Abstract
Excretion of amino acids by yeast cells was reported long ago but has not been characterized in molecular terms. It is typically favored by overproduction of the amino acid and/or impairment of its uptake. Here we describe the construction of a yeast strain excreting threonine and homoserine. Using this excretor strain, we then applied a reverse-genetics approach and found that the transporter encoded by the YNL065w/AQR1 gene, a protein thought to mediate H+ antiport, is involved in homoserine and threonine excretion. Furthermore, overexpression of AQR1 led to increased excretion of several amino acids (alanine, aspartate, and glutamate) known to be relatively abundant in the cytosol. Transcription of the AQR1 gene is induced severalfold by a number of amino acids and appears to be under the negative control of Gcn4. An Aqr1-green fluorescent protein fusion protein is located in multiple internal membrane structures and appears to cycle continuously between these compartments and the plasma membrane. The Aqr1 sequence is significantly similar to the vesicular amine transporters of secretory vesicles of neuronal cells. We propose that Aqr1 catalyzes transport of excess amino acids into vesicles, which then release them in the extracellular space by exocytosis.
Uptake of amino acids across the plasma membrane occurs in all living cells. Many genes encoding amino acid transporters have been cloned from a wide variety of organisms, and on the basis of primary sequence analyses, they have been classified in just a few families (58, 66). Amino acid uptake systems are generally used by cells to scavenge amino acids from the environment for use in protein synthesis and/or as sources of nitrogen, carbon, and/or energy. In animals, several amino acids also act as neurotransmitters whose re-uptake from the synaptic cleft involves specific transporters (47, 55). While amino acid import has been investigated extensively, much less is known today about systems involved in amino acid excretion. The existence of proteins involved in this excretion may at first sight be surprising, given the crucial role played by amino acids in cell metabolism. However, several studies suggest that amino acid excretion may be a more widespread process than anticipated. For instance, studies of bacteria have shown that excretion of amino acids can occur when the central metabolism is imbalanced: e.g., when carbon and energy sources are present in excess and massively used by the cells but growth is limited by lack of an essential nutrient or other compound. Metabolic overflow is, for instance, a prerequisite to effective l-glutamate acid production by Corynebacterium glutamicum. It is induced in biotechnological processes by limiting the formation of biotin, by adding antibiotics or inhibitors of fatty acid biosynthesis, or by introducing a fatty acid or glycerol auxotrophy (35). Excretion of amino acids is also observed under conditions of limited peptide catabolism (10). For instance, l-lysine excretion is observed in bacteria using l-lysine-containing peptides as a carbon source but lacking l-lysine-degrading enzymes (19). Deregulated anabolism is yet another situation favoring excretion of amino acids by bacteria (10). In support of the view that export of amino acids is physiologically relevant under at least some conditions, genes encoding membrane proteins specifically involved in amino acid excretion have been cloned from bacteria like C. glutamicum and Escherichia coli (16). These studies have led to defining completely novel transporter families, one of which appears conserved throughout all kingdoms of life (58, 67).
Studies using yeast as a model have shown that this simple eukaryote can also release detectable amounts of amino acids into the medium. This was typically observed with cells producing unusually large amounts of an amino acid as a result of deregulated anabolism and impaired catabolism (30). It has also been reported that mutants auxotrophic for a specific amino acid or for pyrimidine excrete a number of different and unrelated amino acids once the cells are transferred to a medium lacking the required compound, such conditions causing growth arrest (30). These studies have also shown that excretion is generally more pronounced if cells are defective in the permeases that mediate high-affinity uptake of the excreted compound (29, 30, 41, 45). This has led to the notion that high-affinity permeases may play an important role in retention of intracellular compounds (31). Yet the transport mechanisms involved in excretion of amino acids by yeast remain unknown.
In pluricellular organisms, transfer of amino acids between tissues involves rounds of transport out of and into the cell. For instance, root plant cells are known to release amino acids into the xylem sap for subsequent distribution to different organs, but the mechanism of this excretion remains uncharacterized. Similarly, leaf cells excrete amino acids into the apoplast (surrounding cell walls) via mechanisms that remain to be elucidated, after which these amino acids are incorporated into phloem cells for supply to sink organs (40). In mammals, the directional transfer of several amino acids between cells plays an important role in metabolism and signaling (12). One example is the glutamine-glutamate cycle in the nervous system. Glutamate is first loaded into secretory vesicles via the VGLUT vesicular glutamate/H+ antiporter (54). After its release from nerve terminals by regulated exocytosis, glutamate is taken up by astrocytes surrounding the synapse and converted to glutamine. Glutamine is then excreted by the astrocytes and taken up by neurons, which reconvert it to glutamate. Excretion of glutamine by astrocytes is mediated by the system 1 protein (SN1), a glutamine-Na+/H+ antiporter (13) also expressed in liver. This cycle illustrates the importance of the two modes of amino acid release by animal cells: (i) by a cell-surface-associated transporter and (ii) via exocytosis, the latter requiring prior loading of the vesicle catalyzed by an amino acid/H+-antiporter.
In this study, we report the first molecular characterization in yeast of a protein involved in excretion of amino acids. This protein (Aqr1/Ynl065w) (62) is a member of the multidrug resistance transporter family, a category of proteins thought to mediate H+ antiport (28, 57). Aqr1 is apparently involved in excretion of amino acids present at high concentrations in the cytosol. Interestingly, Aqr1 is located in multiple internal-membrane structures and appears to cycle between these compartments and the cell surface. According to one model of Aqr1-dependent amino acid excretion discussed here, the protein would load internal vesicles with amino acids, these being subsequently released into the external medium by exocytosis.
MATERIALS AND METHODS
Strains and growth conditions.
All Saccharomyces cerevisiae strains used in this work (Table 1) are isogenic with Σ1278b (7), except for strain F4. Cells were grown on rich (YPD) or minimal buffered medium (pH 6.1, except when mentioned otherwise) (38), with 3% glucose as the carbon source except when noted otherwise. Nitrogen sources were added as indicated to the following final concentrations: proline, 0.1%; urea, 0.1%; (NH4)2SO4, 0.25%; or l-threonine (Thr), 1 to 10 mM. When necessary, cultures of auxotrophic strains were supplemented with uracil (0.14 mM), Thr (0.25 to 10 mM), or l-homoserine (Hom) (0.025 to 12.5 mM). In the AQR1/YNL065w overexpression experiment, cells were grown on minimal ammonium medium plus 5-μg/ml doxycycline hydrochloride. The sensitivity of strains 23344c (ura3), JA324 (ura3 aqr1Δ::lacZ), JA248 (gap1Δ agp1Δ gnp1Δ), and IVU271 (gap1Δ agp1Δ gnp1Δ aqr1Δ) to hygromycin B (0.01 to 0.1 M), tetramethylammonium (0.02 to 0.2 M), and LiCl (0.01 to 0.1 M) was assessed by comparative growth tests on YPD solid medium.
TABLE 1.
Yeast srains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| 23344c | MATα ura3 | Laboratory collection |
| 30629c | MATaura3 gap1Δ::kanMX2 | 36 |
| 30633c | MATaura3 gap1Δ::kanMX2 agp1Δ::kanMX2 | 36 |
| EK041 | MATα ura3 gap1Δ gnp1Δ::kanMX4 | This study |
| JA248 | MATα ura3 gap1Δ gnp1Δ agp1Δ | This study |
| FA010 | MATα ura3 gap1Δ ssy1Δ::kanMX4 | F. Abdel-Sater and B. André (unpublished data) |
| F4 | MATathr4 | S. A. Cruzcampo (Seville, Spain) |
| ∑-A3hu | MATα ura3 hom3Δ | I. Velasco et al. (unpublished data) |
| IVU41 | MATα ura3 thr1Δ::kanMX4 | This study |
| IVU61-1B | MATaura3 gap1Δ gnp1 Δ agp1 Δ thr1Δ::kanMX4 | This study |
| IVU441 | MATα ura3 gap1Δ agp1Δ gnp1Δ hom3Δ::kanMX4 | This study |
| JA324 | MATα ura3 aqr1Δ::lacZ-kanMX4 | This study |
| JA316 | MATα ura3 qdr3Δ::lacZ-kanMX4 | This study |
| JA310 | MATα ura3 qdr1Δ::lacZ-kanMX4 | This study |
| IVU271 | MATα ura3 gap1Δ gnp1Δ agp1Δ aqr1Δ::kanMX4 | This study |
| IVU11 | MATα ura3 gap1Δ gnp1Δ agp1Δ qdr3Δ::kanMX4 | This study |
| IVU281 | MATα ura3 gap1Δ gnp1Δ agp1Δ qdr1Δ::kanMX4 | This study |
| IHI120 | Mataura3 gcn4Δ::LEU2 | 37 |
| 30629a | MATα ura3 gap1Δ | 36 |
| JA551 | MATα ura3 gap1Δ aqr1-GFP::kanMX | This study |
| JA618 | MATα ura3 npi1 aqr1-GFP::kanMX | This study |
| JA629 | MATα ura3 gap1Δ end3Δ aqr1-GFP::kanMX | This study |
| 32600a | MATα ura3 gap1Δ gnp1Δ agp1Δ npi1 | This study |
Construction of mutant strains.
