Abstract
Sequencing the actinomycin (acm) biosynthetic gene cluster of Streptomyces antibioticus IMRU 3720, which produces actinomycin X (Acm X), revealed 20 genes organized into a highly similar framework as in the bi-armed acm C biosynthetic gene cluster of Streptomyces chrysomallus but without an attached additional extra arm of orthologues as in the latter. Curiously, the extra arm of the S. chrysomallus gene cluster turned out to perfectly match the single arm of the S. antibioticus gene cluster in the same order of orthologues including the the presence of two pseudogenes, scacmM and scacmN, encoding a cytochrome P450 and its ferredoxin, respectively. Orthologues of the latter genes were both missing in the principal arm of the S. chrysomallus acm C gene cluster. All orthologues of the extra arm showed a G +C-contents different from that of their counterparts in the principal arm. Moreover, the similarities of translation products from the extra arm were all higher to the corresponding translation products of orthologue genes from the S. antibioticus acm X gene cluster than to those encoded by the principal arm of their own gene cluster. This suggests that the duplicated structure of the S. chrysomallus acm C biosynthetic gene cluster evolved from previous fusion between two one-armed acm gene clusters each from a different genetic background. However, while scacmM and scacmN in the extra arm of the S. chrysomallus acm C gene cluster are mutated and therefore are non-functional, their orthologues saacmM and saacmN in the S. antibioticus acm C gene cluster show no defects seemingly encoding active enzymes with functions specific for Acm X biosynthesis. Both acm biosynthetic gene clusters lack a kynurenine-3-monooxygenase gene necessary for biosynthesis of 3-hydroxy-4-methylanthranilic acid, the building block of the Acm chromophore, which suggests participation of a genome-encoded relevant monooxygenase during Acm biosynthesis in both S. chrysomallus and S. antibioticus.
Keywords: actinomycin, actinomycin halves, biosynthesis, Streptomyces chrysomallus, Streptomyces anulatus Streptomyces antibioticus, genomes, 3-hydroxy-4-methylanthranilic acid (4-MHA), evolution of biosynthetic gene cluster, genetic transmission of biosynthetic gene cluster, actinomycin C, actinomycin X
Introduction
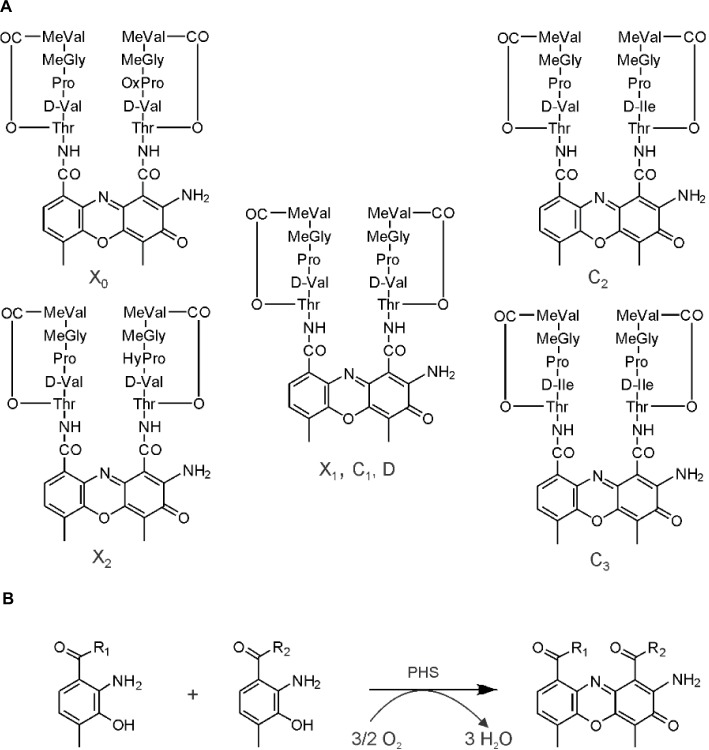
Streptomyces antibioticus IMRU 3720 produces a mixture of actinomycins (Acms), designated as actinomycin X (Acm X).1 Acms are chromopeptides consisting of two pentapeptide lactone rings attached in amide linkage to 2-amino-phenoxazine-3-on-4,6-dimethyl-1,9-dicarboxylic acid (actinocin; Figure 1). Members of the Acm X mixture differ from each other by substitutions in the “proline” site of their β-pentapeptide lactone rings.2–4 Thus, Acm X2 contains a 4-oxo-proline residue whereas Acm X1 and Acm X0 contain a proline or a 4-trans-hydroxyproline, respectively.5 In contrast, members of the Acm C complex produced by Streptomyces anulatus var. chrysomallus ATCC 11523 (designated Streptomyces chrysomallus) and its relatives vary exclusively in the “d-valine” sites of their pentapeptide lactone rings by substitution of one or both d-valine residue with d-allo-isoleucine giving Acm C2 and Acm C3, respectively (Figure 1).5–7 Acm X and Acm C (and also Acm C1 [syn. D] produced by Streptomyces parvulus as an alone-standing Acm) are the preferred types of Acms formed in streptomycetes.5 Strains producing Acms of the Z, F, G and Y type are less frequent.6,8,9 Typically, these latter mixtures are produced in low yield in line with their more complicated structures compared to Acm X and Acm C.9
Figure 1.
(A) Structures of actinomycins of the X complex and the C complex. There are different designations in the literature for the various actinomycins such as for actinomycin D, which is synonymous with actinomycin C1 or actinomycin X1. (B) Phenoxazinone formation reaction. This reaction can be spontaneous or catalyzed by phenoxazinone synthase (PHS) for both in an oxygen dependency.
Abbreviations: Thr, threonine; Val, valine; Pro, proline; HyPro, 4-trans-hydroxyproline, OxoPro, 4-oxo-proline; MeGly, N-methylglycine, Sarcosine; MeVal, N-methyl-l-valine; d-Ile, d-allo-isoleucine.
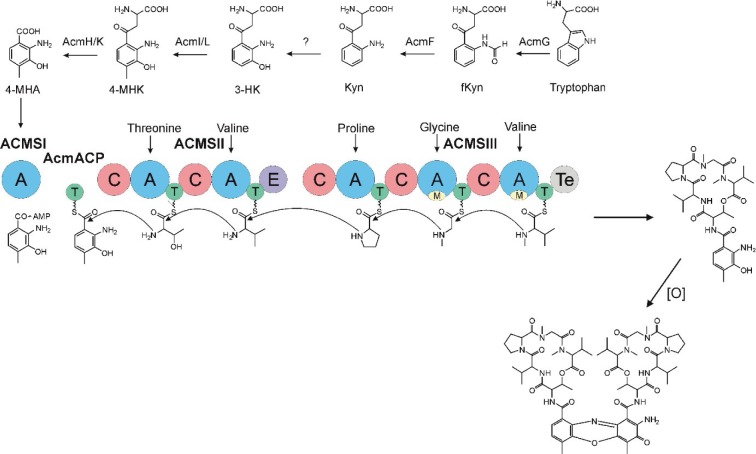
Initial in vivo studies of Acm biosynthesis in S. antibioticus and S. chrysomallus indicated differences in the physiology of formation.5,10–12 Later enzymatic studies performed in S. antibioticus revealed the presence of an enzyme phenoxazinone synthase (PHS), which is not expressed in S. chrysomallus (or S. parvulus).13–17 Its coordinate expression and regulation with Acm X biosynthesis in S. antibioticus indicated its role in the formation of the phenoxazinone chromophore of Acm.13–15 Since S. antibioticus proved to be a difficult source to isolate new Acm biosynthetic enzymes, the focus of Acm biosynthesis research turned onto S. chrysomallus (and to a lesser extent to S. parvulus), from which enzymes were characterized involved in formation of the chromophore precursor 3-hydroxy-4-methylanthranilic acid (4-MHA) or the actinomycin synthetases (ACMSs) involved in the nonribosomal assembly of the half molecule precursors of Acms (Acm halves).18–26 Based on their protein sequences, the acm C biosynthetic gene cluster of S. chrysomallus was cloned.27 Its genetic and biochemical analysis together with the previous enzymatic data allowed to draw a detailed picture of Acm biosynthesis (Figure 2).
Figure 2.
