Abstract
Nicotine withdrawal symptoms contribute to relapse in smokers, thereby prolonging the harm caused by smoking. To investigate the molecular basis for this phenomenon, we conducted a genomewide association study (GWAS) of DSM-IV nicotine withdrawal in a sample of African American (AA) and European American (EA) smokers. A combined AA and EA meta-analysis (n=8,021) identified three highly correlated SNPs in the protocadherin (PCDH)-α, -β and -γ gene cluster on chromosome 5 that were associated with nicotine withdrawal (p < 5×10−8). We then studied one of the SNPs, rs31746, in an independent sample of smokers who participated in an intravenous nicotine infusion study that followed overnight smoking abstinence. After nicotine infusion, abstinent smokers with the withdrawal risk allele experienced greater alleviation of their urges to smoke, as assessed by the Brief Questionnaire on Smoking Urges (BQSU). Prior work has shown that the PCDH-α, -β and -γ genes are expressed in neurons in a highly organized manner. We found that rs31746 mapped to a long-range neuron-specific enhancer element shown previously to regulate PCDH-α, -β and -γ gene expression. Using Braincloud mRNA expression data, we identified a robust and specific association between rs31746 and PCDH-β8 mRNA expression in frontal cortex tissue (p < 1×10−5). We conclude that PCDH-α, -β and -γ gene cluster regulatory variation influences the severity of nicotine withdrawal. Further studies on the PCDH-α, -β and -γ genes and their role in nicotine withdrawal may inform the development of novel smoking cessation treatments and reduce the harm caused by tobacco smoking.
Keywords: smoking, relapse, nicotine, eQTL, GWAS
Introduction
Tobacco smoking is a persistent global public health problem that is responsible for over 5 million deaths annually.1, 2 Because there are clear health benefits of quitting smoking irrespective of smoking history, current age, or presence of smoking-induced diseases,1–3 efforts to promote smoking cessation are a high public health priority. More than 95 percent of smoking quit attempts fail within one year,4 and symptoms of nicotine withdrawal (NicW) are a major precipitating factor for relapse.5, 6 It has been argued that the severity of NicW symptoms, rather than the number of cigarettes smoked or the severity of nicotine dependence, best predicts the outcome of smoking cessation attempts.7 Thus, understanding the genetics of NicW could help to improve tobacco smoking cessation treatment efforts and significantly reduce the harm caused by smoking.
NicW has both an affective/cognitive component (e.g., irritability, frustration, anxiety, depressed mood, restlessness, and difficulty concentrating) and a somatic component (e.g., insomnia, decreased heart rate, increased appetite, constipation, cough, and dizziness).8 NicW symptoms are thought to be mediated by neuroadaptations that affect the function of nicotinic acetylcholine receptors (nAChR), notably α2, β2, α5, α6, and β4.9–11 For example, single-photon emission computed tomography (SPECT) studies indicate that β2*-nAChR availability increases during early abstinence and remains at elevated levels for ~1 month before returning to baseline.12, 13 Changes in the firing of dopaminergic neurons in the ventral tegmental area (VTA) that reduce the amount of dopamine released into the nucleus accumbens (NA) may also contribute to the development of symptoms of NicW,14, 15 as well as symptoms of withdrawal from opioids, cocaine and alcohol.16 Human fMRI neuroimaging studies have provided additional insight into the neuroadaptations that may be responsible for NicW, and several studies have shown that NicW disrupts resting state functional connectivity in the brain.17 These cessation-induced disruptions in functional connectivity are associated with cognitive impairments and craving to smoke.18
Although genetic variation influences the development of NicW, few risk genes have been identified.19–22 The identification of risk genes could provide insight into the molecular mechanism of NicW in humans. We conducted a genome-wide association study (GWAS) of DSM-IV NicW in a combined sample of 8021 European American (EA) and African American (AA) tobacco smokers. The sample was recruited for studies on the genetics of cocaine, opioid, and alcohol dependence (Yale-Penn study),23–25 and was augmented with GWAS data from subjects in the Study of Addiction: Genetics and Environment (SAGE), which are available to researchers through dbGAP (Database of Genotypes and Phenotypes).26–28 We previously published a GWAS of FTND score that was based partially on this dataset.29
Based on the GWAS findings, we investigated the association of an identified NicW risk allele in the protocadherin (PCDH)-α, -β and -γ gene cluster with characteristics of NicW in an independent intravenous (IV) nicotine infusion study. The IV nicotine infusion study included tobacco smokers who had completed a ~10 hour period of smoking abstinence. We also characterized the effects of the NicW-associated risk allele on gene regulatory mechanisms and PCDH-α, -β and -γ gene expression using publically available transcriptomic and epigenomic data from postmortem human brain.
Methods
Subjects
The EA and AA participants in the NicW GWAS are described in Table 1. The Yale–Penn-1 and Yale–Penn-2 subjects were recruited at multiple Eastern US sites for genetic studies of drug and alcohol dependence, as described elsewhere.23–25 These subjects were phenotyped using the polydiagnostic Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA).30 Yale-Penn study participants provided written informed consent prior to their participation and the institutional review board at each participating site approved the studies. The National Institute on Drug Abuse and the National Institute on Alcohol Abuse and Alcoholism issued Certificates of Confidentiality to protect Yale-Penn study participants. The GWAS also included data from the Study of Addiction: Genetics and Environment (SAGE),26 obtained via the database of Genotypes and Phenotypes (accession: phs000092.v1.p1). The SAGE dataset includes subjects from COGA (the Collaborative Study on the Genetics of Alcoholism),28 FSCD (the Family Study of Cocaine Dependence) 27 and COGEND (the Collaborative Genetic Study of Nicotine Dependence).26 Only subjects who smoked at least 100 cigarettes in their lifetime were included in the GWAS.
Table 1.
Distribution of subjects by population and nicotine withdrawal prevalence.
| Study | Total N | Nicotine withdrawal1 | FTND score | ND % | Former smokers % | Age (SEM) | Male % | |
|---|---|---|---|---|---|---|---|---|
| Negative N | Positive N (%) | |||||||
| Yale-Penn-1: | ||||||||
| European American | 1575 | 418 | 1157 (74) | 5.42(0.05) | 87 | 14 | 37.8 (0.3) | 61 |
| African American | 2083 | 813 | 1270 (61) | 4.42(0.05) | 74 | 9 | 42.2 (0.2) | 58 |
| Yale-Penn-2: | ||||||||
| European American | 861 | 254 | 607 (71) | 4.88(0.07) | 81 | 21 | 43.4 (0.4) | 66 |
| African American | 672 | 261 | 411 (61) | 4.18(0.08) | 71 | 19 | 37.9 (0.4) | 65 |
| SAGE: | ||||||||
| European American | 1953 | 1081 | 872 (45) | 3.10(0.08) | 58 | 35* | 38.3 (0.3) | 51 |
| African American | 877 | 384 | 493 (56) | 3.96(0.10) | 56 | 17* | 39.5 (0.3) | 53 |
| Total | 8021 | |||||||
DSM-IV nicotine withdrawal symptom; FTND, Fagerstrom Test for Nicotine Dependence; ND, DSM-IV nicotine dependent;
Estimated based on non-missing age of use data, which was 98% of the total data; SEM, standard error of the mean.
