Abstract
Aging is one of the greatest challenges for biomedical research in the developed world. The continuing increase in lifespan without accompanying increases in health span has extreme economic and societal implications for an aging society. The majority of adult‐onset diseases such as cancer, cardiovascular disease, and Alzheimer's are a direct consequence of aging, and with changing demographics, more and more people are afflicted. If we knew how to slow down the negative effects of aging, we could delay or prevent all of these diseases at once. We therefore need to understand the underlying molecular mechanisms of aging and longevity so as to develop applications that ultimately improve health span.
Subject Categories: Ageing
During the past two decades, the field of aging biology has made spectacular advances in finding and explaining important aging‐regulating mechanisms, such as insulin/Igf1 signaling, TOR, and telomere maintenance. This progress would have been impossible without model organisms, the workhorses of genetics, and molecular biology. Most of our knowledge about the mechanisms of aging has been obtained thanks to the yeast Saccharomyces cerevisiae, the nematode worm Caenorhabditis elegans, the fruit fly Drosophila melanogaster, and the house mouse Mus musculus. These organisms have been extensively used by biologists, are very well characterized, and easy to maintain. Indeed, with the exception of C. elegans, these species have been welcome or unwelcome human companions long before the era of modern science. The genomes of these organisms were among the first to be sequenced, and there is an impressive arsenal of precise tools for genetic manipulation available for each species.
However, these “canonical” model organisms have their limitations. Yeasts are tremendous genetic systems and have provided fundamental insights on cellular aging, but they prevent studies on multicellular and systemic aging. Nematode worms and flies, whose lifespan is extremely short—2–3 weeks for the worm and about 2 months for the fly—are extremely practical invertebrate model organisms. However, adults are mostly composed of post‐mitotic cells, which prevent the study of stem cell function and cancer in the context of aging. Additionally, they entirely lack an adaptive immune system. While the mouse is the shortest‐lived vertebrate used widely for aging studies, its maximum lifespan of more than 3 years commands considerable time and resources for lifespan studies. Hence, a vertebrate even shorter‐lived than mice with active adult stem cell compartments would be a welcome addition.
Moreover, short‐lived species rely on rapid life cycles and prolific reproduction to achieve their evolutionary success. This does not reflect many aspects of human aging, as Homo sapiens is characterized by slow sexual maturation, prolonged parental care, a few progeny, and a long lifespan. Since species with rapid life cycles are not selected to survive for extended time, they may be missing the mechanisms that promote extended longevity, which is what many aging researchers are looking for. In other words, the biology of a mouse or a fly can help us reveal the causes of aging and disease, but may be of limited value for discovering the basis of long lifespan and disease prevention. Therefore, it is also important to include long‐lived species in the aging research bestiary.
Finally, relying on a small number of established model organisms neglects the diversity of adaptations that have evolved in nature and that may be of great value for human health. For example, some species maintain a unique ability to regenerate their organs, others are highly resistant to a variety of stresses or diseases, while various species just live extremely long. Once we can understand and compare the mechanisms behind these unique “talents”, we could adapt this knowledge to benefit human health.
In this Commentary, we discuss three relatively new model organisms that are of interest to aging research: the African turquoise killifish, the planarian flatworm, and the naked mole rat. Advances in genome sequencing and new technologies for genetic manipulation have made it possible to develop genetic tools for these and other species within reasonable time. Genomes can be rapidly and affordably sequenced; cross‐reacting commercially available antibodies can be identified based on protein sequence from close relatives; genes can be targeted using CRISPR/Cas9 or RNAi; and a wide variety of in vivo and in vitro expression technologies are available. Thanks to these technological advances, we can now venture beyond the established model organisms and explore the great diversity of species for biomedical research.
