Abstract
The WNT signaling pathway is a critical mediator of tissue homeostasis and repair, and frequently co-opted during tumor development. Almost all colorectal cancers (CRC) demonstrate hyperactivation of the WNT pathway, which in many cases is believed to be the initiating and driving event. In this short review, we provide a focused overview of recent developments in our understanding of the WNT pathway in CRC, describe new research tools that are enabling a deeper understanding of WNT biology, and outline ongoing efforts to target this pathway therapeutically.
Keywords: colorectal cancer, CRC, Wnt, APC, beta-catenin, RSPO
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer-related death in the United States and accounts for almost 600,000 deaths worldwide, annually [1]. The majority of CRCs arise sporadically in patients with no family history of disease, and while colonoscopic removal of premalignant tumors has led to an overall reduction in morbidity, patients that progress to advanced disease have few effective treatment options and a dismal prognosis [2]. In the age of rational drug design and precision medicine, CRC research and treatment is somewhat lagging. It has been more than 25 years since Vogelstein and colleagues proposed the key genetic drivers of CRC progression, and almost a decade since next generation genomic technologies began to define the full genetic landscape of CRC. To date, this vast knowledge has not translated into effective targeted therapies.
Mutational inactivation of the Adenomatous Polyposis Coli (APC) tumor suppressor is thought to be the initiating event in most sporadic and familial CRCs. Disruption of APC drives activation of the WNT signaling pathway, and a wealth of evidence suggests that WNT hyperactivation is the key oncogenic driver in most, if not all, CRCs. In the past 5 years, extensive genomic analyses and the development of sophisticated model systems to study normal and transformed intestine have expanded our understanding of WNT signaling in intestinal biology and CRC pathogenesis.
The WNT Pathway
WNT, a portmanteau of Wingless and Int, was first identified for its role regulating segment polarity in Drosophila, and its proto-oncogene function in breast tumors induced by mouse mammary tumor virus. The WNT family consists of 19 secreted, cysteine-rich glycoproteins that have been implicated in diverse biological processes, including cell fate specification, cell proliferation, cell migration, dorsal axis formation, and asymmetric cell division [3, 4]. The canonical, or β-catenin–dependent, signaling cascade is a multistep process that involves the relocalization, phosphorylation, and degradation of multiple proteins, culminating in a coordinated transcriptional response; detailed mechanistic models for Wnt signal transduction have been covered extensively elsewhere [5, 6]. While the full, specific complement of factors involved is often cell and tissue dependent, the core signal transduction cascade is constant (Fig 1). Briefly, WNT ligands bind FRIZZLED (FZD) and LRP receptor complexes, initiating membrane recruitment of key scaffold proteins (AXIN, DVL), and disruption of the β-catenin destruction complex (minimally composed of AXIN, APC, CK1, GSK3β). In the absence of this complex, β-catenin accumulates in the cytosol, and through poorly understood mechanisms, translocates into the nucleus where it associates with TCF family transcription factors and a host of co-activators to drive transcription of target genes. The direct transcriptional output of Wnt activation is context dependent. For instance, in the mammalian colon, Wnt activation drives proliferation and stem-like phenotypes [7–9], while in embryonic stem cells β-catenin can promote pluripotency or stem cell differentiation [10, 11].
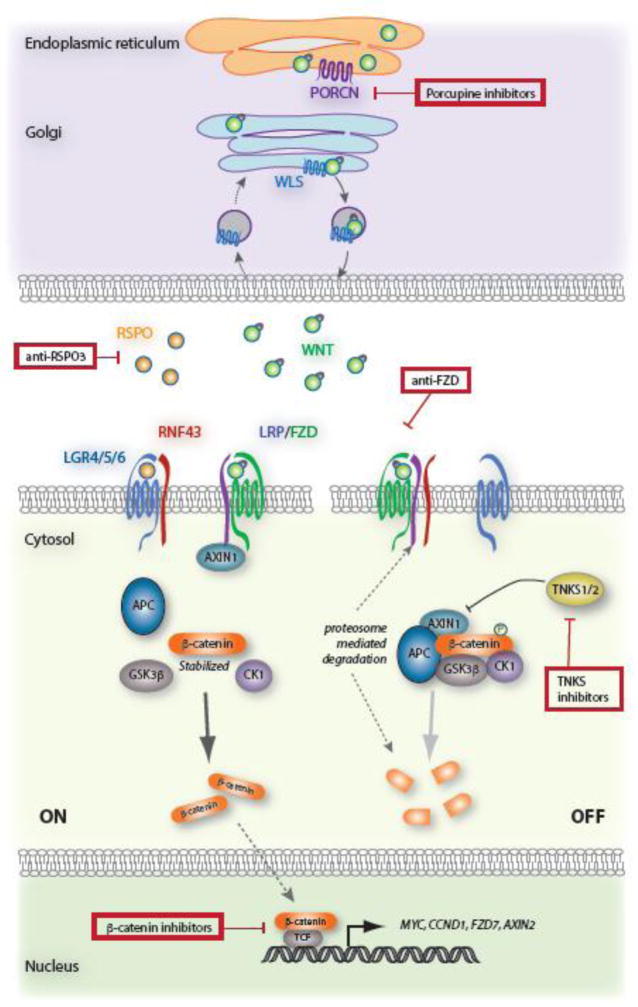
Figure 1. WNT Pathway Overview.