The NicW diagnosis was based on DSM-IV, which requires that four NicW symptoms be experienced within the first 24 hours after either quitting or smoking less than usual, or smoking (or using other nicotine containing products) to avoid problems caused by quitting or smoking less than usual. The NicW symptoms included: 1) Were you irritable, angry, or frustrated? 2) Were you nervous or anxious? 3) Were you restless? 4) Did you have trouble concentrating? 5) Did your heart slow down? 6) Did you feel down or depressed? 7) Did you have such a strong desire for cigarettes that you couldn’t think of anything else? 8) Did your appetite increase or did you gain weight? 9) Did you have trouble sleeping? Table 1 shows the number of subjects and NicW rates for each study group and demographic and smoking-related characteristics.
The laboratory study, which was conducted independently from the GWAS, included 179 non-treatment-seeking EA (50%) and AA (50%) smokers recruited from the New Haven, Connecticut area. Seventy-three percent of subjects were male and the mean age was 36.2 (s.d.=8.9) years. Some subjects from this sample have been included in previous studies, including genetic studies related to smoking and nicotine.31–34 All laboratory subjects smoked > 10 cigarettes/day for the past year and had expired carbon monoxide levels of ≥10 ppm at the initial screening. Based on a structured interview (Structured Clinical Interview for DSM-IV),35 the subjects were free of major medical problems and current psychiatric disorders, including dependence on alcohol or drugs (other than nicotine). Plasma nicotine, cotinine, and 3′-hydroxycotinine concentrations were measured at baseline (prior to nicotine infusion) using HPLC interfaced with tandem mass spectrometry (LC/MS/MS), as previously described.31, 36 Baseline Minnesota Nicotine Withdrawal Scale (MNWS)37 and Brief Questionnaire of Smoking Urges (BQSU) 38 were completed after IV lines were established. After baseline assessments, subjects were administered IV doses of saline, followed in uniform order by escalating doses of nicotine (0.5 mg per 70 kg of body weight and 1 mg per 70 kg of body weight). Each infusion lasted 30 seconds and was separated from the preceding one by 30 minutes. The first dose of nicotine, 0.5 mg per 70 kg of body weight, is approximately equal to the dose of nicotine delivered by smoking one-half of a cigarette, while the second dose of nicotine, 1.0 mg per 70 kg of body weight, is approximately equal to the dose of nicotine delivered by smoking 1 cigarette.39 Subjects repeated the MNWS and BQSU 20 minutes after the final infusion of nicotine. One hundred seventy-nine subjects completed the MNWS and 177 subjects completed the BQSU. The distribution of MNWS and BQSU outcomes is shown in Fig S2. DSM-IV nicotine withdrawal was not measured in the laboratory subjects. All laboratory study subjects provided written informed consent prior to participation, and were paid for their participation. Yale University and the VA Connecticut Healthcare System institutional review boards approved the laboratory study.
Genotyping and Genotype Imputation
The array-genotyped SNP data were acquired with the following Illumina (San Diego, CA, USA) arrays: Yale-Penn-1 by the HumanOmni1-Quad v1.0; Yale-Penn-2 by the HumanCore Exome BeadChip; and SAGE by the Human 1M. IMPUTEv2 software40 was used to impute additional SNPs for each array. The 1000 Genomes June 2011 (http://www.1000genomes.org/) release was used as the reference for imputation. The 1000 Genomes reference included phased haplotypes for 1094 individuals of diverse ancestries, including samples of European descent, Asian descent, African descent, and an admixed American sample.41 SNPs with imputed information scores < 0.80 and minor allele frequencies < 2.5 percent (for both imputed and directly genotyped SNPs) were excluded from the GWAS analysis. To evaluate our GWAS finding of association between NicW and PCDH-α, -β and –γ gene cluster variation, the laboratory subjects were genotyped for rs31746 with a 2 μl TaqMan allelic discrimination assay (Applied Biosystems, Foster City, CA, USA). We considered rs31746 to be the “best” SNP to select for follow-up studies in the laboratory sample because in contrast to the other two SNPS, rs31746 was directly genotyped by all arrays and it was positioned in a regulatory element. The majority of laboratory subjects (n=124) were assigned to either EA or AA ancestry groups based on a previously described ancestry informative genetic marker method.32,42 Subjects not assigned by a genetic ancestry were assigned based on their self-reported ancestry. Four subjects with a genetic assignment that was discordant with the self-report were re-assigned to the ancestry group defined by the genetic markers.
Analysis
Quality control procedures were performed with PLINK43 and EIGENSOFT v4.2.44, 45 using array-based genotype calls. EA and AA subjects were analyzed separately. One subject was retained from each group of related subjects (identity-by-descent > 0.25). Subjects were excluded from the present study if they were ever more than six standard deviations from the mean of principal component (PC) 1, 2 or 3 during five iterations of PC analysis (PCA) in EIGENSOFT v4.2.44, 45 Association testing was performed with PLINK.43 The SNP was coded as the allele dosage, and age, sex and the first 10 PCs (to control for population stratification) were included as covariates. We observed acceptable genomic inflation factor (λ) values for our analyses that included 10 PCs and no additional steps were taken to include or exclude covariates from the model. The results from each array were combined by meta-analysis. SNPs were excluded from the meta-analyses if they were not present (either directly genotyped or well-imputed) in all studies. There were 7,445,053 SNPs for the AA sample, 5,675,219 SNPs for the EA sample, and 4,795,232 SNPs for the EA+AA sample. Regional associations were evaluated with plots generated using LocusZoom (http://locuszoom.sph.umich.edu/locuszoom).46 For laboratory participants, BQSU and MNWS effects were analyzed with linear mixed models using JMP Pro (v10.0.0) software (SAS Institute Inc.). The following independent variables were included in the models as fixed effects: rs31746 genotype (with number of risk allele copies coded additively as 0, 1 and 2), sex, population (EA or AA), time point (pre- or post-nicotine) and the interaction of time point with rs31746 genotype. A random effect was included to account for correlations between the repeated measures from each subject. We performed additional analyses to control for the potential confounding effects of differences in baseline smoking behavior, evaluating the fold-change in BQSU with models that included the following independent variables: sex, race, baseline nicotine plasma concentration, baseline cotinine plasma concentration, and baseline nicotine metabolite ratio.
