Abstract
We report the identification and disruption of the Cryptococcus neoformans var. grubii UGD1 gene encoding the UDP-glucose dehydrogenase, which catalyzes the conversion of UDP-glucose into UDP-glucuronic acid. Deletion of UGD1 led to modifications in the cell wall, as revealed by changes in the sensitivity of ugd1Δ cells to sodium dodecyl sulfate, NaCl, and sorbitol. Moreover, two of the yeast's major virulence factors—capsule biosynthesis and the ability to grow at 37°C—were impaired in ugd1Δ strains. These results suggest that the UDP-dehydrogenase represents the major, and maybe only, biosynthetic pathway for UDP-glucuronic acid in C. neoformans. Consequently, deletion of UGD1 blocked not only the synthesis of UDP-glucuronic acid but also that of UDP-xylose. To differentiate the phenotype(s) associated with the UDP-glucuronic acid defect alone from those linked to the UDP-xylose defect, ugd1Δ mutants were phenotypically compared to strains from which the gene encoding UDP-xylose synthase (i.e., that required for synthesis of UDP-xylose) had been deleted. Finally, studies of strains from which one of the four CAP genes (CAP10, CAP59, CAP60, or CAP64) had been deleted revealed common cell wall phenotypes associated with the acapsular state.
Glucuronic acid is an essential component in the synthesis of many bacterial and eukaryotic polysaccharides (33, 39). In eukaryotic cells, the biosynthesis of such polysaccharides begins with the cytoplasmic synthesis of nucleotide sugar precursors (UDP-xylose, UDP-glucuronic acid, GDP-mannose, CMP-sialic acid, etc.). The precursors are then transported inside specific organelles (Golgi, endoplasmic reticulum, etc.) and finally transferred to the nascent polysaccharide via the action of glycosyltransferases. The key enzyme in UDP-glucuronic acid biosynthesis is a UDP-glucose dehydrogenase which converts UDP-glucose to UDP-glucuronic acid. It provides the main pathway for producing this nucleotide sugar—a mutation in the gene encoding this protein results in a complete lack of UDP-glucuronic acid in fly (3). However, in plants a salvage pathway exists as part of the inositol oxidation pathway (35).
Cryptococcus neoformans is an encapsulated yeast which is responsible for very serious infections in immunocompromised subjects (6). Present in the environment, the yeast is thought to infect people very early in life: it then remains in a dormant state (putatively in alveolar macrophages) until an immune defect occurs (16). The yeast then multiplies and spreads into a variety of organs and most noticeably into the brain, where it provokes severe (and lethal if untreated) meningoencephalitis. It has been demonstrated that a number of virulence factors are essential for virulence in C. neoformans: production of a polysaccharide capsule, synthesis of melanin, and the ability to grow at 37°C are the most notable (5). Certain other factors (like the production of urease or phospholipase) have been shown to influence a given strain's degree of virulence but do not appear to be as essential as the three cited above. The main capsule component is a very-high-molecular-weight polysaccharide called glucuronoxylomannan (GXM) which represents 88% of the capsular mass (see reference 4). GXM is an α-1,3-mannose polymer, branched with xylose, glucuronic acid, and O-acetyl residues. Recent work on the C. neoformans polysaccharide capsule has begun to unveil different aspects of its biosynthesis. A high proportion of the proteins identified to date have been found to be highly conserved in evolution. Cas1p, Uxs1p, Cap59p/Cap60p, Cap10p, and Man1p thus have clear orthologues in the human genome (1, 24, 25, 43). C. neoformans is an easy-to-manipulate microorganism, and a great number of tools for studying its genetics now exist (21). Moreover, a large number of anti-GXM monoclonal antibodies (MAb) have been purified and represent unique tools for probing polysaccharide structure (6). In the present study, we identified the C. neoformans var. grubii gene encoding UDP-glucose dehydrogenase. We then demonstrated that this enzyme plays a central role in the biology and the virulence of C. neoformans, since it is necessary for capsule biosynthesis, normal cell wall structure, and growth at 37°C.
MATERIALS AND METHODS
Strains and culture media.
The C. neoformans strains used in this study are listed in Table 1. The strains were routinely cultured on yeast extract-peptone-dextrose (YPD) medium at 30°C (36). Synthetic dextrose (SD) medium was prepared as described previously (36). The bacterial strain Escherichia coli XL1-Blue (Stratagene, La Jolla, Calif.) was used for the propagation of all plasmids.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| KNH99 | MATα | 32 |
| NE303 | MATα uxs1Δ::nat1 | This study |
| NE305 | MATα cap10Δ::nat1 | This study |
| NE309 | MATα cap60Δ::nat1 | This study |
| NE321 | MATα ugd1Δ::nat1 | This study |
| NE333 | MATα cap64Δ::nat1 | This study |
| NE367 | MATα cap59Δ::nat1 | This study |
| NE280 | MATα ugd1Δ::nat1 UGD1 | This study |
MAb.
The anticapsular MAb E1 (14), CRND-8 (kindly provided by T. Shinoda, Tokyo, Japan) (22), and 4H3, 2H1, and 5E4 (kindly provided by A. Casadevall, Albert Einstein College of Medicine, New York, N.Y.) (7) were used in immunoblotting and immunofluorescence experiments as previously described (13, 25).
