Abstract
A 2.4-Å-resolution x-ray crystal structure of the carrier-protein independent halogenase, WelO5, in complex with its welwitindolinone precursor substrate, 12-epi-fischerindole U, reveals that the C13 chlorination target is proximal to the anticipated site of the oxo group in a presumptive cis-halo-oxo-iron(IV) (haloferryl) intermediate. Prior study of related halogenases forecasts substrate hydroxylation in this active-site configuration, but x-ray crystallographic verification of C13 halogenation in single crystals mandates that ligand dynamics must reposition the oxygen ligand to enable the observed outcome. Ser189Ala WelO5 effects a mixture of halogenation and hydroxylation products, showing that an outer sphere hydrogen bonding group orchestrates ligand movements to achieve a configuration that promotes halogen transfer.
INTRODUCTION
Oxygen-dependent Fe/2OG halogenases target unactivated sp3-hybridized carbon centers for installation of chlorine or bromine1,2–9, transformations that are of broad synthetic utility10. The initial steps of the halogenase reaction – combination of the enzyme’s iron(II) cofactor with 2-oxo-glutarate (2OG) and O2 to produce CO2, succinate and an iron(IV)-oxo (ferryl) intermediate that abstracts a hydrogen atom (H•) from the substrate5,11 – are conserved with those of the more extensively studied Fe/2OG hydroxylases12–15. The two reactions diverge in the fate of the resulting substrate (carbon) radical (C•), which can couple either with the hydroxo ligand of the Fe(III)–OH intermediate produced by the H•-abstraction step or, in the halogenases, with a cis-coordinated chloride/bromide ligand. The key structural feature that enables this alternative radical coupling is an abbreviated protein ligand set comprising a pair of histidine residues16,17. Whereas the hydroxylases have, additionally, a carboxylate ligand from an Asp or Glu residue to complete the 2-His-1-carboxylate facial-triad18, the halogenases have a glycine or alanine residue at the corresponding sequence position16. The effective carboxylate deletion vacates an iron coordination site that accommodates the halide ligand, which is ultimately transferred as a radical to the substrate C• in the final step of the reaction. Elucidation of this underlying mechanistic strategy raised the question of how these enzymes direct coupling of the substrate radical with the halogen while avoiding the transfer of the HO• that prevails in the related hydroxylases.
SyrB2 from Pseudomonas syringae, the founding member of the Fe/2OG halogenase subclass3, chlorinates L-threonine covalently tethered as a thioester to the phosphopantetheine arm of its carrier protein, SyrB1, and has served as the primary subject for mechanistic analysis11,19–21. Comparison of SyrB2 reactions with the native L-threoninyl-SyrB1 substrate to analogs that present different amino acids implies that positioning of the substrate far from the oxygen ligand and closer to the halogen is the primary structural feature ensuring selective halogenation19. These studies reveal a strong inverse correlation between the halogenation selectivity and the rate of H• abstraction, with ferryl decay ~ 130 times slower with the primarily chlorinated native substrate than with an analog designed to project the target carbon closer to the FeIV=O intermediate that is primarily hydroxylated11,19. Pulse electron paramagnetic resonance (EPR) experiments verified the expected trend in substrate positioning relative to an EPR active iron-nitrosyl cofactor designed to mimic reaction intermediates21. Although this study provided mechanistically relevant structural data, and several computational studies have afforded global but speculative structural models for the SyrB2 complex with its substrates20,22–24, no high-resolution experimental structure of the O2-reactive complex of any Fe/2OG halogenase has yet been reported.
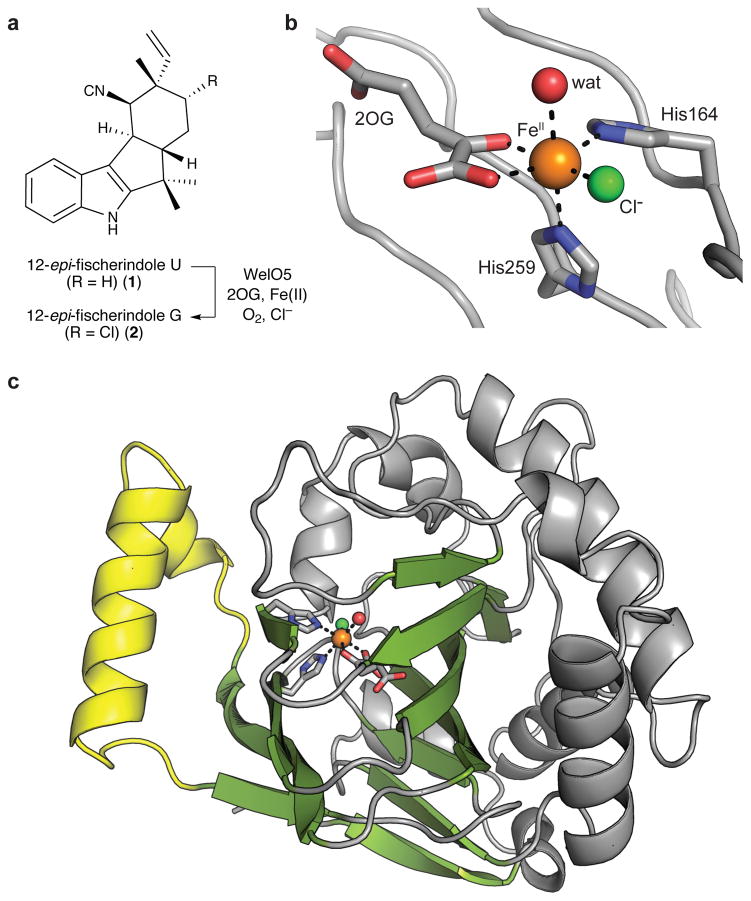
The recently discovered halogenase, WelO5, chlorinates C13 of 12-epi-fischerindole U (1) to yield 12-epi-fischerindole G (2) (Fig. 1a)25,26. This enzymatic transformation constitutes the first step in the oxidative maturation of 1 to welwitindolinones in stigonematalean cyanobacterium Hapalosiphon welwischii UTEX B1830. As the first example of an Fe/2OG halogenase that acts on a free, small-molecule substrate, the enzyme obviates the barrier to structural characterization imposed by the hydrolytically labile aminoacyl-carrier protein substrates of other halogenases. Here we report a series of x-ray crystal structures of WelO5. In these structures, we found the C13 chlorination target of 1 oriented away from the halogen ligand and proximal to the predicted location of the oxo group in the FeIV=O intermediate. Because the substrate must reside far from the oxygen ligand in the reactive haloferryl state, ligand rearrangement must occur during the catalytic cycle, promoted by a distinctive cosubstrate conformation and second-sphere hydrogen-bonding group.
