Abstract
Background
This study aimed to uncover the molecular mechanisms underlying mild and severe pneumonia by use of mRNA sequencing (RNA-seq).
Material/Methods
RNA was extracted from the peripheral blood of patients with mild pneumonia, severe pneumonia, and healthy controls. Sequencing was performed on the HiSeq4000 platform. After filtering, clean reads were mapped to the human reference genome hg19. Differentially expressed genes (DEGs) were identified between the control group and the mild or severe group. A transcription factor-gene network was constructed for each group. Biological process (BP) terms enriched by DEGs in the network were analyzed and these genes were also mapped to the Connectivity map to search for small-molecule drugs.
Results
A total of 199 and 560 DEGs were identified from the mild group and severe group, respectively. A transcription factor-gene network consisting of 215 nodes and another network consisting of 451 nodes were constructed in the mild group and severe group, respectively, and 54 DEGs (e.g., S100A9 and S100A12) were found to be common, with consistent differential expression changes in the 2 groups. Genes in the transcription factor-gene network for the mild group were mainly enriched in 13 BP terms, especially defense and inflammatory response (e.g., S100A8) and spermatogenesis, while the top BP terms enriched by genes in the severe group include response to oxidative stress (CCL5), wound healing, and regulation of cell differentiation (CCL5), and of the cellular protein metabolic process.
Conclusions
S100A9 and S100A12 may have a role in the pathogenesis of pneumonia: S100A9 and CXCL1 may contribute solely in mild pneumonia, and CCL5 and CXCL11 may contribute in severe pneumonia.
MeSH Keywords: Genes, vif; Pneumonia, Aspiration; Sequence Analysis, RNA; Small Molecule Libraries
Background
Pneumonia, especially community-acquired pneumonia (CAP), is the leading reason for adult hospitalization in low- and middle-income countries [1]; Streptococcus pneumoniae (pneumococcus) is believed to be the main cause [2]. According to Said et al., the actual burden of bacteremic pneumococcal pneumonia in adults is significantly underestimated [3].
Expression profiling under different physiological conditions has been employed to investigate the molecular mechanisms underlying various diseases [4,5] and to provide potential biomarkers for targeted therapy [6,7]. Microarrays and RNA sequencing (RNA-seq) are both available for genomic profiling. Notably, RNA-seq allows the detection of new transcripts [8]. Additionally, RNA-seq avoids the introduction of related biases during hybridization of microarrays. Thus, we used this technique to detect gene expression profiles associated with mild and severe pneumonia in order to deepen our insights into the molecular mechanisms underlying these 2 diseases.
Material and Methods
Patient enrollment and sampling
This study was approved by the Medical Ethics Committee of the General Hospital of the People’s Liberation Army (301 Hospital). From June 2013 to December 2013, 18 adult patients with pneumonia were included in this study, including 9 cases with mild pneumonia and 9 cases with severe pneumonia. In addition, 9 healthy adult volunteers were recruited as normal controls. Patients meeting any of the following symptoms were considered to have severe pneumonia: (1) disturbance of consciousness; (2) respiratory rate ≥30 times/min, (3) PaO2 <60 mm Hg, PaO2/FiO2 <300, requiring mechanical ventilation; (4) systolic blood pressure <90 mm Hg; (5) concurrent septic shock; (6) X-ray showing bilateral or multi-lobe involvement, or pulmonary involvement expanding ≥50% within 48 h after hospitalized; and (7) oliguria (urine volume <20 ml/h, or <80 ml/4 h, or concurrent acute renal failure and requiring dialysis.
Peripheral blood samples were collected from each patient and volunteer. Blood samples from 3 randomly selected patients were mixed at 1: 1: 1 as a final sequencing sample. Thus, there were 3 samples for severe pneumonia patients (numbered WLL1, WLL2, WLL3), 3 samples for patients with mild pneumonia (WLL4, WLL5, WLL6), and 3 for normal controls (WLL7, WLL8, WLL9). Written informed consent was provided by each patient and volunteer before sampling.
Total RNA extraction, library construction, and sequencing
Total RNA was extracted from the plasma of these 9 samples using the miRNeasy Serum/Plasma Kit (QIAGEN, Hilden, Germany). Then, rRNA was removed using Epicenter Ribo-ZeroTM kit (Illumina Inc., San Diego, CA) and the remaining RNA (polyA+, polyA−) was recovered and purified. Afterwards, the purified RNA was broken into short segments using random fragmentation reagent (Fragmentation Buffer). Next, reverse transcription was performed to construct a cDNA library. RNA concentration was measured with a Qubit® 2.0 Fluorometer and the RNA integrity number (RIN) was measured by use of Bioanalyzer 2100 (Agilent, CA, USA). Sequencing was performed on the HiSeq4000 platform to generate paired-end reads (150 bp in length).
Sequence quality control and alignment
First, reads were filtered by removing the bases with continuous quality value <10 at both ends, and reads including less than 80% bases with quality value >Q20, and reads shorter than 50 nt, as well as rRNA sequences. Sequence quality control was done with the Fastx toolkit. Next, clean reads were mapped to the human reference genome hg19 using TopHat 2.1.1 software (download site: http://ccb.jhu.edu/software/tophat/index.shtml).
Identification of differentially expressed genes correlated with mild and severe pneumonia
Gene differential expression analysis was performed between the normal control and the mild or severe group using the edgeR package of R (version 3.1.0) [9]. | logFC | >1 and p<0.05 were used as cutoffs for a differentially expressed gene (DEG). Volcanic plots were used to visualize gene expression differences, and a heat map was also drawn to display the gene expression profile of differentially expressed genes based on the hierarchical clustering results using Euclidean distance [10] with the pheatmap package of R [11].
The correlation coefficient of any 2 genes based on their gene expression values in each group was calculated, and only gene pairs with absolute correlation coefficients >0.9 were retained. A heat map was used to visualize the expression correlation matrix.