The African turquoise killifish, a naturally short‐lived vertebrate
The turquoise killifish (Nothobranchius furzeri Jubb; Fig 1) is a freshwater teleost inhabiting seasonal water bodies that form during the brief rainy season in the arid Mozambican and Zimbabwean savannah woodland. The intermittent availability of water exerts a strong selective pressure on both survival and reproduction, ultimately shaping the life cycle of this species with a set of unique adaptations. Turquoise killifish undergo explosive growth to reach sexual maturation within 3–4 weeks. They mate for 2–3 months and, depending on the strain, survive a total of 4–8 months (Valenzano et al, 2015). After reproduction, killifish undergo a broad spectrum of age‐related transformations, which include loss of pigmentation, increased risk for cancer, motor and cognitive decline, cellular senescence, and loss of fecundity (Kim et al, 2016).
Figure 1. New model organisms for aging research.
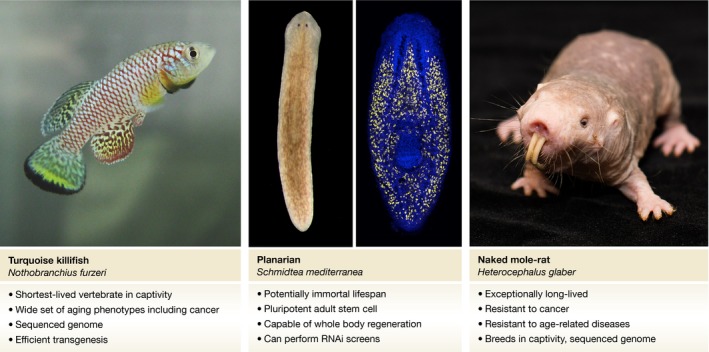
Representatitve male adult turquoise killifish (left). Planarian (center); the right panel shows neoblasts labeled with an RNA fluorescent in situ hybridization probe to the smedwi‐1 transcript, a PIWI protein family homolog. Naked mole rat (right).
Rapid sexual maturation ensures that the fish can reproduce before the onset of the incoming dry season. Once fertilized, embryos enter a developmental‐arrested state called diapause, during which they can sustain life for several months up to years, in the absence of water. This sophisticated adaptation—equivalent to plant seeds—enables embryos in diapause to survive throughout the dry seasons and during prolonged periods of drought. It enables turquoise killifish—as well as other oviparous Cyprinodontiformes—to outcompete all other teleosts in environments where water is absent for prolonged lengths of time.
Compared to mice and zebrafish, whose median captive lifespan exceeds 2.5 years, the killifish's captive median lifespan of only 4 months in the shorter‐lived strains makes it an extremely convenient model organism to test interventions aimed at modulating aging and ultimately longevity. Unlike the widely adopted non‐vertebrate aging models, such as yeast, roundworms (C. elegans), and flies, the turquoise killifish is a vertebrate, and it is therefore equipped with a prominent adult stem cell compartment. Additionally, although naturally short‐lived, turquoise killifish spontaneously develop different types of cancer, the onset of which is age‐dependent (Kim et al, 2016). Since it is a teleost, killifish has an adaptive immune system similar to mammals, including both humoral and cellular immunity, which is entirely missing in invertebrates. For these reasons, killifish can become a powerful system to study the role of stem cell function, cancer, and adaptive immunity in the context of aging. The possibility to raise them in a laboratory setting, a recently sequenced and assembled genome, and the successful development of transgenesis via Tol2 and CRISPR/Cas9 technology make this species a promising new system to experimentally study the biological mechanisms underlying several aspects of the vertebrate aging process (Kim et al, 2016).
Several established turquoise killifish laboratory strains exist, one of which, the “GRZ”—derived from wild‐caught fish collected in 1968—is highly inbred and has been used as a reference species for the sequencing of the genome. The availability of a highly inbred line constitutes a significant advantage over other already established laboratory model organisms, such as zebrafish, and provides an optimal platform to develop transgenic and mutant lines.
