Structured Abstract
Background
Despite extensive efforts to monitor and prevent surgical site infections (SSIs), real-time surveillance of clinical practice has been sparse and expensive or non-existent. However, natural language processing (NLP) and machine learning (i.e., Bayesian network analysis) may provide the methodology necessary to approach this issue in a new way. We investigated the ability to identify SSIs following colorectal surgery (CRS) through an automated detection system using a Bayesian network.
Materials and Methods
Patients who underwent CRS from 2010 to 2012 and were captured in our institutional American College of Surgeons National Surgical Quality Improvement Program (ACS-NSQIP) comprised our cohort. A Bayesian network was applied to detect SSIs using risk factors from ACS-NSQIP data and keywords extracted from clinical notes by NLP. Two surgeons provided expertise informing the Bayesian network to identify clinically meaningful SSIs (CM-SSIs) occurring within 30 days after surgery.
Results
We utilized data from 751 CRS cases experiencing 67 (8.9%) SSIs and 78 (10.4%) CM-SSIs. Our Bayesian network detected ACS-NSQIP-captured SSIs with an ROC area under the curve of 0.827, but this value increased to 0.892 when using surgeon-identified CM-SSIs.
Conclusions
A Bayesian network coupled with NLP has the potential to be used in real-time SSI surveillance. Moreover, surgeons identified CM-SSI not captured under current NSQIP definitions. Future efforts to expand CM-SSI identification may lead to improved and potentially automated approaches to survey for post-operative SSI in clinical practice.
Keywords: surgical site infection, colorectal surgery, natural language processing, Bayesian network
Introduction
Surgical site infection (SSI) is the most common healthcare-associated infection, accounting for 31% of all hospital inpatient infections (1) and resulting in increased length of hospital stay and resource utilization. Colorectal surgery (CRS) is a broad specialty with a large distribution of indications for surgery and patient risk factors (2) and defining CRS SSI through medical records is not an easy task (3). SSI following CRS can be as high as 20% and has been deemed an important quality metric (4).
Current measurement and reporting of CRS SSI either depends on imperfect administrative data (Agency for Healthcare Research and Quality Patient Safety Indicator, AHRQ PSI, 13 covers only postoperative sepsis), expensive clinical surveillance (National Healthcare Safety Network, NHSN, SSI) or retrospective chart review of a sample of surgical cases (American College of Surgeons National Surgical Quality Improvement Program, ACS-NSQIP). Over the past decade, accurate identification and monitoring of surgical complications have been emphasized to improve health care quality and to decrease financial costs. The ACS-NSQIP is one such effort. Previous studies have retrospectively examined patient characteristics identified in NSQIP data to assess SSI risk factors (2, 5, 6). These patient characteristics can be regarded as potential factors in predicting SSI, but are not conclusive. Also, lagged abstraction of SSIs in ACS-NSQIP and further delay from capture to release of the national data makes it impractical to use this information for real-time SSI surveillance in clinical practice. Although NHSN methods have been developed for clinical monitoring, hospital infection control efforts are employee intensive and expensive. In addition, the effort and time required to review the medical record and evaluate potential SSI is a real economic burden for many healthcare centers.
Natural language processing (NLP) and machine learning techniques could automate the chart review and be applied to all patients, enabling faster identification of emerging changes in infections or the risks thereof. NLP is a technique that enables computers to derive meaning from human language (i.e., natural language). It can convert unstructured free text in electronic medical records to a structured form, providing important detailed information that is unavailable in structured data. A Bayesian network is a graphical model-based machine learning technique that encodes probabilistic relationships among variables. It can learn causal relationships to understand a problem domain and provide an ideal representation for combining prior knowledge (7, 8). These approaches represent promising alternatives to gain efficiencies in identifying SSI.