DNA handling.
Genomic DNA purification was carried out as previously described (16). Southern blotting and colony hybridization were carried out using standard protocols (34). Probe labeling, hybridization, washing, and detection of hybridized bands were performed using a DIG nonradioactive nucleic acid labeling and detection system (Roche Diagnostic, Meylan, France) according to the manufacturer's instruction. Software from the University of Wisconsin Genetics Computer Group (Madison, Wis.) was used for nucleic acid sequence analysis (12). Restriction endonuclease digestion and ligation were carried out using standard methods, as recommended by the suppliers.
Gene disruption.
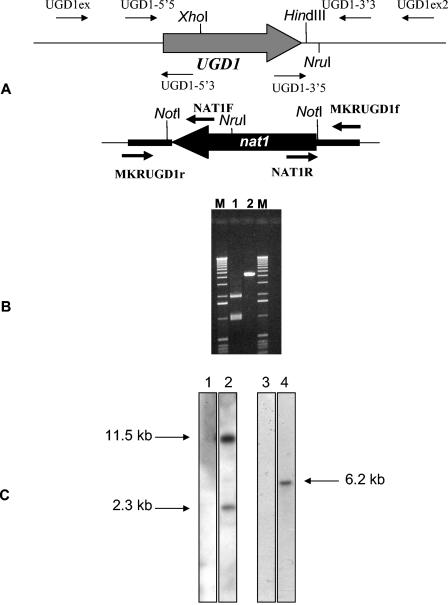
The disruption cassette was constructed by PCR fusion with a strategy similar to that used by Kuwayama and colleagues for investigation of the bacteria Clostridium difficile (26). For UGD1, upstream and downstream gene fragments and the nat1 marker (28) were PCR amplified using an HFPCR kit from Clontech (Palo Alto, Calif.). The primers used for these amplifications are listed elsewhere (see Table SA in the supplemental material), and their positions are shown in Fig. 1. UGD1-5′3 and UGD1-3′5 contain sequences recognized by the M13R and M13F primers, respectively. Similarly, the MKRUGD1f and MKRUGD1r primers, as well as UGD1-5′3 and UGD1-3′5, were designed to anneal to M13R and M13F, respectively. Consequently, UGD1-3′5 and the UGD1-5′3 contain the reverse complements of MKRUGD1f and MKRUGD1r, respectively. In addition, 5 ng of each of the three gel-purified, amplified fragments was used as a substrate for PCR fusion with the primers UGD1-5′5 and UGD1-3′3 via the following program: 94°C for 30 s followed by 35 cycles of 94°C for 15 s and 68°C for 4 min. The final PCR fragment represented the ugd1Δ::nat1 allele.
FIG. 1.
Disruption of UGD1. (A) Positions of the different primers used to construct the disruption cassettes and to screen the transformants. (B) Agarose gel electrophoresis of the UGD1ex-UGD1ex2 PCR products after NotI digestion. Lane 1, ugd1Δ strains; lane 2, KNH99 strains; lanes M, 1-kb ladder molecular mass marker. (C) Southern blot analysis. A total of 5 μg of strain KNH99 (lanes 1 and 3) and strain ugd1Δ (lanes 2 and 4) genomic DNA was digested with NruI (lanes 1 and 2) or XhoI (lanes 3 and 4), transferred onto a nylon membrane, and probed with a nat1 open reading frame-specific probe.
The PCR-amplified fragment was used to transform the KNH99 strains by biolistic DNA delivery, and transformants were selected on YPD medium containing 100 μg of nourseothricin (Werner BioAgents)/ml. Disruption of the other genes (UXS1, CAP10, CAP59, CAP60, and CAP64) was performed using the same strategy and the appropriate primers (see Table SA in the supplemental material).
The ugd1Δ strain was reconstituted (using the primers UGD1-5′5 and UGD1-3′3) by cloning a 3.8-kbp PCR-amplified fragment into the NotI site of a plasmid containing a hygromycin resistance cassette (20). pNE331, the resulting plasmid, was HindIII digested and used to transform the NE321 strain (MATα ugd1Δ) by biolistic DNA delivery. Transformants were selected on YPD medium containing 200 U of hygromycin (Calbiochem)/ml. A total of 10 hygromycin-resistant strains were obtained, all of them capsulated. Two were stored at −80°C for further studies.
Nucleotide sequence accession numbers.
The GenBank accession number for UGD1 is AY530214.
RESULTS
Identification of UGD1.