Figure 1. Structure of WelO5 in the absence of substrate.
(a) Schematic of the reaction catalyzed by WelO5. (b) Selected amino acid residues and cosubstrate molecules in the active site of WelO5 are shown in stick format. FeII, Cl, and water ligands illustrated as spheres. (c) Core Fe/2OG β-sandwich fold shown in green and external helix shown in yellow.
RESULTS
X-ray crystal structure of apo WelO5
WelO5 was crystallized anaerobically with FeII and its 2OG cosubstrate, initially in the absence of 1 (Fig. 1b). Although a monomer in solution (Supplementary Results, Supplementary Fig. 1), the x-ray structure of WelO5, solved at 2.0-Å resolution by single-wavelength anomalous dispersion methods (Supplementary Table 1), contains three WelO5 molecules in the asymmetric unit (ASU), all of which adopt the eight-stranded β-sandwich topology (Fig. 1c) characteristic of Fe/2OG enzymes27. Each monomer contains a single FeII ion at full occupancy in its active site, and all three sites have identical coordination geometry (Fig. 1b and Supplementary Fig. 2). WelO5 contains a glycine (Gly166) at the sequence position that would provide the Asp/Glu ligand in standard Fe/2OG hydroxylases. The structure confirmed that WelO5 does not supply a coordinating carboxylate from an alternative sequence position and the absence of the side chain allows for direct coordination of a chloride ion, consistent with use of the same underlying mechanistic strategy as the carrier-protein dependent Fe/2OG halogenases16.
X-ray crystal structure of substrate-bound WelO5
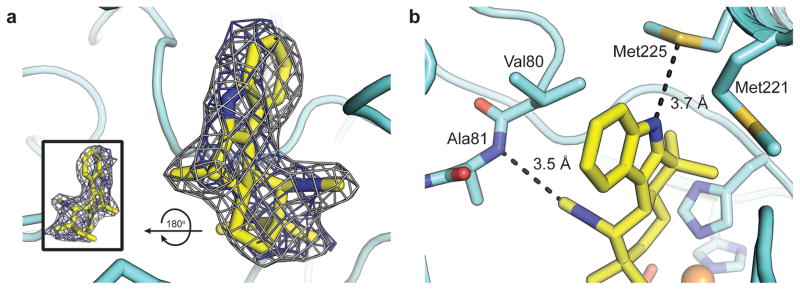
A soak of 1 into apo FeII/2OG-WelO5 crystals yielded a 2.4-Å-resolution structure of the enzyme-substrate complex (Supplementary Table 2). Clear electron density for the substrate is observed in one molecule of the ASU, with a lesser degree of occupancy in the other two monomers (Supplementary Fig. 3). The isocyanide and tertiary isoprenoid substituents are readily discerned (Fig. 2a), allowing for unequivocal orientation of the substrate in the WelO5 active site. The binding pocket is predominantly hydrophobic (Supplementary Fig. 4), in keeping with the large and hydrophobic nature of the substrate, but two regions of the enzyme undergo conformational change to contribute key hydrogen-bonding contacts to the indole nitrogen and isocyanide moieties of 1 (Fig. 2b). The WelO5 active site is embedded between the two β-sheets that constitute the core fold and flanked by connecting loop structures. A short loop (residues 75-82) in WelO5 shifts 3.4 Å to allow the backbone amide nitrogen of Ala82 to engage in an unusual carbon-mediated hydrogen bond with the isocyanide moiety of 1 (Fig. 2b). Another connecting segment near the active site (Fig. 1c) forms an α-helical structure that sits ajar from the rest of the protein in the absence of substrate. The open position of this external helix in the apo structure exposes the active site to solvent (Supplementary Fig. 5), distinct to WelO5 among structurally characterized Fe/2OG halogenases (Supplementary Fig. 6)16,17,28. Upon binding of 1, the external helix shifts 9 Å to close over the active site (Supplementary Fig. 7a), locked in place by hydrophobic interactions with the core fold and an extended hydrogen-bonding network that involves side chains and backbone atoms immediately downstream of the H-X-G sequence in the active site (Supplementary Fig. 7b). This observation, coupled with the fact that the segment of the protein diverges most in sequence from the equivalent region in known WelO5 homolog AmbO5 (Supplementary Fig. 8)29,30, suggests the external helix may dictate substrate preference. Closure of the motif is observed in the single WelO5 molecule in the ASU that displays full occupancy of 1. Lattice interactions with adjacent symmetry mates preclude movement in the other two monomers (Supplementary Fig. 3), providing a rationale for diminished substrate occupancy in those chains. The chlorination target, C13, of 1 is just 4.5 Å from the FeII center, closest to the cofactor among all its carbon atoms. The pro-R hydrogen of C13 is directed toward the FeII (Fig. 3a), consistent with the known stereoselectivity of the reaction25. Surprisingly, C13 is farther from the chlorine ligand, 5.7 Å away (Fig. 3a). Assuming that the oxo/hydroxo ligand of the FeIV=O/FeIII-OH unit would occupy the open coordination position axial to His259, the binding site is typical of Fe/2OG hydroxylases (Supplementary Figs. 9–11)
Figure 2. The substrate binding site in WelO5.
(a) A 2Fo-Fc composite electron density map (gray mesh, contoured at 1.0σ) and Fo-Fc omit map (blue mesh, contoured at 3.0σ) for 1 (yellow sticks) with rotated view in inset. (b) Zoomed-in view of hydrogen bonding interactions (black dashed lines) with 1. Selected amino acid side chains shown in stick format.
