Abstract
The small-subunit (SSU) processome is a large ribonucleoprotein required for the biogenesis of the 18S rRNA and likely corresponds to the terminal knobs visualized by electron microscopy on the 5′ end of nascent rRNAs. The original purification of the SSU processome of Saccharomyces cerevisiae resulted in the identification of 28 proteins. Here, we characterize 12 additional protein components, including five small-ribosomal-subunit proteins (Rps4, Rps6, Rps7, Rps9, and Rps14) that had previously been copurified. Our multiple criteria for including a component as a bona fide SSU processome component included coimmunoprecipitation with Mpp10 (an SSU processome component), the U3 snoRNA, and the anticipated pre-rRNAs. Importantly, the association of specific ribosomal proteins with the SSU processome suggests that the SSU processome has roles in both pre-rRNA processing and ribosome assembly. These ribosomal proteins may be analogous to the primary or secondary RNA binding proteins first described in bacterial in vitro ribosome assembly maps. In addition to the ribosomal proteins and based on the same experimental approach, we found seven other proteins (Utp18, Noc4, Utp20, Utp21, Utp22, Emg1, and Krr1) to be bona fide SSU processome proteins.
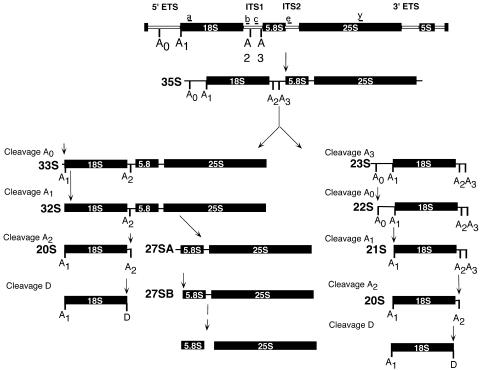
Ribosomes are essential for the translation of mRNA into protein. Ribosome biogenesis in Saccharomyces cerevisiae begins with the transcription of the 35S pre-rRNA, which is then cleaved and processed at more than 10 different processing sites to give rise to the mature 18S, 25S, and 5.8S rRNAs (Fig. 1).
FIG. 1.
Schematic diagram of pre-rRNA processing in S. cerevisiae. The 35S pre-rRNA is transcribed as a single transcript that is subsequently cleaved at the A0, A1, and A2 sites by the SSU processome. Cleavage at A2 or A3 separates precursors to the small-subunit (18S) and large-subunit (5.8S and 25S) rRNAs, whereas cleavage at A0 or A1 matures the 5′ end of the 18S rRNA. The broken line represents multiple processing steps. The locations of oligonucleotides a, b, c, e, and y, used to probe for precursor and mature rRNAs in subsequent experiments, are shown.
Small nucleolar ribonucleoproteins (snoRNPs) are required for many of the different processing steps and modifications that occur relative to the pre-rRNA (16). There are three classes of snoRNPs (H/ACA box, C/D box, and RNase mitochondrial RNA processing) that are required for ribosome biogenesis, each of which contains a small nucleolar RNA (snoRNA). H/ACA box snoRNAs are required for site-specific pseudouridylation of rRNA, while C/D box snoRNAs are required for 2′-O-ribose methylation of specific nucleotides in rRNA.
The U3 snoRNA and its associated proteins are required for the processing of the small ribosomal subunit at cleavage sites A0, A1, and A2 (Fig. 1). Cleavages at A0 and A1 in the 5′ external transcribed spacer mature the 5′ end of the pre-rRNA. Cleavage at A2 or A3 in internal transcribed spacer 1 separates the small-ribosomal-subunit precursor rRNA from the large-ribosomal-subunit precursors. Defects in cleavage at the A0, A1, and A2 sites lead to a reduction in the levels of the 18S rRNA. This reduction causes accumulation of the 35S and 23S pre-rRNAs and a reduction in the levels of the 27SA2 and 20S pre-rRNAs (39).
A large RNP required for the processing of the small-ribosomal-subunit rRNA, called the small-subunit (SSU) processome, has recently been purified (4). This preribosomal complex contains the U3 snoRNA and at least 28 proteins. We defined the SSU processome components as having the following properties. (i) They are nucleolar. (ii) They are able to coimmunoprecipitate with the U3 snoRNA and Mpp10 (a protein specific to the SSU processome). (iii) They are required for 18S rRNA biogenesis. Subsequent large-scale tandem affinity purification studies have also purified several 80 to 90S preribosomal complexes that contain many SSU processome components (9, 33). Additional proteomic studies have also revealed similar proteins required for ribosome biogenesis and subcomplexes, including SSU processome proteins (17, 29). Collectively, these studies have purified an 80S or 90S preribosome which serves as a precursor to the 43S preribosome and is required for cleavages at A0, A1, and A2. Here, we present results that expand upon these studies and validate the role of seven nonribosomal proteins in pre-rRNA processing.
During the original purification, analysis was limited to proteins whose peptide sequences were present more than once in the mass spectrometric analysis (4). Due to this stringent criterion, we hypothesized that there may be some additional SSU processome components that had been eliminated in the original purification. In addition, the initial purification included the presence of five ribosomal proteins (Rps4, Rps6, Rps7, Rps14, and Rps28), but it was not known at that time whether they were components of the SSU processome or contaminants. Therefore, we aimed to determine whether these five ribosomal proteins are SSU processome components by testing (i) their association via coimmunoprecipitation with known SSU processome components, i.e., Mpp10 and the U3 snoRNA, and (ii) their association with precursors to the 18S rRNA. Since there is no in vitro ribosome assembly system in yeast, the order of assembly of ribosomal proteins in eukaryotes is currently unknown. We have found a subset of ribosomal proteins to be associated with the SSU processome, suggesting that these proteins are able to associate with early precursors to the 18S rRNA. Therefore, the association of specific ribosomal proteins with the SSU processome suggests that it has roles in both pre-rRNA processing and ribosome assembly.