The biosynthesis of UDP-glucuronic acid involves the dehydrogenation of UDP-glucose by a UDP-glucose dehydrogenase (Fig. 2). We identified the UDP-glucose dehydrogenase gene in the C. neoformans genome (http://www.broad.mit.edu/annotation/fungi/cryptococcus_neoformans/index.html) by looking for sequence homologies with the corresponding bovine gene (19). Moreover, two traces of cDNA sequences specific for this gene were identified at the Oklahoma Sequencing Center (http://www.genome.ou.edu/cneo.html), and these enabled us to determine the 3′ and 5′ extremity sequences. During the course of our experiments, the C. neoformans UGD1 cDNA was cloned independently by Doering's group; they demonstrated, by means of enzyme assays, that it indeed encoded a protein with UDP-glucose dehydrogenase activity (2). The C. neoformans var. grubii UGD1 gene (UDP-glucose dehydrogenase) contains 14 introns of 66.6 nucleotides on average and encodes a protein of 468 amino acids sharing 99% identity with its C. neoformans var. neoformans orthologue. The sequence of the UDP-glucose dehydrogenase is highly conserved in evolution: the closest homologue is the human gene, which shares 74% similarity in its amino acid sequence with the gene encoding Ugd1p (38). As expected for a protein of this family, Ugd1p contains an N-terminal NAD binding domain (pfam03721.9; UDPG_MGDP_dh_N), a central UDP-glucose/GDP-mannose dehydrogenase family domain (pfam00984.11; UDPG_MGDP_dh), and a C-terminal UDP binding domain (pfam03720.9; UDPG_MGDP_dh_C) (see Fig. SA in the supplemental material).
FIG. 2.
UDP-glucuronic acid biosynthetic pathway. The UGD1 gene encodes a UDP-glucose dehydrogenase, which catalyzes the conversion of UDP-glucose into UDP-glucuronic acid (2). The UPD-glucuronic acid is then transformed into UDP-glucose by a UDP-xylose synthase encoded by the UXS1 gene (1).
Disruption of UGD1.
We disrupted UGD1 with the nat1 marker. Correct integration of the cassette was determined by PCR using a primer that annealed to a region outside the disruption cassette (UGD1ex) and a primer that annealed to a sequence within the marker (NAT1F) (Fig. 1A) (see Table SA in the supplemental material). We screened 20 colonies and identified five putative, homologous integrants. A second pair of primers was used to check for correct integration of the cassette (UGD1ex2-NAT1R) (Fig. 1A) (see Table SA in the supplemental material). PCR amplification and restriction of the UGD1ex-UGD1ex2 region were used to verify the knockout of the wild-type gene in each putative deletion strain (Fig. 1B) (see Table SA in the supplemental material). Furthermore, DNA hybridization analysis was used to confirm that additional, ectopic integration of the cassette had not occurred in the transformant genomes (Fig. 1C). Reintegration of the gene into the disruptant strain was performed using hygromycin as a selective marker (see Materials and Methods).
ugd1Δ strains are acapsular and do not grow on SD medium or at 37°C.
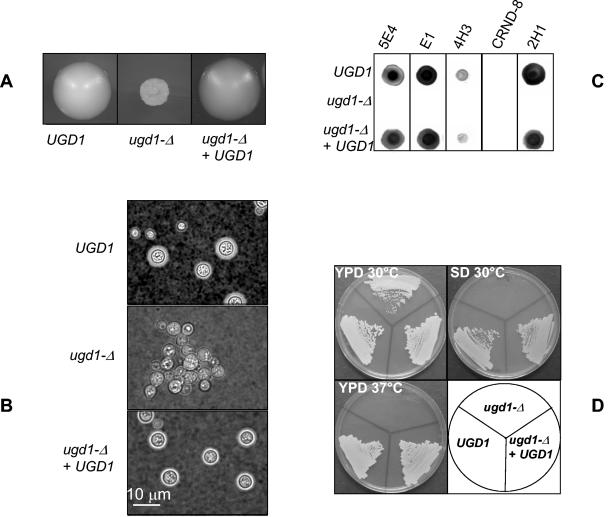
Colonies from ugd1Δ strains displayed a dry phenotype when grown on YPD medium (Fig. 3A). Moreover, examination under the microscope showed that the ugd1Δ cells formed large clusters. These phenotypes have been previously associated with the absence of capsule in the acapsular cap mutant strains (8-11). Indeed, microscope examination after negative staining with India ink confirmed that the ugd1Δ cells did not possess a capsule at all (Fig. 3B). Moreover, the ugd1Δ cells gave completely negative results in a dot blot assay using five different anti-GXM antibodies (Fig. 3C). Although we cannot completely exclude the possibility that these cells synthesize a very small capsule with a modified structure not recognized by the different MAb used here, this hypothesis seems to be unlikely, as all our efforts to purify some polysaccharides from a ugd1Δ cell culture supernatant have failed (data not shown). Thus, UGD1 appears to be necessary for capsule biosynthesis.
FIG. 3.
Cellular and capsular phenotypes associated with UGD1 disruption. (A) Colony morphologies of the original strain (UGD1), the mutant strain (ugd1Δ), and a reconstituted strain (ugd1Δ UGD1) after incubation on YPD plates for 3 days at 30°C. (B) India ink negative staining of the capsule. (C) Dot blot analysis of the capsule structure. Cell suspensions for each strain were spotted (3 × 104 cells per spot) onto nitrocellulose membranes and probed with different anti-GXM MAb. (D) Growth defects of a ugd1Δ strain at 37°C on SD medium after incubation for 3 days.