WNT ligand is palmitoylated by Porcupine (PORCN) in the endoplasmic reticulum, and transported to the plasma membrane via Wntless (WLS). Once secreted and binds receptors FZD and LRP5/6 and this interaction recruits AXIN to the cell membrane and destabilizes the destruction complex, composed of APC, AXIN1, CK1, and GSK3β. Reduced β-catenin phosphorylation by GSK3β and CK1 decreases protein turnover, enabling it to translocate into the nucleus, where it can associate with co-activators and promote the transcription of a plethora of target genes, including MYC, CCND1, FZD, AXIN2. The secreted ligand, RSPO, forms complex with LGR4/5/6 and RNF43, preventing RNF43-dependent degradation of FZD receptors, and potentiating WNT signaling. In the absence of WNT or RSPO ligand, the destruction complex efficiently phosphorylates β-catenin, tagging it for subsequent degradation by the proteasome. Red boxes indicate various nodes of existing small molecule inhibitors designed for WNT-targeted therapies.
WNT in the Intestine
Wnt signaling is an essential factor in normal intestinal function, and in particular, for the maintenance and self-renewal of epithelial stem cells located at the base of intestinal crypts [12, 13]. Using engineered animal models, several groups have shown that either blocking upstream Wnt ligand-mediated signaling [14, 15], disrupting the TCF transcriptional co-activator [16, 17] or deleting β-catenin itself [18] drives loss of intestinal crypts and breakdown of the tissue. Of note, epithelial-specific deletion of the Wnt ligand itself blocks expansion of intestinal epithelium cultured ex vivo but does not disrupt the intestinal architecture or expression of stem cell markers in vivo [19], implying that there are redundant sources of Wnt production in the intestine.
WNTs are secreted glycoproteins that must undergo a series of modifications before they are physiologically active. In the endoplasmic reticulum (ER), the transmembrane O-acyltransferase Porcupine (PORCN) catalyzes the lipidation (palmitoylation) of nascent WNT proteins at two distinct sites [reviewed in [20]]. Next, lipid-bound WNTs are glycosylated in the ER before transitioning to the Golgi, where they interact with a second transmembrane protein, Wntless (WLS). WLS is essential for the delivery and secretion of WNT ligands at the cell surface [21] and following secretion, WI protein is recycled back to the Golgi, via retromer endosome transport [22–24]. Inhibiting either the lipidation or transport of WNT via PORCN or WLS is sufficient to completely block WNT ligand secretion, and thus, offers two potential therapeutic nodes for WNT-ligand driven disease.
Recent work, led primarily by Hans Clevers’ laboratory, has further refined our understanding of how Wnt signaling is regulated in the adult intestine. Beginning with the identification of Leucine-rich G-protein coupled receptor 5 (LGR5) as a key marker of stem cells in the gut [25], subsequent studies implicated this ‘marker’ as a direct regulator of Wnt activity, identified the ligand/s for LGR5 as R-Spondin/s (RSPOs), and produced a structure-based model to explain the control of Wnt signaling. In this model, Wnt ligand-mediated signaling is limited via two transmembrane E3-ligases, RNF43 and ZNRF3, which polyubiquitinate active WNT-FZD complexes and target them for degradation in the lysosome [26]. In LGR5+ intestinal stem cells, however, Wnt-mediated signaling is maintained at high levels by R-Spondins (RSPO1-4), secreted ligands that bridge a physical interaction between Lgr4/5 and RNF43/ZNRF3, thus sequestering the ligases, and enabling continued signal activation from the WNT-FZD complex [27].
WNT pathway disruptions in CRC
Since the initial identification of APC alterations in human CRC, and the recognition that the APC protein controls WNT activity, it was clear that the majority of colon tumors carried high levels of WNT pathway activity. In fact, the 2012 report from The Cancer Genome Atlas (TCGA) consortium estimated that up to 92% of sporadic CRCs contained at least one alteration in a known WNT regulator [28]. In addition to the WNT regulators described by the TCGA, several studies have since expanded the range of potential WNT-driving genetic alterations.
In 2012, de Sauvage and colleagues identified the first recurrent genomic translocations in CRC, involving RSPO family members (RSPO2 and RSPO3) [29]. Interestingly, although relatively few cases have been identified, in both their study and in follow up work [30–32], RSPO translocations were mutually exclusive with APC mutations. Though correlative, this pattern suggests either synthetic lethality, as has been shown for coincident RAS-MAPK alterations [33], or that these events are functionally redundant. Indeed, enforced expression of RSPO1 or RSPO3 can drive proliferation in the intestine in animal models, but in both cases seems to be due to a paracrine effect of the secreted RSPO ligand [34, 35]. Further work will be required to determine whether RSPO rearrangements alone are sufficient to drive tumor development in the intestine. Similar to RSPO fusions, RNF43 mutations were identified in a significant proportion of human CRCs (up to 18%), and were found to be largely exclusive with APC (and RSPO) alterations [36].
Thus, accumulated evidence suggests that the vast majority (i.e., close to 100%) of CRCs carry WNT signaling alterations. Still, there are many unknowns regarding the strong selection for mutations in particular WNT pathway genes, and this may significantly impact our ability to exploit such alterations for therapeutic gain. For instance, CRCs show a remarkable bias toward truncating mutations in APC within a central 200 amino acid region of the protein, known as the mutation cluster region. Such a clustering of non-sense and frameshift mutations is not typical of classic tumor suppressor genes, and implies that these specific disruptions provide a selective advantage for tumor growth. To explain this phenomenon, many have proposed the ‘just right’ hypothesis [37], which infers that pre-cancerous cells must receive ‘not too little, and not too much’ WNT activation to drive transformation. Indeed the level of Wnt induction is dependent on the position of the APC truncation [38], but there is little experimental evidence to explain why this is important for CRC development.
A related, but distinct and unexplained mystery, is the dominance of APC alterations in CRC at the expense of other WNT activating mutations. In hepatocellular carcinoma (HCC), up to 25–35% of tumors show heightened WNT pathway activation, driven through disruption of AXIN1 or activating mutations in CTNNB1 (cbioportal.org) yet they do not show recurrent mutations in APC [28]. In contrast, AXIN and β-catenin mutations make up only a small fraction (5–6%) of CRC cases. Defining the molecular underpinnings of this remarkable tumor-specific mutational profile may provide critical insights into how Wnt pathway deregulation drives tumor growth in different tissues. For this, the development of sophisticated model systems with tight cell and tissue-specific control over gene activation or disruption will be essential.