We have also tested whether seven other nonribosomal proteins (Utp18, Noc4, Utp20, Utp21, Utp22, Emg1, and Krr1), present only once in the purifications or subsequently found by others to coimmunoprecipitate with SSU processome components, were additional components of the SSU processome (7, 8, 32). A subset of these proteins has been partially characterized as being involved in ribosome biogenesis (3, 7, 9, 17, 25, 29, 33). For example, Utp18 was first identified (9), localized to the nucleolus (12), and shown to coimmunoprecipitate with the 5′ external transcribed spacer and the U3 snoRNA (17). Here we show that Utp18 is required for pre-18S rRNA processing by Northern blot analysis of pre-rRNAs; we also show that Utp18 coimmunoprecipitates with the SSU processome protein, Mpp10, by Western blot analysis. Similarly, Noc4 was previously identified (9, 29) and localized (9, 26), and the pre-rRNA processing phenotype of the temperature-sensitive Noc4 mutant was analyzed (26). However, Noc4 was found not to coimmunoprecipitate with the U3 snoRNA (9). Here, we determined the resulting defects in pre-rRNA processing of cells depleted of Noc4 and report that, contrary to previously published results, Noc4 does indeed coimmunoprecipitate with both the U3 snoRNA and the SSU processome protein, Mpp10. In this study, we determined that these proteins are components of the SSU processome, which therefore places them in a specific preribosomal complex. Much to our surprise, we found that the SSU processome associates with the 23S pre-rRNA, a precursor that has been cleaved at A3 but not at A0, A1, or A2. Furthermore, we are able to unify, support, and expand upon previous studies. In addition to further characterizing the nonribosomal proteins, we have validated that five ribosomal proteins, previously not known to be components of pre-rRNA complexes, are components of the SSU processome.
Collectively, we have validated that these 12 proteins are indeed bona fide SSU processome components and are essential for 18S rRNA biogenesis. Together, these results suggest that the SSU processome, in addition to its role in pre-rRNA processing, also has a role as a ribosome assembly intermediate.
MATERIALS AND METHODS
Yeast strains and media.
All yeast strains were derived from YPH499 (MATa ura3-52 lys2-80 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1). Yeast strains were grown in rich medium, either YEPD (1% yeast extract, 2% peptone, 2% glucose) or YPG/R (1% yeast extract, 2% peptone, 2% galactose, 2% raffinose) as specified below.
Expression of proteins from a conditional promoter.
Strains which expressed N termini that were tagged with a triple hemagglutinin epitope tag (3×HA) from a galactose-inducible and glucose-repressible promoter were created as described previously (22) with plasmid pFA6a-kanMX6-PGAL1-3×HA and with primers with 50 nucleotides of complementarity to the gene of interest (the Utp1, Utp7, Utp18, Noc4, Utp20, Emg1, Bfr2, Enp1, Rps9B, or Rps14A gene).
C-terminal 3×HA tagging.
Yeasts expressing proteins with 3×HA tags were constructed as described previously (15) with plasmid pYM1 (kanMX6 selectable marker) and 50 nucleotides with complementarity to the gene of interest (the Utp1 to Utp10, Utp12 to Utp17, Utp21, Utp22, Krr1, Enp1, Enp2, Rpf2, Imp4, Rps4A, Rps6B, Rps7B, Rps27A, Rps28A, Rpl33A, Nop7, or Sof1 genes).
Analysis of pre-rRNA processing by Northern blotting.
Strains expressing an N-terminal 3×HA tag were grown in YPG/R and then washed and resuspended in YEPD. RNA was extracted from 10 ml of cells grown to an optical density at 600 nm of 0.4 to 0.5 in YPG/R and of cells grown in YEPD (24 h). RNA extraction and Northern blotting were carried out as previously described (20). Equal amounts of RNA (5 μg) were loaded in each lane.
Immunoprecipitations.
Immunoprecipitations were carried out with N- and C-terminally 3×HA-tagged strains. Protein-protein coimmunoprecipitations were carried out with 200 μl each of anti-HA (12CA5 hybridoma culture supernatant) with glass bead protein extracts and blotted with anti-Mpp10 antibodies (20). Immunoprecipitations for protein, RNA, and pre-rRNA were carried out with extracts from strains that had been N-terminally tagged with GAL-3×HA and C-terminally tagged with 3×HA as previously described.
Protein-snoRNA coimmunoprecipitations were carried out with 200 μl of anti-HA (12CA5) on tagged protein extracts made by glass bead disruption, and RNA was extracted and analyzed as previously described (20). Protein-pre-rRNA coimmunoprecipitations were carried out as previously described (40), except that Rps4A and Rps6B pre-rRNA coimmunoprecipitates were probed with oligonucleotides e and b.
Immunofluorescence.
Yeast strains expressing 3×HA-tagged proteins were used in indirect immunofluorescence assays to determine subcellular localization as described previously (1, 5). Mouse anti-HA (12CA5 hybridoma culture supernatant; dilution, 1:1,000) and rabbit anti-Mpp10 polyclonal antibodies (dilution, 1:2,000) (6) were detected with tetramethylrhodamine B isothiocyanate-conjugated goat anti-mouse immunoglobulin G (dilution, 1:100) and fluorescein isothiocyanate-conjugated donkey anti-rabbit immunoglobulin G (dilution, 1:300) secondary antibodies (Jackson ImmunoResearch). The localization of tagged Utp4, Utp18, Noc4, Utp20, Bfr2, and Emg1 was carried out with N-terminally 3×HA-tagged strains, whereas the localization of Utp21, Utp22, and Enp2 was carried out with C-terminally 3×HA-tagged strains.
RESULTS
Previous work identified 28 protein components and the U3 snoRNA as components of the SSU processome (4). Here, we identify a number of additional proteins which, based on a combination of factors, are SSU processome candidates.