More surprisingly, ugd1Δ mutant strains were not able to grow on SD medium (Fig. 3D). This growth defect was not complemented by the addition of glucuronic acid, UDP-glucuronic acid, nicotinic acid, amino acids, or 1 M sorbitol to the medium (data not shown). The cells were able to grow on yeast extract-dextrose and peptone-dextrose media, albeit less well than on YPD medium (data not shown). The ugd1Δ mutants were also temperature sensitive and did not grow on YPD medium at 37°C (Fig. 3D). In contrast, no modification of melanin production was observed, as tested on Niger seed agar (data not shown). All phenotypes associated with UGD1 disruption were common to the five ugd1Δ mutant strains isolated and were restored by reintroduction of the gene. Thus, ugd1Δ UGD1 strains displayed smooth colony morphology, synthesized a capsule, and grew similarly to the wild-type strain on SD medium and on YPD medium at 37°C (Fig. 3). We tried to measure the intracellular concentration of UDP-glucuronic acid in the wild-type and mutated C. neoformans cells by a high-pressure liquid chromatography procedure (27). However, the wild-type concentration was at the limit of detection of the system and we did not obtain conclusive results (data not shown). On the other hand, the strongly marked phenotypes associated with the UGD1 deletion (i.e., the complete absence of capsule and the strong growth defects at 37°C and on SD medium) suggest that the UDP-glucose dehydrogenase pathway represents the major, if not the only, means for C. neoformans to produce UDP-glucuronic acid.
Cell wall phenotypes in ugd1Δ mutants.
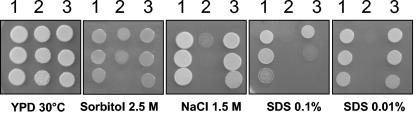
We hypothesized that growth defects in the ugd1Δ mutant strains are associated with a change in the properties of the cell wall. Indeed, ugd1Δ strains were not able to grow on YPD medium supplemented with sodium dodecyl sulfate (SDS) (0.01% wt/vol) or containing a high NaCl concentration (1.5 M): additionally, the mutants showed reduced growth when the medium was supplemented with 2.5 M sorbitol (Fig. 4). In contrast, ugd1Δ strains were not affected by the addition of CaCl2 (50 mM) to the medium or the replacement of glucose with glycerol (YPG) (data not shown).
FIG. 4.
Growth defects associated with UGD1 disruption. The C. neoformans cells were grown in liquid YPD medium overnight and washed with sterile water, and serial dilutions (106, 105, and 104 cells) of each strain (KNH99 [lanes 1], ugd1Δ [lanes 2], and ugd1Δ UGD1 [lanes 3]) were spotted onto different media and observed after incubation for 3 days at 30°C.
Phenotype linkage.
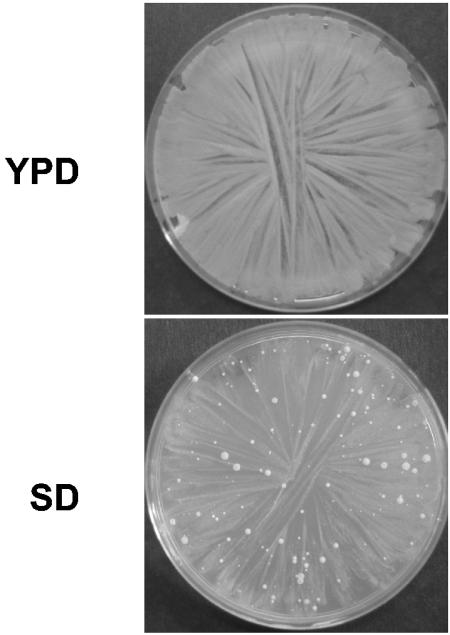
After several days' incubation, a number of spontaneous revertant mutants bearing a suppressor mutation of the ugd1Δ-associated SD growth defect appeared on the agar plates (Fig. 5). We recovered two of these mutant strains from each of the five ugd1Δ strains originally obtained (named Sup1 to Sup10). We then tested whether the phenotypes associated with the ugd1Δ mutation were suppressed in these strains. All 10 strains were able to grow at 37°C and on YPD medium containing 0.01% SDS, although they were unable to grow when the SDS concentration was increased to 0.1% (Table 2). In contrast, none of the revertant mutant strains were encapsulated: when a large number of ugd1Δ cells were plated, all the colonies which appeared on SD plates displayed a dry phenotype indicative of an acapsular structure (data not shown). Similarly, all strains were still sorbitol sensitive. However, the mutants differed in their sensitivity to NaCl: some (Sup1) were as sensitive as the ugd1Δ strains, whereas others (Sup2, Sup6 to Sup8) were completely resistant and grew as well as the wild type on YPD NaCl (1.5 M) medium. A last group of mutant strains (Sup3 to Sup5, Sup9, and Sup10) was characterized by partial restoration of NaCl resistance (Table 2).
FIG. 5.
ugd1Δ suppressor mutant strains. Spontaneous appearance of suppressor mutant strains on SD medium. A total of 107 cells were plated on YPD and SD media and incubated for 3 days at 30°C.
TABLE 2.
