Abstract
Background
Over the past 20 years, many marine seabird populations have been gradually declining and the factors driving this ongoing deterioration are not always well understood. Avipoxvirus infections have been found in a wide range of bird species worldwide, however, very little is known about the disease ecology of avian poxviruses in seabirds. Here we present two novel avipoxviruses from pacific shearwaters (Ardenna spp), one from a Flesh-footed Shearwater (A. carneipes) (SWPV-1) and the other from a Wedge-tailed Shearwater (A. pacificus) (SWPV-2).
Results
Epidermal pox lesions, liver, and blood samples were examined from A. carneipes and A. pacificus of breeding colonies in eastern Australia. After histopathological confirmation of the disease, PCR screening was conducted for avipoxvirus, circovirus, reticuloendotheliosis virus, and fungal agents. Two samples that were PCR positive for poxvirus were further assessed by next generation sequencing, which yielded complete Shearwaterpox virus (SWPV) genomes from A. pacificus and A. carneipes, both showing the highest degree of similarity with Canarypox virus (98% and 67%, respectively). The novel SWPV-1 complete genome from A. carneipes is missing 43 genes compared to CNPV and contains 4 predicted genes which are not found in any other poxvirus, whilst, SWPV-2 complete genome was deemed to be missing 18 genes compared to CNPV and a further 15 genes significantly fragmented as to probably cause them to be non-functional.
Conclusion
These are the first avipoxvirus complete genome sequences that infect marine seabirds. In the comparison of SWPV-1 and −2 to existing avipoxvirus sequences, our results indicate that the SWPV complete genome from A. carneipes (SWPV-1) described here is not closely related to any other avipoxvirus genome isolated from avian or other natural host species, and that it likely should be considered a separate species.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-017-3680-z) contains supplementary material, which is available to authorized users.
Keywords: Avipoxvirus, Poxvirus, Next generation sequencing, dermatitis, Ardenna, Shearwater
Background
The Avipoxvirus genus includes a divergent group of viruses that cause diseases in more than 278 species of wild and domestic birds in terrestrial and marine environments worldwide [1, 2]. Relatively little is known about the origins, worldwide host distribution and genetic diversity of avipoxviruses [3]. In affected birds, avipoxviruses typically cause proliferative ‘wart-like’ growths that are most commonly restricted to the eyes, beak or unfeathered skin of the body (so-called ‘dry’ pox), but infections can also develop in the upper alimentary and respiratory tracts (‘wet’ or ‘diptheritic’ pox) [2]. The incubation period and magnitude of avipoxvirus infection is variable, and is rarely fatal although secondary bacterial or fungal infections are common and cause increased mortality [2]. Such conditions in naïve populations can reach a much higher prevalence with substantial fatality [4, 5].
Avipoxviruses belong to the subfamily Chordopoxvirinae (ChPV) of the Poxviridae family, which are relatively large double-stranded DNA (dsDNA) viruses that replicate in the cytoplasm of infected cells [6]. Although poxviruses have evolved to infect a wide range of host species, to date only six avipoxvirus genomes have been published; a pathogenic American strain of Fowlpox virus (FPVUS) [7], an attenuated European strain of Fowlpox virus (FP9) [8], a virulent Canarypox virus (CNPV) [9], a pathogenic South African strain of Pigeonpox virus (FeP2), a Penguinpox virus (PEPV) [3], and a pathogenic Hungarian strain of Turkeypox virus (TKPV) [10]. Although these genome sequences demonstrate that avipoxviruses have diverged considerably from the other chordopoxviruses (ChPVs), approximately 80 genes have been found to be conserved amongst all ChPVs and to comprise the minimum essential poxvirus genome [11]. These genes tend to be present in the central core of the linear genome with the remainder presumed to be immunomodulatory and host specific genes located towards the terminal regions of the genome [3]. With the exception of TKPV (188 kb), avipoxvirus genomes (266–360 kb) tend to be bigger than those of other ChPVs due in part to multiple families of genes.
Over the past two decades, the status of the world’s bird populations have deteriorated with seabirds declining faster than any other group of birds [12]. On Lord Howe Island in eastern Australia, the Flesh-footed Shearwater Ardenna carneipes has been declining for many years and is therefore listed as Vulnerable in the state of New South Wales [13]. The ongoing threat of plastic pollution, and toxicity from the elevated concentration of trace elements such as mercury could be confounding drivers of this declining species [14]. Infectious diseases, including those caused by avipoxviruses, have also been identified as an important risk factor in the conservation of small and endangered populations, particularly in island species [15–18]. The impact of the introduction of avipoxviruses has been severe for the avifauna of various archipelagos [19]. The emergence of distinctive avipoxvirus with a high prevalence (88%) in Hawaiian Laysan Albatross (Phoebastria immutabilis) enabled one of the first detailed studies of the epidemiology and population-level impact of the disease in the seabirds [20]. However, relatively little is known about the general prevalence or effects of poxviruses in seabird species, including for shearwaters (Ardenna or Puffinus spp.). Therefore, the aim of the present study was to identify and characterize pathogens associated with clinical disease in breeding colonies of Flesh-footed Shearwater and Wedge-tailed Shearwater sourced from Lord Howe Island in 2015.
Results
Identification of fungal pathogens
In the sample from A. pacificus (15–1526, and 15–1527), there were multifocal areas of inflammation and exudation associated with serocellular surface crust that contained abundant branching fungal hyphae and aggregations of bacteria (Fig. 1c). A PCR screening was conducted for the presence of fungal pathogen using the ITS region to amplify a segment of approximately 550 bp. Two samples (out of 6) were positive for fungal pathogens, and direct Sanger sequencing of the purified gel bands resulted in a 550 bp sequence after trimming off primer sequences (data not shown). These sequences were further verified using high-throughput NGS, and generated con tigs of 3,430 bp (15–1526; GenBank accession KX857213) and 5,188 bp (15–1527; GenBank accession KX857212). A BLASTn search for the bird coinfected with fungal pathogen (15–1526) returned multiple hits to various fungal species, all with very similar scores; however, the best match (88%) was to the Phaeosphaeria nodorum (GenBank Accession EU053989.1, and value ≤ e-153), a major necrotrophic fungal pathogen of wheat [21]. Similar search model for the fungal pathogen of bird 15–1527, demonstrated a highest hit (96%) to the Metarhizium anisopliae var. anisopliae (GenBank Accession AY884128.1, and value ≤ e-173), an entomopathogenic fungus [22].
Fig. 1.
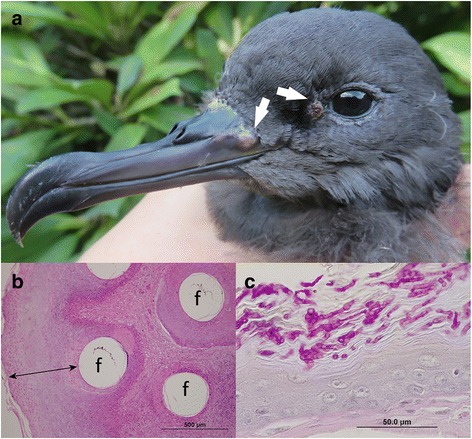
Pathological evidence of characteristic pox and fungal lesions. a Grossly well circumscribed, popular, crusting pox lesions across the featherless skins (white arrows). b Feather skin demonstrating diffuse proliferation of the epidermis and follicular infundibula with keratinocytes containing eosinophilic intracytoplasmic inclusions (Bollinger bodies) and serocellular surface crust (double head arrow). c Inflammatory exudates associated with serocellular surface crust that contained abundant branching fungal hyphae and aggregations of bacteria
Identification of virus
Samples from six shearwater chicks of two different species, A. carneipes and A. pacificus, with evidence of gross well circumscribed, popular, crusting lesions across the feather skins (Fig. 1a), were biopsied, with blood and liver samples also collected. Histological examinations of the skin demonstrated focal to diffuse full thickness necrosis of the epidermis and a thick serocellular surface crust. A marked heterophilic rich inflammatory cellular response and exudation was present alongside abundant macrophages and perifollicular fibroplasia. In some areas there was focal proliferation of the adjacent epidermis associated with ballooning degeneration of keratinocytes with eosinophilic intracytoplasmic inclusions (Fig. 1b). A PCR screening was conducted for the presence of poxvirus, circovirus and reticuloendotheliosis virus, which are likely to cause this type of skin lesions. Two birds (A. pacificus 15–1526 and A. carneipes 15–1528) were positive by PCR targeting the 4b gene that encodes a core protein of ChPV, however, there were no evidence of either circovirus or reticuloendotheliosis for any of the samples used in this study. Direct Sanger sequencing of the purified gel bands resulted in a 578 bp sequence after trimming off primer sequences (data not shown). A BLASTn search with these sequences returned multiple hits to the 4b core gene from a variety of poxviruses, all with very similar scores; however, the best match was to the Canarypox virus 4b core protein gene ((bird 15–1526; similarity with AY318871 was 99% and identity score ≤ e-162), and bird 15–1528; similarity with LK021654 was 99% and identity score ≤ e-157)).
Genome sequence and annotation of viruses
The Shearwaterpox virus complete genomes were assembled using CLC Genomics workbench 9.5.2 under La Trobe University Genomics Platform. The assembled complete genomes of SWPV-1 and −2 were 326,929 and 351,108 nt, respectively. The SWPV-1 and −2 complete genomes were annotated as described in the methods using CNPV as a reference genome (Additional file 1: Table S1 and Additional file 2: Table S2). We took a conservative approach to the annotation in order to minimize the inclusion of ORFs that were unlikely to represent functional genes. Table 1 lists the 310 and 312 genes annotated for SWPV-1 and −2, respectively. For the most part, these two new complete genomes are collinear to CNPV although there are a number of rearrangements of blocks of 1–6 genes in addition to insertions and deletions with respect to CNPV (Table 1). Comparison of the predicted proteins of SWPV-2 to orthologs in CNPV reveal the vast majority are >98% identical (aa), with more than 80 being completely conserved. In contrast, the orthologs of SWPV-1 only have an average aa identity of 67% to CNPV. However, with the lower average identity, greater genetic distance, comes a much greater range of variation in the level of identity and a significant number of predicted proteins are 80 – 90% identical (aa) to CNPV orthologs.
Table 1.
