Abstract
Background
Somatic copy number alterations (SCNAs) occurring in tumors can provide information about tumor classification, patient's outcome or treatment targets. Liquid biopsies, incl. plasma samples containing circulating cell-free tumor DNA (ccfDNA) can be used to assess SCNAs for clinical purposes, however specify and reliability of methods have to be tested.
Methods
SNP microarrays (Affymetrix) were used to generate whole-genome copy number profiles from plasma ccfDNA (OncoScan) and paired tumor biopsies (CytoScan) from ten patients with metastatic cancers. Numerical, segmental and focal SCNAs were assessed using ASCAT/TuScan and SNP-FASST2.
Results
Aberrations in ccfDNA in 4 patients resembled numerical (76%) and segmental (80%) aberrations in tDNA. Three patients represented low correlation due to postponed sampling time, ccfDNA quality and possible treatment interference. Breakpoints of high-amplitude amplification were assessed with high accuracy and relative breakpoints difference of only 7% (0.02–37%). Similarly, biallelic losses were reliably detected. Array was 100% successful in detection of SCNAs on clinically relevant genes compared to SCNAs in tumor biopsies. Tracking of SCNAs changes during the treatment course of one patient also indicated that apoptosis/necrosis of non-cancerous cells presumably induced by treatment can influence ccfDNA composition and introduce false-negative findings into the analysis of liquid biopsies.
Conclusions
Genomic alterations detected in ccfDNA from liquid biopsies by comprehensive SNP array are reliable source for information for stratification of patients for targeted treatment.
General significance
Clinically relevant SCNAs can be detected in ccfDNA with high resolution and can therefore serve as an alternative to tumor biopsy in defining treatment targets.
Keywords: Copy number alterations, Circulating cell-free tumor DNA, Array profiling, Diagnostics
Highlights
-
•
Genome-wide copy number alterations can be detected in tumor ccfDNA isolated from blood.
-
•
Alterations in ccfDNA reflect alterations in corresponding tumor.
-
•
Array-based technology assigns alterations break-points with high precision.
-
•
Composition of ccfDNA can be altered by treatment.
1. Introduction
Molecular tumor profiling is essential for identification of patients that may be eligible for targeted therapy. Tumor tissue is normally obtained from biopsies that not always reflect tumor heterogeneity. Moreover, it may not be possible to obtain serial biopsies, restricting the possibility to track tumor evolution and treatment response. Therefore, liquid biopsies have emerged as an attractive alternative for cancer diagnostics and their significance dramatically increased with the introduction of next-generation sequencing (NGS) into analysis of circulating cell-free DNA (ccfDNA) released from the tumor.
The ccfDNA is released into the blood stream from necrotic and/or apoptotic cells as longer size or shorter 160–180 bp fragments, respectively [13]. The amount of ccfDNA is by itself a prognostic marker and patients with increased levels of ccfDNA exhibit an unfavorable prognosis [1], [9]. PCR-based methods have been employed to identify tumor-specific mutations and DNA methylation patterns during disease progression and treatment (reviewed in [17]). Recently, advances in next-generation sequencing (NGS) have enabled whole-genome/exome sequencing (WGS, WES) of ccfDNA and depicted complex tumor profiles from ccfDNA [4], [14]. NGS has also been applied for detection of particular somatic copy number alterations (SCNAs) [11], [21] or genome-wide SCNA profiles [19], [26] in tumor samples, as well as, in plasma [10]. Even though advances in bioinformatics in principle allow for transformation of sequencing reads into quantitative assessment of copy numbers, the derived SCNAs are sensitive to the quality of the ccfDNA and the choice of processing algorithms and thresholds [10]. NGS based analyses have usually longer turn-around time and are still relatively labor intensive and expensive for diagnostic purposes, i.e. if individual samples have to be processed to meet patient report's deadline and standards. Considering these limitations, array-based approaches for detection of SCNAs in ccfDNA might have several benefits in the clinical settings.
Arrays comprising a weighted distribution of SNPs on cancer relevant genes were recently employed for SCNA analysis of ccfDNA present in the urine from patients with urothelial bladder cancer [22]. The assay utilizes molecular inversion probe (MIP) technology [8] and was initially designed for FFPE material. The technology is independent of DNA fragment size, requires minimal DNA input and can partly compensate for suboptimal DNA quality. In the present study, we examined ccfDNA isolated from blood samples collected from patients with various cancer types, and systematically compared whole-genome copy number profiles obtained from ccfDNA with the corresponding tumor biopsies. Specifically, we have focused on SCNAs occurring on the genes with treatment relevance. We also show that the ccfDNA-derived SCNA profiles can be used to monitor treatment response and point out specific risks/challenges related to ccfDNA analysis.
2. Materials and methods
2.1. Patients and cfDNA samples
Table 1.
Patient cohort overview.
| Patient # | Diagnosis | Age at inclusion | Sex | Biopsy site | Days between cfDNA & tDNA sampling | cfDNA concentration (ng/ml plasma) | Days treatment-cfDNA sampling | tDNA status |
|---|---|---|---|---|---|---|---|---|
| Patient 1 | Stomach c. | 65 | M | Liver | 0 | 58.03 | Known | |
| Patient 2 | Cholangiocarcinoma | 34 | M | Liver | 0 | 202.13 | Known | |
| Patient 3 | Pancreas c. | 58 | F | Liver | 0 | 29.01 | Known | |
| Patient 4 | Oesophagus c. | 83 | M | Liver | 0 | 191.13 | Known | |
| Patient 5 | Lung c. | 60 | M | Lymph node | 96 | 10.63 | Known | |
| Patient 6 | Colon c. | 50 | F | Stomach | 0 | 65.31 | In treatment | Known |
| Patient 7 | Breast c. | 60 | F | Skin | 0 | 5.6 | Known | |
| Patient 8 | Colon c. | 74 | F | Liver | 0 | 41.39 | Unknown | |
| Patient 9 | Cervix c. | 35 | F | Lymph node | 0 | 8.31 | Unknown | |
| Patient 10 | Colon c. | 64 | M | Liver | 0 | 27.23 | Unknown |
Age: Age at the inclusion to CoPPO study.
