Abstract
Background
Some studties reported that the polymorphism of TM6SF2 gene E167K affects the occurrence and the progression of hepatocytes carcinoma (hepatocellular, HCC). In oeder to investigate the effects of the polymorphism of TM6SF2 gene E167K in the pathogenesis of HCC, we explored its influence on the cell cycle in hepatocellular carcinoma cell HEPA1-6.
Methods
HEPA 1–6 cells which could respectively overexpress TM6SF2 wild type and E167K variant were cultured and HEPA 1–6 cells with zero load plasmids were used as matched control. Flow cytometry was used to detect the cell cycles of these 3 type of HEPA 1–6 cells. Realtime fluores-cence quantitative PCR and western blot were used to analyzed the expression of regulatory factors (Cyclin D1、p53、P16、P27、P21 and Rb) of cell cycle. T-test was used in statistical analysis.
Results
Cell cycle phase distribution was presented by the proportion of cells in each phases (%). Compared with the control group, the cell cycle phase distribution (G1 phase 57.36 ± 0.21%, G2/M phase 25.61 ± 0.36%,S phases 19.31 ± 0.25%) had no differences in wild type group (G1 phase 57.63 ± 0.28%, G2/M phase 25.77 ± 0.51%, S phases 19.54 ± 0.25%; P < 0.05). Between variant type group and wild type group,G1 phase was significantly decreased (variant type group G1 phase 36.26 ± 0.31%, P < 0.05),S phase and G2/M phase were increased(variant type group S phase 28.41 ± 0.31%, P < 0.05;G2/M phase 35.23 ± 0.14%, P < 0.05), respectively. Compared with control group,the relative expression of CyclinD1、P53 and Rb mRNA in variant type group was significantly upregulated (2.03 ± 0.01 VS 1.04 ± 0.06, 1.88 ± 0.05 VS 1.37 ± 0.03, 1.29 ± 0.06 VS 1.15 ± 0.03, P < 0.05) and P27 mRNA in variant type group was significantly downregulated (0.56 ± 0.02 VS 0.85 ± 0.05, P < 0.05).
Compared with wild type group, the relative expression of CyclinD1、P53 and Rb mRNA in variant type group was significantly upregulated (wild type group 1.00 ± 0.00, 1.48 ± 0.09, 1.18 ± 0.01, P < 0.05) and P27 mRNA in variant type group was significantly downregulated (variant type group 0.82 ± 0.05,P < 0.05). There was no statistical significance between wild type group and control group (P > 0.05). P16 and P21 expression showed no statistical sigtfificance in any of these three groups (P > 0.05).
Conclusion
E167K polymorphism of TM6SF2 gene affects cell cycles of HEPA1–6 cells via up-regulating CyclinD1、P53 and Rb and down-regulating P27.
Keywords: Hepatocellular carcinoma cell (HCC), Cell cycle, TM6SF2 E167K polymorphism, Western blot, Pcr
Background
The Strictly cell cycle is the guarantee of proliferation and division, which is very important for maintaining the normal function of the organism. A key function of the cell cycle is to ensure accurate replication and segregation of the genome, because errors in genetic transmission can cause mutations and chromosomal rearrangements that may lead to cell death or disease [1]. Many studies have reported that deregulated cell proliferation and suppressed cell death together provide the underlying platform for neoplastic progression [2–4]. Exploring the changes of cell cycle, which has the great significance to reveal the pathogenesis of the disease. Many studies have reported that the transmembrane 6 superfamily member 2 (TM6SF2) is one of the most important genes of lipid metabolism in the liver and the E167K polymorphism can independently lead to liver fat accumulation, which has an important role in chronic liver disease [5–7]. While some studies reported that the polymorphism of E167K affects the occurrence and the progression of hepatocytes Carcinoma (hepatocellular, HCC) [5, 8, 9]. In order to investigate the effects of the polymorphism of TM6SF2 gene E167K in then pathogenesis of HCC, we explored its influence on the cell cycle in hepatocellular carcinoma cell HEPA 1–6.