Prediction of transcription factors and construction of transcription factor-gene network
Transcriptional regulators are responsible for the transcriptional regulation of gene expression [12]. To find the key transcriptional regulators of the DEGs we identified above, we searched the TRED database (Transcriptional Regulation Element Database, http://rulai.cshl.edu/TRED), which includes both cis- and trans-regulatory elements and provides both promoter sequences and transcription factor binding information [13]. Only experimentally validated data were used in our study. Next, a transcription factor-gene network containing transcription factors and their target genes was constructed, which was further visualized using software Cytoscape2.8.0 [14].
Functional annotation of genes in the transcription factor-gene network
Genes in the transcription factor-gene network were mapped to the GO functional nodes, and the biological process (BP) terms enriched by these genes were predicted using GOstat (P value <0.05 as cutoff) [15].
Screening of small-molecule drugs
DEGs in the transcription factor-gene network were also mapped to the Connectivity map (Cmap) to search for small-molecule drugs [16,17]. Only drugs with | score | >0.8 were retained.
Results
Quality control of reads and statistics
A total of 346G data were generated from the 9 samples. After quality control, all the clean reads had one end containing >99.97% Q20 bases and the other end >96.38% Q20 bases (Table 1). The proportion of clean reads from the 2 ends was larger than 95% and 75%.
Table 1.
Quality statistics of reads.
| Sample | Raw reads | Raw bases | Q20 value | Clean reads | Clean base | Clean rate |
|---|---|---|---|---|---|---|
| WLL1 | 52831743 | 7928532000 | 99.99% | 50735720 | 7609666306 | 0.960326446 |
| WLL1 | 58572433 | 7928532000 | 97.10% | 44421351 | 6662121716 | 0.758400304 |
| WLL2 | 103035876 | 15455381400 | 99.98% | 98747743 | 14810817868 | 0.958382137 |
| WLL2 | 103035876 | 15455381400 | 97.50% | 87943761 | 13189434520 | 0.85352563 |
| WLL3 | 47936143 | 7190421450 | 99.97% | 45842678 | 6875831673 | 0.956328047 |
| WLL3 | 47936143 | 7190421450 | 96.67% | 39203922 | 5879662949 | 0.817836387 |
| WLL4 | 55117713 | 8267656950 | 99.97% | 52810671 | 7920931057 | 0.958143365 |
| WLL4 | 55117713 | 8267656950 | 96.86% | 45772099 | 6864690206 | 0.830442638 |
| WLL5 | 42414859 | 6362228850 | 99.97% | 40625314 | 6093303572 | 0.957808536 |
| WLL5 | 42414859 | 6362228850 | 96.49% | 34754331 | 5212336104 | 0.819390464 |
| WLL6 | 52831743 | 7924761450 | 99.97% | 50675720 | 7600736984 | 0.959190765 |
| WLL6 | 52831743 | 7924761450 | 96.38% | 43274947 | 6490230720 | 0.819108826 |
| WLL7 | 54142181 | 8121327150 | 99.99% | 51811388 | 7771187924 | 0.956950515 |
| WLL7 | 54142181 | 8121327150 | 97.28% | 44077521 | 6610710884 | 0.814106861 |
| WLL8 | 58572433 | 8785864950 | 99.99% | 56266066 | 8439365160 | 0.960623678 |
| WLL8 | 58572433 | 8785864950 | 97.30% | 48447586 | 7266131771 | 0.827139723 |
| WLL9 | 42047930 | 6307189500 | 99.99% | 40469826 | 6070080949 | 0.962468925 |
| WLL9 | 42047930 | 6307189500 | 97.23% | 34410808 | 5160914737 | 0.818371035 |
With reference to the human reference genome hg19, more than 70% clean reads were aligned in each sample, each with coverage rate of >70%, mostly above 80%, and sequencing depth of >3.4, mostly 4–5.5 (Table 2).
Table 2.
Statistics of aligned reads.
| Sample | Aligned reads | Unique reads | Alignment rate | Specific alignment rate | Coverage | Depth |
|---|---|---|---|---|---|---|
| WLL1 | 66658130 | 65814534 | 0.700506324 | 0.691641024 | 0.8124469 | 4.656258953 |
| WLL2 | 106477220 | 105906423 | 0.570337791 | 0.567280357 | 0.8897298 | 7.498857573 |
| WLL3 | 60484089 | 59601427 | 0.711187619 | 0.700809051 | 0.7997421 | 4.401589422 |
| WLL4 | 66873642 | 65760610 | 0.678350203 | 0.667059873 | 0.8028072 | 5.141637117 |
| WLL5 | 55225190 | 54350513 | 0.732627356 | 0.721023733 | 0.705912 | 4.090939095 |
| WLL6 | 67240507 | 66192609 | 0.715700156 | 0.704546451 | 0.8202879 | 5.359257339 |
| WLL7 | 62030490 | 61253756 | 0.646899528 | 0.638799175 | 0.7911858 | 4.366865004 |
| WLL8 | 75880654 | 74793902 | 0.724649103 | 0.714270781 | 0.828035 | 5.46437753 |
| WLL9 | 52286836 | 51342443 | 0.698269141 | 0.685657162 | 0.7566747 | 3.453845744 |
DEGs associated with mild and severe pneumonia and screening of co-expressed genes
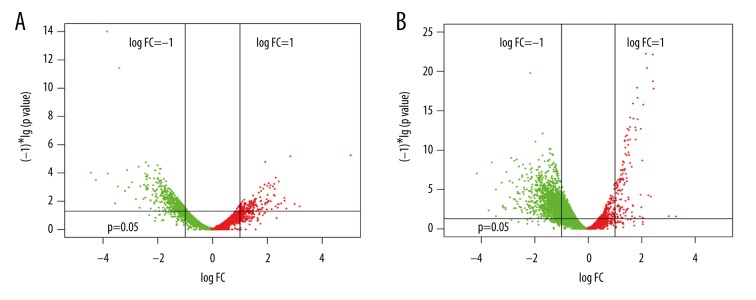
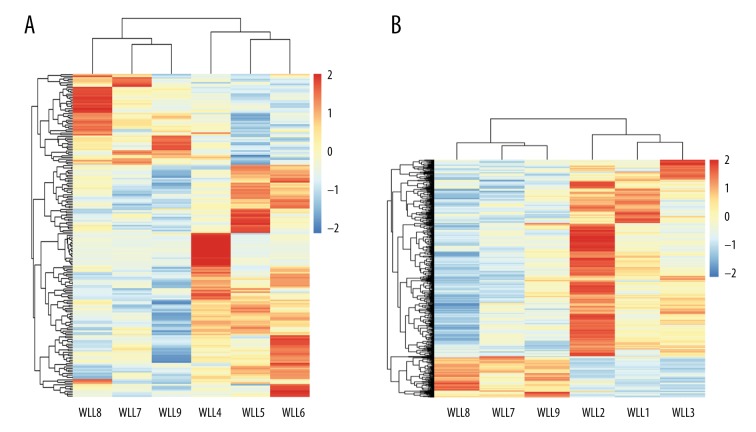
A total of 199 and 560 DEGs were identified from the mild group and severe group, respectively (Figure 1). Overall, the identified DEGs could distinguish the pneumonia samples from the control sample (Figure 2).