Models of Colon Biology & CRC
Much of our understanding of the WNT pathway in CRC comes from model systems. Human CRC cell lines have been extensively used to characterize many aspects of CRC biology, and while they are readily accessible and grow rapidly, thus facilitating biochemistry and cell biology experiments, the genetic complexity and variability of such lines often confounds detailed genetic analysis. While cell lines are still heavily used today, the coming years will see a major shift toward new in vitro and in vivo tools to study normal intestinal function and CRC biology.
Organoids
A recent advance that has revolutionized the modeling of CRC is the development of techniques to isolate and maintain normal and transformed intestinal ‘organoids’. In 2009, Clevers and colleagues reported that mouse small intestinal stem cells are able to proliferate in 3D Matrigel culture, spontaneously self-organize into crypt-villus units, self-renew, and differentiate into all intestinal lineages [39]. The relative ease in establishing and manipulating organoids has sparked a wave of studies to characterize the genetics, signaling, metabolism, and therapeutic response of normal and transformed stem cells [40–44]. In one example, Farin et al. recently used organoids to develop a model of how Wnt gradients in the intestine are established, and maintain the geography of stem and progenitor cells within the crypt [45]. First, Wnt is delivered to the Lgr5+ stem cell via direct contact with the Wnt-producing Paneth cell. Second, low level, diffuse Wnt dispersal is generated as a consequence of cell division. This elegant work was made possible by the unique three-dimensional architecture and heterotypic cell-cell interactions enabled by organoid culture. While the precise molecular mechanism by which Wnt ligand is delivered from the Paneth cell to the intestinal stem cell remains to be shown, organoids will likely play a key role in defining this process.
Adding to the flexibility of organoid culture systems, advances in CRISPR/Cas9-based genome editing provide a means to efficiently and strategically introduce CRC-relevant mutations in vitro. Two independent studies highlight the power of this approach, recapitulating the proposed sequence of oncogenic disruptions observed in sporadic CRC [40, 42]. In addition to the culture and manipulation of ‘normal’ colonic tissue, numerous institutes, including ours, have invested in the generation of patient-derived organoid banks that include both normal and cancer-derived tissue [41]. This resource provides an unprecedented opportunity to study Wnt pathway alterations and a multitude of other cancer associated genetic events, and it provides a platform to interrogate therapeutic responses in normal and cancer-derived tissue. Indeed, the first large-scale example of this showed how medium throughput drug screens on a human colon organoid bank can identify rare drug sensitivities and/or drug combinations that have potent mutation-specific effects on individual organoids [41].
Finally, to bridge the gap between in vitro and in vivo analysis of CRC, we and others have developed strategies for the orthotopic engraftment of normal [46] and transformed mouse and human organoids (O’Rourke et al, in press; J. Roper et al, in press). Platforms such as this provide a unique, functional way to study the mechanisms of disease progression in the colon in situ, and/or develop tailored pre-clinical models for drug testing.
Animal models
The earliest genetic model of CRC arose by chance from an ENU mutagenesis screen performed in the late 1980s in the laboratory of William Dove. The multiple intestinal neoplasia (Min) phenotype was subsequently mapped and identified as a single nonsense (truncating) mutation in Apc (ApcMin) [47]. ApcMin mice served as the backbone of genetic and pre-clinical research in CRC for many years, and are still used today. However, they are often deemed an imperfect model of human CRC because they develop tumors predominantly in the small intestine, rather than in the colon. The generation of increasingly sophisticated genetic engineering technologies has dramatically improved our ability to model human CRC in the mouse. Not surprisingly, most efforts have focused on strategies to induce deletion of Apc in the colon, as this provides the most robust induction of tumor development of any genetic lesion tested.
One of the earliest strategies to localize tumor development to the colon focused on administering Cre-expressing adenovirus to the colonic lumen of mice carrying a Cre-conditional allele of Apc [48]. Though this approach proved difficult for many in the field to reproduce, more than 10 years after the initial description, two groups published similar methods to enable the investigation of colon cancer in the mouse [49, 50]. In parallel, many groups have developed tissue-specific Cre mouse strains. These transgenic and knock-in alleles including Fabp-Cre [51], Cdx2-Cre [52], Cdx2-CreER [53], CAC [54], and Car1-CreER [55], all enable reproducible, colon-restricted gene inactivation or activation, depending on the genetic context. One recent publication describes the development of the Car1-CreER strain, which outlines an extremely complex model to generate oncogenic mutations in the four most common CRC-associated alterations (Apc, Kras, Trp53 and Smad4) [55]. This is an impressive example of what can be accomplished with traditional genetically engineered mouse models (GEMMs), but also a cautionary tale. Due to the extra-colonic complications of p53 loss, and the difficulty in breeding so many alleles, the authors could only analyze a single mouse with this genetic configuration. When developing complex models to interrogate multiple-hit CRC, there is a clear need to consider alternate strategies.
To accelerate the generation of tailored and complex animal models, we recently described the generation of inducible CRISPR/Cas9-based mice, which allows the simultaneous induction of multiple mutations from a single targeted allele [56]. In addition to providing a powerful platform for multiplexed mutagenesis in vivo, we believe this technology creates a specific opportunity for investigating Apc and Wnt biology. Due to the unusual genomic structure of Apc, in which 75% of its coding sequence is contained within a single exon, it has been challenging to produce Cre-conditional alleles that represent the most frequent CRC-associated mutations. As mentioned above, the majority of Apc mutations in human CRC lie between amino acids 1250 and 1450 in the “Mutation Cluster Region”, and there is a strong correlation between the type of APC truncation, level of Wnt signaling and disease severity in familial cases [57–59]. Inducible CRISPR/Cas9-based genome editing now provides an avenue to assess the impact of specific Apc alterations on tumor initiation, progression and response.