Biosynthesis of Acm C1 (X1, D) in Streptomyces chrysomallus. Upper row: reaction sequence leading from tryptophan to 3-hydroxy-4-methylanthranilic acid (4-MHA), enzyme designations (Keller et al27; still non-identified 3-kynurenine-monooxygenase is indicated by a question mark). Lower row: nonribosomal ACMS assembly line of Acm half molecule (shown Acm X1 half) and oxidative condensation of Acm X1 halves to Acm X1.
Abbreviation: ACMS, actinomycin synthetase.
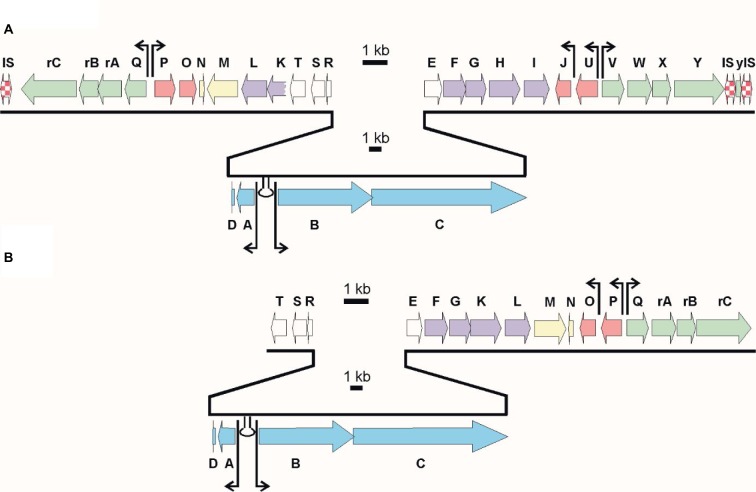
Remarkably, the acm C biosynthetic gene cluster of S. chrysomallus has a palindromic appearance by the presence of two invertedly oriented arms flanking the peptide synthetase genes and their cohorts in the center. Each arm contained orthologous sets of genes (but not all) from the other arm and in the same arrangement. It was assumed that this was the result of a previous duplication of a simpler version (minimal) acm gene cluster with subsequent rearrangements and deletions, which also may explain loss of a 3-kynurenine monooxygenase gene necessary for 4-MHA synthesis or the gene encoding a phenoloxidase responsible for condensation of Acm halves in the last step of Acm biosynthesis (Figure 2).28 However, the duplication model was hampered by the presence of two additional genes embedded in the extra arm but both absent in the principal arm, which precluded to deduce how the minimal acm cluster looked like. In view of the considerable amount of data available on Acm X biosynthesis in S. antibioticus, we decided to clone the acm X biosynthetic gene cluster and compare its structure with the acm C biosynthetic gene cluster in S. chrysomallus. To this end, we sequenced the genome of both S. antibioticus and S. chrysomallus.
Materials and methods
General materials and methods
S. anulatus var. chrysomallus ATCC 11523 and S. antibioticus IMRU 3720 were derived from the American Type Culture Collection (ATCC, Manassas, VA, USA). For historical reasons instead of S. anulatus the name of the original isolate S. chrysomallus is used throughout this study.7 Growth and maintenance of both strains and their derivatives were as described.11–13,29 All other methods for microbiological handling of streptomycetes and for genetic manipulations were according to Hopwood et al.30 For preparation of large genomic DNA fragments, protoplasts of S. chrysomallus and S. antibioticus were prepared and lysed in a protocol as described in Supplementary materials. Polymerase chain reaction (PCR) using chromosomal DNA of S. antibioticus was performed to close gaps in the genes saacmB and saacmC as described in the SI section. Common biochemicals and chemicals were from standard commercial sources.
Genome sequencing and analyses
These procedures were according to state of the art techniques and are described in “Results and discussion” section.
Bioinformatics analyses
Programs used for sequence alignments were BLASTP, Clustal Omega,31 Genedoc,32 BProm (Softberry),33 Pepper34 and ISfinder.35 Phylogenetic tree construction was done by using the phylogeny.fr website.36 Analysis of NRPS sequences was done using online tools such as NRPSpredictor2.37
Accession numbers
Accessions for the draft genome projects of S. antibioticus IMRU 3720 and S. anulatus var. chrysomallus ATCC11523 are given in “Results and discussion” section.
Results and discussion
Genome sequencing of S. antibioticus
Purified genomic DNA of S. antibioticus IMRU 3720 was used as input for the construction of a DNA paired end whole-genome sequencing (WGS) library using the Nextera DNA Library Preparation Kit (Illumina). Sequencing of the library in a paired-end run using the MiSeq desktop sequencer (Illumina) yielded 2,409,697 reads (410.9 Mb). An initial assembly performed with the Roche GS de novo Assembler software (release 2.8) resulted in 162 contigs in 56 scaffolds, with 167 contigs larger than 500 bp in total. Manual inspection of the contig ends revealed lack of coverage at the end of most of the contigs as the cause for the contig breaks. To overcome this problem, a second WGS library was constructed using the TruSeq DNA PCR-Free Library Preparation Kit (Illumina) for paired end library construction and the MiSeq reagent kit v3 (600 cycles) for sequencing, adding 2,654,170 reads (532.8 Mb) to the assembly. The final automatic draft assembly contained 42 contigs in 20 scaffolds with 49 contigs larger than 500 bp in total. Comparison to several complete sequenced Streptomyces spp. revealed a high degree of synteny between the scaffolds and the respective reference genomes. Using Streptomyces collinus Tü 365 [GenBank: CP006259.1; PMID: 24140291] as a reference, the linear genome could be assembled in one single scaffold consisting of 14 contigs (although the orientation of 4 contigs is unknown).
The S. antibioticus draft genome consists of a single, linear chromosome of ~8,480.1 kbp with a G+C content of 71.78%. The NCBI Prokaryotic Genome Annotation Pipeline (PGAP) pipeline (PMID: 18416670) was used for gene prediction and annotation, resulting in a total of 7,536 predicted genes (7,154 CDS, 69 tRNAs, 18 rRNAs in 6 operons, and 5 ncRNAs). The draft genome of S. antibioticus was subjected to analysis using antiSMASH.38 From this, it was deduced that it contains at least 28 different secondary metabolite gene clusters encoding the biosynthesis of secondary metabolites such as nonribosomal peptides, polyketides, siderophores and pigments (Table 1). They are distributed over the whole genome with no preference for distinguished regions. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession LHQL00000000. The version described in this paper is version LHQL01000000.
Table 1.
Genomic data for Streptomyces chrysomallus and Streptomyces antibioticus
| Genomic data | S. chrysomallus | Plasmid scrP1 | S. antibioticus |
|---|---|---|---|
| Genome size | 8,759 Mb | 87.5 kB | 8,480 Mb |
| Contigs | 7 | 1 | 42 |
| G+C mole% content | 71.74% | 70.87% | 71.78% |
| Gene count | 7637 | 59 | 7536 |
| Protein coding genes | 7359 | 66 | 7154 |
| RNA genes | 68 tRNAs, 18 rRNAs, 1 ncRNA | – | 69 tRNAs, 18 rRNAs, 5 ncRNAs |
| Biosynthetic gene clusters | 36 | – | 28 |
| NRPS gene clusters | 12 | – | 6 |
| NRPS-PKSI gene clusters | 1 | ||
| Phosphopantetheinyl-transferase genes | 1 | 1 | |
| Acyl carrier protein synthase genes | 3 | 1 | |
| MbtH-like protein genes | 6 | 2 |
Genome structure of S. chrysomallus (S. anulatus var. chrysomallus)
Purified genomic DNA of S. chrysomallus ATCC 11523 was used as input for the construction of a DNA paired end WGS library using the Nextera DNA Library Preparation Kit (Illumina). Sequencing of the library in a paired-end run using the MiSeq desktop sequencer (Illumina) yielded 2,245,616 reads (319.7 Mb). An initial assembly performed with the Roche GS de novo Assembler software (release 2.8) resulted in 364 contigs in 70 scaffolds, with 379 contigs larger than 500 bp in total. Manual inspection of the contig ends revealed lack of coverage at the end of most of the contigs as the cause for the contig breaks. To overcome this problem, a second WGS library was constructed using the TruSeq DNA PCR-Free Library Preparation Kit (Illumina) for paired end library construction and the MiSeq reagent kit v3 (600 cycles) for sequencing, adding 2,879,576 reads (591.7 Mb) to the assembly. The final automatic draft assembly contained 57 contigs in 28 scaffolds with 67 contigs larger than 500 bp in total.