SNP functional annotation and characterization
A literature search led us to previous work identifying DNase I hypersensitivity sites and CTCF binding sites important in regulating the expression of genes in the PCDH gene cluster.47–49 The positions of NicW-associated SNPs relative to these regulatory regions were examined using the following UCSC Genome Browser tracks: “DNaseI HS Density Signal from ENCODE/Duke” track for ventromedial prefrontal cortex and fibroblast tissue, and “Transcription Factor ChIP-seq (161 factors) from ENCODE” track for CCCTC-binding factor (CTCF).50–52 HS5-1 sites were mapped to the genome using primers that had been previously used to generate electrophoretic mobility shift assay probes for HS5-1a and HS5-1b.47 UCSC Genome Browser51 screenshots containing the indicated track data were used to illustrate the relative position of the SNPs and regulatory elements. The P-values for the association of rs31746 with PCDH-α, -β and -γ gene cluster mRNA expression were extracted from BrainCloud (http://braincloud.jhmi.edu/; dbGaP Study Accession: phs000417.v2.p1).53 BrainCloud includes data from human prefrontal cortex (PFC) tissue and an association with gene expression in other brain regions was not tested. Fifty-five different PCDH-α, -β and -γ gene mRNAs were targeted by 63 probes. All gene names and corresponding expression P-values are shown in Table S3.
Results
Genomewide association study of nicotine withdrawal symptoms: rs31746 associated with NicW across populations
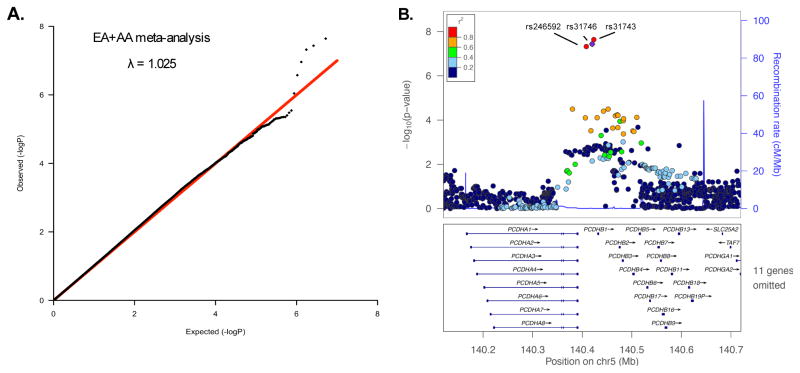
The demographic characteristics of GWAS subjects are shown in Table 1. There was little evidence of systemic P-value inflation (genomic inflation factor λ < 1.029) for the EA, AA or EA+AA GWAS meta-analyses (Figure 1A and Supplemental Figure S1). There were no NicW associations (P > 5×10−8) observed in the meta-analyzed EA or AA samples. In the meta-analyzed EA+AA sample, three closely mapped SNPs (rs246592, rs31746, rs31743) in an intergenic position within the PCDH-α, -β and -γ gene cluster on chromosome 5 (Figure 1B) were associated with NicW after a genomewide test correction (Pmeta < 5×10−8). No heterogeneity was detected in the meta-analysis (Cochran’s Q, p > 0.5, I2 =0). The statistical strength of the association was nearly indistinguishable among the three SNPs (P=2.3×10−8 – 4.8×10−8) and, as indicated by the 1000 Genome Project data,41 the pairwise correlation between all SNPs was high in populations of European (r2=1, D′=1) and African (r2>0.66, D′>0.98) ancestry. Rs31746 was directly genotyped by all arrays used in the study. The effect of rs31746, as indicated by the magnitude and direction of the odds ratio, was consistent across the six studies that were included in the meta-analysis (Figure 2). The rs31746*A allele (EA frequency = 68%; AA frequency=47%) was associated with elevated risk for NicW (ORmeta =1.21 [95%CI 1.13–1.30]; P=3.67×10−8). The association of rs31746 to each individual DSM-IV NicW criterion for Yale-Penn 1 and Yale-Penn 2 subjects is shown in Table S2. Based on the EA+AA meta-analysis, the strongest associations were observed for “Did you feel down or depressed?” (Pmeta= 5×10−4), “Were you irritable, angry, or frustrated?” (Pmeta= 5×10−4) and “Were you restless?” (Pmeta= 1×10−3). The individual DSM NicW criterion data were unavailable for SAGE subjects.
Figure 1.
A genomewide association study of nicotine withdrawal. A. A quantile-quantile plot of observed and expected P-values for a meta-analysis of nicotine withdrawal that included European American (EA) and African American (AA) subjects (n=8,021). The genomic inflation factor, λ, is the observed median test statistic divided by the expected median test statistic. B. A regional plot generated with LocusZoom (http://locuszoom.sph.umich.edu/locuszoom/) showing nicotine withdrawal associated SNPs in the protocadherin-α, -β and -γ gene cluster. The linkage disequilibrium (r2) is based on the 1000 Genomes Ad-mixed American sample (AMR).
Figure 2.
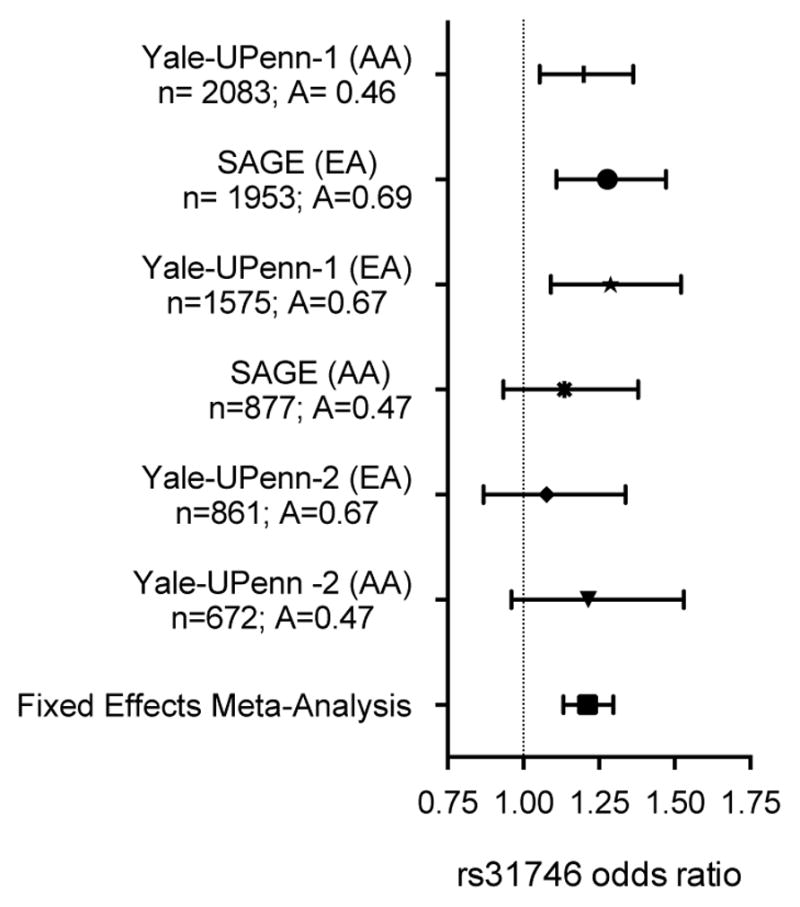
Rs31746 is associated with nicotine withdrawal in European American (EA) and African American (AA) smokers. The odds ratio (± 95% confidence interval) for rs31746*A observed for six studies and for the combined fixed effects meta-analysis (Pmeta = 3.67 × 10−8; OR=1.2 [95% confidence interval=1.13–1.30]). For each sample, the size (n) and rs31746 A-allele frequency are shown.