The gnp1Δ, thr1Δ, hom3Δ, aqr1Δ, qdr1Δ, and thr1Δ null mutations were introduced in yeast strains by the PCR-based gene deletion method and the disruption cassette of plasmid pUG6 (33). The same procedure was used to replace with lacZ the coding sequences of the AQR1, QDR3, and QDR1 genes in strain 23344c, except that the template DNA for PCR amplification was plasmid pUG6-lacZ (8). The qdr3Δ null mutation was introduced into strain JA248 by transformation with the NotI restriction fragment of the pYORC-YBR043c plasmid containing the kanMX marker gene flanked by the 5′ and 3′ regions of the open reading frame YBR043c/QDR3 (62). The JA551 strain expressing Aqr1 fused at its extreme C terminus to the GFP preceded by a five-glycine-alanine linker was isolated by integration of a DNA fragment generated by PCR with plasmid GA5-GFP65T-KanMX6 as the template DNA. Yeast cells were transformed by the method of Gietz et al. (27). All gene modifications were checked by PCR tests. All oligonucleotides used for the construction of strains are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Construct | Oligonucleotide | Sequence (5′ to 3′) |
|---|---|---|
| gnp1Δ::kanMX4 | D5-GNP1 | TCGCTTTCTCAAGTAGCTTATATAATATCAAATATTGCACGCGGCCGCCAGCTGAAGCTT |
| D3-GNP1 | GCTGTAGGAAGGCTGGGATTTGAAATAATATGCTAATTGAGCGGCCGCATAGGCCACTAG | |
| aqr1Δ::lacZ-kanMX4 | S1-YNL065w | TATTTTTTTGAGAATCCAAGCTAGATTCAGAAAGTCGAATCAGCAATGTTCGTACGCTGCAGGTCGAC |
| S2-YNL065w | TTATAAAAAAAATATGCAGGATAAGTGTCTATCAAATTGAAGAATGCCAAGCATAGGCCACTAGTGGATCTG | |
| qdr3Δ::lacZ-kanMX4 | S1-YBR043c | ACTTGTTAACGTCTATGCTAGGACCGAAGTCAGAAAGCGATAAACATGTTCGTACGCTGCAGGTCGAC |
| S2-YBR043c | AAAAGTCTACAATGAAAATATTTACGCTTCCGTCTTATTAACCGATATGAGCATAGGCCACTAGTGGATCTG | |
| qdr1Δ::lacZ-kanMX4 | S1-YIL120w | AAAAAAATAACAGATAGCTCATGAAGAGACTTCTATAAGTAAATCATGTTCGTACGCTGCAGGTCGAC |
| S2-YIL120w | GTTTCTGGAAAGTGGGGGCAGAGACTTTTTAGTTTTACGACTTTTTTTCTGCATAGGCCACTAGTGGATCTG | |
| thr1Δ::kanMX4 | D5-THR1 | CAAGTGTACTTCTAACCTGCCTAATGGTTATAACAGTAGCGCGGCCGCCAGCTGAAGCTTCGTACGC |
| D3-THR1 | ACAGTATACTAGGGGTAAAGGACATTTCATTGCTGTTCGAGCGGCCGCATAGGCCACTAGTGGATCTG | |
| hom3Δ::kanMX4 | D5-HOM3 | CCAATACTCTCTCCATCGCTTAAGCTCACATAGCTATCGCGCGGCCGCCAGCTGAAGCTTCGTACGC |
| D3-HOM3 | CCCTGACATTACATTTAGGGAATAATCGCCCCAATATAGGGCGGCCGCATAGGCCACTAGTGGATCTG | |
| Plasmid pCJ115 | pGal-AQR1 | TACCTCTATACTTTAACGTCAAGGAGAAAAAACTATAATGCTACGAAGTAACAGTATATA |
| pGFP-AQR1 | TACTGTTAATTGCTCCAGCACCAGCACCAGCACCTGCTCCATTATGATTATCGTTCTGGT | |
| AQR1-GFP fusion | 5-AQR1-GFP | CAAAAAGGGAACAAAAAGAGACCAGAACGATAATCATAATGGAGCAGGTGCTGGTGCTGG |
| 3-AQR1-GFP | GCAGGGATTATATGCTGAAATTTCCACTGTATATATTTGCGCGGCCGCATAGGCCACTAG | |
| Plasmid plVU13 | C5-YNL065w | CGCGGATCCGCGATGTCACGAAGTAACAGTATATAC |
| C3-YNL065w | CGCGATATCGCGAATTATGATTATCGTTCTGGTCTC |
Plasmid constructions.
All procedures for DNA manipulation were standard (5, 59). The centromere-based (pMACR7) and 2μm-based (pYH3-R2) plasmids containing the mutant HOM3-R allele have been described elsewhere (21; Velasco et al., unpublished data). Plasmid pIVU13 derives from the yeast expression vector pCM190 (24) and contains the AQR1 coding region under control of the tetO promoter. The centromere-based plasmid (pCJ115) expressing AQR1-GFP under the control of the inducible GAL promoter was isolated by recombination in yeast between a linearized YCp-GAL-GAP1-GFP plasmid (14) and the AQR1 coding region obtained by PCR with genomic DNA of wild-type yeast as template. The recombinant plasmid was purified by cloning into E. coli. The YCpAQR1-lacZ plasmid contains a lacZ fusion with the promoter region, the translation initiation codon, and a short portion of the coding region of the AQR1 gene, from bp −1000 to +33. This construction has been described elsewhere (62).
Amino acid and methylamine uptake assays.
Threonine uptake activities were determined by measuring incorporation of [14C]Thr in growing cells as described by Grenson et al. (30, 32). A similar procedure was used to determine uptake of [14C]methylamine (0.2 mM) in cells growing on proline medium.
HPLC experiments.
Cells were grown in minimal liquid medium and harvested by filtration with a Millipore MF membrane filter (0.45 μM) while the medium was collected. The filters were rinsed and cells were resuspended in distilled water. Cell suspensions and collected medium were boiled for 15 min and centrifuged for 10 min at 10,000 × g to eliminate cellular debris. Supernatants were then collected, and the concentrations of amino acids were analyzed by high-performance liquid chromatography (HPLC) as fluorescent derivatives following the Waters AccQ-Tag Instruction Manual (Millipore Corporation, Milford, Mass.). A chromatographer equipped with a Waters AccQ-Tag column, an automatic injector (Waters 715 Ultra Wisp), and a scanning fluorescence detector (Waters 474) were used. Data integration and processing were performed with the Waters Millennium32 software.
β-Galactosidase assays.
β-Galactosidase activities were measured as described earlier (4) and are expressed in nanomoles of o-nitrophenol formed per minute per milligram of protein. Protein concentrations were measured with the Folin reagent and the standard used was bovine serum albumin. In the experiment of Fig. 7A, the influence of the presence of several other amino acids (2 mM final concentration) was tested. These amino acid were arginine, alanine, aspartate, cysteine, glycine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, serine, threonine, tryptophan, tyrosine, valine, asparagine, citrulline, and ornithine. Cytosine and ammonium were also tested.
FIG. 7.
Expression of an AQR1-lacZ fusion gene. (A) Cells of the wild-type strain (23344c) transformed with the YCpAQR1-lacZ plasmid were grown on minimal medium with urea as the sole nitrogen source. At time zero of the experiment, the indicated amino acids were added at a 2 mM final concentration. After 2 h, cells were collected and β-galactosidase activity was assayed as described in Materials and Methods. (B) Cells of the wild type (23344c) and gnc4 mutant (IHI120) transformed with the YCpAQR1-lacZ plasmid were grown on minimal medium containing urea as the sole nitrogen source. β-Galactosidase activity was assayed as described in Materials and Methods.
Fluorescence microscopy.
Cells were grown on minimal buffered glucose medium (pH 6.1) with glutamate as the sole nitrogen source and viewed with a Nikon Eclipse E600 microscope equipped with appropriate fluorescence light filter sets. Images were captured with a Nikon DXM1200 digital camera and processed with Adobe Photoshop 5.0 (Adobe Systems, Mountain View, Calif.). The FL4-64 fluorescent lipid was used to label the vacuolar membrane.
RESULTS
Yeast cells overproducing Thr and Hom do not excrete detectable amounts of either amino acid.
