescues the cse4 null phenotype, high levels of either protein appear to have deleterious consequences to cells, manifested by lengthened generation times and increased rates of mitotic chromosome loss. The negative effects of Cse4 overexpression are likely caused by imbalances in nucleosome assembly pathways. Nonlethal mutations in core histone genes or in genes regulating histone expression are known to have pleiotropic effects on gene transcription, silencing, and chromosome segregation (17, 18, 21, 28, 43, 46). Likewise, Smith and coworkers have shown that histone H3 and H4 levels affect Cse4 function and vice versa: overexpression of Cse4 suppresses a specific H4 mutation, hhf1-20, and overexpression of H3 enhances the phenotype of some cse4 mutations (14, 52). Similarly, overexpression of H4 suppresses mutational defects in Cnp1, the Schizosaccharomyces pombe Cse4 homologue (8). In the case of Cse4 overexpression, the flood of Cse4 protein resulting from GAL1-driven expression might significantly reduce the supply of H3-H4 tetramers available for bulk chromatin assembly by competing with H3 for binding to H4, while increasing the concentration of Cse4-H4 tetramers to abnormally high levels. An increased concentration of Cse4-H4 tetramers might promote incorporation of Cse4 nucleosomes into noncentromeric loci and cause the formation of aberrant chromatin structures, as appears to be the case in human cells, where overexpression of CENP-A causes mistargeting of the CENP-A protein to noncentromeric loci (58). ChIP assays of strains overexpressing Cse4 and Cse4Δ129 reproducibly show higher recoveries of non-CEN sequences in Cse4 immunoprecipitates than observed with wild-type strains (L. Morey, unpublished data), suggesting that mistargeting of Cse4 occurs. As either H3-H4 tetramer deficit or Cse4-H4 tetramer excess is expected to be deleterious, we propose that the primary role of the Cse4 N terminus in wild-type cells is to facilitate the assembly and centromere targeting of Cse4-H4 tetramers at low cellular concentrations of Cse4, allowing the cell to maintain low Cse4 levels to protect the pathway of conventional (i.e., H3-containing) nucleosome assembly from interference by excess Cse4.
The steady-state level of Cse4Δ129 is consistently higher than that of the other Cse4ΔNT proteins when expression is driven by the GAL1 promoter. This difference is not observed when Cse4ΔNT proteins are expressed from their endogenous promoters (e.g., Fig. 1B), where the low levels of protein detected by Western blotting are variable and influenced by strain background and growth conditions. Nonetheless, the high level of GAL1-driven Cse4Δ129 protein is attributable to its increased stability, implying that the Cse4 N terminus contains one or more elements that act to enhance Cse4p turnover. Incremental deletion of Cse4 N-terminal amino acids does not result in significant stabilization of the protein until the final 37 residues are removed (amino acids 93 to 129). The observed Cse4 stabilization could be due to the loss of a solitary destabilizing element located between residues 93 and 129, or this region (residues 93 to 129) may contain one of several redundant destabilizing elements located throughout the N terminus. The C-terminal half of the region containing residues 93 to 129 is rich in charged amino acids, including six lysine residues that might be targets of ubiquitination. While there is at present no strong argument to posit a functional significance to the protein destabilizing function of the Cse4 N terminus, under conditions where it would be advantageous for the cell to regulate the level of Cse4 protein, the stability determinant(s) in the N terminus could provide the means.
The phenotype of strains (over)expressing Cse4Δ129 as their only Cse4 protein is surprisingly benign. The 40% increase in doubling time observed for the cse4Δ129 strain is not significantly different from that observed for a similar strain also grown on galactose but overexpressing wild-type Cse4. S. cerevisiae strains experiencing relatively high rates of chromosome missegregation often grow at near wild-type rates (22); however, in this case, the cse4Δ129 strain exhibits a 40% increase in doubling time but only a sixfold increase in mitotic chromosome loss. No significant differences in sporulation or spore viability were observed (data not shown), suggesting no major impairment of meiotic chromosome segregation in cse4Δ129 cells. Cells overexpressing wild-type Cse4 exhibit a larger increase in mitotic loss of CFs than do cells of the cse4Δ129 strain, suggesting that it is the overexpression of the HFD and not the lack of the Cse4 N terminus that causes the increase in chromosome missegregation events.
Minichromosome (i.e., CEN plasmid) loss rates are often used as a measure of centromere function (23). In contrast to the results observed with CFs, the cse4Δ129 allele caused decreased mitotic stability of the 10.9-kbp minichromosome, while overexpression of wild-type Cse4 caused no statistically significant change in plasmid segregation. The magnitude of the effect, 50% plasmid retention versus 83% in the control, is not as large as that observed with the mcm21Δ and mcm22Δ mutants (6 and 13% plasmid retention, respectively). The mcm mutants were of interest, because several Mcm gene products (Mcm16p, Mcm17p [Chl4p], Mcm21p, and Mcm22p) are kinetochore components (11), and genetic interactions have been detected between cse4 END mutations and both mcm21 and mcm22 (9). While it appears that the END performs a nonessential Mcm-like function that is disproportionately more important for minichromosome segregation than CF segregation, the results argue against the simplest model in which the Cse4 END interacts with Mcm21p or Mcm22p to perform this function. Otherwise, we would expect the minichromosome loss phenotype of cse4Δ129 to be as severe as that exhibited by mcm21Δ and mcm22Δ cells.