Phenotypes of the suppressor ugd1Δ mutant strains
| Strain | Resulta
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| YPD (30°C) | YPD (37°C) | SD | CaCl2 (50 mM) | SDS (0.01%) | SDS (0.1%) | Melanin | NaCl (1.5 M) | YPG | Sorbitol (2.5 M) | Capsule | |
| KNH99 | + | + | + | + | + | + | + | + | + | + | + |
| ugd1Δ | ± | − | − | ± | − | − | + | − | ± | ± | − |
| Sup1 | + | + | + | + | + | − | + | − | + | ± | − |
| Sup2 | + | + | + | + | + | − | + | + | + | ± | − |
| Sup3 | + | + | + | + | + | − | + | ± | + | ± | − |
| Sup4 | + | + | + | + | + | − | + | ± | + | ± | − |
| Sup5 | + | + | + | + | + | − | + | ± | + | ± | − |
| Sup6 | + | + | + | + | + | − | + | + | + | ± | − |
| Sup7 | + | + | + | + | + | − | + | + | + | ± | − |
| Sup8 | + | + | + | + | + | − | + | + | + | ± | − |
| Sup9 | + | + | + | + | + | − | + | ± | + | ± | − |
| Sup10 | + | + | + | + | + | − | + | ± | + | ± | − |
+, growth similar to that seen with the wild-type strain; ±, growth reduced compared to that seen with the wild-type strain; −, no growth.
Study of the uxs1 and cap mutant strains.
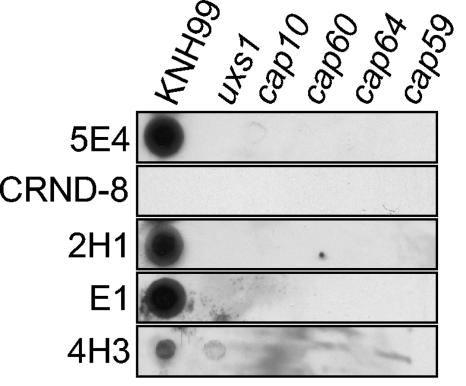
As shown in Fig. 2, UDP-glucuronic acid is the substrate of UDP-xylose synthase. Thus, cells from which the UGD1 gene has been deleted are not only unable to synthesize UDP-glucuronic acid or able to synthesize it only poorly but are also completely or partially devoid of UDP-xylose. The UXS1 gene encoding the xylose synthase in C. neoformans has been previously identified and studied with a C. neoformans var. neoformans strain (1, 30). To differentiate phenotypes directly associated with the lack of the UDP-glucuronic acid from those purely associated with the lack of UDP-xylose, we deleted the UXS1 gene from a C. neoformans var. grubii strain. As with C. neoformans var. neoformans usx1Δ strains, the C. neoformans var. grubii uxs1Δ strains synthesized a capsule which differs from the wild-type structure, as shown by our antibody binding analysis (Fig. 6). It is noteworthy that the phenotypes of the uxs1Δ strains were identical in both C. neoformans var. neoformans and C. neoformans var. grubii, since only the 4H3 antibody recognized the altered polysaccharide capsule structure (30) (Fig. 6).
FIG. 6.
Capsular phenotypes of the cap and uxs1 mutant strains. The results of dot blot analysis of the capsule structure are shown. Cell suspensions for each strain were spotted (3 × 104 cells per spot) onto nitrocellulose membranes and probed with different anti-GXM MAb.
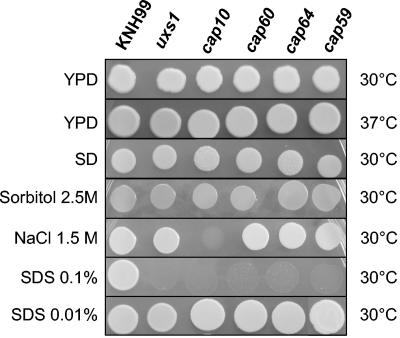
Furthermore, to see whether certain ugd1Δ mutant strain phenotypes were a common feature of the acapsular mutants, we deleted the CAP10, CAP59, CAP60, and CAP64 genes from C. neoformans var. grubii. As with the C. neoformans var. neoformans mutant strains, the cap10Δ, the cap59Δ, the cap60Δ, and the cap64Δ var. grubii strains were all acapsular, as revealed by India ink negative staining and dot blot analysis (Fig. 6 and data not shown). The growth phenotypes of these mutant strains were compared to those of the ugd1Δ strains (Fig. 7): all were able to grow on SD medium and did not show any temperature-dependent growth defects. As for the ugd1Δ strains, growth of the cap and uxs1 mutant strains was not affected by the presence of CaCl2 (50 mM) in the medium or by replacement of glucose with glycerol (YPG) (data not shown). These strains were also sensitive to 0.1% SDS but grew perfectly well when the concentration of SDS was reduced to 0.01%. In the presence of 1.5 M NaCl, the cap10Δ mutant strain was the only strain affected—the other cap mutant strains and the uxs1Δ strain were comparable to the original strain KNH99. Finally, the cap and uxs1Δ strains did not show any defects in melanin production when tested on Niger seed agar plates (data not shown). We studied the phenotypes of one further cap10Δ mutant, three more cap60Δ mutants, three more cap64Δ mutants, five more cap59Δ strains, and three more uxs1Δ strains (all independently isolated) and obtained identical results, thus confirming the linkage between the different phenotypes and the corresponding gene disruptions (data not shown).
FIG. 7.
Phenotypes of the cap and uxs1 strains. The C. neoformans cells were grown in liquid YPD medium overnight and washed with sterile water, and 106 cells were spotted onto different media and observed after incubation for 3 days.