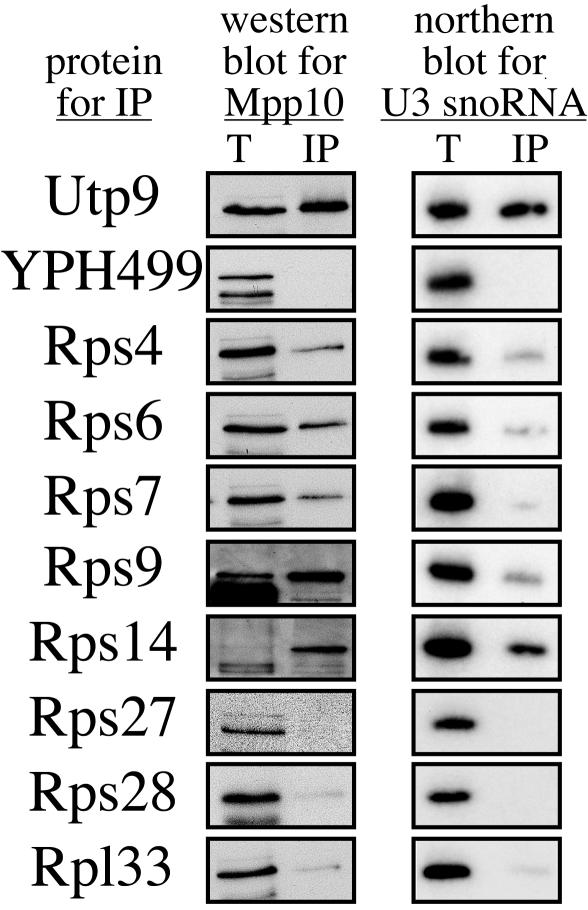
During the original SSU processome purification, five copurifying ribosomal proteins, Rps4, Rps6, Rps7, Rps14, and Rps28, were identified (4). In order to assess whether these and other ribosomal proteins might be components of the SSU processome, 3×HA-tagged ribosomal proteins Rps4, Rps6, Rps7, Rps9, Rps14, Rps27, Rps28, and Rpl33 were tested for their ability to coimmunoprecipitate with Mpp10 and the U3 snoRNA, two components of the SSU processome (Fig. 2). Four of the five SSU processome-copurifying ribosomal proteins (Rps4, Rps6, Rps7, and Rps14) were able to coimmunoprecipitate with Mpp10 and the U3 snoRNA, though to different degrees. Rps9 was also able to coimmunoprecipitate with Mpp10 and the U3 snoRNA. Relative to the other ribosomal proteins, Rps7 was able to coimmunoprecipitate only with small amounts of Mpp10 and the U3 snoRNA. In contrast, ribosomal proteins Rps27, Rps28, and Rpl33 did not coimmunoprecipitate with Mpp10 or the U3 snoRNA (Fig. 2). As expected, no coimmunoprecipitation was observed with the untagged parental strain, YPH499, while Utp9 (a known SSU processome component) did coimmunoprecipitate with both Mpp10 and the U3 snoRNA (4). We verified that each ribosomal protein (both positive and negative) was enriched by immunoprecipitation by stripping the blot and reprobing it for the HA-tagged protein (data not shown).
FIG. 2.
A subset of ribosomal proteins is found in the SSU processome. 3×HA-tagged ribosomal proteins Rps4, Rps6, Rps7, Rps9, Rps14, Rps27, Rps28, and Rpl33 were immunoprecipitated with beads conjugated with HA antibodies. 3×HA-tagged Utp9 (a bona fide SSU processome component) and YPH499 (the untagged parental strain) were used as positive and negative controls, respectively. Ribosomal proteins were tested with Western blotting for their abilities to coimmunoprecipitate Mpp10. Ribosomal proteins were also tested with Northern blotting for their abilities to coimmunoprecipitate the U3 snoRNA. Results for totals (lanes T), representing 5% (Western blot) and 10% (Northern blot) of the proteins extracted, and immunoprecipitates (lanes IP) are shown.
We examined whether proteins represented as single peptides by mass spectrophotometry were SSU processome components (YJL069c and Enp1). In addition, we tested whether several proteins identified in tandem affinity purification-tagged SSU processome protein coimmunoprecipitates (8, 9) were also members of the SSU processome (YJL069c, YBL004w, YLR409c, YGR090w, Noc4, Emg1, Krr1, Bfr2, Enp1, and Enp2) (Tables 1 and 2). Proteins would be considered components of the SSU processome (i) if they were nucleolar, (ii) if they could coimmunoprecipitate with Mpp10 and the U3 snoRNA, both known SSU processome components, and (iii) if their depletion led to a decrease in the levels of the 18S rRNA.
TABLE 1.
Additional nonribosomal components of the SSU processome
| Protein | Alias | Gene product | Mol wt (103) | Essential? | Homolog (GenBank no. and/or % homology) | Motif and/or commentb | References |
|---|---|---|---|---|---|---|---|
| Utp18 | YJL069c | 66.3 | Yes | CGI-48 (25) | WD repeats, interacts with Utp21; one peptide in purification | 4, 12 | |
| Noc4 | YPR144c | 63.5 | Yes | MGC3162 (31) | Noc domain and ribosome biogenesis | 8, 9, 12, 24, 29 | |
| Utp20 | YBL004w | 287 | Yes | DRIMa (NP05531 8.1; 23) | HEAT repeats and homolog of DRIM | 8, 9, 12, 33, 34 | |
| Utp21 | YLR409c | 105 | Yes | TA-WDRP (NP644810.1; 32) | WD repeats, coiled-coil domains, interacts with Utp18 | 8, 9, 12, 13, 33 | |
| Utp22 | YGR090w | 141 | Yes | NOL6 (NRAP; 24) | Ribosome biogenesis and homolog of human Nrap | 8, 9, 12, 33, 38 | |
| Emg1 | Nep1 | YLR186w | 27.3 | Yes | C2Fc (53) | Required for 40S biogenesis and interacts with Nop14; one peptide in purification | 4, 9, 21 |
| Krr1 | YCL059c | 37.2 | Yes | HRB2 (NP008974.3; 61) | Required for 18S, KH domain, KRR-R motif | 8, 9, 13, 14, 32, 37 |
DRIM, down-regulated in metastasis.