Shearwaterpox virus (SWPV) genome annotations and comparative analysis of ORFs relative to CNPV genomes
| SWPV1 synteny | SWPV2 synteny | CNPV synteny | CNPV BLAST hits | SWPV1 % identity | SWPV2 % identity | SWPV1 AA size | SWPV2 AA size | Reference AA size | notes |
|---|---|---|---|---|---|---|---|---|---|
| CNPV001 | CNPV001 hypothetical protein | 72 | |||||||
| SWPV2-001 | CNPV002 | CNPV002 hypothetical protein | 92.941 | 171 | 171 | ||||
| SWPV1-001 | SWPV2-002 | CNPV003 | CNPV003 C-type lectin-like protein | 32.044 | 85.99 | 181 | 208 | 204 | |
| SWPV1-002 | CNPV004 | CNPV004 ankyrin repeat protein | 56.458 | 468 | 514 | ||||
| SWPV1-003 | SWPV2-003 | CNPV005 | CNPV005 conserved hypothetical protein | 87.387 | 99.55 | 220 | 222 | 222 | |
| SWPV2-004 | CNPV006 | CNPV006 hypothetical protein | 88.71 | 134 | 182 | SWPV2: C-terminus fragment, not likely translated | |||
| CNPV007 | CNPV007 ankyrin repeat protein | 674 | |||||||
| SWPV1-004 | SWPV2-005 | CNPV008 | CNPV008 C-type lectin-like protein | 50 | 98.225 | 174 | 169 | 169 | |
| SWPV2-006 | CNPV009 | CNPV009 ankyrin repeat protein | 99.564 | 688 | 688 | ||||
| CNPV010 | CNPV010 ankyrin repeat protein | 734 | |||||||
| SWPV2-007 | CNPV011 | CNPV011 ankyrin repeat protein | 99.147 | 586 | 586 | ||||
| SWPV2-008 | CNPV012 | CNPV012 hypothetical protein | 100 | 189 | 189 | ||||
| SWPV2-009 | CNPV013 | CNPV013 hypothetical protein | 98.81 | 168 | 168 | ||||
| SWPV2-010 | CNPV014 | CNPV014 immunoglobulin-like domain protein | 99.184 | 490 | 490 | ||||
| SWPV2-011 | CNPV015 | CNPV015 ankyrin repeat protein | 97.538 | 528 | 528 | ||||
| SWPV1-005 | CNPV035 C-type lectin-like protein | 35.556 | 138 | 134 | |||||
| SWPV1-006 | CNPV318 ankyrin repeat protein | 58.932 | 487 | 514 | |||||
| SWPV1-007 | SWPV2-012 | CNPV016 | CNPV016 C-type lectin-like protein | 52.128 | 98.81 | 117 | 168 | 168 | |
| SWPV1-008 | SWPV2-013 | CNPV017 | CNPV017 ankyrin repeat protein | 64.471 | 97.912 | 425 | 479 | 486 | |
| SWPV1-009 | CNPV295 ankyrin repeat protein | 56.41 | 277 | 396 | |||||
| SWPV2-014 | CNPV018 | CNPV018 IL-10-like protein | 90.805 | 190 | 191 | ||||
| SWPV2-015 | CNPV019 | CNPV019 ankyrin repeat protein | 99.083 | 436 | 436 | ||||
| SWPV1-010 | SWPV2-016 | CNPV020 | CNPV020 ankyrin repeat protein | 56.311 | 99.761 | 412 | 419 | 419 | |
| SWPV1-011 | CNPV320 Ig-like domain protein | 31.656 | 483 | 469 | |||||
| SWPV1-012 | SWPV2-017 | CNPV021 | CNPV021 ankyrin repeat protein | 62.313 | 99.626 | 528 | 535 | 535 | |
| SWPV1-013 | SWPV2-018 | CNPV022 | CNPV022 putative serpin | 65.642 | 98.324 | 356 | 358 | 358 | |
| SWPV1-014 | PEPV260 ankyrin repeat protein | 53.158 | 190 | 192 | |||||
| SWPV1-015 | CNPV011 ankyrin repeat protein | 34 | 530 | 586 | |||||
| SWPV2-019 | CNPV023 | CNPV023 vaccinia C4L/C10L-like protein | 98.595 | 424 | 427 | ||||
| SWPV2-020 | CNPV024 | CNPV024 hypothetical protein | 96.629 | 178 | 178 | ||||
| SWPV1-016 | SWPV2-021 | CNPV025 | CNPV025 alpha-SNAP-like protein | 57.491 | 98.667 | 304 | 300 | 300 | |
| SWPV1-017 | SWPV2-022 | CNPV026 | CNPV026 ankyrin repeat protein | 54.271 | 98.953 | 397 | 382 | 382 | |
| SWPV1-018 | SWPV2-023 | CNPV027 | CNPV027 ankyrin repeat protein | 59.375 | 98.722 | 646 | 626 | 626 | |
| SWPV1-019 | SWPV2-024 | CNPV028 | CNPV028 ankyrin repeat protein | 57.618 | 99.164 | 408 | 365 | 362 | |
| SWPV1-020 | SWPV2-025 | CNPV029 | CNPV029 C-type lectin-like protein | 50.35 | 99.296 | 142 | 142 | 142 | |
| SWPV1-021 | SWPV2-026 | CNPV030 | CNPV030 ankyrin repeat protein | 63.72 | 98.529 | 345 | 340 | 340 | |
| SWPV1-022 | SWPV2-027 | CNPV031 | CNPV031 hypothetical protein | 60.331 | 97.479 | 120 | 119 | 119 | |
| SWPV1-023 | CNPV013 conserved hypothetical protein | 44.048 | 168 | 168 | |||||
| SWPV1-024 | SWPV2-028 | CNPV032 | CNPV032 Ig-like domain putative IFN-gamma binding protein | 51.837 | 92.149 | 242 | 242 | 242 | |
| SWPV1-025 | SWPV2-029 | CNPV033 | CNPV033 Ig-like domain protein | 48.095 | 93.496 | 238 | 246 | 246 | |
| SWPV2-030 | CNPV034 | CNPV034 ankyrin repeat protein | 99.848 | 659 | 659 | ||||
| SWPV2-031 | CNPV035 | CNPV035 C-type lectin-like protein | 94.776 | 133 | 134 | ||||
| SWPV1-026 | SWPV2-032 | CNPV036 | CNPV036 conserved hypothetical protein | 48.235 | 98.947 | 88 | 95 | 95 | |
| SWPV1-027 | SWPV2-033 | CNPV037 | CNPV037 conserved hypothetical protein | 63.068 | 99.441 | 178 | 179 | 179 | |
| SWPV1-028 | SWPV2-034 | CNPV038 | CNPV038 vaccinia C4L/C10L-like protein | 54.523 | 99.516 | 411 | 413 | 413 | |
| SWPV1-029 | SWPV2-035 | CNPV039 | CNPV039 G protein-coupled receptor-like protein | 67.284 | 97.859 | 323 | 327 | 327 | |
| SWPV1-030 | SWPV2-036 | CNPV040 | CNPV040 ankyrin repeat protein | 57.36 | 93.401 | 589 | 591 | 591 | SWPV2: High SNP Density |
| SWPV1-031 | SWPV2-037 | CNPV041 | CNPV041 ankyrin repeat protein | 66.284 | 98.605 | 432 | 430 | 430 | |
| SWPV1-032 | SWPV2-038 | CNPV042 | CNPV042 ankyrin repeat protein | 72.712 | 99.339 | 608 | 605 | 605 | |
| SWPV1-033 | SWPV2-039 | CNPV043 | CNPV043 conserved hypothetical protein | 74.627 | 99.005 | 202 | 201 | 201 | |
| SWPV1-034 | SWPV2-040 | CNPV044 | CNPV044 ankyrin repeat protein | 67.316 | 99.583 | 470 | 480 | 480 | |
| SWPV1-035 | SWPV2-041 | CNPV045 | CNPV045 G protein-coupled receptor-like protein | 65.231 | 100 | 331 | 332 | 332 | |
| SWPV1-036 | SWPV2-042 | CNPV046 | CNPV046 ankyrin repeat protein | 68.08 | 98.667 | 452 | 450 | 450 | |
| SWPV1-037 | SWPV2-043 | CNPV047 | CNPV047 conserved hypothetical protein | 65.6 | 99.194 | 125 | 124 | 124 | |
| SWPV1-038 | SWPV2-044 | CNPV048 | CNPV048 alkaline phosphodiesterase-like protein | 68.238 | 98.502 | 804 | 801 | 801 | |
| SWPV1-039 | SWPV2-045 | CNPV049 | CNPV049 hypothetical protein | 72.667 | 100 | 148 | 150 | 150 | |
| SWPV1-040 | SWPV2-046 | CNPV050 | CNPV050 ankyrin repeat protein | 67.422 | 98.864 | 352 | 352 | 352 | |
| SWPV1-041 | SWPV2-047 | CNPV051 | CNPV051 DNase II-like protein | 63.683 | 96.75 | 398 | 408 | 401 | |
| SWPV1-042 | SWPV2-048 | CNPV052 | CNPV052 C-type lectin-like protein | 50 | 100 | 182 | 171 | 171 | |
| SWPV1-043 | FWPV ankyrin repeat protein | 45 | 329 | 406 | |||||
| SWPV1-044 | SWPV2-049 | CNPV053 | CNPV053 conserved hypothetical protein | 68.148 | 100 | 135 | 146 | 146 | |
| SWPV1-045 | SWPV2-050 | CNPV054 | CNPV054 conserved hypothetical protein | 62.59 | 99.286 | 141 | 140 | 140 | |
| SWPV1-046 | SWPV2-051 | CNPV055 | CNPV055 conserved hypothetical protein | 74.534 | 100 | 162 | 163 | 163 | |
| SWPV1-047 | SWPV2-052 | CNPV056 | CNPV056 dUTPase | 80.986 | 98.621 | 155 | 145 | 145 | |
| SWPV1-048 | SWPV2-053 | CNPV057 | CNPV057 putative serpin | 63.107 | 99.02 | 301 | 306 | 306 | |
| SWPV1-049 | SWPV2-054 | CNPV058 | CNPV058 bcl-2 like protein | 51.744 | 98.857 | 174 | 180 | 175 | |
| SWPV1-050 | SWPV2-055 | CNPV059 | CNPV059 putative serpin | 71.302 | 99.704 | 338 | 338 | 338 | |
| SWPV1-051 | SWPV2-056 | CNPV060 | CNPV060 conserved hypothetical protein | 46.939 | 95.098 | 236 | 206 | 316 | SWPV2: Large internal deletion, Translated but not likely functional |
| SWPV1-052 | SWPV2-057 | CNPV061 | CNPV061 DNA ligase | 80.995 | 98.761 | 567 | 565 | 565 | |
| SWPV1-053 | SWPV2-058 | CNPV062 | CNPV062 putative serpin | 70.94 | 100 | 349 | 350 | 350 | |
| SWPV1-054 | SWPV2-059 | CNPV063 | CNPV063 hydroxysteroid dehydrogenase-like protein | 71.348 | 99.441 | 359 | 358 | 358 | |
| SWPV1-055 | SWPV2-060 | CNPV064 | CNPV064 TGF-beta-like protein | 56.897 | 98.587 | 272 | 283 | 282 | |
| SWPV1-056 | SWPV2-061 | CNPV065 | CNPV065 semaphorin-like protein | 69.735 | 99.485 | 573 | 583 | 583 | |
| SWPV1-057 | SWPV2-062 | CNPV066 | CNPV066 hypothetical protein | 37.349 | 98.519 | 139 | 399 | 405 | SWPV1: Low BLAST hits, possible unique ORF |
| SWPV2-063 | CNPV067 | CNPV067 hypothetical protein | 100 | 57 | 57 | ||||
| SWPV1-058 | no significant BLAST hits | 239 | SWPV1: Possible Unique ORF | ||||||
| SWPV1-059 | SWPV2-064 | CNPV068 | CNPV068 GNS1/SUR4-like protein | 84.825 | 99.611 | 257 | 257 | 257 | |
| SWPV1-060 | SWPV2-065 | CNPV069 | CNPV069 late transcription factor VLTF-2 | 87.5 | 100 | 154 | 155 | 155 | |
| SWPV1-061 | SWPV2-066 | CNPV070 | CNPV070 putative rifampicin resistance protein, IMV assembly | 88.065 | 100 | 553 | 551 | 551 | |
| SWPV1-062 | SWPV2-067 | CNPV071 | CNPV071 mRNA capping enzyme small subunit | 89.273 | 100 | 289 | 289 | 289 | |
| SWPV2-068 | CNPV072 | CNPV072 CC chemokine-like protein | 96.262 | 132 | 312 | SWPV2: N-terminus fragment | |||
| SWPV1-063 | SWPV2-069 | CNPV073 | CNPV073 hypothetical protein | 45.263 | 100 | 110 | 109 | 109 | |
| SWPV1-064 | SWPV2-070 | CNPV074 | CNPV074 NPH-I, transcription termination factor | 92.756 | 99.685 | 635 | 635 | 635 | |
| SWPV1-065 | SWPV2-071 | CNPV075 | CNPV075 mutT motif putative gene expression regulator | 79.295 | 100 | 226 | 228 | 230 | |
| SWPV1-066 | SWPV2-072 | CNPV076 | CNPV076 mutT motif | 84.549 | 99.569 | 233 | 232 | 232 | |
| CNPV077 | CNPV077 hypothetical protein | 78 | |||||||
| SWPV1-067 | CNPV011 ankyrin repeat protein | 29.806 | 435 | 586 | |||||
| SWPV1-068 | SWPV2-073 | CNPV078 | CNPV078 RNA polymerase subunit RPO18 | 82.39 | 100 | 161 | 160 | 160 | |
| SWPV2-074 | CNPV079 | CNPV079 Ig-like domain protein | 94.161 | 274 | 272 | ||||
| SWPV1-069 | SWPV2-075 | CNPV080 | CNPV080 early transcription factor small subunit VETFS | 96.682 | 100 | 633 | 633 | 633 | |
| SWPV2-076 | CNPV081 | CNPV081 Ig-like domain protein | 97.006 | 334 | 333 | ||||
| SWPV1-070 | SWPV2-077 | CNPV082 | CNPV082 NTPase, DNA replication | 88.818 | 99.748 | 790 | 794 | 794 | |
| SWPV1-071 | SWPV2-078 | CNPV083 | CNPV083 CC chemokine-like protein | 60.352 | 91.855 | 223 | 221 | 221 | |
| SWPV1-072 | CNPV215 CC chemokine-like protein | 30.994 | 195 | 204 | |||||
| SWPV1-073 | SWPV2-079 | CNPV084 | CNPV084 uracil DNA glycosylase | 86.364 | 97.706 | 220 | 218 | 218 | |
| SWPV1-074 | SWPV2-080 | CNPV085 | CNPV085 putative RNA phosphatase | 67.895 | 74.312 | 245 | 303 | 403 | SWPV2: High SNP Density |
| SWPV1-075 | CNPV216 conserved hypothetical protein | 39.225 | 398 | 404 | |||||