Comparison to several completely sequenced Streptomyces spp. revealed a high degree of synteny between the scaffolds and the respective reference genomes. Using S. collinus Tü 365 9 (GenBank: CP006259.1; PMID: 24140291) as a reference, the linear genome could be assembled in two scaffolds (representing the chromosome and one plasmid) consisting of 7 and 1 contigs, respectively.
The genome consists of a linear chromosome of ~8,759.6 kbp with a G+C content of 71.74% and a linear plasmid of 87.5 kbp with a G+C content of 70.87%. The PGAP pipeline (PMID: 18416670) was used for gene prediction and annotation, resulting in a total of 7,637 predicted genes (7,359 CDS, 68 tRNAs, 18 rRNAs in 6 operons, and 1 ncRNA). According to the antiSMASH38 results, the draft genome of S. chrysomallus contains at least 36 different secondary metabolite gene clusters (Table 1). Similar to S. antibioticus, these are distributed over the whole genome. No secondary metabolite gene clusters were located on the S. chrysomallus linear plasmid. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession JPZP01000000. The version described in this paper is version JPZP01000000.
Structure comparison of actinomycin (acm) biosynthetic gene clusters
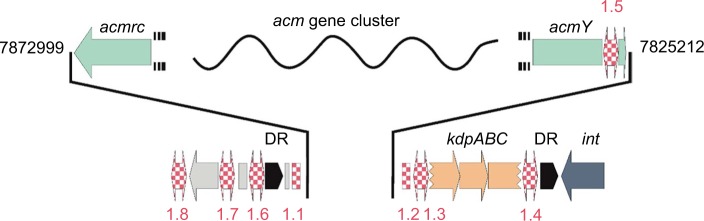
The acm X biosynthetic gene cluster of S. antibioticus IMRU 3720 (accession LHQL00000000) is located between nt positions 8178370 and 8215920 of the draft genome. In S. chrysomallus, the acm C gene cluster is located between nt positions 7825212 to 7872999 of the draft genome (accession NZ_CM003601.1). Both gene clusters thus lie near the end of their chromosomes. In S. chrysomallus, this stands in agreement with the location close to the silent region of the acm biosynthetic gene locus (acm class I/III locus) on the previous genetically established chromosomal map of S. chrysomallus.22,29 In addition, we saw coincidence of the locations of most of the auxotrophic markers with the locations of the corresponding genes on the draft genome.22,29
In S. antibioticus, the acm gene cluster comprises 20 individual CDSs spanning 37.5 kb. Each of these CDSs has an orthologue situated in S. chrysomallus acm C gene cluster, which as a length of 47.8 kb comprising 28 CDSs due to the presence of eight doubly occurring orthologues in left arm (Table 2, Figure 3). The framework formed by the genes in the S. antibioticus acm X gene cluster corresponds to the major region of the S. chrysomallus acm C gene cluster, which spans the center and right arm of the latter except the two orthologues, scacmM and scacmN, that are located in the opposite left arm. This, however, clearly shows perfect identity in the arrangement of all orthologues in the left arm to that of their counterparts in the S. antibioticus acm X gene cluster (Figure 3). As can be seen from Figure 3, both gene clusters are organized into four sets of tandemly arranged genes (apart from two additional ones in the left arm of the S. chrysomallus acm C gene cluster). These are pairwise arranged in opposite orientation to each other. From the inverted orientations of genes at least two promoter regions could be inferred. The first is situated between the operons saacmADRST (scacmADRST) and saacmBCEFG-KLM (scacmBCEFGHI) containing biosynthetic genes and the genes sa(sc)acmE, sa(sc)acmS and sa(sc)acmT, which encode conserved proteins of unknown function (Table 2). Immediately downstream the last gene of each operon is located inverted repeat (IR) structures most probably serving as transcriptional terminators.
Table 2.
Genes of the Streptomyces antibioticus acm X biosynthetic gene cluster and similarities to corresponding protein sequences encoded by orthologues in the Streptomyces chrysomallus acm C biosynthetic gene cluster
| Genes in S. antibioticus | Encoded function | Gene orthologues in S. chrysomallus acm gene cluster left arm/right arm | Protein identity to S. chrysomallus orthologue
|
|
|---|---|---|---|---|
| % Identity | % Similarity | |||
| AFM16_36345 | α/β hydrolase | |||
| AFM16_36350 | Ser/Thr protein kinase | |||
| saacmrC | UvrA-like protein | scacmrC/scacmY | 87/83 | 92/90 |
| saacmrB | ABC 2-type transporter | scacmrB/scacmX | 89/85 | 95/92 |
| saacmrA | ABC transporter ATPase subunit | scacmrA/scacmW | 86/83 | 92/89 |
| saacmQ | Siderophore-interacting protein | scacmQ/scacmV | 82/79 | 89/90 |
| saacmP | TetR family transcriptional regulator | scacmP/scacmU | 77/66 | 83/72 |
| saacmO | LmbU-like protein | scacmO/scacmJ | 77/71 | 84/78 |
| saacmN | Ferredoxin | scacmN | 73 | 83 |
| saacmM | Cytochrome P450 | scacmM | 86 | 90 |
| saacmL | Methyltransferase | scacmL/scacmI | 88/82 | 93/88 |
| saacmK | Kynureninase | scacmK/scacmH | 84/82 | 89/92 |
| saacmG | Tryptophan 2,3-dioxygenase | scacmG | 78 | 82 |
| saacmF | Aryl formamidase | scacmF | 82 | 86 |
| saacmE | Hypothetical protein | scacmE | 82 | 89 |
| saacmC | Peptide synthetase ACMS III | scacmC | 76 | 83 |
| saacmB | Peptide synthetase ACMS II | scacmB | 72 | 81 |
| saacmA | Peptide synthetase ACMS I | scacmA | 75 | 84 |
| saacmD | 4-MHA carrier protein AcmACP | scacmD | 75 | 87 |
| saacmR | MbtH-like protein | scacmR | 80 | 90 |
| saacmS | Hypothetical protein | scacmS | 74 | 85 |
| saacmT | Hypothetical protein | scacmT | 73 | 84 |
| AFM16_36455 | Putative alpha-1,2-mannosidase | |||
| AFM16_36460 | Threonine transporter RhtB | |||
Abbreviations: ACMS, actinomycin synthetase; 4-MHA, 3-hydroxy-4-methylanthranilic acid.
Figure 3.
Alignment of the actinomycin biosynthetic gene clusters of Streptomyces chrysomallus (A) and Streptomyces antibioticus (B). Arrows designate putative start points of transcription. Stem-loop symbols indicate large inverted repeats. Gene designations Keller et al.27
The second promoter region (different in sequence from the first one) is located between the regulatory genes saacmP or (scacmP/U) and the self-resistance genes saacmQ (scacmQ/V). The promoter regions in each gene cluster share high similarity with their counterparts in the other, which is surprising in view of the known different regulation of Acm biosynthesis in Acm X and Acm C producers.10–13,39 Different search programs for promoter sequences indicate an additional promoter upstream saacmO and scacmJ, respectively, which encode a transcriptional regulator (Figure 3). However, as long as these latter promoters are not mapped, their nature remains speculative.
In S. antibioticus, the borders of the acm biosynthetic gene cluster are defined by saacmT on the left side and saacmrC on the right side (Figure 3; Table 2). Beyond these genes, no genes with Acm-biosynthetic relevance were identified (Table 2). In S. chrysomallus, the acm gene cluster ranges from scacmrC to scacmY (as revealed by previous cosmid sequencing) and is surrounded by various insertion-sequence (IS) elements and gene fragments not existent in the flanking regions of the acm gene cluster of S. antibioticus (revealed by genome sequencing in this paper). This indicates lack of synteny between the two genomes in respect of the integration locus of the acm gene clusters.
