Abstract
Klinefelter syndrome (KS) (47,XXY) is the most common male sex chromosome aneuploidy. Diagnosis and clinical supervision remain a challenge due to varying phenotypic presentation and insufficient characterization of the syndrome. Here we combine health data-driven epidemiology and molecular level systems biology to improve the understanding of KS and the molecular interplay influencing its comorbidities. In total, 78 overrepresented KS comorbidities were identified using in- and out-patient registry data from the entire Danish population covering 6.8 million individuals. The comorbidities extracted included both clinically well-known (e.g. infertility and osteoporosis) and still less established KS comorbidities (e.g. pituitary gland hypofunction and dental caries). Several systems biology approaches were applied to identify key molecular players underlying KS comorbidities: Identification of co-expressed modules as well as central hubs and gene dosage perturbed protein complexes in a KS comorbidity network build from known disease proteins and their protein–protein interactions. The systems biology approaches together pointed to novel aspects of KS disease phenotypes including perturbed Jak-STAT pathway, dysregulated genes important for disturbed immune system (IL4), energy balance (POMC and LEP) and erythropoietin signalling in KS. We present an extended epidemiological study that links KS comorbidities to the molecular level and identify potential causal players in the disease biology underlying the identified comorbidities.
Introduction
Klinefelter syndrome (KS) (47,XXY) is the most common male sex chromosome aneuploidy with a prevalence of approximately 1 in 650 males. Diagnosis of KS and prevention and treatment of its comorbidities keep being a challenge. It is estimated that only 25% of KS males are ever diagnosed due to variable phenotype and possibly insufficient characterisation of the syndrome. Many KS males are only diagnosed in adult life due to infertility (1).
The presence of an extra chromosome can cause higher levels of gene expression and gene products in amounts affected by regulation at different levels, protein degradation and modification (2). Also the interaction pattern of the proteins encoded by the additional chromosome can influence the severity of a trisomy (3). In the case of KS, X chromosome inactivation (XCI) counterweights the extra dosage from the many X chromosome genes (n 2000). Still, roughly 15% of X-linked genes escape XCI including genes in the two pseudoautosomal regions (PARs) (4).
KS males present certain shared phenotypes such as small testes, tall stature, narrow shoulders, broad hips, sparse body hair, gynecomastia and decreased verbal intelligence. KS males also have increased risk of a wide range of additional disorders compared to the background population, i.e. comorbidities. Testicular dysfunction is prevalent in KS. It is estimated that KS causes 1–3% of all male infertility cases (5). In fact, 10% of all male patients with azoospermia are likely to be caused by KS. Numerous KS comorbidities are related to hypogonadism: osteoporosis, cognitive disorders, metabolic syndrome and diabetes and cardiac disease. Consequently, KS males are allocated lifelong testosterone replacement therapy from puberty. Yet, the therapy does not counteract or prevent all comorbidities. It is still unclear whether the chromosome abnormality adds an extra testosterone-independent facet to the complexity of KS comorbidities (6).
Comorbidities have been of interest for many years in epidemiology studies. In recent years, more data-driven approaches have linked molecular aetiology to observed disease correlations extracted from electronic patient records. Studying disease comorbidities using a systems biology approach potentially enhances the understanding of disease origin and progression and the longitudinal interplay between comorbidities (7,8). Genes associated with the same disease tend to be co-expressed and work together in protein complexes (9–12). Thus, studying co-expressed genes and protein complexes can give further functional insight into the biological transformation between healthy and diseased individuals and potentially further improve disease diagnosis, treatment and target discovery (13).
Here we present a population-wide study of KS comorbidities extracted from data in the Danish National Patient Registry (14) collected over a period of 18.5 years. Gene expression data was generated from KS patients and controls from peripheral blood and co-expressed modules of high relevance for the KS phenotype were identified. A KS comorbidity network was built based on the extracted disease-pair frequencies, known disease-associated proteins and their protein–protein interactions (PPIs). Disease distances and nodes central for the interplay between diseases were subsequently explored in the network. The various analytical approaches used in this study point out the same disturbed functionalities and thereby strengthen their relevance for KS.
Results
Klinefelter syndrome comorbidities
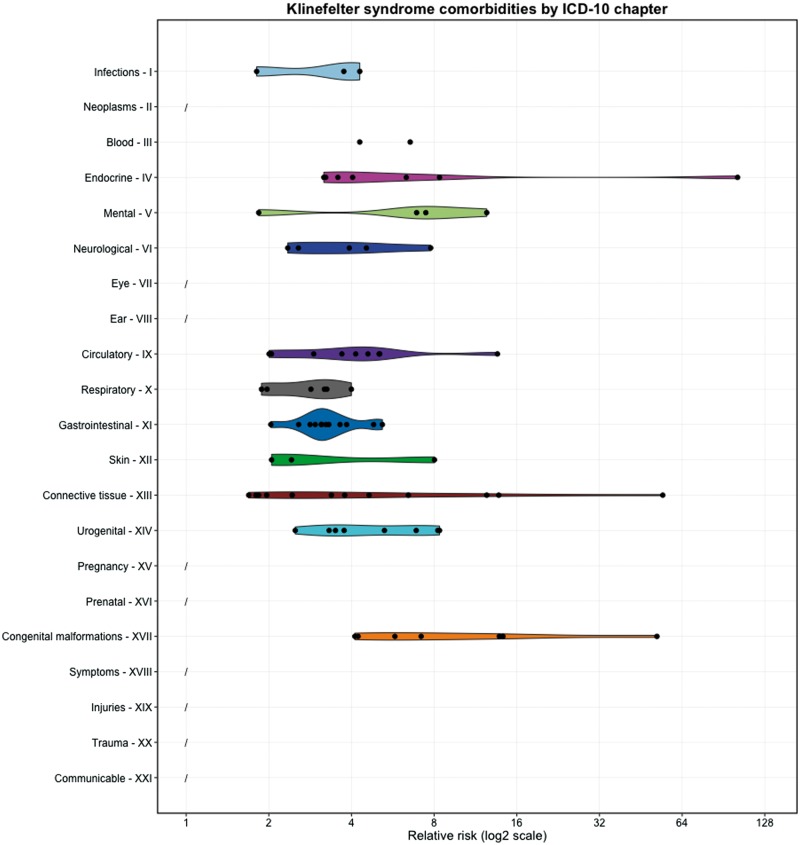
KS comorbidities were systematically extracted from the Danish National Patient Registry (14) using the International Classification of Diseases version 10 (ICD-10) level-3 codes checking for co-occurrence of the KS code Q98 with all other disease codes (see ‘Materials and Methods’ for details). Comorbidities were defined as diseases significantly diagnosed a minimum of 50% more frequently in men with KS compared to controls, i.e. relative risk (RR) ≥ 1.5. P-values were obtained using the Fisher’s test and Benjamini-Hochberg correction was applied to obtain a false discovery rate (FDR) of 5% corresponding to P ≤ 0.0082. A total of 78 significant KS comorbidities was identified with a minimum of five KS patients diagnosed (Supplementary Material, Table S1). Figure 1 displays the KS comorbidities by ICD-10 chapters (short abbreviations for chapter titles in Supplementary Material, Table S2). Many well-known KS comorbidities were as expected observed in the patient records including E29 ‘testicular dysfunction’ (RR = 101.88), M81 ‘osteoporosis without pathological fraction’ (RR = 12.433), N62 ‘hypertrophy of the breast’ (RR = 8.252), F79 ‘unspecified mental retardation’ (RR = 6.907), E14 ‘unspecified diabetes mellitus’ (RR = 6.332), Q53 ‘undescended testicle’ (RR = 4.227), E66 ‘obesity’ (RR = 4.038) and G40 ‘epilepsy’ (RR = 3.928) (15–17). Less established KS comorbidities were also observed e.g. E23 ‘hypofunction and other disorders of the pituitary gland’ (RR = 8.369), K02 ‘dental caries’ (RR = 4.816) and K07 ‘dentofacial anomalities [including malocclusion]’ (RR = 3.84). All comorbid level-3 codes were checked for observations representing phenotypes diverging in opposite directions as level-4 codes. Only one such case was identified: E29 ‘testicular dysfunction’ (RR = 101.88) that includes both E29.0 ‘testicular hyperfunction’ and E29.1 ‘testicular hypofunction’ of which only the latter was found comorbid with KS (RR = 117.67).