Rs31746 is associated with smoking urges in smokers following overnight abstinence and intravenous nicotine infusion
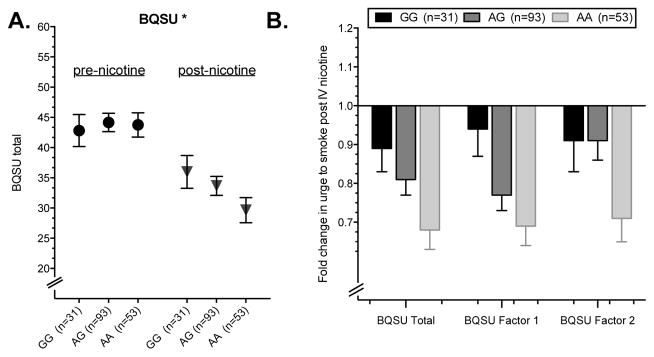
To extend our GWAS findings and gain insight into the potential behavioral basis for the association of PCDH-α, -β and -γ gene SNPs with NicW, we investigated one of the genome-wide significant SNPs, rs31746, in relation to withdrawal characteristics in a sample of current smokers who participated in an IV nicotine infusion laboratory study (n=179). Because this SNP was directly genotyped by all GWAS arrays, we considered it the strongest association signal and the best SNP in the region to select for follow-up analysis. The laboratory procedure was initiated after the subjects completed an overnight (~10 h) period of smoking abstinence. This time period is sufficient for the development of withdrawal symptoms in dependent smokers.54, 55 Following the IV nicotine infusion session, features of NicW were lower than at the pre-infusion baseline (main effect of time point: BQSU:F(1,175)= 105.89, P < 0.001; MNWS: F(1,177)=63.01, P < 0.001). The change in BQSU, but not MNWS, in response to the nicotine infusion session differed by rs31746 genotype (genotype by time point interaction: BQSU: F(1,175)= 5.37, P = 0.022) (Figure 3A). There was no significant main effect of rs31746 genotype on BQSU or MNWS. The interactive effect of genotype was similar for BQSU Factor 1, the urge to smoke for reward (F(1,175) = 4.40, P = 0.037), and BQSU Factor 2, the urge to smoke to relieve negative affect (F(1,175) = 4.68, P = 0.032). The withdrawal risk allele, rs31746*A, was associated with a greater percent reduction in the urge to smoking following the nicotine infusion session. The average percent reduction in total BQSU score by rs31746 genotype was 32% for AA, 19% for AG, and 11% for GG (Figure 3B). The association of genotype to change in BQSU score was significant after adjusting for baseline plasma cotinine and nicotine levels, and NMR (main effect of rs31746, P < 0.02). The effect was also consistent among EA and AA subjects when the two groups were analyzed separately (EA genotype by time point interaction for BQSU: F(1,85)= 2.87, P = 0.094; AA genotype by time point interaction for BQSU: F(1,88)= 2.88, P = 0.093). The average percent reduction in total BQSU score by rs31746 genotype was similar for EA subjects (30% for AA, 19% for AG, and 16% for GG) and AA subjects (34% for AA, 19% for AG, and 10% for GG).
Figure 3.
The nicotine withdrawal risk allele, rs31746*A, is associated with a greater reduction in the urge to smoke following an intravenous nicotine infusion session. A. Mean Brief Questionnaire on Smoking Urges (BQSU) score pre- and post-administration of nicotine to smokers following overnight smoking abstinence. B. The average change in BQSU total, BQSU Factor 1 and BQSU Factor 2 scores for each genotype group. * p<0.05 for the genotype by treatment interaction. Error bars = ± standard error of the mean (SEM).
Rs31746 maps to a long-range neuron-specific enhancer element and is associated with gene expression in human brain
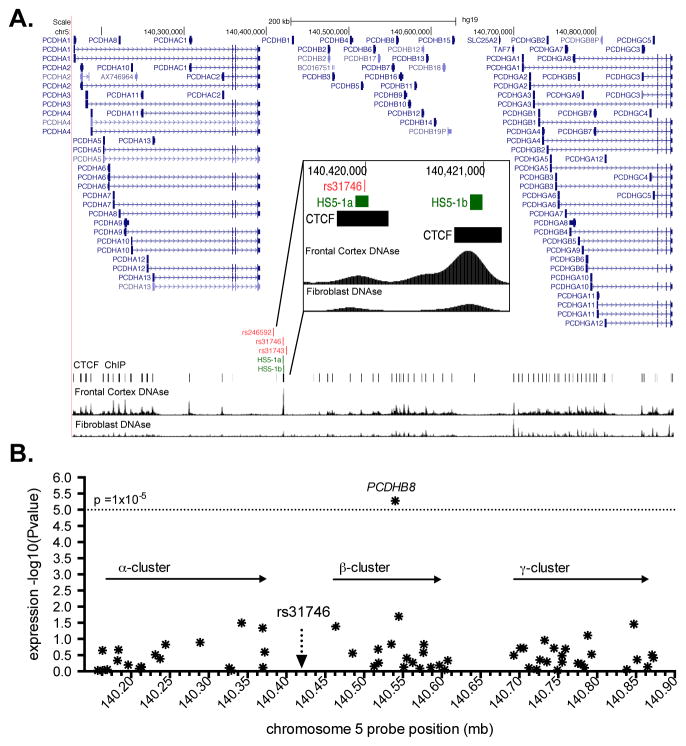
To gain insight into the potential molecular mechanisms linking PCDH-α, -β and -γ gene cluster SNPs to NicW risk, we surveyed the three NicW-associated SNPs and their flanking sequences for evidence of potential regulatory effects. As shown in Figure 4A, rs31746 mapped to a region with marked DNAse1 hypersensitivity (HS), while rs31743 and rs246592 did not. The DNAse1 HS site presented in Figure 4A contains HS5-1a and HS5-1b, components of a previously characterized neuron-specific enhancer element (HS5-1) that regulates the expression of PCDH-α, -β and -γ genes.48,56 Rs31746 mapped to within a 113 base-pair element that corresponds to HS5-1a.47 CCCTC-binding factor (CTCF), which is required for the regulatory function of HS5-1, binds to HS5-1a (Figure 4A). Based on the position of rs31746 within this regulatory element, we hypothesized that rs31746 could affect PCDH-α, -β and -γ gene expression. To test this, we used BrainCloud human PFC gene expression data to query the association of rs31746 with the expression of 55 different PCDH-α, -β and -γ gene cluster mRNAs that were targeted by 63 probes. We observed a robust association between rs31746 and PCDHβ8 mRNA expression (Figure 4B). The withdrawal risk allele, rs31746*A, was associated with lower PCDHβ8 mRNA expression (p=5.32×10−6).