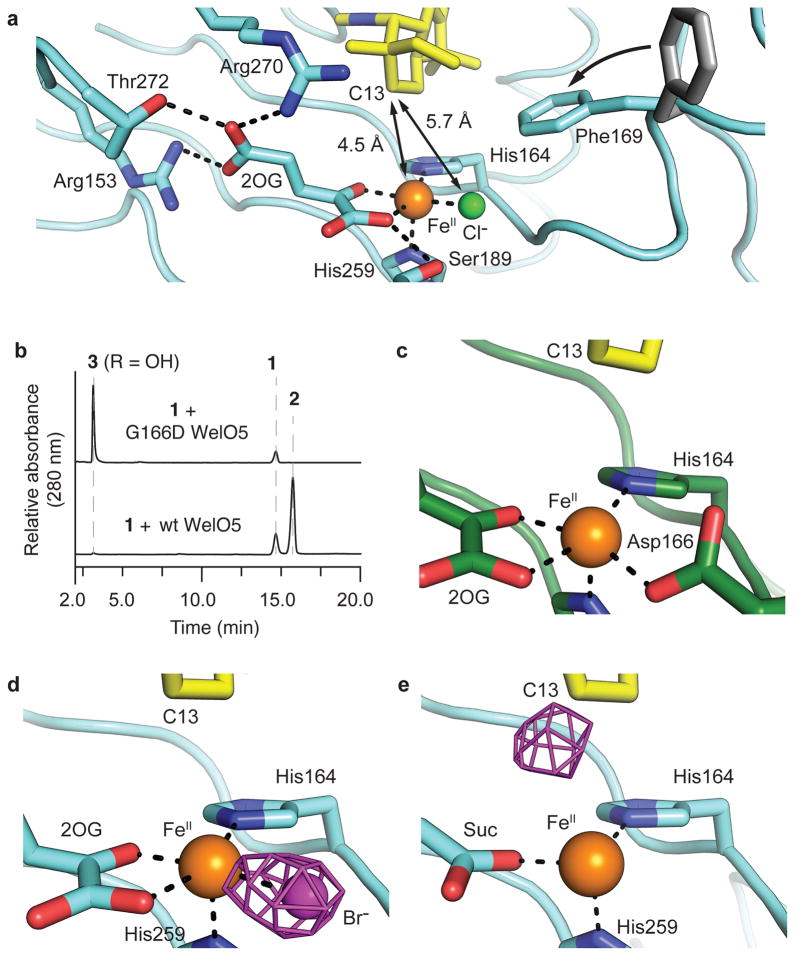
Figure 3. Functional characterization of the WelO5 substrate binding site.
(a) Positioning of substrate (yellow) proximal to the active site FeII (orange) and distal to the –Cl (green) ligands in 1-bound WelO5 (cyan) is facilitated by conformational change of Phe169, as compared to apo WelO5 (gray), to occlude a pocket near the –Cl binding site. An unusual conformation of 2OG is enabled by a WelO5-specific hydrogen bonding network. (b) G166D WelO5 exclusively hydroxylates 1 to give 3. (c) G166D WelO5 restores the His-X-Asp-Xn-His FeII binding motif. Assays were performed in triplicate and representative HPLC traces are shown. (d) Anomalous Fourier density (purple mesh, contoured at 4.0σ), illustrating -Br coordination to FeII prior to reaction initiation, is consistent with the assigned halogen binding site. (e) Migration of Br-associated density to the substrate C13 position after reaction with O2.
Crystal structure and activity of the G166D WelO5 variant
From the crystallographic data, the active site of wt WelO5, when poised for O2 activation, appeared primed to hydroxylate the substrate. In support of this observation, Gly166 → Asp substitution in WelO5 results in exclusive hydroxylation of 1 to give 3 (Fig. 3b and Supplementary Figs. 12–16). Interestingly, when we performed this analysis with wt WelO5, we observed trace amounts of 3 (< 2% of 2) (Supplementary Fig. 17), consistent with the reactivity patterns of carrier-protein dependent halogenases19. A 2.5-Å-resolution structure of G166D WelO5 in complex with 1 shows that Asp166 coordinates FeII and the substrate location observed in wt WelO5 is retained in the variant enzyme (Fig. 3c and Supplementary Fig. 18).
Bromination by wild-type WelO5 in single crystals
To confirm that the substrate binding site observed in the wt enzyme is also competent for halogenation, we repeated the soak of 1 into wt WelO5 co-crystallized with NaBr31. Collection of x-ray diffraction datasets at the Br K edge (13. 5 keV) (Supplementary Table 3) in anaerobic crystals and those exposed to O2 allowed for direct visualization of the reaction outcome via tracking of the location of the halide. Anomalous Fourier density consistent with an FeII-coordinating bromide in 1-bound WelO5 (Fig. 3d) and for a brominated analog of 2 in WelO5 after O2 exposure (Fig. 3e) validated that the substrate binding site allows for halogenation.
Mechanism of halogenation by WelO5
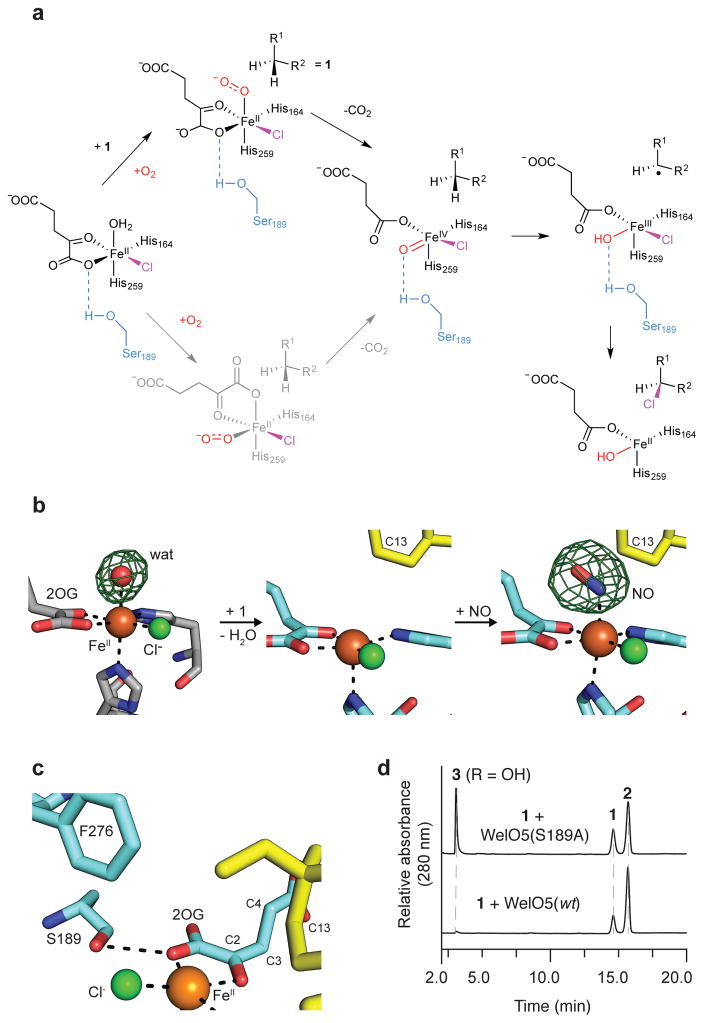
To accomplish selective halogenation, the structure of WelO5 mandates that some of the ligands in the haloferryl intermediate will occupy different positions relative to their location in the reactant complex (Fig. 4a). Migration of the oxo unit to the position currently occupied by the C1 carboxylate of 2OG (Fig. 4a, bottom route in gray) would be consistent with the known reactivity patterns of wt WelO5.25,26 One route to an alternative configuration of ligands would involve 2OG rearrangement promoted by O2 binding, as has been observed upon interaction with O2 surrogate NO and FeII in the x-ray structure of clavaminate synthase32, an Fe/2OG enzyme reported to catalyze oxacyclization and desaturation reactions. An NO-bound structure of WelO5 (Fig. 4b) showed that the stable O2 analog instead adds in the open coordination position in this system, trans to the distal His ligand, without accompanying rearrangement of 2OG.