Figure 1.
Volcanic plots showing gene expression status of the differentially expressed genes in each group. (A) Mild pneumonia group; (B) Severe pneumonia group.
Figure 2.
Heat maps showing the gene expression profile of differentially expressed genes based on hierarchical clustering. (A) Mild pneumonia group; (B) Severe pneumonia group.
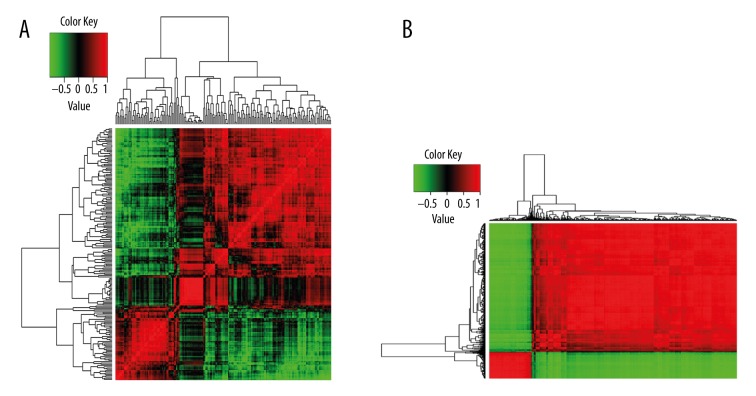
The correlation coefficient of any 2 DEGs based on their gene expression values was calculated in each group, and a 199×199 matrix and a 560×560 correlation coefficient matrix were obtained, respectively, as shown in Figure 3. Using a cutoff of 0.9, 1128 gene pairs and 1170 gene pairs were retained in the mild group and severe group, respectively.
Figure 3.
Correlation coefficient matrix based on correlation coefficient of any 2 DEGs based on their gene expression values. (A) Mild pneumonia group; (B) Severe pneumonia group.
Construction of transcription factor-gene network
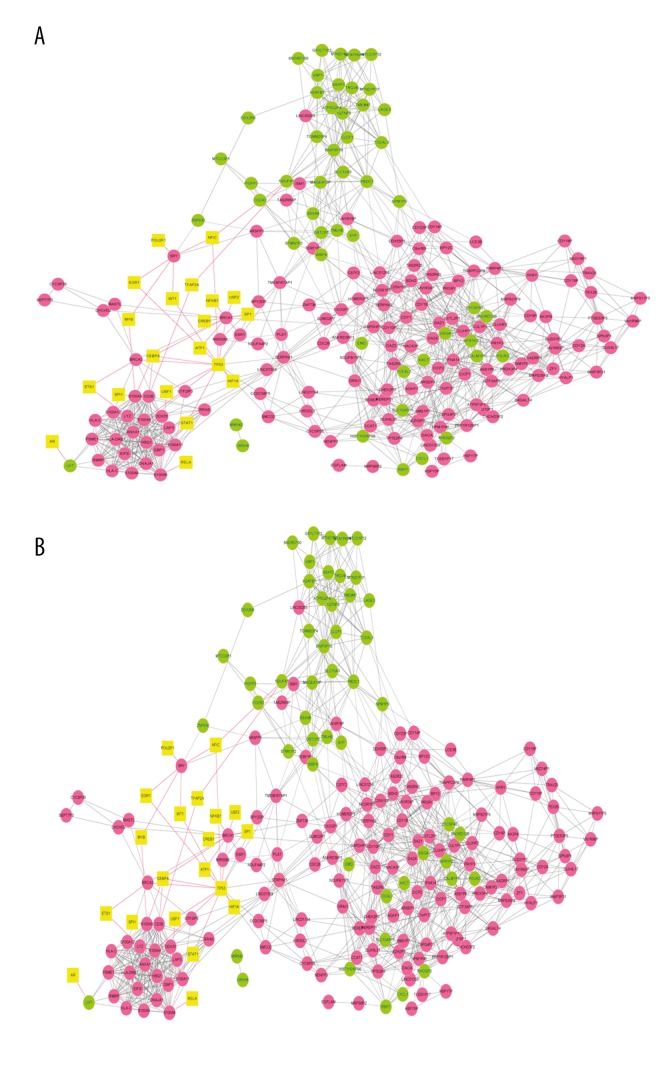
A total of 36 and 93 of transcription factor-gene pairs were obtained, respectively. Taking into account the co-expressed gene pairs identified above, a transcription factor-gene network consisting of 215 nodes and another network consisting of 451 nodes were constructed in the mild group and severe group, respectively (Figure 4). In the former, S100A9 (24), S100A8 (20), S100A12 (20), DAZ1 (24), DAZ4 (23), and DAZ3 (18) were found to be co-expressed with more DEG-encoded proteins; in the latter network, PSMA1 (8), CCL5 (6), and CXCL11 (2) were co-expressed with more proteins.