Figure 1.
Klinefelter syndrome (KS) comorbidities extracted from Danish patient records (n = 78). Comorbidities are summarised by disease chapter and each dot represents a KS comorbidity with a certain relative risk (RR). The disease chapter titles are shortened (see Supplementary Material, Table S2). There were no comorbidities between KS and disorders from chapters 2, 7, 8, 16 and 18–21. See Supplementary Material, Table S1 for the full list of KS comorbidities and RRs.
Differentially expressed genes in peripheral blood
Blood samples from eight KS patients and eight controls were expression profiled using microarrays. A total of 12,947 genes were found to be expressed in at least one of the 16 samples (intensity cut off ≥7.27). In this gene set, we found an enrichment of genes encoded by chromosomes 4 (P = 0.0232), X (P = 0.017) and Y (P = 2.483e-05) (Fisher’s exact test, P ≤ 0.05) (Supplementary Material, Fig. S1). Statistical testing identified 363 differentially expressed genes (146 up- and 217 down-regulated) considering an absolute log2 fold change (lfc) ≥ 1.5 and a Benjamini–Hochberg-corrected FDR of 5% corresponding to P ≤ 0.0069 (Supplementary Material, Table S3).
The most significant differentially expressed and upregulated KS gene was the long non-coding RNA X inactive specific transcript (XIST) (lfc = 8.41). A total of 22 X chromosome genes was deregulated in KS compared to controls: 16 were up-regulated (AKAP17A, ASMTL, CSF2RA, EIF1AX, EIF2S3, GPR82, GTPBP6, IL3RA, PLCXD1, PPP2R3B, PRKX, RP11-EPO6O15.3, SEPT6, SLC25A6, TMSB4X and XIST) and six were down-regulated (BEND2, BEX1, COX7B, FOXO4, NHS and TFE3). Some of these genes were expressed from pseudoautosomal region 1 (PAR1): AKAP17A, ASMTL, CSF2RA, GTPBP6, IL3RA and PLCXD1, but no differentially expressed genes were observed encoded from pseudoautosomal region 2. An interesting observation was the up-regulation of another long non-coding RNA, RP11-706O15.3. The function of this gene is still unknown, but it is located near PAR1 at Xp22.33 and, thus, potentially escapes XCI and/or might be involved in the XCI process itself.
Gene set enrichment analysis (GSEA) was performed to gain functional understanding of the deregulated genes (n = 363). The most enriched biological process terms were ‘immune response’ and ‘response to bacterium’ and the most enriched pathways were ‘Cytokine–cytokine receptor interaction’ and ‘Jak-STAT signalling pathway’ (full list in Supplementary Material, Table S4). The ‘Cytokine–cytokine receptor interaction’ pathway included 18 deregulated genes (seven up-regulated: CCR7, CSF2RA, IFNA4, IL3RA, IL6ST, IL7R and TNFRSF25; and 11 down-regulated: CXCL1, CXCL16, CXCL2, CXCR1, EPOR, IL1R1, IL4, INHBB, LEP, LEPR and OSM). Multiple genes are involved in both the ‘Cytokine–cytokine receptor interaction’ and the ‘Jak-STAT signalling pathway’ (CSF2RA, EPOR, IFNA4, IL3RA, IL4, IL6ST, IL7R, LEP, LEPR and OSM).
Co-expressed gene modules correlating with Klinefelter syndrome status
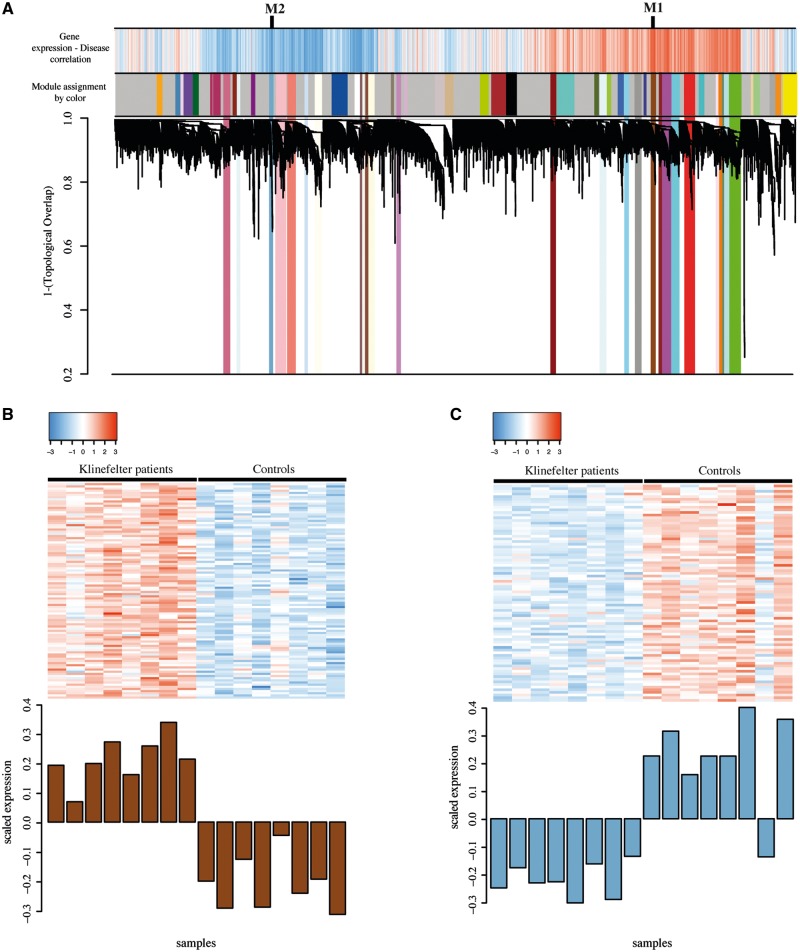
A total of 54 co-expressed modules containing genes with similar expression patterns was identified by applying weighted gene co-expression network analysis (WGCNA) (18) (see ‘Materials and Methods’ for details). Of these, 27 modules had expression signatures that correlated with KS status either positively (higher expressed in KS than controls, n = 14) or negatively (lower expressed in KS than controls, n = 13). All modules were assigned a colour and labelled numerically according to their degree of significant correlation to KS status (Fig. 2A and Supplementary Material, Table S5).