The animal models described above clearly demonstrate that Apc loss is sufficient for tumorigenesis in the intestine. But, once established, is WNT hyperactivation necessary for tumor maintenance and progression? Various distinct animal models suggest the answer is yes. For example, doxycycline-mediated activation of β-catenin, or suppression of Apc, drives hyperplasia in the intestinal crypts, whereas withdrawal of β-catenin, or restoration of Apc, reverses these acute effects [9, 60]. More importantly, restoration of Apc is sufficient to induce sustained tumor regression, even in tumors carrying additional oncogenic alterations in Kras and p53 [9]. This work provides compelling evidence that viable therapeutic targets for the Wnt pathway could have a profound impact on the treatment of CRC.
WNT signaling as a therapeutic target in CRC
Despite the overwhelming evidence that WNT pathway hyperactivation drives CRC, targeted WNT therapies have not made headway in the clinic. While the overall survival (OS) for patients with CRC has progressively lengthened over the last 30 years, it is largely attributed to advances in surgery, chemotherapy, adenoma detection and removal by colonoscopy, the use of aspirin and NSAIDs for other clinical indications, and more recently, development of agents targeting VEGF and EGFR signaling. Hence, the question remains, why have WNT-targeted agents failed to have a clinical impact, and where do we head from here? We will not attempt to detail all WNT-targeted therapeutics, but rather provide some key examples of current strategies to control WNT pathway hyperactivation, and where future efforts might lead. Conceptually, WNT-targeting approaches can be divided into three categories: 1) Targeting the WNT ligand-receptor interface, 2) Regulation of the endogenous destruction complex, and 3) Direct interference with β-catenin-mediated transcription.
The WNT ligand/receptor interface
Although direct WNT pathway agents are not yet in clinical practice, a number are in various stages of preclinical development, some are in early Phase clinical trials. Perhaps the most mature of all efforts to target hyperactive WNT signaling are small molecules and antibodies designed to block WNT-FZD ligand-receptor interaction at the cell surface. For example, Vantictumab (OMP-18R5) is an antibody targeting FZD receptors [61], and Ipafricept is a decoy-receptor, comprised of the FZD8 extracellular domain and the Fc domain of human immunoglobulin [62]. While both drugs have shown acceptable safety profiles and progressed to Phase 1b evaluation for breast, ovarian and pancreatic cancers, neither is under investigation for treatment of CRC. This is not surprising, as the majority of CRCs activate WNT signaling independent of actual WNT ligand. A similar paradigm is expected to hold true for strategies that aim to prevent the secretion of Wnt ligands (i.e., Porcupine inhibitors) or neutralize Wnt ligand directly, thus reinforcing the challenge of targeting activated WNT signaling in CRC.
While drugs tested to date that block ligand-receptor signaling have been ineffective in the majority of CRCs, tumors carrying RSPO rearrangements or RNF43 mutations, which require ligand-mediated activation to initiate the WNT response, are potential candidates for this approach [31, 41, 63–65]. Indeed, an ongoing basket trial for patients with treatment-refractory metastatic CRC carrying RSPO/RNF43 and BRAF mutations combines a PORCN inhibitor (WNT974), a BRAF inhibitor (LGX818), and the EGFR-targeted antibody Cetuximab. Preliminary work with PORCN inhibitors in CRC organoids [41], xenograft models [31] or GEMMs [66] appears promising, but it is still unclear whether these inhibitors, alone or in combination, will provide an effective and safe therapeutic window for CRC treatment.
In a related, and perhaps more direct approach, Genentech and OncoMed Pharmaceuticals have developed RSPO3 neutralizing antibodies to target tumors with RSPO3 chromosome rearrangements. This approach has shown efficacy in patient-derived xenografts [32], and the OncoMed drug (OMP-131R10) has now entered a Phase 1a/b study for patients with various advanced solid tumors. An important aspect to ensure success of these clinical studies will be incorporating sensitive biomarker assays to prospectively identify the small fraction of CRC cases that are predicted to benefit from this approach.
Targeting the Destruction Complex
The destruction complex, composed of APC, AXIN, GSK3b, CK1, is the core network that regulates β-catenin degradation, and thereby serves as a cytosolic gatekeeper for β-catenin-mediated transcription. As the destruction complex enforces endogenous tumor suppression, it represents an attractive target to attenuate WNT signaling. In theory, there are numerous paths to stabilize the destruction complex, but the best explored is the regulation of AXIN function via Tankyrase activity.
Tankyrase enzymes (TNKS and TNKS2) are members of the Poly ADP-Ribose Polymerase (PARP) family of proteins that act to PARsylate AXIN, marking it for degradation, thus destabilizing the destruction complex. In the past 5–6 years, numerous academic groups and commercial entities have identified independent TNKS/2 inhibitors as potent suppressors of WNT pathway activity. Most importantly, several studies showed that TNKS blockade can block the proliferation of APC-mutant CRC cell lines in vitro, and some have demonstrated efficacy in vivo in GEMMs with Apc-mutant adenomas. Moreover, three recent studies showed that TNKS inhibition synergizes with approved chemotherapies [67] and drugs targeting RAS-MAPK and PI3K-AKT pathways [68, 69]. Despite the excitement of TNKS inhibition as a potential therapy to treat the majority of APC-mutant CRCs, the field is clouded by uncertainty, as multiple studies have reported conflicting data on the toxicity of TNKS blockade. In addition to toxicity associated with WNT suppression in normal intestine and bone marrow, TNKS modulates the stability of proteins involved in cell division (e.g., PLK1) and telomere elongation (e.g., TERF1), among others [70]. Thus, there may be multiple confounding variables that influence the effectiveness of TNKS-based approaches. Further genetic and pharmacologic studies will be required to address the safety and efficacy of TNKS inhibition as a therapeutic strategy in CRC.