Figure 4.
Transcription factor-gene network consisting of transcription factor and co-expressed genes. (A) Mild pneumonia group; (B) Severe pneumonia group.
We further compared the DEGs in the transcription factor-gene network between the mild group and severe group and found 54 common genes that showed consistent differential expression changes in the 2 groups (Table 3).
Table 3.
The common differentially expressed genes shared by mild pneumonia and severe pneumonia.
| Common differentially expressed genes differentially expressed genes | Mild group | Severe group | ||
|---|---|---|---|---|
| logFC | P value | logFC | P value | |
| AGKP1 | 1.17 | 3.01E-02 | 1.50 | 7.48E-05 |
| ARSFP1 | 1.26 | 3.59E-02 | 1.88 | 1.14E-05 |
| ATP5F1P1 | 1.00 | 4.87E-02 | 1.09 | 4.47E-04 |
| BDH2 | 1.16 | 2.41E-02 | 1.24 | 2.76E-04 |
| BPY2 | 1.26 | 8.97E-03 | 1.39 | 8.45E-07 |
| BPY2C | 1.35 | 1.76E-02 | 1.62 | 2.51E-04 |
| BRCA1 | 3.37 | 3.72E-12 | 1.67 | 7.64E-13 |
| BRCA2 | 3.82 | 9.92E-15 | 2.14 | 1.66E-20 |
| C1orf137 | 1.11 | 2.81E-02 | 1.32 | 3.36E-05 |
| CCAT1 | 1.06 | 2.53E-02 | 1.18 | 6.74E-06 |
| CCDC58P5 | 1.18 | 3.68E-02 | 2.13 | 4.65E-07 |
| CDC26 | 1.36 | 2.31E-02 | 1.52 | 2.65E-04 |
| CDY1 | 1.34 | 6.00E-03 | 1.14 | 8.35E-05 |
| CDY10P | 2.09 | 2.40E-04 | 1.80 | 1.20E-05 |
| CDY18P | 1.44 | 5.01E-03 | 1.39 | 2.08E-05 |
| CDY19P | 1.47 | 4.18E-03 | 1.38 | 2.49E-05 |
| CDY1B | 1.34 | 6.00E-03 | 1.09 | 2.20E-04 |
| CDY2A | 1.08 | 2.28E-02 | 1.11 | 4.07E-05 |
| CLUHP1 | 1.35 | 4.03E-03 | 1.30 | 4.51E-07 |
| CLUHP2 | 1.18 | 1.14E-02 | 1.32 | 2.01E-07 |
| CYCSP39 | 1.18 | 4.50E-02 | 1.42 | 9.89E-04 |
| CYCSP55 | 1.17 | 2.63E-02 | 1.27 | 2.70E-04 |
| DAOA | 1.25 | 1.37E-02 | 1.66 | 3.06E-07 |
| DAZ3 | 1.26 | 5.43E-03 | 1.13 | 1.20E-06 |
| DDX3Y | 1.39 | 3.11E-02 | 1.32 | 8.88E-03 |
| DNM1P24 | 1.13 | 2.63E-02 | 1.22 | 2.27E-04 |
| DUX4L31 | 1.52 | 3.93E-03 | 1.43 | 3.22E-05 |
| EXTL2P1 | 1.44 | 3.80E-02 | 1.63 | 4.93E-03 |
| EZH2P1 | 1.41 | 1.19E-02 | 1.42 | 1.33E-03 |
| GAPDHP17 | 2.01 | 3.06E-03 | 2.84 | 9.80E-10 |
| GTF3AP5 | 1.74 | 5.70E-03 | 1.97 | 1.26E-04 |
| HOMER2P1 | 1.09 | 2.79E-02 | 1.35 | 3.48E-06 |
| KIR3DL3 | 1.03 | 2.91E-02 | 1.03 | 1.47E-04 |
| MED14P1 | 2.05 | 4.70E-02 | 2.74 | 2.37E-03 |
| MRPS17P5 | 2.08 | 1.13E-02 | 2.26 | 3.87E-03 |
| MRPS35P2 | 1.26 | 4.71E-02 | 2.34 | 7.88E-07 |
| MRPS6P2 | 1.11 | 4.57E-02 | 2.40 | 6.68E-09 |
| NACA3P | 1.46 | 5.06E-03 | 1.23 | 3.39E-04 |
| NCOR1P2 | 1.63 | 1.20E-02 | 2.02 | 6.70E-05 |
| NDUFB11P1 | 1.20 | 2.04E-02 | 1.05 | 2.88E-03 |
| PRDX3P4 | 1.81 | 2.44E-03 | 1.60 | 3.90E-04 |
| PRYP3 | 1.26 | 1.25E-02 | 1.08 | 7.27E-04 |
| RMRP | 1.03 | 2.91E-02 | -1.48 | 9.21E-07 |
| S100A12 | 2.26 | 2.36E-04 | 2.56 | 1.19E-08 |
| S100A9 | 1.86 | 5.29E-05 | 1.27 | 2.92E-08 |
| SLC25A15P1 | 2.04 | 4.71E-03 | 2.60 | 2.00E-05 |
| SMCO2 | 1.03 | 3.93E-02 | 1.24 | 1.91E-04 |
| SRY | 1.13 | 3.18E-02 | 1.34 | 2.48E-04 |
| TAS2R43 | 2.66 | 3.62E-05 | 2.55 | 5.32E-07 |
| TAS2R8 | 1.62 | 1.94E-03 | 1.14 | 1.27E-03 |
| TCEAL7 | −1.18 | 2.64E-02 | -1.00 | 3.79E-03 |
| TEX26 | 1.56 | 5.94E-03 | 1.21 | 4.55E-03 |
| TMEM167AP1 | 3.16 | 1.89E-03 | 3.15 | 1.97E-03 |
| TPTE2P4 | 1.42 | 3.24E-03 | 1.94 | 8.38E-12 |
GO functional annotation of genes in the transcription factor-gene network
The genes in the transcription factor-gene network for the mild group were mainly enriched in 13 biological process terms, especially defense and inflammatory response (e.g., CXCL1, CD36, S100A8, S100A9, ANXA1, LYZ, VSIG4, S100A12) and spermatogenesis (e.g., DAZ3, DAZ4, DAZ1, BPY2C, CDY1B, DNAJA1, BRCA2, BPY2, CDY2A, CDY1) (Table 4A). In contrast, the top BP terms enriched by genes in the severe group were much different, including response to oxidative stress (GPX2, PRDX6, PTGS1, SNCA, CLU, PDLIM1, CCL5, ETV5), wound healing (GP1BB, KLKB1, F13A1, APOH, SERPINB2, PF4, ITGB3, NRG1), and regulation of cell differentiation (PF4, ITGB3, MBNL3, CNTF, DLX5, CLU, GNAS, PF4, NRG1, CCL5, CD74), and of cellular protein metabolic process (DAZ3, UBE2C, BRCA1PSMA1, PSMA5, SOCS1, SNCA, SERPINB10, ITGB3, CDC26, UBE2C, TIMP1) (Table 4B).