Figure 2.
Gene co-expression modules correlating with Klinefelter syndrome (KS) status. (A) Gene co-expression analysis dendrogram showing co-expressed gene modules identified using Weighted gene co-expression network analysis (WGCNA) (n = 54). The top colour bar states expression correlation with KS status (red: positive correlation; blue: negative correlation). The bar below informs about module membership. In total, 27 modules correlated significantly with KS status and reported here as coloured across the dendrogram. (B) Scaled expression per individual of the 99 genes in the most positively correlated module with KS status, M1/saddlebrown. (C) Scaled expression per individual of the 81 genes in the most negatively correlated module with KS status, M2/skyblue3.
The most significant and positively correlated module was M1/saddlebrown (WGCNA student asymptotic t-test P = 1.12e−07) (Fig. 2B). This module included 99 genes including many X-linked and PAR1 genes: AKAP17A, CA5B, EIF1AX, IL3RA, SCML1, SEPT6, XIST and ZBED1. An interesting gene in the module was DDX17 that recently was discovered as protein interactor of XIST (19). M1 contained 32 of the significantly up-regulated genes in KS identified in the statistical test described previously (Supplementary Material, Table S3). GSEA showed that the M1 genes were involved in the KEGG pathways ‘neuroactive ligand-receptor interaction pathway’ and ‘calcium signalling pathway’, and several Online Mendelian Inheritance in Man (OMIM) diseases such as ‘abnormal thyroid hormone metabolism’, ‘epilepsy’ and ‘deafness’ (Supplementary Material, Table S5 for complete list).
The most significant negatively correlated module was M2/skyblue3 (WGCNA student asymptotic t-test P = 5.21e−06) (Fig. 2C). This module included 81 genes; none of them located in the PARs. M2 contained 38 of the down-regulated genes in KS and GSEA revealed enriched biological processes such as ‘immune system process’ and ‘response to bacteria’ and the KEGG pathway ‘platelet activation’ (Supplementary Material, Table S5). Interestingly, both WGCNA and the differential expression analysis showed significantly enriched biological terms related to the immune system. These terms contained mainly downregulated genes. In order to evaluate the overlap between the genes involved in these terms within M2 and the deregulated genes we computed the hypergeometric distribution obtaining a significant overlap (P = 3.35e−47). This result showed that the GSEA analysis and the consequent interpretation of the results were consistent, that is, the down-regulation of genes involved in the immune system could explain some diseases associated with KS and its comorbidities.
Supplementary Material, Figure S2 displays the chromosomal distribution of genes in each of the 27 modules correlating significantly with KS status. M1 was one of the modules with the highest X chromosome gene content. Yet, overall the modules contained genes spread out genome-wide underlining that having an extra chromosome changes gene expression globally.
Klinefelter syndrome comorbidity network
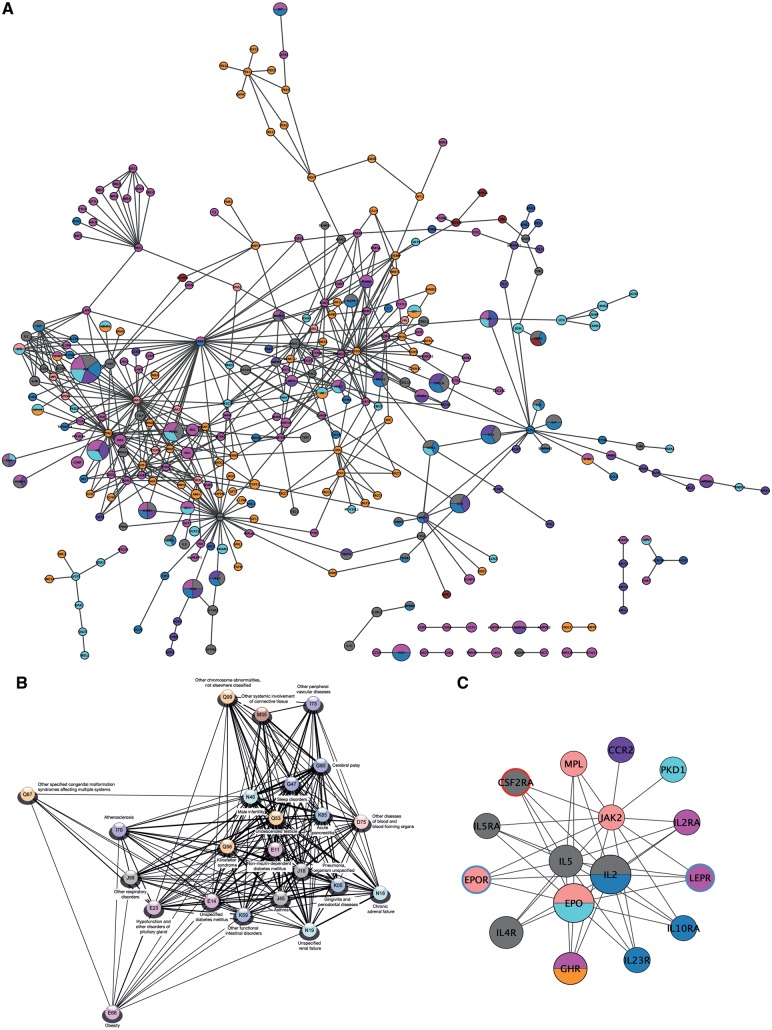
One of the main aims of this study was to investigate the molecular relationship between KS and its comorbidities and to identify key players in their crosstalk. For this purpose, a KS comorbidity network was constructed consisting of disease sub-networks each covering KS or one of its comorbidities. Each disease sub-network was built of associated proteins extracted from disease-gene databases and following connecting with observed PPIs from InWeb_InBioMap (InWeb_IM) (11). The disease sub-networks were following merged into the KS comorbidity network (see ‘Materials and Methods’ and Supplementary Material, Fig. S3 for further details). Each of the sub-networks were tested for increased PPI connectivity compared to random networks of the same size and node degree distribution, thereby testing their functional relevance under the hypothesis that functionally related proteins are more likely to interact than unrelated proteins (20). It was possible to build disease sub-networks for KS and 29 of its comorbidities, and the sub-networks for KS and 22 comorbidity subsequently showed significantly high connectivity (P-value cut off ≤ 0.05) (Supplementary Material, Table S6). Accordingly, the KS comorbidity network covered 23 diseases and consisted of 299 nodes and 528 edges (Fig. 3A).