Remarkably, turquoise killifish populations from different habitats—characterized by more or less dry conditions, and longer or shorter rainy seasons—display significant differences in captive lifespan: Killifish from more arid areas are shorter‐lived than those derived from more wet areas (Valenzano et al, 2015; Blazek et al, 2017). Genetic mapping enabled the identification of the genomic regions associated with these differences in life expectancy. The results showed that although a large number of genomic regions are associated with differential survival—the region with the largest association with strain‐specific survival is located on the sex chromosome, in proximity with the sex‐determining locus. Future studies will determine which specific genetic elements are causally responsible for differences in adult life expectancy. Genetic mapping, population genetics, and mutagenesis screening will become key tools to dissect the genetic architecture of killifish‐specific natural traits, including embryonic diapause, explosive maturation, and the wide spectrum of age‐related transformations, including cancer, and motor and cognitive decline.
This organism will also help to address a fundamental question related to the role of natural selection, random drift, and demography in shaping life history in nature. The unique evolutionary trajectory of this clade of teleosts will provide a concrete example of how different life‐history strategies evolved in nature under different ecological conditions. These results might help shed light on the general mechanisms involved in the regulation of lifespan even in longer‐lived species.
An ever‐growing number of research groups are adopting the turquoise killifish as an experimental model organism, and molecular resources for this species are rapidly increasing, making it more available to a broader research community. The development of a large collection of benchmarked antibodies, an improved genome annotation, and the availability of transgenic line stock centers will become key resources to further develop this species and make it competitive with the more established and canonical vertebrate model organisms, such as zebrafish and mice.
Planarians and the basis of an immortal life history
Planarian flatworms (Fig 1), members of the phylum Platyhelminthes, have long been the subject of research, including by one of the founders of modern genetics, Thomas Hunt Morgan before his illuminating work with fruit flies (Reddien & Sánchez Alvarado, 2004). These freshwater‐dwelling worms, like the freshwater cnidarian hydra, have the amazing capacity to regrow all tissues and organs from nearly any amputated starting fragment. Allied to this regenerative capacity is an equally startling developmental plasticity that allows animals to indefinitely grow and shrink in response to nutritional status. After regeneration and during growth/starvation, all tissues and organs achieve and maintain perfect scaling. These traits are driven by a population of pluripotent adult stem cells called neoblasts (Fig 1). During regeneration, these cells can collectively interpret signals from wounds, the 3D polarity and positional information in remaining tissues and then proliferate and differentiate to replace all missing structures. During growth and starvation, neoblasts can characteristically adjust both the rate at which they divide and the relative levels of symmetric and asymmetric division to respond to nutritional status. Even in the absence of any tissue damage, planarians constantly replace their differentiated cells with new ones provided by the stem cell population. Moreover, some species of this group of animals have evolved to be somatically immortal, meaning that they do not show any age‐associated decline in any aspect of their physiological functions.
The conclusion that some planarians must be somatically immortal is deduced from the fact that some species have no functional germline or gonads and reproduce by splitting in half to produce an anterior fragment that regenerates a tail and a posterior fragment that regenerates a brain and all other missing organs. The somatic adult stem cells have also adopted the role of the germline, and for these species to persist over any evolutionarily significant time, these somatic cells must avoid the aging process. This means that the physiological, cellular, and molecular processes that we think cause aging in most other animals must somehow be avoided or counteracted in planarians, in particular in the pluripotent stem cell population. So how are these stem cells protected from aging and can these processes be studied in a tractable way?
Molecular genetic research with planarians has blossomed over the past two decades, with two main model species, Schmidtea mediterranea and Dugesia japonica. The number of laboratories dedicated to studying regenerative mechanisms and stem cell biology in Schmidtea in particular has continually increased driven by a growing molecular toolkit. This includes the ability to relatively easily knock down genes, an array of cell‐specific markers that has begun to reveal heterogeneity in the stem cell population and extensive publically available genomic and transcriptomic resources. These tools have led to advances in understanding the mechanisms underpinning the amazing regenerative process and stem cell function. In the broadest sense, all these studies help to explain the key life‐history trait of planarians: their endless regenerative capacity that can be considered as the antithesis of the aging process.