In our study, HEPA 1–6 cells which could respectively overexpress TM6SF2 wild type and E167K variant were cultured and HEPA 1–6 cells with zero load plasmids were used as matched control. To explore the cell cycle of HEPA 1–6 cells affected by E167K polymorphism of TM6SF2 gene and the possible mechanisms.
Methods
Culture of HEPA 1–6 cell lines
The HEPA 1–6 cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) medium containing 100 U/mL penicillin, 10% fetal bovine serum (FBS; Hyclone, USA) and 100 μg/mL streptomycin (Gibco®, USA), and incubated at temperature 37 °C in a humidified atmosphere with 5% CO2. The cells were treated at the time of approximately 80% confluence.
Construction of Lentiviral vectors
The present study had three groups, including TM6SF2 wild type, TM6SF2 variant type and zero load plasmids as matched control. The lentiviral plasmids (Shanghai Genechem Co., LTD) transfected into 293 T cells, after 24 h culture in DMEM with 10% FBS. 293 T cells were incubated with transfection complexes (expression plasmid, packaging plasmid and transfection reagent) for 48–72 h. We then concentrated the lentivirus and transfected it into HEPA 1–6 cells. The success of the transfection process was validated by performing western blot, real time-polymerase chain reaction (RT-PCR) as well as the percentage of HEPA 1–6 cells with green fluorescent protein (GFP).
Biochemical indicator assay
Cyclin D1、p53、P16、P27、P21 and Rb monoclonal antibody were purchased from the Biotechnology Corporation, USA. The primer of Cyclin D1、p53、P16、P27、P21 and Rb were designed by Invitrogen Corporation, USA. While the primer of β-actin was synthesized by Nanjing Kingsy Biotechnology Co., ltd.
Cell cycle determination
The logarithmic growth phase of cells were digested into suspension in a centrifuge tube of 15 ml, centrifugaled at room temperature for 5 min with 800 r/min, centrifugaled and washed 2 times repeatly with PBS, and then fixed at temperature 4 °C with 2 ml 70% absolute alcoholo vernight.
The fixed cells were centrifugaled at room temperature for 5 min with 800 r/min and then retained 0.5 ml PBS to percuss the cells into suspension, and then add 1 mg/ml PI and RNase A, and water bath at temperature 37 °C for 30 min. Flow cytometry was used to detect the cell cycle.
Western blotting
Total protein of the HEPA 1–6 cells was extracted by RIPA buffer (Sigma-Aldrich, USA). Bradford method was performed to determine the protein concentration following the manufacturer’s protocols, and proteins were frozen at −70 °C until analysis. Antibodies against Cyclin D1(JIANGSU KEYGEN BIOTECH. CO., LTD, KG22272)、p53(JIANGSU KEYGEN BIOTECH. CO., LTD, KG22606)、P16(JIANGSU KEYGEN BIOTECH. CO., LTD, KG22602)、P27(JIANGSU KEYGEN BIOTECH. CO., LTD, KG30242)、P21(JIANGSU KEYGEN BIOTECH. CO., LTD, KG30240)、Rb(JIANGSU KEYGEN BIOTECH. CO., LTD, KG21109) and GAPDH (KGAA002) were used. The expression level of Cyclin D1 or the other protein was normalized relative to the corresponding GAPDH (endogenous reference) level in each lane. Images of Western Blotting were analyzed using Gel-Pro Analyzer Version 4.5 Software (Media Cybernetics, USA).
Quantitative real time PCR
Total RNA was isolated from the HEPA 1–6 cells using Trizol reagent (Invitrogen: 15,596–026, USA) following the manufacturer’s protocols. Complementary DNA (cDNA) synthesis was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher: K1622, USA).