Days after last treatment: Days after the last treatment has been given.
2.2. CytoScan and OncoScan assay
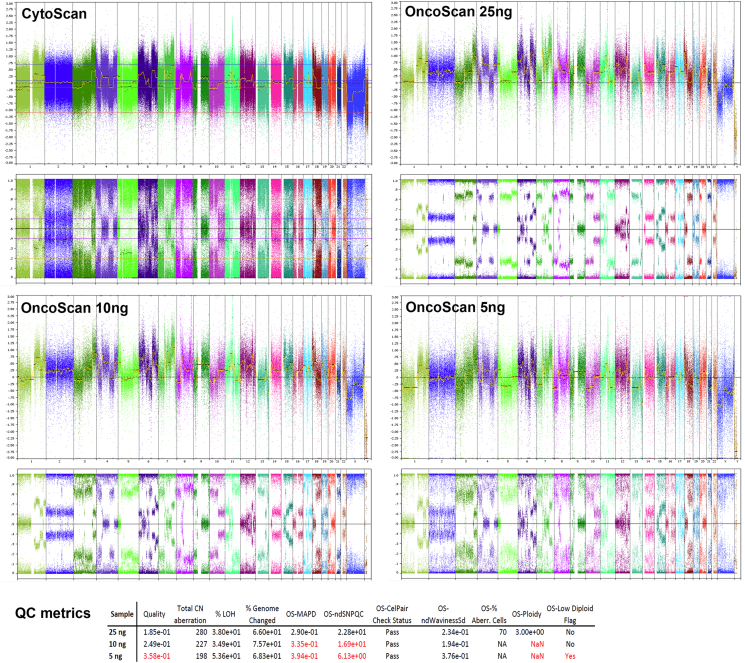
CytoScan assay (Affymetrix, Santa Clara, USA) was performed on fresh-frozen specimens from tumor biopsies according to the manufactures instructions. Tumor samples were analyzed as a part of the diagnostics in the CoPPO study where CytoScan is the pre-defined platform. OncoScan assay (Affymetrix, Santa Clara, USA) for analysis of ccfDNA was also performed according to the manufactures instructions. The ccfDNA samples were analyzed retrospectively. To test the minimal input of ccfDNA (recommended total DNA 80 ng), various amounts of DNA with known SCNAs profile were used as starting material. Briefly, 25, 10, and 5 ng of tDNA with known SCNAs profiles were analyzed and assessed by standard QC parameters. The results are summarized in Fig. S1 and were used to estimate minimal input of ccfDNA used in the study. SCNAs profile of the testing sample was identical if using CytoScan or OncoScan platform indicating that the applied platform will have presumably minimal impact on the analysis.
Fig. S1.
Titration experiment. Whole genome views on tDNA analyzed on CytoScan arrays with recommended concentration and analyzed on OncoScan arrays with total input of 25, 10 and 5 ng. Table shows QC parameters of samples analyzed on OncoScan.
2.3. Data analysis
OSCHP files from OncoScan and CEL files from the CytoScan assay were imported into NEXUS software (BioDiscovery) and used for the analysis and visualization of SCNAs and loss of heterozygosity (LOH). Files are available at the GEO database under accession number GSE85937. CytoScan CEL files were processes by ASCAT 2.1 (quadratic correction, Gamma 0.55, removal of 3% outliers) and by pre-defined NEXUS settings (quadratic correction, median recenter probes, SNP-FASST2 segmentation, removal of 3% outliers, and minimum of 50 probes per segment). OncoScan OSCHP files were processed similarly by TuScan (Affymetrix implementation of ASCAT) and by pre-defined NEXUS settings (ratio column as Log2Ratio, median recenter probes, SNP-FASST2 segmentation, removal of 3% outliers, and minimum of 50 probes per segment). Overall, both approaches performed similarly on OncoScan data (correlation in parameter “% of genome changed” was R = 0.96), and had insignificant effect on assessment of SCNAs of interest. Both of the platforms require at least 10% tumor burden in order to detect the somatic copy number aberrations. The SM Viewer Software (Affymetrix) was applied for detection of somatic mutations in each sample. The platform requires at least 20–30% of mutation's frequency in order to detect/assign its presence. Out of the 74 hot-spot mutations detectable by the OncoScan (BRAF, KRAS, EGFR, IDH1, IDH2, PTEN, PIK3CA, NRAS, TP53), one mutation (PIK3CA, c.3140A > G, p.H1047R) was detected in Patient 8, what was in concordance with the finding from whole-exome sequencing (routine in CoPPO patients analysis; data not shown).
SCNAs (loss, gain, biallelic loss, or high amplification) and LOH calls for each sample were confirmed by visual inspection and followed by manual interpretation of whole-genome profiles. In general, presence of numerical (involving whole chromosome) and segmental (involving at least two chromosomal bands) aberrations was assigned for each chromosome. Subsequently, clinically relevant deletions/biallelic losses and local amplification (> 4 copies relative to average ploidy on the particular chromosome) on 55 genes, selected based on their treatment relevance [25], were examined.
3. Results
3.1. Performance and limitations of ccfDNA analysis on arrays
In order to define the minimum input of ccfDNA (recommended 80 ng total DNA), different total amounts of tDNA were used as a starting material. Briefly, 25, 10 and 5 ng of tDNA with known SCNAs profile was analyzed by OncoScan and assessed by standard QC parameters as described above (Fig. S1). The lowest input (5 ng) failed the most of the QC parameters, although displaying a visually recognizable SCNAs profile. 10 ng tDNA was border-line with respect to the QC parameters as MAPD and WavinessSd, and showed a distinct SCNA profile, but algorithms failed to assign sample characteristics as e.g. ploidy. The input of 25 ng passed all QC parameters and the sample was correctly assessed for SCNAs, ploidy, and aberrant cell fraction. The majority of the samples contained at least 20 ng of ccfDNA. Hence if possible, a minimum input of 20 ng ccfDNA was used in the study. This resulted in acceptable QC metrics in all but one sample.