Comparison of biosynthetic genes and their translation products in S. antibioticus and S. chrysomallus: nonribosomal assembly genes
The overall genetic composition of the two gene clusters did not reveal a conspicuous difference in the underlying basic biochemistry of Acm formation in the two strains. Moreover, searches in the database revealed that the arrangement of biosynthetic genes of peptide backbone assembly (ACMS genes and cohorts) acmABCDR and their immediate neighbors acmST and acmE, respectively, is not only conserved in S. antibioticus and S. chrysomallus but also in the genomes of other streptomycetes, which may therefore be Acm producers, too (e.g., Streptomyces mutabilis strain TRM45540, JNFQ00000000.1). A duplicated ACMS gene assembly (mutually missing acmA/D or acmR in their repeats) is present in the acm gene clusters of Streptomyces iakyrus, a producer of Acm G or of Streptomyces fradiae, a producer of AcmY (unpublished materials; Crnovčić et al. The genome sequence of the actinomycin Y producing Streptomyces fradiae).40
Comparison of translation products of the S. antibioticus and S. chrysomallus ACMS genes revealed that they shared high similarity in sequence and length (between 72% and 88% identity). The only one notable difference was in the sequence of the substrate specificity-determining amino acid residues of A-domains of module 2 of the ACMS IIs (Table 3).
Table 3.
Substrate specificity determining residues of ACMS modules
| Nonribosomal codons in
| ||
|---|---|---|
| Module | Streptomyces chrysomallus | Streptomyces antibioticus |
| ACMS II | FWNVGMVH (threonine) | FWNVGMVH (threonine) |
| ACMS II | AYFWGVTF (isoleucine) | AYWWGVTF (valine) |
| ACMS III | VQFAAHVV (proline) | VQFAAHVV (proline) |
| ACMS III | ILQLGLIW (glycine) | ILQLGLIW (glycine) |
| ACMS III | AYWWGVTF (valine) | AYWWGVTF (valine) |
Abbreviation: ACMS, actinomycin synthetase.
The slight deviation in their nonribosomal code stands in agreement with the previously experimentally determined substrate specificities of the ACMS IIs from S. chrysomallus and S. antibioticus, which prefer l-isoleucine or l-valine, respectively, as substrates.16,39 In fact, only d-valine is present in position 2 of the peptide chains of all members of the Acm X complex in S. antibioticus but not d-allo-isoleucine, which on the other hand can be incorporated in the peptide chains of the Acm C complex from S. chrysomallus (Figure 1).
4-MHA biosynthesis in S. chrysomallus and S. antibioticus
Similar to the ACMSs genes and their cohorts, strict orthology was noticed also for their downstream genes in both acm gene clusters. The presence of saacmF, saacmG, saacmK and saacmL, which encode tryptophan dioxygenase, kynurenine formamidase, hydroxykynureninase and 3-hydroxykynurenine-4-methyltransferase, respectively, strongly suggests that the biosynthesis of 4-MHA in both S. antibioticus and S. chrysomallus is the same (Figure 2; Table 2). The role of the corresponding orthologues of these genes in 4-MHA synthesis in S. chrysomallus and S. parvulus was clearly proven.19–22,27,39,41 Importantly, the data show that the methyl group of 4-MHA similar to S. chrysomallus stems from the methylation of 3-hydroxykynurenine (3-HK) by 3-hydroxykynurenine-4-methyltransferase encoded by saacmL (scacmL) delivering 3-hydroxy-4-methylkynurenine (4-MHK). It was shown recently that the S. chrysomallus orthologue scAcmL does not methylate 3-hydroxyanthranilic acid.41 This most likely excludes that its S. antibioticus orthologue saAcmL is identical with the previously isolated 3-HA methyltransferase (HAMT) of S. antibioticus, which can methylate 3-hydroxyanthranilic acid to give 4-MHA (Figure 4).42 HAMT appears to be a nonpathway-specific enzyme whose sequence and genomic location in S. antibioticus remains unknown.
Figure 4.
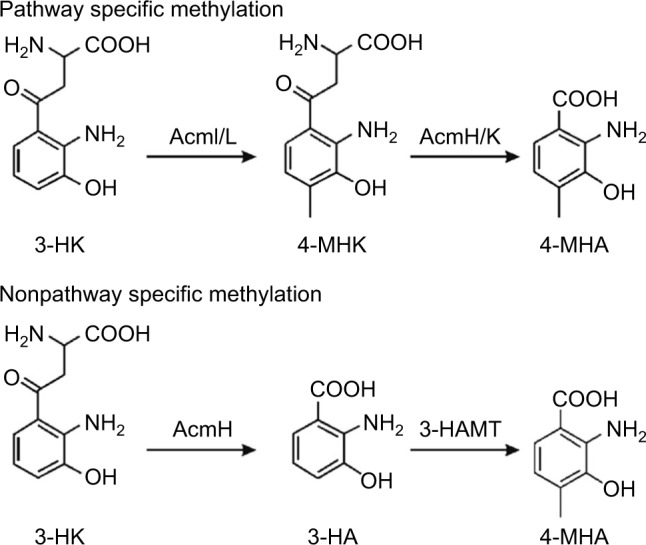
Reaction schemes of 3-HK-4-methyltransferase (AcmI or AcmL) and of 3-HA-4-methyltransferase (HAMT) in the formation of 4-MHA. The upper scheme includes the pathway specific conversion of 4-MHK to 4-MHA, whereas the lower scheme includes nonpathway specific methylation step prior conversion of 3-HK to 3-HA.
Abbreviations: 4-MHA, 3-hydroxy-4-methylanthranilic acid; 3-HK, 3-hydroxykynurenine; 4-MHK, 3-hydroxy-4-methylkynurenine.
Interestingly, similar to S. chrysomallus, the acm gene cluster of S. antibioticus does not contain a gene encoding a kynurenine-3-monooxygenase (KMO) necessary for synthesis of 3-HK (Figures 2 and 3). This indicates that the earlier noted absence of this gene in the acm C biosynthetic gene cluster is for natural reasons.27 3-HK is an obligatory intermediate in 4-MHA formation and can be formed from kynurenine only enzymatically (Figure 2). Remarkably enough, the biosynthetic gene clusters for the antitumor compounds anthramycin and sibiromycin from Streptomyces refuineus and Streptosporangium sibiricum, respectively, besides orthologues of saacmF, saacmG, saacmK and saacmL indeed both contain a KMO gene (orf23 and sibC, respectively).43,44 Similar to the case of the Acm chromophore, 4-MHA is a building block of anthramycin and sibiromycin structures. Therefore, the presence of a KMO gene situated somewhere outside the acm gene clusters on the genomes of S. antibioticus and S. chrysomallus is necessary. BLASTP searches using as queries the sequences of Orf23, SibC and in addition of QbsG, a KMO from the gram-negative Pseudomonas fluorescens involved in the biosynthesis of the siderophore quinolobactin,45 indeed revealed a single KMO gene, JI76_02815, in the S. chrysomallus genome. Its translated sequence has significant similarity and comparable length to Orf23, SibC and QbsG and other KMOs in the database (35%, 30% and 40% identity, respectively). It is located in the biosynthetic gene cluster of a secondary metabolite of unknown structure (nt positions 663951–661838). This gene could be a possible candidate for gene sharing with the acm C biosynthetic gene cluster.
Searches for a KMO gene in S. antibioticus did not reveal an orthologue encoding a monooxygenase with significant similarity to ORF23, SibC or QbsG (Figure S1). The existence of non-orthologous bacterial KMO has been postulated for the tryptophan degradation pathway in Burkholderia cepacia.46 However, since the corresponding gene in Burkholderia sp. is not yet known, the presence of a non-orthologous KMO gene in S. antibioticus remains to be verified. In any case, sequence comparisons of those flavine-dependent monooxygenases from S. antibioticus with >25% identity did not allow to determine a suitable candidate for the KMO gene involved in Acm X biosynthesis (Figure S1).
Cytochrome P450 monooxygenase gene acmM and ferredoxin gene acmN mark a biochemical difference between acm C and acm X biosynthetic gene clusters
saacmM and saacmN of S. antibioticus acm X gene cluster like their orthologues scacmM and scacmN in the left arm of the S. chrysomallus acm C gene cluster lie immediately downstream of the 4-MHA biosynthesis genes saacmL and scacmL, respectively (Table 2; Figure 3). They encode a cytochrome P450 and a ferredoxin (Table 2).27 Previous determination of scacmM and scacmN sequences in S. chrysomallus had revealed that both genes are pseudogenes encoding nonfunctional proteins.27 Remarkably, the sequences of their orthologues in S. antibioticus do not show mutations in their sequences and therefore seem to encode functional full-length proteins. The defect of scacmM and scacmN in S. chrysomallus indicates the only relevant biochemical difference between Acm biosynthesis of S. antibioticus and S. chrysomallus and suggests their specific role in Acm X biosynthesis and not in that of Acm C. These two families of Acms differ in their structures solely in the presence of some rare amino acid residues in their peptide chains (Figure 1).