Figure 4.
The position and regulatory effects of nicotine-withdrawal-associated variants in the protocadherin (PCDH)-α, -β, and -γ gene cluster. A. A modified UCSC Genome Browser (http://genome.ucsc.edu) image showing the position of the three nicotine-withdrawal-associated risk variants relative to a neuron-specific enhancer element (HS5-1a,b; green bars) and the DNA binding sites for CCCTC-binding factor (CTCF; black bars). ENCODE generated DNAseI hypersensitivity regions for frontal cortex (FC) DNA and fibroblast DNA are also shown. B. The association of rs31746 with PCDH-α, -β, and -γ gene mRNA expression in prefrontal cortex based on BrainCloud (http://braincloud.jhmi.edu). Each point represents an mRNA probe.
Discussion
Nicotine withdrawal (NicW) is common among tobacco smokers and its symptoms contribute to smoking relapse following quit attempts. Using GWAS, we identified three highly correlated SNPs in the PCDH-α, -β and -γ gene cluster that were associated with DSM-IV NicW. Subsequently, we evaluated the influence of one of these SNPs, rs31746, on smoking withdrawal behavior in an independent sample of current tobacco smokers who had abstained from smoking for ~10 hours. Following IV nicotine infusion, the NicW risk allele was associated with a greater reduction in the urge to smoke, a clinically relevant characteristic of NicW.6 That is, individuals with the NicW risk allele experienced greater alleviation of their withdrawal symptoms in a laboratory setting following nicotine infusion. Rs31746 is located within a previously characterized PCDH-α, -β and –γ gene cluster neuron-specific enhancer element, HS5-1. Using publically available data, we observed a robust association between rs31746 and PCDHβ8 mRNA expression in postmortem human prefrontal cortex tissue. Our results indicate that NicW risk is influenced by regulatory variation in the PCDH-α, -β and -γ gene cluster.
Our initial association finding emerged from a GWAS of six separately analyzed samples derived from three independent cohorts containing nearly 4,400 EA subjects and more than 3,600 AA subjects. The results of the nicotine infusion laboratory study, which used a different study design from the GWAS, supported the GWAS finding by showing an association of the rs31746*A allele with a greater change in the urge to smoke, a clinically important characteristic of NicW. This critical insight suggests one potential mechanism, regulation of smoking urges and craving, by which rs31746 might influence NicW risk. However, the laboratory study subjects had a ~10 hour period of smoking abstinence and there was no association of rs31746 to NicW characteristics at the smoking abstinence baseline prior to nicotine infusion. Evaluating genotype effects in subjects after a longer period of abstinence might also be informative given prior work suggesting that some withdrawal symptoms peak 2–3 days post-cessation57. The limited abstinence period may be why we observed effects for QSU but not MNWS. Although withdrawal symptoms and urges are similar, and often correlated, constructs, they follow different time courses during abstinence; indeed some research suggests that BQSU may have greater sensitivity to early abstinence (24 h) compared to the MNWS.58–60 Real-time behavioral assessments of NicW in abstinent smokers over an extended period could provide important additional insight into the role of rs31746 in NicW. Alternatively, the size of the laboratory sample may have limited the statistical power to detect some effects during early abstinence.
The NicW-associated SNPs mapped to the PCDH-α, -β, and -γ gene cluster. Genes expressed from this ~800-kilobase region on human chromosome 5 encode structurally similar transmembrane proteins that are enriched at the synapses of neurons.61 The expression of PCDH-α, -β, and -γ genes in the nervous system is tightly controlled.62, 63 The specific position of rs31746 within the gene cluster is noteworthy; it maps to HS5-1, a previously characterized long-range, neuron-specific enhancer element.47–49 The critical function of this regulatory element is supported by several studies, including data compiled by ENCODE on DNAse hypersensitivity and transcription factor binding. Rs31746 is within HS5-1a, a component of HS5-1 that interacts directly with CTCF.47 HS5-1 recruits CTCF and additional proteins that control the chromatin structure of the PCDH-α, -β, and -γ gene region. Through protein partners, HS5-1 and other cis-regulatory elements in the region direct long-range DNA looping and interactions that facilitate the expression of specific PCDH-α, -β, and -γ gene mRNAs via alternative promoter usage.47–49, 56, 64 Targeted deletion of HS5-1 in mice reduced the expression of PCDHβ8 mRNA, which we observed to be associated with the rs31746*A risk allele.56 The position of rs31746 in a previously characterized and important regulatory element identifies it as a strong candidate for follow-up studies on molecular mechanisms linking the PCDH-α, -β, and -γ genes to NicW risk.
PCDH-α, -β, and -γ genes are important for many neuronal processes,65–67 but there have been few behavioral studies of the function of specific PCDH-α, -β, and -γ genes. Mice genetically engineered to express low levels of PCDH-α had abnormal fear-related contextual learning and working memory.68 Using RNA-seq to investigate changes in the mouse nucleus accumbens, Eipper-Mains et al. observed dynamic alterations in PCDH-α, -β, and -γ gene mRNA expression in response to cocaine and cocaine withdrawal.69 McGowan et al. observed that the PCDH-α, -β, and -γ gene cluster was differentially regulated by epigenetic mechanisms in the brains of adult rats following a low versus high early-life maternal care paradigm.70 This well established early-life stress paradigm induces later-life differences in hypothalamic-pituitary-adrenal axis function.70, 71 Interestingly. Suderman et al. found that syntenic regions in the human PCDH-α, -β, and -γ gene cluster were differentially regulated in subjects who had experienced severe abuse during childhood, suggesting a potentially conserved mechanism for responding to early life stressors.72 These findings are concordant with the important role of stress regulation and stress response pathways in nicotine withdrawal and smoking relapse.34, 73–78 Combined, these studies indicate that PCDH-α, -β, and -γ mRNA expression is altered in the mammalian brain in response to stimuli. Alterations that affect brain functional connectivity are thought to be associated with many neuropsychiatric disorders, including nicotine addiction.79 Future research is warranted to examine the specific effects of PCDH-α, -β, and -γ genes on brain functional connectivity.