The primers for PCR amplification of the fragments containing Cyclin D1、p53、P16、P27、P21 and Rb were synthesized by Nanjing Sipu Kim Technology Co. Ltd.: 5- CAGATCATCCGCAAACACGC -3 and 5- AAGTTGTTGGGGCTCCTCAG -3 for Cyclin D1, 5-CTGGATTGGCAGCCAGACT-3 and 5- GCTCGACGCTAGGATCTGAC-3 for P53, 5- TGTGCCACACATCTTTGACCT-3 and 5- AGGACCTTCGGTGACTGATGA-3 for P16, 5- CCCTGAACGGAGCTGAAGTC-3 and 5- TAACCGCGCAGCAGATAGTC-3 for P27, 5- CGTTCACAGGTGTTTCTGCG-3 and 5- CATTAGCGCATCACAGTCGC-3 for P21, 5- TGCAGTATGCTTCCACCAGG-3 and 5- TGTTGGTGTTGGCAGACCTT-3 for Rb, 5-CTCCATCCTGGCCTCGCTGT-3 and 5-GCTGTCACCTTCACCGTTCC-3 for β-actin. The PCR amplification profile was as follows: pre-denaturation at 95 °C for 10 min, 35 cycles, denaturation at 95 °C for 15 s, annealing at 58 °C for 60 s, extending at 72 °C for 30 s, and finally extending at 72 °C for 10 min to terminate the reaction. We used the comparative threshold cycle (CT) method to obtain the above genes [10].
Statistical analysis
Data are expressed as the mean ± standard deviation (S.D.) and T-test was used in statistical analysis. These statistical analyses were performed using SPSS 17.0 statistical software (SPSS Inc., Chicago, IL, USA). P < 0.05 were considered statistically significant.
Results
Constructed HEPA 1–6 cell lines with overexpress TM6SF2 successfully
The lentiviral plasmids transfected into HEPA 1–6 after 48 h, the positive rate of GFP >95% was observed, which prompted we have constructed stable strains successfully.
Detected cell cycle by flow cytometry
Table 1 showed the proportion of cells in each cell cycle stage of the three groups. Compared with the wild type group, the cell cycle phase distribution (G1,S and G2/M) had no differences in control group (t was 1/336, 1.127, 0.444, respectively, P all >0.05.); Compared with the wild type group and the control group, G1 phase was significantly decreased and S phase, G2/M phase were increased in variant type group (p all < 0.001.).
Table 1.
Comparison of the percentage of cell cycle in three groups (n = 3)
| groups | Cell cycle stage (%) | ||
|---|---|---|---|
| G1 | S | G2/M | |
| variant type group | 36.26 ± 0.31 | 28.41 ± 0.31 | 35.23 ± 0.14 |
| wild type group | 57.63 ± 0.28 | 19.54 ± 0.25 | 25.77 ± 0.51 |
| the control group | 57.36 ± 0.21 | 19.31 ± 0.25 | 25.61 ± 0.36 |
| t1 | −88.607 | 38.577 | 30.982 |
| p1 | <0.001 | <0.001 | <0.001 |
| t2 | −97.595 | 39.577 | 29.201 |
| p2 | <0.001 | <0.001 | <0.001 |
| t3 | 1.336 | 1.127 | 0.444 |
| p3 | >0.05 | >0.05 | >0.05 |
t1:Between variant type group and wild type group; t2: Between variant type group and the control group; t3: Between wild type group and the control group;
Detection of Cyclin D1、p53、P16、P27、P21 and Rb protein expression by western blot
Compared with control group and wild type group,the relative expression of CyclinD1、P53 and Rb in variant type group was significantly upregulated and P27 mRNA in variant type group was significantly downregulated (P < 0.05), there was no statistical significance between wild type group and control group (P > 0.05). P16 and P21 expression showed no statistical sigtfificance in any of these three groups (P > 0.05). (Fig. 1).
Fig. 1.
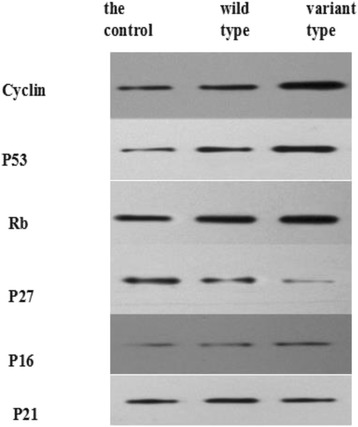
Detection of Cyclin D1、p53、P16、P27、P21 and Rb protein expression by Western blot
Detection of Cyclin D1、p53、P16、P27、P21 and Rb mRNA expression by quantitative real time PCR
Compared with control group, the relative expression of CyclinD1、P53 and Rb mRNA in variant type group was significantly upregulated (t was 28.190, 15.152, 3.615, P all <0.05) and P27 mRNA in variant type group was significantly downregulated (t was 9.328, P < 0.05). Compared with wild type group, the relative expression of CyclinD1、P53 and Rb mRNA in variant type group was significantly upregulated (t was 178.532, 6.729, 3.132, P all <0.05) and P27 mRNA in variant type group was significantly downregulated (t was 8.363, P < 0.05). There was no statistical significance between wild type group and control group (P > 0.05). P16 and P21 expression showed no statistical sigtfificance in any of these three groups (P > 0.05). (Table 2).