AcmE and AcmT each belongs to subfamilies of conserved proteins of unknown function
saacmT and saacmE like their S. chrysomallus orthologues scacmT and scacmE flank the peptide synthetase gene ensembles in both gene clusters. They were noted previously to encode conserved proteins of unknown functions.27 The translated sequences of scacmE and scacmT shared 46% identity with each other, whereas their similarity to their orthologues saacmE and saacmT in S. antibioticus is 88% and 82% identity, respectively. Surprisingly, despite the similarity between the acmEs and acmTs BLASTP searches using translated sequences of sc(sa)acmE or sc(sa)acmT as queries revealed that each of them belongs to a distinct subfamily of conserved proteins (Figure S2). In fact, acmE and acmT are also both present in the acm G gene clusters of S. iakyrus40 the acm Y gene cluster of S. fradiae (unpublished materials; Crnovčić et al. The genome sequence of the actinomycin Y producing Streptomyces fradiae), the acm gene cluster of S. mutabilis (acc. no. JNFQ00000000.1) and several as yet not annotated gene clusters from different streptomycetes in the database. The simultaneous presence of both acmT and acmE in all of these gene clusters may not be accidental and suggests for each of the two homologues a distinct role in Acm biosynthesis (Figure S3). Moreover, the analogy in the location of saacmT and scacmT in the two acm biosynthetic gene clusters of S. antibioticus and S. chrysomallus indicates that scacmT most probably marks the end of the original framework of the S. chrysomallus acm C gene cluster, to which the left extra arm once became attached (see Genetic origin of the S. chrysomallus acm biosynthetic gene cluster section).
Regulatory gene sequences
The acm X biosynthetic gene cluster carries in its right side (Figure 3) two invertedly oriented block of genes each containing the same regulatory genes and self-resistance genes as in the acm C gene cluster of S. chrysomallus.27 saacmO is orthologous to lmbU, the representative of a small family of transcriptional activator genes first detected in the lincomycin biosynthetic gene cluster from Streptomyces lincolnensis.47 Other members of the family are novE of the novobiocin biosynthetic gene cluster from Streptomyces spheroides or hrmB of the hormaomycin biosynthetic gene cluster.48,49 NovE – in conjunction with an unknown protein factor encoded by the genome of S. spheroides – was shown to activate the transcription of the pathway-specific regulatory genes novG.50,51 NovG, a homologue of the well-known transcriptional activator StrR,52 then in turn activates the transcription of all synthetic genes of the novobiocin gene cluster in a single large transcript.53 Importantly, however, NovE can positively regulate novobiocin biosynthesis alone when NovG is absent.53 Neither a novG homologue is missing in the acm biosynthetic gene clusters of both S. antibioticus and S. chrysomallus nor is present any target sequences for NovG-or StrR-like transcriptional activators, it may be assumed that saAcmO or scAcmO(J) can positively regulate acm biosynthetic genes on their own in conjunction with unknown factors provided by their genomes. In fact, it was previously reported that lincomycin biosynthesis in S. lincolnensis most likely is directly regulated by LmbU, too.54
scacmP, such as its orthologues scacmU and saacmP all lying upstream of saacmO and scacmO or scacmJ, respectively, has similarity to TetR-like repressors from a variety of streptomycetes.55 The role of these repressors in the regulation of Acm biosynthesis or Acm self-resistance is currently investigated in this laboratory.
Further search for regulatory sequences in the acm biosynthetic gene clusters revealed the presence of a TTA leu codon in the gene scacmC encoding ACMS III in S. chrysomallus. It is located in codon position 19 (of 4248 codons total length), whereas the orthologue saascmC from S. antibioticus has a phe TTC codon in that position. The TTA codon is the rarest codon in streptomycetes and is decoded by the tRNA BldA.56 Interestingly, comparison of the gene sequences of scacmB and saacmB encoding ACMS II revealed a TTG start codon for the S. antibioticus gene, whereas in case of S. chrysomallus that gene has a GTG start codon. The TTG start codon is a rare start codon in prokaryotes and is present in ~3% Escherichia coli genes or 4% of genes of Streptomyces coelicolor.57 Its significance in the expression of Acm X biosynthesis is not clear.
Selfresistance genes
From all genes of the S. antibioticus acm gene cluster, the four genes saacmQ, saacmrA, scacmrB, scacmrC are the most similar to genes in the acm C gene cluster of S. chrysomallus (80%–92% identity; Table 2). saacmQ encodes a siderophore interacting protein, whereas saacmrA and saacmrB are typical resistance genes encoding the subunits of an ABC transporter (AcmrA, AcmrB) involved in drug export.27 AcmrC similarly encodes resistance protein, which is an excinuclease with similarity to UvrA from E. coli most probably involved in DNA repair.27 Homologues of the latter genes have been found in the biosynthetic gene clusters of compounds such as daunomycin, which like actinomycin intercalate into DNA.58 While saacmrA, scacmrB, scacmrC clearly encode known self-resistance mechanisms against drugs, the role of the siderophore interacting protein encoded by saacmQ (or scacmQ/V) is still unclear. A role of iron in the mechanism of actinomycin is not known.
PHS genes in S. antibioticus and S. chrysomallus
Phenoxazinone formation as final step in actinomycin biosynthesis in S. antibioticus has for long been attributed to the action of PHS, a 650 aa residues two copper centers-containing phenoloxidase.13,15,59 However, later it was found that Acm X production persisted in a ΔphsA mutant of S. antibioticus IMRU 3720, which indicated the dispensability of the enzyme for Acm biosynthesis.60 No substitute for PHS was identified because protein extracts of that ΔphsA mutant did not contain any PHS activity.60 The PHS gene phsA (AFM16_29460) is not contained in the acm biosynthetic gene cluster of S. antibioticus and is located to the core region of the genome directly downstream of the geosmin synthase gene cyc2 (AFM16_29455). No orthologue of phsA was detected in the S. chrysomallus genome. Instead, BLASTP searches revealed as most similar enzyme a laccase gene, encoding a protein of 611 aa residues (JI76_35910) and showing 46% identity to PHS in the carboxyterminal 400 aa. The laccase gene is situated in a biosynthetic gene cluster of an unknown secondary metabolite (nt position 826570). Like PHS, lac-cases can catalyze formation of phenoxazinones in vitro and in vivo.61,62 However, no laccase enzyme activity – using 3-HA as substrate – converting 3-HA to colored products was detectable in protein extracts from S. chrysomallus thierefore excluding involvement of the laccase gene in phenoxazinone formation of Acm biosynthesis.
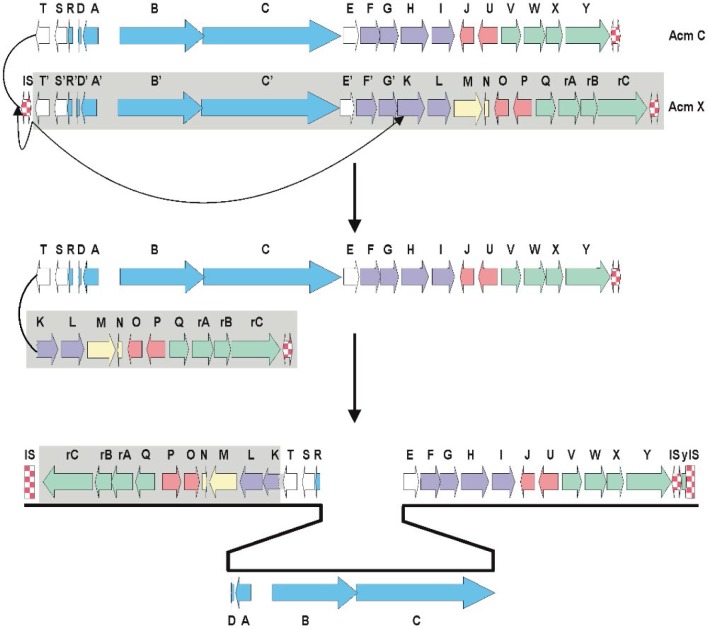
Genetic origin of the S. chrysomallus acm biosynthetic gene cluster
Frame plot analysis of genes in the left and right arm of the S. chrysomallus acm biosynthetic gene cluster (Figure 3) showed different G+C mole% contents. Each orthologue in the left arm (from scacmK to scacmrC) had a 2.5% lower G+C-contents than their counterparts in the right arm (average 69.4 G+C mole % vs. 71.9 mole%; Table S1). This indicates that the entire left arm ranging from acmK until acmrC stems from a different genetic background. In view of the additional presence of the pseudogenes scacmM and scacmN in that left arm, which is a perfect copy of the single arm of the acm X biosynthetic gene cluster of S. antibioticus (Figure 3), it can be argued that this arm was a previous part of an acm X biosynthetic cluster donated by a foreign streptomycete such as S. antibioticus. In fact, calculations of G+C mole%-contents of genes and in third codon-positions along with sequence alignments of protein sequences encoded by the orthologues in all three arms of the S. chrysomallus and S. antibioticus acm biosynthetic gene clusters showed that encoded protein sequences from the left arm were more similar to their orthologues in S. antibioticus than to their orthologues from the right arm of their own gene cluster (Table 2). From this we infer that the S. chrysomallus acm C biosynthetic gene cluster arose from an original minimal acm C gene cluster framework ranging from scacmT to scacmY to which was fused the left arm during its evolution. Meanwhile, further searches in the data bank revealed indeed the existence of such one-armed acm biosynthetic gene cluster not having orthologues of acmM and acmN (S. mutabilis strain TRM45540, JNFQ00000000.1).