| SWPV1-076 | SWPV2-081 | CNPV086 | CNPV086 TNFR-like protein | 67.327 | 71.569 | 103 | 112 | 117 | |
| SWPV2-082 | CNPV087 | CNPV087 putative glutathione peroxidase | 98.473 | 131 | 198 | SWPV2: C-terminus fragment, not likely translated | |||
| SWPV1-077 | CNPV227 N1R/p28-like protein | 74.638 | 256 | 359 | |||||
| SWPV1-078 | SWPV2-083 | CNPV088 | CNPV088 conserved hypothetical protein | 55.769 | 97 | 104 | 100 | 100 | |
| SWPV1-079 | SWPV2-084 | CNPV089 | CNPV089 conserved hypothetical protein | 64.935 | 100 | 164 | 159 | 159 | |
| SWPV1-080 | SWPV2-085 | CNPV090 | CNPV090 conserved hypothetical protein | 62.393 | 100 | 124 | 127 | 127 | |
| SWPV1-081 | SWPV2-086 | CNPV091 | CNPV091 HT motif protein | 64.634 | 100 | 77 | 83 | 83 | |
| SWPV1-082 | SWPV2-087 | CNPV092 | CNPV092 conserved hypothetical protein | 64.901 | 97.945 | 140 | 146 | 146 | |
| SWPV1-083 | SWPV2-088 | CNPV093 | CNPV093 virion protein | 60.37 | 99.625 | 270 | 267 | 267 | |
| SWPV1-084 | SWPV2-089 | CNPV094 | CNPV094 T10-like protein | 75 | 98.909 | 282 | 275 | 275 | |
| SWPV1-085 | SWPV2-090 | CNPV095 | CNPV095 conserved hypothetical protein | 71.111 | 100 | 47 | 45 | 45 | |
| SWPV1-086 | SWPV2-091 | CNPV096 | CNPV096 ubiquitin | 100 | 100 | 77 | 85 | 85 | |
| SWPV1-087 | SWPV2-092 | CNPV097 | CNPV097 conserved hypothetical protein | 70.031 | 99.705 | 298 | 339 | 339 | |
| SWPV1-088 | SWPV2-093 | CNPV098 | CNPV098 hypothetical protein | 67.442 | 98.75 | 61 | 80 | 80 | |
| SWPV1-089 | SWPV2-094 | CNPV099 | CNPV099 beta-NGF-like protein | 62.162 | 97.949 | 186 | 195 | 195 | |
| SWPV1-090 | SWPV2-095 | CNPV100 | CNPV100 putative interleukin binding protein | 51.176 | 98.225 | 211 | 168 | 169 | |
| SWPV2-096 | CNPV101 | CNPV101 hypothetical protein | 98.824 | 85 | 85 | ||||
| SWPV1-091 | SWPV2-097 | CNPV102 | CNPV102 conserved hypothetical protein | 54.167 | 99.048 | 102 | 105 | 105 | |
| SWPV1-092 | SWPV2-098 | CNPV103 | CNPV103 N1R/p28-like protein | 62.304 | 98.947 | 188 | 190 | 190 | |
| SWPV1-093 | SWPV2-099 | CNPV104 | CNPV104 putative glutaredoxin 2, virion morphogenesis | 86.4 | 99.2 | 125 | 125 | 125 | |
| SWPV1-094 | SWPV2-100 | CNPV105 | CNPV105 conserved hypothetical protein | 77.35 | 98.718 | 234 | 234 | 234 | |
| SWPV1-095 | SWPV2-101 | CNPV106 | CNPV106 putative elongation factor | 76.829 | 98.039 | 103 | 102 | 102 | |
| SWPV2-102 | CNPV107 | CNPV107 hypothetical protein | 100 | 77 | 77 | ||||
| SWPV1-096 | PEPV083 transforming growth factor B | 64 | 444 | 336 | |||||
| SWPV1-097 | SWPV2-103 | CNPV108 | CNPV108 putative metalloprotease, virion morphogenesis | 85.489 | 100 | 633 | 632 | 632 | |
| SWPV1-098 | SWPV2-104 | CNPV109 | CNPV109 NPH-II, RNA helicase | 86.05 | 99.706 | 681 | 681 | 681 | |
| SWPV1-099 | SWPV2-105 | CNPV110 | CNPV110 virion core proteinase | 87.441 | 99.763 | 421 | 422 | 422 | |
| SWPV1-100 | SWPV2-106 | CNPV111 | CNPV111 DNA-binding protein | 80.612 | 99.488 | 391 | 391 | 391 | |
| SWPV1-101 | SWPV2-107 | CNPV112 | CNPV112 putative IMV membrane protein | 81.481 | 100 | 81 | 81 | 81 | |
| SWPV1-102 | SWPV2-108 | CNPV113 | CNPV113 thymidine kinase | 75.978 | 99.441 | 181 | 179 | 179 | |
| SWPV1-103 | SWPV2-109 | CNPV114 | CNPV114 HT motif protein | 69.62 | 100 | 79 | 82 | 82 | |
| SWPV1-104 | SWPV2-110 | CNPV115 | CNPV115 DNA-binding phosphoprotein | 71.429 | 82.353 | 282 | 289 | 289 | SWPV2: High SNP density |
| SWPV1-105 | SWPV2-111 | CNPV116 | CNPV116 unnamed protein product | 73.913 | 98.551 | 66 | 69 | 69 | |
| SWPV1-106 | SWPV2-112 | CNPV117 | CNPV117 DNA-binding virion protein | 88.854 | 99.677 | 314 | 310 | 310 | |
| SWPV1-107 | SWPV2-113 | CNPV118 | CNPV118 conserved hypothetical protein | 75.762 | 99.387 | 656 | 652 | 653 | |
| SWPV1-108 | SWPV2-114 | CNPV119 | CNPV119 virion core protein | 83.969 | 100 | 131 | 131 | 131 | |
| SWPV1-109 | SWPV2-115 | CNPV120 | CNPV120 putative IMV redox protein, virus assembly | 80.851 | 100 | 94 | 93 | 93 | |
| SWPV1-110 | SWPV2-116 | CNPV121 | CNPV121 DNA polymerase | 89.17 | 99.899 | 988 | 988 | 988 | |
| SWPV1-111 | CNPV122 | CNPV122 putative membrane protein | 83.088 | 273 | 274 | ||||
| SWPV1-112 | SWPV2-117 | CNPV123 | CNPV123 conserved hypothetical protein | 82.312 | 85.336 | 571 | 502 | 571 | SWPV2: High SNP density |
| SWPV1-113 | SWPV2-118 | CNPV124 | CNPV124 variola B22R-like protein | 67 | 98.957 | 1906 | 1916 | 1918 | |
| SWPV1-114 | SWPV2-119 | CNPV125 | CNPV125 variola B22R-like protein | 71.669 | 99.66 | 1742 | 1767 | 1767 | |
| SWPV1-115 | SWPV2-120 | CNPV126 | CNPV126 variola B22R-like protein | 64.456 | 98.847 | 1902 | 1839 | 1951 | SWPV2: N-terminus fragment |
| SWPV2-121 | CNPV126 variola B22R-like protein | 96 | 153 | 1951 | SWPV2: C-terminus fragment, not likely translated | ||||
| SWPV1-116 | SWPV2-122 | CNPV127 | CNPV127 RNA polymerase subunit RPO30 | 96.154 | 100 | 182 | 182 | 182 | |
| SWPV1-117 | SWPV2-123 | CNPV128 | CNPV128 conserved hypothetical protein | 77.072 | 98.752 | 742 | 721 | 721 | SWPV2: High SNP Density |
| SWPV1-118 | SWPV2-124 | CNPV129 | CNPV129 poly(A) polymerase large subunit PAPL | 83.898 | 99.788 | 472 | 472 | 472 | |
| SWPV1-119 | SWPV2-125 | CNPV130 | CNPV130 DNA-binding virion core protein | 76.471 | 100 | 114 | 119 | 119 | |
| SWPV1-120 | SWPV2-126 | CNPV131 | CNPV131 conserved hypothetical protein | 64.115 | 99.517 | 212 | 207 | 207 | |
| SWPV1-121 | SWPV2-127 | CNPV132 | CNPV132 conserved hypothetical protein | 81.081 | 99.324 | 151 | 148 | 148 | |
| SWPV1-122 | SWPV2-128 | CNPV133 | CNPV133 conserved hypothetical protein | 73.737 | 100 | 90 | 99 | 99 | |
| SWPV1-123 | SWPV2-129 | CNPV134 | CNPV134 variola B22R-like protein | 65.517 | 99.001 | 1774 | 1801 | 1801 | |
| SWPV1-124 | SWPV2-130 | CNPV135 | CNPV135 putative palmitylated EEV envelope lipase | 89.418 | 99.735 | 378 | 378 | 378 | |
| SWPV1-125 | SWPV2-131 | CNPV136 | CNPV136 putative EEV maturation protein | 75.602 | 99.68 | 622 | 625 | 625 | |
| SWPV1-126 | SWPV2-132 | CNPV137 | CNPV137 conserved hypothetical protein | 62.26 | 98.925 | 467 | 462 | 465 | |
| SWPV1-127 | SWPV2-133 | CNPV138 | CNPV138 putative serine/threonine protein kinase, virus assembly | 83.632 | 100 | 445 | 444 | 444 | |
| SWPV1-128 | SWPV2-134 | CNPV139 | CNPV139 conserved hypothetical protein | 81.69 | 100 | 213 | 213 | 213 | |
| SWPV1-129 | SWPV2-135 | CNPV140 | CNPV140 conserved hypothetical protein | 78.788 | 100 | 65 | 66 | 66 | |
| SWPV1-130 | SWPV2-136 | CNPV141 | CNPV141 HAL3-like domain protein | 88.333 | 100 | 182 | 184 | 184 | |
| SWPV1-131 | no significant BLAST hits | 28 | 101 | 571 | SWPV1: Possible Unique ORF | ||||
| SWPV1-132 | SWPV2-137 | CNPV142 | CNPV142 N1R/p28-like protein | 48.266 | 98.442 | 314 | 321 | 321 | |
| SWPV1-133 | SWPV2-138 | CNPV143 | CNPV143 ankyrin repeat protein | 54.103 | 98.361 | 634 | 671 | 671 | |
| SWPV1-134 | SWPV2-139 | CNPV144 | CNPV144 ankyrin repeat protein | 59.011 | 99.281 | 562 | 556 | 556 | |
| SWPV1-135 | SWPV2-140 | CNPV145 | CNPV145 conserved hypothetical protein | 75.814 | 100 | 439 | 440 | 440 | |
| SWPV1-136 | SWPV2-141 | CNPV146 | CNPV146 RNA polymerase subunit RPO7 | 88.525 | 100 | 66 | 62 | 62 | |
| SWPV1-137 | SWPV2-142 | CNPV147 | CNPV147 conserved hypothetical protein | 80.851 | 100 | 188 | 188 | 188 | |
| SWPV1-138 | SWPV2-143 | CNPV148 | CNPV148 virion core protein | 86.533 | 100 | 347 | 348 | 348 | |
| SWPV2-144 | CNPV149 | CNPV149 putative thioredoxin binding protein | 99.673 | 306 | 306 | ||||
| CNPV150 | CNPV150 ankyrin repeat protein | 351 | |||||||
| SWPV2-145 | CNPV151 | CNPV151 ankyrin repeat protein | 99.029 | 412 | 412 | ||||
| SWPV2-146 | CNPV152 | CNPV152 hypothetical protein | 98 | 149 | 187 | SWPV2: C-terminus fragment, not likely translated | |||
| SWPV2-147 | CNPV153 | CNPV153 Rep-like protein | 99.359 | 312 | 312 | ||||
| SWPV1-139 | CNPV159 N1R/p28-like protein | 78.488 | 333 | 337 | |||||
| SWPV1-140 | FWPV121 CC chemokine-like protein | 46 | 93 | 121 | |||||
| SWPV1-141 | SWPV2-148 | CNPV154 | CNPV154 variola B22R-like protein | 90.067 | 98.286 | 1939 | 875 | 1928 | SWPV2: N-terminus fragment/SWPV1: Low SNP Density |
| SWPV1-142 | SWPV2-149 | CNPV155 | CNPV155 variola B22R-like protein | 82.427 | 99.454 | 1810 | 1831 | 1830 | |
| SWPV2-150 | CNPV156 | CNPV156 hypothetical protein | 96.287 | 834 | 832 | ||||
| SWPV2-151 | CNPV157 | CNPV157 TGF-beta-like protein | 87.679 | 343 | 349 | ||||
| CNPV158 | CNPV158 TGF-beta-like protein | 172 | |||||||
| CNPV159 | CNPV159 N1R/p28-like protein | 337 | |||||||
| CNPV160 | CNPV160 N1R/p28-like protein | 396 | |||||||
| SWPV2-152 | CNPV161 | CNPV161 TGF-beta-like protein | 99.441 | 358 | 358 | ||||
| SWPV2-153 | CNPV162 | CNPV162 TGF-beta-like protein | 97.987 | 149 | 149 | ||||
| CNPV163 | CNPV163 hypothetical protein | 92 | |||||||
| CNPV164 | CNPV164 hypothetical protein | 98 | |||||||
| SWPV2-154 | CNPV165 | CNPV165 N1R/p28-like protein | 98.75 | 320 | 346 | SWPV2: C-terminus fragment, not likely translated | |||
| SWPV1-143 | SWPV2-155 | CNPV166 | CNPV166 Ig-like domain protein | 96.812 | 95.652 | 345 | 345 | 345 | SWPV1: Low SNP Density |
| SWPV1-144 | SWPV2-156 | CNPV167 | CNPV167 Ig-like domain protein | 94.767 | 88.372 | 172 | 168 | 171 | SWPV1: Low SNP Density |
| SWPV2-157 | CNPV168 | CNPV168 N1R/p28-like protein | 96 | 350 | 358 | ||||
| SWPV1-145 | CNPV169 | CNPV169 N1R/p28-like protein | 83.578 | 337 | 332 | SWPV1: CNPV-168/169 Fusion | |||
| SWPV1-146 | SWPV2-158 | CNPV170 | CNPV170 thymidylate kinase | 100 | 100 | 121 | 212 | 212 | SWPV1: N-terminus fragment |
| SWPV1-147 | SWPV2-159 | CNPV171 | CNPV171 late transcription factor VLTF-1 | 96.923 | 100 | 260 | 260 | 260 | |
| SWPV1-148 | SWPV2-160 | CNPV172 | CNPV172 putative myristylated protein | 83.125 | 99.403 | 336 | 335 | 335 | |
| SWPV1-149 | SWPV2-161 | CNPV173 | CNPV173 putative myristylated IMV envelope protein | 91.358 | 98.354 | 243 | 243 | 243 | |
| SWPV1-150 | SWPV2-162 | CNPV174 | CNPV174 conserved hypothetical protein | 47.917 | 100 | 96 | 96 | 96 | |
| SWPV1-151 | SWPV2-163 | CNPV175 | CNPV175 conserved hypothetical protein | 84.158 | 100 | 303 | 303 | 303 | |