In this study, we aim to demonstrate that the application of a Bayesian network coupled with NLP can be used to detect NSQIP SSIs. Moreover, we demonstrate that a properly trained Bayesian network is able to identify clinically meaningful SSIs (CM-SSIs) currently not captured under NSQIP definitions. Collectively this automated system may lead to improved and potentially real-time SSI surveillance for post-operative CM-SSI in clinical practice.
Materials and Methods
This study was approved by the Mayo Clinic institutional review board. We performed a retrospective review of all adult patients who underwent colorectal surgery at Mayo Clinic Hospital, Methodist Campus, from 2010 to 2012 who were sampled for ACS-NSQIP institutional data.
A Bayesian network was applied to detect thirty-day SSIs identified by ACS-NSQIP. Discrepancies between the results of the Bayesian network and ACS-NSQIP were independently reviewed by two attending surgeons through retrospective chart review, where only patient identification and operation date were given to them (i.e., the SSI case status of the Bayesian network and ACS-NSQIP were not available to them). Upon review of cases, these surgeons further identified CM-SSIs. Agreement in determining CM-SSIs between two surgeons was 82% with a Cohen’s kappa (9) of 0.44, which indicates moderate agreement. The Bayesian network was retrained using CM-SSI to examine the capability to detect clinically important SSIs (i.e., CM-SSIs).
Study Outcome
The main outcome of this study was to assess the performance of Bayesian network in abstracting ACS-NSQIP SSI and further evaluate the potential to identify CM-SSIs from electronic medical records. The surgeons defined CM-SSI as any SSI that required intervention, which is a broader than ACS-NSQIP criteria.
The key points of NSQIP definitions of SSI that lead to excluding some CM-SSIs are as follows:
Cellulitis alone does not count as an SSI.
Anastomotic leaks were first implemented as an organ/space SSI in January 2013
Before 2012, a subsequent procedure could be captured as an index procedure if the first procedure was not captured. Since January 2012, NSQIP has excluded a return to the operating room as the principal operative procedure, if it is related to an occurrence or complication from another procedure within 30 days.
Dirty/infected wounds at the index procedure are not assigned as postop SSIs, despite the potential that clinically relevant SSI can and do develop in these patients. The POA (present on admission) was introduced in June 2010 as an optional field capture and has been implemented since January 2011 as PATOS (present at time of surgery).
In NSQIP, a SSI must meet every criterion of the definition. If not, it cannot be assigned. Lack of documentation in particular can be problematic. A phone call and putting the patient on antibiotics is not enough without full symptom documentation or supporting evidence that this is in fact an infection and not cellulitis. If there is any doubt, it is not assigned.
Detection of SSIs using Bayesian Network
A Bayesian network was developed to detect SSIs using the following strategies: 1) pre- and intra-operation ACS-NSQIP-based risk factors; 2) pre-, intra-, and post-operation ACS-NSQIP-based risk factors; 3) keywords extracted from clinical notes by NLP; 4) both 2) and 3). The keywords represent current medical comorbidities and conditions related to SSI represented by presence or absence during 30-day postoperative period. We used an open-source NLP pipeline, MedTagger, which was developed by Mayo Clinic (10, 11), to extract keywords from clinical notes. The keywords have been compiled by subject matter experts (e.g., surgeons) on 1,856 colorectal surgical cases at Mayo Clinic Hospital, Methodist Campus from 2005 to 2013 in our previous study (12) and expanded through personal communication with NSQIP annotators. Detailed descriptions of the variables are found in Table 1.
Table 1.