Some study limitations should be noted. A genomewide significant effect was only observed with a meta-analysis of six studies (n=8021), which may reflect limited power. The top GWAS SNP remained significant (p < 5×10−8) after correction for genomic inflation (λ = 1.025); however, a spurious finding due to underlying ancestry differences within samples or to chance cannot be completely excluded. The observed effects related to NicW in an independent laboratory sample support the validity of the GWAS finding. Also, the GWAS sample included subjects recruited for genetic studies of alcohol, cocaine and opioid use disorder, while the laboratory study recruited just for tobacco use disorder. Rs31746 was associated only with nicotine dependence (pmeta = 5×10−4) in the combined GWAS sample and not alcohol, cocaine or opioid dependence (pmeta > 0.05), but the high prevalence of these co-occurring disorders in the GWAS sample could limit the generalizability of the results. Although, prior work has linked NicW symptoms to smoking relapse following a quit attempt 6, 7, 80, we could not evaluate a potential moderating effect of the NicW risk variant on relapse rate after smoking cessation. Additional studies will be required to determine how the NicW risk SNPs influence withdrawal symptoms and relapse in a treatment context.
In summary, using GWAS we identify a regulatory SNP in the PCDH-α, -β, and -γ gene cluster that was associated with NicW and smoking urges. Additional studies of these genes in response to stress, nicotine, and NicW are warranted. Understanding and potentially alleviating NicW symptoms could facilitate smoking cessation, thereby significantly reducing the harm associated with smoking.
Supplementary Material
Acknowledgments
This research was supported by the Veterans Administration (VA) Mental Illness Research, Education and Clinical Center (MIRECC) and a VA VISN1 Career Development Award; NIH grants R03 DA027474, R01 AA017535, R01 DA030976, R01 DA12690, MSTP 5T32GM007205-38, CTSA 8UL1TR000142 TL1, F30 DA037665; and Army STARRS, which is sponsored by the Department of the Army and funded under cooperative agreement number U01MH087981 with the US Department of Health and Human Services, National Institutes of Health, National Institute of Mental Health (NIH/NIMH). The Yale Biomedical Supercomputer, funded by NIH/NCRR (1 S10 RR19895-01), was used for some analyses. The publically available datasets used for the analyses described in this manuscript were obtained from dbGaP (accession number phs000092.v1.p and phs000417.v2.p1). Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative (GEI) (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning and with general study coordination was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C).
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Conflict of interest statement
Dr. Sofuoglu has served as an expert witness on behalf of Pfizer in lawsuits related to varenicline. Dr. Kranzler has served as a consultant or advisory board member for Alkermes, Indivior, Lundbeck, and Otsuka. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which is supported by AbbVie, Ethypharm, Lilly, Lundbeck, and Pfizer. The other authors declare no biomedical financial interests or potential conflicts of interest.
References
- 1.Banks E, Joshy G, Weber M, Liu B, Grenfell R, Egger S, et al. Tobacco smoking and all-cause mortality in a large Australian cohort study: findings from a mature epidemic with current low smoking prevalence. BMC Med. 2015;13(38) doi: 10.1186/s12916-015-0281-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter BD, Abnet CC, Feskanich D, Freedman ND, Hartge P, Lewis CE, et al. Smoking and mortality--beyond established causes. N Engl J Med. 2015;372(7):631–640. doi: 10.1056/NEJMsa1407211. [DOI] [PubMed] [Google Scholar]
- 3.Jha P, Ramasundarahettige C, Landsman V, Rostron B, Thun M, Anderson RN, et al. 21st-century hazards of smoking and benefits of cessation in the United States. N Engl J Med. 2013;368(4):341–350. doi: 10.1056/NEJMsa1211128. [DOI] [PubMed] [Google Scholar]
- 4.Fiore MC, Novotny TE, Pierce JP, Giovino GA, Hatziandreu EJ, Newcomb PA, et al. Methods used to quit smoking in the United States. Do cessation programs help? JAMA. 1990;263(20):2760–2765. [PubMed] [Google Scholar]
- 5.Zhou X, Nonnemaker J, Sherrill B, Gilsenan AW, Coste F, West R. Attempts to quit smoking and relapse: factors associated with success or failure from the ATTEMPT cohort study. Addict Behav. 2009;34(4):365–373. doi: 10.1016/j.addbeh.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Allen SS, Bade T, Hatsukami D, Center B. Craving, withdrawal, and smoking urges on days immediately prior to smoking relapse. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2008;10(1):35–45. doi: 10.1080/14622200701705076. [DOI] [PubMed] [Google Scholar]
- 7.West RJ, Hajek P, Belcher M. Severity of withdrawal symptoms as a predictor of outcome of an attempt to quit smoking. Psychol Med. 1989;19(4):981–985. doi: 10.1017/s0033291700005705. [DOI] [PubMed] [Google Scholar]
- 8.Hughes JR. Effects of abstinence from tobacco: valid symptoms and time course. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2007;9(3):315–327. doi: 10.1080/14622200701188919. [DOI] [PubMed] [Google Scholar]
- 9.Changeux JP. Nicotine addiction and nicotinic receptors: lessons from genetically modified mice. Nature Reviews Neuroscience. 2010;11(6):389–401. doi: 10.1038/nrn2849. [DOI] [PubMed] [Google Scholar]
- 10.Dani JA, Bertrand D. Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annu Rev Pharmacol Toxicol. 2007;47:699–729. doi: 10.1146/annurev.pharmtox.47.120505.105214. [DOI] [PubMed] [Google Scholar]
- 11.Gotti C, Clementi F, Fornari A, Gaimarri A, Guiducci S, Manfredi I, et al. Structural and functional diversity of native brain neuronal nicotinic receptors. Biochem Pharmacol. 2009;78(7):703–711. doi: 10.1016/j.bcp.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 12.Cosgrove KP, Batis J, Bois F, Maciejewski PK, Esterlis I, Kloczynski T, et al. beta2-Nicotinic acetylcholine receptor availability during acute and prolonged abstinence from tobacco smoking. Arch Gen Psychiatry. 2009;66(6):666–676. doi: 10.1001/archgenpsychiatry.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mamede M, Ishizu K, Ueda M, Mukai T, Iida Y, Kawashima H, et al. Temporal change in human nicotinic acetylcholine receptor after smoking cessation: 5IA SPECT study. J Nucl Med. 2007;48(11):1829–1835. doi: 10.2967/jnumed.107.043471. [DOI] [PubMed] [Google Scholar]