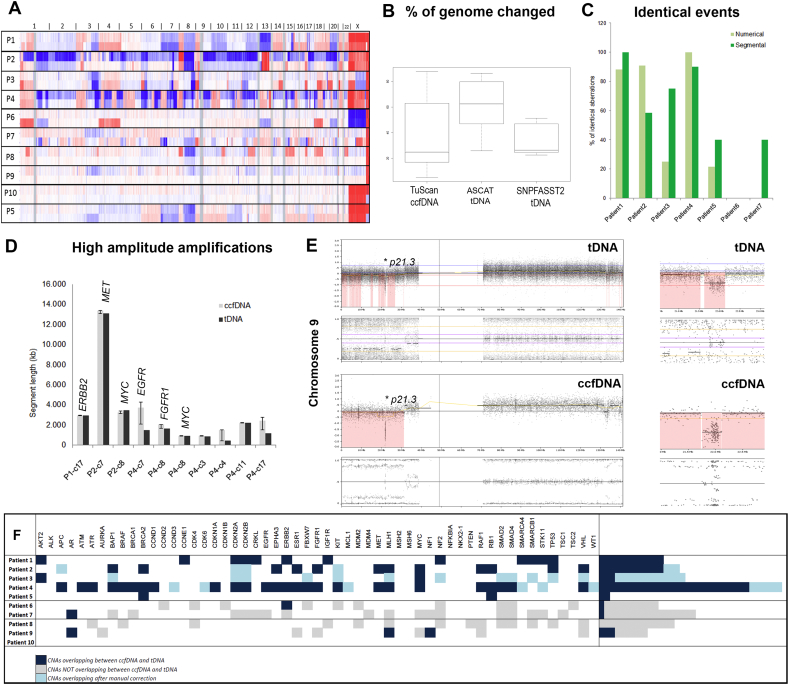
When looking on the percentage of the genome subjected to SCNAs (% of genome changed) as calculated by TuScan (ccfDNA) and by ASCAT (tDNA), the similar trend was only seen in some sample pairs (Patient 1, 2, and 4). Overall, there was a tendency towards underestimation of percentage of altered genome in ccfDNA by TuScan algorithm compared to tDNA processed by ASCAT (Fig. 1B). This was reflected by the low correlation between ccfDNA and tDNA (R = 0.58). Application of SNP-FASST2 instead of ASCAT, improved the correlation between the ccfDNA-tDNA pairs (R = 0.74), thus indicating need not only for an algorithmic adjustment but also a possible biological phenomenon, as e.g. presence of various tumor clones. Even though the above assessed parameters are interesting in the large cohort studies, they will have only minimal influence on the single patient diagnostics and for clinical purposes, sensitivity in detection of particular SCNAs will be more important.
Fig. 1.
Comparison of genomic profiles of ccfDNA and corresponding tDNA. (A) Overview plots showing probe densities on chromosomes for patient's paired samples. Upper plot for each patient represents probe densities distribution in ccfDNA and below probe densities in tDNA. Numbers above the plot indicates chromosome numbers. (B) Boxplots for percentages of genome subjected to aberrations in 7 patients with known genomic profile as assigned in ccfDNA by TuScan, and in tDNA assigned by ASCAT and SNPFASST2. (C) Plot showing percentual overlap of numerical (light-grey) and segmental (dark-grey) aberrations between tDNA and ccfDNA in individual patients. (D) Graph representing length of high amplitude amplifications detected in ccfDNA (light-grey) and in corresponding tDNA (dark-grey). “P” on x-axis indicates patient's number and “c” chromosome. Error bars on ccfDNA columns correspond to differences between individual break-points between tDNA and ccfDNA. (E) Chromosome view (NEXUS, BioDiscovery) on chromosome 9 in tDNA and ccfDNA. Arrows mark biallelic deletion in 9p21.3 involving CDKN2A/B and is enlarged is section to the right. (F) Aberrations detected on clinically relevant cancer genes in tDNA and ccfDNA. Dark-blue indicates alterations found in ccfDNA and tDNA. Light-grey marks alterations detected only in tDNA or only in ccfDNA. Light-blue marks corresponding alterations after performance of manual correction.