Table 4A.
GO functional annotation of genes in the transcription factor-gene network for the mild pneumonia group.
| Biological process term | Gene number | P value | Gene |
|---|---|---|---|
| GO: 0009611~response to wounding | 8 | 0.007259044 | CXCL1, CD36, S100A8, S100A9, ANXA1, LYZ, VSIG4, S100A12 |
| GO: 0006952~defense response | 8 | 0.015612576 | CXCL1, S100A8, S100A9, ANXA1, LYZ, IFNA14, VSIG4, S100A12 |
| GO: 0006954~inflammatory response | 7 | 0.002598886 | CXCL1, S100A8, S100A9, ANXA1, LYZ, VSIG4, S100A12 |
| GO: 0019953~sexual reproduction | 7 | 0.013338106 | DAZ3, DAZ4, DAZ1, BPY2C, CDY1B, DNAJA1, XKRY, BRCA2, BPY2, CDY2A, CDY1 |
| GO: 0048232~male gamete generation | 6 | 0.010136281 | DAZ3, DAZ4, DAZ1, BPY2C, CDY1B, DNAJA1, BRCA2, BPY2, CDY2A, CDY1 |
| GO: 0007283~spermatogenesis | 6 | 0.010136281 | DAZ3, DAZ4, DAZ1, BPY2C, CDY1B, DNAJA1, BRCA2, BPY2, CDY2A, CDY1 |
| GO: 0007276~gamete generation | 6 | 0.026768941 | DAZ3, DAZ4, DAZ1, BPY2C, CDY1B, DNAJA1, BRCA2, BPY2, CDY2A, CDY1 |
| GO: 0007010~cytoskeleton organization | 6 | 0.03855316 | CXCL1, UXT, RHOQP2, S100A9, BRCA2, BRCA1 |
| GO: 0032270~positive regulation of cellular protein metabolic process | 5 | 0.017614315 | DAZ3, DAZ4, DAZ1, PSME1, CLCF1, CDC26, BRCA1 |
| GO: 0051247~positive regulation of protein metabolic process | 5 | 0.020217214 | DAZ3, DAZ4, DAZ1, PSME1, CLCF1, CDC26, BRCA1 |
| GO: 0051052~regulation of DNA metabolic process | 4 | 0.012850438 | S100A11, TP53, BRCA2, BRCA1 |
| GO: 0030155~regulation of cell adhesion | 4 | 0.020911374 | EGFLAM, CD36, SERPINI1, SERPINI2 |
| GO: 0031401~positive regulation of protein modification process | 4 | 0.046097288 | PSME1, CLCF1, CDC26, BRCA1 |
Table 4B.
GO functional annotation of genes in the transcription factor-gene network for the severe pneumonia group.
| Biological process term | Gene number | P value | Genes |
|---|---|---|---|
| GO: 0006979~response to oxidative stress | 8 | 0.00498357 | GPX2, PRDX6, PTGS1, SNCA, CLU, PDLIM1, CCL5, ETV5 |
| GO: 0042060~wound healing | 8 | 0.011168559 | GP1BB, KLKB1, F13A1, APOH, SERPINB2, PF4, ITGB3, NRG1 |
| GO: 0045596~negative regulation of cell differentiation | 8 | 0.02053092 | PTHLH, CNTF, PF4, ITGB3, MBNL3, HIST1H4I, OMG, CD74 |
| GO: 0045597~positive regulation of cell differentiation | 8 | 0.02716175 | CNTF, DLX5, CLU, GNAS, PF4, NRG1, CCL5, CD74 |
| GO: 0032270~positive regulation of cellular protein metabolic process | 8 | 0.02945066 | DAZ3, PSMA1, CNTF, PSMA5, KLKB1, CDC26, UBE2C, BRCA1 |
| GO: 0051247~positive regulation of protein metabolic process | 8 | 0.035793321 | DAZ3, PSMA1, CNTF, PSMA5, KLKB1, CDC26, UBE2C, BRCA1 |
| GO: 0032269~negative regulation of cellular protein metabolic process | 9 | 0.002127619 | PSMA1, PSMA5, SOCS1, SNCA, SERPINB10, ITGB3, CDC26, UBE2C, TIMP1 |
| GO: 0009617~response to bacterium | 9 | 0.003271572 | GPX2, PPBP, CCL20, DEFB113, SOCS1, SNCA, CCL5, S100A12, B2M |
| GO: 0007017~microtubule-based process | 9 | 0.015645909 | TUBBP5, OPA1, CKS2, BRCA2, TUBB1, UBE2C, BRCA1, SPAST, KIF2A |
| GO: 0007626~locomotory behavior | 9 | 0.023984998 | PPBP, CCL20, SNCA, S100A9, PF4, CXCL11, CCL5, XCL2, NOVA1 |
| GO: 0051248~negative regulation of protein metabolic process | 10 | 6.45E-04 | PSMA1, PSMA5, SOCS1, SNCA, SERPINB10, ITGB3, CDC26, UBE2C, FLNA, TIMP1 |
| GO: 0031399~regulation of protein modification process | 10 | 0.013193547 | PRKAR2B, PSMA1, CNTF, PSMA5, SOCS1, SNCA, CDC26, UBE2C, PDCD4, BRCA1 |