HEAT, Huntington-elongation-A subunit-TOR; KH, lysine homology; KRR-R, lysine arginine arginine arginine.
C2F is a possible homolog.
TABLE 2.
Nucleolar proteins that are not components of the SSU processome
| Protein | Alias | Gene product | Mol wt (103) | Essential? | Homolog (GenBank no. and/or % homology) | Motif and/or commentb | References |
|---|---|---|---|---|---|---|---|
| Enp1 | Meg1 | YBR247c | 55 | Yes | BYSLa (42) | NLS, coiled-coil domains, interacts with Nop1; one peptide in purification | 3, 4, 9, 31-33 |
| Enp2 | YGR145w | 81.7 | Yes | FLJ14075 (38) | WD repeats, interacts with Mpp10 and Bfr2, and has homology to Spb1 | 8, 13 | |
| Bfr2 | YDR299w | 61.2 | Yes | AATF (NP036270.1; 25) | Interacts with Lcp5, Crm1, and Enp2 | 8, 12-14, 37 |
BYSL is a possible homolog.
NLS, nuclear localization sequence; WD, tryptophan-aspartate repeat.
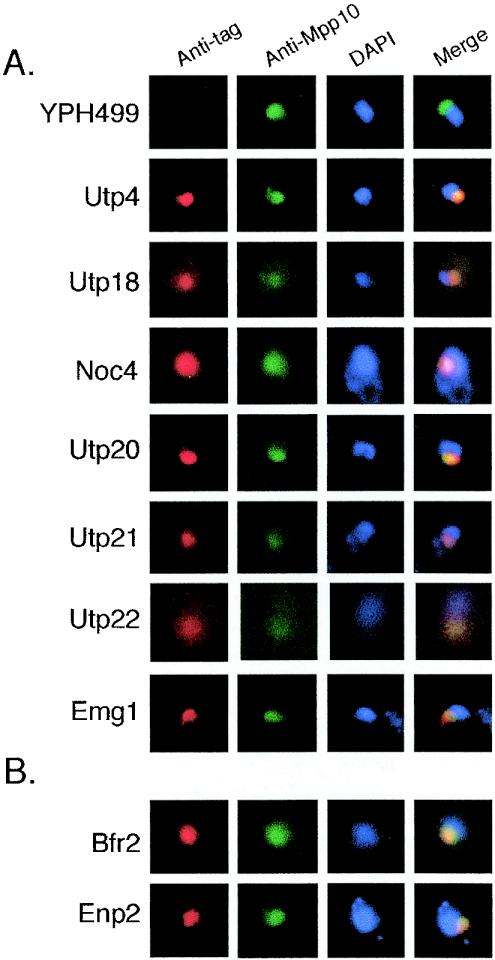
Since ribosome biogenesis occurs within the nucleolus, we reasoned that proteins that were part of the SSU processome would also be nucleolar and colocalize with another nucleolar protein, Mpp10. Therefore, we first determined the subcellular localization of the candidate SSU processome proteins. We localized HA-tagged YJL069c, YBL004w, YLR409c, YGR090w, Noc4, Emg1, Bfr2, and Enp2 (Fig. 3). An untagged parental strain, YPH499, and a known SSU processome component, Utp4-3×HA, were used as negative and positive controls, respectively. All of these proteins colocalized with Mpp10 and are therefore localized primarily to the nucleolus. Enp1, Krr1, and Noc4 have previously been localized to the nucleolus (3, 9, 25, 32). While the present work was in progress, YJL069c, YGR090w, and Bfr2 were localized to the nucleolus, whereas YBL004w, YLR409c, and Noc4 were localized to other cellular compartments (12). When overexpressed, Enp2 appears to accumulate predominantly in the nucleoplasm; however, we found that it localizes primarily to the nucleolus when expressed from its own promoter (19). Further analysis (see below) indicates that only seven of these proteins are part of the SSU processome. Bfr2 and Enp2 were localized to the nucleolus but are not SSU processome components, since they did not coimmunoprecipitate with Mpp10 and the U3 snoRNA (Fig. 3B and data not shown). Therefore, four of these proteins have subsequently been named to reflect their function in the biogenesis of the small ribosomal subunit (Utp18 for YJL069c, Utp20 for YBL004w, Utp21 for YLR409c, and Utp22 for YGR090w) (Fig. 3A).
FIG. 3.
Subcellular localization of new SSU processome proteins and two other related proteins. All proteins were 3×HA tagged and colocalized using anti-HA(red) or nucleolar protein Mpp10 (green) with anti-Mpp10 by indirect immunofluorescence. DAPI (4′,6′-diamidino-2-phenylindole) (blue) was used to stain the nuclear DNA. Merge, merge of tagged protein, Mpp10, and DAPI. (A) Nucleolar localization of Utp18, Noc4, Utp20, Utp21, Utp22, and Emg1. The parental strain YPH499 (untagged) and Utp4 (a bona fide SSU processome component) were used as negative and positive controls, respectively. (B) Nucleolar localization of Bfr2 and Enp2.