3.2. Resolution in SCNAs detected in ccfDNA and in tumor biopsies
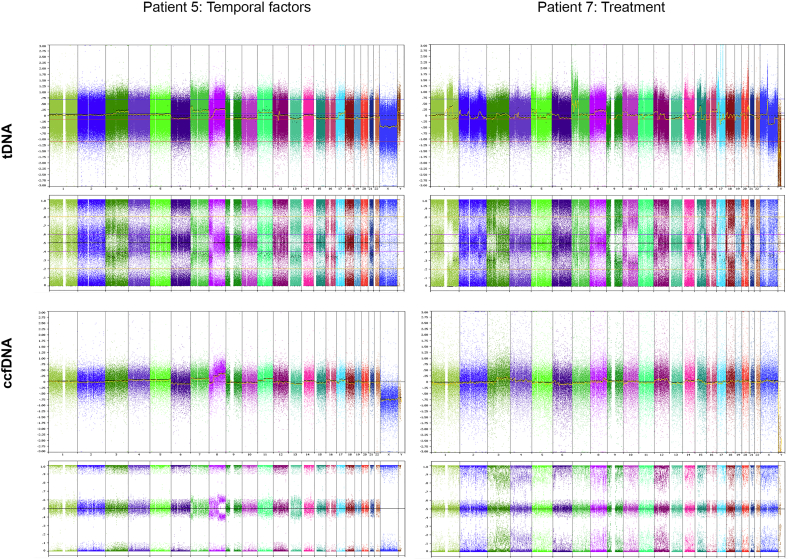
To assess the degree of correspondence between alterations detected in ccfDNA and tumor biopsies, we investigated raw copy number changes; i.e. whole chromosome gains or losses (numerical aberrations) and subchromosomal gains and losses (segmental aberrations). The percentage of identical aberrations in the seven patients with known SCNAs profile in tDNA was remarkably uneven (Fig. 1C). On average, the analyzed patients had 53% (numerical aberrations) and 58% (segmental aberrations) of identical events in ccfDNA and tDNA, varying from 0 to 100%. Whereas aberrations in ccfDNA from patients 1–4 resembled to a high degree aberrations in tDNA (76% for numerical and 80% for segmental aberrations, respectively), the three remaining patients represented likeness only in 33% (Patient 5), 0% (Patient 6) and 15% (Patient 7) of the events. Patient 5 was the only patient whose biopsy was taken 95 days after the sampling of ccfDNA. Hence, the observed discrepancy in the genomic profiles could reflect tumor evolution. Indeed, tDNA was more altered than ccfDNA obtained earlier, showing an overall shift in the ploidy (2.00 in ccfDNA and 4.63 in tDNA), as well as the presence of new segmental aberrations (Fig. S3). Patient 7 had suboptimal ccfDNA input of 10 ng and failed most of the array QC (Table S1). Patient 6 had ccfDNA showing genomic profile without SCNAs (Fig. S3). The difference to the other patients was that Patient 6 received regorafenib treatment at the time of ccfDNA sampling (issue investigated below), though hereby not excluding other possible causes. There was also marginal discrepancy between detection of numerical and segmental aberrations in the Patients 1–4 (Fig. 1C). While segmental aberrations were identical in 80% of events with standard deviation of 18.2%, numerical aberrations displayed deviation of 34.5%. This was due to Patient 3, whose ccfDNA had borderline QC parameters and contained substantial amount of ccfDNA from normal cells; making the assessment of numerical aberrations more challenging. Segmental aberrations, on the other hand, were in general more pronounced and TuScan was able to detect differences between segments with different copy numbers.
Fig. S3.
Examples of biological bias in ccfDNA analysis. Whole genome views of tDNA and ccfDNA of Patient 5 and Patient 7. Patient 5 displays a less altered genomic profile in ccfDNA collected 95 days before the tumor biopsy. Patient 7, exposed to regorafenib treatment under sampling of ccfDNA, shows ccfDNA genomic profile without aberrations, i.e. corresponding to genomic profile of normal cells
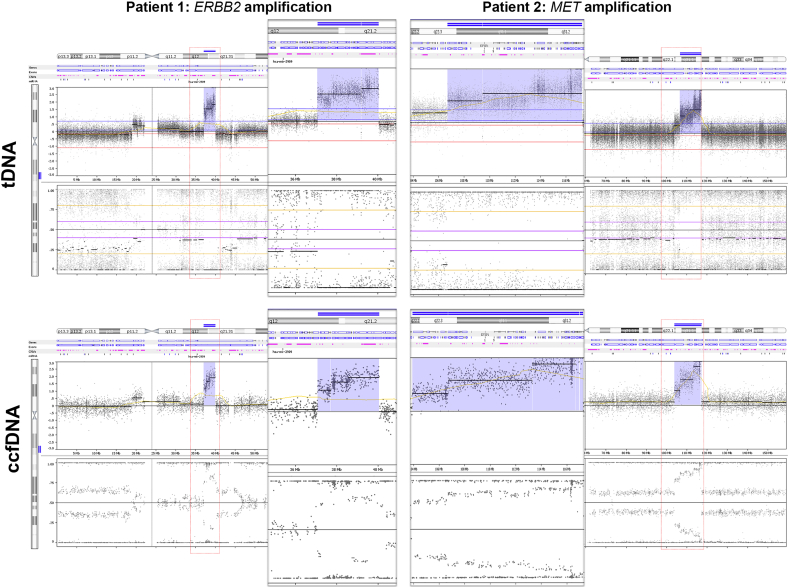
In order to assess array resolution for detection of events in ccfDNA, break-points of high amplitude focal amplifications and of biallelic losses were analyzed. Ten high amplitude focal amplifications were detected among the patients, and their size was defined as a simple difference (kb) between the break-points delineating the amplified segment (Fig. 1D). Subsequently, difference between ccfDNA and tDNA break-points position was calculated as percentual difference relative to the size of amplified segment in tDNA. The average difference was 26.75% though including three distinct outliers. Exclusion of outliers decreased the relative difference to 7%, ranging from 0.02% to 37.1% indicating a high precision in array-based analysis of focal events in ccfDNA. Furthermore, there was no correlation between segment length and detected relative difference (R = − 0.242). This suggests that smaller or larger focal events are detected with equal resolution of break-points and high accuracy. Interestingly in all cases, where clinically relevant gene was detected as amplified in tDNA, the gene was also amplified in ccfDNA (for examples see Fig. S2).
Fig. S2.
Examples of high-amplitude amplification. Examples of high amplitude amplifications detected in tumor DNA and in corresponding ccfDNA occurring in patient 1 (amplification of ERBB2) and in patient 2 (large amplification including e.g. MET).
Detection of biallelic losses by DNA sequencing of ccfDNA remains biased and particularly dependent on tumor content in the sample. Noticeably, the three biallelic losses identified in our cohort were detected in tDNA as well as in the ccfDNA counterparts; i.e. Patient 1: 15q22.2 (CCNB2), Patient 3: 9p21.3-p21.1 (CDKN2A/B), and Patient 4: 9p21.3 (CDKN2A/B) (Fig. 1E).
Taken together, the results show that array-based whole genome analysis represents a suitable tool for SCNAs analysis of blood ccfDNA. The genomic aberrations in tDNA are to a high degree reflected in ccfDNA, however, the representative power of ccfDNA can be conditioned by quality and quantity of ccfDNA as well as temporal and biological factors.