| GO: 0007610~behavior | 13 | 0.0167272 | TAS2R1, TAS2R5, IL18, S100A9, SNCA, PF4, CXCL11, CCL5, PRKAR2B, CCL20, PPBP, XCL2, NOVA1 |
| GO: 0032268~regulation of cellular protein metabolic process | 16 | 0.001064045 | DAZ3, PRKAR2B, PSMA1, CNTF, PSMA5, KLKB1, SOCS1, SNCA, SERPINB10, EIF1, ITGB3, CDC26, UBE2C, PDCD4, BRCA1, TIMP1 |
| GO: 0009611~response to wounding | 16 | 0.003144496 | F13A1, S100A9, CLU, PF4, ITGB3, CXCL11, CCL5, S100A12, CCL20, GP1BB, FCN2, KLKB1, SERPINB2, APOH, VCAN, NRG1 |
| GO: 0006952~defense response | 18 | 0.002159946 | IFNA21, CLU, SNCA, S100A9, HLA-B, CXCL11, CCL5, CD74, S100A12, PAGE1, PPBP, IFNA7, CCL20, DEFB113, FCN2, KLKB1, BNIP3L, HLA-DRA |
| GO: 0010605~negative regulation of macromolecule metabolic process | 18 | 0.01223182 | SNCA, SOCS1, BRCA2, PF4, ITGB3, UBE2C, CDC26, PDCD4, BRCA1, FLNA, TIMP1, PSMA1, PSMA5, BNIP3L, HBZ, SERPINB10, NRG1, ENO1 |
| GO: 0042127~regulation of cell proliferation | 19 | 0.011317448 | IL18, CLU, PTGS1, MMP7, NAP1L1, BRCA2, SPARC, PRRX2, BRCA1, FTH1, TIMP1, PTHLH, CTH, CNTF, DLX5, APOH, GLMN, EMP3, NRG1 |
| GO: 0006955~immune response | 23 | 6.37E-05 | HLA-DRB1, IL18, ENPP3, CLU, SNCA, TNFRSF17, PF4, HLA-B, PF4V1, CXCL11, CCL5, FTH1, CD74, B2M, PPBP, CCL20, FCN2, BNIP3L, HLA-DRB5, NFIL3, FCGR3B, XCL2, HLA-DRA |
Prediction of relevant small-molecule drugs
Using | score | >0.8 as cutoff, 9 small-molecule drugs were found to be negatively correlated with DEGs in the mild group, of which mevalolactone had the maximum correlation coefficient, and 3 were found in the severe group, of which alsterpaullone had the maximum correlation coefficient (Table 5).
Table 5.
Small-molecule drugs were found negatively correlated with differentially expressed genes with coefficient <−0.8.
| Mild | ||
|---|---|---|
| Cmap | Correlation | P value |
| Mevalolactone | −0.988 | 0.01935 |
| Vincamine | −0.98 | 0.0002 |
| Dipivefrine | −0.969 | 0.00571 |
| Lycorine | −0.86 | 0.00146 |
| Sulmazole | −0.858 | 0.0291 |
| Etacrynic Acid | −0.829 | 0.04122 |
| Pentamidine | −0.828 | 0.00312 |
| Prestwick-691 | −0.81 | 0.04994 |
| Vanoxerine | −0.801 | 0.01675 |
| Fenoterol | 0.823 | 0.01094 |
| Depudecin | 0.857 | 0.04119 |
| Sanguinarine | 0.867 | 0.03599 |
| Rifabutin | 0.906 | 0.00174 |
| Severe pneumonia group | ||
| Cmap | Correlation | P value |
| Alsterpaullone | −0.997 | 0 |
| Valdecoxib | −0.899 | 0.00192 |
| Clofibrate | −0.861 | 0.03845 |
| Lycorine | 0.82 | 0.0004 |
| Riboflavin | 0.842 | 0.00101 |
| Carmustine | 0.848 | 0.00663 |
| Docosahexaenoic acid ethyl ester | 0.862 | 0.03839 |
| Atracurium besilate | 0.884 | 0.00316 |
| Retrorsine | 0.907 | 0.00008 |
| Anisomycin | 0.931 | 0.00004 |
| Emetine | 0.963 | 0 |
| Cephaeline | 0.996 | 0 |
Discussion
Based on the transcriptome data from RNA-seq, we first identified DEGs associated with mild and severe pneumonia, and also the co-expressed ones in each group, and then constructed a transcription factor-gene network based on the predicted transcription factors. Furthermore, we tried to uncover which biological pathways the DEGs in the network are involved. Finally, we predicted 2 potential small-molecule drugs for the treatment of mild pneumonia and severe pneumonia.
The transcription factor-gene network for the mild group and that for the severe group consisted of 215 and 451 nodes, respectively, which shared 54 common DEGs (e.g., BRCA1, BRCA2, CDY1B, CDY2A, S100A12, S100A9) with consistent differential expression change in the 2 groups, indicating the similarity in molecular mechanisms between mild pneumonia and severe pneumonia.