Figure 4. Proposed mechanism for WelO5-mediated halogenation and its structural basis.
(a) Pathways for localization of the FeIV=O moiety into the plane defined by 2OG, -Cl, and the proximal His164 ligand in the reactant complex. Bottom pathway in gray illustrates a possible route to the same complex that is not operant in WelO5 but may apply more generally to other Fe/2OG enzymes, including carrier-protein dependent halogenases. (b) A structure of WelO5 bound to the stable oxygen mimic nitric oxide (NO) (far right, NO/1-bound WelO5), in which NO binds at the open coordination site (middle, 1-bound WelO5) occupied by a water molecule prior to substrate binding (left, apo WelO5). Fo-Fc omit maps (green mesh, contoured at 3.0σ) are shown for the apo and NO-bound structures. Modeling of a water molecule into the analogous position in the 1-bound structure yields no 2Fo-Fc electron density in maps contoured at 1.0σ after refinement. (c) Ser189 in WelO5, coupled with substitution of a conserved Arg by Phe276, could stabilize the FeIV=O unit in the site vacated by the 2OG C1 carboxylate group upon conformational change or dissociation of CO2. (d) WelO5 Ser189Ala yields a mixture of –OH (3) and –Cl (2) modified products. Assays were performed in triplicate and representative HPLC traces are shown.
Inspection of the structure of WelO5 revealed a distinctive hydrogen-bonding group in the FeII secondary sphere, potentially positioned appropriately to promote ligand dynamics (Fig. 4a, top pathway, in black). Ser189 is located near the C1 carboxylate of 2OG and the halogen ligand, within hydrogen bonding distance of the former ligand (Figs. 4a–c). In this position, Ser189 could interact directly with the oxygenic ligand in the peroxysuccinate, FeIV=O and/or FeIII-OH states (Fig. 4a, top pathway and Supplementary Fig. 19) – but only after the proposed ligand rearrangement has occurred. The hydrogen-bonding capacity of the side chain would constrain the oxygenic group away from C13 so as to suppress hydroxylation and permit transfer of the cis halogen ligand. Consistent with such a role for Ser189, its substitution by Ala relaxes the selectivity of WelO5, resulting in an equal proportion of –OH (3) and –Cl (2) modified products (Fig. 4d).
DISCUSSION
Based on proposals for substrate positioning in SyrB219,20, we predicted that the WelO5 target carbon would reside adjacent to the halogen. In other Fe/2OG enzymes, the position occupied by the coordinated water in the FeII•2OG complex is presumed to represent the location of the oxo ligand in the FeIV=O intermediate. We thus anticipated a binding site for 1 in the quadrant defined by the open coordination position and the -Cl ligand, likely very close to the latter. In many other crystallographically characterized Fe/2OG enzymes, the analogous site, defined by the positions of the water ligand and carboxylate residue in the H-X-D/E motif, is employed for substrate binding (Supplementary Fig. 9)27. In WelO5, this pocket is accessible to solvent and sufficiently large to accommodate 1 in the more open state of the active site that we observe in the substrate-free structure. But in the substrate-bound protein, the site is definitively occluded by a conformational change in Phe169, and the substrate instead resides in the opposite quadrant, over His164 (Supplementary Fig. 9). The observed location is, however, consistent with the endpoint of a proposed substrate binding channel in SyrB216 (Supplementary Fig. 10).
The experimentally validated substrate-positioning hypothesis put forth for control of alternative group transfer in carrier-protein dependent halogenases11,19,21 dictates that the target C-H bond be held away from the FeIV=O unit with the bond vectors close to perpendicular to ensure H• abstraction through a π-pathway20,21. To square this requirement with the observed substrate-binding site in WelO5, we considered ligand configurations for the haloferryl intermediate state distinct from that implied by the x-ray crystal structure of the reactant complex. Location of the oxo unit in the plane defined by His164, 2OG, and the –Cl ligand (Fig. 4a) would reconcile the crystallographically observed location of the substrate in WelO5 and the known reactivity trends in the subfamily of Fe/2OG halogenases as a whole11. Indeed, corresponding geometries were recently proposed for the haloferryl complex in SyrB220,21. In one of these analyses20, location of the oxo trans to the His164 equivalent is proposed to occur in the context of a trigonal bipyramidal complex (axis defined by the O=FeIV-His164 vector). In WelO5, such a complex could arise via slight migration of the –Cl ligand to bring it closer to C13, perhaps an additional feature that would promote selective halogen transfer. Although that study predicts a similar reactive intermediate structure, it relied entirely on intuition and computational analysis to deduce the disposition of the halogenation target within the active site.