Table 2.
Detection of Cyclin D1、p53、P16、P27、P21 and Rb mRNA expression by Quantitative Real Time PCR
| groups | Quantitative Real Time PCR | |||||
|---|---|---|---|---|---|---|
| Cyclin D1 | p53 | Rb | P27 | P21 | P16 | |
| variant type group | 2.03 ± 0.01 | 1.88 ± 0.05 | 1.29 ± 0.06 | 0.56 ± 0.02 | 0.78 ± 0.06 | 1.00 ± 0.04 |
| wild type group | 1.00 ± 0.00 | 1.48 ± 0.09 | 1.18 ± 0.01 | 0.82 ± 0.05 | 0.77 ± 0.02 | 1.01 ± 0.06 |
| the control group | 1.04 ± 0.06 | 1.37 ± 0.03 | 1.15 ± 0.03 | 0.85 ± 0.05 | 0.78 ± 0.04 | 1.00 ± 0.06 |
| t1 | 178.532 | 6.729 | 3.132 | 8.363 | 0.158 | 0.240 |
| P1 | <0.001 | 0.0016 | 0.041 | <0.001 | >0.05 | >0.05 |
| t2 | 28.190 | 15.152 | 3.615 | 9.328 | 0 | 0 |
| P2 | <0.001 | <0.001 | 0.014 | <0.001 | >0.05 | >0.05 |
| t3 | 1.156 | 2.008 | 1.643 | 0.735 | 0.387 | 0.204 |
| P3 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 | >0.05 |
t1:between variant type group and wild type group; t2:between variant type group and the control group; t3:between wild type group and the control group;
Discussion
Recent genome-wide association studies have identified that variant in TM6SF2 was significantly associated with liver dieases in multiple ethnic groups. Some studies assessed the interaction between TM6SF2 E167K variant in the conditioning of HCC development had come to the conclusion that TM6SF2 E167K variant may be potential genetic risk factors for developing HCC [11–13]. Our study was to explore the cell cycle of HEPA 1–6 cells affected by E167K polymorphism of TM6SF2 gene and the possible mechanisms.
Eukaryotic DNA replication is regulated to ensure all chromosomes replicate once and only once per cell cycle [14]. Errors that result in underreplication or overeplication of the genome in any cell cycle have disastrous consequences and can produce a large array of human genetic diseases, including cancer, birth defects, and many developmental abnormalities [15]. Cell cycle included G1, S, G2 and M phases. G1/S phase and G2/M phase were the important “Check” for cell cycle and were to maintain the normal operation of the cell cycle. Cell cycle regulation by protein phosphorylation ensures that pre-RC assembly can only occur in G1 phase, whereas helicase activation and loading can only occur in S phase [14]. Once the cell passed through the G1/S phase, it will no longer depend on the exogenous proliferation and division signal and complete cell cycle independently [14]. Therefore, the G1/S phase is the most critical period of cell cycle regulation. In our study, G1 phase was significantly decreased and S phase and G2/M phase were increased in variant type group than wild type group and the control group. The result suggested that TM6SF2 gene mutation of E167K may promote DNA replication and accelerate cell cycle of human hepatocellular carcinoma cell line HEPA 1–6 and thereby promote the development of liver cancer cells. Therefore, the acceleration of cell cycle may has the important influence in malignant progression of HCC cells.