GO functional annotation revealed that genes involved in defense and inflammatory responses and spermatogenesis may have important roles in the pathogenesis of mild pneumonia. Among them, 3 S100 gene family members (S100A8, S100A9, and S100A12) showed upregulated expression in patients with mild pneumonia (the latter 2 were also upregulated in severe pneumonia), suggesting minor difference in the roles of this family between different pneumonia types. The proteins encoded by this gene family are also known as migration inhibitory-related proteins (MRP), which are mainly expressed in granulocytes, macrophages, activated endothelial cells, and epithelial cells. S100A8 and S100A9 in the form of heteromeric dimer calprotectin (S100A8/A9) can chelate Zn+ to inhibit the growth of a wide variety of microorganisms [18–20]. Raquil et al. reported high expression of S100A8 and S100A9 proteins in the alveolar walls of lungs of mice infected with S. pneumoniae, and confirmed that both proteins have an important role in leukocyte migration, strongly suggesting their involvement in the transepithelial migration of macrophages and neutrophils [21]. S100A12 can activate the receptor for advanced glycation end-products (RAGE), which is expressed ubiquitously in the lungs, mainly on endothelial and respiratory epithelial cells [22], and activation of RAGE triggers NF-κB signaling pathway [23,24], resulting in the transcription of proinflammatory factors. Elevated S100A12 level has been detected in patients with bacterial pneumonia [25], in patients with sepsis due to pneumonia [26], and in patients with acute respiratory distress syndrome [23]. Taken together, these 3 S100 gene family members have critical roles in the pathogenesis of pneumonia, although they may function in pneumonia of varying severity.
Raquil et al. also observed an opposite trend in the expression of S100A8 and S100A9 and CXCL1, a member of the CXC (C-X-C motif) chemokine family [27]. In the present study we also found CXCL1 expression was downregulated. According to the functional annotation, CXCL1 may play a role in mild pneumonia via BP terms in response to wounding, defense response, inflammatory response, and cytoskeleton organization. Despite these findings, the role of CXCL1 needs to be further investigated.
Interestingly, the expression of CXCL11, another CXC chemokine family member, was overexpressed in patients with severe pneumonia in the present study. McAllister et al. found that CXCL11 were undetectable at day 0 but was detectable at days 7 and 14 after Pneumocystis infection in mice with pneumonia caused by this strain [28], showing that the expression of this chemokine may be related to pneumonia severity. However, we found another C-C chemokine, CCL5 [29], showing a downregulated expression in severe pneumonia. Palaniappan et al. reported that CCL5 is an essential factor for the induction and maintenance of protective pneumococcal immunity [30]. Singh et al. further pointed out that CCL5 blockade altered humoral and cellular pneumococcal immunity via modulating PspA (Pneumococcal surface protein A)-specific T helper cells during S. pneumonia-induced carriage [31]. According to functional annotation, CCL5 was thought to contribute to the pathogenesis of severe pneumonia via response to oxidative stress, positive regulation of cell differentiation, response to bacterium, locomotory behavior, response to wounding, and defense response. In addition, genes involved in the regulation of cellular protein metabolic process and positive regulation of protein metabolic process (e.g., DAZ3, PSMA1, CNTF, PSMA5, KLKB1, CDC26, UBE2C, and BRCA1) are also speculated to have a role in the pathogenesis of severe pneumonia, although their involvement in this disease has not been reported yet.
Finally, we predicated that mevalolactone and alsterpaullone may be used as potential drugs for the treatment of mild pneumonia and severe pneumonia, respectively. In fact, alsterpaullone, which is a GSK3 (glycogen synthase kinase-3) inhibitor, can inhibit the replication of influenza virus [29], and it has been used for the prevention and treatment of pneumonia caused by influenza virus [30], further validating the credibility of our prediction. Thus, the use of mevalolactone in the therapy of mild pneumonia may be feasible, although there is no report on the role of mevalolactone in pneumonia treatment (mevalolactone is the precursor of in vivo synthesis of terpene compounds and steroids [31]). However, this needs more clinical evidence.
Conclusions
By use of RNA-seq, we found some genes may contribute to the pathogenesis of both pneumonias (e.g., S100A9 and S100A12) and some may have more important roles in the pathogenesis of mild pneumonia (e.g., S100A9 and CXCL1) or severe pneumonia (e.g., CCL5 and CXCL11). Additionally, we predicated 2 small-molecule drugs, mevalolactone and alsterpaullone, that may have potential for the treatment of mild pneumonia and severe pneumonia, respectively. However, since most of our findings were drawn by bioinformatics analyses, more evidence is needed.
Footnotes
Conflicts of interest
None.