Threonine (Thr)-overproducing mutants of the yeast S. cerevisiae, accumulating up to 40 times more Thr than the wild-type strain, have been previously isolated. In all cases studied, overproduction is linked to mutations in the HOM3 gene causing expression of a feedback-insensitive aspartate kinase, a key enzyme in the regulation of the Thr biosynthetic pathway (20, 22, 46). Homoserine (Hom), a precursor of Thr biosynthesis, is also overproduced in these HOM3-R mutants (20). We have used a HOM3-R allele cloned into a centromere-based vector and tested whether cells transformed with this plasmid excrete detectable amounts of Thr and Hom. For this, we tested whether HOM3-R cells in drops of high-density cell suspension (5-μl drops of a suspension of ∼108 cells ml−1) are able to cross-feed Thr (thr4) or Hom (hom3Δ) auxotrophs spread at lower density (150 μl of a suspension of ∼107 cells ml−1) on the same petri dish. The results in Fig. 1 show that neither the thr4 nor the hom3Δ auxotroph was efficiently cross-fed by wild-type cells transformed with HOM3-R, irrespective of whether the nitrogen source in the medium was proline, NH4+, urea, or glutamate. Similarly, HOM3-R-transformed cells growing exponentially in liquid medium (with urea, proline, or NH4+ as the sole nitrogen source) did not excrete Thr or Hom in detectable amounts (data not shown). On the other hand, wild-type cells transformed with the HOM3-R gene inserted into a high-copy-number plasmid excreted low but detectable levels of Thr into the medium (Table 3). In contrast Hom, present at a much lower intracellular concentration, was not detected in the external medium (Table 3).
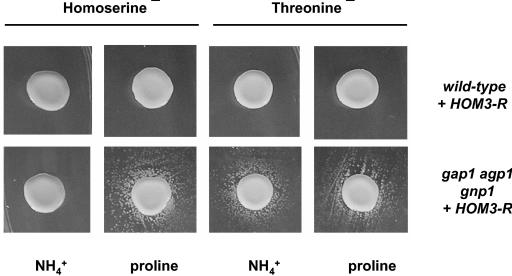
FIG. 1.
Excretion of Hom and Thr by yeast cells grown on solid medium. Cell suspensions of wild-type (23344c) or gap1 agp1 gnp1 (JA248) strains transformed with the pMACR7 plasmid (HOM3-R) were dropped over a lawn of Thr (thr4; strain F4) or Hom (hom3; strain Σ-A3hu) auxotroph mutants, and the petri dishes were incubated for 4 days at 29°C. Proline or NH4+ was the sole source of nitrogen in the medium.
TABLE 3.
Impact of mutations in GAP1, AGP1, GNP1, and AQR1 genes on intracellular and external Thr and Hom concentrationsa
| Strain type | Concn (nmol mg−1 [dry wt])
|
|||
|---|---|---|---|---|
| Thr
|
Hom
|
|||
| Intracellular | External | Intracellular | External | |
| Wild type | 210.3 ± 18.2 | 3.2 ± 0.5 | 39.9 ± 4.4 | BDb |
| gap1Δ agp1Δ gnp1Δ | 157.1 ± 10.7 | 45.3 ± 6.4 | 36.4 ± 4.4 | 4.4 ± 0.7 |
| aqr1Δ | 196.5 ± 19.9 | 2.2 ± 0.2 | 30.7 ± 4.9 | BD |
| gap1Δ agp1Δ gnp1Δ aqr1Δ | 185.9 ± 9.4 | 37.0 ± 6.4 | 29.5 ± 1.9 | BD |
Cells transformed with the high-copy-number plasmid containing the HOM3-R2 allele (pYH3-R2) were grown on minimal medium with proline as the sole nitrogen source. Intracellular and external concentrations of Thr and Hom were quantified by HPLC (see Materials and Methods). The average values and standard deviations obtained from three independent transformed clones are shown. The Strains were 23344c (wild type), JA324 (aqr1Δ), JA248 (gap1Δ agp1Δ gnp1Δ), and IVU271 (gap1Δ agp1Δ gnp1Δ aqr1Δ).
BD, below detection.
Previous studies have shown that excretion of amino acids (30) and nucleobases (29) is more pronounced if the cells are defective in the uptake of these compounds. We thus sought to identify the main Thr and Hom permeases in yeast in order to repeat the above excretion assays with HOM3-R-transformed cells lacking these permeases.
Identification of Gap1, Agp1, and Gnp1 as the main Thr and Hom uptake systems in yeast.
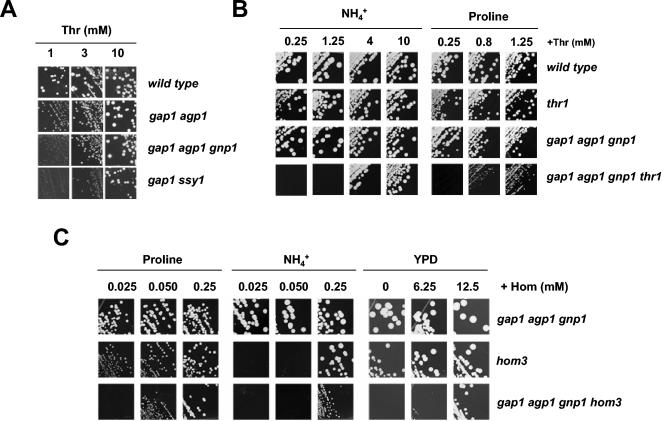
It has been previously reported that the general amino acid permease Gap1 and the broad-specificity amino acid permease Agp1 are involved in Thr uptake (36, 53). Yet one or more other Thr uptake systems must exist in yeast, since a gap1Δ agp1Δ mutant strain can still grow on 1 mM Thr as the sole nitrogen source, albeit more slowly than the wild type (Fig. 2A). Furthermore, the gap1Δ agp1Δ and wild-type strains grow similarly on higher Thr concentrations (Fig. 2A). Like Agp1, the additional Thr permeases sustaining growth on Thr medium appear to be under the positive control of the Ssy1 sensor for external amino acids, as the gap1Δ ssy1Δ strain grows more slowly on Thr at low concentration (1 to 3 mM) than the gap1Δ agp1Δ strain (Fig. 2A). Further experiments identified Gnp1 as an additional permease able to mediate Thr uptake and showed that a triple-mutant gap1Δ agp1Δ gnp1Δ strain is largely defective in incorporation of this amino acid. Namely, a gap1Δ agp1Δ additionally defective for Gnp1 grew more slowly on a low concentration of Thr (1 mM) as the sole nitrogen source (Fig. 2A). Furthermore, a low external concentration of Thr was not able to compensate for a thr1Δ auxotrophy if this mutation was present in a gap1Δ agp1Δ gnp1Δ strain (Fig. 2B). Finally, incorporation of Thr (0.1 mM) in the gap1Δ agp1Δ gnp1Δ strain was largely defective (Fig. 3). Other experiments indicated that besides Gap1, Agp1, and Gnp1, other, lower-affinity, Thr uptake systems likely exist in yeast. First, slow but significant residual incorporation of Thr (0.1 mM) was detected in the gap1Δ agp1Δ gnp1Δ triple-mutant strain (Fig. 3). Second, growth of the gap1Δ agp1Δ gnp1Δ strain on Thr as the sole nitrogen source was almost normal when the amino acid was present at a concentration of 3 mM or more (Fig. 2A). Third, it remained possible to compensate for a thr1Δ auxotrophy introduced into the gap1Δ agp1Δ gnp1Δ strain by adding Thr to the medium, although it took a higher Thr concentration to do so than with the thr1Δ single mutant (Fig. 2B).
FIG. 2.
The Gap1, Agp1, and Gnp1 permeases are involved in Thr and Hom utilization. (A) Growth test on increasing concentrations of Thr as the sole nitrogen source. The strains were 23344c (wild type), 30633c (gap1 agp1), JA248 (gap1 agp1 gnp1), and FA010 (gap1 ssy1). (B) The gap1 agp1 gnp1 mutations reduce the ability of external Thr to complement the growth defect of a Thr auxotroph strain. Cells were grown on minimal media containing proline or NH4+ as nitrogen source and increasing concentrations of Thr. The strains were 23344c (wild type), IVU41 (thr1), JA248 (gap1 agp1 gnp1), and IVU61-1B (gap1 agp1 gnp1 thr1). (C) The gap1 agp1 gnp1 mutations reduce the ability of external Hom to complement the growth defect of a Hom auxotroph strain. Cells were grown on minimal media containing proline or NH4+ as nitrogen source or on YPD medium. Hom was eventually added to the media at the indicated final concentrations. The strains were JA248 (gap1 agp1 gnp1), Σ-A3hu (hom3), and IVU441 (gap1 agp1 gnp1 hom3).