It has also made possible to look specifically at mechanisms associated with aging, for example telomere shortening, which is associated with cellular senescence and the loss of proliferative ability in adult stem cell populations. In most eukaryotes, telomeres are maintained by the reverse transcriptase enzyme telomerase that serially adds repeat sequences to the chromosome ends when activated. In order to explain an endless regenerative capacity, planarians' stem cells must have mechanisms to maintain their telomeres, leading to the hypothesis that they must have the capacity to activate telomerase or some other chromosome end maintenance mechanism as required. In fact, asexual planarians can upregulate telomerase activity whenever they regenerate or reproduce by fission to restore telomere length in response to signals that require increases in stem cell proliferation (Tan et al, 2012). This activity is in part controlled at the levels of expression and splicing of the gene encoding the Schmidtea protein subunit of telomerase. The study also showed that the sexually reproducing species of Schmidtea, from which the asexual species evolved, does not maintain telomere length in the same way, despite also having an apparent indefinite capacity to regenerate. The question of whether sexually reproducing planarians are also somatically immortal and, if so, how they maintain their chromosome ends is still open (Tan et al, 2012).
We would also expect that planarian stem cells exhibit high levels of genome stability to avoid the accumulation of mutations despite all the proliferative activity they must undertake, even in homeostatic conditions. The observation that these cells can withstand large amounts of damage from exogenous sources, for example up to 20 Gray of ionizing radiation, without affecting subsequent regeneration suggests they must have robust genome surveillance and repair mechanisms. While currently little is known about canonical DNA damage response pathways, it has been shown that PIWI proteins and piRNAs, normally associated with protecting germline cells from DNA damage induced by retrotransposons, are an important feature of planarian stem cell biology (Shibata et al, 2016). Indeed, one general strategy by which planarian adult stem cells achieve immortality may have been using many of the same features that ensure immortality of the germline across all metazoans.
Future work will look at more detail into how planarians stem cells avoid the aging process and whether individual stem cells show any signatures of aging within the population or whether damaged or corrupted cells are rapidly eliminated. Another possibility is that some neoblasts in any individual worm are given a transient special status and are specifically protected somewhat like a germline. Whatever the answers, studying the tractable planarian system will lead to an explanation about how any animal could have evolved to avoid the aging process entirely.
Naked mole rat, the longest‐lived rodent
Naked mole rat Heterocephalus glaber (Fig 1) is a small rodent that lives in large underground colonies in East Africa. As the name suggests, these animals are naked, albeit this is not the most interesting aspect of their biology. Naked mole rats are true eusocial animals similar to bees or ants. Each colony has one breeding female, the queen, who mates with one or two breeder males. The queen rules her colony by suppressing reproductive functions in other animals that work as cleaners, nannies, and soldiers. Naked mole rats feed on roots and tubers and rarely if ever emerge above ground. Hence, predators cannot easily get to them.
The protected environment of underground borrows together with eusocial lifestyle likely contributed to the evolution of exceptional longevity with a maximum lifespan of 31 years that makes the naked mole rat the longest‐lived rodent known today. As the longer‐lived mammalian species tend to be larger, the naked mole rat is particularly striking considering its small body size. In addition, the animal has an extreme resistance to cancer and other age‐related diseases. As naked mole rats age, they do not show a gradual decline of physiological functions, which led them to be classified as one of a few species displaying negligible senescence (Buffenstein, 2008) and makes them a very valuable model for studying mechanisms of longevity and disease resistance.
The naked mole rat is also a practical model organism. Provided with a constant‐temperature housing and a diet of fresh fruit and vegetables, they thrive and breed in captivity. The genome has been sequenced and two independent, high‐quality genome assemblies are available. Many of the antibodies available to mouse or rat proteins cross‐react with the naked mole rat, and their cells can be cultured in vitro with small modifications from mouse protocols. Importantly, the naked mole rat is a distant relative of two well‐characterized laboratory rodents, the house mouse and Norway rat, and a closer relative of the guinea pig. Both mouse and rat are very short‐lived, while the guinea pig has an intermediate lifespan. This enables comparative approaches to study the naked mole rat's longevity mechanisms in relation to shorter‐lived relatives.