3.3. Detection of clinically relevant SCNAs in ccfDNA
From a clinical point of view, numerical or segmental aberrations in adult, non-CNS, solid tumors have rather prognostic than treatment relevant impact. On contrary, amplification (activation) or deletions/LOH (loss-of-function) of a specific gene can represent a possible treatment target. To verify the feasibility of ccfDNA for detection of clinically relevant SCNAs, we analyzed 55 clinically relevant genes in ccfDNA and in corresponding tumor biopsies. In the first 4 patients with comparable ccfDNA-tDNA pairs, a total number of 76 SCNAs were detected in tumor biopsies. From those, 54 (70%) were also recognized in ccfDNA (Fig. 1F). Discrepancies between the SCNAs calls were caused either by missing LOH calls or by the over/under-estimation of SCNA, i.e. gain interpreted as amplification and vice versa, or biallelic loss was assessed as monoallelic deletion. In all cases, the differences could be explained by normal cell contamination and could be corrected after visual inspection of genomic profiles (Fig. 1F). In patients 8, 9, and 10, where the tumor biopsies only contained normal cells, we identified 14 (Patient 8) and 9 (Patient 9) clinically relevant SCNAs in ccfDNA (Fig. 1F). In Patient 10, ccfDNA did not possess sufficient tumor burden. Hence, clinically relevant SCNAs can be reliably detected in ccfDNA. The liquid biopsies can therefore be used as an alternative source of SCNA information for defining of treatment targets where tumor biopsy is not an option.
3.4. Presumptive non-cancerous cell apoptosis can bias the diagnostic power of ccfDNA
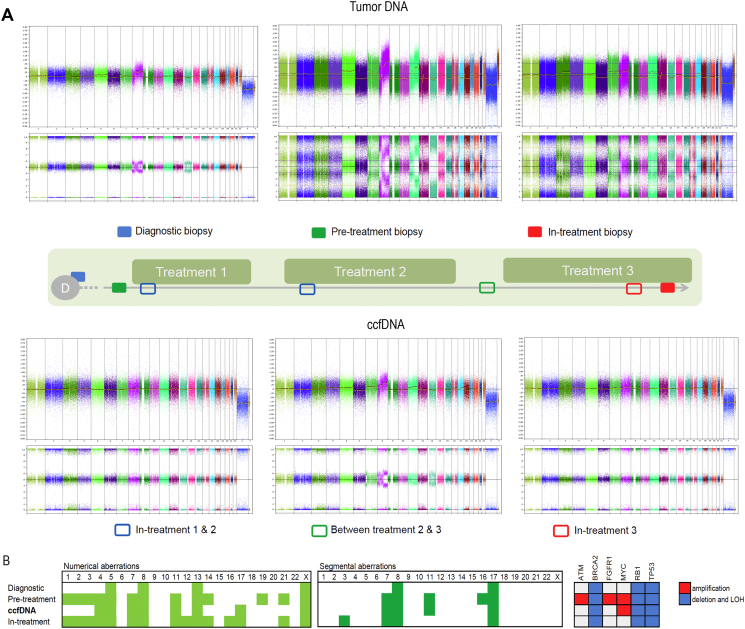
The diagnostic biopsy showed a relatively stable genomic profile characterized by unbalanced gain of 8q and LOH on chromosome 13. At CoPPO inclusion, metastatic tDNA was more affected and displayed aneuploidy on several chromosomes, pronounced amplification 8q, LOH on chromosome 13, and in addition, unbalanced gain/amplification of 11q, local gain 12q13 and 17p deletion. After the third treatment regime, tDNA showed less prominent aberrations, as simple gain 8q and LOH on chromosome 13, and lacked 17p deletion or high gain 11q observed in the pre-treatment biopsy (Fig. 2A,B). Interestingly, despite having comparable plasma concentrations of ccfDNA (31, 14, 11 and 20 ng/ml, respectively), none of the ccfDNA samples, taken under the treatments, contained sufficient amount of tumor ccfDNA. None of the aberrations observed in tDNA could be detected in ccfDNA. The only ccfDNA sample with recognizable SCNAs was obtained between treatments, i.e. between treatment 2 and 3. The SCNAs resembled to a large extend SCNAs from pre- and in-treatment biopsies; e.g. high gain 8q, deletion 17p, LOH on chromosome 13 and gain 11q. These results indicate that apoptosis/necrosis of non-cancerous cells induced by chemotherapy can influence ccfDNA composition and introduce false-negative findings into the whole-genome SCNAs analysis of liquid biopsies. Indeed, used array platform and subsequent data processing require at least 10–30% of tumor burden in order to detect the deviations from normal cell profile. Moreover, OncoScan detects mutations with frequency of at least 20–30%. In agreement with these observations, patients BRAF driver mutation, was detected in ccfDNA by ddPCR with frequency of app. 1% for in-treatment samples and of 10.8% in between-treatment sample (unpublished results), hence far below the array sensitivity. However, we cannot exclude other technical factors affecting DNA composition [23], nor the tumor behavior acquiring more quiescent state during the treatment.
Fig. 2.