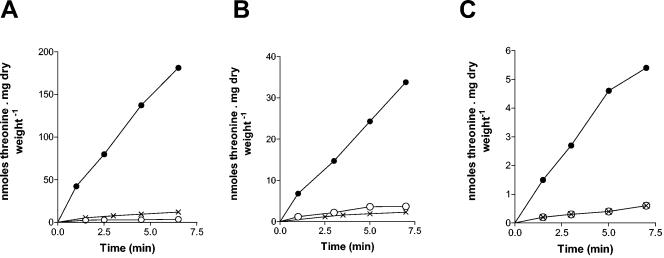
FIG. 3.
Gap1, Agp1 and Gnp1 are the main Thr uptake systems. Time course accumulation of [14C]Thr added at time zero at a 0.1 mM initial concentration to cells growing on minimal medium with proline (A) or NH4+ (B) as the sole nitrogen source. The strains were gap1 (•, strain 30629c), gap1 agp1 (♦, strain 30633c), gap1 gnp1 (○, strain EK041), and gap1 agp1 gnp1 (⋄, strain JA248).
We also tested whether Gap1, Agp1, and Gnp1 mediate Hom uptake. Experiments to directly measure Hom uptake were not carried out because of the high cost of commercially available radioactively labeled Hom. We therefore tested whether Hom could inhibit Thr uptake specifically mediated by Gap1, Agp1, or Gnp1. In each case, the initial rate of Thr uptake was markedly reduced if excess Hom was present (Fig. 4) and the kinetic data were consistent with competitive inhibition (not shown). That gap1Δ agp1Δ gnp1Δ cells are largely defective in Hom uptake was supported by another observation: a higher external concentration of Hom was needed to compensate for the hom3Δ auxotrophy when this mutation was introduced into the gap1Δ agp1Δ gnp1Δ strain (Fig. 2C).
FIG. 4.
Hom inhibits Gap1, Agp1, and Gnp1 uptake activities. (A) Initial velocity of 0.1 mM [14C]Thr uptake in wild-type (○, •) and gap1 (×) strains. The cells were grown on minimal proline medium, and Hom (10 mM final concentration) was added (○) or not (•, ×) to the medium at time zero of the experiment. (B) Initial velocity of 0.1 mM [14C]Thr uptake in gap1 gnp1 (○, •) and gap1 agp1 gnp1 (×) strains. The cells were grown on minimal proline medium to which citrulline (5 mM final concentration) was added 30 min before the uptake assay to induce AGP1 transcription. Hom (10 mM final concentration) was added (○) or not (•, ×) to the medium at time zero of the experiment. (C) Initial velocity of 0.1 mM [14C]Thr uptake in gap1 agp1 (○, •) and gap1 agp1 gnp1 (×) strains. The cells were grown on minimal NH4+ medium to which citrulline was added (5 mM final concentration) 30 min before the uptake assay to induce GNP1 transcription. Hom (10 mM final concentration) was added (○) or not (•, ×) to the medium at time zero of the experiment. The strains were 23344c (wild type), 30629c (gap1), 30633c (gap1 agp1), EK041 (gap1 gnp1), and JA248 (gap1 agp1 gnp1).
Yeast gap1 agp1 gnp1 mutant cells overproducing Thr and Hom excrete both amino acids.
As mentioned above, a yeast mutant defective in high-affinity uptake of a given compound tends to excrete this molecule into the external medium. This is particularly true under conditions favoring internal accumulation of the compound (29, 30, 41, 45). We thus tested excretion of Thr and Hom by the gap1Δ agp1Δ gnp1Δ strain defective in Thr and Hom uptake. The triple-mutant strain was transformed with the centromeric plasmid bearing the HOM3-R allele. Feeding of the thr4 strain by the gap1Δ agp1Δ gnp1Δ strain was readily detectable (Fig. 1). This means that a strain overproducing endogenously synthesized Thr excretes detectable amounts of Thr into the external medium, provided the strain is additionally deficient in high-affinity Thr uptake. This excretion was observed with cells growing on different nitrogen sources (urea, proline, glutamate, and ammonium) (Fig. 1) (data not shown). In the case of Hom, the result was different. Cross-feeding was observed if the cells were grown on proline, urea, or glutamate medium, but not if they were grown on ammonium (Fig. 1) (data not shown). The exact cause of this nitrogen source effect is unknown. A likely explanation is that Hom excreted by gap1Δ agp1Δ gnp1Δ HOM3-R cells is taken up by surrounding hom3Δ cells mainly via Gap1, which is inactive on ammonium but highly active on the other tested nitrogen sources. Excretion from cells growing on proline medium was confirmed by HPLC amino acid quantification (Table 3) (data not shown). In this experiment, HOM3-R was carried on a high-copy vector to further increase the endogenous production of Thr and Hom. The data show that Thr excretion by gap1Δ agp1Δ gnp1Δ cells is more pronounced than that in wild-type cells. Hom was also overproduced in HOM3-R cells but to lower levels than Thr. In this case, only cultures of gap1Δ agp1Δ gnp1Δ cells showed some slight excretion of Hom (Table 3). These data corroborate the growth test data and further illustrate that amino acid permeases have the capability to limit excretion of amino acids, most likely by catalyzing their re-uptake once the amino acids have leaked out of the cell.
Although the cross-feeding tests shown in Fig. 1 have been carried out using growth media buffered at pH 6.1, net excretion of Hom by the gap1 agp1 gnp1 strain was also observed at pH 4.5 and 7.0 (data not shown). Curiously, the cross-fed auxotrophs grew slightly faster around the donor strain on media at pH 4.5 and 7.0 compared to pH 6.1. Whether these faster growth phenotypes at pH 4.5 and 7.0 are due to higher excretion of amino acids by the donor cells, more efficient incorporation of the excreted amino acids, improved utilization of other specific nutrients of the medium, or a combination of these putative effects will require further investigation.
The YNL065w/AQR1 gene is involved in Hom and Thr excretion.
Although the excretion of metabolites by yeast cells has been illustrated in several previous studies, the transport mechanisms involved remain poorly known. On the other hand, the functions of about 120 of the 282 membrane-transport proteins thus far inventoried in the yeast proteome remain unknown (3, 64). One might thus speculate that some of these uncharacterized transporters are in fact involved in excretion, a transport process that is hard to tackle by the classical genetic methods usually applied to yeast. Among the proteins potentially involved in the efflux of compounds are the multidrug transporters of the major facilitator superfamily (28). The multidrug transporter family includes no less than 23 proteins, usually viewed as H+ antiporters because they share high sequence similarity with bacterial antibiotic resistance proteins known to act by this mechanism (57). Several yeast proteins in this family, such as Flr1p, are clearly involved in drug resistance (1, 9, 39, 50, 61, 63), and the same function has been proposed for the other members of the family (28, 57). Yet there is growing evidence that at least some members of this family are not involved in drug resistance but rather in excretion of natural compounds. For instance, the Dtr1 protein was recently reported to be specifically expressed during sporulation. It is targeted to the plasma membrane where it mediates excretion of di-tyrosine, a constituent of the spore cell wall (23). Similarly, the Tpo1 protein of the same family was recently found to be located at the plasma membrane and to be involved in the detoxification of excess polyamines (2). Among the 23 multidrug transporters of the major facilitator superfamily, some are closely related in sequence to the di-tyrosine/H+ exchanger (28, 57). Interestingly, the promoters of some of these genes (e.g., YBR043C/QDR3) contain an unusually high number of 5′-GATA-3′ sequences, a feature typical of genes sensitive to nitrogen catabolite repression (44). Furthermore, it was recently reported that another gene encoding a close homologue of Dtr1 (YNL065w/AQR1) is induced by glutamate (43). These observations prompted us to test whether these putative transporters could play a role in Thr and Hom excretion. The genes encoding the putative transporters (AQR1, QDR3, and YIL120w/QDR1) were deleted from the wild-type strain and from the gap1Δ agp1Δ gnp1Δ triple mutant. The derived strains were transformed with a centrometric plasmid bearing the HOM3-R allele and used as described above in cross-feeding tests. Deletion of QDR1 and QDR3 had no detectable effect on Thr or Hom excretion (not shown), but deletion of AQR1 completely suppressed the ability of the gap1Δ agp1Δ gnp1Δ strain to cross-feed hom3Δ cells (Fig. 5). This suggests that Aqr1 is essential to Hom excretion. Consistently, HPLC measurements in liquid cultures showed that the aqr1Δ mutation prevents excretion of Hom by the gap1Δ agp1Δ gnp1Δ strain (Table 3). Furthermore, the effect of the aqr1Δ mutation does not seem to be due to impaired endogenous synthesis of Hom (Table 3). A lack of Aqr1 also reduced cross-feeding of the thr4 auxotroph, but this effect was not as pronounced as with hom3Δ cells and varied in intensity from one experiment to the other (data not shown) (see Fig. 8B). In conclusion, our data indicate that Aqr1 is an essential component of Hom excretion. Although it appears also to also contribute to Thr excretion, other excretion systems for this amino acid likely exist.