SSI risk factors and indication concepts for colorectal surgery.
| Potential risk factors | SSI indications in clinical notes | ||
|---|---|---|---|
| Pre-operation | Intra-operation | Post-operation | Post-operation Medical Concept‡ |
| age gender current smoker pre-op hospital stay diabetes anemia† BMI (>=30) prior operation§ steroid usage blood transfusion |
ASA score* operation type wound classification duration of operation MIS** vs. others |
anemia† ascites wound disruption abdominal tenderness† antibiotics† |
wound infection cellulitis abdominal contamination purulent drainage fascial/wound dehiscence abdominal infection purposeful reopening of the incision pain/swelling/redness at the incision abscess infected fluid antibiotic treatment for infection |
extracted from clinical notes using NLP;
each concept consists of a set of the corresponding keywords; the other variables were from ACS-NSQIP
surgical procedure within 30 days prior to the assessed operation
American Society of Anesthesiologists (ASA) physical status classification
Minimally Invasive Surgery
Clinical Review of Discrepant Cases
Medical records of discrepant cases between the Bayesian network outcomes and NSQIP-SSI were independently reviewed by two surgeons (JYA, DWL) to determine whether the patient had CM-SSI. When the surgeons disagreed, the final determination of whether or not a CM-SSI existed was based on consolidation through discussion of surgeons. Once CM-SSIs were identified, the Bayesian network was recalculated to identify CM-SSI.
Statistical Methods
The performance of SSI detection using a Bayesian network is reported in receiver operating characteristic (ROC) area under the curve (AUC). The evaluation was performed in a 10-fold cross-validation framework to produce reliable test results—i.e., divide the data randomly into 10 sets, train on 9 datasets and test on the remaining one dataset, repeat 10 times, and combine the results to compute AUC. A logistic regression model using a ridge estimator (13) to predict SSI was also developed for comparison.
Results