One pathway to the proposed intermediate configuration would involve migration of the 2OG C1 carboxylate to the position axial to the distal His ligand prior to O2 binding, followed by FeIV=O formation by the standard mechanism (Fig. 4a, bottom route, in gray). C1-carboxylate migration to accommodate a distinct oxygen-binding and oxo location may well be operant in carrier-protein-dependent halogenases and perhaps other Fe/2OG enzymes mediating non-hydroxylation reactions, as observed in NO-bound CAS32. A distinctive 2OG conformation in WelO5, in which the C3-C4 bond projects out of the plane defined by the halogen ligand and the FeII-2OG chelate motif and towards the substrate binding site (Fig. 4b), would yield a highly unfavorable 1,3-diaxial interaction between the C1 carboxylate and the C3-C4 bond on the trajectory to such a rearrangement. Migration, instead, of the oxygen-derived moiety in the FeII-peroxysuccinate (Supplementary Fig. 19) or haloferryl state would obviate this clash. The NO-bound structure of WelO5 (Fig. 4b) also dictates that relocation of the oxo unit away from the C13 halogenation target must occur post-O2 addition. Importantly, the arrangement is fully distinct from the findings of an analogous experiment in SyrB2 that shows NO already distal to the substrate chlorination site upon formation of the FeII-2OG-Cl complex21.
WelO5 exhibits another unusual feature that may serve to promote residency of the haloferryl oxo ligand in the position vacated by the departing C1 functional group. Ser189 is located near the carboxylate and halogen, within hydrogen bonding distance of the former ligand (Fig. 4c). A second notable feature is the absence in the active site of a generally conserved positively charged residue27, typically an Arg, and its replacement by Phe276. In other Fe/2OG enzymes, the side chain of this Arg residue is positioned optimally to hydrogen bond with the oxo moiety of the FeIV=O intermediate. Ser189 and, perhaps, the succinate carboxylate ligand could functionally replace the Arg and the protein carboxylate ligand (Supplementary Fig. 20) to direct the oxo of the FeIV=O unit to the position necessary to enable halogenation. The observation that Ser189Ala exhibits relaxed halogenation selectivity further supports the idea that a second-sphere hydrogen-bonding group can exert catalytic control in this scaffold.
The carrier-protein dependent halogenases, as well as other enzymes in the larger Fe/2OG family with non-hydroxylation outcomes (e.g. desaturation, oxacyclization, etc.), may employ strategies similar to those proposed here for WelO5. Exploitation of the 2OG cosubstrate and outer sphere hydrogen bonding groups in localization of the reactive intermediate distal to the substrate C-H target might, in a more general sense, facilitate reactions that demand suppression of –OH transfer. Indeed, the mechanisms of known Fe/2OG enzymes that catalyze non-canonical reactions12, and in particular how they avoid potentially competing hydroxylation outcomes, remain largely unknown. In the few examples characterized, residues in peripheral structural elements and/or the intrinsic properties of the substrate are proposed to impede hydroxylation to allow for a different type of reactivity to occur33. The structure of WelO5 suggests a new and potentially more versatile strategy, relocation of the reactive FeIV=O moiety, to promote a different reaction outcome in this scaffold.
ONLINE METHODS
General methods
All PCR reactions were carried out on a C1000 thermal cycler (BioRad). DNA sequencing was performed by Elim BioPharm Inc. Preparative scale reverse-phase analytical and semipreparative HPLC was performed using a Dionex UHPLC with a photo-diode array UV/Vis detector (Thermo Fisher Scientific). HRMS analysis was conducted using a Q Exactive Benchtop Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a Dionex RSLC (Thermo Fisher Scientific). Conventional low-resolution LC-MS analysis was conducted with a Shimadzu LCMS-2020 system. The NMR spectrum was recorded on a Bruker AV600 (600 MHz) spectrometer or a Bruker Avance III 700 MHz spectrometer equipped with a 1H/13C/15N triple-resonance inverse probe (1.7 mm microprobe).
WelO5-G166D, WelO5-S189A and WelO5-G166D-S189A protein purification
Single amino acid variants of WelO5 (G166D or S189A) were generated according to the protocol described in the QuikChange site-directed mutagenesis kit (Agilent Technologies) using plasmid pQTEV::welO5 as the template and the following primer sequences.
-
G166D WelO5
WelO5-G166D-5′: GCGAAATCGCAGTCCATGTTGATAATGATTTCCTGTTAATGCC
WelO5-G166D-3′: GGCATTAACAGGAAATCATTATCAACATGGACTGCGATTTCGC
S189A WelO5 WelO5-S189A-5′:TGATTTATCTGATCAGCTTGCTTACTTCATCCCCCTTACTGT WelO5-S189A-3′:ACAGTAAGGGGGATGAAGTAAGCAAGCTGATCAGATAAATCAA
A double variant of WelO5 (G166D-S189A) was generated using plasmid pQTEV::welO5-G166D as the template and the primers for S189A WelO5 generation following an otherwise identical protocol compared to that described for the single variants. The sequences of transformants were verified by Sanger methods and introduced into C43 (DE3) E. coli cells for overexpression and purification via the procedure described for wild-type (wt) WelO525.
WelO5-G166D, WelO5-S189A and WelO5-G166D-S189A in vitro activity assays
Individual assays for each WelO5 variant with 12-epi-fisherindole U (1, isolated from H. welwitschii B1830 as described previously25 and purified to homogeneity) as the substrate were carried out in HEPES buffer (20 mM), pH=7.4 on a 100-μL scale containing the following components: enzyme (20 μM), 1 (500 μM), 2OG (1 mM), NaCl (2 mM), and (NH4)2Fe(SO4)2 (500 μM). In a typical assay, all components except (NH4)2Fe(SO4)2 and the protein were mixed under ambient conditions and then purged with nitrogen gas. Protein samples were mixed with an aliquot of freshly prepared (NH4)2Fe(SO4)2, pre-purged with nitrogen gas, and the resulting solution was incubated briefly (~10 min) under a nitrogen atmosphere followed by purge with air. The samples were incubated under aerobic conditions at 30 °C for 30 min. The assay was quenched by extraction with ethyl acetate and centrifugation to isolate the supernatant. The extracts were combined, dried under a stream of nitrogen gas, and redissolved in methanol (50 μL). A portion of the methanol solution was injected into a Dionex UHPLC over a Kinetex 2.6um XB-C18 column (2.1 × 150 mm, 100Å, Phenomenex). Analyses were carried out using a 50% acetonitrile/50% water mobile phase under isocratic conditions using a 0.4 mL/min flow rate with monitoring by UV absorption at 278 nm. For accurate mass determination of the enzymatic product(s) derived from each assay, the methanol solution was injected to a Dionex RSLC paired with a Thermo Scientific Q Exactive Benchtop Quadrupole-Orbitrap mass spectrometer. All analytical-scale assays (100 μl) were performed in triplicate and representative HPLC traces are shown.