Patient case. (A) Longitudinal study of genomic aberrations in tDNA and ccfDNA obtained from Patient 5 before and during treatment. The top part of the figure shows whole genome profiles of tDNA, analyzed by CytoScan, originating from diagnostic (blue rectangle), pre- (green rectangle) and in-treatment (red rectangle) biopsies. Below are whole genome profiles of ccfDNA analyzed by OncoScan, and sampled after the initiation of treatment 1 and 2 (blue empty rectangle), between treatments 2 and 3 (green empty rectangle) and after 73 days of treatment 3 (red empty rectangle). Notice the absence of aberrations in cfDNA obtained under treatment. (B) Distribution of numerical and segmental aberrations and of focal alterations in cancer related genes in diagnostic, pre-treatment and in-treatment biopsies and chronologically placed ccfDNA obtained between treatments 2 and 3. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
SCNAs including oncogenes or tumor suppressor genes can be deeply involved in tumor progression. For clinical purposes, a number of the amplified genes may be targeted by specific drugs such as ERBB2 ([18] or MET [15]. The alteration in genomic profile can also pint-point the tumors with defects in homologous recombination and hereby define patients sensitive to PARP inhibitors [12]. Consequently, fast and reliable SCNAs analysis is becoming an important part of the clinical cancer diagnostics. In parallel, molecular profiling of cancer biomarkers circulating in the blood is an emerging companion diagnostic. However, clinical implementation is still precautious and limited by the lack of knowledge about the origin and function of ccfDNA [7], one-angle molecular characterization [17] and absence of analytical consensus [3]. In this study, we have validated an array-based copy number analysis of plasma ccfDNA from metastatic cancer patients eligible for inclusion into clinical trials, and hereby supplied a reliable tool required for complex molecular profiling of ccfDNA.
Systematic comparison of ccfDNA and tDNA obtained from tumor biopsy showed that genomic SCNAs profile assembled from ccfDNA to a high degree reflects SCNAs in a tumor. Though, the representative power of ccfDNA can be temporally conditioned, as illustrated by the case where tumor DNA was obtained > 2 months after the ccfDNA sample. In some occasions, genomic profiles of ccfDNA can also marginally differ from those of tDNA obtained from simultaneous biopsies. In view of that, the overlap of numerical and segmental aberrations was never 100%. Considering that a biopsy represents only a temporal and spatial snap-shot of a tumor, there is a risk for misled discovery of potential targets. Hence, ccfDNA can be more representative of the tumor burden than a single biopsy [11]. Therefore, complex analysis of ccfDNA including sequencing and SCNAs profiling can contribute to exploration of tumor heterogeneity [2], [5], major clone characterization and disclose the presence of other lesions.
It was noteworthy that treatment was also found to have a profound effect on the ccfDNA composition. This phenomenon was previously reported as an accidental finding in a lung cancer patient who received neoadjuvant treatment during ccfDNA sampling [26]. Here, the finding was hypothetically explained by rapid tumor shrinkage leading to de-attachment of tumor from adjacent blood supply, and thus to limited release of ccfDNA into the blood stream. However, ccfDNA in our study was taken not only 1–2 days after the treatment initiation but also more than a month in a treatment. The possible explanation can be then based on the fact that the majority of ccfDNA is of non-cancerous origin and originates from apoptotic and stressed cells [6]. Chemotherapy and radiotherapy can stress normal cells and increase non-cancerous ccfDNA release. The SCNAs profile of tumor ccfDNA is then diluted by normal cell ccfDNA and alters the results of ccfDNA analysis. This was clearly manifested on the case study where all ccfDNA samples taken under the treatments showed genomic profiles without detectable SCNAs. Hereby, we raise the consideration that apoptosis/necrosis of normal cells induced by chemotherapy/targeted therapy can influence ccfDNA composition and therefore introduce false-negative findings in the SCNAs analysis of liquid biopsies. OncoScan can only detect SCNAs if the tumor burden is above min.10%.
The sensitivity issue of the arrays and related algorithms can be on the other hand compensated by NGS techniques (detection of mutations with 0.5–1% frequency) or by specialized methods such as ddPCR, which can detect mutations with frequency of 0.005% [16]. However, reliable estimation of tumor burden cannot be detected by low-coverage NGS technology (< 50 ×) and requires reliable SCNA analytical platform as the one demonstrated here. On the other hand, advances in bioinformatics (e.g. ABSOLUTE) will probably soon overcome this obstacle and allow for estimation of high amplitude amplification also in diagnostic settings. Nevertheless, techniques allowing for the assessment of aberrant cell fractions appear to be an essential supplement for proper interpretation of ccfDNA analysis results.
Even though numerical and segmental aberrations can vary between tDNA and ccfDNA as described above, driver events including local amplifications or deletions/biallelic losses remain consistent during tumor evolution [20]. The analysis of 55 genes confirmed this view, since every SCNA detected in tDNA was also present in ccfDNA. SCNAs detected in ccfDNA can therefore be directly used for patient stratification. Yet, the majority of SCNAs on clinically relevant genes is only indicative for treatment and requires further assessment of e.g. mutations in the particular gene. Deletions/LOHs are a typical examples when in order to achieve biallelic inactivation and fulfill the criteria for targeted treatment; the remaining allele has to contain an inactivating mutation. Targeted DNA sequencing [21] or massively multiplexed PCR [11] of ccfDNA still meet the difficulties of estimation of deletions/biallelic losses. Therefore, a supplement of array-based SCNAs analysis to targeted DNA sequencing/WES for genomic screening of cancer patients appears to be an optimal solution for ccfDNA analysis in diagnostic settings.
5. Conclusion
Our results validate a new method for precise detection of copy number alterations in ccfDNA isolated from liquid biopsies (blood) with a great potential to be implemented in the cancer diagnostics within a short time. The genomic aberrations detected in ccfDNA resemble to a high degree alterations in corresponding tumor biopsies. Moreover, array-based genomic profiling provides high resolution and precision in detection of clinically relevant alterations. For proper interpretation of ccfDNA profiles, it is however essential to consider certain technical and biological pitfalls (Table 2). The major concern raised by our study is treatment, which can significantly alter ccfDNA composition by increasing the release of ccfDNA from non-cancerous cells.
Table 2.