A total of 751 colorectal surgery cases developing 67 (8.9%) NSQIP-identified SSIs (i.e., 36 superficial incisional SSIs, 5 deep incisional SSIs, and 26 organ/space SSIs) at a tertiary hospital were evaluated with the Bayesian network classifier using various variables to detect SSIs.
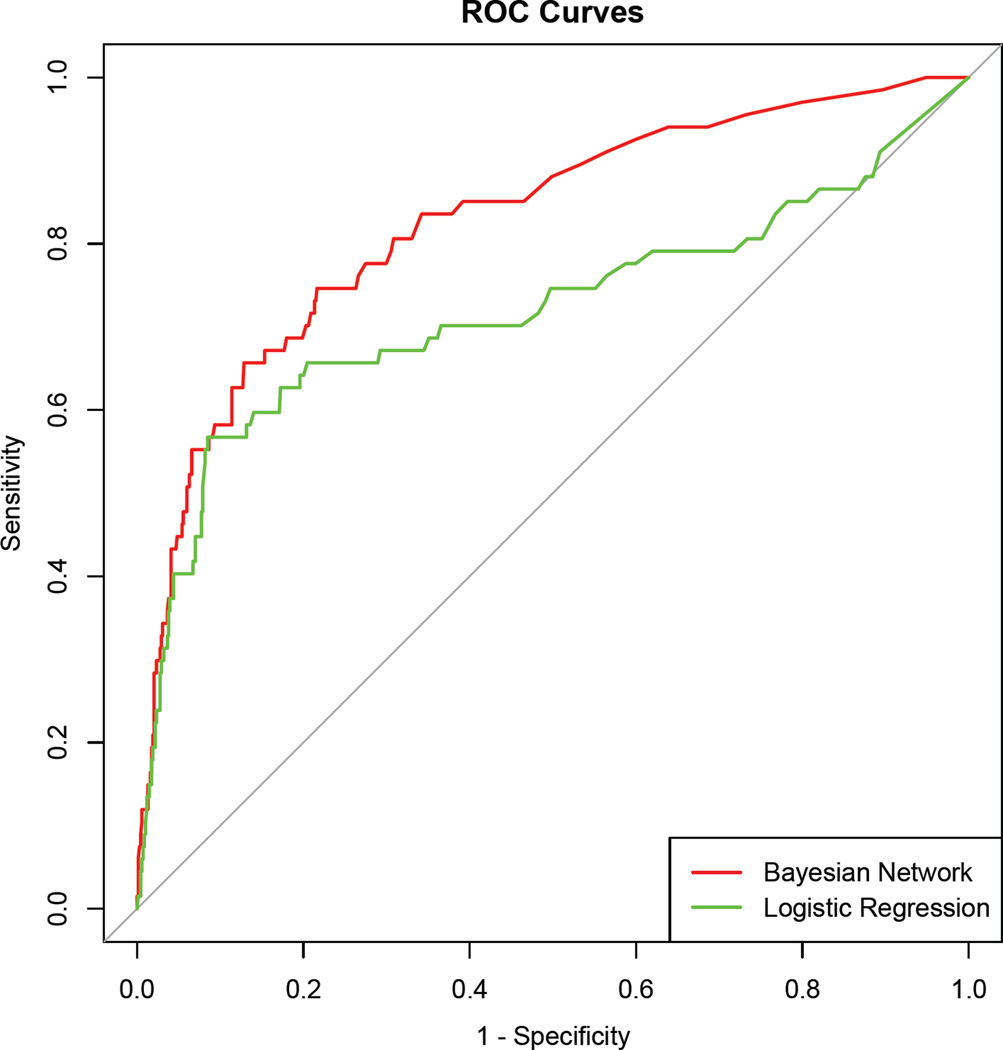
Table 2 shows AUC of the Bayesian network classifier using different sets of variables with 10-fold cross validation. The best performance (AUC=0.827) was achieved when using both risk factors and keywords from clinical narratives extracted by NLP. For comparison, a logistic regression was also employed using the same variables as the best performed Bayesian network (Appendix includes the coefficients and odd ratios in the logistic regression). The corresponding ROC curves are shown in Figure 1. The Bayesian network produced higher performance than the logistic regression (AUC=0.719, p=0.002) that is commonly used in a clinical domain.
Table 2.
Bayesian network AUC for ACS-NSQIP SSI identification.
| Feature used | AUC | 95% | p-value† |
|---|---|---|---|
| a. pre/intra-op risk factors | 0.643 | 0.5767 – 0.7075 | NA |
| b. risk factors | 0.721 | 0.6526 – 0.7880 | 0.002 (a vs. b) |
| c. keywords | 0.799 | 0.7349 – 0.8638 | 0.071 (b vs. c) |
| d. risk factors & keywords | 0.827 | 0.7695 – 0.8823 | 0.305 (c vs. d) |
p-value of the difference between two AUCs
Figure 1.
ROC AUC for detection of ACS-NSQIP SSI (using risk factors & keywords)
top curve: Bayesian network (AUC=0.827)
bottom curve: logistic regression (AUC=0.719)
Surgeon record review found that 44% of Bayes network false positives (i.e., SSIs according to the system but non-SSIs in NSQIP) are CM-SSIs in clinical practice even though they are not captured under ACS-NSQIP’s definitions. Table 3 includes a summary of the surgeons’ justifications for the eleven non-ACS-NSQIP CM-SSIs. One case was due to NSQIP abstraction error. The rest did not fit into NSQIP SSI criteria. As a measure of the importance of focusing on CM-SSI, the median hospital stay for CM-SSI cases was 7 (IQR = 9.25) days while the median stay for ACS-NSQIP SSIs was 5 (IQR = 7) days.
Table 3.