Cell cycle regulation is a hot topic in the field of oncology and it is a very complex and delicate process. A variety of internal and external factors involved in the process of cell cycle regulation. These factors included cells cyclin (cyclin), cyclin dependent-kinase (CDK) and some tumor suppressor genes such as Rb, p16, p21, p27, p53 protein products [16, 17]. A large number of studies have showed that cyclin, Rb, p16, p21, p27 and p53 played an important role in G1/S phase of cell cycle [18–22]. Uncontrolled cell proliferation is the hallmark of cancer, and tumor cells have typically acquired damage to genes that directly regulate their cell cycles [23]. In our study, our results showed that CyclinD1、P53 and Rb were increased and P27 was decreased in variant type group, which suggested that E167K polymorphism of TM6SF2 gene may change the cell cycle of hepatocellular carcinoma cell HEPA 1–6 through up-regulatating CyclinD1、P53 and Rb and down-regulatating P27.
Conclusions
In conclusion, this study elucidated that E167K polymorphism of TM6SF2 gene can affect cell cycles of HEPA1–6 cells and the possible mechanisms may up-regulate CyclinD1、P53 and Rb and down-regulate P27. Cell cycle disorder further promoted the deterioration of cell energy metabolism, which thereby formed the vicious cycle to promote progression of HCC.
Acknowledgements
We thank Qingdao Medical University, Qingdao Municipal Hospital, Digestive Disease Key Laboratory of Qingdao and all the participants in the our study.
Funding
This study was supported by the Key Research Project of Shandong Province (2016GSF201217)、Medical and Health Technology Development Project of Shandong Province (2015WS0321)、Qingdao, Shinan District Science and Technology Development Project Fund (2015–6-014-YY、2016–3-016-YY).
Availability of data and materials
No.
Authors’ contributions
Study concept and design: SXD, LLL. acquisition of the data: SXD, YXM. analysis and interpretation of the data: SXD, CFL. drafting of the manuscript: SXD, LLL. critical revision of the manuscript for important intellectual content: SYX, YNX. statistical analysis: SXD, WWJ, LLL. administrative technical and material support: YNX. supervision: SYX,YNX. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
No.
Ethics approval and consent to participate
This study was approved by the ethics committee on human research of Qingdao municipal hospital (Qingdao, China). This study was performed in accordance with the principles of the declaration of Helsinki and its appendices [24].
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CDK
Cyclin dependent-kinase
- Cyclin
Cells cyclin
- HCC
Hepatocellular carcinoma cell
- PCR
Polymerase chain reaction
- RT-PCR
Real time-polymerase chain reaction
- TM6SF2
The transmembrane 6 superfamily member 2
Contributor Information
Shuixian Du, Phone: +86-532-88905508, Email: 15866813623@163.com.
Linlin Lu, Email: lulili@163.com.
Yingxia Miao, Email: miaoyxua@163.com.
Wenwen Jin, Email: jinwwsw@163.com.
Changfei Li, Email: lichnagdei@163.com.
Yongning Xin, Phone: +86-532-82789463, Email: xinyongning1@163.com.
Shiying Xuan, Phone: +86-532-88905289, Email: xuansydxy1@163.com.