Targeting β-catenin-mediated transcription
The ultimate effector of WNT pathway activity is β-catenin mediated gene transcription. Hence, it follows that the most direct path to blocking WNT hyperactivation is by inhibiting this transcriptional response. However, it is clear from genetic studies that complete ablation of β-catenin is overtly toxic to normal intestinal epithelium [18]. As an alternative, several groups have attacked this issue by developing strategies to block β-catenin association with specific transcriptional co-activators. The first example of this was published more than 10 years ago, with the description of ICG-001, a small molecule that specifically blocks the interaction of β-catenin with CREB binding protein (CBP), but not p300 [71]. ICG-001, now known as PRI-724, was developed by Prism Pharma for treatment of CRC, pancreatic adenocarcinoma, myeloid malignancies and HCV-induced liver cirrhosis (clinicaltrials.gov). A number of Phase 1 trials for these indications are ongoing, and Prism has registered a Phase 2 randomized study for initial treatment of metastatic CRC in combination with FOLFOX/Bevacizumab. The results of this study are widely anticipated by both oncologists and scientists studying WNT hyperactivation in CRC and other cancers.
Beyond this early example, various screens have identified a number of small molecule inhibitors, such as iCRT14 [72] and CCT036477 [60]. Promising in vitro data shows their capacity to reduce levels of Wnt reporter gene activity, decrease expression of β-catenin transcriptional targets, and block binding to the co-activator TCF [60, 73–75]. Despite this potential, strong in vivo data demonstrating the efficacy of these compounds in colorectal cancer is lacking. Thus, further studies are required to validate β-catenin as a therapeutic target and to ensure anti-tumor activity in a mechanism that minimizes intestinal toxicity.
WNT and immunotherapy - toward the unknown
The most promising cancer treatments to emerge over the past 5–10 years are immune checkpoint inhibitors, such as anti-CTLA4, anti-PD1, and anti-PDL1 blocking antibodies. These immunotherapies suppress the endogenous mechanism that restrains autoimmunity, and permit adaptive immune response to ablate tumors cells. In contrast to many targeted therapies, immunotherapies often induce sustained anti-tumor responses, leading to long-term regression of advanced cancers. DNA mismatch repair (MMR) deficient, hypermutated CRCs demonstrated promising responses to checkpoint blockade [76]; however, these tumors represent only 10–15% of CRCs. The mechanism underlying the lack of response in non-hypermutated CRCs and other solid tumors is not clearly defined. Some evidence from colon and lung cancers suggests it is due to the reduced burden of neoantigens [76, 77], while other models imply active T cell exclusion from the tumor mass [78]. Interestingly, a recent analysis of melanoma non-responders concluded that activated WNT/β-catenin signaling mediates resistance to anti-CTLA4 and anti-PD-L1 treatment by preventing recruitment of antigen presenting dendritic cells (DCs) [79]. Additional work suggests that suppressing WNT activity may decrease DC-mediated tolerance [80–83], and drive differentiation of cytotoxic T lymphocytes, thus bolstering anti-tumor immunity [84]. It is tempting to speculate that part of the reason CRCs show a poor response to checkpoint blockade is due to high WNT activity, and that inhibition of WNT signaling would provide both a cell intrinsic and non-cell intrinsic anti-tumor effect, in combination with immunotherapy. Of course, it is possible WNT inhibition may have other unexpected consequences for effective immune-based approaches. Regardless, our first priority is to generate safe and effective WNT-targeted small molecules. The identification of such drugs will likely have far reaching applications beyond the treatment of CRC.
Conclusion
In the 30 years since its discovery, WNT signaling has emerged one of the most important biological pathways in development and disease. In CRC, WNT pathway hyperactivation is arguably the most critical cancer driver, and represents an exciting avenue for targeted therapy. As new technologies pave the way for a more refined understanding of WNT function in normal and transformed cells, we expect the identification and development of WNT-targeted therapeutics to accelerate, and hope that these efforts translate into a significant clinical benefit.
Table 1.
WNT-targeted therapeutics
| Node | Target | Drug | CRC Model | Efficacy in vivo | Stage | Source |
|---|---|---|---|---|---|---|
| Ligand/Receptor | Porcupine | WNT974 | Human CRC organoids, GEMMs | Yes* | Phase 1 | [41, 63, 64] |
| ETC-159 | PDX | Yes | Pre-clinical | [31] | ||
| C59 | GEMM | Yes | [65] | |||
| RSPO | OMP-131R10 | Xenografts, GEMMs | Yes | Phase 1 | [32]# | |
| FZD receptor | OMP-18R5 | PDX | Yes | Phase 1 | [61] | |
| OMP-54F28 | Cell lines, PDX | Yes | Phase 1 | [85] | ||
| LRP6 | GSK3178022 | Cell lines, xenograft, PDX | Yes | Pre-clinical | [86] | |
| Destruction Complex | Tankyrase | XAV939 | CRC lines | N/A | Pre-clinical | [87] |
| JW55 | CRC lines | Yes | Pre-clinical | [88] | ||
| NVP-TNKS656 | CRC lines, PDX | Yes | Pre-clinical | [69] | ||
| G007-LK | Cell lines, xenografts, GEMMS | Dose-limiting toxicity | Pre-clinical | [89] | ||
| G631 | Xenografts | Dose-limiting toxicity | Pre-clinical | [90] | ||
| Transcription | β-catenin/TCF | ICG-001 (PRI-724) | CRC lines, xenograft, Human trial | Yes | Phase 1/2 | [71] |
| 2,4-diamino-quinazoline | CRC lines, xenograft | No | Pre-clinical | [91] | ||
| PNU-74654 | CRC lines | N/A | Pre-clinical | [92] | ||
| iCRT3, iCRT5, iCRT14 | CRC lines, GEMMS | Yes* | Pre-clinical | [72, 74, 75] | ||
| CCT036477 | CRC lines, β-catenin mutant mice | No | Pre-clinical | [60] |
Reference describes an RSPO3 Antibody developed by Genentech, OMP-131R10 has not been published
Non-CRC tumors
PDX– Patient-derived xenograft
Xenografts– Refers to CRC lines subcutaneously engrafted into immunocompromised mice
Acknowledgments
EMS is supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM07739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program. LED is supported by a K22 Career Development Award from the NCI/NIH (CA 181280-01), with funding from the Starr Cancer Consortium (I8-A8-030) and NIH/NCI (5R01CA195787-02).