FIG. 5.
AQR1 is required for Hom excretion. Cell suspensions of wild-type (23344c), gap1 agp1 gnp1 (JA248), and gap1 agp1 gnp1 aqr1 (IVU271) strains transformed with the pMACR7 plasmid (HOM3-R) were dropped over a lawn of Hom (hom3, strain Σ-A3-hu) auxotroph mutants and the petri dishes were incubated for 4 days at 29°C. Proline was the sole source of nitrogen in the medium.
FIG. 8.
Mutations impairing endocytosis affect Aqr1 subcellular location and Aqr1-mediated excretion of amino acids. (A) Subcellular location of an Aqr1-GFP fusion protein. Cells of the JA551 (AQR1-GFP ura3), JA618 (AQR1-GFP npi1 ura3), and JA629 (AQR1-GFP end3Δ ura3) strains were grown on minimal glutamate medium and examined by fluorescence microscopy. Cells were labeled with FM4-64 to stain the vacuolar membrane and endosomal compartments (not shown). (B) Cell suspensions of wild-type (23344c), gap1 agp1 gnp1 (JA248), gap1 agp1 gnp1 aqr1 (IVU271), and gap1 agp1 gnp1 npi1 (32600a) strains transformed with the pMACR7 plasmid (HOM3-R) were dropped over a lawn of Thr (thr4, strain F4) or Hom (hom3, strain Σ-A3hu) auxotroph mutants and the petri dishes were incubated for 4 days at 29°C. Glutamate was the sole source of nitrogen in the medium.
The apparent role of Aqr1 in Hom and Thr excretion could be indirect: e.g., lack of Aqr1 could affect the electrical plasma membrane potential. However, we could not see any effect of the aqr1Δ mutation on the ability of cells to incorporate methylammonium neither on their sensitivity to toxic concentrations of hygromycin B, tetramethylammonium, or lithium chloride (data not shown) (see Materials and Methods). These observations make it unlikely that Aqr1 affects amino acid excretion by altering the potential of the plasma membrane.
Overexpression of Aqr1 leads to excretion of amino acids, including aspartate, glutamate, and alanine.
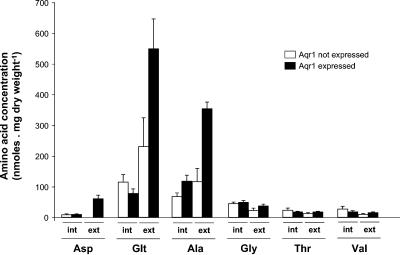
The above data show that Aqr1 is involved in excretion of Hom. We next addressed the following question: can Aqr1 mediate efflux of other amino acids? We overexpressed the AQR1 gene in a gap1Δ agp1Δ gnp1Δ aqr1Δ strain producing normal levels of Thr and Hom (not transformed with the HOM3-R plasmid) and measured the release of amino acids into the growth medium. For this, we placed the AQR1 behind the regulatable Tet promoter in a high-copy-number vector and used the resulting plasmid to transform gap1Δ agp1Δ gnp1Δ aqr1Δ cells. The cells were grown on a minimal medium devoid of amino acids and containing NH4+ as the sole nitrogen source. To this medium, doxycycline, an antibiotic relieving repression of Tet promoter-dependent expression of AQR1, was added or not. The growth media were collected 22 h after derepression of AQR1 expression and subjected to HPLC analysis (Fig. 6). Many amino acids (e.g., arginine, leucine, phenylalanine, glutamine, lysine, and isoleucine) were undetectable in the external medium, regardless of whether AQR1 was expressed or not (not shown). Aspartate, remarkably, was detected only in the medium of cells overexpressing AQR1. Hence, Aqr1 would appear to mediate excretion of this amino acid also. Many other amino acids were detected in the medium of cells that were not expressing Aqr1, but AQR1 overexpression caused a net increase in the external concentration of several of them. The effect was more pronounced for glutamate and alanine (Fig. 6). Interestingly, these amino acids (Glu, Ala, and Asp) are precisely those reported to be present at the highest concentrations in the cytosol of cells growing on glucose-ammonium medium (48). Other amino acids (e.g., arginine, ornithine, histidine, and glutamine) are present at high levels in cells growing under these conditions, but these are found mostly in the vacuole.
FIG. 6.
AQR1 is involved in excretion of aspartate, alanine, and glutamate. Cells of the gap1 agp1 gnp1 aqr1 strain (IV271) transformed with the 2μm plasmid pIVU13 (Tet-AQR1) were grown on minimal medium containing NH4+ as the sole nitrogen source plus doxycycline to repress AQR1 transcription. At time zero of the experiment (early exponential phase, 0.2 to 0.4 mg of dry weight ml−1), the culture was filtered and cells were resuspended in the same medium with (Aqr1 not expressed) or without (Aqr1 expressed) doxycycline. Cells were harvested after 22 h, i.e., at late-exponential/early-stationary phase (1 to 2 mg of dry weight ml−1), and the concentrations of free amino acids inside cells (int.) and in the external medium (ext.) were measured by HPLC as detailed in Materials and Methods. Data are shown for aspartate (Asp), glutamate (Glt), alanine (Ala), glycine (Gly), Thr, and valine (Val).
These results thus show that in cells where no amino acid is overproduced, overexpression of the AQR1 gene leads to a significant increase in Asp, Glu, and Ala excretion. Aqr1-dependent excretion was most pronounced for Glu and Ala, the two amino acids present at the highest concentrations in the cytosol (48). Yet both amino acids were also excreted by cells not expressing Aqr1. This suggests the involvement of other export systems or mechanisms. In the case of Asp, no excretion was detected when Aqr1 was not expressed, suggesting that Aqr1 is the main Asp excretion system under the conditions tested. These results, together with those obtained with cells overproducing Thr and Hom, suggest that the role of Aqr1 could be to mediate excretion of amino acids present at high concentrations in the cytosol.
Analysis of AQR1 gene expression.
Given the role of Aqr1 in the excretion of several amino acids, we sought to determine whether expression of the AQR1 gene is influenced by levels of amino acids in the medium. To test this, we constructed a plasmid bearing the lacZ gene under the control of the AQR1 gene's promoter. We introduced the plasmid into wild-type cells and monitored β-galactosidase activity in transformed cells growing on minimal medium devoid of amino acids or to which a single amino acid was added (see Materials and Methods). A significant level of AQR1-lacZ expression was observed in urea-grown cells, and expression was higher if γ-amino-n-butyric acid (GABA; approximately four- to fivefold), glutamate (five- to sixfold), or proline (three- to fourfold) was also present in the medium (Fig. 7A). Other amino acids had no significant effect on AQR1-lacZ expression (not shown). The higher expression of AQR1 on urea-plus-glutamate medium corroborates previous whole-genome transcription analysis data showing that AQR1 is one of the many genes that are induced by glutamate (43).
Analysis of the AQR1 promoter revealed that it contains several perfect matches to the consensus binding site for the Gcn4 transcription factor. We thus tested whether AQR1 is under Gcn4 control. The plasmid bearing the AQR1-lacZ gene was introduced into wild-type and gcn4Δ strains, and transformed cells were grown on urea or ammonium medium. The results of β-galactosidase assays unexpectedly revealed that AQR1 expression is three- to fourfold higher in the gcn4Δ mutant than in the wild type (Fig. 7B), thus suggesting that Gcn4 exerts a negative control on AQR1 expression. Gcn4 is known to be more active when the internal level of any one of several amino acids is reduced (34). That an AQR1-lacZ gene is more highly expressed in a gcn4Δ strain might thus be rationalized as a mechanism limiting amino acid excretion under conditions of amino acid privation.
Aqr1 is mainly intracellular and transits constantly through the plasma membrane.