Structure determination of WelO5-G166D enzymatic product 3
To determine the structure of the WelO5-G166D enzymatic product 3, the reactions were scaled proportionally to 12 mL from a 100-μL scale reaction and incubated at 30 °C for 16 h. The reaction mixture was extracted with ethyl acetate (12 mL × 3), and combined ethyl acetate extracts were dried under nitrogen gas. The crude material was dissolved in methanol and purified by a Dionex HPLC equipped with a Luna 5um C18 column (250 × 4.6 mm, 100 Å, Phenomenex). The mobile phase gradient ran from 50–80% acetonitrile in water over 35 min at a 1 mL/min flow rate with monitoring by UV absorption at 278 nm. The target peak eluted with a retention time of 11.0 min and was collected and dried. The regiochemistry of the hydroxyl substituent on 3 was determined by analyzing its 1H NMR spectrum (Supplementary Fig. 12) in comparison with those of 1 and 2 (Supplementary Fig. 15), and its 1H-1H COSY spectrum (Supplementary Fig. 13). The relative stereochemistry of the hydroxyl substituent on 3 was determined by analyzing its NOESY spectrum (Supplementary Fig. 14). The spectral data are tabulated in Supplementary Table 4.
Purification of WelO5 for crystallographic trials
WelO5 containing an N-terminal hexahistidine tag was overexpressed and purified by Ni-NTA affinity chromatography as described25. To remove metal ions incorporated during overexpression, the protein sample was dialyzed at 4°C against 20 mM HEPES pH 7.5, 10 mM EDTA, followed by two rounds of dialysis in 20 mM HEPES pH 7.5 to remove EDTA. WelO5 was further purified via anion exchange chromatography using a HiPrep DEAE FF column in 20 mM HEPES pH 7.5 and a gradient of 0–50% 1 M NaCl over 10 column volumes. Purified WelO5 eluted at 200 mM NaCl. Size exclusion chromatography with a Superdex 200 pg column (20 mM HEPES pH 7.5, 200 mM NaCl) yielded a single monodisperse peak that elutes as an apparent monomer. Fractions containing WelO5 were pooled, exchanged into 20 mM HEPES pH 7.5, and concentrated with an Amicon MWCO 10K centrifugal filter. Samples intended for co-crystallization in the presence of bromide, generated to assign the location of the halogen ligand via anomalous diffraction methods, were produced as described. LC-MS grade chloride-free water used in all of the chromatography procedures and no further purification was performed after the dialysis step.
Crystallization and x-ray structure determination
Purified protein samples were rendered anaerobic by purging for 30 minutes with argon gas and transferred to an anoxic chamber (Coy Laboratory Products). Protein samples were diluted to 15 mg/mL in 20 mM HEPES pH 7.5, 50 mM NaCl (or NaBr) and cocrystallized with 1 equivalent of Fe(NH4)2(SO4)2 and 5 equivalents of 2OG. The samples were mixed in a 1:1 ratio with a precipitant solution consisting of 20% PEG 3350, 0.1 M sodium acetate pH 4.5, and 0.1 M Bis-tris pH 5.5. Rod-shaped crystals were obtained in hanging drop vapor diffusion trials within a week of setup. Substrates were incorporated by overlay of crystal drops with 2 μL of a 2 mM solution of 1 (diluted from a 10 mM stock in DMSO) prepared in the crystallization solution. The soaks were allowed to proceed for 6–8 hours at room temperature. Crystals were prepared for data collection by mounting on rayon loops after cryoprotection in crystallization solution supplemented with 24% glycerol, followed by flash freezing in liquid nitrogen for data collection. LC-MS grade water was used to prepare all solutions for bromine co-crystallization experiments. In datasets collected after reaction initiation with O2, the crystal trays were removed from the anaerobic chamber immediately before harvest and exposed to ambient O2 during cryoprotection and harvest. In NO-bound structures, 50–100 μL of a 64 mM diethylamine NONOate solution prepared in 20 mM NaOH was added to the reservoir solution in the anaerobic chamber prior to crystal harvest. The well was quickly resealed and, after 30 minutes incubation in the NO-enriched atmosphere, the crystals were harvested and flash frozen as previously described.
WelO5 crystallizes in the P212121 space group with three molecules in the asymmetric unit. G166D WelO5 crystals belong to the C2 space group with a single molecule in the asymmetric unit. All crystallographic datasets were collected at beamlines 23ID-D, 23ID-B, and 21ID-F of the Advanced Photon Source at Argonne National Lab. Resulting diffraction images were processed with the software package HKL200034. Phase information was obtained using iron single-wavelength anomalous diffraction methods. A dataset collected at 1.74 Å yielded usable anomalous signal to 3.9 Å resolution. The program autoSHARP35 was used to locate three heavy atom sites (phasing power of 0.58) that correspond to the native iron cofactor in each WelO5 monomer in the ASU. Solvent flattening and density modification procedures generated interpretable electron density maps that were subjected to an autobuilding procedure using ARP/wARP36. The initial model was then refined against a native dataset collected to 2.0 Å resolution. All subsequent structures were solved by molecular replacement using PHASER37 with the substrate-free WelO5 model as the search model. Refinement and model building steps were performed with the programs Refmac538 and COOT39, respectively. Bromine anomalous Fourier maps were generated using datasets collected at 0.92 Å with the programs CAD and FFT40, both part of the CCP4 software suite41. Coordinates and restraints for 1 were generated with the PRODRG server42. A summary of the processing and refinement statistics for all structures can be found in Supplementary Tables 1–3. Ramachandran outlier analysis and other validation procedures were carried out using the Molprobity server43. Figures were generated using the PyMOL molecular graphics software package (Schrödinger LLC).