Figure 3.
Klinefelter syndrome (KS) comorbidity network, disease separation and protein complex with disturbed functionality. (A) The global KS comorbidity network created from disease-associated proteins from knowledge databases linked by observed protein–protein interactions (PPIs). The network represents the sub-networks for KS and the 22 comorbidities with significant high connectivity. Nodes are coloured according to their associated disease chapter similarly to Figure 1. Nodes associated to diseases belonging to multiple chapters are multi-coloured. Node size is scaled to the number of KS comorbidities each node is associated with. (B) Disease separations of the 23 diseases represented in the comorbidity network clustered after similarity and only including edges between diseases with overlapping network topology. (C) The most statistical significant cluster in the network. The size and colouring of the nodes are similar to in the full network. Node border colour represents significant up- (red) and down-regulation (blue) in KS compared to controls.
Network distances between each of the disease pairs in the KS comorbidity network were determined by comparing the network-based distances between the proteins involved in the two diseases (21). As expected, most diseases had a negative disease separation indicating overlapping network topology. The comorbidities with most PPI topological overlap (TO) to KS was Q53 ‘undescended testicle’, N46 ‘male infertility’ and E14 ‘unspecified diabetes mellitus’ (Fig. 3B and Supplementary Material, Table S7). The comorbidity network had the common topological characteristics of networks from the molecular level biological domain and the KS sub-network did not stand out in any topological measures (see Supplementary Material, Text S1 and Text S2 for more details and Supplementary Material, Table S8).
Numerous nodes in the network were associated to multiple KS comorbidities. In this study, we defined two categories of such nodes: comorbidity nodes and comorbidity-linking hubs. Comorbidity nodes are associated in disease-gene databases to numerous comorbidities themselves. Comorbidity-linking hubs are central to multiple comorbidities through their first-order interaction partners, which are associated with multiple comorbidities. Thus, the term ‘hub’ here refers to the centrality of the node to numerous diseases and is not referring to the PPI connectivity of the node. The KS comorbidity network can be considered as the ‘backbone’ for tissue-agnostic network and applying gene expression data from different tissues will give tissue-specific information. In this study, we applied the gene expression data from blood presented above. This data set covered 174 nodes in the network (Supplementary Material, Table S9).
Some comorbidity nodes were associated with numerous codes from the same or disease-related ICD-10 chapters, e.g. chapters I and Q both featuring heart diseases. Of more interest were comorbidity nodes linking diseases affecting diverse pathophysiology across tissues. Numerous such nodes were present in the network and involved in up to twelve diseases. Three nodes linking minimum five diseases were significantly down-regulated in KS: POMC (lfc = −0.762), LEP (lfc = −0.785) and IL4 (lfc = −0.814) linking seven, six and five diseases, respectively.
Comorbidity-linking hubs were also abundant in the network, i.e. nodes that connect multiple comorbidities through their first order interactions partners. The biggest deregulated comorbidity-linking hubs were: LEP, LEPR (lfc = −0.600), POMC, EPOR (lfc = −0.603), CSF2RA (lfc = 0.648), and IL4 connecting nine, eight, eight, seven, seven and six diseases, respectively. Thus, LEP, POMC and IL4 were both comorbidity nodes and comorbidity-linking hubs, i.e. both associated to multiple diseases themselves and also linking multiple diseases through their first-order interaction partners.
Protein complexes with perturbed functionality in KS
Protein complexes, i.e. clusters, were identified in the comorbidity network. In total 18 significant clusters (P ≤ 0.05) were identified with the most significant cluster, C1, containing 16 proteins: CCR2, CSF2RA, EPO, EPOR, GHR, IL2, IL23R, IL2RA, IL4R, IL5, IL5RA, IL10RA, JAK2, LEPR, MPL and PKD1 (Fig. 3C and Supplementary Material, Table S10). These proteins are involved in three main KEGG pathways: ‘Cytokine–cytokine receptor interaction’, ‘Jak-STAT signalling pathway’ and ‘hematopoietic cell lineage’ and also related to numerous KS comorbidities such as obesity, diabetes mellitus, atherosclerosis and respiratory disorders. In this cluster, CSF2RA, LEP and EPOR were deregulated as before mentioned, which might cause this complex to have disturbed functionality in KS.
Discussion
KS is the most common male sex chromosome aneuploidy. KS males have a 70% increased risk of being hospitalized and have a reduced life expectancy by two years compared to normal karyotype males due to increased disease susceptibility (22). Still, KS is under diagnosed and treatment is most often limited to testosterone therapy, which overcomes some but by far all comorbidities. In this study, we identified 78 significant KS comorbidities using population-wide patient record data. Up to half of the diseases were not previously reported in comorbidity studies on KS comorbidities (15–17). Among the novel and unexpected comorbidity findings was E23 ‘hypofunction and other disorders of the pituitary gland’ (RR = 5.42). Usually KS males present a normal growth hormone-IGF-I axis (23) and increased luteinizing hormone and follicle-stimulating hormone levels due to their androgen deficiency (17,24). Our observation could in theory result from inappropriate ICD-10 registration of luteinizing hormone/follicle-stimulating hormone hyposecretion observed during androgen replacement therapy, which occasionally suppresses gonadotropins to subnormal levels due to negative feedback. Another still less established comorbidity of KS is dental anomalies although several case studies of observed late eruption and dental problems in KS males have been published (25–28). Consequently, a conclusion from this study is to prioritise dental observation of KS males.