DISCUSSION
GXM is composed of mannose, xylose, glucuronic acid, and O-acetyl residues. In our previous studies, we isolated two genes (CAS1 and UXS1) necessary for O-acetylation and xylosylation of the capsule, respectively (25, 30). Here, we constructed a strain lacking the UDP-glucose dehydrogenase. Although a salvage pathway exists in plants which are able to synthesize UDP-glucuronic acid via the oxidation of inositol to glucuronic acid and subsequent activation to the nucleotide sugar (35), phenotypic analysis of the udg1Δ strain suggests strongly that most if not all of the UDP-glucuronic acid is synthesized through the conversion of UDP-glucose. If a major salvage pathway existed in C. neoformans, one can predict that the residual UDP-glucuronic acid would compensate at least partly for the UDP-glucose dehydrogenase. UGD1 appeared completely required for expression of two of the three major attributes necessary for virulence in C. neoformans, i.e., the presence of a capsule and the ability to grow at 37°C. These results thus suggest that the ugd1Δ cells are largely devoid of UDP-glucuronic acid. Moreover, after this paper had been submitted and was under review, a related one appeared that provides further support for the findings presented here. Thus, Griffith and colleagues studied the consequences of the UGD1 deletion with C. neoformans var. neoformans and demonstrated using mass spectrometry analysis that the ugd1Δ strains were complexly devoid of UDP-glucuronic acid (18).
As UDP-glucuronic acid is the precursor of UDP-xylose, it was important to differentiate the phenotypes associated with the defect of UDP-xylose alone from those associated with the UDP-glucuronic acid/UDP-xylose double defect. We were able to link certain phenotypes to defects in one or the other of these UDP sugars, whereas others seem to be related to both defects. Thus, the acapsularity and the temperature sensitivity phenotypes were specific to ugd1Δ strains (i.e., default in UDP-glucuronic acid only), since the uxs1Δ strains were capsulated and grew at 37°C. Similar deductions can be made for the cell wall phenotype of the ugd1Δ mutant strains, as revealed by the inability to grow in the presence of high NaCl or sorbitol concentrations. The study of the mutants' sensitivity to SDS revealed a cumulative effect of the two UDP sugar defects: uxs1Δ strains were sensitive to SDS at 0.1% but not at 0.01%, whereas ugd1Δ strains were not even able to grow at the latter concentration. This result suggests that both UDP sugars play a role in SDS resistance in C. neoformans. None of the phenotypes studied here could be considered the consequence of a UDP-xylose defect alone, since none was common to the ugd1Δ and the uxs1Δ strains.
After several days of incubation of the ugd1 mutant strains on SD medium, a number of colonies appeared on the plates. Each of these spontaneous revertant strains bears one or several mutations bypassing the SD growth defect associated with the UGD1 deletion. The results of the study of these strains are indicative of the diversity of the metabolic pathways affected by the UDP-glucuronic acid defect in C. neoformans cells. For example, none of the 10 revertant strains studied was encapsulated. Similarly, all the revertant mutants had the same sorbitol sensitivity as the ugd1Δ strains. Thus, the sorbitol sensitivity and capsular phenotypes of the ugd1Δ strains and the growth defect on SD of the ugd1Δ strains seem to be consequences of independent changes in cell metabolism. In contrast, all the revertant mutant strains were able to grow at 37°C and on 0.01% SDS. This result indicates a clear linkage between these phenotypes and strongly suggests that they are consequences of the same defect in C. neoformans metabolism. The picture is more complex for NaCl sensitivity, since not all the revertant mutants had the same phenotype. This indicates that the metabolic defect causing NaCl sensitivity partly overlaps with the one causing temperature and 0.01% SDS sensitivities or the growth defect on SD.
One can imagine at least two different (but not necessarily exclusive) hypotheses to explain why so many phenotypes are affected by the absence of one nucleotide sugar. Firstly, although the C. neoformans cell wall does not contain glucuronic acid residues, it has been demonstrated previously that glycoproteins can indeed contain these moieties (23, 42). This type of posttranslational modification is often necessary for the full enzymatic activity of the corresponding proteins. For Candida albicans, for example, the deletion of the PMT1 gene (encoding a mannosyl transferase) has multiphenotypic consequences and affects the virulence of the resulting strains (41). With C. neoformans, it is possible that glucuronic acid residues are necessary for the functionality of various proteins involved in capsule and cell wall biosynthesis or of those necessary for growth at 37°C.
Alternatively, the addition of glucuronic acid residues to a nascent polysaccharide chain could be an early biosynthetic step influencing GXM polymerization. The absence of such residues would thus impair the synthesis of this polysaccharide. Similarly, in higher eukaryotes, sulfated glycosaminoglycans are synthesized as proteoglycans on specific serine residues in the so-called glycosamino-protein linkage region [glucuronic acid(β-1,3)galactose(β-1,3)galactose(β-1,4)xylose-β-1-O-Ser] common to heparan sulfate, heparin, chondroitin sulfate, and dermatan sulfate chains: a mutation in the UDP-glucose dehydrogenase gene impairs the synthesis of all these polysaccharides (39). In support of this hypothesis, it was previously demonstrated that the deletion of either the CAS1 or UXS1 gene (required for capsule O-acetylation or xylosylation, respectively) did not modify the position or the number of glucuronic acid residues on GXM—probably because xylose and O-acetyl residues are added to the nascent GXM chain after the glucuronic acid moieties (25, 30).