Source of support: This work was supported by grants from the Welfare Industry Research Program of the Ministry of Health (No. 201302017, 201502019), the National Natural Science Fund (No.81272060, 81371561), the Hai Nan Natural Science Fund (20158315), the Youth Training Program of the PLA (No.13QNP171), the Beijing Scientific and Technologic Supernova Supportive Project (Z15111000030000/XXJH2015B100), the PLA General Hospital Science and Technology Innovation Nursery Fund Project (16KMM56), and the PLA Logistic Major Science and Technology Project (14CXZ005, AWS15J004, BWS14J041)
References
- 1.Zar H, Madhi S, Aston S, Gordon S. Pneumonia in low and middle income countries: Progress and challenges. Thorax. 2013;68(11):1052–56. doi: 10.1136/thoraxjnl-2013-204247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Örtqvist Å, Hedlund J, Kalin M. Seminars in respiratory and critical care medicine. Copyright© 2005 by Thieme Medical Publishers, Inc; 333 Seventh Avenue, New York, NY 10001, USA: 2005. Streptococcus pneumoniae: Epidemiology, risk factors, and clinical features; pp. 563–74. [DOI] [PubMed] [Google Scholar]
- 3.Said MA, Johnson HL, Nonyane BA, et al. AGEDD Adult Pneumococcal Burden Study Team. Estimating the burden of pneumococcal pneumonia among adults: A systematic review and meta-analysis of diagnostic techniques. PLoS One. 2013;8:e60273. doi: 10.1371/journal.pone.0060273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solga AC, Pong WW, Walker J, et al. RNA-sequencing reveals oligodendrocyte and neuronal transcripts in microglia relevant to central nervous system disease. Glia. 2015;63:531–48. doi: 10.1002/glia.22754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akat KM, Moore-McGriff DV, Morozov P, et al. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc Natl Acad Sci USA. 2014;111:11151–56. doi: 10.1073/pnas.1401724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craciun FL, Bijol V, Ajay AK, et al. RNA sequencing identifies novel translational biomarkers of kidney fibrosis. J Am Soc Nephrol. 2016;27:1702–13. doi: 10.1681/ASN.2015020225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao S, Fung-Leung W-P, Bittner A, et al. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9:e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson MD, Mccarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression dataSMotn. Bioinformatics. 2009;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deza E, Deza MM. Encyclopedia of Distances. Springer; 2013. [Google Scholar]
- 11.Wang L, Cao C, Ma Q, et al. RNA-seq analyses of multiple meristems of soybean: Novel and alternative transcripts, evolutionary and functional implications. BMC Plant Biology. 2013;14:1–19. doi: 10.1186/1471-2229-14-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blancafort P, Segal DJ. Designing transcription factor architectures for drug discovery. Mol Pharmacol. 2004;66:1361–71. doi: 10.1124/mol.104.002758. [DOI] [PubMed] [Google Scholar]
- 13.Jiang C, Xuan Z, Zhao F, Zhang MQ. TRED: A transcriptional regulatory element database, new entries and other development. Nucleic Acids Res. 2007;35:D137–40. doi: 10.1093/nar/gkl1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odelius K, Finne A, Albertsson AC. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–32. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beißbarth T, Speed TP. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–65. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 16.Lamb J, Crawford ED, Peck D, et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 17.Flynn C, Zheng S, Yan L, et al. Connectivity map analysis of nonsense-mediated decay-positive BMPR2-related hereditary pulmonary arterial hypertension provides insights into disease penetrance. Am J Respir Cell Mol Biol. 2012;47:20–27. doi: 10.1165/rcmb.2011-0251OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohnle PG, Hunter MJ, Hahn B, Chazin WJ. Zinc-reversible antimicrobial activity of recombinant calprotectin (migration inhibitory factor – related proteins 8 and 14) J Infect Dis. 2000;182:1272–75. doi: 10.1086/315810. [DOI] [PubMed] [Google Scholar]
- 19.Clohessy P, Golden B. Calprotectin-mediated zinc chelation as a biostatic mechanism in host defence. Scand J Immunol. 1995;42:551–56. doi: 10.1111/j.1365-3083.1995.tb03695.x. [DOI] [PubMed] [Google Scholar]
- 20.Nisapakultorn K, Ross KF, Herzberg MC. Calprotectin expression inhibits bacterial binding to mucosal epithelial cells. Infec Immun. 2001;69:3692–96. doi: 10.1128/IAI.69.6.3692-3696.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raquil M-A, Anceriz N, Rouleau P, Tessier PA. Blockade of antimicrobial proteins S100A8 and S100A9 inhibits phagocyte migration to the alveoli in streptococcal pneumonia. J Immunol. 2008;180:3366–74. doi: 10.4049/jimmunol.180.5.3366. [DOI] [PubMed] [Google Scholar]
- 22.Uchida T, Shirasawa M, Ware LB, et al. Receptor for advanced glycation end-products is a marker of type I cell injury in acute lung injury. Am J Respir Crit Care Med. 2006;173:1008–15. doi: 10.1164/rccm.200509-1477OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittkowski H, Sturrock A, Van Zoelen MA, et al. Neutrophil-derived S100A12 in acute lung injury and respiratory distress syndrome. Crit Care Med. 2007;35:1369–75. doi: 10.1097/01.CCM.0000262386.32287.29. [DOI] [PubMed] [Google Scholar]
- 24.Moroz O, Antson A, Dodson E, et al. The structure of S100A12 in a hexameric form and its proposed role in receptor signalling. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 3):407–13. doi: 10.1107/s0907444901021278. [DOI] [PubMed] [Google Scholar]
- 25.Hou F, Wang L, Wang H, et al. Elevated gene expression of S100A12 is correlated with the predominant clinical inflammatory factors in patients with bacterial pneumonia. Mol Med Rep. 2015;11:4345–52. doi: 10.3892/mmr.2015.3295. [DOI] [PubMed] [Google Scholar]
- 26.Achouiti A, Föll D, Vogl T, et al. S100A12 and soluble receptor for advanced glycation end products levels during human severe sepsis. Shock. 2013;40:188–94. doi: 10.1097/SHK.0b013e31829fbc38. [DOI] [PubMed] [Google Scholar]
- 27.Raquil MA, Anceriz N, Rouleau P, Tessier PA. Blockade of antimicrobial proteins S100A8 and S100A9 inhibits phagocyte migration to the alveoli in streptococcal pneumonia. J Immunol. 2008;180:3366–74. doi: 10.4049/jimmunol.180.5.3366. [DOI] [PubMed] [Google Scholar]
- 28.Mcallister F, Ruan S, Steele C, et al. CXCR3 and IFN protein-10 in Pneumocystis pneumonia. J Immunol. 2006;177:1846–54. doi: 10.4049/jimmunol.177.3.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zlotnik A, Yoshie O. Chemokines: A new classification system and their role in immunity. Immunity. 2000;12:121–27. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 30.Palaniappan R, Singh S, Singh UP, et al. CCL5 modulates pneumococcal immunity and carriage. J Immunol. 2006;176:2346–56. doi: 10.4049/jimmunol.176.4.2346. [DOI] [PubMed] [Google Scholar]
- 31.Singh R, Singh S, Singh UP, et al. CCL5 modulates pneumococcal surface protein A (PspA) peptide-specific T helper cell responses. FASEB J. 2008;22(Suppl 1):853.15. [Google Scholar]