XCI compensates extensively for the extra X chromosome gene dosage in KS males and probably increases their chances of survival compared to other aneuploidies. The master effector of XCI, the non-coding RNA XIST, was the most significantly upregulated gene in KS consistent with previous findings (29,30). Still, it is estimated that approximately 15% of the X encoded genes escape XCI (31) prompting the characteristic KS phenotypes and comorbidities. In this study, we identified the genes escaping XCI. We found that the majority of the 363 significantly deregulated genes were not X encoded. Thus, having an extra X chromosome influences gene expression across the genome and underlines the trans-regulatory effect of the escapees. Interestingly, it has been shown that the mere presence of extra chromosome leads to genomic instability and changes in gene expression in cancer cell lines (32–34). These studies showed that gain of chromosomes triggers replication stress promoting genomic instability and possibly contributing to tumorigenesis in cell lines. Moreover, the authors showed a remarkable change in the transcriptional regulation (33). Other studies showed that excess expression of one or more X chromosome genes influences brain development and it has been hypothesized that specific brain-expressed genes that are known to escape XCI could be responsible for cognitive disorders of KS patients (35,36). Future work with in vivo analyses is needed to better understand this phenomenon, but the gain of extra chromosomes seems to be responsible for the observed changes in the global gene expression.
The most enriched biological process term for the 363 differential expressed genes was ‘immune response’. This is consistent with that the lack of testosterone in KS patients enhances cellular and humoral immunity and androgen replacement treatment may suppress this process (37). Moreover, it has been reported that infectious diseases cause increased mortality of KS patients (38).
Previously focus has mostly been on single protein–disease relationships. Yet, proteins work in concert and we are only starting to fully understand how protein subunits work in a network perspective. The KS comorbidity network emphasises knowledge that can be further studied as disease interplay at the PPI level. The three downregulated comorbidity nodes and comorbidity-linking hubs, POMC, LEP and IL4, are involved in the regulation of energy balance and the immune system, both known to be affected by KS. POMC is a hormone precursor for multiple peptides active across tissues including adrenocorticotropic hormone that regulates cortisol release, and also the three melanocyte stimulating hormones that balance food intake and energy consumption amongst others. LEP is also a long-term regulator of the energy balance by suppressing food intake and works through POMC-containing neurons (39). IL4 is produced by T-cells and plays an essential role in inducing adaptive immune response (40). Possibly, IL4 also plays a role in autoimmune diseases, which KS patients have increased risk of, including triggering self-antigen presentation in pancreatic islet cells driving the development of autoimmune diabetes (41). The comorbidity-linking hub EPOR is the receptor for EPO. Testosterone stimulates erythropoiesis through EPO stimulation (42). Thus, the observed hypoandrogenism in KS might explain the down-regulation of EPOR.
The combined network biology approaches used in this study emphasised perturbation of ‘cytokine–cytokine receptor interaction’ and ‘Jak-STAT pathways’, immune system alterations and disturbed EPO signalling and energy balance through POMC and LEP in KS. LEP and EPO appear relevant to the increased prevalence of at least diabetes and obesity in KS patients (43) and might be used as biomarkers for the clinical management of KS in the future. An interesting observation was that both LEP and LEPR were down-regulated. Another possible future treatment of KS might be to add additional XIST molecules edited to complement the escapees like it was done recently for Trisomy 21 (44). This could lead to silencing of the complete extra X chromosome. Administering such treatment in foetal life has the potential of KS males being symptom free, because it obstructs the primary inducers of the KS-associated comorbidities.
A potential weakness of this study is that the use of registry data solely consisting of hospital diagnosis data for each patient. There are several factors such as socio-economic factors, lifestyle and other drug prescriptions that affect the risk of diseases. However, it is difficult to obtain such data for the entire population of a full country. Thorough follow-up studies should include as many as these as possible.
As already mentioned a significant fraction of KS patients are never diagnosed. The comorbidity profile of KS described in this study could potentially be used to identify undiagnosed patients. However, the use of a disease trajectory approach similar to the one published earlier (45,46) where the temporal order of diseases is incorporated would presumably be more powerful compared to the comorbidity profile described here. This will be the target of further work.
Although the expression data from different individuals in general was in good agreement the cohort is admittedly small. Nevertheless, we pointed out the importance and power of data integration approaches, in particular the integration of comorbidities with other biological data types. In future studies we will increase the number of samples, in order to further confirm the results. In this work, a first attempt has been done providing relevant results consistent with previous studies and adding novel biological aspects of KS.
In this study, several novel and less clinically established KS comorbidities were identified in an unbiased, data-driven way. Several genes in the identified co-expressed modules, network hubs and clusters may be promising biomarkers for prevention or treatment of common KS comorbidities. The data-driven epidemiological approach presented here can be applied to any aneuploidy and disease in general and shed light on their dosage-provoked comorbidities, established as well as non-established ones pointing at their aetiology and molecular interplay.
Materials and Methods
Klinefelter syndrome comorbidities
Comorbidities were identified from the over-occurrence of KS with another disease, X, compared to individuals not having KS. In this study, we used diagnoses from electronic patient records in the Danish National Patient Registry (14) for 6.8 million patients hospitalized between 1996 and 2014 comprising 90 million hospital encounters and 87 million unique patient-diagnosis associations at the ICD-10 level-3 (45,47).
ICD-10 has a hierarchical structure divided into chapters, blocks, level-3 codes and finally level-4 codes, which are the most specific. Each level-4 code can be ‘rounded’ to its parent level-3 code and further to the block or chapter. To get larger counts and thereby better statistics, we have used level-3 codes. This also reduces a misclassification where an inappropriate level-4 code was selected among the level-4 codes under a specific level-3 code.
KS comorbidities were extracted using the level-3 disease code Q98 ‘other sex chromosome abnormalities, male phenotype, not elsewhere classified’, but only including patients diagnosed with level-4 codes Q98.0 ‘KS karyotype 47,XXY’, Q98.1 ‘KS, male with more than two X chromosomes’, Q98.2 ‘KS, male with 46,XX karyotype’ or Q98.4 ‘KS, unspecified’ (n = 595). Comorbidities were defined as significantly increased RR of Q98 co-occurring with other level-3 ICD-10 codes when compared to a control population. Fisher’s Exact test was used to assign a P-value to the association using uncorrected counts. We defined the full control population as all patients in the Danish National Patient Registry without a KS diagnosis. Including all patients from this population as the non-exposed comparison group in the statistical analysis will give a very skewed number of exposed (KS) patients versus non-exposed patients. Therefore, we sampled a matched background (BG) population including 20 controls per KS patient. The choice of 20 control patients per KS patient is a compromise between having enough controls to pick up rare diseases, but not too many to increase the statistical power of the test. The controls were matched to have same gender and year of birth as the KS patient.