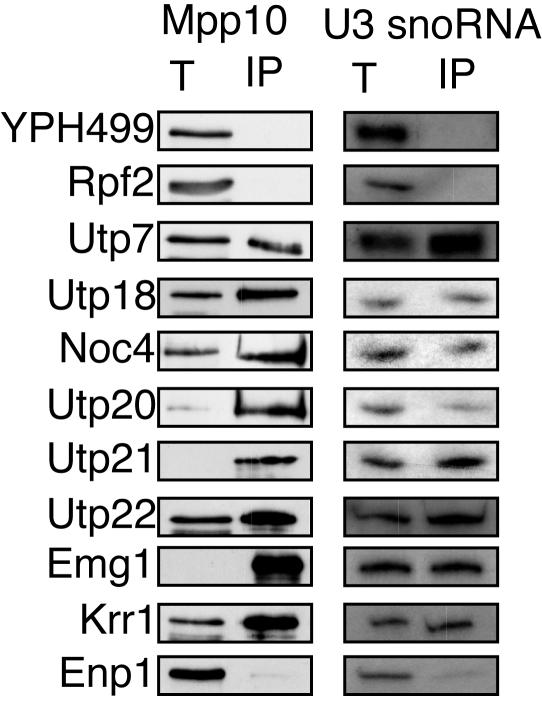
We determined whether candidate SSU processome proteins were able to coimmunoprecipitate with Mpp10 and the U3 snoRNA. HA-tagged proteins were immunoprecipitated and analyzed for the presence of the U3 snoRNA and Mpp10 by Northern and Western blotting, respectively. Utp18, Noc4, Utp20, Utp21, Utp22, Emg1, and Krr1 all coimmunoprecipitated with Mpp10 and the U3 snoRNA (Fig. 4). As expected, no coimmunoprecipitation was observed for an untagged strain (YPH499). Similarly, Rpf2-3×HA (a nucleolar protein involved in large-subunit biogenesis) did not coimmunoprecipitate with Mpp10 or the U3 snoRNA, while Utp7-3×HA, an SSU processome component, did (Fig. 4).
FIG. 4.
Coimmunoprecipitation experiments define new proteins as SSU processome components. Immunoprecipitations of 3×HA-tagged Utp18, Noc4, Utp20, Utp21, Utp22, Emg1, Krr1, and Enp1 were carried out using anti-HA antibodies. 3×HA-tagged proteins were immunoprecipitated and tested for their ability to coimmunoprecipitate with Mpp10 as determined by Western blot analysis. U3 snoRNA that coimmunoprecipitated with 3×HA-tagged proteins were analyzed by Northern blotting. Results for totals (lanes T), representing 5% (Mpp10) or 10% (U3 snoRNA) of the total protein extracted, and for immunoprecipitates (lanes IP) are shown. The parental strain, YPH499 (untagged), and 3×HA-tagged Rpf2 (a protein involved in large-subunit biogenesis) (40) were used as negative controls. 3×HA-tagged Utp7 (a bona fide SSU processome component) was used as a positive control.
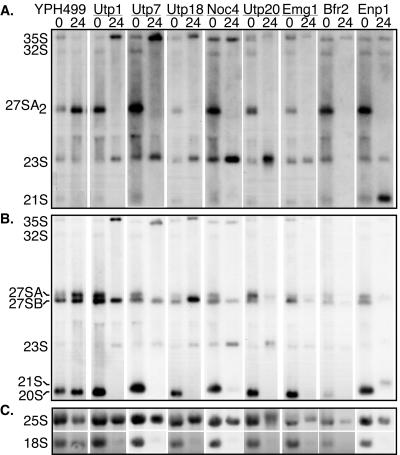
Collectively, our results suggest that these proteins are involved in ribosome biogenesis. To substantiate this finding, we determined whether the new nonribosomal SSU processome components have functions in ribosome biogenesis. Since all of the proteins tested are encoded by essential genes, we fused a galactose-inducible promoter and a 3×HA tag to each gene of interest in the chromosome. When these strains are grown in medium with galactose and raffinose, the gene is transcribed and the protein is expressed. However, when the yeast strains are grown in medium containing dextrose, the promoter is unable to induce gene expression, and its protein levels are depleted over time. RNA was extracted from strains grown to early log phase in medium containing galactose and raffinose (undepleted) and from strains grown for 24 h in dextrose (depleted). Upon RNA analysis by Northern blotting, Utp1, Utp7, Utp18, Noc4, Utp20, and Emg1 revealed similar pre-rRNA processing defects (Fig. 5). In all of these strains, 35S and 23S pre-rRNAs accumulated, and 27SA2, 20S, and 18S rRNAs were no longer present. Accumulation of these precursors suggests defects in pre-rRNA processing at cleavage sites A0, A1, and A2 (Fig. 5). Different processing defects were also observed in cells depleted of Bfr2 and Enp1. Strikingly, when the Bfr2 protein was depleted, the levels of all the pre-rRNAs were reduced; however, 18S rRNA levels were more affected than those of 25S rRNA. In Enp1 depletion, accumulation of the processing intermediates 35S, 23S, and 21S pre-rRNA (A1 to A3) was observed, and levels of the 27SA and 18S rRNAs were reduced. The 21S pre-rRNA has previously been noted to be a precursor that is normally present in the strains we studied (20). The observed increase in the accumulation of the 21S pre-rRNA precursor suggests a defect in cleavage at A2. While this work was in progress, pre-rRNA processing defects were reported in Noc4, Utp20, Utp22, Enp1, and Emg1 (3, 7, 25, 29). Collectively, depletion of each of these proteins led to a significant reduction in the levels of the mature 18S rRNA but not in that of the 25S rRNA. In addition, protein depletion did not affect the levels of U3, U14, 5.8S rRNA, suggesting that the observed processing defects were not due to general RNA degradation (data not shown).
FIG. 5.
Depletion of new SSU processome proteins leads to defects in pre-18S rRNA processing. Strains expressing Utp1, Utp7, Utp18, Noc4, Utp20, Emg1, Bfr2, and Enp1 from galactose-inducible and glucose-repressible promoters were grown in early log phase (time point 0) in galactose- and raffinose-supplemented media (undepleted) and then shifted into glucose (depleted) for 24 h. YPH499, the untagged parental strain, was used as a control. RNA from undepleted or depleted yeast strains was analyzed for the presence of pre-rRNAs by Northern blotting. Equal amounts of RNA were separated on formaldehyde-1.2% agarose gels, transferred to membranes, and hybridized with the specific oligonucleotide probes shown in Fig. 1. (A) Northern blot using oligonucleotide c, which hybridizes to 35S, 32S, 27SA2, 23S and 21S pre-rRNAs; (B) Northern blot using oligonucleotides b and e, which hybridize to the 35S, 32S, 27SA, 27SB, 23S, 20S pre-rRNAs; (C) Northern blot using oligonucleotides a and y, which hybridize to the 18S and 25S rRNAs, respectively.