Model of formation of the acm C gene cluster and factors contributing to its stability in S. chrysomallus
The opposite orientation of the arms of the S. chrysomallus acm C biosynthetic gene cluster implies a previous head-to-head orientation of two different one-armed acm biosynthetic gene clusters (Figure 5). Possibly the two clusters could have lain side-by-side as result of matings between different Acm producers or by transmission of a second gene cluster located on a mobile genetic element.
Figure 5.
Model of fusion of two different single-armed biosynthetic gene clusters from an Acm X producer (lower gene cluster) and an Acm C producer, leading to the present double-armed acm C gene cluster of Streptomyces chrysomallus.
Subsequent recombination between the long inverted arms of the resultant huge palindrome seems unlikely in view of the high conservation of the different G+C mole% contents in full length of the two arms. Therefore, we propose unsymmetrical excision of the largest part of the putative acm C cluster portion in the primary fusion product, that is from acmT’ to acmG’ as depicted in Figure 5. This could have been catalyzed by the same IS elements initially responsible for side by side combination sitting in the flanks and between the two acm gene clusters. In fact, remnants of IS-elements (ISSc1.1 and ISSc1.2) are still visible in the direct neighborhood of the scacmrC- and scacmY-ends of the present S. chrysomallus acm biosynthetic gene cluster (Figures 3 and 6). Imprecise excision may be indicated by the fact that scacmK, the first gene of the left arm has lost the first 107 codons of its original sequence by comparison with the sequence of its orthologue scacmH in the right arm (Figure 5). No coding sequences, promoters or remnants of an IS-element are contained in the 108 nt long stretch between the first detectable codon (108) of scacmK and the stop codon of the preceding scacmT which suggests that the truncated scacmK and its downstream genes including scacmM most probably will not be transcribed. A 22 nt IR downstream of scacmT may mark the previous transcriptional terminator at the end of the right acm gene cluster. In fact, a similar IR is situated at 37 nt after the stop codon of saacmT in the S. antibioticus acm gene cluster (Figure 5). On the other hand, inspection of the promoter regions of the self-resistance genes scacmQrArBrC and of the regulatory genes scacmO and scacmP transcription of these genes cannot be disregarded. Remarkably, the gene scacmY located at the opposite end of the acm C biosynthetic gene cluster encoding a self-resistance gene most probably involved in DNA repair is disrupted by integration of an IS element. Its function could be overtaken by its orthologue scAcmrC as a kind of functional complementation between the two arms in the S. chrysomallus acm C gene cluster, which may select for maintenance of its two-armed structure.
Figure 6.
Physical map of flanking regions of the acm biosynthetic gene cluster of Streptomyces chrysomallus. The borders of the gene cluster are indicated by their genomic coordinates. The various IS elements or IS element fragments are symbolized by red and white checks. IS elements with inverted repeats are with arrows. Hypothetical proteins are in gray. IS elements are hatched. Terminal genes of the acm gene cluster are in blue, disrupted kdpABC is in beige, and DR are in black. Physical map is not at scale.
Abbreviation: DR, directed repeat.
IS elements and directed repeats (DRs) flank the acm biosynthetic gene cluster in S. chrysomallus
Sequence analysis of the flanking regions of the acm gene cluster of S. chrysomallus revealed two 165 nt DRs and several IS-elements to both sides (Figure 6). The DRs share 69% identity and possess in their central part three purine-rich stretches each of 22 nt length with 90% and 95% identity. They cover four tandemly arranged motifs with the consensus TGGGGAG and at some distance two tandem motifs with the consensus GAAAGA (Figure S4). The significance of these motifs is not knownl; however, highly similar DRs at more than 50 kb distance and their uniqueness in the S. chrysomallus genome may not be accidental. DRs flanking a biosynthetic gene cluster has been reported in case of the mithramycin biosynthetic gene cluster Streptomyces argillaceus, which indicated previous Campbell-type integration of the gene cluster into the genome from a phage or plasmid.63 The presence of various IS elements within and at the periphery of the acm biosynthetic gene cluster indicates transpositional events in the evolution of the gene cluster and/or its transmission to its present host. The presence of IS-elements in secondary metabolite gene cluster has been described as an indication of transposition as a tool to generate biosynthetic and structural diversity.64 Moreover, transposition of antibiotic or other secondary metabolite gene clusters within bacterial species in the form of composite IS elements (transposons) has been shown as a means to spread genetic diversity in virulence and defense mechanisms among bacterial species.65
All IS elements and fragments flanking the two-armed acm gene cluster are listed in Table S2. BLAST searches using IS finder35 revealed that they belong to the IS3 and IS5 families of IS elements. The IS elements, ISSc1.1 and ISSc1.2 are fragments, which may be older than the next-coming IS elements ISSc1.3 and ISSc1.4, which both possess IRs and DRs. ISSc1.3 and ISSc1.4 most probably were part of a composite transposon carrying a truncated kdpABC operon that was integrated to the right side of the acm biosynthetic gene cluster into the flank of ISsc1.2. A single alone-standing kdpED, possibly the rest of the previous kdpABCED was found to still reside at nt position 6672757 (i.e., one Mb from the acm biosynthetic gene cluster) neighboring a trkA operon (nt position 6678844). Nevertheless, a complete kdpABCED operon is still present in the genome at nt position 5415894. Interestingly, the gene scacmY is truncated by an IS element (ISSc1.5), which is highly similar to ISSc1.3 and like the latter has CTAG as DR. There are further CTAG sequences within the gene cluster as well as in the flanking regions such as one flanking the IS element ISSc1.7 (Figure 6). A single non-targeted CTAG lies between the right DR and an integrase gene (intJI76_33900) to its right side. Two other IS elements (ISSc 1.6 and 1.8) had no recognizable DR but IRs. However, no IRs could be seen flanking the acm C gene cluster of S. chrysomallus, which excludes that the gene cluster is still transmissible as it is the case for the gene cluster encoding the nonribosomal peptide cereulide in some pathogenic bacilli.65
Conclusion
S. antibioticus and S. chrysomallus are representatives of the two main groups of Acm-producing streptomycetes distinguished by the production of Acm X and Acm C, respectively. Acm X and Acm C are mixtures of Acms which differ from each other by specific amino acid substitutions in the “proline” and “d-valine” site, respectively, of their two pentapeptide chains (Figure 1). The acm X biosynthetic gene cluster from S. antibioticus – presented here – has only one single arm lying to the acmE-side of the peptide synthetase genes ensemble, whereas the bi-armed acm C biosynthetic gene cluster of S. chrysomallus has an additional extra arm containing a number of orthologues also present in the principal arm of the gene cluster. Nevertheless, both gene clusters have in common a framework defined by the genes scacmT (resp. saacmT) and scacmY (resp. acmrC) comprising 20 genes in the acm X gene cluster of S. antibioticus and 18 genes in the S. chrysomallus acm C gene cluster (Figure 3). The clue to the significance of the extra arm of the S. chrysomallus acm C gene cluster came from the observation that the orthologues scacmM and scacmN, which encode a cytochrome P450 monooxygenase and its ferredoxin (Figure 3), are sitting in the left (extra) arm, which turned out to be identical in all ortholgues to the arm of the S. antibioticus acm X gene cluster. Further analyses revealed indeed differences between the extra and principal arm such as different G+C contents of orthologues in each arm and – importantly enough – the higher similarities of extra arm-gene products with those of their counterparts in the S. antibioticus acm X gene cluster rather than to those encoded by the principal arm. These data suggested, that the extra arm was derived from a gene cluster originating from a foreign streptomycete, which most probably could have been a producer of Acm X or related Acms.