Footnotes
Compliance with Ethics Guidelines
Conflict of Interest
Emma M. Schatoff, Benjamin I. Leach, and Lukas E. Dow declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 4.McCartney BM, I, Näthke S. Cell regulation by the Apc protein Apc as master regulator of epithelia. Current opinion in cell biology. 2008;20(2):186–193. doi: 10.1016/j.ceb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Stamos JL, Weis WI. The beta-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5(1):a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17(1):9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker N, Ridgway R, van Es J, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2008 doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 8.Sansom OJ, Reed KR, Hayes AJ, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18(12):1385–90. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Dow LE, O’Rourke KP, Simon J, et al. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell. 2015;161(7):1539–52. doi: 10.1016/j.cell.2015.05.033. This paper defined the importance of sustained Apc loss for driving tumorigenesis in the colon. Specifically, that restoring normal levels of Apc is sufficient to revert even advanced carcinomas to normal epithelium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyashenko N, Winter M, Migliorini D, et al. Differential requirement for the dual functions of beta-catenin in embryonic stem cell self-renewal and germ layer formation. Nat Cell Biol. 2011;13(7):753–61. doi: 10.1038/ncb2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marucci L, Pedone E, Di Vicino U, et al. beta-catenin fluctuates in mouse ESCs and is essential for Nanog-mediated reprogramming of somatic cells to pluripotency. Cell Rep. 2014;8(6):1686–96. doi: 10.1016/j.celrep.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15(1):19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 13.Schepers A, Clevers H. Wnt signaling, stem cells, and cancer of the gastrointestinal tract. Cold Spring Harb Perspect Biol. 2012;4(4):a007989. doi: 10.1101/cshperspect.a007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhnert F, Davis CR, Wang HT, et al. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2004;101(1):266–71. doi: 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flanagan DJ, Phesse TJ, Barker N, et al. Frizzled7 functions as a Wnt receptor in intestinal epithelial Lgr5(+) stem cells. Stem Cell Reports. 2015;4(5):759–67. doi: 10.1016/j.stemcr.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korinek V, Barker N, Moerer P, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19(4):379–83. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 17.van Es JH, Haegebarth A, Kujala P, et al. A critical role for the Wnt effector Tcf4 in adult intestinal homeostatic self-renewal. Mol Cell Biol. 2012;32(10):1918–27. doi: 10.1128/MCB.06288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fevr T, Robine S, Louvard D, et al. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol. 2007;27(21):7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farin HF, Van Es JH, Clevers H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143(6):1518–1529 e7. doi: 10.1053/j.gastro.2012.08.031. [DOI] [PubMed] [Google Scholar]
- 20.Das S, Yu S, Sakamori R, et al. Wntless in Wnt secretion: molecular, cellular and genetic aspects. Front Biol (Beijing) 2012;7(6):587–593. doi: 10.1007/s11515-012-1200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banziger C, Soldini D, Schutt C, et al. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006;125(3):509–22. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 22.Belenkaya TY, Wu Y, Tang X, et al. The retromer complex influences Wnt secretion by recycling wntless from endosomes to the trans-Golgi network. Dev Cell. 2008;14(1):120–31. doi: 10.1016/j.devcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Zhang P, Wu Y, Belenkaya TY, et al. SNX3 controls Wingless/Wnt secretion through regulating retromer-dependent recycling of Wntless. Cell Res. 2011;21(12):1677–90. doi: 10.1038/cr.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harterink M, Port F, Lorenowicz MJ, et al. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat Cell Biol. 2011;13(8):914–23. doi: 10.1038/ncb2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 26.Koo BK, Spit M, Jordens I, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488(7413):665–9. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 27.de Lau W, Peng WC, Gros P, et al. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 2014;28(4):305–16. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seshagiri S, Stawiski EW, Durinck S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–4. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinmura K, Kahyo T, Kato H, et al. RSPO fusion transcripts in colorectal cancer in Japanese population. Mol Biol Rep. 2014;41(8):5375–84. doi: 10.1007/s11033-014-3409-x. [DOI] [PubMed] [Google Scholar]
- 31.Madan B, Ke Z, Harmston N, et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene. 2016;35(17):2197–207. doi: 10.1038/onc.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Storm EE, Durinck S, de Sousa e Melo F, et al. Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature. 2016;529(7584):97–100. doi: 10.1038/nature16466. This paper demonstrated that antibodies targeting RSPO3 were sufficient to halt tumor growth in xenografts of CRCs carrying PTPRK-RSPO3 fusions. This example implies RSPO fusions are a driving event in CRC and highlights a potential therapeutic approach. [DOI] [PubMed] [Google Scholar]