Centromere identity across the eukaryotic world exhibits an epigenetic character with little or no dependence on unique centromere DNA sequence identifiers except in budding yeast (20, 56). In multicellular eukaryotes, euchromatic sequences can acquire centromere function (1), while functional centromere DNA can become inactive (13). In S. pombe, centromere function is influenced by factors that affect chromatin structure without changing the underlying primary DNA sequence (4, 42). Recently, it has been shown that S. cerevisiae point centromeres can switch between segregation-competent and -incompetent states. Establishment of the active, segregation-competent state requires the CHL4 (MCM17) gene product (39). The experiments reported here do not address the question of whether the Cse4 N terminus is required to establish an active centromere on CEN DNA. In our experiments, the cse4ΔNT allele was introduced into cells before the wild-type CSE4 allele was eliminated. Subsequent growth through dozens of generations resulted in loss of the preexisting wild-type Cse4 by dilution, but at no time were the cells faced with the need to build an active centromere de novo. We can conclude with some certainty that the HFD of Cse4 is sufficient to propagate active centromeres in S. cerevisiae under conditions of Cse4Δ129 protein overexpression. Elucidation of the role of the Cse4 N terminus in establishing active centromeres de novo awaits further investigation.
Acknowledgments
We thank Sam Stoler for helpful advice throughout this project and Diana Wentworth for technical assistance with Northern and Western blots.
This work was supported in part by grants from the National Institutes of Health to M.F.-H. (GM54766) and R.E.B. (GM61120).
REFERENCES
- 1.Amor, D. J., K. Bentley, J. Ryan, J. Perry, L. Wong, H. Slater, and K. H. Choo. 2004. Human centromere repositioning “in progress.” Proc. Natl. Acad. Sci. USA 101:6542-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, R. E., K. Harris, and K. Zhang. 1998. Mutations synthetically lethal with cep1 target Saccharomyces cerevisiae kinetochore components. Genetics 149:73-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, R. E., and D. C. Masison. 1990. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol. Cell. Biol. 10:2458-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell, C., K. A. Martin, A. Greenall, A. Pidoux, R. C. Allshire, and S. K. Whitehall. 2004. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol. Cell. Biol. 24:4309-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 6.Cheeseman, I. M., C. Brew, M. Wolyniak, A. Desai, S. Anderson, N. Muster, J. R. Yates, T. C. Huffaker, D. G. Drubin, and G. Barnes. 2001. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheeseman, I. M., D. G. Drubin, and G. Barnes. 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, E. S., S. Saitoh, M. Yanagida, and K. Takahashi. 2003. A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol. Cell 11:175-187. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Y., R. E. Baker, K. C. Keith, K. Harris, S. Stoler, and M. Fitzgerald-Hayes. 2000. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 20:7037-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 11.De Wulf, P., A. D. McAinsh, and P. K. Sorger. 2003. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17:2902-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong, F., and K. E. van Holde. 1991. Nucleosome positioning is determined by the (H3-H4)2 tetramer. Proc. Natl. Acad. Sci. USA 88:10596-10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earnshaw, W. C., and B. R. Migeon. 1985. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma 92:290-296. [DOI] [PubMed] [Google Scholar]
- 14.Glowczewski, L., P. Yang, T. Kalashnikova, M. S. Santisteban, and M. M. Smith. 2000. Histone-histone interactions and centromere function. Mol. Cell. Biol. 20:5700-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes, J. J., D. J. Clark, and A. P. Wolffe. 1991. Histone contributions to the structure of DNA in the nucleosome. Proc. Natl. Acad. Sci. USA 88:6829-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henikoff, S., K. Ahmad, J. S. Platero, and B. van Steensel. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirschhorn, J. N., A. L. Bortvin, S. L. Ricupero-Hovasse, and F. Winston. 1995. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 15:1999-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, L., W. Zhang, and S. Y. Roth. 1997. Amino termini of histones H3 and H4 are required for a1-α2 repression in yeast. Mol. Cell. Biol. 17:6555-6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janke, C., J. Ortiz, T. U. Tanaka, J. Lechner, and E. Schiebel. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karpen, G. H., and R. C. Allshire. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13:489-496. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman, P. D., J. L. Cohen, and M. A. Osley. 1998. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol. Cell. Biol. 18:4793-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keith, K. C., R. E. Baker, Y. Chen, K. Harris, S. Stoler, and M. Fitzgerald-Hayes. 1999. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell. Biol. 19:6130-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koshland, D., J. C. Kent, and L. H. Hartwell. 1985. Genetic analysis of the mitotic transmission of minichromosomes. Cell 40:393-403. [DOI] [PubMed] [Google Scholar]