- 14.Fung YK, Schmid MJ, Anderson TM, Lau YS. Effects of nicotine withdrawal on central dopaminergic systems. Pharmacology Biochemistry and Behavior. 1996;53(3):635–640. doi: 10.1016/0091-3057(95)02063-2. [DOI] [PubMed] [Google Scholar]
- 15.Hildebrand BE, Nomikos GG, Hertel P, Schilstrom B, Svensson TH. Reduced dopamine output in the nucleus accumbens but not in the medial prefrontal cortex in rats displaying a mecamylamine-precipitated nicotine withdrawal syndrome. Brain Res. 1998;779(1–2):214–225. doi: 10.1016/s0006-8993(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 16.Rossetti ZL, Hmaidan Y, Gessa GL. Marked inhibition of mesolimbic dopamine release - a common feature of ethanol, morphine, cocaine and amphetamine abstinence in rats. Eur J Pharmacol. 1992;221(2–3):227–234. doi: 10.1016/0014-2999(92)90706-a. [DOI] [PubMed] [Google Scholar]
- 17.Sutherland MT, McHugh MJ, Pariyadath V, Stein EA. Resting state functional connectivity in addiction: Lessons learned and a road ahead. Neuroimage. 2012;62(4):2281–2295. doi: 10.1016/j.neuroimage.2012.01.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lerman C, Gu H, Loughead J, Ruparel K, Yang Y, Stein EA. Large-scale brain network coupling predicts acute nicotine abstinence effects on craving and cognitive function. JAMA Psychiatry. 2014;71(5):523–530. doi: 10.1001/jamapsychiatry.2013.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pergadia ML, Agrawal A, Heath AC, Martin NG, Bucholz KK, Madden PA. Nicotine withdrawal symptoms in adolescent and adult twins. Twin Res Hum Genet. 2010;13(4):359–369. doi: 10.1375/twin.13.4.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pergadia ML, Heath AC, Martin NG, Madden PA. Genetic analyses of DSM-IV nicotine withdrawal in adult twins. Psychol Med. 2006;36(7):963–972. doi: 10.1017/S0033291706007495. [DOI] [PubMed] [Google Scholar]
- 21.Xian H, Scherrer JF, Madden PA, Lyons MJ, Tsuang M, True WR, et al. The heritability of failed smoking cessation and nicotine withdrawal in twins who smoked and attempted to quit. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2003;5(2):245–254. [PubMed] [Google Scholar]
- 22.Xian H, Scherrer JF, Madden PA, Lyons MJ, Tsuang M, True WR, et al. Latent class typology of nicotine withdrawal: genetic contributions and association with failed smoking cessation and psychiatric disorders. Psychol Med. 2005;35(3):409–419. doi: 10.1017/s0033291704003289. [DOI] [PubMed] [Google Scholar]
- 23.Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, et al. Genome-wide association study of alcohol dependence: significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014;19(1):41–49. doi: 10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H, et al. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry. 2014;76(1):66–74. doi: 10.1016/j.biopsych.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, et al. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry. 2014;19(6):717–723. doi: 10.1038/mp.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bierut LJ, Strickland JR, Thompson JR, Afful SE, Cottler LB. Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend. 2008;95(1–2):14–22. doi: 10.1016/j.drugalcdep.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edenberg HJ. The collaborative study on the genetics of alcoholism: an update. Alcohol Res Health. 2002;26(3):214–218. [PMC free article] [PubMed] [Google Scholar]
- 29.Gelernter J, Kranzler HR, Sherva R, Almasy L, Herman AI, Koesterer R, et al. Genome-wide association study of nicotine dependence in American populations: identification of novel risk loci in both African-Americans and European-Americans. Biol Psychiatry. 2015;77(5):493–503. doi: 10.1016/j.biopsych.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80(3):303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Sofuoglu M, Herman AI, Nadim H, Jatlow P. Rapid nicotine clearance is associated with greater reward and heart rate increases from intravenous nicotine. Neuropsychopharmacology. 2012;37(6):1509–1516. doi: 10.1038/npp.2011.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herman AI, Jatlow PI, Gelernter J, Listman JB, Sofuoglu M. COMT Val158Met modulates subjective responses to intravenous nicotine and cognitive performance in abstinent smokers. Pharmacogenomics J. 2013;13(6):490–497. doi: 10.1038/tpj.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeVito EE, Herman AI, Waters AJ, Valentine GW, Sofuoglu M. Subjective, physiological, and cognitive responses to intravenous nicotine: effects of sex and menstrual cycle phase. Neuropsychopharmacology. 2014;39(6):1431–1440. doi: 10.1038/npp.2013.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen KP, Herman AI, Morean ME, Kranzler HR, Gelernter J, Sofuoglu M. FKBP5 variation is associated with the acute and chronic effects of nicotine. Pharmacogenomics J. 2015;15(4):340–346. doi: 10.1038/tpj.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.First MB, Spitzer RL, Gibbon M, Williams JBW. The Structured Clinical Interview for Dsm-Iii-R Personality-Disorders (Scid-Ii) .1. Description. J Pers Disord. 1995;9(2):83–91. [Google Scholar]
- 36.Dempsey D, Tutka P, Jacob P, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76(1):64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 37.Hughes JR, Hatsukami D. Signs and symptoms of tobacco withdrawal. Arch Gen Psychiatry. 1986;43(3):289–294. doi: 10.1001/archpsyc.1986.01800030107013. [DOI] [PubMed] [Google Scholar]
- 38.Cox LS, Tiffany ST, Christen AG. Evaluation of the brief questionnaire of smoking urges (QSU-brief) in laboratory and clinical settings. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2001;3(1):7–16. doi: 10.1080/14622200020032051. [DOI] [PubMed] [Google Scholar]
- 39.Djordjevic MV, Stellman SD, Zang E. Doses of nicotine and lung carcinogens delivered to cigarette smokers. J Natl Cancer Inst. 2000;92(2):106–111. doi: 10.1093/jnci/92.2.106. [DOI] [PubMed] [Google Scholar]
- 40.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Genomes Project C. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang BZ, Zhao H, Kranzler HR, Gelernter J. Practical population group assignment with selected informative markers: characteristics and properties of Bayesian clustering via STRUCTURE. Genet Epidemiol. 2005;28(4):302–312. doi: 10.1002/gepi.20070. [DOI] [PubMed] [Google Scholar]