RR is normally defined as:
With this definition, RR overestimates the association of rare diseases. This can be addressed by adding, a pseudo count (or continuity correction) to each of the four patient groups in the calculation of the RR. For a pseudo count of k, RR is defined as:
A study found a pseudo count of 1/2 to be recommended for a generalization of Fisher’s Exact test to N groups (the Mantel–Haenszel procedure) (48). We rounded this to an integer of 1. For the comorbidities, we required a minimum of five KS patients diagnosed with disease X and a RR ≥ 1.5. We utilized Benjamini–Hochberg to limit the FDR to 5%. The level-4 ICD-10 subcategories of the extracted KS comorbidities were checked for observations representing phenotypes diverging in opposite directions.
Three level-3 codes had phenotypic opposite directed level-4 codes: E23 ‘Hypofunction and other disorders of pituitary gland’, E29 ‘Testicular dysfunction’ and E34 ‘Other endocrine disorders’. For these three level-3 codes, we used the level-4 codes to get more detailed information after seeing that the level-3 code was significant.
Gene expression data and analysis
Blood samples were available from eight KS males and eight normal karyotype male controls. Total RNA was extracted with QIAamp RNA Blood Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. RNA quantity and quality was determined using NanoDrop (Thermo Scientific, Wilmington, Delaware, US) with Bioanalyzer Nano Kit (Agilent Technologies, Santa Clara, California, US). The samples were amplified (one round) using the MessageAmp II aRNA Amplification Kit (Applied Biosystems, Carlsbad, California, US) and the aRNA was applied and run on Agilent Human Genome Microarrays 44K (Agilent Technologies, Santa Clara, California, US). Hybridization and scanning of the one-color arrays were performed according to instructions from the manufacturer (Agilent Technologies, Santa Clara, California, US).
Processing of the gene expression microarray data was performed using the R software suite version 3.3.1 (www.r-project.org). gProcessedSignals were loaded into the limma R/Bioconductor package (49,50) and normalized between arrays using the quantile normalization procedure (51). Probes were collapsed for each systematic gene ID by taken the median expression value. The age of the KS patients and controls varied (Supplementary Material, Table S11), which was corrected for using the ComBat method (52) implemented in the Surrogate Variable Analysis package (53). A Gaussian mixture model was employed to determine a threshold of genuine gene expression using the mixtools package (54). The expectation maximization algorithm was used to model the log2-transformed intensity signals of the probes using a mixture of three Gaussian distributions. Genes with mean of log2-transformed expression values over the 95% percentile of two distributions of lowest expressed genes were considered expressed (cut off ≥ 7.27) and included in the further analysis. The expectation maximization algorithm was implemented using the normalmixEM function from the mixtools R package. The limma package (50) was used to perform differential expression analysis. Genes were considered differentially expressed if having an absolute fold change ≥1.5 and an Benjamini–Hochberg-corrected P ≤ 0.05. The raw microarray data is deposited in ArrayExpress with accession number E-MTAB-4922. Description of the arrays are available in Supplementary Material, Table S11.
Co-expressed gene modules correlating with KS status
The WGCNA package in R was used to define co-expressed gene modules (55). Correlations were estimated using the biweight mid-correlation and a signed weighted correlation network was used to identify co-expression modules with high TO (18). The TO is a useful approach to exclude spurious or isolated connections during network construction. TO considers each pair of genes in relation to all other genes in the network. Genes have high TO if they are connected to roughly the same group of genes in the network (i.e. they share the same neighbourhood). The TO captures relationships among neighbourhoods of genes, and is therefore more robust than pairwise correlation alone for clustering genes by similarity (18).
Modules were defined as branches of a hierarchical cluster tree using the top-down dynamic tree cut method (56). The expression patterns of each module were summarized by the module eigengene, defined as the first principal component of a given module. Pairs of modules with high module eigengene correlations (r > 0.9) were merged. A weighted signed network was computed based on a fit to scale-free topology. A thresholding power of 26 was chosen (lowest threshold resulting in a scale-free R2 fit of 0.8), and the pairwise TO between genes was calculated, which converted pairwise correlation values [−1,1] to TO co-expression values [0,1] where values close to 1 represented highly shared co-expression neighbourhoods. The TO dendrogram was used to define modules using the dynamic tree cut method function in WGCNA (55) with a minimum module size set to 40 genes and the deepSplit parameter set to 2 (57). Finally, a measure of module-disease and gene-disease correlations to KS were calculated, Student asymptotic P-value, using the R function corPvalueStudent of the WGCNA method.
The gProfileR package (58) was used to compute the GSEA of the differentially expressed genes and the genes in modules. The background was set to the total list of expressed genes defined above. The hypergeometric distribution was used to estimate the significance of enriched pathways and processes using a threshold of FDR ≤ 5%.
Klinefelter syndrome comorbidity associated proteins and network
In order to identify proteins associated with KS comorbidities we created a mapping between the Disease Ontology Database (59) and ICD-10 codes, and then used the associations between diseases and proteins extracted from the Diseases database (60), Genetics home reference (61), OMIM (62) and the UniProt Knowledgebase (63). Disease sub-networks were built for KS and all extracted comorbidities with associated proteins connected with observed physical PPIs. We used high confidence PPIs from InWeb_IM (11) using the recommended cut off of confidence score ≥ 0.1. A common hypothesis is that proteins associated to the same disease and, thus, having some functional relationship, are more likely to interact than randomly selected proteins (20). Therefore, each disease sub-network was statistically tested for this hypothesis by comparing the observed number of PPIs between the disease proteins compared to 1,000 random sub-networks of the same size and node degree distribution picked randomly in the whole InWeb_IM network of high confidence interactions. P-values were calculated as the proportion of random networks with equal or more PPIs than the true network (workflow illustrated in Supplementary Material, Fig. S3). A significance level of P ≤ 0.05 was set as cut off for a functional network. Disease separations were calculated as previously published (21) and network topology was investigated (see Supplementary Material, Text S1). Subsequently, the ClusterONE algorithm (64) implemented in Cytoscape (65) was used to extract protein clusters using the default parameters and the InWeb_IM confidence score as edge weights.
Supplementary Material
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
Funding
FP7 grant SyBoSS EU 7th Framework (G.A. N° 242129); Novo Nordisk Foundation (grant agreement NNF14CC0001); MedBioinformatics EU Horizon 2020 grant (agreement no 634143); BioMedBridges EU FP7 Capacities Specific Programme grant (agreement number 284209); International Research and Research Training Centre in Endocrine Disruption of Male Reproduction and Child Health (EDMaRC). Funding to pay the Open Access publication charges for this article was provided by Novo Nordisk Foundation (grant agreement NNF14CC0001).