Overview over advantages and disadvantages in use of OncoScan for genome-wide analysis of SCNAs in circulating cell-free tumor DNA.
| Characteristics | Comments | Contras | |
|---|---|---|---|
| Input material | 20 ng ccfDNA | 5 ng is possible for recognizable SCNAs profile | Patient with low tumor mass excluded |
| Processing algorithm | TuScan | Freely available via NEXUS (BioDiscovery) | |
| Ploidy assessment | Yes | Manual correction required | |
| Estimation of altered genome | Yes | Only informative, not for diagnostic purposes | |
| Aberrant cell fraction | Yes | Best performance with > 50% of aberrant cell fraction | Composition of ccfDNA might cause false-negative findings |
| Resolution and sensitivity of SCNAs detection | |||
| SCNAs | Precision | ||
| Numerical SCNAs | 20–100% | Visual inspection and correction required | Information about possible biological bias is required; e.g. treatment, heterogeneity, time of sampling |
| Segmental SCNAs | 40–100% | Visual inspection and correction required | |
| High amplitude amplification | 100% | Deviation in assignment of break-points is 7% of segment length | |
| Biallelic losses | Yes | ||
| Clinically relevant aberration | 70(100)% | Precision is 100% after manual correction of LOH | Need for manual interference |
The following are the supplementary data related to this article.
QC parameters of the cfDNA analysis on OncoScan and general characteristics of cfDNA and tDNA
Acknowledgments
Maria Guschina, Julie Fisker Nielsen and Susanne Smed are thanked for their excellent technical assistance.
References
- 1.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M., Antonarakis E.S., Azad N.S., Bardelli A., Brem H., Cameron J.L., Lee C.C., Fecher L.A., Gallia G.L., Gibbs P., Le D., Giuntoli R.L., Goggins M., Hogarty M.D., Holdhoff M., Hong S.M., Jiao Y., Juhl H.H., Kim J.J., Siravegna G., Laheru D.A., Lauricella C., Lim M., Lipson E.J., Marie S.K., Netto G.J., Oliner K.S., Olivi A., Olsson L., Riggins G.J., Sartore-Bianchi A., Schmidt K., Shih I.M., Oba-Shinjo S.M., Siena S., Theodorescu D., Tie J., Harkins T.T., Veronese S., Wang T.L., Weingart J.D., Wolfgang C.L., Wood L.D., Xing D., Hruban R.H., Wu J., Allen P.J., Schmidt C.M., Choti M.A., Velculescu V.E., Kinzler K.W., Vogelstein B., Papadopoulos N., Diaz L.A., Jr. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolli N., Avet-Loiseau H., Wedge D.C., Van Loo P., Alexandrov L.B., Martincorena I., Dawson K.J., Iorio F., Nik-Zainal S., Bignell G.R., Hinton J.W., Li Y., Tubio J.M., McLaren S., S O.M., Butler A.P., Teague J.W., Mudie L., Anderson E., Rashid N., Tai Y.T., Shammas M.A., Sperling A.S., Fulciniti M., Richardson P.G., Parmigiani G., Magrangeas F., Minvielle S., Moreau P., Attal M., Facon T., Futreal P.A., Anderson K.C., Campbell P.J., Munshi N.C. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bronkhorst A.J., Aucamp J., Pretorius P.J. Cell-free DNA: preanalytical variables. Clin. Chim. Acta. 2015;450:243–253. doi: 10.1016/j.cca.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 4.Chan K.C., Jiang P., Zheng Y.W., Liao G.J., Sun H., Wong J., Siu S.S., Chan W.C., Chan S.L., Chan A.T., Lai P.B., Chiu R.W., Lo Y.M. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin. Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 5.Chicard M., Boyault S., Colmet Daage L., Richer W., Gentien D., Pierron G., Lapouble E., Bellini A., Clement N., Iacono I., Brejon S., Carrere M., Reyes C., Hocking T., Bernard V., Peuchmaur M., Corradini N., Faure-Conter C., Coze C., Plantaz D., Defachelles A.S., Thebaud E., Gambart M., Millot F., Valteau-Couanet D., Michon J., Puisieux A., Delattre O., Combaret V., Schleiermacher G. Genomic copy number profiling using circulating free tumor DNA highlights heterogeneity in neuroblastoma. Clin. Cancer Res. 2016;22:5564–5573. doi: 10.1158/1078-0432.CCR-16-0500. [DOI] [PubMed] [Google Scholar]
- 6.Glebova K., Veiko N., Kostyuk S., Izhevskaya V., Baranova A. Oxidized extracellular DNA as a stress signal that may modify response to anticancer therapy. Cancer Lett. 2015;356:22–33. doi: 10.1016/j.canlet.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Gravina S., Sedivy J.M., Vijg J. The dark side of circulating nucleic acids. Aging Cell. 2016;15:398–399. doi: 10.1111/acel.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardenbol P., Baner J., Jain M., Nilsson M., Namsaraev E.A., Karlin-Neumann G.A., Fakhrai-Rad H., Ronaghi M., Willis T.D., Landegren U., Davis R.W. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 9.Hashad D., Sorour A., Ghazal A., Talaat I. Free circulating tumor DNA as a diagnostic marker for breast cancer. J. Clin. Lab. Anal. 2012;26:467–472. doi: 10.1002/jcla.21548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heitzer E., Auer M., Ulz P., Geigl J.B., Speicher M.R. Circulating tumor cells and DNA as liquid biopsies. Genome Med. 2013;5:73. doi: 10.1186/gm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkizlar E., Zimmermann B., Constantin T., Swenerton R., Hoang B., Wayham N., Babiarz J.E., Demko Z., Pelham R.J., Kareht S., Simon A.L., Jinnett K.N., Rabinowitz M., Sigurjonsson S., Hill M. Detection of clonal and subclonal copy-number variants in cell-free DNA from patients with breast cancer using a massively multiplexed PCR methodology. Transl. Oncol. 2015;8:407–416. doi: 10.1016/j.tranon.