- 33.Unni AM, Lockwood WW, Zejnullahu K, et al. Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. Elife. 2015;4:e06907. doi: 10.7554/eLife.06907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhanja P, Saha S, Kabarriti R, et al. Protective role of R-spondin1, an intestinal stem cell growth factor, against radiation-induced gastrointestinal syndrome in mice. PLoS One. 2009;4(11):e8014. doi: 10.1371/journal.pone.0008014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilkens J, Timmer NC, Boer M, et al. RSPO3 expands intestinal stem cell and niche compartments and drives tumorigenesis. Gut. 2016 doi: 10.1136/gutjnl-2016-311606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giannakis M, Hodis E, Jasmine Mu X, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46(12):1264–6. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albuquerque C, Breukel C, van der Luijt R, et al. The ‘just-right’ signaling model: APC somatic mutations are selected based on a specific level of activation of the beta-catenin signaling cascade. Hum Mol Genet. 2002;11(13):1549–60. doi: 10.1093/hmg/11.13.1549. [DOI] [PubMed] [Google Scholar]
- 38.Gaspar C, Franken P, Molenaar L, et al. A targeted constitutive mutation in the APC tumor suppressor gene underlies mammary but not intestinal tumorigenesis. PLoS genetics. 2009;5(7):e1000547. doi: 10.1371/journal.pgen.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 40••.Drost J, van Jaarsveld RH, Ponsioen B, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521(7550):43–47. doi: 10.1038/nature14415. This study, along with Matano et al (Ref 36) demonstrated the ability to sequencially and specifically manipulate genomic loci in cultured human organoids. This enabled the authors of both studies to recapitulate the proposed sequence of oncogenesis in the colon (The Vogelgram), and highlight the stepwise transition to CRC. [DOI] [PubMed] [Google Scholar]
- 41•.van de Wetering M, Francies HE, Francis JM, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–45. doi: 10.1016/j.cell.2015.03.053. This paper demonstrated the potential to establish living repositories of patient-derived organoid cultures, and use them to prospectively identify drug sensitivities and new driver mutations in CRC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matano M, Date S, Shimokawa M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nature medicine. 2015;21(3):256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 43.Yilmaz OH, Katajisto P, Lamming DW, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486(7404):490–5. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grun D, Lyubimova A, Kester L, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525(7568):251–5. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 45.Farin HF, Jordens I, Mosa MH, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530(7590):340–3. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- 46.Yui S, Nakamura T, Sato T, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat Med. 2012;18(4):618–23. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- 47.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247(4940):322–4. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 48.Shibata H, Toyama K, Shioya H, et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science. 1997;278(5335):120–3. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 49.Hung KE, Maricevich MA, Richard LG, et al. Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(4):1565–70. doi: 10.1073/pnas.0908682107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hadac JN, Leystra AA, Paul Olson TJ, et al. Colon Tumors with the Simultaneous Induction of Driver Mutations in APC, KRAS, and PIK3CA Still Progress through the Adenoma-to-carcinoma Sequence. Cancer Prev Res (Phila) 2015;8(10):952–61. doi: 10.1158/1940-6207.CAPR-15-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saam JR, Gordon JI. Inducible gene knockouts in the small intestinal and colonic epithelium. J Biol Chem. 1999;274(53):38071–82. doi: 10.1074/jbc.274.53.38071. [DOI] [PubMed] [Google Scholar]
- 52.Hinoi T, Akyol A, Theisen BK, et al. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 2007;67(20):9721–9730. doi: 10.1158/0008-5472.CAN-07-2735. [DOI] [PubMed] [Google Scholar]
- 53.Feng Y, Sentani K, Wiese A, et al. Sox9 induction, ectopic Paneth cells, and mitotic spindle axis defects in mouse colon adenomatous epithelium arising from conditional biallelic Apc inactivation. Am J Pathol. 2013;183(2):493–503. doi: 10.1016/j.ajpath.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xue Y, Johnson R, Desmet M, et al. Generation of a transgenic mouse for colorectal cancer research with intestinal cre expression limited to the large intestine. Mol Cancer Res. 2010;8(8):1095–104. doi: 10.1158/1541-7786.MCR-10-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tetteh PW, Kretzschmar K, Begthel H, et al. Generation of an inducible colon-specific Cre enzyme mouse line for colon cancer research. Proc Natl Acad Sci U S A. 2016;113(42):11859–11864. doi: 10.1073/pnas.1614057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dow LE, Fisher J, O’Rourke KP, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol. 2015;33(4):390–4. doi: 10.1038/nbt.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowan AJ, Lamlum H, Ilyas M, et al. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci U S A. 2000;97(7):3352–7. doi: 10.1073/pnas.97.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sieber OM, Segditsas S, Knudsen AL, et al. Disease severity and genetic pathways in attenuated familial adenomatous polyposis vary greatly but depend on the site of the germline mutation. Gut. 2006;55(10):1440–8. doi: 10.1136/gut.2005.087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaspar C, Fodde R. APC dosage effects in tumorigenesis and stem cell differentiation. Int J Dev Biol. 2004;48(5–6):377–86. doi: 10.1387/ijdb.041807cg. [DOI] [PubMed] [Google Scholar]