We next examined the subcellular location of Aqr1. For this, the GFP preceded by a linker was fused in frame to the extreme C terminus of Aqr1. Remarkably, in cells expressing the fusion gene under the control of the natural AQR1 promoter, the fluorescence was weak in intensity and mainly present in multiple small punctate structures distributed in the whole cytoplasm (Fig. 8A) In at least some cells, a weak fluorescent signal was also detectable at the cell surface. The internal subcellular location pattern of Aqr1 differed from the one reported for proteins located in the endoplasmic reticulum, Golgi, or endosomes and is more compatible with a location in multiple internal vesicles. As a small fraction of the protein appears also located at the cell surface in some cells, we addressed the question of whether Aqr1 cycles between the internal and the plasma membranes. We thus examined the location of Aqr1-GFP in mutant strains defective in endocytosis. We first used the npi1/rsp5 strain in which endocytosis of multiple plasma membrane proteins was shown to be defective. This endocytosis defect is caused by a mutation reducing the level of the HECT-type ubiquitin ligase Rsp5, a protein involved in ubiquitylation and subsequent endocytosis of a wide variety of plasma membrane transporters (56). In the npi1/rsp5 mutant expressing Aqr1-GFP, the fluorescence was mainly redistributed to the cell surface (Fig. 8A). A similar location was observed in the end3Δ strain, another classically used mutant defective in endocytosis (52). These data strongly suggest that Aqr1 constantly cycles between multiple internal membranes and the cell surface. Furthermore, endocytosis of Aqr1 is apparently dependent on a normal Rsp5 ubiquitin ligase function. Other yeast proteins were reported to cycle constantly between the cell surface and internal membranes: e.g., the Snc1 SNARE protein (42). When expressed above normal levels, Snc1 tends to accumulate mostly at the cell surface. Interestingly, the same was observed when Aqr1-GFP was overexpressed (e.g., by placing its gene under the control of the GAL promoter), with a fraction of the protein being targeted to the endosome/vacuolar degradation pathway (62; our unpublished data). Finally, we tested whether the npi1 mutation provoking accumulation of Aqr1 at the plasma membrane affects Aqr1's ability to mediate excretion of Hom and Thr. The results revealed that the npi1 mutation markedly reduced the ability of the gap1Δ agp1Δ gnp1Δ excretor strain to cross-feed the Hom and Thr auxotroph strains (Fig. 8B). This suggests that Aqr1 is most active as an amino acid excretion protein when located in internal membranes. As discussed below, this observation supports the model according to which Aqr1 functions by loading internal vesicles with amino acids, which are then released into the external medium by exocytosis.
DISCUSSION
Yeast cells can excrete metabolites such as amino acids. This occurs under particular conditions, e.g., when the amino acid is produced at abnormally high levels as a result of deregulated anabolism or when amino acid metabolism rapidly shifts to an imbalanced situation (30). The mechanisms of this excretion remain unknown. It is highly interesting to investigate the molecular details of this cell process in yeast, because amino acid excretion exists in other eukaryotes as well. For instance, plants often associate with mycorrhizal fungi whose expanding mycelium scavenges nitrogen from the surrounding soil. This nitrogen is then transmitted to the plant in the form of amino acids through export mechanisms which remain unknown (11). Transfer of amino acids between tissues of pluricellular organisms such as plants involves rounds of transport out of and into the cell, but the proteins involved in amino acid export remain uncharacterized (51). Yeast mutants defective in excretion of specific amino acids might thus be used as recipient strains to characterize genes involved in amino acid excretion in other species.
Efflux of an amino acid may be catalyzed by an energy-driven carrier located at the plasma membrane. Several amino acid exporters of this type have indeed been characterized in bacteria (16). In eukaryotes, excretion of metabolites can in principle also be mediated by exocytosis. In this case, the compound must first be incorporated into vesicles via transporters located in the vesicle membranes. For instance, several vesicular transporters responsible for the loading of synaptic vesicles with neurotransmitters have been characterized in animals (17). These proteins act as H+ antiporters, the H+ gradient being established by V-type H+-ATPases responsible for the acidification of internal membrane compartments (65).
In this study, we have defined a methodology for investigating the excretion of specific amino acids in yeast. We used cells artificially overproducing Thr and Hom as a result of a dominant HOM3-R allele. As these cells growing in standard growth media did not excrete much Hom or Thr into the medium, we sought to identify the genes coding for the main uptake systems for these amino acids in yeast, since reports suggest that excretion of a given compound is often more pronounced in cells defective in high-affinity uptake of that compound. This is probably because influx systems catalyze re-uptake of the extruded compound (31). We thus found that three amino acid permeases, Gap1, Agp1, and Gnp1, are the main Thr and Hom uptake systems in yeast. When Hom and Thr were overproduced in the triple gap1 agp1 gnp1 mutant strain, we detected net excretion of both amino acids into the medium. This excretion was observed on solid media in cross-feeding tests and in liquid media by HPLC measurements. Starting with this strain, we then applied a reverse-genetics approach to test candidate genes possibly involved in Thr and Hom excretion. A similar strategy could be applied to identify genes involved in other metabolite excretion processes. For instance, mutant alleles or genetic strategies causing abnormal accumulation of many different metabolites have been described, and the main yeast permeases involved in the uptake of most natural nutritional compounds (sugars, amino acids, phosphate, nucleobases, vitamins, etc.) are known (64). It should thus be possible to construct yeast strains overproducing and excreting a large set of specific metabolites. These strains could then be used to search for genes involved in metabolite excretion, such genes having largely eluded characterization by standard methods.
Using this approach, we found that the AQR1 gene is involved in excretion of amino acids. Several results support this conclusion. First, Hom and Thr excretion by the overproducing strain is impaired if the AQR1 gene is deleted, and this loss of excretion is not caused by reduced endogenous synthesis of the amino acids. Second, if Aqr1 is overexpressed in cells defective in the uptake of multiple amino acids, aspartate is excreted into the medium; this phenomenon is not observed if Aqr1 is not expressed. Furthermore, a net increase in excretion of two other amino acids (i.e., glutamate and alanine) occurs when Aqr1 is overexpressed. Interestingly aspartate, glutamate, and alanine are the most abundant amino acids in the cytosol of cells growing under the tested conditions. Our results thus suggest that the normal role of Aqr1 might be to catalyze excretion of amino acids present at relatively high levels in the cytosol. This process could (e.g., by providing a mechanism for protecting cells against the accumulation of amino acids to toxic levels). That the normal function of Aqr1 is indeed to mediate amino acid excretion is supported by the observation that transcription of the AQR1 gene is influenced by amino acids. Hence, as previously reported on the basis of a genome-wide transcriptome analysis (43), AQR1 transcription is induced severalfold by glutamate. Two other amino acids, proline and GABA, also promote higher-level expression of AQR1. Why these amino acids (and not other ones) increase AQR1 transcription, however, requires further investigation. The AQR1 gene is also expressed to a higher level in a gcn4Δ strain characterized by defective general control of amino acid biosynthesis control. Although this might in principle be an indirect effect of the internal pool of amino acids, the presence of several potential binding sites for Gcn4 in the AQR1 gene promoter suggests that AQR1 could be a direct target gene of Gcn4. Many yeast genes are reported to be under the negative control of Gcn4 (49). Lower expression of AQR1 in cells deprived of one or several amino acids (conditions under which Gcn4 is active) is consistent with a role of Aqr1 amino acid excretion. Yet our data do not rule out another possibility: that the main physiological role of Aqr1 might be to mediate excretion of compounds other than amino acids, e.g., amino acid derivatives, recognizing amino acids also but less efficiently and only when they are present at unusually high levels. It was also previously reported that Aqr1 confers a moderate increase of yeast tolerance to toxic concentrations of ketoconazole, the cationic dye crystal violet, and short-chain monocarboxylic acids (62). We favor the view that the potential recognition by Aqr1 of these toxic compounds corresponds to opportunistic side effects of Aqr1's natural function, which is to mediate excretion of amino acids (and possibly amino acid derivatives).
Our data also show that Aqr1 is not the sole yeast protein involved in amino acid excretion. For instance, cells lacking Aqr1 are able to excrete glutamate and alanine. Furthermore, excretion of Thr still occurs in the aqr1 mutant, and cross-feeding tests have even shown that a cell lacking Aqr1 can excrete some Hom when the level of this amino acid is very high: i.e., when the HOM3-R gene is introduced on a high-copy vector (our unpublished data). The Aqr1 protein is part of the large MFS protein family (57). Several members of this family (e.g., Flr1) are clearly involved in drug resistance, and consistently, their genes are under the control of the PDR transcriptional network (15). Yet the functions of many other proteins of this family remain unknown, and their genes are apparently not under PDR control (15, 57). It is thus tempting to speculate that close homologues of Aqr1 might be involved in excretion of amino acids or derivatives. In support of this view, one such protein (Dtr1) is specifically expressed during sporulation, and catalyzes excretion of di-tyrosine, a component of the spore cell wall (23). Furthermore, a gene encoding another close homologue of Aqr1 (YBR043c/QDR3) is under nitrogen control (our unpublished observations). Several of these genes could be singly deleted without detectably altering Thr or Hom excretion in cross-feeding tests. As many transporters in yeast display partial functional redundancy, we are currently investigating the impact of deleting several genes closely related to AQR1 on the excretion of Hom, Thr, and other amino acids as well.