| SWPV1-152 | SWPV2-164 | CNPV176 | CNPV176 DNA-binding virion core protein | 87.747 | 100 | 253 | 252 | 252 | |
| SWPV1-153 | SWPV2-165 | CNPV177 | CNPV177 conserved hypothetical protein | 84.733 | 100 | 131 | 130 | 130 | |
| SWPV1-154 | SWPV2-166 | CNPV178 | CNPV178 putative IMV membrane protein | 85.135 | 100 | 148 | 148 | 148 | |
| SWPV1-155 | SWPV2-167 | CNPV179 | CNPV179 poly(A) polymerase small subunit PAPS | 88.667 | 100 | 300 | 302 | 302 | |
| SWPV1-156 | SWPV2-168 | CNPV180 | CNPV180 RNA polymerase subunit RPO22 | 87.634 | 99.462 | 186 | 186 | 186 | |
| SWPV1-157 | SWPV2-169 | CNPV181 | CNPV181 conserved hypothetical protein | 82.353 | 100 | 136 | 136 | 136 | |
| SWPV1-158 | SWPV2-170 | CNPV182 | CNPV182 RNA polymerase subunit RPO147 | 93.866 | 99.922 | 1288 | 1288 | 1288 | |
| SWPV1-159 | SWPV2-171 | CNPV183 | CNPV183 putative protein-tyrosine phosphatase, virus assembly | 85.542 | 100 | 166 | 166 | 166 | |
| SWPV1-160 | SWPV2-172 | CNPV184 | CNPV184 conserved hypothetical protein | 91.534 | 100 | 190 | 189 | 189 | |
| SWPV1-161 | SWPV2-173 | CNPV185 | CNPV185 ankyrin repeat protein | 32.632 | 96.341 | 337 | 328 | 328 | |
| SWPV1-162 | SWPV2-174 | CNPV186 | CNPV186 IMV envelope protein | 100 | 100 | 329 | 330 | 330 | |
| SWPV1-163 | SWPV2-175 | CNPV187 | CNPV187 RNA polymerase associated protein RAP94 | 91.114 | 99.75 | 799 | 799 | 799 | |
| SWPV1-164 | SWPV2-176 | CNPV188 | CNPV188 late transcription factor VLTF-4 | 70.115 | 92.941 | 170 | 170 | 170 | |
| SWPV1-165 | SWPV2-177 | CNPV189 | CNPV189 DNA topoisomerase | 88.608 | 99.684 | 316 | 316 | 316 | |
| SWPV1-166 | SWPV2-178 | CNPV190 | CNPV190 conserved hypothetical protein | 77.124 | 99.346 | 153 | 153 | 153 | |
| SWPV1-167 | SWPV2-179 | CNPV191 | CNPV191 conserved hypothetical protein | 70.874 | 99.029 | 103 | 103 | 103 | |
| SWPV1-168 | SWPV2-180 | CNPV192 | CNPV192 mRNA capping enzyme large subunit | 88.221 | 99.764 | 848 | 846 | 846 | |
| SWPV1-169 | SWPV2-181 | CNPV193 | CNPV193 HT motif protein | 72.619 | 100 | 104 | 106 | 106 | |
| SWPV1-170 | SWPV2-182 | CNPV194 | CNPV194 virion protein | 71.223 | 100 | 139 | 140 | 140 | |
| SWPV1-171 | SWPV2-183 | CNPV195 | CNPV195 hypothetical protein | 51.2 | 98.611 | 139 | 144 | 144 | |
| SWPV1-172 | SWPV2-184 | CNPV196 | CNPV196 conserved hypothetical protein | 62.963 | 100 | 189 | 190 | 190 | |
| SWPV1-173 | SWPV2-185 | CNPV197 | CNPV197 N1R/p28-like protein | 61.679 | 97.818 | 279 | 275 | 275 | |
| SWPV1-174 | SWPV2-186 | CNPV198 | CNPV198 C-type lectin-like protein | 55.844 | 99.359 | 159 | 156 | 156 | |
| SWPV1-175 | SWPV2-187 | CNPV199 | CNPV199 deoxycytidine kinase-like protein | 79.111 | 100 | 222 | 225 | 225 | |
| SWPV1-176 | SWPV2-188 | CNPV200 | CNPV200 Rep-like protein | 72.903 | 97.59 | 152 | 166 | 166 | |
| SWPV1-177 | SWPV2-189 | CNPV201 | CNPV201 conserved hypothetical protein | 60 | 97.661 | 197 | 167 | 192 | |
| SWPV1-178 | SWPV2-190 | CNPV202 | CNPV202 N1R/p28-like protein | 69.203 | 99.638 | 275 | 276 | 276 | |
| SWPV1-179 | SWPV2-191 | CNPV203 | CNPV203 N1R/p28-like protein | 64.935 | 99.738 | 380 | 382 | 382 | |
| SWPV1-180 | SWPV2-192 | CNPV204 | CNPV204 conserved hypothetical protein | 53.226 | 100 | 53 | 61 | 61 | |
| SWPV1-181 | SWPV2-193 | CNPV205 | CNPV205 N1R/p28-like protein | 71.885 | 99.371 | 317 | 318 | 318 | |
| SWPV1-182 | SWPV2-194 | CNPV206 | CNPV206 putative photolyase | 84.989 | 99.364 | 464 | 472 | 472 | |
| SWPV1-183 | CNPV081 Ig-like domain protein | 53.988 | 332 | 333 | |||||
| SWPV1-184 | SWPV2-195 | CNPV207 | CNPV207 N1R/p28-like protein | 64.535 | 98.235 | 193 | 173 | 183 | |
| SWPV1-185 | SWPV2-196 | CNPV208 | CNPV208 conserved hypothetical protein | 52.239 | 97.5 | 172 | 200 | 200 | |
| SWPV1-186 | SWPV2-197 | CNPV209 | CNPV209 N1R/p28-like protein | 65.686 | 100 | 311 | 310 | 310 | |
| SWPV1-187 | SWPV2-198 | CNPV210 | CNPV210 N1R/p28-like protein | 74.419 | 99.237 | 130 | 131 | 131 | |
| SWPV1-188 | SWPV2-199 | CNPV211 | CNPV211 conserved hypothetical protein | 49.02 | 98.148 | 54 | 54 | 54 | |
| SWPV1-189 | SWPV2-200 | CNPV212 | CNPV212 N1R/p28-like protein | 76.136 | 98.295 | 175 | 176 | 176 | |
| SWPV1-190 | no significant BLAST hits | 70 | SWPV1: Possible Unique ORF | ||||||
| SWPV1-191 | SWPV2-201 | CNPV213 | CNPV213 deoxycytidine kinase-like protein | 58.768 | 99.539 | 216 | 216 | 217 | |
| SWPV2-202 | CNPV214 | CNPV214 vaccinia C4L/C10L-like protein | 99.438 | 356 | 356 | ||||
| SWPV1-192 | CNPV012 conserved hypothetical protein | 37.41 | 165 | 189 | |||||
| SWPV1-193 | CNPV223 ankyrin repeat protein | 31.579 | 674 | 847 | |||||
| SWPV1-194 | SWPV2-203 | CNPV215 | CNPV215 CC chemokine-like protein | 49.751 | 96.078 | 202 | 204 | 204 | |
| SWPV2-204 | CNPV216 | CNPV216 conserved hypothetical protein | 98.762 | 401 | 404 | ||||
| SWPV2-205 | CNPV217 | CNPV217 N1R/p28-like protein | 95.152 | 330 | 330 | ||||
| SWPV1-195 | CNPV223 ankyrin repeat protein | 38.474 | 729 | 847 | SWPV1: N-terminus fragment | ||||
| SWPV1-196 | SWPV2-206 | CNPV218 | CNPV218 N1R/p28-like protein | 66.667 | 99.522 | 318 | 223 | 437 | SWPV2: N-terminus fragment |
| SWPV1-197 | CNPV228 N1R/p28-like protein | 53 | 161 | 371 | SWPV1: N-terminus fragment | ||||
| SWPV1-198 | CNPV160 N1R/p28-like protein | 79.293 | 367 | 396 | SWPV1: Fragment/CNPV-220/221 Fusion | ||||
| SWPV1-199 | CNPV160 N1R/p28-like protein | 66.582 | 360 | 396 | SWPV1: Paralog to SWPV1-198? | ||||
| SWPV1-200 | CNPV161 TGF-beta-like protein | 36.882 | 256 | 358 | |||||
| SWPV1-201 | CNPV162 TGF-beta-like protein | 50 | 141 | 149 | |||||
| SWPV1-202 | no significant BLAST hits | 98 | SWPV1: Possible Unique ORF | ||||||
| SWPV2-207 | CNPV219 | CNPV219 N1R/p28-like protein | 99.713 | 349 | 349 | ||||
| SWPV2-208 | CNPV220 | CNPV220 N1R/p28-like protein | 80.263 | 85 | 178 | SWPV2: N-terminus fragment | |||
| SWPV2-209 | CNPV221 | CNPV221 N1R/p28-like protein | 94.231 | 213 | 281 | SWPV2: N-terminus fragment | |||
| SWPV2-210 | CNPV222 | CNPV222 N1R/p28-like protein | 99.649 | 285 | 285 | ||||
| SWPV2-211 | CNPV223 | CNPV223 ankyrin repeat protein | 98.819 | 847 | 847 | ||||
| SWPV1-203 | SWPV2-212 | CNPV224 | CNPV224 hypothetical protein | 50.382 | 100 | 126 | 239 | 239 | |
| SWPV2-213 | CNPV225 | CNPV225 N1R/p28-like protein | 74.038 | 94 | 159 | SWPV2: N-terminus fragment | |||
| SWPV2-214 | CNPV226 | CNPV226 N1R/p28-like protein | 96.825 | 126 | 134 | ||||
| CNPV227 | CNPV227 N1R/p28-like protein | 359 | |||||||
| CNPV228 | CNPV228 N1R/p28-like protein | 371 | |||||||
| SWPV1-204 | SWPV2-215 | CNPV229 | CNPV229 ankyrin repeat protein | 44.498 | 97.926 | 423 | 434 | 434 | |
| SWPV2-216 | CNPV230 | CNPV230 hypothetical protein | 98.462 | 65 | 65 | ||||
| SWPV1-205 | SWPV2-217 | CNPV231 | CNPV231 MyD116-like domain protein | 72.222 | 98.101 | 100 | 158 | 158 | SWPV1: large in-frame deletions |
| SWPV1-206 | SWPV2-218 | CNPV232 | CNPV232 CC chemokine-like protein | 59.024 | 93.137 | 205 | 204 | 204 | |
| SWPV1-207 | SWPV2-219 | CNPV233 | CNPV233 ankyrin repeat protein | 56.936 | 99.788 | 476 | 471 | 471 | |
| SWPV2-220 | CNPV234 | CNPV234 ankyrin repeat protein | 100 | 508 | 508 | SWPV2: High SNP Density | |||
| SWPV1-208 | PEPV008 vaccinia C4L/C10L-like protein | 55 | 420 | 411 | |||||
| SWPV2-221 | CNPV235 | CNPV235 conserved hypothetical protein | 88.426 | 432 | 432 | ||||
| SWPV1-209 | SWPV2-222 | CNPV236 | CNPV236 ribonucleotide reductase small subunit | 83.282 | 95.666 | 324 | 323 | 323 | |
| SWPV2-223 | CNPV237 | CNPV237 ankyrin repeat protein | 97.732 | 441 | 441 | ||||
| SWPV1-210 | CNPV234 ankyrin repeat protein | 30.545 | 559 | 508 | |||||
| SWPV1-211 | SWPV2-224 | CNPV238 | CNPV238 late transcription factor VLTF-3 | 95.111 | 100 | 225 | 225 | 225 | |
| SWPV1-212 | SWPV2-225 | CNPV239 | CNPV239 virion redox protein | 80.282 | 100 | 72 | 75 | 75 | |
| SWPV1-213 | SWPV2-226 | CNPV240 | CNPV240 virion core protein P4b | 88.788 | 99.848 | 660 | 659 | 659 | |
| SWPV1-214 | SWPV2-227 | CNPV241 | CNPV241 immunodominant virion protein | 47.368 | 99.07 | 242 | 215 | 215 | |
| SWPV1-215 | SWPV2-228 | CNPV242 | CNPV242 RNA polymerase subunit RPO19 | 88.166 | 98.817 | 169 | 169 | 169 | |
| SWPV1-216 | SWPV2-229 | CNPV243 | CNPV243 conserved hypothetical protein | 81.501 | 98.928 | 373 | 373 | 373 | |
| SWPV1-217 | SWPV2-230 | CNPV244 | CNPV244 early transcription factor large subunit VETFL | 95.91 | 100 | 709 | 709 | 709 | |
| SWPV1-218 | SWPV2-231 | CNPV245 | CNPV245 intermediate transcription factor VITF-3 | 90.667 | 99.667 | 300 | 300 | 300 | |
| SWPV1-219 | SWPV2-232 | CNPV246 | CNPV246 putative IMV membrane protein | 80 | 98.667 | 76 | 75 | 75 | |
| SWPV1-220 | SWPV2-233 | CNPV247 | CNPV247 virion core protein P4a | 81.494 | 99.664 | 897 | 893 | 893 | |
| SWPV1-221 | SWPV2-234 | CNPV248 | CNPV248 conserved hypothetical protein | 78.723 | 100 | 281 | 279 | 279 | |
| SWPV1-222 | SWPV2-235 | CNPV249 | CNPV249 virion protein | 74.269 | 99.405 | 167 | 168 | 168 | |
| SWPV1-223 | SWPV2-236 | CNPV250 | CNPV250 conserved hypothetical protein | 36.082 | 94.595 | 73 | 56 | 99 | SWPV2: N-terminus fragment |
| SWPV1-224 | SWPV2-237 | CNPV251 | CNPV251 putative IMV membrane protein | 69.565 | 100 | 69 | 69 | 69 | |
| SWPV1-225 | SWPV2-238 | CNPV252 | CNPV252 putative IMV membrane protein | 68.478 | 98.913 | 92 | 92 | 92 | |
| SWPV1-226 | SWPV2-239 | CNPV253 | CNPV253 putative IMV membrane virulence factor | 73.585 | 98.113 | 53 | 53 | 53 | |
| SWPV1-227 | SWPV2-240 | CNPV254 | CNPV254 conserved hypothetical protein | 75 | 98.958 | 96 | 96 | 96 | |
| SWPV1-228 | SWPV2-241 | CNPV255 | CNPV255 predicted myristylated protein | 84.282 | 99.728 | 368 | 368 | 368 | |
| SWPV1-229 | SWPV2-242 | CNPV256 | CNPV256 putative phosphorylated IMV membrane protein | 81.006 | 100 | 188 | 192 | 192 | |
| SWPV1-230 | SWPV2-243 | CNPV257 | CNPV257 DNA helicase, transcriptional elongation | 87.229 | 99.784 | 462 | 462 | 462 | |
| SWPV1-231 | SWPV2-244 | CNPV258 | CNPV258 conserved hypothetical protein | 77.647 | 100 | 86 | 89 | 89 | |
| SWPV1-232 | SWPV2-245 | CNPV259 | CNPV259 DNA polymerase processivity factor | 81.86 | 100 | 432 | 112 | 434 | |