In the absence of substrate, the model for each WelO5 molecule in the ASU contains the full sequence of amino acids, apart from an affinity tag and the ten residues at the N-terminus of the protein that immediately follow the tag. The model consists of residues 11-290 in chains A and C, residues 10-290 in chain B, three FeII ions, three chlorine atoms, three molecules of 2OG, and 456 water molecules. Ramachandran and Molprobity analyses indicate 100% of the residues in preferred and allowed regions with a clashscore of 0.61 and three rotamer outliers. Soak of WelO5 crystals with 1 yielded electron density for the molecule in all three WelO5 monomers in the ASU (Supplementary Fig. 5) but the molecule was only modeled into two of the active sites chains A and B. Only chain B displays full occupancy of 1 and substrate binding is associated with cleary evidence for a conformational change in the external helix motif and rotamer change in the side chain of Phe169. These structural changes are located at the periphery of the protein structure and are not observed in the other two molecules in the ASU due to more extensive engagement of the corresponding regions in lattice contacts (Supplementary Fig. 3). Irrespective of these peripheral conformational changes, a short loop region in the core fold (residues 79-82) containing a motif that interacts with the isocyanide moiety in 1 shifts inwards towards the substrate binding site in all three molecules, consistent with partial occupancy of these sites by 1. In chain B, residues 11-290 can be modeled, although the density for residues 215-219 is weak compared to the surrounding peptide chain. In chain C, residues 11-215 and 220-290 can be modeled. Residues 216-219 are disordered and were not included in the final model. In chain A, residues 10-290 can be modeled but no movement of the external helix is observed, consistent with observation of more extensive lattice contacts in this monomer. The model for substrate-bound WelO5 also contains three FeII ions, three chlorine atoms, three molecules of 2OG, and 251 water molecules. Ramachandran and Molprobity analyses indicate 99.5% of the residues in preferred and allowed regions with a clashscore of 1.44 and no rotamer outliers.
The substrate-bound structure of G166D WelO5 exhibits some disorder in the loop region following residue 166 (residues 169-173) as well as in a neighboring region (residues 90-97) (Supplementary Fig. 18). Residues 10-89, 98-168, and 174-290 are included in the final model in addition to a single FeII ion and 2OG molecule. The external helix does not undergo conformational change but clear density for 1 is observed and the substrate is modeled at full occupancy. Residues in the 167-176 loop facilitate external-helix closure in the wt enzyme (Supplementary Fig. 8), providing a rationale for failure to observe the conformational change in G166D WelO5, likely a consequence of the disorder in that region. The loop containing residues 79-82 moves closer to the active site, as observed in wt WelO5 + 1 monomers, even those that fail to undergo lid loop closure due to lattice contacts. Ramachandran and Molprobity analyses indicate 99.5% of the residues in preferred and allowed regions with a clashscore of 2.38 and two rotamer outliers.
The 1-/NO-bound WelO5 model consists of residues 11-214, 218-290 in chain A, residues 11-214, 219-290 in chain B, and residues 11-217, 224-290 in chain C. The final model also consists of three Fe(II), three chloride ions, three 2OG molecules, three molecules of 1, two NO molecules, and 190 water molecules. Ramachandran and Molprobity analyses indicate 100% of the residues in preferred and allowed regions with a clashscore of 1.15 and a single rotamer outlier. Compared to the 1-bound WelO5 structure in the absence of NO, more significant occupancy of substrate 1 is observed in all three WelO5 chains. The external helical motif in chains A and B closes over the active site while the active site of chain C remains exposed. The lattice contacts in each chain, as described above, are consistent with the observed trends. Two NO molecules are modeled into the active sites of chains A and B, both of which exhibited strong difference density for the molecule. Chain C displayed much weaker difference density, which we elected to leave unmodeled.
Supplementary Material
Acknowledgments
We thank Ryan Martinie, Carsten Krebs, and Marty Bollinger for critical discussions and reading of the manuscript. We acknowledge David Smith, Joseph Brunzelle, and the staff at the CCP4/APS School for Macromolecular Crystallography for assistance with x-ray data collection and analysis. This work has been supported by NIH grant GM100011 (A.K.B.), the Searle Scholars Program (A.K.B.), and the Department of Chemistry at the University of Pittsburgh (X.L.). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104).
Footnotes
Accession codes
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5IQS (WelO5), 5IQT (WelO5 + 1), 5IQU (G166D WelO5 + 1), and 5IQV (WelO5 + 1 + NO).
Author contributions
A.J.M., A.O.M., and N.A. purified and crystallized protein samples and collected x-ray datasets. A.J.M. and A.O.M. processed x-ray datasets and solved structures. Q.Z. and M.L.H. prepared samples and performed biochemical experiments. A.J.M, X.L., and A.K.B. designed the experimental approach, analyzed data, and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available in the online version of the paper.