A central question emerging from this work is through which mechanism does Aqr1 favor amino acid excretion? The protein (586 amino acids) is predicted to be made of 12 transmembrane domains flanked by N- and C-terminal hydrophilic tails and shares highly significant sequence similarity with several MFS drug resistance proteins (from bacteria and fungi) most likely acting as H+ antiport systems. Interestingly, when compared against the entire set of human proteins, Aqr1 exhibits the highest degree of similarity with members of the VAT family (18). The VAT proteins are in fact similar to many bacterial and fungal MFS proteins of the multidrug resistance family (60). These proteins are expressed in secretory vesicles in neuronal and endocrine cells. They function as H+ antiporters and catalyze the transport of acetylcholine and biogenic amines into the secretory vesicles which then release them into the extracellular space by exocytosis. When expressed under the natural AQR1 gene promoter, an Aqr1-GFP fusion is present in multiple membrane compartments dispersed throughout the cytoplasm. Furthermore, we have provided evidence that, under these conditions, Aqr1 constantly cycles between these internal membranes and the cell surface. These observations suggest two possible models of Aqr1 function. According to one model, Aqr1 would be active at the plasma membrane as an amino acid/H+ exchanger. Its constant cycling between the plasma and internal membranes could provide a means of controlling its activity. Yet we have studied the subcellular location of Aqr1 under different growth conditions (e.g., by varying amino acid availability) and have found no conditions favoring higher accumulation of Aqr1 at the cell surface. According to the second model, Aqr1 would be naturally present in the membrane of intracellular vesicles and would act mainly as an amino acid/H+ antiporter to load these vesicles with amino acids present at relatively high concentration in the cytosol. The amino acids would then be released into the external medium by exocytosis. The Aqr1 protein having reached the plasma membrane through exocytosis would then be rapidly internalized back to internal vesicles, and this would require prior ubiquitylation of the transporter, as shown for endocytosis of many plasma membrane proteins. In support of this model, we found that accumulation of Aqr1 at the cell surface by means of the npi1/rsp5 mutation also reduces the ability of cells to mediate Aqr1-dependent excretion of amino acids. This second model thus implies that exocytosis would be the main mechanism through which amino acids are excreted in yeast. A similar model has recently been proposed for auxin transport in plants. Auxin is a derivative of tryptophan acting as a key plant growth hormone. Polar cell-to-cell transport of auxin requires an auxin-H+ antiporter (Pin1). Although Pin1 was initially viewed as a carrier mediating active export of auxin across the plasma membrane, current data (25, 26) are also consistent with a role of Pin1 in loading auxin into plasma-derived endosomal and endosome-recycling vesicles. In other words, auxin might be released from plant cells by exocytosis (6).
In yeast, bulk excretion of several compounds present in excess concentration in the cytosol (e.g., primary and secondary metabolites, ions, etc.) might thus be at least partially mediated by the concerted action of vesicular H+ antiporters and exocytosis. Plasma membrane uptake systems could then possibly catalyze the selective re-uptake of compounds useful for cell metabolism. Under specific nutrient supply conditions, for instance, the high-affinity Gap1 permease efficiently limits excretion of several amino acids (our unpublished data) and the high-affinity Mep1 and Mep2 permeases limit that of ammonium (45). Release of molecules via exocytosis provides an obvious means of preventing excess accumulation of compounds in the cytosol. This cell function might have then evolved into systems mediating cell-to-cell communication, as for neurotransmitter in the case of animals, possibly for auxin in the case of plants, and why not also in fungi when growing as multicellular hyphae or pseudohyphae?
Acknowledgments
We are grateful to Catherine Jauniaux for skillful technical contribution at several stages of this work. We thank Anne-Marie Marini for critical reading of the manuscript and suggestions. We also thank members of Isabel Calderon's laboratory for many discussions and encouragements.
This study was supported by the European Community (EUROFAN project, FPIV); the Communauté Française de Belgique (ARC project no. 98/03-223); grant FRSM 3.4597.00 F for Medical Scientific Research, Belgium; Ministerio de Educación y Cultura, España (fellowship to Isabel Velasco and grant no. ALI99-0496); and a postdoctoral grant to S. Tenreiro (BPD/5649/01 from Fundação para a Ciência e a Tecnologia, Portugal).
REFERENCES
- 1.Alarco, A. M., I. Balan, D. Talibi, N. Mainville, and M. Raymond. 1997. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J. Biol. Chem. 272:19304-19313. [DOI] [PubMed] [Google Scholar]
- 2.Albertsen, M., I. Bellahn, R. Kramer, and S. Waffenschmidt. 2003. Localization and function of the yeast multidrug transporter Tpo1p. J. Biol. Chem. 278:12820-12825. [DOI] [PubMed] [Google Scholar]
- 3.André, B. 1995. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast 11:1575-1611. [DOI] [PubMed] [Google Scholar]
- 4.André, B., C. Hein, M. Grenson, and J. C. Jauniaux. 1993. Cloning and expression of the UGA4 gene coding for the inducible. Mol. Gen. Genet. 237:17-25. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 6.Baluska, F., J. Samaj, and D. Menzel. 2003. Polar transport of auxin: carrier-mediated flux across the plasma membrane or neurotransmitter-like secretion? Trends Cell Biol. 13:282-285. [DOI] [PubMed] [Google Scholar]
- 7.Béchet, J., M. Grenson, and J. M. Wiame. 1970. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur. J. Biochem. 12:31-39. [DOI] [PubMed] [Google Scholar]
- 8.Boles, E., P. de Jong-Gubbels, and J. T. Pronk. 1998. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J. Bacteriol. 180:2875-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broco, N., S. Tenreiro, C. A. Viegas, and I. Sa-Correia. 1999. FLR1 gene (ORF YBR008c) is required for benomyl and methotrexate resistance in Saccharomyces cerevisiae and its benomyl-induced expression is dependent on pdr3 transcriptional regulator. Yeast 15:1595-1608. [DOI] [PubMed] [Google Scholar]
- 10.Burkovski, A., and R. Kramer. 2002. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl. Microbiol. Biotechnol. 58:265-274. [DOI] [PubMed] [Google Scholar]
- 11.Chalot, M., and A. Brun. 1998. Physiology of organic nitrogen acquisition by ectomycorrhizal fungi and ectomycorrhizas. FEMS Microbiol. Rev. 22:21-44. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhry, F. A., R. J. Reimer, and R. H. Edwards. 2002. The glutamine commute: take the N line and transfer to the A. J. Cell Biol. 157:349-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhry, F. A., R. J. Reimer, D. Krizaj, D. Barber, J. Storm-Mathisen, D. R. Copenhagen, and R. H. Edwards. 1999. Molecular analysis of system N suggests novel physiological roles in nitrogen metabolism and synaptic transmission. Cell 99:769-780. [DOI] [PubMed] [Google Scholar]
- 14.de Craene, J. O., O. Soetens, and B. André. 2001. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276:43939-43948. [DOI] [PubMed] [Google Scholar]
- 15.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 16.Eggeling, L., and H. Sahm. 2003. New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch. Microbiol. 180:155-160. [DOI] [PubMed] [Google Scholar]
- 17.Eiden, L. E. 2000. The vesicular neurotransmitter transporters: current perspectives and future prospects. FASEB J. 14:2396-2400. [DOI] [PubMed] [Google Scholar]
- 18.Eiden, L. E., M. K. Schafer, E. Weihe, and B. Schutz. 2004. The vesicular amine transporter family (SLC18): amine/proton antiporters required for vesicular accumulation and regulated exocytotic secretion of monoamines and acetylcholine. Pfluegers Arch. 447:636-640. [DOI] [PubMed] [Google Scholar]
- 19.Erdmann, A., B. Weil, and R. Kramer. 1993. Lysine secretion by wild-type Corynebacterium glutamicum triggered by dipeptide uptake. J. Gen. Microbiol. 139:3115-3122. [Google Scholar]
- 20.Farfan, M., and I. L. Calderon. 2000. Enrichment of threonine content in Saccharomyces cerevisiae by pathway engineering. Enzyme Microb. Technol. 26:763-770. [DOI] [PubMed] [Google Scholar]
- 21.Farfan, M., E. Martin-Rendon, and I. L. Calderon. 1996. Effect of gene amplification on threonine production by yeast. Biotechnol. Bioeng. 49:667-674. [DOI] [PubMed] [Google Scholar]
- 22.Farfán, M.-J., L. Aparicio, and I. L. Calderón. 1999. Threonine overproduction in yeast strains carrying the HOM3-R2 mutant allele under the control of different inducible promoters. Appl. Environ. Microbiol. 65:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felder, T., E. Bogengruber, S. Tenreiro, A. Ellinger, I. Sa-Correia, and P. Briza. 2002. Dtrlp, a multidrug resistance transporter of the major facilitator superfamily, plays an essential role in spore wall maturation in Saccharomyces cerevisiae. Eukaryot. Cell 1:799-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gari, E., L. Piedrafita, M. Aldea, and E. Herrero. 1997. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13:837-848. [DOI] [PubMed] [Google Scholar]
- 25.Geldner, N., N. Anders, H. Wolters, J. Keicher, W. Kornberger, P. Muller, A. Delbarre, T. Ueda, A. Nakano, and G. Jurgens. 2003. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112:219-230. [DOI] [PubMed] [Google Scholar]
- 26.Geldner, N., J. Friml, Y. D. Stierhof, G. Jurgens, and K. Palme. 2001. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413:425-428. [DOI] [PubMed] [Google Scholar]
- 27.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 28.Goffeau, A., J. Park, I. T. Paulsen, J. L. Jonniaux, T. Dinh, P. Mordant, and M. H. Saier, Jr. 1997. Multidrug-resistant transport proteins in yeast: complete inventory and phylogenetic characterization of yeast open reading frames with the major facilitator superfamily. Yeast 13:43-54. [DOI] [PubMed] [Google Scholar]
- 29.Grenson, M. 1969. The utilization of exogenous pyrimidines and the recycling of uridine-5′-phosphate derivatives in Saccharomyces cerevisiae, as studied by means of mutants affected in pyrimidine uptake and metabolism. Eur. J. Biochem. 11:249-260. [DOI] [PubMed] [Google Scholar]
- 30.Grenson, M. 1973. Specificity and regulation of the uptake and retention of amino acids and pyrimidines in yeast, p. 179-193. In Z. Vanek, Z. Hostalek, and J. Cudlin (ed.), Genetics of industrial microorganisms. Academia, Prague, Czech Republic.