| SWPV1-233 | SWPV2-246 | CNPV260 | CNPV260 conserved hypothetical protein | 91.071 | 99.77 | 112 | 434 | 112 | |
| SWPV1-234 | SWPV2-247 | CNPV261 | CNPV261 Holliday junction resolvase protein | 80.405 | 100 | 151 | 152 | 152 | |
| SWPV1-235 | SWPV2-248 | CNPV262 | CNPV262 intermediate transcription factor VITF-3 | 86.126 | 100 | 383 | 383 | 383 | |
| SWPV1-236 | SWPV2-249 | CNPV263 | CNPV263 RNA polymerase subunit RPO132 | 94.301 | 100 | 1158 | 1157 | 1157 | |
| SWPV1-237 | SWPV2-250 | CNPV264 | CNPV264 A type inclusion-like protein | 81.015 | 99.502 | 602 | 601 | 603 | |
| SWPV1-238 | SWPV2-251 | CNPV265 | CNPV265 A type inclusion-like/fusion protein | 67.015 | 99.789 | 471 | 475 | 475 | |
| SWPV1-239 | SWPV2-252 | CNPV266 | CNPV266 conserved hypothetical protein | 89.286 | 99.286 | 140 | 140 | 140 | |
| SWPV1-240 | SWPV2-253 | CNPV267 | CNPV267 RNA polymerase subunit RPO35 | 77.558 | 99.016 | 303 | 305 | 305 | |
| SWPV1-241 | SWPV2-254 | CNPV268 | CNPV268 conserved hypothetical protein | 73.529 | 100 | 72 | 75 | 75 | |
| SWPV1-242 | SWPV2-255 | CNPV269 | CNPV269 conserved hypothetical protein | 70.796 | 100 | 113 | 113 | 113 | |
| SWPV1-243 | SWPV2-256 | CNPV270 | CNPV270 conserved hypothetical protein | 70.588 | 100 | 119 | 120 | 120 | |
| SWPV1-244 | SWPV2-257 | CNPV271 | CNPV271 DNA packaging protein | 89.963 | 99.648 | 272 | 284 | 284 | |
| SWPV1-245 | SWPV2-258 | CNPV272 | CNPV272 C-type lectin-like EEV protein | 76.136 | 99.448 | 182 | 181 | 181 | |
| SWPV1-246 | CNPV012 conserved hypothetical protein | 30.147 | 172 | 189 | |||||
| SWPV1-247 | SWPV2-259 | CNPV273 | CNPV273 conserved hypothetical protein | 62.816 | 99.635 | 276 | 274 | 274 | |
| SWPV1-248 | SWPV2-260 | CNPV274 | CNPV274 putative tyrosine protein kinase | 63.197 | 99.628 | 286 | 269 | 269 | |
| SWPV1-249 | SWPV2-261 | CNPV275 | CNPV275 putative serpin | 72.271 | 99.408 | 340 | 338 | 338 | |
| SWPV1-250 | SWPV2-262 | CNPV276 | CNPV276 conserved hypothetical protein | 56.667 | 100 | 227 | 252 | 252 | |
| SWPV1-251 | SWPV2-263 | CNPV277 | CNPV277 G protein-coupled receptor-like protein | 90 | 99.677 | 310 | 310 | 310 | |
| SWPV1-252 | SWPV2-264 | CNPV278 | CNPV278 conserved hypothetical protein | 89.691 | 98.958 | 97 | 96 | 96 | |
| SWPV1-253 | SWPV2-265 | CNPV279 | CNPV279 beta-NGF-like protein | 63.415 | 100 | 167 | 169 | 169 | |
| SWPV1-254 | SWPV2-266 | CNPV280 | CNPV280 HT motif protein | 67.692 | 99.231 | 134 | 130 | 130 | |
| SWPV1-255 | SWPV2-267 | CNPV281 | CNPV281 conserved hypothetical protein | 71.728 | 99.533 | 192 | 214 | 214 | |
| SWPV1-256 | SWPV2-268 | CNPV282 | CNPV282 HT motif protein | 71.552 | 100 | 118 | 120 | 120 | |
| SWPV1-257 | SWPV2-269 | CNPV283 | CNPV283 CC chemokine-like protein | 63.208 | 100 | 110 | 111 | 111 | |
| SWPV1-258 | SWPV2-270 | CNPV284 | CNPV284 putative interleukin binding protein | 37.405 | 90.769 | 192 | 193 | 195 | |
| SWPV1-259 | SWPV2-271 | CNPV285 | CNPV285 EGF-like protein | 62.992 | 99.206 | 123 | 126 | 126 | |
| SWPV1-260 | SWPV2-272 | CNPV286 | CNPV286 putative serine/threonine protein kinase | 76.744 | 99.672 | 303 | 305 | 305 | |
| SWPV1-261 | SWPV2-273 | CNPV287 | CNPV287 conserved hypothetical protein | 73.248 | 98.758 | 165 | 160 | 161 | |
| SWPV1-262 | SWPV2-274 | CNPV288 | CNPV288 C-type lectin-like protein | 52.414 | 88.435 | 163 | 147 | 147 | |
| SWPV1-263 | SWPV2-275 | CNPV289 | CNPV289 putative interleukin binding protein | 58.993 | 99.281 | 132 | 139 | 139 | |
| SWPV1-264 | SWPV2-276 | CNPV290 | CNPV290 conserved hypothetical protein | 84 | 83.784 | 75 | 75 | 75 | |
| SWPV1-265 | SWPV2-277 | CNPV291 | CNPV291 ankyrin repeat protein | 48.067 | 98.99 | 613 | 594 | 594 | |
| SWPV1-266 | SWPV2-278 | CNPV292 | CNPV292 hypothetical protein | 37.209 | 100 | 101 | 74 | 74 | |
| SWPV1-267 | SWPV2-279 | CNPV293 | CNPV293 ankyrin repeat protein | 55.634 | 99.648 | 305 | 284 | 284 | |
| SWPV1-268 | SWPV2-280 | CNPV294 | CNPV294 ankyrin repeat protein | 68.447 | 99.07 | 424 | 430 | 430 | |
| SWPV1-269 | PIPV223 host range protein | 51 | 138 | 143 | |||||
| SWPV1-270 | FWPV217 hypothetical protein | 50 | 330 | 328 | |||||
| SWPV1-271 | SWPV2-281 | CNPV295 | CNPV295 ankyrin repeat protein | 57.736 | 100 | 264 | 396 | 396 | |
| SWPV1-272 | SWPV2-282 | CNPV296 | CNPV296 ankyrin repeat protein | 67.195 | 99.127 | 438 | 458 | 458 | |
| SWPV1-273 | SWPV2-283 | CNPV297 | CNPV297 ankyrin repeat protein | 54.972 | 99.457 | 717 | 737 | 737 | |
| SWPV1-274 | SWPV2-284 | CNPV298 | CNPV298 ankyrin repeat protein | 64.591 | 99.825 | 573 | 571 | 571 | |
| SWPV1-275 | SWPV2-285 | CNPV299 | CNPV299 putative serine/threonine protein kinase | 67.893 | 99.333 | 303 | 300 | 300 | |
| SWPV1-276 | SWPV2-286 | CNPV300 | CNPV300 ankyrin repeat protein | 75.82 | 98.77 | 253 | 244 | 244 | |
| SWPV1-277 | CNPV219 N1R/p28-like protein | 28.467 | 142 | 349 | |||||
| SWPV1-278 | CNPV228 N1R/p28-like protein | 43.038 | 87 | 371 | |||||
| SWPV1-279 | TKPV163 ankyrin repeat protein | 40 | 432 | 434 | |||||
| SWPV1-280 | SWPV2-287 | CNPV301 | CNPV301 ankyrin repeat protein | 59.546 | 99.241 | 510 | 527 | 527 | |
| SWPV1-281 | SWPV2-288 | CNPV302 | CNPV302 conserved hypothetical protein | 45.026 | 100 | 175 | 193 | 193 | |
| SWPV1-282 | SWPV2-289 | CNPV303 | CNPV303 ankyrin repeat protein | 68.938 | 99.4 | 499 | 500 | 500 | |
| SWPV1-283 | SWPV2-290 | CNPV304 | CNPV304 ankyrin repeat protein | 62.105 | 99.785 | 476 | 466 | 466 | |
| SWPV1-284 | SWPV2-291 | CNPV305 | CNPV305 N1R/p28-like protein | 54.545 | 100 | 261 | 262 | 262 | |
| SWPV1-285 | SWPV2-292 | CNPV306 | CNPV306 hypothetical protein | 30.769 | 98.611 | 73 | 72 | 72 | |
| SWPV1-286 | SWPV2-293 | CNPV307 | CNPV307 C-type lectin-like protein | 55.828 | 100 | 165 | 154 | 154 | |
| SWPV1-287 | SWPV2-294 | CNPV308 | CNPV308 ankyrin repeat protein | 58.757 | 99.44 | 359 | 357 | 357 | |
| SWPV1-288 | SWPV2-295 | CNPV309 | CNPV309 ankyrin repeat protein | 69.388 | 100 | 195 | 196 | 196 | |
| SWPV1-289 | SWPV2-296 | CNPV310 | CNPV310 ankyrin repeat protein | 47.359 | 99.255 | 540 | 537 | 537 | |
| SWPV1-290 | SWPV2-297 | CNPV311 | CNPV311 EFc-like protein | 54.4 | 99.194 | 125 | 124 | 124 | |
| SWPV1-291 | SWPV2-298 | CNPV312 | CNPV312 conserved hypothetical protein | 53.704 | 98.795 | 168 | 166 | 166 | |
| SWPV1-292 | SWPV2-299 | CNPV313 | CNPV313 Ig-like domain protein | 69.43 | 98.165 | 213 | 218 | 218 | |
| SWPV1-293 | SWPV2-300 | CNPV314 | CNPV314 ankyrin repeat protein | 71.552 | 99.829 | 580 | 629 | 584 | |
| SWPV1-294 | CNPV011 ankyrin repeat protein | 32 | 513 | 586 | |||||
| SWPV1-295 | SWPV2-301 | CNPV315 | CNPV315 G protein-coupled receptor-like protein | 59.17 | 99.365 | 315 | 315 | 315 | |
| SWPV1-296 | CNPV014 Ig-like domain protein | 59.624 | 230 | 490 | |||||
| SWPV1-297 | CNPV014 Ig-like domain protein | 59.641 | 240 | 490 | |||||
| SWPV1-298 | CNPV015 ankyrin repeat protein | 45.455 | 74 | 528 | |||||
| SWPV1-299 | CNPV150 ankyrin repeat protein | 36.364 | 84 | 351 | |||||
| SWPV1-300 | SWPV2-302 | CNPV316 | CNPV316 ankyrin repeat protein | 35.294 | 99.632 | 162 | 544 | 544 | |
| SWPV2-303 | CNPV317 | CNPV317 hypothetical protein | 100 | 55 | 55 | ||||
| SWPV2-304 | CNPV318 | CNPV318 ankyrin repeat protein | 98.054 | 514 | 514 | ||||
| SWPV2-305 | CNPV319 | CNPV319 ankyrin repeat protein | 97.638 | 637 | 739 | SWPV2: C-terminus fragment, not likely translated | |||
| SWPV1-301 | PIPV253 EFc-like protein | 69 | 124 | 124 | |||||
| SWPV1-302 | CNPV015 ankyrin repeat protein | 45.276 | 520 | 528 | |||||
| SWPV1-303 | CNPV223 ankyrin repeat protein | 40 | 480 | 847 | |||||
| SWPV1-304 | SWPV2-306 | CNPV320 | CNPV320 Ig-like domain protein | 76.858 | 99.787 | 468 | 469 | 469 | |
| SWPV2-307 | CNPV321 | CNPV321 EFc-like protein | 99.194 | 124 | 124 | ||||
| SWPV2-308 | CNPV322 | CNPV322 ankyrin repeat protein | 98.408 | 689 | 690 | ||||
| SWPV1-305 | CNPV035 C-type lectin-like protein | 35.556 | 138 | 134 | |||||
| SWPV1-306 | CNPV008 C-type lectin-like protein | 50 | 174 | 169 | |||||
| SWPV1-307 | SWPV2-309 | CNPV323 | CNPV323 conserved hypothetical protein | 75.61 | 93.651 | 84 | 186 | 182 | |
| SWPV1-308 | SWPV2-310 | CNPV324 | CNPV324 conserved hypothetical protein | 87.387 | 99.55 | 220 | 222 | 222 | |
| SWPV1-309 | CNPV325 | CNPV325 ankyrin repeat protein | 56.458 | 468 | 514 | ||||
| SWPV1-310 | SWPV2-311 | CNPV326 | CNPV326 C-type lectin-like protein | 32.044 | 85.99 | 181 | 208 | 204 | |
| SWPV2-312 | CNPV327 | CNPV327 hypothetical protein | 92.941 | 171 | 171 | ||||
| CNPV328 | CNPV328 hypothetical protein | 72 |
This difference in similarity between the new viruses and CNPV is easily visualized in complete genome dotplots (Fig. 2a and b). Significantly more indels are present in the SWPV-1 vs CNPV dotplot (Fig. 2a). However, when the phylogenetic relationships of these viruses were examined together with the other available complete genomes, SWPV-1 was still part of the CNPV clade (Fig. 3a). From this alignment, CNPV is 99.2%, 78.7%, 69.4%, 69.5%, 68.8% and 66.5% identical (nt) to SWPV-2, SWPV-1, FeP2, PEPV, FWPV and TKPV, respectively. A greater selection of viruses was included in the phylogenetic tree by using other fragments of incompletely sequenced avipoxvirus genomes. For example, Vultur gryphus poxvirus (VGPV), Flamingopox virus (FGPV) and Hawaiian goose poxvirus (HGPV) are all more similar to SWPV-2 and CNPV than SWPV-1 (Fig. 3b), this confirms that other poxviruses are as closely related to CNPV as SWPV-2. By also building phylogenetic trees with partial nucleotide sequences from the p4b gene (Fig. 4) and DNA polymerase gene (Fig. 5), we discovered that several other viruses are within the SWPV-1, SWPV-2 and CNPV clade. This includes a poxvirus isolated from Houbara Bustards (Chlamydotis undulata) in captive-breeding programs in Morocco [23], but named CNPV-morocco, and avipoxviruses isolated from American crow (Corvus brachyrhynchos) and American robin (Turdus migratorius) [24], which is almost identical to CPNV-1 within this relatively small fragment of the genome.