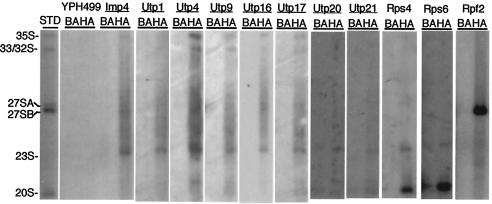
Since the SSU processome proteins affect the processing of RNA precursors to the small-ribosomal-subunit rRNA, we determined with which specific precursor rRNAs they were associated. 3×HA-tagged SSU processome proteins were immunoprecipitated, RNA was extracted, and Northern blots were probed with oligonucleotides specific for pre-rRNAs. Utp1, Utp4, Utp9, Utp16, Utp17, Utp20, Utp21, Rps4, and Rps6 were all tested for their ability to coimmunoprecipitate with pre-rRNA species. Utp1, Utp4, Utp9, Utp16, and Utp17 were all able to coimmunoprecipitate with the 35S, 33/32S, and 23S rRNA precursors (Fig. 6). Utp20 and Utp21 were able to efficiently coimmunoprecipitate with the 23S pre-rRNA (Fig. 6). Pre-rRNA coimmunoprecipitation experiments demonstrate that SSU processome proteins efficiently associate with the 23S pre-rRNA. None of these proteins were able to coimmunoprecipitate with the 20S pre-rRNA. Rps4 and Rps6 also coimmunoprecipitated with the 23S pre-rRNA (Fig. 6). This suggests that a subset of small-ribosomal-subunit proteins is found on pre-rRNA precursors with the SSU processome. As expected, Rps4 and Rps6 also coimmunoprecipitated with the 20S pre-rRNA, the RNA that is exported from the nucleolus to the cytoplasm, where the small ribosomal subunit undergoes a final cleavage step. No coimmunoprecipitation of pre-rRNA was observed in the untagged strain, YPH499. In contrast, an SSU processome component, Imp4, was able to coimmunoprecipitate with the 35S, 33/32S, and 23S pre-rRNAs. Rpf2, a protein essential for large-subunit biogenesis, was able to coimmunoprecipitate with the 27SB pre-rRNA specie, but not with precursors to the 18S rRNA, as previously described (40). Together, these data suggest that SSU processome components are associated with the 35S, 33/32S, and 23S pre-rRNA species. In addition, Rps4 and Rps6 are also associated with the 23S and 20S pre-rRNA, the latter of which represents the fully assembled 40S preribosome.
FIG. 6.
SSU processome and small-ribosomal-subunit proteins associate with precursor rRNAs. 3×HA-tagged Imp4, Utp1, Utp4, Utp9, Utp16, Utp17, Utp20, Utp21, Rps4, Rps6, and Rpf2 were immunoprecipitated with anti-HA antibodies bound to Sepharose beads and analyzed by Northern blotting for their ability to coimmunoprecipitate with pre-rRNAs (lanes HA). An untagged strain, YPH499, and 3×HA-tagged Rpf2, a protein involved in large-subunit biogenesis, were used as controls. Imp4, a known SSU processome component, was used as a positive control. All experiments were also done in parallel without antibody, i.e., with beads alone (lanes BA), and pre-rRNA precursors that immunoprecipitated were determined by comparison to a standard (lane STD). Northern blots were probed with oligonucleotides b, c, and e, which hybridize to the 35S, 32S, 27SA2, 27SB, 23S, 21S, and 20S pre-rRNAs (as shown in Fig. 1). Rps4 and Rps6 Northern blots were probed with oligonucleotides b and e, which hybridize to the 35S, 32S, 23S, 21S, and 20S pre-rRNAs (see Fig. 1).
DISCUSSION
During the original SSU processome purification, 28 components that are required for 18S rRNA biogenesis were identified (4). Upon further analysis, we have identified 12 additional components of the SSU processome. These components include seven nonribosomal proteins (Utp18, Noc4, Utp20, Utp21, Utp22, Emg1, and Krr1) (Table 1) and five ribosomal proteins (Rps4, Rps6, Rps7, Rps9, and Rps14). All of the nonribosomal proteins are nucleolar, required for the processing of the small ribosomal subunit, and coimmunoprecipitate with Mpp10, U3, and the 35S, 33/32S, and 23S pre-rRNAs. The ribosomal proteins are able to coimmunoprecipitate with Mpp10 and the U3 snoRNA, suggesting that this subset of ribosomal proteins is made up of bona fide SSU processome components. In addition, Rps4 and Rps6 both coimmunoprecipitate with the 23S and 20S pre-rRNAs. Other proteins identified but subsequently found not to be components of the SSU processome are described in Table 2.
The SSU processome is thus a preribosomal complex of at least 40 proteins and the U3 snoRNA. As might be expected based on the large number and sizes of the proteins, this complex sediments at 80S on a sucrose gradient (4). Surprisingly, pre-rRNA coimmunoprecipitation experiments demonstrate that SSU processome proteins efficiently associate with the 23S pre-rRNA, a precursor which is often labeled as an aberrant pre-rRNA (2, 39). However, this precursor is normally present in the strain in our study (10). Therefore, the SSU processome remains associated with rRNA precursors of the mature 18S rRNA that have been separated from rRNA precursors to the large ribosomal subunit by cleavage at A3.
Although we tested a large number of ribosomal and nonribosomal proteins to find additional components of the SSU processome, we may not have exhausted all possible candidates. We tested a subset of small-ribosomal-subunit proteins based upon their identification in our original purification of the SSU processome and their identification in other purifications (4, 8, 9). Although we found a distinct set of ribosomal proteins to be components of the SSU processome, there may be additional ribosomal protein components that were not tested. In addition, we may not have identified some nonribosomal protein components because we relied on the existing copurifications and cannot rule out the possibility that the conditions used did not disrupt the integrity of this complex. Currently, the SSU processome is thought to be composed of the U3 snoRNA, 35 nonribosomal proteins (Utp1 to Utp18, Utp20 to Utp22, Noc4, Nop1, Nop56, Nop5/58, Snu13, Mpp10, Imp3, Imp4, Dhr1, Rrp9, Rrp5, Emg1, and Krr1), and 5 ribosomal proteins (Rps4, Rps6, Rps7, Rps9, and Rps14) (4).