- 31.Grenson, M. 1992. Amino acid transporters in yeast: structure, function and regulation, p. 219-245. In J. J. L. L. M. De Pont (ed.), Molecular aspects of transport proteins. Elsevier Science, Amsterdam, The Netherlands.
- 32.Grenson, M., M. Mousset, J. M. Wiame, and J. Bechet. 1966. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae. Biochim. Biophys. Acta 127:325-338. [DOI] [PubMed] [Google Scholar]
- 33.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikeda, M. 2003. Amino acid production processes. Adv. Biochem. Eng. Biotechnol. 79:1-35. [DOI] [PubMed] [Google Scholar]
- 36.Iraqui, I., S. Vissers, F. Bernard, J.-O. de Craene, E. Boles, A. Urrestarazu, and B. André. 1999. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19:989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iraqui, I., S. Vissers, M. Cartiaux, and A. Urrestarazu. 1998. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol. Gen. Genet. 257:238-248. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs, P., J. C. Jauniaux, and M. Grenson. 1980. A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. J. Mol. Biol. 139:691-704. [DOI] [PubMed] [Google Scholar]
- 39.Jungwirth, H., F. Wendler, B. Platzer, H. Bergler, and G. Hogenauer. 2000. Diazaborine resistance in yeast involves the efflux pumps Ycf1p and Flr1p and is enhanced by a gain-of-function allele of gene YAP1. Eur. J. Biochem. 267:4809-4816. [DOI] [PubMed] [Google Scholar]
- 40.Koch, W., M. Kwart, M. Laubner, D. Heineke, H. Stransky, W. B. Frommer, and M. Tegeder. 2003. Reduced amino acid content in transgenic potato tubers due to antisense inhibition of the leaf H+/amino acid symporter StAAP1. Plant J. 33:211-220. [DOI] [PubMed] [Google Scholar]
- 41.Lacroute, F., and P. P. Slonimski. 1964. Etude physiologique des mutants résistant au 5-fluorouracile chez la levure. Crit. Rev. Acad. Sci. Paris 258:2172-2174. [PubMed] [Google Scholar]
- 42.Lewis, M. J., B. J. Nichols, C. Prescianotto-Baschong, H. Riezman, and H. R. Pelham. 2000. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell 11:23-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, Z., T. Sekito, C. B. Epstein, and R. A. Butow. 2001. RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J. 20:7209-7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magasanik, B., and C. A. Kaiser. 2002. Nitrogen regulation in Saccharomyces cerevisiae 475. Gene 290:1-18. [DOI] [PubMed] [Google Scholar]
- 45.Marini, A.-M., S. Soussi-Boudekou, S. Vissers, and B. André. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4282-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin-Rendon, E., M. J. Farfan, C. Ramos, and I. L. Calderon. 1993. Isolation of a mutant allele that deregulates the threonine biosynthesis in Saccharomyces cerevisiae. Curr. Genet. 24:465-471. [DOI] [PubMed] [Google Scholar]
- 47.Masson, J., C. Sagne, M. Hamon, and S. El Mestikawy. 1999. Neurotransmitter transporters in the central nervous system. Pharmacol. Rev. 51:439-464. [PubMed] [Google Scholar]
- 48.Messenguy, F., D. Colin, and J. P. ten Have. 1980. Regulation of compartmentation of amino acid pools in Saccharomyces cerevisiae and its effects on metabolic control. Eur. J. Biochem. 108:439-447. [DOI] [PubMed] [Google Scholar]
- 49.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nunes, P. A., S. Tenreiro, and I. Sá-Correia. 2001. Resistance and adaptation to quinidine in Saccharomyces cerevisiae: role of QDR1 (YIL120w), encoding a plasma membrane transporter of the major facilitator superfamily required for multidrug resistance. Antimicrob. Agents Chemother. 45:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pilot, G., H. Stransky, D. F. Bushey, R. Pratelli, U. Ludewig, V. P. Wingate, and W. B. Frommer. 2004. Overexpression of GLUTAMINE DUMPER1 leads to hypersecretion of glutamine from Hydathodes of Arabidopsis leaves. Plant Cell 16:1827-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raths, S., J. Rohrer, F. Crausaz, and H. Riezman. 1993. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 120:55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Regenberg, B., L. During-Olsen, M. C. Kielland-Brandt, and S. Holmberg. 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet. 36:317-328. [DOI] [PubMed] [Google Scholar]
- 54.Reimer, R. J., and R. H. Edwards. 2004. Organic anion transport is the primary function of the SLC17/type I phosphate transporter family. Pfluegers Arch. 447:629-635. [DOI] [PubMed] [Google Scholar]
- 55.Roettger, V. R., and S. G. Amara. 1999. GABA and glutamate transporters: therapeutic and etiologic implications for epilepsy. Adv. Neurol. 79:551-560. [PubMed] [Google Scholar]
- 56.Rotin, D., O. Staub, and R. Haguenauer-Tsapis. 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176:1-17. [DOI] [PubMed] [Google Scholar]
- 57.Sa-Correia, I., and S. Tenreiro. 2002. The multidrug resistance transporters of the major facilitator superfamily, 6 years after disclosure of Saccharomyces cerevisiae genome sequence. J. Biotechnol. 98:215-226. [DOI] [PubMed] [Google Scholar]
- 58.Saier, M. H., Jr. 2000. Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology 146:1775-1795. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Schuldiner, S., A. Shirvan, and M. Linial. 1995. Vesicular neurotransmitter transporters: from bacteria to humans. Physiol. Rev. 75:369-392. [DOI] [PubMed] [Google Scholar]
- 61.Tenreiro, S., A. R. Fernandes, and I. Sa-Correia. 2001. Transcriptional activation of FLR1 gene during Saccharomyces cerevisiae adaptation to growth with benomyl: role of Yap1p and Pdr3p. Biochem. Biophys. Res. Commun. 280:216-222. [DOI] [PubMed] [Google Scholar]
- 62.Tenreiro, S., P. A. Nunes, C. A. Viegas, M. S. Neves, M. C. Teixeira, M. G. Cabral, and I. Sa-Correia. 2002. AQR1 gene (ORF YNL065w) encodes a plasma membrane transporter of the major facilitator superfamily that confers resistance to short-chain monocarboxylic acids and quinidine in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 292:741-748. [DOI] [PubMed] [Google Scholar]
- 63.Tenreiro, S., P. C. Rosa, C. A. Viegas, and I. Sa-Correia. 2000. Expression of the AZR1 gene (ORF YGR224w), encoding a plasma membrane transporter of the major facilitator superfamily, is required for adaptation to acetic acid and resistance to azoles in Saccharomyces cerevisiae. Yeast 16:1469-1481. [DOI] [PubMed] [Google Scholar]
- 64.Van Belle, D., and B. André. 2001. A genomic view of yeast membrane transporters. Curr. Opin. Cell Biol. 13:389-398. [DOI] [PubMed] [Google Scholar]
- 65.Van Dyke, R. W. 1996. Acidification of lysosomes and endosomes. Subcell. Biochem. 27:331-360. [DOI] [PubMed] [Google Scholar]
- 66.Wipf, D., U. Ludewig, M. Tegeder, D. Rentsch, W. Koch, and W. B. Frommer. 2002. Conservation of amino acid transporters in fungi, plants and animals. Trends Biochem. Sci. 27:139-147. [DOI] [PubMed] [Google Scholar]
- 67.Yen, M. R., Y. H. Tseng, P. Simic, H. Sahm, L. Eggeling, and M. H. Saier, Jr. 2002. The ubiquitous ThrE family of putative transmembrane amino acid efflux transporters. Res. Microbiol. 153:19-25. [DOI] [PubMed] [Google Scholar]