Cases of clinically meaningful SSI that are not NSQIP SSI
| Clinically meaningful SSI (by Surgeons) | ACS-NSQIP | |||
|---|---|---|---|---|
| Level of intervention |
Problem | SSI type | Reason not SSI | No. patients |
| Opened wound; Abx treatment |
Wound infection | Superficial | NSQIP Abstraction Error | 1 |
| Emergency operation |
Anastomotic leak | Deep space | NSQIP definition limitation | 5 |
| Opened wound; Abx treatment |
Wound infection | Superficial | NSQIP definition limitation | 1 |
| Opened wound; Abx treatment; Reoperation |
Wound infection; Re-operation for Small bowel fistula |
Organ/Space | NSQIP definition limitation | 1 |
| Opened wound; Abx treatment; Reoperation |
UTI; Wound infection; Aspiration pneumonitis |
Superficial | NSQIP definition limitation | 1 |
| Emergency operation |
Anastomotic leak | Organ/Space | NSQIP definition limitation | 1 |
| Antibiotic treatment for wound cellulitis |
Cellulitis | Superficial | NSQIP definition limitation | 1 |
In order to evaluate the capability of detecting CM-SSI, the data set was relabeled with CM-SSIs identified by surgeons (78 CM-SSIs: 44 superficial incisional SSIs, 6 deep incisional SSIs, and 28 organ/space SSIs). The Bayesian network retrained with CM-SSIs produced AUC of 0.892, which is higher than the one trained with ACS-NSQIP SSIs (AUC=0.827). This higher AUC on CM-SSI might be partially due to the consistent CM-SSI definition as opposed to changes in NSQIP SSI definitions over time.
Discussion
Identification of CM-SSI provides an important opportunity to improve surgical quality of care. Moreover, rapid response to developing infections can efficiently minimize their adverse effects to patients. Real time stratification of patients who require additional healthcare resources enables, not just quality improvement efforts, but enhances the care of current patients. Our Bayesian network analysis showed promising performance for colorectal SSI surveillance in clinical practice. In the Bayesian network, ACS-NSQIP-based risk factors were not sufficient to accurately detect SSIs. The Bayesian network produced the strongest performance when using both risk factors from ACS-NSQIP data and keywords extracted from clinical narratives by NLP.
Further refinement of keywords, not only from clinical notes but also from other types of electronic medical records (e.g., operation notes, radiology reports, microbiology), and ongoing assessment of risk factors are necessary to better detect CM-SSIs. Automatic extraction of risk factors to implement real-time SSI surveillance should be the next step. Informatics techniques such as NLP, which allows for the automatic extraction of patients’ medical conditions and statistical machine learning techniques such as a Bayesian network to detect the corresponding outcomes, are essential components in the realization of efficient SSI surveillance. The variables used in our Bayesian network (Table 1) are common concepts that can be extracted from any electronic medical record system, which allows the system to be transportable and generalizable to other institutions.
Several previous studies have explored automated detection of SSI. Hu et al. built a logistic regression model using claim data and electronic medical records to identify SSIs and produced high specificity (from 0.788 to 0.988) (14). Soguero-Ruiz et al. used a machine learning model to predict deep incisional SSIs. They used only blood test data and utilized a Gaussian process regression to deal with missing values and produced prediction accuracy of 0.69 to 0.91 depending on the blood test categories (15). Also, there has been a study to automatically identify keywords related to post-surgical complications through sublanguage analysis instead of manual process by subject matter experts (12). In other studies, recent advances in NLP have produced promising results in text analytics from clinical text (16) and have been successfully applied in various clinical applications including medication information extraction (17), patient medical status identification (18–20), sentiment analysis (21), decision support (22, 23), genomewide association studies (24, 25), and diagnosis code assignment (26, 27).
In this study, we have discovered remaining potential to improve our understanding and detection of CM-SSIs and the resulting clinical impact on patients. From our surgeons’ perspectives, a CM-SSI is any infectious complication that occurred as a direct result of surgery and required any intervention above that required for a patient without an SSI. ACS-NSQIP definitions for SSI, however, have been modified multiple times and are very strict, as clear consensus across institutions is required. NSQIP SSIs do not capture all CM-SSI, which results in prolonged hospital stays and consumption of healthcare resources. In order to improve surgical outcomes and better monitor patients after surgery, it will be necessary to identify SSIs that are clinically useful.