Fig. 2.
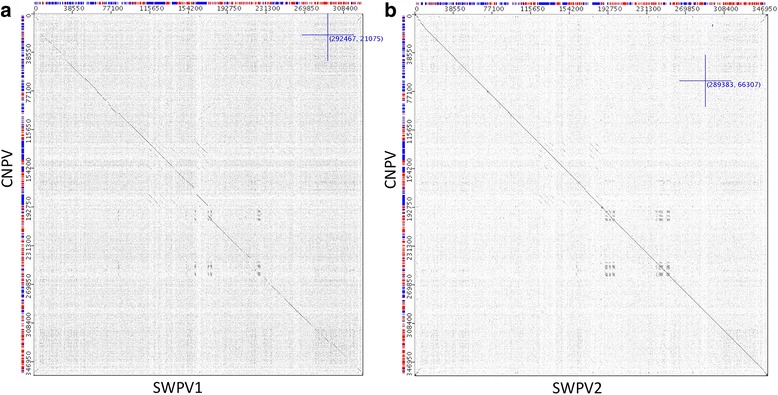
Dotplots of Shearwaterpox viruses (SWPV-1 and 2) vs CNPV genomes. Horizontal sequence: SWPV-1 (a) and SWPV-2 (b), vertical sequence CNPV. Red and blue boxes represent genes transcribed to the right and left of the genome, respectively
Fig. 3.
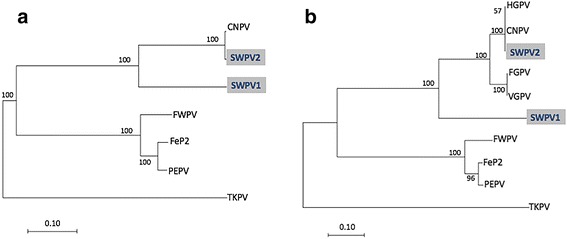
Phylogenetic relationship between Shearwaterpox viruses (SWPV-1 and 2) and other avipoxviruses. a Phylogenetic tree of 173 kbp core region (large gaps removed) from available complete avipoxvirus genomes. b Phylogenetic tree highlighting viruses closely related to CNPV. The sequences were aligned with ClustalO and MEGA7 was used to create a maximum likelihood tree based on the Tamura-Nei method and tested by bootstrapping with 1000 replicates. The abbreviations and GenBank accession details for poxviruses strains were used: Canarypox virus (CNPV; AY318871), Pigeonpox virus (FeP2; KJ801920), Penguinpox virus (PEPV; KJ859677) Fowlpox virus (FWPV; AF198100), Shearwaterpox virus 1 (SWPV-1; KX857216), Shearwaterpox virus 2 (SWPV-2; KX857215), Turkeypox virus (TKPV; NC_028238), Vultur Gryphus poxvirus (VGPV; AY246559), Flamingopox virus (FGPV; HQ875129 and KM974726), Hawaiian goose poxvirus (HGPV; AY255628)
Fig. 4.
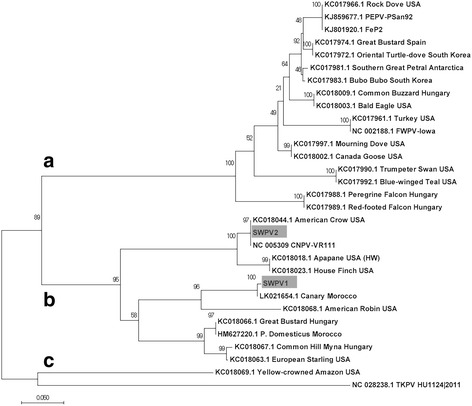
Maximum likelihood phylogenetic tree from partial DNA sequences of p4b gene of avipoxviruses. Novel Shearwaterpox viruses (SWPV-1 and SWPV-2) are highlighted by gray background
Fig. 5.
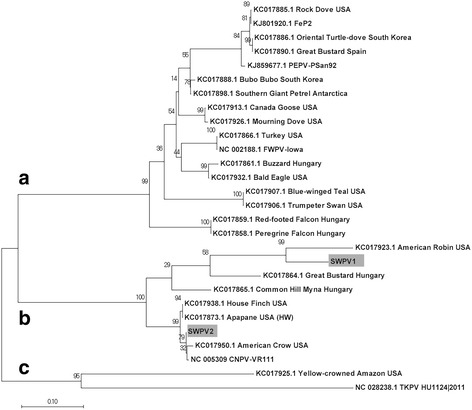
Maximum likelihood phylogenetic tree from partial DNA sequences of DNA polymerase gene of avipoxviruses. Novel Shearwaterpox viruses (SWPV-1 and SWPV-2) are highlighted by gray background
Features of SWPV-2
As noted above, and displayed in the Dotplot (Fig. 2b), SWPV-2 is very similar to CNPV with almost 98% nt identity. However, a 1% difference still gives approximately 10 mutations in an average sized gene any of which could have drastic effects if an early STOP codon is introduced to the gene sequence. Similarly, small changes to promoter regions can significantly alter gene expressions that are impossible to predict in these viruses. With this annotation strategy, 18 CNPV genes were deemed to be missing from the SWPV-2 complete genome and a further 15 genes significantly fragmented as to probably cause them to be non-functional (Table 1). No novel genes were predicted in SWPV-2, and no rearrangement of genes compared to CNPV was observed.
Features of SWPV-1
As expected from the much lower percent nt identity, SWPV-1 was found to be considerably more different to CNPV than SWPV-2 when compared at the level of genes present or absent. (Table 1). 43 CNPV genes are absent from SWPV-1 and a further 6 are significantly fragmented. There are 4 predicted genes in SWPV-1 that are not present in any other poxvirus, nor do they match any sequences in the NR protein database using BLASTP. However, they are all relatively short ORFs and it is possible that they are not functional genes. Additionally, SWPV-1 encodes nine polypeptides that do not match CNPV proteins, but do match proteins from other avipoxviruses (penguinpox, turkeypox, pigeonpox and fowlpox). This could be due to recombination among ancestral viruses, but could also result from the loss of the corresponding ortholog in CNPV leaving another virus to provide the “best match”.
As might be expected given the greater distance between SWPV-1 and CPNV than between SWPV-2 and CNPV, there are more instances of minor rearrangements that created a loss of synteny (Table 1). However, since most of these involve the families of repeated genes, it is also possible that divergence of these sequences has led to the inability to distinguish between the orthologous and paralogous genes.
Evidence of recombination among avipoxviruses
When we reviewed a graph of nt identity between the 2 new complete genomes and CNPV using BBB (not shown), there were several relatively short syntenic regions where 1) SWPV-1 matched CNPV significantly better than the majority of the genome, and 2) SWPV-2 matched CNPV significantly worse than the majority of the genome. To examine these regions in more detail, the Visual Summary feature of BBB was used to display individual SNPs for these genome comparisons (Fig. 6a and b). This analysis revealed that SWPV-1 and SWPV-2 were unique in these regions and confirmed that the genome sequences of SWPV-1and SWPV-2 were not contaminated during their assembly. However, when these regions were used as query sequences in BLASTN searches of all poxvirus sequences the best match remained CNPV suggesting that these sequences originated from avipoxvirus genomes that are not represented in the public databases.
Fig. 6.
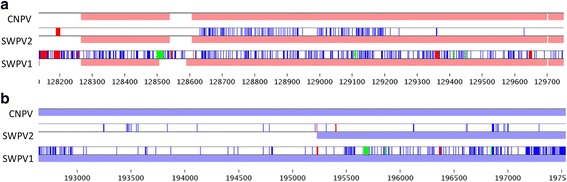
Region of recombination in Shearwaterpox viruses (SWPV) detected in A. carneipes and A. pacificus. Nucleotide differences to CNPV are shown in blue (SNPs), green/red (indels). Figure 6a. Region of recombination in SWPV-2. On the middle track, SWPV-2 has very few differences to CNPV except for highly divergent block in the middle of this region. Figure 6b. Region of recombination in SWPV-1. On the bottom track, SWPV-1 is very different to CNPV except for highly similar block between nt 193,000 and 195,500
Discussion
This paper describes the detection and characterization of two novel avipoxvirus complete genome sequences in a naturally occurring infections of avian pox in a naïve population of shearwaters. The DNA sequences of SWPV-1 and SWPV-2 are significantly different than each other but nevertheless had closest similarity with Canarypox virus (67% and 98%, respectively). Furthermore, the genetic distance and novel genome structure of SWPV-1 from A. carneipes considered to be missing 43 genes likened to CNPV and contained 4 predicted genes which are not found in any other poxvirus and is overall sufficiently genetically different to be considered a separate virus species. Whilst, the SWPV-2 complete genome was missing 18 genes compared to CNPV, with a further 15 genes significantly fragmented as to probably cause them to be non-functional. Furthermore, the phylogenetic distribution of SWPV-1 indicates that shearwaters and perhaps other long-lived, vagile marine birds could be important hosts for avipoxvirus dispersal around the globe. The natural hosts of these avipoxviruses maybe this population of shearwaters, other migratory birds that use Lord Howe Island for breeding or resident avian host reservoir species. Species such as the Lord Howe White-eye (Zosterops tephropleura) and Lord Howe Golden Whistler (Pachcephala petoralis contempta) are candidate passerine birds that might provide such function.
Examining the phylogenetic relationship between the Shearwaterpox viruses and other avipoxviruses, it is evident that the SWPV-2 is most closely related to Canarypox virus. The SWPV-1 and SWPV-2 complete genomes both contain several genes that are more closely related to CNPV throughout their entire genome. As shown in Fig. 3 it is reasonable to postulate that these viruses originated from a common ancestor that diverged from a CNPV-like progenitor related to fowlpox, penguinpox and pigeonpox viruses. Finer resolution of the phylogenetic relationship using partial nucleotide sequences of p4b and DNA polymerase genes of avipoxviruses revealed that SWPV isolated from seabirds also clustered in global clade B consisting of avipoxviruses originating from Canary Morocco, Canarypox and poxviruses from American crow and American robin. Given their genetic diversity, it is perhaps not surprising that Shearwater species can be exposed to multiple avipoxviral infections. Studies such as those by Barnett et al. [25] suggest that the species specificity of poxviruses is variable. Some genera, such as Suipoxvirus are highly restricted to individual vertebrate hosts, swinepox for instance, whereas others, such as avipoxviruses demonstrate some evidence of cross-species infection within a predator–prey system [24]. This suggests that the avipoxviruses can infect a diverse range of bird species if they are within a close enough proximity to each other [26]. Thus far, there were no clear patterns regarding species-specificity in the Shearwaterpox viruses described here.
While overt and systemic lesions and fatal disease can occur, avian pox tends to be a self-limiting localized infection of apterial skin with full recovery possible. Many bird species experience life-long immunity if the immune system is not weakened and or the birds are not infected by different strains [27, 28]. As shown in our example, secondary infections can occur and these may contribute to morbidity and mortality [29–31]. Similar to the example in shearwaters, Shivaprasad et al. [30] reported evidence of poxvirus infection and secondary fungal pathogens in canaries (Serinus canaria). Stressful conditions, poor nutrition, overt environmental contamination and other underlying causes of immunosuppression and ill health may contribute to the pathogenesis of such lesions. This was the primary reason we tested for avian circovirus and other potential pathogens.
Avian pox has not been previously reported in shearwaters (Ardenna spp.) from Lord Howe Island, nor has it been documented for any other bird species in this region. So it is difficult to attribute the causality of this unique event in these species. The value of complete genome characterization and analysis is highlighted since a phylogenetic relationship based on single gene studies such as the polymerase gene may have falsely implicated Canarypox virus as a potential exotic introduced emerging disease from domesticated birds. Although we cannot trace the actual source of infection in the shearwater chicks, it is more likely that the infection in the birds resulted from parental feeding or arthropod mediated transmission from other island bird species [32]. While, the reservoir host of these novel Shearwaterpox viruses is unknown, mosquitoes are suspected to play a part in transmission within the island. Avipoxvirus infection appears to be relatively rare in seabirds, but it has been reported in several species when they occur on human-inhabited islands that harbor mosquito vectors [33]. According to the Lord Howe Island Board, ship rats, mice, cats, humans and other invasive pest species such as owls are implicated in the extinction of at least five endemic birds, two reptiles, 49 flowering plants, 12 vegetation communities and numerous threatened invertebrates [34]. These rodents and invasive pests have also been highlighted for the potential reservoir of poxvirus infections [3, 35]. Transmission of avipoxvirus by prey–predator and other migratory seabirds likely plays a prominent role; however, the mode of avipoxvirus transmission on Lord Howe Island is not completely understood. Studies by Gyuranecz et al. [24], for example, postulated that raptors may acquire poxvirus infection from their avian prey. This suggests that the poxvirus in shearwaters is likely to be transmitted from other island species such as other migratory seabirds and/or prey–predator, although, it is difficult to be certain without further studies.