The order of assembly of ribosomal proteins with rRNA was first described for Escherichia coli during the early 1970s (27, 35). The first steps in analysis of the ribosomal pattern of assembly came in 1966, when Staehelin and Meselson observed that 30 to 40% of the small-ribosomal-subunit proteins partially disassembled during density gradient centrifugation in 5 M cesium chloride (35). This discovery enabled the establishment of a system for reconstituting ribosomes, which facilitated the elucidation of a detailed pathway for in vitro ribosome assembly, termed the “30S assembly map” (11, 27, 28, 36). Ribosomal proteins were grouped according to their abilities to bind to rRNA and to each other. Primary binders (i.e., S4, S7, S8, S15, S17, and S20) are ribosomal proteins that bind to rRNA directly, whereas secondary and tertiary binders are ribosomal proteins that require the presence of one or more ribosomal proteins (28). Many of the bacterial primary binding proteins (for example, S7, S8, S15, and S20) do not have yeast homologues, making it difficult to extrapolate the bacterial data to S. cerevisiae (23, 30). Only Rps9 and Rps14 yeast ribosomal proteins have bacterial homologues, i.e., the primary and tertiary binding proteins S4 and S11, respectively (23, 30). Since S. cerevisiae rRNAs and bacterial rRNAs are different, and since not all ribosomal proteins are conserved in both organisms, S. cerevisiae may have a set of primary binding proteins that is distinct from that in bacteria.
We propose that the ribosomal proteins associated with the SSU processome may be analogous to the primary or secondary binding proteins described for bacteria, since cleavages by the SSU processome represent early pre-rRNA maturation steps for the small-ribosomal-subunit rRNA. We found that the yeast ribosomal proteins Rps4, Rps6, Rps7, Rps9, and Rps14 were bona fide components of the SSU processome and may therefore represent a distinct set of yeast ribosomal proteins involved in the early stages of ribosome assembly. Because there is no in vitro ribosomal assembly system for eukaryotic ribosomes, we can only hypothesize which ribosomal proteins bind first on the basis of their association with pre-rRNAs. For example, Rps4 and Rps6 were both able to coimmunoprecipitate the 23S pre-rRNA, suggesting that they may be involved in ribosome assembly prior to cleavage at sites A0, A1, and A2 (Fig. 6). These results are consistent with those of Kruiswijk et al., who hypothesized that a specific set of ribosomal proteins (Rps23, Rps18, Rps2, Rps30, Rps5, Rps11, Rps19, Rps4, Rps21, Rps9, Rps22, and Rps3 [in new nomenclature]) were involved in the early stages of ribosomal assembly (18). In agreement with this hypothesis, we found that Rps4 and Rps9 may be required for the early steps of ribosome assembly. However, we were unable to confirm the current ribosomal protein counterpart for 10 ribosomal proteins (S19, S12, S22, S20, S21, S11, S6, S17, S5, and S29) that were found by Kruiswijk et al. to be associated with an early step of assembly of the small ribosomal subunit (18, 23, 30). In addition, two ribosomal proteins that we have found to be SSU processome components, Rps7 and Rps14, were not analyzed by Kruiswijk et al.
Our results are consistent with those of Grandi et al. and Schäfer et al., who reported the identification of a 90S RNP that contains 35 nonribosomal proteins and the U3 snoRNA (9, 33). They identified 27 SSU processome proteins that we had identified previously (Nop1, Noc4, Nop56, Mpp10, Imp3, Imp4, Sof1, Rrp5, Rrp9, Utp1, Utp2, Utp4, Utp5, Utp6, Utp7 Utp8, Utp9, Utp10, Utp11, Utp12, Utp13, Utp15, Utp16, Utp17, Utp18, Utp21, Utp22, and Emg1) but did not identify 2 other SSU processome components (Utp3 and Utp14). Due to the large overlap of protein components, it seems highly probable that we have independently characterized the same complex. Our results are also consistent with those of Krogan et al. and Peng et al., who used genomics and proteomic experiments to identify additional proteins required for ribosome biogenesis (17, 29).
One important difference between the interpretation of our results and those previously published is whether Enp1 is a component of the SSU processome-90S preribosome (3, 9, 33). We have shown here that tagged Enp1 does not detectably coimmunoprecipitate with other SSU processome components or the U3 snoRNA, compared to positive and negative controls, because Enp1 coimmunoprecipitated with the same amounts of Mpp10 and the U3 snoRNA as did the negative controls (Rpf2 and the untagged parental strain, YPH499). In addition, genetic depletion of Enp1 led to the accumulation of the 21S pre-rRNA, a phenotype different from that caused by the depletion of any other SSU processome component. The appearance of the 21S pre-rRNA upon Enp1 depletion suggests that it may participate in a later complex, as has been suggested by Milkereit et al. (25, 33). However, sedimentation of Enp1 on sucrose density gradients shows that Enp1 sediments at both 40S and 90S (33). These results therefore suggest that Enp1 may have multiple roles in ribosome biogenesis, and we cannot rule out the possibility that it may also be present in substoichiometric amounts in the SSU processome (3).
In addition to the SSU processome's role in pre-rRNA processing, it also likely has a role in RNA folding, as has been suggested previously (4, 39). Furthermore, the specific association of a subset of ribosomal proteins with the SSU processome suggests that the SSU processome is also an assembly intermediate for ribosome biogenesis. Therefore, folding of the pre-rRNA for processing is intertwined with ribosome assembly.
Acknowledgments
K.A.B. and J.E.G.G. were supported by predoctoral fellowships from the National Institutes of Health (GM67564 and GM20905). K.A.B. was previously supported by a Research Service Award (GM07499) from the National Institute of General Medical Sciences (NIGMS). S.G. was supported by a Leslie H. Warner fellowship in cancer research. This work was supported by NIH grant GM52581 to S.J.B.