The critical ability of surgeons to assimilate information and make diagnosis is in fact largely based on the lack of or limited documentation of information. This leads one to the conclusion that novel approaches to the semantics and pragmatics of language are needed to adequately reflect what an expert surgeon would conclude as a CM-SSI. In addition the resources used to collect and validate this critical information must be replaced over time by tools which can automate this process or enhance abstractors’ ability to identify and confirm additional patient information over their current work flow limitations.
Our study has several limitations. This is a retrospective study in one institution based on clinical documents, not real-time assessment and therefore the system performance depends on the quality of documentation. The system performance has not been validated yet in other institutions and through actual surveillance in clinical setting. SSIs annotated in ACS-NSQIP might not be fully generalizable to current definitions. For example, anastomotic leaks, important SSIs from a clinical perspective, were first implemented as an organ/space SSI in January 2013, after our study period. Additionally, we have not fully reviewed true negatives (i.e., non-SSIs in both the system and NSQIP, N=663) and may have missed potential CM-SSIs not captured by ACS-NSQIP definitions or our machine learning process. Alternatively, we reviewed one hundred random true negatives and observed five CM-SSI cases (5%), which shows a reasonable estimation of missing CM-SSIs.
In conclusion, our Bayesian network coupled with NLP produced a well-performing model to detect CM-SSIs. Although it did not completely solve the case ascertainment of SSI, our approach may be able to reduce the burden of chart review at the point of care with the good potential to support real-time SSI surveillance and management.
Supplementary Material
Acknowledgments
This study was made possible by NIBIB R01 EB19403.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: S.S., H.L., and D.L. conceived and designed the study. All authors analyzed and interpreted the data. S.S. and H.L. drafted the manuscript. All authors provided critical revisions of the manuscript.
Disclosure: The authors have no conflicts of interest to declare.
References
- 1.Magill SS, Hellinger W, Cohen J, Kay R, Bailey C, et al. Prevalence of healthcare-associated infections in acute care hospitals in Jacksonville, Florida. Infection Control. 2012;33:283–291. doi: 10.1086/664048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirchhoff P, Clavien P-A, Hahnloser D. Complications in colorectal surgery: risk factors and preventive strategies. Patient Saf Surg. 2010;4:5. doi: 10.1186/1754-9493-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedrick TL, Sawyer RG, Hennessy SA, Turrentine FE, Friel CM. Can we define surgical site infection accurately in colorectal surgery? Surgical infections. 2014;15:372–376. doi: 10.1089/sur.2013.013. [DOI] [PubMed] [Google Scholar]
- 4.Hübner M, Diana M, Zanetti G, Eisenring M-C, Demartines N, et al. Surgical site infections in colon surgery: the patient, the procedure, the hospital, and the surgeon. Archives of Surgery. 2011;146:1240–1245. doi: 10.1001/archsurg.2011.176. [DOI] [PubMed] [Google Scholar]
- 5.Smith RL, Bohl JK, McElearney ST, Friel CM, Barclay MM, et al. Wound infection after elective colorectal resection. Annals of surgery. 2004;239:599. doi: 10.1097/01.sla.0000124292.21605.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malone DL, Genuit T, Tracy JK, Gannon C, Napolitano LM. Surgical site infections: reanalysis of risk factors. Journal of Surgical Research. 2002;103:89–95. doi: 10.1006/jsre.2001.6343. [DOI] [PubMed] [Google Scholar]
- 7.Heckerman D. A tutorial on learning with Bayesian networks. Springer; 1998. [Google Scholar]
- 8.Nielsen TD, Jensen FV. Bayesian networks and decision graphs. Springer Science & Business Media; 2009. [Google Scholar]
- 9.Cohen J. A coefficient of agreement for nominal scales. Educational and psychological measurement. 1960;20:37–46. [Google Scholar]