The acapsular phenotype of ugd1Δ mutants is similar to that seen with strains from which one of the five following genes have been deleted: the four CAP genes (CAP10, CAP59, CAP60, and CAP64) (8-11) and VPH1 (15). VPH1 encodes a protein sharing sequence homologies with the “a” subunit of vacuolar (H+)-ATPase complexes from diverse organisms. This protein is involved in the control of vesicular acidification and is necessary for melanin formation and for growth at 37°C (15). In contrast, very little information has been published on the potential role(s) of the Cap proteins. It has been suggested that they might play a role in the regulation of capsule formation (29) and in polysaccharide secretion (17). The results of the present study support a different hypothesis, in which the CAP genes constitute a direct part of the capsule biosynthetic pathway and are particularly involved in glucuronic acid metabolism. For example, a mutation in a glucuronosyltransferase-encoding gene would theoretically result in an acapsular phenotype of the corresponding strain. We have no direct proof that one or several of the Cap proteins have glycosyltransferase activity, but this hypothesis is supported by the recent demonstration that a CAP59-homologous gene (CMT1) encodes a α-1,3-mannosyl transferase (37).
The C. neoformans polysaccharide capsule is a major virulence factor of this yeast and also constitutes a fascinating structure to study. This work revealed the central role of glucuronic acid residues in C. neoformans biology and established a link between this moiety and various C. neoformans virulence factors.
Supplementary Material
Acknowledgments
We are grateful to the members of the C. neoformans H99 sequencing project at the Duke Center for Genome Technology (http://cgt.duke.edu/), Durham, N.C., and the Genome Sequence Centre at the BC Cancer Research Centre (http://www.bcgsc.bc.ca/), Vancouver, British Columbia, Canada. We also thank the members of the C. neoformans cDNA sequencing project at the University of Oklahoma (http://www.genome.ou.edu/cneo.html) funded by the National Institutes of Health-National Institute of Allergy and Infectious Diseases (grant no. AI47079). This work was supported by a grant from SIDACTION (AO-13) and the Pasteur Institute.
We are grateful to T. Shinoda (Tokyo, Japan) for the generous gift of the MAb CRND-8 and to A. Casadevall (New York, N.Y.) for the MAb 4H3, 5E4, and 2H1. We thank T. Fontaine (Institut Pasteur, Paris, France) for his help in the tentative measurement of the intracellular concentration of UDP-glucuronic acid.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org.
REFERENCES
- 1.Bar-Peled, M., C. L. Griffith, and T. L. Doering. 2001. Functional cloning and characterization of a UDP-glucuronic acid decarboxylase: the pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc. Natl. Acad. Sci. USA 98:12003-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar-Peled, M., C. L. Griffith, J. J. Ory, and T. L. Doering. 2004. Biosynthesis of UDP-GlcA, a key metabolite for capsular polysaccharide synthesis in the pathogenic fungus Cryptococcus neoformans. Biochem. J. 381:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binari, R. C., B. E. Staveley, W. A. Johnson, R. Godavarti, R. Sasisekharan, and A. S. Manoukian. 1997. Genetic evidence that heparin-like glycosaminoglycans are involved in wingless signaling. Development 124:2623-2632. [DOI] [PubMed] [Google Scholar]
- 4.Bose, I., A. J. Reese, J. J. Ory, G. Janbon, and T. L. Doering. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchanan, K. L., and J. W. Murphy. 1998. What makes Cryptococcus neoformans a pathogen? Emerg. Infect. Dis. 4:71-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. American Society for Microbiology, Washington, D.C.