Supplementary Material
References
- 1. Bojesen A., Juul S., Gravholt C.H. (2003) Prenatal and postnatal prevalence of Klinefelter syndrome: a national registry study. J. Clin. Endocrinol. Metab., 88, 622–626. [DOI] [PubMed] [Google Scholar]
- 2. Dürrbaum M., Storchova Z. (2016) Effects of aneuploidy on gene expression: implications for cancer. FEBS J., 283, 791–802. [DOI] [PubMed] [Google Scholar]
- 3.Kirk, I. K., Weinhold, N., Belling, K., Skakkebæk, N. E., Jensen, T. S., Leffers, H., Juul, A. and Brunak, S. (2017) Chromosome-wise protein interaction patterns and their impact on functional implications of large-scale genomic aberrations. Cell Systems, 4, 1–8. [DOI] [PubMed] [Google Scholar]
- 4. Deng X., Berletch J.B., Nguyen D.K., Disteche C.M. (2014) X chromosome regulation: diverse patterns in development, tissues and disease. Nat. Rev. Genet., 15, 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lanfranco F., Kamischke A., Zitzmann M., Nieschlag E. (2004) Klinefelter's syndrome. Lancet, 364, 273–283. [DOI] [PubMed] [Google Scholar]
- 6. Høst C., Skakkebaek A., Groth K.A., Bojesen A. (2014) The role of hypogonadism in Klinefelter Syndrome. Asian J. Androl., 16, 185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barabási A.L., Gulbahce N., Loscalzo J. (2011) Network medicine: a network-based approach to human disease. Nat. Rev. Genet., 12, 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu J.X., Thomas C.E., Brunak S. (2016) Network biology concepts in complex disease comorbidities. Nat. Rev. Genet., 17, 615–629. [DOI] [PubMed] [Google Scholar]
- 9. Taylor I.W., Linding R., Warde-Farley D., Liu Y., Pesquita C., Faria D., Bull S., Pawson T., Morris Q., Wrana J.L. (2009) Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat. Biotechnol., 27, 199–204. [DOI] [PubMed] [Google Scholar]
- 10. Lage K. (2014) Protein-protein interactions and genetic diseases: the interactome. Biochim. Biophys. Acta, 1842, 1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li T., Wernersson R., Hansen R.B., Horn H., Mercer J.M., Slodkowicz G., Workman C., Regina O., Rapacki K., Staerfeldt H.H., et al. (2016) A scored human protein-protein interaction network to catalyze genomic interpretation. Nat. Methods,14, 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ideker T., Sharan R. (2008) Protein networks in disease. Genome Res., 18, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun K., Gonçalves J.P., Larminie C., Pržulj N. (2014) Predicting disease associations via biological network analysis. BMC Bioinformatics, 15, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lynge E., Sandegaard J.L., Rebolj M. (2011) The Danish National Patient Register. Scand. J. Public Health, 39, 30–33. [DOI] [PubMed] [Google Scholar]
- 15. Bourke E., Herlihy A., Snow P., Metcalfe S., Amor D. (2014) Klinefelter syndrome: a general practice perspective. Austr. Family Physician, 43, 38–41. [PubMed] [Google Scholar]
- 16. Bojesen A., Gravholt C.H. (2007) Klinefelter syndrome in clinical practice. Nat. Rev. Urol., 4, 192–204. [DOI] [PubMed] [Google Scholar]
- 17. Bojesen A., Gravholt C.H. (2011) Morbidity and mortality in Klinefelter syndrome (47,XXY). Acta Paediatr., 100, 807–813. [DOI] [PubMed] [Google Scholar]
- 18. Zhang B., Horvath S. (2005) A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol., 4, Article17. [DOI] [PubMed] [Google Scholar]
- 19. Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y. (2015) Systematic discovery of Xist RNA binding proteins. Cell, 161, 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goh K.I., Cusick M.E., Valle D., Childs B., Vidal M., Barabási A.L. (2007) The human disease network. Proc. Natl. Acad. Sci. USA, 104, 8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Menche J., Sharma A., Kitsak M., Ghiassian S.D., Vidal M., Loscalzo J., Barabási A.L. (2015) Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science, 347, 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bojesen A., Stochholm K., Juul S., Gravholt C.H. (2011) Socioeconomic trajectories affect mortality in Klinefelter syndrome. J. Clin. Endocrinol. Metab., 96, 2098–2104. [DOI] [PubMed] [Google Scholar]
- 23. Aksglaede L., Skakkebaek N.E., Juul A. (2008) Abnormal sex chromosome constitution and longitudinal growth: serum levels of insulin-like growth factor (IGF)-I, IGF binding protein-3, luteinizing hormone, and testosterone in 109 males with 47,XXY, 47,XYY, or sex-determining region of the Y chromosome (SRY)-positive 46,XX karyotypes. J. Clin. Endocrinol. Metab., 93, 169–176. [DOI] [PubMed] [Google Scholar]
- 24. Aksglaede L., Skakkebaek N.E., Almstrup K., Juul A. (2011) Clinical and biological parameters in 166 boys, adolescents and adults with nonmosaic Klinefelter syndrome: a Copenhagen experience. Acta Paediatr., 100, 793–806. [DOI] [PubMed] [Google Scholar]
- 25. Hunter M.L., Collard M.M., Razavi T., Hunter B. (2003) Increased primary tooth size in a 47,XXY male: a first case report. Int. J. Paediatr. Dent., 13, 271–273. [DOI] [PubMed] [Google Scholar]
- 26. Schulman G.S., Redford-Badwal D., Poole A., Mathieu G., Burleson J., Dauser D. (2005) Taurodontism and learning disabilities in patients with Klinefelter syndrome. Pediatr. Dent., 27, 389–394. [PubMed] [Google Scholar]
- 27. Marques-da-Silva B., Baratto-Filho F., Abuabara A., Moura P., Losso E.M., Moro A. (2010) Multiple taurodontism: the challenge of endodontic treatment. J. Oral Sci., 52, 653–658. [DOI] [PubMed] [Google Scholar]
- 28. D'Alessandro G., Armuzzi L., Cocchi G., Piana G. (2012) Eruption delay in a 47 XXY male: a case report. Eur. J. Paediatr. Dent., 13, 159–160. [PubMed] [Google Scholar]
- 29. Vawter M.P., Harvey P.D., DeLisi L.E. (2007) Dysregulation of X-linked gene expression in Klinefelter's syndrome and association with verbal cognition. Am. J. Med. Genet. A, 144B, 728–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma Y., Li C., Gu J., Tang F., Li C., Li P., Ping P., Yang S., Li Z., Jin Y. (2012) Aberrant gene expression profiles in pluripotent stem cells induced from fibroblasts of a Klinefelter syndrome patient. J. Biol. Chem., 287, 38970–38979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tüttelmann F., Gromoll J. (2010) Novel genetic aspects of Klinefelter's syndrome. Mol. Hum. Reprod., 16, 386–395. [DOI] [PubMed] [Google Scholar]