- 24.Kunitoku, N., T. Sasayama, T. Marumoto, D. Zhang, S. Honda, O. Kobayashi, K. Hatakeyama, Y. Ushio, H. Saya, and T. Hirota. 2003. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev. Cell 5:853-864. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lea, D. E., and C. A. Coulson. 1949. The distribution of the number of mutants in bacterial populations. J. Genet. 49:264-285. [DOI] [PubMed] [Google Scholar]
- 27.Lechner, J., and J. Carbon. 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64:717-725. [DOI] [PubMed] [Google Scholar]
- 28.Lenfant, F., R. K. Mann, B. Thomsen, X. Ling, and M. Grunstein. 1996. All four core histone N-termini contain sequences required for the repression of basal transcription in yeast. EMBO J. 15:3974-3985. [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Y., J. Bachant, A. A. Alcasabas, Y. Wang, J. Qin, and S. J. Elledge. 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16:183-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 31.Malik, H. S., and S. Henikoff. 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157:1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik, H. S., and S. Henikoff. 2003. Phylogenomics of the nucleosome. Nat. Struct. Biol. 10:882-891. [DOI] [PubMed] [Google Scholar]
- 33.Malik, H. S., D. Vermaak, and S. Henikoff. 2002. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc. Natl. Acad. Sci. USA 99:1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McAinsh, A. D., J. D. Tytell, and P. K. Sorger. 2003. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19:519-539. [DOI] [PubMed] [Google Scholar]
- 35.Measday, V., and P. Hieter. 2004. Kinetochore substructure comes to MIND. Nat. Cell Biol. 6:94-95. [DOI] [PubMed] [Google Scholar]
- 36.Meluh, P. B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland, and M. M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94:607-613. [DOI] [PubMed] [Google Scholar]
- 38.Moqtaderi, Z., Y. Bai, D. Poon, P. A. Weil, and K. Struhl. 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383:188-191. [DOI] [PubMed] [Google Scholar]
- 39.Mythreye, K., and K. S. Bloom. 2003. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J. Cell Biol. 160:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Connell, K. F., Y. Surdin-Kerjan, and R. E. Baker. 1995. Role of the Saccharomyces cerevisiae general regulatory factor CP1 in methionine biosynthetic gene transcription. Mol. Cell. Biol. 15:1879-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortiz, J., O. Stemmann, S. Rank, and J. Lechner. 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13:1140-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pidoux, A. L., W. Richardson, and R. C. Allshire. 2003. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 161:295-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinto, I., and F. Winston. 2000. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 19:1598-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poddar, A., N. Roy, and P. Sinha. 1999. MCM21 and MCM22, two novel genes of the yeast Saccharomyces cerevisiae are required for chromosome transmission. Mol. Microbiol. 31:349-360. [DOI] [PubMed] [Google Scholar]
- 45.Saccharomyces Genome Deletion Project. http://www-sequence.stanford.edu/group/yeast_deletion_project.
- 46.Sharp, J. A., A. A. Franco, M. A. Osley, and P. D. Kaufman. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shelby, R. D., O. Vafa, and K. F. Sullivan. 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136:501-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sherman, F., G. Fink, and J. B. Hicks. 1983. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 49.Shero, J. H., M. Koval, F. Spencer, R. E. Palmer, P. Hieter, and D. Koshland. 1991. Analysis of chromosome segregation in Saccharomyces cerevisiae. Methods Enzymol. 194:749-773. [DOI] [PubMed] [Google Scholar]
- 50.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, M. M. 2002. Centromeres and variant histones: what, where, when and why? Curr. Opin. Cell Biol. 14:279-285. [DOI] [PubMed] [Google Scholar]
- 52.Smith, M. M., P. Yang, M. S. Santisteban, P. W. Boone, A. T. Goldstein, and P. C. Megee. 1996. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoler, S., K. C. Keith, K. E. Curnick, and M. Fitzgerald-Hayes. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573-586. [DOI] [PubMed] [Google Scholar]
- 54.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 55.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 56.Sullivan, K. F. 2001. A solid foundation: functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 11:182-188. [DOI] [PubMed] [Google Scholar]
- 57.Van Hooser, A. A., and R. Heald. 2001. Kinetochore function: the complications of becoming attached. Curr. Biol. 11:R855-R857. [DOI] [PubMed] [Google Scholar]
- 58.Van Hooser, A. A., I. I. Ouspenski, H. C. Gregson, D. A. Starr, T. J. Yen, M. L. Goldberg, K. Yokomori, W. C. Earnshaw, K. F. Sullivan, and B. R. Brinkley. 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114:3529-3542. [DOI] [PubMed] [Google Scholar]
- 59.Vermaak, D., H. S. Hayden, and S. Henikoff. 2002. Centromere targeting element within the histone fold domain of Cid. Mol. Cell. Biol. 22:7553-7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westermann, S., I. M. Cheeseman, S. Anderson, J. R. Yates III, D. G. Drubin, and G. Barnes. 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163:215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wigge, P. A., and J. V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeitlin, S. G., R. D. Shelby, and K. F. Sullivan. 2001. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155:1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]