- 43.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 46.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, et al. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci U S A. 2012;109(51):21081–21086. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kehayova P, Monahan K, Chen W, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. 2011;108(41):17195–17200. doi: 10.1073/pnas.1114357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. 2006;103(52):19719–19724. doi: 10.1073/pnas.0609445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42(Database issue):D764–770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raney BJ, Dreszer TR, Barber GP, Clawson H, Fujita PA, Wang T, et al. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014;30(7):1003–1005. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478(7370):519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hendricks PS, Ditre JW, Drobes DJ, Brandon TH. The early time course of smoking withdrawal effects. Psychopharmacology (Berl) 2006;187(3):385–396. doi: 10.1007/s00213-006-0429-9. [DOI] [PubMed] [Google Scholar]
- 55.Tiffany ST, Drobes DJ. The development and initial validation of a questionnaire on smoking urges. Br J Addict. 1991;86(11):1467–1476. doi: 10.1111/j.1360-0443.1991.tb01732.x. [DOI] [PubMed] [Google Scholar]
- 56.Yokota S, Hirayama T, Hirano K, Kaneko R, Toyoda S, Kawamura Y, et al. Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster. J Biol Chem. 2011;286(36):31885–31895. doi: 10.1074/jbc.M111.245605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hughes JR, Higgins ST, Bickel WK. Nicotine withdrawal versus other drug withdrawal syndromes: similarities and dissimilarities. Addiction. 1994;89(11):1461–1470. doi: 10.1111/j.1360-0443.1994.tb03744.x. [DOI] [PubMed] [Google Scholar]
- 58.West R, Ussher M. Is the ten-item Questionnaire of Smoking Urges (QSU-brief) more sensitive to abstinence than shorter craving measures? Psychopharmacology (Berl) 2010;208(3):427–432. doi: 10.1007/s00213-009-1742-x. [DOI] [PubMed] [Google Scholar]
- 59.Bujarski S, Roche DJ, Sheets ES, Krull JL, Guzman I, Ray LA. Modeling naturalistic craving, withdrawal, and affect during early nicotine abstinence: A pilot ecological momentary assessment study. Exp Clin Psychopharmacol. 2015;23(2):81–89. doi: 10.1037/a0038861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teneggi V, Tiffany ST, Squassante L, Milleri S, Ziviani L, Bye A. Smokers deprived of cigarettes for 72 h: effect of nicotine patches on craving and withdrawal. Psychopharmacology (Berl) 2002;164(2):177–187. doi: 10.1007/s00213-002-1176-1. [DOI] [PubMed] [Google Scholar]
- 61.Zipursky SL, Sanes JR. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell. 2010;143(3):343–353. doi: 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 62.Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, et al. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat Genet. 2005;37(2):171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- 63.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97(6):779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 64.Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, et al. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10(1):21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 65.Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488(7412):517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen WV, Alvarez FJ, Lefebvre JL, Friedman B, Nwakeze C, Geiman E, et al. Functional significance of isoform diversification in the protocadherin gamma gene cluster. Neuron. 2012;75(3):402–409. doi: 10.1016/j.neuron.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katori S, Hamada S, Noguchi Y, Fukuda E, Yamamoto T, Yamamoto H, et al. Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J Neurosci. 2009;29(29):9137–9147. doi: 10.1523/JNEUROSCI.5478-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fukuda E, Hamada S, Hasegawa S, Katori S, Sanbo M, Miyakawa T, et al. Down-regulation of protocadherin-alpha A isoforms in mice changes contextual fear conditioning and spatial working memory. Eur J Neurosci. 2008;28(7):1362–1376. doi: 10.1111/j.1460-9568.2008.06428.x. [DOI] [PubMed] [Google Scholar]
- 69.Eipper-Mains JE, Kiraly DD, Duff MO, Horowitz MJ, McManus CJ, Eipper BA, et al. Effects of cocaine and withdrawal on the mouse nucleus accumbens transcriptome. Genes Brain Behav. 2013;12(1):21–33. doi: 10.1111/j.1601-183X.2012.00873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McGowan PO, Suderman M, Sasaki A, Huang TC, Hallett M, Meaney MJ, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS One. 2011;6(2):e14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, Freedman A, et al. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science. 1997;277(5332):1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- 72.Suderman M, McGowan PO, Sasaki A, Huang TC, Hallett MT, Meaney MJ, et al. Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc Natl Acad Sci U S A. 2012;109(Suppl 2):17266–17272. doi: 10.1073/pnas.1121260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guillot CR, Pang RD, Leventhal AM. Anxiety sensitivity and negative urgency: a pathway to negative reinforcement-related smoking expectancies. J Addict Med. 2014;8(3):189–194. doi: 10.1097/ADM.0000000000000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langdon KJ, Leventhal AM, Stewart S, Rosenfield D, Steeves D, Zvolensky MJ. Anhedonia and anxiety sensitivity: prospective relationships to nicotine withdrawal symptoms during smoking cessation. J Stud Alcohol Drugs. 2013;74(3):469–478. doi: 10.15288/jsad.2013.74.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beckham JC, Lytle BL, Vrana SR, Hertzberg MA, Feldman ME, Shipley RH. Smoking withdrawal symptoms in response to a trauma-related stressor among Vietnam combat veterans with posttraumatic stress disorder. Addict Behav. 1996;21(1):93–101. doi: 10.1016/0306-4603(95)00038-0. [DOI] [PubMed] [Google Scholar]
- 76.Brown RA, Lejuez CW, Kahler CW, Strong DR. Distress tolerance and duration of past smoking cessation attempts. J Abnorm Psychol. 2002;111(1):180–185. [PubMed] [Google Scholar]
- 77.Brown RA, Lejuez CW, Kahler CW, Strong DR, Zvolensky MJ. Distress tolerance and early smoking lapse. Clin Psychol Rev. 2005;25(6):713–733. doi: 10.1016/j.cpr.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith PH, Homish GG, Giovino GA, Kozlowski LT. Cigarette smoking and mental illness: a study of nicotine withdrawal. Am J Public Health. 2014;104(2):e127–133. doi: 10.2105/AJPH.2013.301502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu H, Stein EA. Resting state functional connectivity: its physiological basis and application in neuropharmacology. Neuropharmacology. 2014;84:79–89. doi: 10.1016/j.neuropharm.2013.08.023. [DOI] [PubMed] [Google Scholar]
- 80.Piasecki TM, Jorenby DE, Smith SS, Fiore MC, Baker TB. Smoking withdrawal dynamics: I. Abstinence distress in lapsers and abstainers. J Abnorm Psychol. 2003;112(1):3–13. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.