The fact that S. chrysomallus does not produce Acm X led to inspect the sequences of scacmM and scacmN in the acm C biosynthetic gene cluster, which revealed that they are pseudogenes due to mutations in their sequences. Their orthologues in S. antibioticus showed no mutations and most probably encode normal full-length proteins. Therefore, they may encode a biosynthetic step typical for Acm X biosynthesis but not Acm C biosynthesis. The only differences between the AcmCs and AcmXs lie in the presence of rare amino acids such as 4-hydroxy- or 4-oxoproline in the peptide chains of Acm X. Whether the inability of S. chrysomallus to produce Acms with 4-hydroxyproline and 4-oxoproline residues in their peptide chains is due to its inability to express functional translation products of scacmM and scacmN, remains to be seen.
The comparative analyses of the acm X and acm C biosynthetic gene clusters prompted to derive a model of formation of the bi-armed acm C gene cluster in S. chrysomallus. From the orientation of the extra arm and also from the truncated scacmK gene at the front of the extra arm an imprecise excision event may have removed a large part of a previous acm biosynthetic gene cluster situated in head-to-head orientation left to an original acm C gene cluster (Figure 5). The excision resulted in connecting the remaining left arm directly to the downstream region of scacmT as depicted in Figure 5. The presence of various IS-elements in the flanking regions of the acm C gene cluster together with the presence of long DRs indicates its later transmission and insertion into a hot spot region of transposition on the S. chrysomallus most probably with the involvement of a mobile genetic element. Transpositional fusions of gene clusters with involvement of plasmid or phages may be a means to generate higher diversity of compound structures. Such fusions may also have happened in case of the more complex acm biosynthetic gene clusters such as those for acm G40 or acm Y (unpublished materials; Crnovčić et al. The genome sequence of the actinomycin Y producing Streptomyces fradiae), which also show duplicated gene sets in their sequences. The increasing number of known composite Acm biosynthetic gene clusters stands in agreement with the wide-spread occurrence of Acm production among members of the genus Streptomyces.66
Supplementary materials
Description of experimental procedures
Preparation of large genomic DNA fragments
Streptomyces chrysomallus and Streptomyces antibioticus were grown in liquid CM as described previously.1–3 The cultures contained in addition 0.5% glycine. After 36 hours of growth, mycelium was harvested by suction on a Buechner funnel and washed with distilled water. A total of 1.6 g wet weight myceli of each strain were protoplasted in medium P.4 The protoplasts were washed with the same medium and finally concentrated into ~0.5 mL medium P. The suspension was then portionwise (50 µL) added to 10 mL 0.2 M ethylenediaminetetraacetic acid (EDTA), pH 8, 0.2% sodium dodecyl sulfate (SDS). After each addition of protoplasts, the solution was gently inverted several times to ensure complete lysis of protoplasts. RNase (50 µg f.c.) was added and the solution was incubated for 1 hour at 37°C. DNA was extracted and reextracted by gentle shaking with phenol and chloroform as described in reference.4 After addition of 0.3 M NaOAc, the DNA was spooled onto a glass rod from the interphase overlayered with absolute ethanol and after drying redissolved in tris EDTA (TE) buffer. Repeated precipitation of the DNA sample with ethanol and/or polyethylene glycol (PEG) 6000 yielded 3.8 mg S. chrysomallus DNA and 2.8 mg of S. antibioticus DNA (260/280=1.67 and 1.58, respectively). Agarose gel electrophoresis (0.6% in Tris/Borate/EDTA [TBE]) revealed bands of both chromosomal DNAs corresponding to approximate sizes of ~50 kb (standards used: λ DNA, λ DNA PstI digest). Quantitation of the DNA was photometrically at 260 nm. The concentration of the DNA was adjusted to 50 ng µL−1 for genome sequencing.
Gap filling by polymerase chain reaction (PCR)
Due to non-overlapping ends of respective contigs the sequences of peptide synthetases genes acmB and acmC in both S. antibioticus and S. chrysomallus each had a gap in their central portions. In case of S. chrysomallus, the total sequences of these genes were already known by previous cosmid sequencing.5 In case of the S. antibioticus genes, the gap was closed by PCR using S. antibioticus chromosomal DNA as template. The primers used were for acmB acmB_f: CGCACGAACTCACGTAGATGTTCC and acmB_r: CTGCACGACACCATCACCACTCAG. For acmC, the primers were acmC_f: TCCC GAGTACAGGGAGTCGTAGAG and acmC_r: CAGCTCCCTCCTCAACCTCATCAC.
>acmB gap sequence
CTGCACGACACCATCACCACTCAGCGCGTCACGGCGCTCTGGCTCACCAGCTCCCTCCTCAACCTCGTCACCGACCACACCCCCGAAACACTCGCCCACGTACACCAGGTCTGGACCGGCGGCGAAGCCGTCTCCGGCGCGACCGTCCAACAACTCCAGCGCGTTTGCCCGGCGTTGACCGTCATCGACGGGTACGGCCCCACCGAGACCACCACCTTCGCCACCCACCACCCCGTCCCCCGCCCTTACACCGGCAACCCCACCGTCCCCATCGGCCGCCCCATGGCCAACACCCGCACCTACGTACTCGACCCCCACCTCCAACCTGTCCCGCCCGGCGCCACGGGCGAACTGCACATCGGCGGCGCCGGTCTCGCCCGCGGTTATCTGAACCGGCCGGGACTGACCGCGGAACGCTTCATCGCCGACCCCTACGCCAGCGAACCCGGCGCCCGCATGTACCGCACCGGCGACCTGGTGCGCTGGAACGCCGACGGCGATCTCGAGTACCTGGGCCGCAGCGACCACCAGCTCAAGATCCGCGGCTTCCGCATCGAACCCGGCGAGATCGAGAACACCCTCACCGACCACCCCGACATCACCCAAGCCGCCGTCATCGCACACCACGACGAGCCCGGCAACCCTCGACGCCTCACCGCATACGTCGTCGCCGACACCGCCGTAACGCCGGAACATCTACGTGAGTTCGTGCG
>acmC gap sequence
CAGCTCCCTCCTCAACCTCATCACCGACCACACCCCCGAAACACTCACCCACGTACACCAGGTCTGGACCGGCGGCGAAGCCGTCTCCGGCGCGACCGTCCAACAACTCCAGCGCGCTTGCCCGGCGTTGACCGTCATCGACGGCTACGGCCCCACCGAGACCACCACCTTCGCCACCCACCACCCCGTCCCCCGCCCCTACACCGGCAACCCCACCGTCCCCATCGGCCGCCCCATGGCCAACACCCGCACCTACGTACTCGACACACAACTCCGCCCCGTACCGCCCGGCGTGACCGGAGAGCTGTACATCGCCGGCGCCCATCTGGCCCGCGGCTACCTCGGCAAGCCCCATCTCACCGCCGAACGCTTCGTCGCCGACCCCTACGCCACCGAACCCGGCGCCCGCATGTACCGCACAGGCGACCTCGCACGCCACAACCCCGACGGCGACCTCGAATACCTCGGCCGCACCGACCACCAGCTCAAGATCCGTGGCTTCCGCATCGAACCCGGCGAGATCGAGAACACCCTCACCGACCACCCCGACATCGCCCAGGCCGCCGTCGACCAGCCCGGCGACACCCGCCTCACCGCCTACATCGTGCCCGACACCACCGACACGGCGAAGGACGACGAGGAGCGAAGCCAGATCGGCGAGTGGCAGGATCTCTACGACTCCCTGTACTCGGGA
Alignments and nearest neighbor tree of different flavine monooxygenase protein sequences from Streptomyces antibioticus (AFM16) and Streptomyces chrysomallus (JI76) with similarity to kynurenine-3-monooxygenase. Included are bacterial KMO sequences Orf23 from Streptomyces refuineus (ABW71854.1), SibC from Streptosporangium sibiricum (ACN39726.1), QbsG from Stenotrophomonas maltophilia (CRX68935.1). The tree consists of two main clades, of which the upper clade has a subclade (shaded) containing conserved KMO sequences. Sequences in the other subclade are less similar to KMOs having 30% identity and less to SibC or Orf23. Red numbers denote branch values representing a measure of support for the node. The bar at the bottom denotes phylogenetic distance.