- 32. Passerini V., Ozeri-Galai E., de Pagter M.S., Donnelly N., Schmalbrock S., Kloosterman W.P., Kerem B., Storchova Z. (2016) The presence of extra chromosomes leads to genomic instability. Nat. Commun., 7, 10754.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sheltzer J.M., Torres E.M., Dunham M.J., Amon A. (2012) Transcriptional consequences of aneuploidy. Proc. Natl. Acad. Sci. USA, 109, 12644–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dürrbaum M., Kuznetsova A.Y., Passerini V., Stingele S., Stoehr G., Storchova Z. (2014) Unique features of the transcriptional response to model aneuploidy in human cells. BMC Genomics, 15, 139.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wallentin M., Skakkebaek A., Bojesen A., Fedder J., Laurberg P., Østergaard J.R., Hertz J.M., Pedersen A.D., Gravholt C.H. (2016) Klinefelter syndrome has increased brain responses to auditory stimuli and motor output, but not to visual stimuli or Stroop adaptation. Neuroimage Clin., 11, 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DeLisi L.E., Maurizio A.M., Svetina C., Ardekani B., Szulc K., Nierenberg J., Leonard J., Harvey P.D. (2005) Klinefelter's syndrome (XXY) as a genetic model for psychotic disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet., 135B, 15–23. [DOI] [PubMed] [Google Scholar]
- 37. Koçar I.H., Yesilova Z., Ozata M., Turan M., Sengül A., Ozdemir I. (2000) The effect of testosterone replacement treatment on immunological features of patients with Klinefelter's syndrome. Clin. Exp. Immunol., 121, 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bojesen A., Juul S., Birkebaek N., Gravholt C.H. (2004) Increased mortality in Klinefelter syndrome. J. Clin. Endocrinol. Metab., 89, 3830–3834. [DOI] [PubMed] [Google Scholar]
- 39. Ehrlich S., Weiss D., Burghardt R., Infante-Duarte C., Brockhaus S., Muschler M.A., Bleich S., Lehmkuhl U., Frieling H. (2010) Promoter specific DNA methylation and gene expression of POMC in acutely underweight and recovered patients with anorexia nervosa. J. Psychiatr. Res., 44, 827–833. [DOI] [PubMed] [Google Scholar]
- 40. Choi P., Reiser H. (1998) IL-4: role in disease and regulation of production. Clin. Exp. Immunol., 113, 317–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mueller R., Bradley L.M., Krahl T., Sarvetnick N. (1997) Mechanism underlying counterregulation of autoimmune diabetes by IL-4. Immunity, 7, 411–418. [DOI] [PubMed] [Google Scholar]
- 42. Bachman E., Travison T.G., Basaria S., Davda M.N., Guo W., Li M., Westfall J.C., Bae H., Gordeuk V., Bhasin S. (2014) Testosterone induces erythrocytosis via increased erythropoietin and suppressed hepcidin: evidence for a new erythropoietin/hemoglobin set point. J. Gerontol. A Biol. Sci. Med. Sci., 69, 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rocca M.S., Pecile V., Cleva L., Speltra E., Selice R., Di Mambro A., Foresta C., Ferlin A. (2016) The Klinefelter syndrome is associated with high recurrence of copy number variations on the X chromosome with a potential role in the clinical phenotype. Andrology, 4, 328–334. [DOI] [PubMed] [Google Scholar]
- 44. Jiang J., Jing Y., Cost G.J., Chiang J.C., Kolpa H.J., Cotton A.M., Carone D.M., Carone B.R., Shivak D.A., Guschin D.Y., et al. (2013) Translating dosage compensation to trisomy 21. Nature, 500, 296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jensen A.B., Moseley P.L., Oprea T.I., Ellesøe S.G., Eriksson R., Schmock H., Jensen P.B., Jensen L.J., Brunak S. (2014) Temporal disease trajectories condensed from population-wide registry data covering 6.2 million patients. Nat. Commun., 5, 4022.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beck M.K., Jensen A.B., Nielsen A.B., Perner A., Moseley P.L., Brunak S. (2016) Diagnosis trajectories of prior multi-morbidity predict sepsis mortality. Sci. Rep, 6, 36624.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roque F.S., Jensen P.B., Schmock H., Dalgaard M., Andreatta M., Hansen T., Søeby K., Bredkjær S., Juul A., Werge T., et al. (2011) Using electronic patient records to discover disease correlations and stratify patient cohorts. PLoS Comput. Biol., 7, e1002141.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sweeting M.J., Sutton A.J., Lambert P.C. (2004) What to add to nothing? Use and avoidance of continuity corrections in meta-analysis of sparse data. Stat. Med., 23, 1351–1375. [DOI] [PubMed] [Google Scholar]
- 49. Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol., 5, R80.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 43, e47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smyth G.K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol., 3, Article 3. [DOI] [PubMed] [Google Scholar]
- 52. Johnson W.E., Li C., Rabinovic A. (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8, 118–127. [DOI] [PubMed] [Google Scholar]
- 53. Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics, 28, 882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Benaglia T., Chauveau D., Hunter D.R., Young D.S. (2009) mixtools: an R Package for Analyzing Finite Mixture Models. J. Stat. Softw., 32, 1–29. [Google Scholar]
- 55. Langfelder P., Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Langfelder P., Horvath S. (2012) Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw., 46, pii: i11. [PMC free article] [PubMed] [Google Scholar]
- 57. Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S., Mill J., Cantor R.M., Blencowe B.J., Geschwind D.H. (2011) Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature, 474, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Reimand J., Kull M., Peterson H., Hansen J., Vilo J. (2007) g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res., 35, W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kibbe W.A., Arze C., Felix V., Mitraka E., Bolton E., Fu G., Mungall C.J., Binder J.X., Malone J., Vasant D., et al. (2015) Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res., 43, D1071–D1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pletscher-Frankild S., Pallejà A., Tsafou K., Binder J.X., Jensen L.J. (2015) DISEASES: text mining and data integration of disease-gene associations. Methods, 74, 83–89. [DOI] [PubMed] [Google Scholar]
- 61. Mitchell J.A., McCray A.T. (2003) The Genetics Home Reference: a new NLM consumer health resource. AMIA Annu. Symp. Proc., 2003, 936.. [PMC free article] [PubMed] [Google Scholar]
- 62.Online Mendelian Inheritance in Man, OMIM®- McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD) Online Mendelian Inheritance in Man, OMIM®- McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD).
- 63. UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res., 43, D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nepusz T., Yu H., Paccanaro A. (2012) Detecting overlapping protein complexes in protein-protein interaction networks. Nat. Methods, 9, 471–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.