Although it is a relatively new research model, several molecular mechanisms contributing to longevity have already been identified. As mentioned above, naked mole rats are highly resistant to cancer. Only a few cases of spontaneous tumors have been found in thousand of animals monitored over two decades in research colonies and zoos. Investigating the mechanism of cancer resistance revealed a very unique adaptation that gave the naked mole rat the title of the “vertebrate of the year” in 2013. Their cells secrete a high molecular weight glycosaminoglycan polymer, hyaluronan, a main non‐protein component of the extracellular matrix (Tian et al, 2013). The biological properties of hyaluronan depend on its molecular weight: Short polymers promote cell division and are associated with wounds and inflammation, while long polymers are antiproliferative and anti‐inflammatory. While animals, including humans, also produce hyaluronan, the naked mole rat's hyaluronan molecules are up to 10 times longer. High molecular weight hyaluronan binds to cell receptors, promotes cell quiescence, and restricts the growth of premalignant cells. In addition, hyaluronan has antioxidant and cytoprotective properties that may also contribute to longevity. Since shorter versions of hyaluronan are already used in the clinic, the discovery of high molecular weight hyaluronan in a cancer‐resistant animal opens new avenues for clinical applications.
Other mechanisms contributing to cancer resistance in the naked mole rat include a unique structure of the INK4 locus. In human and mouse, the INK4 locus encodes three important tumor suppressors, p15, p16, and ARF. Remarkably, the naked mole rat's INK4 locus encodes a fourth isoform pALT that is even more efficient at arresting the cell cycle than p15 or p16.
One more unique molecular feature that may contribute to long lifespan is the naked mole rat's ribosome structure (Azpurua et al, 2013). The 28S rRNA is split into two fragments, resulting in four separate rRNAs. In addition, naked mole rat ribosomes are very accurate: Their fidelity of protein synthesis is 10‐fold higher than in the mouse resulting in fewer aberrant proteins being made. It also shows the potential importance of proteostasis for longevity.
Remarkably, the naked mole rat does not use a single strategy to achieve long lifespan. It has evolved multiple adaptations that affect different cellular and physiological systems. Clearly, to extend the lifespan by almost an order of magnitude when compared to the house mouse, more than a single improvement is required. Thus, what we have learned from the naked mole rat is exciting, but there is a lot more that we do not know. Moreover, while the naked mole rat possesses some conserved mechanisms of longevity, others, such as high molecular weight hyaluronan and split ribosomal RNA, are unique to this species. This illustrates that evolution does not choose the same path, and animals evolve unique mechanisms of longevity that are shaped by the ecology and genetics of each species. This underscores the importance of studying a wide range of animal models.
Discussion
The increasing number and use of non‐canonical model organisms is expanding our knowledge about the molecular basis of biological aging and providing novel experimental opportunities to tackle scientific questions that were unapproachable using the classical model organisms. For instance, long‐lived model organisms, such as the naked mole rat, help us to reveal the basis of extreme longevity and organisms with negligible senescence, such as planarians, help us to understand how to achieve optimal tissue maintenance and an immortal somatic cell line. We can use these fundamental discoveries for applications to fight the aging process and associated age‐related disorders in humans.
On the other side of the spectrum, short‐lived vertebrate model organisms provide experimental platforms to rapidly test the effect of potential interventions on longevity and age‐related diseases, including cancer and neurodegenerative diseases. For example, genes from long‐lived organisms can be introduced into the short‐lived organisms to test their effect on lifespan and fitness. Alternatively, short‐lived organisms could be manipulated using small molecules to shift their molecular pathways toward those discovered in long‐lived organisms. This approach will also serve as a platform to translate basic discoveries into practical applications.