- 7.Casadevall, A., and M. D. Scharff. 1991. The mouse antibody response to infection with Cryptococcus neoformans: VH and VL usage in polysaccharide binding antibodies. J. Exp. Med. 174:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y. C., and K. J. Kwon-Chung. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14:4912-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. C., and K. J. Kwon-Chung. 1998. Isolation of the third capsule-associated gene CAP60, required for virulence in Cryptococcus neoformans. Infect. Immun. 66:2230-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. C., and K. J. Kwon-Chung. 1999. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 181:5636-5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 64:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devereux, J. P., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acid Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dromer, F., E. Guého, O. Ronin, and B. Dupont. 1993. Serotyping of Cryptococcus neoformans by using a monoclonal antibody specific for capsular polysaccharide. J. Clin. Microbiol. 31:359-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dromer, F., J. Salamero, A. Contrepois, C. Carbon, and P. Yeni. 1987. Production, characterization, and antibody specificity of a mouse antibody reactive with Cryptococcus neoformans capsular polysaccharide. Infect. Immun. 55:742-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson, T., T. Liu, A. Gueyikian, X. Zhu, J. Gibbons, and P. R. Williamson. 2001. Multiple virulence factors of Cryptococcus neoformans are dependent on VPH1. Mol. Microbiol. 42:1121-1131. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Hermoso, D., G. Janbon, and F. Dromer. 1999. Epidemiological evidence for dormant Cryptococcus neoformans infection. J. Clin. Microbiol. 37:3204-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García-Rivera, J., Y. C. Chang, K. J. Kwon-Chung, and A. Casadevall. 2004. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffith, C. L., J. S. Klutts, L. Zhang, S. B. Levery, and T. L. Doering. UDP-glucose dehydrogenase plays multiple roles in the biology of the pathogenic fungus Cryptococcus neoformans. J. Biol. Chem., in press. [DOI] [PubMed]
- 19.Hempel, J., J. Perozich, H. Romovacek, A. Hinich, I. Kuo, and D. S. Feingold. 1994. UDP-glucose dehydrogenase from bovine liver: primary structure and relationship to other dehydrogenases. Protein Sci. 3:1074-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua, J. H., J. D. Meyer, and J. K. Lodge. 2000. Development of positive markers for the fungal pathogen, Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 7:125-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hull, C. M., and J. Heitman. 2002. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36:557-615. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda, R., S. Nishimura, A. Nishikawa, and T. Shinoda. 1996. Production of agglutinating monoclonal antibody against antigen 8 specific for Cryptococcus neoformans serotype D. Clin. Diagn. Lab. Immunol. 3:89-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.James, P. G., R. Cherniak, R. G. Jones, and C. A. Stortz. 1990. Cell-wall glucans of Cryptococcus neoformans CAP 67. Carbohydr. Res. 198:23-38. [DOI] [PubMed] [Google Scholar]
- 24.Janbon, G. 2004. Cryptococcus neoformans capsule biosynthesis and regulation. FEMS Yeast Res. 4:765-771. [DOI] [PubMed] [Google Scholar]
- 25.Janbon, G., U. Himmelreich, F. Moyrand, L. Improvisi, and F. Dromer. 2001. Cas1p is a membrane protein necessary for the O-acetylation of the Cryptococcus neoformans capsular polysaccharide. Mol. Microbiol. 42:453-469. [DOI] [PubMed] [Google Scholar]
- 26.Kuwayama, H., S. Obara, T. Morio, M. Katoh, H. Urushibara, and Y. Tanaka. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, R., D. Monsey, A. Weston, K. Duncan, C. Rithner, and M. McNeil. 1996. Enzymatic synthesis of UDP-galactofuranose and an assay for UDP-galactopyranose mutase based on high-performance liquid chromatography. Anal. Biochem. 242:1-7. [DOI] [PubMed] [Google Scholar]
- 28.McDade, H. C., and G. M. Cox. 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39:151-154. [DOI] [PubMed] [Google Scholar]
- 29.McFadden, D. C., and A. Casadevall. 2001. Capsule and melanine synthesis in Cryptococcus neoformans. Med. Mycol. 39:19-30. [PubMed] [Google Scholar]
- 30.Moyrand, F., B. Klaproth, U. Himmelreich, F. Dromer, and G. Janbon. 2002. Isolation and characterization of capsule structure mutant strains of Cryptococcus neoformans. Mol. Microbiol. 45:837-849. [DOI] [PubMed] [Google Scholar]
- 31.Nicholas, K. B., H. B. J. Nicholas, and D. W. I. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW. NEWS 4:14. [Google Scholar]
- 32.Nielsen, K., G. Cox, P. Wang, D. Toffaletti, J. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts, I. A. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285-315. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Seitz, B., C. Klos, M. Wurm, and R. Tenhaken. 2000. Matrix polysaccharide precursors in Arabidopsis cell walls are synthesized by alternative pathways with organ-specific expression patterns. Plant J. 21:537-546. [DOI] [PubMed] [Google Scholar]
- 36.Sherman, F. 1992. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 37.Sommer, U., H. Liu, and T. L. Doering. 2003. An α-1,3-mannosyltransferase of Cryptococcus neoformans. J. Biol. Chem. 278:47724-47730. [DOI] [PubMed] [Google Scholar]
- 38.Spicer, A. P., L. A. Kaback, T. J. Smith, and M. F. Seldin. 1998. Molecular cloning and characterization of the human and mouse UDP-glucose dehydrogenase genes. J. Biol. Chem. 273:25117-25124. [DOI] [PubMed] [Google Scholar]
- 39.Sugahara, K., and H. Kitagawa. 2000. Recent advances in the study of the biosynthesis and functions of the sulfated glycosaminoglycans. Curr. Opin. Struct. Biol. 10:518-527. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acid Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timpel, C., S. Strahl-Bolsinger, K. Ziegelbauer, and J. Ernst. 1998. Multiple functions of Pmt1p-mediated protein O-mannosylation in the fungal pathogen Candida albicans. J. Biol. Chem. 273:20837-20846. [DOI] [PubMed] [Google Scholar]
- 42.Turner, S. H., and R. Cherniak. 1991. Multiplicity in the structure of the glucuronoxylomannan of Cryptococcus neoformans, p. 123-142. In J. P. Latgé and D. Boucias (ed.), Fungal cell wall and immune response, vol. H 53. NATO ASI Series, Heidelberg, Germany.
- 43.Wills, E. A., I. S. Roberts, M. Del Poeta, J. Rivera, A. Casadevall, G. M. Cox, and J. R. Perfect. 2001. Identification and characterization of the Cryptococcus neoformans phosphomannose isomerase-encoding gene, MAN1, and its impact on pathogenicity. Mol. Microbiol. 40:610-620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.