Interestingly, these new shearwaterpox virus complete genomes also provide evidence that supports the hypothesis that recombination may play an important role in the evolution of avipoxviruses. A number of genes in SWPV-1 appear to be rearranged compared to CNPV and blocks of unusual similarity scores were seen in both SWPVs. Software that is designed to look for gross recombination between two viruses, such as two strains of HIV, fails to detect this level of recombination and it is left to the investigator to observe such small events by eye after visualizing the distribution of SNPs between viruses. Such relatively small exchanges of DNA may still exert important influences on virus evolution, and has been predicted to have been a driver in the evolution of smallpox [36].
Conclusions
These are the first avipoxvirus complete genome sequences that infect marine bird species. The novel complete genome sequences of SWPV-1 and −2 have greatly enhanced the genomic information for the Avipoxvirus genus, which will contribute to our understanding of the avipoxvirus more generally, and track the evolution of poxvirus infection in such a non-model avian species. Together with the sequence similarities observed between SWPV and other avipoxviruses, this study concluded that the SWPV complete genome from A. carneipes (SWPV-1) described here is not closely related to any other avipoxvirus complete genome isolated from avian or other natural host species, and that it likely should be considered a separate species. Further investigations of Shearwaterpox viruses genetic and pathogenesis will provide a unique approach to better assess the risk associated to poxvirus transmission within and between marine bird species.
Methods
Source of sampling
A total of six samples were collected from two different species of shearwater, five were from Flesh-footed Shearwater (ID: 15-1527-31), and other one was from Wedge-tailed Shearwater (ID: 15–1526). Of size birds, two were recoded to have evidence of gross well circumscribed lesions in the beak (Fig. 1a) and ankle, and others had feather defects (fault lines across the vanes of feathers). Samples were collected from fledglings (approximately 80–90 days of age) of both species on Lord Howe Island, New South Wales (32.53̊S, 159.08̊E) located approximately 500 km off the east coast of Australia during April-May 2015. Samples were collected with the permission of the Lord Howe Island Board (permit no. LHIB 02/14) under the approval of the University of Tasmania and Charles Sturt University Animal Ethics Committees (permit no. A0010874, A0011586, and 09/046). Samples from one individual of each shearwater species were collected including skin lesions, liver and skin biopsies, as well as blood for identifying the causative agents. Depending on the samples, either 25 mg of skin tissue were cut out and chopped into small pieces or 50–100 μL of blood were aseptically transferred into clean 1.5 mL microcentrifuge tube (Eppendorf), and genomic DNA was isolated using the Qiagen blood and tissue mini kit (Qiagen, Germany). The extracted DNA has been stored at −20 °C for further testing. Histopathological examination of the skin was performed.
Archived viral and fungal pathogen testing
Initially, the extracted DNA was screened for detecting novel circoviruses [37, 38] and reticuloendotheliosis virus [39]. For poxvirus screening, the primers PoxP1 (5′-CAGCAGGTGCTAAACAACAA-3′) and PoxP2 (5′-CGGTAGCTTAACGCCGAATA-3′) were synthesized from published literature and used to amplify a segment of approximately 578 bp from the 4b core protein gene for all ChPV species [40]. Optimized PCR reactions mixture contained 3 μL of extracted genomic DNA, 25 pmol of each primer (GeneWorks, Australia), 1.5 mM MgCl2, 1.25 mM of each dNTP, 1xGoTaq® Green Flexi Reaction Buffer, 1 U of Go Taq DNA polymerase (Promega Corporation, USA) and DEPC distilled H2O (Invitrogen, USA) was added to a final volume of 25 μL. The PCR amplification was carried out in an iCycler thermal cycler (Bio-Rad) under the following conditions: denaturation at 94 °C for 2 min followed by 35 cycles of 94 °C for 1 min, 60 °C for 1 min and 72 °C for 1 min, and a final extension step of 2 min at 72 °C.
The internal transcribed spacer (ITS) region was chosen for screening and identification of fungal pathogens [41]. A set of fungus-specific primers ITS1 (5′- TCCGTAGGTGAACCTGCGG -3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC -3′) were designed and used to amplify a segment of approximately 550 bp from the fungal ITS gene [42]. The PCR was standardized to amplify ITS genes, and the 25-μL reaction mixture contained 3 μL of extracted genomic DNA, 25 pmol of each primer (GeneWorks, Australia), 1.5 mM MgCl2, 1.25 mM of each dNTP, 1xGoTaq® Green Flexi Reaction Buffer, 1 U of Go Taq DNA polymerase (Promega Corporation, USA). The PCR reaction involved initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 1 min, and with a final step of one cycle extension at 72 °C for 10 min.
Amplified PCR products, together with a standard molecular mass marker (Sigma), were separated by electrophoresis in 2.0% agarose gel and stained with GelRed (Biotium, CA). Selected bands were excised and purified using the Wizard® SV Gel and PCR Clean-Up System (Promega, USA) according to the manufacturer’s instructions. Purified amplicons were sequenced with PCR primers by the Australian Genome Research Facility Ltd (Sydney) using an AB 3730xl unit (Applied Biosystems). For each amplicon, sequences were obtained at least twice in each direction for each isolate. The sequences were trimmed for primers and aligned to construct contigs (minimum overlap of 35 bp, minimum match percentage of 95%) using Geneious Pro (version 10.0.2).
High throughput sequencing
Next-generation sequencing (NGS) was used to sequence the poxvirus genomes. Virion enrichment was performed by centrifugation for 2 min at 800 × g to remove tissue debris, and the supernatants were subsequently filtered through 5 μm centrifuge filters (Millipore) [43]. The filtrates were nuclease treated to remove unprotected nucleic acids using 8 μL RNase Cocktail Enzyme Mix (Invitrogen). Viral nucleic acids were subsequently extracted using QIAamp DNA mini (Qiagen). The genomic libraries were prepared with an insert size of 150 paired-end. DNA sequencing (NGS) was performed on a HiSeq4000 sequencing platform (Illumina) by Novogene, China.
Bioinformatics
Assembly of the viral genome was conducted according to the established pipeline [44] in CLC Genomics workbench 9.5.2 under La Trobe University Genomics Platform. Briefly, the preliminary quality evaluation for each raw read was generated using quality control (QC) report. The raw data were preprocessed to remove ambiguous base calls, and bases or entire reads of poor quality using default parameters. The datasets were trimmed to pass the quality control based on PHRED score or per base sequence quality score. Trimmed sequence reads were mapped against closely available host genome (Albatross) to remove possible remaining host DNA contamination, and post-filtered reads were mapped against reference Canarypox virus complete genome sequence. Consensus sequences were used to generate the complete poxvirus genome. Avipoxvirus complete genome sequences were aligned using MAFFT [45]. Then the poxvirus specific bioinformatics analyses were performed using the Viral Bioinformatics Resource Centre (virology.uvic.ca) [46], and the further analyses were conducted using the following tools: Viral Orthologous Clusters Database for sequence management (VOCs) [11]; Base-By-Base for genome/gene/protein alignments [47, 48]; Viral Genome Organizer for genome organization comparisons (VGO) [11], and Genome Annotation Transfer Utility for annotation (GATU) [49].
Open reading frames (ORFs) longer than 60 amino acids with minimal overlapping (overlaps cannot exceed 25% of one of the genes) to other ORFs were captured using the CLC Genomics Workbench (CLC) ORF analysis tool as well as GATU [49], and other protein coding sequence and annotation software described in Geneious (version 10.0.2, Biomatters, New Zealand). These ORFs were subsequently extracted into a FASTA file, and similarity searches including nucleotide (BLASTN) and protein (BLASTP) were performed on annotated ORFs as potential genes if they shared significant sequence similarity to known viral or cellular genes (BLAST E value ≤ e-5) or contained a putative conserved domain as predicted by BLASTp [50]. The final SWPV annotation was further examined with other poxvirus ortholog alignments to determine the correct methionine start site, correct stop codons, signs of truncation, and validity of overlaps.
Phylogenetic analysis
Phylogenetic analyses were performed using full poxvirus genome sequences for Shearwater species determined in this study with related avipoxvirus genome sequences available in GenBank database. A selection of partial sequences from seven completely sequenced avipoxvirus genomes and fragments of incompletely sequenced avipoxvirus genomes from Vultur Gryphus poxvirus (VGPV), flamingopox virus (FGPV) and Hawaiian goose poxvirus (HGPV) were also used for phylogenetic analysis. To investigate closer evolutionary relationship among avipoxviruses, partial nucleotide sequences of p4b and DNA polymerase genes were selected. The avipoxvirus sequences were aligned using ClustalO, and then manually edited in Base-by-Base. MEGA7 was used to create a maximum likelihood tree based on the Tamura-Nei method and tested by bootstrapping with 1000 replicates. An additional analysis was performed using complete genome nucleotide sequences of Canarypox virus (CNPV; AY318871), Pigeonpox virus (FeP2; KJ801920), Fowlpox virus (FPV; AF198100), Turkeypox virus (TKPV; NC_028238), Shearwaterpox virus strain-1 (SWPV-1; KX857216), and Shearwaterpox virus strain-2 (SWPV-2; KX857215), which were aligned with MAFTT in Base-By-Base for genome/gene/protein alignments [48]. The program jModelTest 2.1.3 favoured a general-time-reversible model with gamma distribution rate variation and a proportion of invariable sites (GTR + I + G4) for the ML analysis [51].
Additional files
Summary of SWPV1 genome annotations (DOCX 52 kb)
Summary of SWPV2 genome annotations (DOCX 145 kb)
Acknowledgments
The authors are extremely grateful to La Trobe University School of Life Science Publication Booster Award for their financial support to SS. Additional funding for this project was generously provided by the Detached Foundation, Trading Consultants Ltd, and L. Bryce. Assistance in the field was provided by the Lord Howe Island community and numerous dedicated volunteers, particularly A. Fidler, P. Lewis, A. Lombal, K. Richards, and V. Wellington. The authors thank Chad Smithson for help with genome assembly.
Funding
Funding for this project was generously provided by the Detached Foundation, Trading Consultants Ltd, and L. Bryce. However, none of these have grant numbers assigned since all are donations from private philanthropists. Additional financial support was provided to SS through the La Trobe University School of Life Science Publication Booster Award. CU and JI were funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant. The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The complete genome sequences of the Shearwaterpox virus 1 from a Flesh-footed Shearwater (Ardenna carneipes) and Shearwaterpox virus 2 from a Wedge-tailed Shearwater (Ardenna pacificus) have been deposited in the NCBI database under GenBank accession numbers: [SWPV-1, GenBank: KX857216] and [SWPV-2, GenBank: KX857215].
Authors’ contributions
Conceived and designed the experiments: SS, SRR. Performed the experiments: SS, SRR. Analyzed the data: SS, CU, JI, SRR. Contributed reagents/materials/analysis tools: SS, SD, JLL, IH, KH, CU, JI, SRR. SS, JLL, CU, JI, SRR wrote the initial manuscript. All authors read, edited and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
Samples were collected with the permission of the Lord Howe Island Board (permit no. LHIB 02/14) under the approval of the University of Tasmania and Charles Sturt University Animal Ethics Committees (permit no. A13836, A0010874, A0011586, and 09/046).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- ChPV
Chordopoxvirinae
- CNPV
Canarypox virus
- dsDNA
double-stranded
- FGPV
Flamingopox virus
- FP9
European strain of Fowlpox virus
- FPVUS
South African strain of Fowlpox virus
- GATU
Genome Annotation Transfer Utility
- HGPV
Hawaiian goose poxvirus
- ITS
internal transcribed spacer
- ML
Maximum likelihood
- NGS
Next-generation sequencing
- ORF
open reading frame
- PCR
polymerase chain reaction
- PEPV
Penguinpox virus
- QC
Quality control
- SWPV-1
Shearwaterpox virus 1
- SWPV-2
Shearwaterpox virus 2
- TKPV
Turkeypox virus
Contributor Information
Subir Sarker, Phone: +61 3 9479 2317, Email: s.sarker@latrobe.edu.au.
Shubhagata Das, Email: sdas@csu.edu.au.
Jennifer L. Lavers, Email: jennifer.lavers@utas.edu.au
Ian Hutton, Email: ian@ianhuttontours.com.
Karla Helbig, Email: k.helbig@latrobe.edu.au.
Jacob Imbery, Email: jimbery@live.ca.
Chris Upton, Email: cupton@uvic.ca.