REFERENCES
- 1.Adams, C. C., J. Jakovlyevic, J. Roman, P. Harnpicharnchai, and J. L. J. Woolford. 2002. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA 8:150-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang, C., P. Mitchell, E. Petfalski, and D. Tollervey. 2000. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 28:1684-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, W., J. Bucaria, D. A. Band, A. Sutton, and R. Sternglanz. 2003. Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res. 31:690-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunbar, D. A., F. Dragon, S. J. Lee, and S. J. Baserga. 2000. A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. USA 97:13027-13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunbar, D. A., S. Wormsley, T. M. Agentis, and S. J. Baserga. 1997. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol. 17:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eschrich, D., M. Buchhaupt, P. Kötter, and K.-D. Entian. 2002. Nep1p (Emg1p), a novel protein conserved in eukaryotes and archaea, is involved in ribosome biogenesis. Curr. Genet. 40:326-338. [DOI] [PubMed] [Google Scholar]
- 8.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 9.Grandi, P., V. Rybin, J. Baβler, E. Petfalski, D. Strauβ, M. Marzioch, T. Schäfer, B. Kuster, H. Tschochner, D. Tollervey, A.-C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 10.Granneman, S., and S. J. Baserga. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43-50. [DOI] [PubMed] [Google Scholar]
- 11.Held, W. A., S. Muizushima, and M. Nomura. 1973. Reconstitution of Escherichia coli 30S ribosomal subunits from purified molecular components. J. Biol. Chem. 248:5720-5730. [PubMed] [Google Scholar]
- 12.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 13.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, T. H., M. Neville, J. C. Rain, T. McCarthy, P. Legrain, and M. Rosbash. 2000. Identification of novel Saccharomyces cerevisiae proteins with nuclear export activity: cell cycle-regulated transcription factor Ace2p shows cell cycle independent nucleocytoplasmic shuttling. Mol. Biol. Cell 20:8047-8058. [Google Scholar]
- 15.Knop, M., K. Siegers, G. Pereira, W. Zachiariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 16.Kressler, D., P. Linder, and J. de la Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 18.Kruiswijk, T., R. J. Planta, and J. M. Krop. 1978. The course of the assembly of ribosomal subunit in yeast. Biochim. Biophys. Acta 517:378-389. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, A., S. Agarwal, J. A. Heyman, S. Matson, M. Heidtman, S. Piccirillo, L. Umansky, A. Drawid, R. Jansen, Y. Liu, K. H. Cheung, P. Miller, M. Gerstein, G. S. Roeder, and M. Snyder. 2002. Subcellular localization of the yeast proteome. Genes Dev. 16:707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, S. J., and S. J. Baserga. 1999. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 19:5441-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, P. C. C., and D. J. Thiele. 2001. Novel stress-responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol. Biol. Cell 12:3644-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longtine, M. S., A. R. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 23.Mager, W. H., R. J. Planta, J. G. Ballesta, J. C. Lee, K. Mizuta, K. Suzuki, J. R. Warner, and J. Woolford. 1997. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 25:4872-4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milkereit, P., O. Gadal, A. Podtelejnikov, S. Trumtel, N. Gas, E. Petfalski, D. Tollervey, M. Mann, E. Hurt, and H. Tschochner. 2001. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 105:499-509. [DOI] [PubMed] [Google Scholar]
- 25.Milkereit, P., H. Kühn, N. Gas, and H. Tschochner. 2003. The pre-ribosomal network. Nucleic Acids Res. 31:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milkereit, P., D. Strauss, J. Bassler, O. Gadal, H. Kuhn, S. Schutz, N. Gas, J. Lechner, E. Hurt, and H. Tschochner. 2003. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 278:4072-4081. [DOI] [PubMed] [Google Scholar]
- 27.Mizushima, S., and M. Nomura. 1980. Assembly mapping of 30S ribosomal proteins from E. coli. Nature 226:1214-1218. [DOI] [PubMed] [Google Scholar]
- 28.Nomura, M. 1973. Assembly of bacterial ribosomes. Science 179:864-873. [DOI] [PubMed] [Google Scholar]
- 29.Peng, W. T., M. D. Robinson, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, J. Vladimir, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919-933. [DOI] [PubMed] [Google Scholar]
- 30.Planta, R. J., and W. H. Mager. 1998. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast 14:471-477. [DOI] [PubMed] [Google Scholar]
- 31.Roos, J., J. M. Luz, S. Centoducati, R. Sternglanz, and W. J. Lennarz. 1997. ENP1, an essential gene encoding a nuclear protein that is highly conserved from yeast to humans. Gene 185:137-146. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki, T., A. Toh-e, and Y. Kikuchi. 2000. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol. Cell. Biol. 20:7971-7979.11027267 [Google Scholar]
- 33.Schäfer, T., D. Straub, E. Petfalski, D. Tollervey, and E. Hurt. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22:1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwirzke, M., A. Gnirke, P. Bork, D. Tarin, and U. H. Weidle. 1998. Differential gene expression in mammary carcinoma cell lines: identification of DRIM, a new gene down-regulated in metastasis. Anticancer Res. 18:1409-1421. [PubMed] [Google Scholar]
- 35.Staehelin, T., and M. Meselson. 1966. In vitro recovery of ribosomes and of synthetic activity from synthetically inactive ribosomal subunits. J. Mol. Biol. 15:245-249. [DOI] [PubMed] [Google Scholar]
- 36.Traub, P., and M. Nomura. 1968. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. USA 59:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. J. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 38.Utama, B., D. Kennedy, K. Ru, and J. S. Mattick. 2002. Isolation and characterization of a new nucleolar protein, Nrap, that is conserved from yeast to humans. Genes Cells 7:115-132. [DOI] [PubMed] [Google Scholar]
- 39.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 40.Wehner, K. A., and S. J. Baserga. 2002. The sigma 70-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9:329-339. [DOI] [PubMed] [Google Scholar]