The increased availability of a larger set of model organisms with known biology and sequenced genomes allows for statistically sound comparative genomics of aging and lifespan. Other long‐lived organisms, such as the Greenland shark, bowhead whale, elephant, quahog clam, and hydra, will provide additional material for research on longevity. More short‐lived organisms, such as shrews, Labord's chameleon, and others, will further help to identify the genomic signature of short lifespan and compressed life cycle. Comparative genomics will enable to detect the shared genomic features that underlie rapid or slow aging, as well as short or long lifespan. However, a key feature of a model organism is the possibility to raise it for the entire life cycle under controlled laboratory conditions, which explains the important role of yeast, worm, flies, and mice as the most prominent aging model systems to date. Of course, this is not practical for extremely long‐lived or large species, such as Greenland sharks, whales, or elephants. However, analyzing cultured cells from these organisms could also help to identify the unique mechanisms contributing to their longevity.
Each model organism will enable research to address different questions and levels of research, spanning from population and life‐history studies to heavily mechanistic studies of function. By comparing how aging trajectories differ, identifying the underlying evolutionary mechanisms that have driven these differences, and understanding how they are reflected in conserved genetic pathways, we will both understand aging and develop the conceptual framework for a future generation of interventions. Given the huge societal impact of increasing lifespans, funding bodies need to be open‐minded about the research they finance. As we have shown in this Commentary, there is an urgent need to expand the bestiary of models for aging research. The study of non‐traditional model organisms leads to highly mechanistic discoveries, and it is no longer an esoteric pursuit. We hope that using these novel models and recruiting young scientists to work on them will lead to groundbreaking discoveries ultimately leading to increases in health span and lifespan.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Azpurua J, Ke Z, Chen IX, Zhang Q, Ermolenko DN, Zhang ZD, Gorbunova V, Seluanov A (2013) Naked mole‐rat has increased translational fidelity compared with the mouse, as well as a unique 28S ribosomal RNA cleavage. Proc Natl Acad Sci USA 110: 17350–17355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek R, Polacik M, Kacer P, Cellerino A, Rezucha R, Methling C, Tomasek O, Syslova K, Terzibasi Tozzini E, Albrecht T, Vrtilek M, Reichard M (2017) Repeated intraspecific divergence in life span and aging of African annual fishes along an aridity gradient. Evolution 71: 386–402 [DOI] [PubMed] [Google Scholar]
- Buffenstein R (2008) Negligible senescence in the longest living rodent, the naked mole‐rat: insights from a successfully aging species. J Comp Physiol [B] 178: 439–445 [DOI] [PubMed] [Google Scholar]
- Kim Y, Nam HG, Valenzano DR (2016) The short‐lived African turquoise killifish: an emerging experimental model for ageing. Dis Model Mech 9: 115–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A (2004) Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol 20: 725–757 [DOI] [PubMed] [Google Scholar]
- Shibata N, Kashima M, Ishiko T, Nishimura O, Rouhana L, Misaki K, Yonemura S, Saito K, Siomi H, Siomi MC, Agata K (2016) Inheritance of a nuclear PIWI from pluripotent stem cells by somatic descendants ensures differentiation by silencing transposons in planarian. Dev Cell 37: 226–237 [DOI] [PubMed] [Google Scholar]
- Tan TC, Rahman R, Jaber‐Hijazi F, Felix DA, Chen C, Louis EJ, Aboobaker A (2012) Telomere maintenance and telomerase activity are differentially regulated in asexual and sexual worms. Proc Natl Acad Sci USA 109: 4209–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Azpurua J, Hine C, Vaidya A, Myakishev‐Rempel M, Ablaeva J, Mao Z, Nevo E, Gorbunova V, Seluanov A (2013) High‐molecular‐mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499: 346–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzano DR, Benayoun BA, Singh PP, Zhang E, Etter PD, Hu CK, Clement‐Ziza M, Willemsen D, Cui R, Harel I, Machado BE, Yee MC, Sharp SC, Bustamante CD, Beyer A, Johnson EA, Brunet A (2015) The African turquoise killifish genome provides insights into evolution and genetic architecture of lifespan. Cell 163: 1539–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]