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., Nava Rodrigues D., Robinson D., Omlin A., Tunariu N., Boysen G., Porta N., Flohr P., Gillman A., Figueiredo I., Paulding C., Seed G., Jain S., Ralph C., Protheroe A., Hussain S., Jones R., Elliott T., McGovern U., Bianchini D., Goodall J., Zafeiriou Z., Williamson C.T., Ferraldeschi R., Riisnaes R., Ebbs B., Fowler G., Roda D., Yuan W., Wu Y.M., Cao X., Brough R., Pemberton H., A'Hern R., Swain A., Kunju L.P., Eeles R., Attard G., Lord C.J., Ashworth A., Rubin M.A., Knudsen K.E., Feng F.Y., Chinnaiyan A.M., Hall E., de Bono J.S. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouliere F., Robert B., Arnau Peyrotte E., Del Rio M., Ychou M., Molina F., Gongora C., Thierry A.R. High fragmentation characterizes tumour-derived circulating DNA. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murtaza M., Dawson S.J., Tsui D.W., Gale D., Forshew T., Piskorz A.M., Parkinson C., Chin S.F., Kingsbury Z., Wong A.S., Marass F., Humphray S., Hadfield J., Bentley D., Chin T.M., Brenton J.D., Caldas C., Rosenfeld N. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 15.Nishio M., Horiike A., Nokihara H., Horinouchi H., Nakamichi S., Wakui H., Ohyanagi F., Kudo K., Yanagitani N., Takahashi S., Kuboki Y., Yamamoto N., Yamada Y., Abe M., Tahata T., Tamura T. Phase I study of the anti-MET antibody onartuzumab in patients with solid tumors and MET-positive lung cancer. Investig. New Drugs. 2015;33:632–640. doi: 10.1007/s10637-015-0227-5. [DOI] [PubMed] [Google Scholar]
- 16.Page K., Guttery D.S., Fernandez-Garcia D., Hills A., Hastings R.K., Luo J., Goddard K., Shahin V., Woodley-Barker L., Rosales B.M., Coombes R.C., Stebbing J., Shaw J.A. Next generation sequencing of circulating cell-free DNA for evaluating mutations and gene amplification in metastatic breast cancer. Clin. Chem. 2017;63:532–541. doi: 10.1373/clinchem.2016.261834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polivka J., Jr., Pesta M., Janku F. Testing for oncogenic molecular aberrations in cell-free DNA-based liquid biopsies in the clinic: are we there yet? Expert. Rev. Mol. Diagn. 2015;15:1631–1644. doi: 10.1586/14737159.2015.1110021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pritchard K.I., Shepherd L.E., O'Malley F.P., Andrulis I.L., Tu D., Bramwell V.H., Levine M.N., National Cancer Institute of Canada Clinical Trials, G HER2 and responsiveness of breast cancer to adjuvant chemotherapy. N. Engl. J. Med. 2006;354:2103–2111. doi: 10.1056/NEJMoa054504. [DOI] [PubMed] [Google Scholar]
- 19.Rothwell D.G., Smith N., Morris D., Leong H.S., Li Y., Hollebecque A., Ayub M., Carter L., Antonello J., Franklin L., Miller C., Blackhall F., Dive C., Brady G. Genetic profiling of tumours using both circulating free DNA and circulating tumour cells isolated from the same preserved whole blood sample. Mol. Oncol. 2016;10:566–574. doi: 10.1016/j.molonc.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sottoriva A., Kang H., Ma Z., Graham T.A., Salomon M.P., Zhao J., Marjoram P., Siegmund K., Press M.F., Shibata D., Curtis C. A Big Bang model of human colorectal tumor growth. Nat. Genet. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takai E., Totoki Y., Nakamura H., Morizane C., Nara S., Hama N., Suzuki M., Furukawa E., Kato M., Hayashi H., Kohno T., Ueno H., Shimada K., Okusaka T., Nakagama H., Shibata T., Yachida S. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci. Rep. 2015;5:18425. doi: 10.1038/srep18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Togneri F.S., Ward D.G., Foster J.M., Devall A.J., Wojtowicz P., Alyas S., Vasques F.R., Oumie A., James N.D., Cheng K.K., Zeegers M.P., Deshmukh N., O'Sullivan B., Taniere P., Spink K.G., McMullan D.J., Griffiths M., Bryan R.T. Genomic complexity of urothelial bladder cancer revealed in urinary cfDNA. Eur. J. Hum. Genet. 2016;24:1167–1174. doi: 10.1038/ejhg.2015.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toro P.V., Erlanger B., Beaver J.A., Cochran R.L., VanDenBerg D.A., Yakim E., Cravero K., Chu D., Zabransky D.J., Wong H.Y., Croessmann S., Parsons H., Hurley P.J., Lauring J., Park B.H. Comparison of cell stabilizing blood collection tubes for circulating plasma tumor DNA. Clin. Biochem. 2015;48:993–998. doi: 10.1016/j.clinbiochem.2015.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuxen I.V., Jonson L., Santoni-Rugiu E., Hasselby J.P., Nielsen F.C., Lassen U. Personalized oncology: genomic screening in phase 1. APMIS. 2014;122:723–733. doi: 10.1111/apm.12293. [DOI] [PubMed] [Google Scholar]
- 25.Van Allen E.M., Wagle N., Stojanov P., Perrin D.L., Cibulskis K., Marlow S., Jane-Valbuena J., Friedrich D.C., Kryukov G., Carter S.L., McKenna A., Sivachenko A., Rosenberg M., Kiezun A., Voet D., Lawrence M., Lichtenstein L.T., Gentry J.G., Huang F.W., Fostel J., Farlow D., Barbie D., Gandhi L., Lander E.S., Gray S.W., Joffe S., Janne P., Garber J., MacConaill L., Lindeman N., Rollins B., Kantoff P., Fisher S.A., Gabriel S., Getz G., Garraway L.A. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat. Med. 2014;20:682–688. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia S., Huang C.C., Le M., Dittmar R., Du M., Yuan T., Guo Y., Wang Y., Wang X., Tsai S., Suster S., Mackinnon A.C., Wang L. Genomic variations in plasma cell free DNA differentiate early stage lung cancers from normal controls. Lung Cancer. 2015;90:78–84. doi: 10.1016/j.lungcan.2015.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
QC parameters of the cfDNA analysis on OncoScan and general characteristics of cfDNA and tDNA