- 10.Torii M, Wagholikar K, Liu H. Using machine learning for concept extraction on clinical documents from multiple data sources. Journal of the American Medical Informatics Association. 2011;18:580–587. doi: 10.1136/amiajnl-2011-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H, Bielinski S, Sohn S, Murphy S, Wagholikar K, et al. AMIA Summits Transl Sci Proc. San Francisco, CA: 2013. An information extraction framework for cohort identification using electronic health records; pp. 149–153. [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Sohn S, Murphy S, Lovely J, Burton M, et al. Facilitating post-surgical complication detection through sublanguage analysis. AMIA Summits on Translational Science Proceedings. 2014;2014:77. [PMC free article] [PubMed] [Google Scholar]
- 13.Le Cessie S, Van Houwelingen JC. Ridge estimators in logistic regression. Applied statistics. 1992:191–201. [Google Scholar]
- 14.Hu Z, Simon GJ, Arsoniadis EG, Wang Y, Kwaan MR, et al. Automated Detection of Postoperative Surgical Site Infections Using Supervised Methods with Electronic Health Record Data. MEDINFO 2015: eHealth-enabled Health. 2015:706–710. [PMC free article] [PubMed] [Google Scholar]
- 15.Soguero-Ruiz C, Wang F, Jenssen R, Augestad KM, Alvarez J-LR, et al. AMIA Annual Symposium. San Francisco, CA: 2015. Data-driven Temporal Prediction of Surgical Site Infection. [PMC free article] [PubMed] [Google Scholar]
- 16.Savova G, Masanz J, Ogren P, Zheng J, Sohn S, et al. Mayo Clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohn S, Clark C, Halgrim S, Murphy S, Chute C, et al. MedXN: an Open Source Medication Extraction and Normalization Tool for Clinical Text. Journal of the American Medical Informatics Association. 2014;21:858–865. doi: 10.1136/amiajnl-2013-002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohn S, Kocher J-PA, Chute CG, Savova GK. Drug side effect extraction from clinical narratives of psychiatry and psychology patients. J Am Med Inform Assoc. 2011;18:144–149. doi: 10.1136/amiajnl-2011-000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sohn S, Savova GK. AMIA Annual Symposium. San Francisco, CA: 2009. Mayo Clinic Smoking Status Classification System: Extensions and Improvements; pp. 619–623. [PMC free article] [PubMed] [Google Scholar]
- 20.Sohn S, Ye Z, Liu H, Chute C, Kullo I. AMIA Summits Transl Sci Proc. San Francisco: 2013. Identifying Abdominal Aortic Aneurysm Cases and Controls using Natural Language Processing of Radiology Reports. [PMC free article] [PubMed] [Google Scholar]
- 21.Sohn S, Torii M, Li D, Wagholikar K, Wu S, et al. A Hybrid Approach to Sentiment Sentence Classification in Suicide Notes. Biomedical informatics insights. 2012:43–50. doi: 10.4137/BII.S8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demner-Fushman D, Chapman W, McDonald C. What can natural language processing do for clinical decision support? Journal of Biomedical Informatics. 2009;42:760–772. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aronsky D, Fiszman M, Chapman WW, Haug PJ. Combining decision support methodologies to diagnose pneumonia. American Medical Informatics Association. 2001:12–16. [PMC free article] [PubMed] [Google Scholar]
- 24.Kullo IJ, Fan J, Pathak J, Savova GK, Ali Z, et al. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. Journal of the American Medical Informatics Association. 2010;17:568–574. doi: 10.1136/jamia.2010.004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kullo IJ, Ding K, Jouni H, Smith CY, Chute CG. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS One. 2010;5:e13011. doi: 10.1371/journal.pone.0013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. Journal of the American Medical Informatics Association. 2004;11:392. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pakhomov SVS, Buntrock JD, Chute CG. Automating the assignment of diagnosis codes to patient encounters using example-based and machine learning techniques. Journal of the American Medical Informatics Association. 2006;13:516–525. doi: 10.1197/jamia.M2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.