References
- 1.Hayles J, Wood V, Jeffery L, Hoe KL, Kim DU, Park HO, et al. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol. 2013;3(5):130053. doi: 10.1098/rsob.130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 3.Brown JS, O'Carrigan B, Jackson SP, Yap TA. Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov. 2017;7(1):20–37. doi: 10.1158/2159-8290.CD-16-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt AM, Garcia JT, Hung T, Flynn RA, Shen Y, Qu K, et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet. 2016;48(11):1370–1376. doi: 10.1038/ng.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen LZ, Xia H-X, Xin YN, Lin ZH, Xuan SY. TM6SF2 E167K variant, a novel genetic susceptibility variant, contributing to nonalcoholic fatty liver disease. J Clin Transl Hepatol. 2015;3(4):265–270. doi: 10.14218/JCTH.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eslam M, Mangia A, Berg T, Chan HL, Irving WL, Dore GJ, et al. Diverse impacts of the rs58542926 E167K variant in TM6SF2 on viral and metabolic liver disease phenotypes. Hepatology. 2016;64(1):34–46. doi: 10.1002/hep.28475. [DOI] [PubMed] [Google Scholar]
- 7.Scorletti E, West AL, Bhatia L, Hoile SP, McCormick KG, Burdge GC, et al. Treating liver fat and serum triglyceride levels in NAFLD, effects of PNPLA3 and TM6SF2 genotypes: results from the WELCOME trial. J Hepatol. 2015;63(6):1476–1483. doi: 10.1016/j.jhep.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 8.Wainwright P, Byrne CD. Bidirectional relationships and disconnects between NAFLD and features of the metabolic syndrome. Int J Mol Sci. 2016;17(3):367. doi: 10.3390/ijms17030367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yki-Järvinen H. Diagnosis of non-alcoholic fatty liver disease (NAFLD) Diabetologia. 2016;59(6):1104–1111. doi: 10.1007/s00125-016-3944-1. [DOI] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Falleti E, Cussigh A, Cmet S, Fabris C, Toniutto P. PNPLA3 rs738409 and TM6SF2 rs58542926 variants increase the risk of hepatocellular carcinoma in alcoholic cirrhosis. Dig Liver Dis. 2016;48(1):69–75. doi: 10.1016/j.dld.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Thabet K, Chan HL, Petta S, Mangia A, Berg T, Boonstra A. The MBOAT7 variant rs641738 increases inflammation and fibrosis in chronic hepatitis B. Hepatology. 2017 doi: 10.1002/hep.29064. [DOI] [PubMed] [Google Scholar]
- 13.Dongiovanni P, Valenti L. Genetics of nonalcoholic fatty liver disease. Metabolism. 2016;65(8):1026–1037. doi: 10.1016/j.metabol.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu Rev Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Depamphilis ML. DNA replication and human disease. Cold Spring Harbor Laboratory Press. 2006;9(4):356. [Google Scholar]
- 16.Xie Z, Tan G, Ding M, et al. Foxm1 transcription factor is required for maintenance of pluripotency of P19 embryonal carcinoma cells. Nucleic Acids Res. 2010;38(22):8027–8038. doi: 10.1093/nar/gkq715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan Y, Xie Z, Ding M, et al. Increased levels of FoxA1 transcription factor in pluripotent P19 embryonal carcinoma cells stimulate neural differentiation. Stem Cells Dev. 2010;19(9):1365–1374. doi: 10.1089/scd.2009.0386. [DOI] [PubMed] [Google Scholar]
- 18.Nazim UM, Md R, Lee YJ, Seol DW, Park SY. Enhancement of TRAIL-induced apoptosis by 5-fluorouracil requires activating Bax and p53 pathways in TRAIL-resistant lung cancers. Oncotarget. 2017;8:18095-105. [DOI] [PMC free article] [PubMed]
- 19.Xie X, Shi X, Guan H, Guo Q, Fan C, Dong W, et al. P21-activated kinase 4 involves TSH induced papillary thyroid cancer cell proliferation. Oncotarget. 2017;8:24882-891. [DOI] [PMC free article] [PubMed]
- 20.Crockford A, Zalmas LP, Grönroos E, Dewhurst SM, McGranahan N, Cuomo ME, et al. Cyclin D mediates tolerance of genome-doubling in cancers with functional p53. Ann Oncol. 2017;28(1):149–156. doi: 10.1093/annonc/mdw612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito Y, Ushiku T, Omura G, Yasuhara K, Yoshida M, Takahashi W, et al. Clinical value of the Epstein-Barr virus and p16 status in patients with nasopharyngeal carcinoma: a single-Centre study in Japan. ORL J Otorhinolaryngol Relat Spec. 2017;78(6):334–343. doi: 10.1159/000455901. [DOI] [PubMed] [Google Scholar]
- 22.Lee JI, Kim IH, Nam TJ. Crude extract and solvent fractions of Calystegia soldanella induce G1 and S phase arrest of the cell cycle in HepG2 cells. Int J Oncol. 2017;50(2):414–420. doi: 10.3892/ijo.2017.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 24.Rickham PP. Human experimentation. Code of ethics of the world Medical association. Declaration of Helsinki. Br Med J. 1964;2(5402):177. doi: 10.1136/bmj.2.5402.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No.