- 60.Jarde T, Evans RJ, McQuillan KL, et al. In vivo and in vitro models for the therapeutic targeting of Wnt signaling using a Tet-ODeltaN89beta-catenin system. Oncogene. 2013;32(7):883–93. doi: 10.1038/onc.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gurney A, Axelrod F, Bond CJ, et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci U S A. 2012;109(29):11717–22. doi: 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le PN, McDermott JD, Jimeno A. Targeting the Wnt pathway in human cancers: therapeutic targeting with a focus on OMP-54F28. Pharmacol Ther. 2015;146:1–11. doi: 10.1016/j.pharmthera.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang X, Hao HX, Growney JD, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110(31):12649–54. doi: 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J, Pan S, Hsieh MH, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci U S A. 2013;110(50):20224–9. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koo BK, van Es JH, van den Born M, et al. Porcupine inhibitor suppresses paracrine Wnt-driven growth of Rnf43;Znrf3-mutant neoplasia. Proc Natl Acad Sci U S A. 2015;112(24):7548–50. doi: 10.1073/pnas.1508113112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Lau W, Barker N, Low TY, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476(7360):293–7. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 67.Wu X, Luo F, Li J, et al. Tankyrase 1 inhibitior XAV939 increases chemosensitivity in colon cancer cell lines via inhibition of the Wnt signaling pathway. Int J Oncol. 2016;48(4):1333–40. doi: 10.3892/ijo.2016.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schoumacher M, Hurov KE, Lehar J, et al. Inhibiting Tankyrases sensitizes KRAS-mutant cancer cells to MEK inhibitors via FGFR2 feedback signaling. Cancer Res. 2014;74(12):3294–305. doi: 10.1158/0008-5472.CAN-14-0138-T. [DOI] [PubMed] [Google Scholar]
- 69.Arques O, Chicote I, Puig I, et al. Tankyrase Inhibition Blocks Wnt/beta-Catenin Pathway and Reverts Resistance to PI3K and AKT Inhibitors in the Treatment of Colorectal Cancer. Clin Cancer Res. 2016;22(3):644–56. doi: 10.1158/1078-0432.CCR-14-3081. [DOI] [PubMed] [Google Scholar]
- 70.Riffell JL, Lord CJ, Ashworth A. Tankyrase-targeted therapeutics: expanding opportunities in the PARP family. Nat Rev Drug Discov. 2012;11(12):923–36. doi: 10.1038/nrd3868. [DOI] [PubMed] [Google Scholar]
- 71.Emami KH, Nguyen C, Ma H, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected] Proceedings of the National Academy of Sciences of the United States of America. 2004;101(34):12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gonsalves FC, Klein K, Carson BB, et al. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc Natl Acad Sci U S A. 2011;108(15):5954–63. doi: 10.1073/pnas.1017496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen ZL, Shao WJ, Xu F, et al. Acute Wnt pathway activation positively regulates leptin gene expression in mature adipocytes. Cell Signal. 2015;27(3):587–97. doi: 10.1016/j.cellsig.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 74.Dandekar S, Romanos-Sirakis E, Pais F, et al. Wnt inhibition leads to improved chemosensitivity in paediatric acute lymphoblastic leukaemia. Br J Haematol. 2014;167(1):87–99. doi: 10.1111/bjh.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mathur R, Sehgal L, Braun FK, et al. Targeting Wnt pathway in mantle cell lymphoma-initiating cells. J Hematol Oncol. 2015;8:63. doi: 10.1186/s13045-015-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372(26):2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feig C, Jones JO, Kraman M, et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110(50):20212–7. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–5. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 80.Oderup C, LaJevic M, Butcher EC. Canonical and noncanonical Wnt proteins program dendritic cell responses for tolerance. J Immunol. 2013;190(12):6126–34. doi: 10.4049/jimmunol.1203002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suryawanshi A, Manoharan I, Hong Y, et al. Canonical wnt signaling in dendritic cells regulates Th1/Th17 responses and suppresses autoimmune neuroinflammation. J Immunol. 2015;194(7):3295–304. doi: 10.4049/jimmunol.1402691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Valencia J, Hernandez-Lopez C, Martinez VG, et al. Wnt5a skews dendritic cell differentiation to an unconventional phenotype with tolerogenic features. J Immunol. 2011;187(8):4129–39. doi: 10.4049/jimmunol.1101243. [DOI] [PubMed] [Google Scholar]
- 83.Swafford D, Manicassamy S. Wnt signaling in dendritic cells: its role in regulation of immunity and tolerance. Discov Med. 2015;19(105):303–10. [PMC free article] [PubMed] [Google Scholar]
- 84.Gattinoni L, Zhong XS, Palmer DC, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat Med. 2009;15(7):808–13. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.DeAlmeida VI, Miao L, Ernst JA, et al. The soluble wnt receptor Frizzled8CRD-hFc inhibits the growth of teratocarcinomas in vivo. Cancer Res. 2007;67(11):5371–9. doi: 10.1158/0008-5472.CAN-07-0266. [DOI] [PubMed] [Google Scholar]
- 86.Jackson H, Granger D, Jones G, et al. Novel Bispecific Domain Antibody to LRP6 Inhibits Wnt and R-spondin Ligand-Induced Wnt Signaling and Tumor Growth. Mol Cancer Res. 2016;14(9):859–68. doi: 10.1158/1541-7786.MCR-16-0088. [DOI] [PubMed] [Google Scholar]
- 87.Huang SMA, Mishina YM, Liu S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461(7264):614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 88.Waaler J, Machon O, von Kries JP, et al. Novel synthetic antagonists of canonical Wnt signaling inhibit colorectal cancer cell growth. Cancer research. 2011;71(1):197–205. doi: 10.1158/0008-5472.CAN-10-1282. [DOI] [PubMed] [Google Scholar]
- 89.Lau T, Chan E, Callow M, et al. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res. 2013;73(10):3132–44. doi: 10.1158/0008-5472.CAN-12-4562. [DOI] [PubMed] [Google Scholar]
- 90.Zhong Y, Katavolos P, Nguyen T, et al. Tankyrase Inhibition Causes Reversible Intestinal Toxicity in Mice with a Therapeutic Index < 1. Toxicol Pathol. 2016;44(2):267–78. doi: 10.1177/0192623315621192. [DOI] [PubMed] [Google Scholar]
- 91.Chen Z, Venkatesan AM, Dehnhardt CM, et al. 2,4-Diamino-quinazolines as inhibitors of beta-catenin/Tcf-4 pathway: Potential treatment for colorectal cancer. Bioorg Med Chem Lett. 2009;19(17):4980–3. doi: 10.1016/j.bmcl.2009.07.070. [DOI] [PubMed] [Google Scholar]
- 92.Trosset JY, Dalvit C, Knapp S, et al. Inhibition of protein-protein interactions: the discovery of druglike beta-catenin inhibitors by combining virtual and biophysical screening. Proteins. 2006;64(1):60–7. doi: 10.1002/prot.20955. [DOI] [PubMed] [Google Scholar]