References
- 1.Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Nature’s inventory of halogenation catalysts: oxidative strategies predominate. Chem Rev. 2006;106:3364–78. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- 2.Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature. 2005;436:1191–4. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 3.Vaillancourt FH, Yin J, Walsh CT. SyrB2 in syringomycin E biosynthesis is a nonheme FeII α-ketoglutarate- and O2-dependent halogenase. Proc Natl Acad Sci U S A. 2005;102:10111–6. doi: 10.1073/pnas.0504412102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang W, et al. Biosynthetic chlorination of the piperazate residue in kutzneride biosynthesis by KthP. Biochemistry. 2011;50:6063–72. doi: 10.1021/bi200656k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galoni DP, Barr EW, Walsh CT, Bollinger JM, Jr, Krebs C. Two interconverting Fe(IV) intermediates in aliphatic chlorination by the halogenase CytC3. Nat Chem Biol. 2007;3:113–6. doi: 10.1038/nchembio856. [DOI] [PubMed] [Google Scholar]
- 6.Galoni DP, Vaillancourt FH, Walsh CT. Halogenation of unactivated carbon centers in natural product biosynthesis: trichlorination of leucine during barbamide biosynthesis. J Am Chem Soc. 2006;128:3900–1. doi: 10.1021/ja060151n. [DOI] [PubMed] [Google Scholar]
- 7.Neumann CS, Walsh CT. Biosynthesis of (−)-(1S,2R)-allocoronamic acyl thioester by an Fe(II)-dependent halogenase and a cyclopropane-forming flavoprotein. J Am Chem Soc. 2008;130:14022–3. doi: 10.1021/ja8064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu L, et al. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009;459:731–5. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pratter SM, et al. More than just a halogenase: modification of fatty acyl moieties by a trifunctional metal enzyme. Chembiochem. 2014;15:567–74. doi: 10.1002/cbic.201300345. [DOI] [PubMed] [Google Scholar]
- 10.Podgorsek A, Zupan M, Iskra J. Oxidative halogenation with green oxidants: oxygen and hydrogen peroxide. Angew Chem Int Ed Engl. 2009;48:8424–50. doi: 10.1002/anie.200901223. [DOI] [PubMed] [Google Scholar]
- 11.Matthews ML, et al. Substrate-triggered formation and remarkable stability of the C-H bond-cleaving chloroferryl intermediate in the aliphatic halogenase, SyrB2. Biochemistry. 2009;48:4331–43. doi: 10.1021/bi900109z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bollinger JM, Jr, et al. Mechanisms of 2-oxoglutarate-dependent oxygenases: the hydroxylation paradigm and beyond. In: Hausinger RP, Schofield CJ, editors. 2-oxoglutarate-dependent oxygenases. The Royal Society of Chemistry; London: 2015. pp. 95–122. [Google Scholar]
- 13.Krebs C, Galoni-Fujimori D, Walsh CT, Bollinger JM., Jr Non-heme Fe(IV)-oxo intermediates. Acc Chem Res. 2007;40:484–92. doi: 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solomon EI, et al. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem Rev. 2000;100:235–350. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 15.Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 16.Blasiak LC, Vaillancourt FH, Walsh CT, Drennan CL. Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis. Nature. 2006;440:368–71. doi: 10.1038/nature04544. [DOI] [PubMed] [Google Scholar]
- 17.Khare D, et al. Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis. Proc Natl Acad Sci U S A. 2010;107:14099–104. doi: 10.1073/pnas.1006738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Que L., Jr One motif--many different reactions. Nat Struct Biol. 2000;7:182–4. doi: 10.1038/73270. [DOI] [PubMed] [Google Scholar]
- 19.Matthews ML, et al. Substrate positioning controls the partition between halogenation and hydroxylation in the aliphatic halogenase, SyrB2. Proc Natl Acad Sci U S A. 2009;106:17723–8. doi: 10.1073/pnas.0909649106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong SD, et al. Elucidation of the Fe(IV)=O intermediate in the catalytic cycle of the halogenase SyrB2. Nature. 2013;499:320–3. doi: 10.1038/nature12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinie RJ, et al. Experimental correlation of substrate position with reaction outcome in the aliphatic halogenase, SyrB2. J Am Chem Soc. 2015;137:6912–6919. doi: 10.1021/jacs.5b03370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulik HJ, Drennan CL. Substrate placement influences reactivity in non-heme Fe(II) halogenases and hydroxylases. J Biol Chem. 2013;288:11233–41. doi: 10.1074/jbc.M112.415570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borowski T, Noack H, Radon M, Zych K, Siegbahn PE. Mechanism of selective halogenation by SyrB2: a computational study. J Am Chem Soc. 2010;132:12887–98. doi: 10.1021/ja101877a. [DOI] [PubMed] [Google Scholar]
- 24.Pandian S, Vincent MA, Hillier IH, Burton NA. Why does the enzyme SyrB2 chlorinate, but does not hydroxylate, saturated hydrocarbons? A density functional theory (DFT) study. Dalton Trans. 2009:6201–7. doi: 10.1039/b906866j. [DOI] [PubMed] [Google Scholar]
- 25.Hillwig ML, Liu X. A new family of iron-dependent halogenases acts on freestanding substrates. Nat Chem Biol. 2014;10:921–3. doi: 10.1038/nchembio.1625. [DOI] [PubMed] [Google Scholar]
- 26.Hillwig ML, et al. Identification and characterization of a welwitindolinone alkaloid biosynthetic gene cluster in the stigonematalean Cyanobacterium Hapalosiphon welwitschii. Chembiochem. 2014;15:665–9. doi: 10.1002/cbic.201300794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aik W, McDonough MA, Thalhammer A, Chowdhury R, Schofield CJ. Role of the jelly-roll fold in substrate binding by 2-oxoglutarate oxygenases. Curr Opin Struct Biol. 2012;22:691–700. doi: 10.1016/j.sbi.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Wong C, Fujimori DG, Walsh CT, Drennan CL. Structural analysis of an open active site conformation of nonheme iron halogenase CytC3. J Am Chem Soc. 2009;131:4872–9. doi: 10.1021/ja8097355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hillwig ML, Zhu Q, Liu X. Biosynthesis of ambiguine indole alkaloids in cyanobacterium Fischerella ambigua. ACS Chem Biol. 2014;9:372–7. doi: 10.1021/cb400681n. [DOI] [PubMed] [Google Scholar]
- 30.Hillwig ML, Zhu Q, Ittiamornkul K, Liu X. Discovery of a promiscuous non-heme iron halogenase in ambiguine alkaloid biogenesis: implication for an evolvable enzyme family for late-stage halogenation of aliphatic carbons in small molecules. Angew Chem Int Ed Engl. 2016 doi: 10.1002/anie.201601447. [DOI] [PubMed] [Google Scholar]
- 31.Zhu Q, Hillwig ML, Doi Y, Liu X. Aliphatic halogenase enables late-stage C-H functionalization: selective synthesis of a brominated Fischerindole alkaloid with enhanced antibacterial activity. Chembiochem. 2016;17:466–70. doi: 10.1002/cbic.201500674. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, et al. Crystal structure of a clavaminate synthase-Fe(II)-2-oxoglutarate-substrate-NO complex: evidence for metal centered rearrangements. FEBS Lett. 2002;517:7–12. doi: 10.1016/s0014-5793(02)02520-6. [DOI] [PubMed] [Google Scholar]
- 33.Chang W-c, et al. Mechanism of the C5 stereoinversion reaction in the biosynthesis of carbapenem antibiotics. Science. 2014;343:1140–4. doi: 10.1126/science.1248000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–30. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 36.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–9. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Ten Eyck LF. Fast Fourier transform calculation of electron density maps. Methods Enzymol. 1985;115:324–37. doi: 10.1016/0076-6879(85)15024-x. [DOI] [PubMed] [Google Scholar]
- 41.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;D67:235–42. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. 2004;D60:1355–63. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 43.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. 2010;D66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.