Shane R. Raidal, Email: shraidal@csu.edu.au
References
- 1.Bolte AL, Meurer J, Kaleta EF. Avian host spectrum of avipoxviruses. Avian Pathol. 1999;28(5):415–432. doi: 10.1080/03079459994434. [DOI] [PubMed] [Google Scholar]
- 2.van Riper C, Forrester DJ. Avian Pox. In: Thomas NJ, Hunter DB, Atkinson CT, editors. Infectious diseases of wild birds. Oxford: Wiley Blackwell Publishing; 2007. [Google Scholar]
- 3.Offerman K, Carulei O, van der Walt AP, Douglass N, Williamson A-L. The complete genome sequences of poxviruses isolated from a penguin and a pigeon in South Africa and comparison to other sequenced avipoxviruses. BMC Genomics. 2014;15(1):1–17. doi: 10.1186/1471-2164-15-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkinson CT, LaPointe DA. Introduced avian diseases, climate change, and the future of Hawaiian Honeycreepers. J Avian Med Surg. 2009;23(1):53–63. doi: 10.1647/2008-059.1. [DOI] [PubMed] [Google Scholar]
- 5.Alley MR, Hale KA, Cash W, Ha HJ, Howe L. Concurrent avian malaria and avipox virus infection in translocated South Island saddlebacks (Philesturnus carunculatus carunculatus) N Z Vet J. 2010;58(4):218–223. doi: 10.1080/00480169.2010.68868. [DOI] [PubMed] [Google Scholar]
- 6.Gubser C, Hué S, Kellam P, Smith GL. Poxvirus genomes: a phylogenetic analysis. J Gen Virol. 2004;85(1):105–117. doi: 10.1099/vir.0.19565-0. [DOI] [PubMed] [Google Scholar]
- 7.Afonso CL, Tulman ER, Lu Z, Zsak L, Kutish GF, Rock DL. The genome of fowlpox virus. J Virol. 2000;74(8):3815–3831. doi: 10.1128/JVI.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laidlaw SM, Skinner MA. Comparison of the genome sequence of FP9, an attenuated, tissue culture-adapted European strain of Fowlpox virus, with those of virulent American and European viruses. J Gen Virol. 2004;85(Pt 2):305–322. doi: 10.1099/vir.0.19568-0. [DOI] [PubMed] [Google Scholar]
- 9.Tulman ER, Afonso CL, Lu Z, Zsak L, Kutish GF, Rock DL. The Genome of Canarypox Virus. J Virol. 2004;78(1):353–366. doi: 10.1128/JVI.78.1.353-366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banyai K, Palya V, Denes B, Glavits R, Ivanics E, Horvath B, Farkas SL, Marton S, Balint A, Gyuranecz M, et al. Unique genomic organization of a novel Avipoxvirus detected in turkey (Meleagris gallopavo) Infect Genet Evol. 2015;35:221–229. doi: 10.1016/j.meegid.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. Poxvirus Orthologous Clusters: toward Defining the Minimum Essential Poxvirus Genome. J Virol. 2003;77(13):7590–7600. doi: 10.1128/JVI.77.13.7590-7600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croxall JP, Butchart SHM, Lascelles BEN, Stattersfield AJ, Sullivan BEN, Symes A, Taylor P. Seabird conservation status, threats and priority actions: a global assessment. Bird Conserv Int. 2012;22(1):1–34. doi: 10.1017/S0959270912000020. [DOI] [Google Scholar]
- 13.Reid T, Hindell M, Lavers JL, Wilcox C. Re-examining mortality sources and population trends in a declining seabird: using Bayesian methods to incorporate existing information and new data. PLoS One. 2013;8(4):e58230. doi: 10.1371/journal.pone.0058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bond AL, Lavers JL. Trace element concentrations in feathers of flesh-footed shearwaters (Puffinus carneipes) from across their breeding range. Arch Environ Contam Toxicol. 2011;61(2):318–326. doi: 10.1007/s00244-010-9605-3. [DOI] [PubMed] [Google Scholar]
- 15.Illera JC, Emerson BC, Richardson DS. Genetic characterization, distribution and prevalence of avian pox and avian malaria in the Berthelot’s pipit (Anthus berthelotii) in Macaronesia. Parasitol Res. 2008;103(6):1435–1443. doi: 10.1007/s00436-008-1153-7. [DOI] [PubMed] [Google Scholar]
- 16.Lecis R, Secci F, Antuofermo E, Nuvoli S, Scagliarini A, Pittau M, Alberti A. Multiple gene typing and phylogeny of avipoxvirus associated with cutaneous lesions in a stone curlew. Vet Res Commun. 2017;4:1–7. doi: 10.1007/s11259-016-9674-5. [DOI] [PubMed] [Google Scholar]
- 17.Woolaver LG, Nichols RK, Morton ES, Stutchbury BJM. Population genetics and relatedness in a critically endangered island raptor, Ridgway’s Hawk Buteo ridgwayi. Conserv Genet. 2013;14(3):559–571. doi: 10.1007/s10592-013-0444-4. [DOI] [Google Scholar]
- 18.Thiel T, Whiteman NK, Tirape A, Baquero MI, Cedeno V, Walsh T, Uzcategui GJ, Parker PG. Characterization of canarypox-like viruses infecting endemic birds in the Galapagos Islands. J Wildl Dis. 2005;41(2):342–353. doi: 10.7589/0090-3558-41.2.342. [DOI] [PubMed] [Google Scholar]
- 19.van Riper C, van Riper SG, Hansen WR. Epizootiology and Effect of Avian Pox on Hawaiian Forest Birds. Auk. 2002;119(4):929–942. doi: 10.1642/0004-8038(2002)119[0929:EAEOAP]2.0.CO;2. [DOI] [Google Scholar]
- 20.Young LC, VanderWerf EA. Prevalence of avian pox virus and effect on the fledging success of Laysan Albatross. J Field Ornithol. 2008;79(1):93–98. doi: 10.1111/j.1557-9263.2008.00149.x. [DOI] [Google Scholar]
- 21.Hane JK, Lowe RG, Solomon PS, Tan KC, Schoch CL, Spatafora JW, Crous PW, Kodira C, Birren BW, Galagan JE, et al. Dothideomycete plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum. Plant Cell. 2007;19(11):3347–3368. doi: 10.1105/tpc.107.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghikas DV, Kouvelis VN, Typas MA. The complete mitochondrial genome of the entomopathogenic fungus Metarhizium anisopliae var. anisopliae: gene order and trn gene clusters reveal a common evolutionary course for all Sordariomycetes, while intergenic regions show variation. Arch Microbiol. 2006;185(5):393–401. doi: 10.1007/s00203-006-0104-x. [DOI] [PubMed] [Google Scholar]
- 23.Le Loc’h G, Ducatez MF, Camus-Bouclainville C, Guérin J-L, Bertagnoli S. Diversity of avipoxviruses in captive-bred Houbara bustard. Vet Res. 2014;45(1):98. doi: 10.1186/s13567-014-0098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gyuranecz M, Foster JT, Dán Á, Ip HS, Egstad KF, Parker PG, Higashiguchi JM, Skinner MA, Höfle U, Kreizinger Z, et al. Worldwide Phylogenetic Relationship of Avian Poxviruses. J Virol. 2013;87(9):4938–4951. doi: 10.1128/JVI.03183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnett J, Dastjerdi A, Davison N, Deaville R, Everest D, Peake J, Finnegan C, Jepson P, Steinbach F. Identification of Novel Cetacean Poxviruses in Cetaceans Stranded in South West England. PLoS One. 2015;10(6):e0124315. doi: 10.1371/journal.pone.0124315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haller SL, Peng C, McFadden G, Rothenburg S. Poxviruses and the Evolution of Host Range and Virulence. Infect Genet Evol. 2014;21:15–40. doi: 10.1016/j.meegid.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.EAZWV Transmissible Disease Fact Sheet. http://c.ymcdn.com/sites/www.eazwv.org/resource/resmgr/Files/Transmissible_Diseases_Handbook/Fact_Sheets/006_Avian_Pox.pdf. Accessed 18 Jan 2016.
- 28.Winterfield RW, Reed W. Avian pox: infection and immunity with quail, psittacine, fowl, and pigeon pox viruses. Poult Sci. 1985;64(1):65–70. doi: 10.3382/ps.0640065. [DOI] [PubMed] [Google Scholar]
- 29.Johnson BJ, Castro AE. Canary pox causing high mortality in an aviary. J Am Vet Med Assoc. 1986;189(10):1345–1347. [PubMed] [Google Scholar]
- 30.Shivaprasad HL, Kim T, Tripathy D, Woolcock PR, Uzal F. Unusual pathology of canary poxvirus infection associated with high mortality in young and adult breeder canaries (Serinus canaria) Avian Pathol. 2009;38(4):311–316. doi: 10.1080/03079450903061643. [DOI] [PubMed] [Google Scholar]
- 31.Reza K, Nasrin A, Mahmoud S. Clinical and pathological findings of concurrent poxvirus lesions and aspergillosis infection in canaries. Asian Pac J Trop Biomed. 2013;3(3):182–185. doi: 10.1016/S2221-1691(13)60046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shearn-Bochsler V, Green DE, Converse KA, Docherty DE, Thiel T, Geisz HN, Fraser WR, Patterson-Fraser DL. Cutaneous and diphtheritic avian poxvirus infection in a nestling Southern Giant Petrel (Macronectes giganteus) from Antarctica. Polar Biol. 2008;31(5):569–573. doi: 10.1007/s00300-007-0390-z. [DOI] [Google Scholar]
- 33.VanderWerf EA, Young LC. Juvenile survival, recruitment, population size, and effects of avian pox virus in Laysan Albatross (Phoebastria immutabilis) on Oahu, Hawaii, USA. The Condor. 2016;118(4):804–814. doi: 10.1650/CONDOR-16-49.1. [DOI] [Google Scholar]
- 34.Lord Howe Island Board. http://www.lhib.nsw.gov.au/environment/biodiversity/research. Accessed 20 Sep 2016.
- 35.Tantawi HH, Zaghloul TM, Zakaria M. Poxvirus infection in a rat (Rattus norvegicus) in Kuwait. Int J Zoonoses. 1983;10(1):28–32. [PubMed] [Google Scholar]
- 36.Smithson C, Purdy A, Verster AJ, Upton C. Prediction of Steps in the Evolution of Variola Virus Host Range. PLoS One. 2014;9(3):e91520. doi: 10.1371/journal.pone.0091520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarker S, Lloyd C, Forwood J, Raidal SR. Forensic genetic evidence of beak and feather disease virus infection in a Powerful Owl. Ninox strenua Emu. 2016;116(1):71–74. doi: 10.1071/MU15063. [DOI] [Google Scholar]
- 38.Sarker S, Moylan KG, Ghorashi SA, Forwood JK, Peters A, Raidal SR. Evidence of a deep viral host switch event with beak and feather disease virus infection in rainbow bee-eaters (Merops ornatus) Sci Rep. 2015;5:14511. doi: 10.1038/srep14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biswas SK, Jana C, Chand K, Rehman W, Mondal B. Detection of fowl poxvirus integrated with reticuloendotheliosis virus sequences from an outbreak in backyard chickens in India. Vet Ital. 2011;47(2):147–153. [PubMed] [Google Scholar]
- 40.Huw Lee L, Hwa Lee K. Application of the polymerase chain reaction for the diagnosis of fowl poxvirus infection. J Virol Methods. 1997;63(1–2):113–119. doi: 10.1016/S0166-0934(96)02119-2. [DOI] [PubMed] [Google Scholar]
- 41.Kumar M, Shukla PK. Use of PCR Targeting of Internal Transcribed Spacer Regions and Single-Stranded Conformation Polymorphism Analysis of Sequence Variation in Different Regions of rRNA Genes in Fungi for Rapid Diagnosis of Mycotic Keratitis. J Clin Microbiol. 2005;43(2):662–668. doi: 10.1128/JCM.43.2.662-668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindsley MD, Hurst SF, Iqbal NJ, Morrison CJ. Rapid Identification of Dimorphic and Yeast-Like Fungal Pathogens Using Specific DNA Probes. J Clin Microbiol. 2001;39(10):3505–3511. doi: 10.1128/JCM.39.10.3505-3511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen RH, Mollerup S, Mourier T, Hansen TA, Fridholm H, Nielsen LP, Willerslev E, Hansen AJ, Vinner L. Target-dependent enrichment of virions determines the reduction of high-throughput sequencing in virus discovery. PLoS One. 2015;10(4):e0122636. doi: 10.1371/journal.pone.0122636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao K, Wohlhueter RM, Li Y. Finishing monkeypox genomes from short reads: assembly analysis and a neural network method. BMC Genomics. 2016;17(Suppl 5):497. doi: 10.1186/s12864-016-2826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehlers A, Osborne J, Slack S, Roper RL, Upton C. Poxvirus Orthologous Clusters (POCs) Bioinformatics. 2002;18(11):1544–1545. doi: 10.1093/bioinformatics/18.11.1544. [DOI] [PubMed] [Google Scholar]
- 47.Brodie R, Smith AJ, Roper RL, Tcherepanov V, Upton C. Base-By-Base: Single nucleotide-level analysis of whole viral genome alignments. BMC Bioinformatics. 2004;5(1):1–9. doi: 10.1186/1471-2105-5-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hillary W, Lin S-H, Upton C. Base-By-Base version 2: single nucleotide-level analysis of whole viral genome alignments. Microb Inf Exp. 2011;1:2–2. doi: 10.1186/2042-5783-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tcherepanov V, Ehlers A, Upton C. Genome Annotation Transfer Utility (GATU): rapid annotation of viral genomes using a closely related reference genome. BMC Genomics. 2006;7(1):1–10. doi: 10.1186/1471-2164-7-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772–2 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of SWPV1 genome annotations (DOCX 52 kb)
Summary of SWPV2 genome annotations (DOCX 145 kb)
Data Availability Statement
The complete genome sequences of the Shearwaterpox virus 1 from a Flesh-footed Shearwater (Ardenna carneipes) and Shearwaterpox virus 2 from a Wedge-tailed Shearwater (Ardenna pacificus) have been deposited in the NCBI database under GenBank accession numbers: [SWPV-1, GenBank: KX857216] and [SWPV-2, GenBank: KX857215].