Abbreviation: KMO, kynurenine-3-monooxygenase.
Alignments of AcmT and AcmE sequences from Streptomyces chrysomallus (sc), Streptomyces antibioticus (sa) and Streptomyces mutabilis (WP_043377479.1 and WP_052412530.1, respectively).
Nearest neighbor tree of AcmE and AcmT sequences from different streptomycetes.
Notes: From the listed strains sa (Streptomyces antibioticus), sc (Streptomyces chrysomallus), Streptomyces mutabilis and Streptomyces iakyrus are known Acm producers. The tree shows two main clades each for AcmE or AcmT sequences. Accession numbers are: Streptomyces flavovariabilis (WP_031142912.1); Streptomyces variegatus (WP_052686596.1); S. iakyrus E (CCO61882.1); S. mutabilis E (WP_052412530.1); Streptomyces sp. CNS654 (WP_032769128.1); Streptomyces sp. NRRL F-5008 (WP_051781089.1); S. mutabilis T (WP_043377479.1); Streptomyces sp.1 (WP_030987180.1); Streptomyces sp.2 (WP_030653030.1); S. iakyrus T (WP_033306586.1); Streptomyces sp. NRRL WC-3742 (WP_051838397.1). Red numbers denote branch values. The bar at the bottom indicates phylogenetic distance.
Sequences of directed repeats (DRs) flanking the acm biosynthetic gene cluster of Streptomyces chrysomallus.
Table S1.
Comparisons of G+C mole% contents of genes in the acm biosynthetic gene clusters of Streptomyces antibioticus and Streptomyces chrysomallus
| Gene in S. antibioticus | % G+C | 3rd-letter % G+C | Gene in S. chrysomallus | Gene in S. chrysomallus
|
|
|---|---|---|---|---|---|
| % G+C left arm/right arm | 3rd-letter % G+C left arm/right arm | ||||
| saacmrC | 68.9 | 94 | scacmrC/scacmY | 67.1/70.1 | 91.1/95.8 |
| saacmrB | 69.4 | 95.3 | scacmrB/scacmX | 68.2/71.5 | 91.4/97.3 |
| saacmrA | 72.1 | 90.8 | scacmrA/scacmW | 71.6/73.5 | 89.3/95.4 |
| saacmQ | 73 | 95.6 | scacmQ/scacmV | 72.2/73.7 | 92.5/98.7 |
| saacmP | 73.8 | 90.7 | scacmP/scacmU | 70.3/74 | 85.1/91.5 |
| saacmO | 71.6 | 92.6 | scacmO/scacmJ | 69.4/72.5 | 83.3/90.5 |
| saacmN | 70.5 | 81.2 | scacmN | 72.5 | 92.8 |
| saacmM | 71.7 | 91 | scacmM | 69.6 | 87 |
| saacmL | 68.1 | 91 | scacmL/scacmI | 69.1/70.5 | 93.4/97.4 |
| saacmK | 72.5 | 89 | scacmH/scacmK | 74.1/72.8 | 95/90.6 |
| saacmG | 70.8 | 92.3 | scacmG | 70.8 | 94.7 |
| saacmF | 74.4 | 90.6 | scacmF | 75.7 | 94.8 |
| saacmE | 70.3 | 92.9 | scacmE (acmT) | 70.1 (68.2) | 90.3 (93.4) |
| saacmC | 70.5 | 89.8 | scacmC | 73.2 | 95.6 |
| saacmB | 71.2 | 88.6 | scacmB | 73.4 | 94.3 |
| saacmA | 70.4 | 88 | scacmA | 71.5 | 90.7 |
| saacmD | 62.4 | 88.6 | scacmD | 66.7 | 97.4 |
| saacmR | 68.2 | 92.5 | scacmR | 69.7 | 97 |
| saacmS | 66 | 88.8 | scacmS | 68.4 | 95.7 |
| saacmT | 69.2 | 93.4 | scacmT (scacmE) | 68.2 (70.1) | 93.4 (90.3) |
Table S2.
IS elements flanking the acm biosynthetic gene cluster of Streptomyces chrysomallus
| Name | Length | Family | Highest similarity to | Length |
|---|---|---|---|---|
| ISSc1.1 | 421 (fragment) | IS5 | ISStu1 (Streptomyces turgidiscabies) AY707081 | 899 bp (4e−05) |
| ISSc1.2 | 189 (fragment) | IS481 | ISsco1 (Streptomyces coelicolor) AF099014 | 1057 bp (3e−20) |
| ISSc1.3 | 882 | IS5 | IS1650 (S. coelicolor) AL079356 | 898 bp (9e−11) |
| ISSc1.4 | 1300 | IS3 | ISTesp3 (Terrabacter sp.) AP008980 | 1373 bp (1e−16) |
| ISSc1.5 | 1071 | IS5 | IS1648 (S. coelicolor) AL023861 | 969 bp (4e−07) |
| ISSc1.6 | 1622 | IS3 | IS1372 (Streptomyces lividans) U50076 | 1304 bp (0) |
| ISSc1.7 | 1397 | IS701 | ISSav3 (Streptomyces avermitilis) NC_003155 | 1400 bp (0) |
| ISSc1.8 | 779 | IS5 | IS1647 (S. coelicolor) AL023702 | 878 bp (8e−48) |
References
- 1.Keller U, Krengel U, Haese A. Genetic analysis in Streptomyces chrysomallus. J Gen Microbiol. 1985;131(5):1181–1191. doi: 10.1099/00221287-131-5-1181. [DOI] [PubMed] [Google Scholar]
- 2.Haese A, Keller U. Genetics of actinomycin C production in Streptomyces chrysomallus. J Bacteriol. 1988;170(3):1360–1368. doi: 10.1128/jb.170.3.1360-1368.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katz E, Pienta P, Sivak A. The role of nutrition in the synthesis of actinomycin. Appl Microbiol. 1958;6(4):236. doi: 10.1128/am.6.4.236-241.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hopwood DA, Bibb MJ, Chater KF, et al. Genetic Manipulation of Streptomyces. A Laboratory Manual. Norwich, UK: John Innes foundation; 1985. [Google Scholar]
- 5.Keller U, Lang M, Crnovčić I, Pfennig F, Schauwecker F. The Actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: a genetic hall of mirrors for synthesis of a molecule with mirror symmetry. J Bacteriol. 2010;192(10):2583–2595. doi: 10.1128/JB.01526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Goss WA, Katz E, Waksman SA. Changes in the composition of an actinomycin complex during growth of a Streptomyces culture. Proc Nat Acad Sci USA. 1956;42(1):10–12. doi: 10.1073/pnas.42.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brockmann H, Linge H, Gröne H. Zur kenntnis des actinomycins X (Knowledge of Actinomycin X) Naturwissenschaften. 1953;40:224–224. [Google Scholar]
- 3.Brockmann H, Gröne H. Darstellung und charakterisierung reiner actinomycine (Isolation and characterization of pure actinomycins), XII. mitteil. über actinomycine; antibiotica aus actinomyceten. Chem Ber. 1954;87:1036–1051. [Google Scholar]
- 4.Martin HH, Pampus G. Untersuchung über die bildung des actinomycins X (Investigation of formation of actinomycin X). Über die beeinflussung des Komponentenverhältnisses beim Actinomycin X durch Variation der mikrobiologischen Kulturbedingungen. Arch Mikrobiol. 1956;25:90–108. [PubMed] [Google Scholar]
- 5.Brockmann H. Die Actinomycine. Angew Chem. 1960;72:939–947. [Google Scholar]
- 6.Brockmann H, Pfennig N. Auftrennung von actinomycin C durch gegenstromverteilung (Separation of actinomycins C) Naturwissenschaften. 1952;39:429. [Google Scholar]
- 7.Lindenbein W. Über einige chemisch interessante Actinomyceten-stämme und ihre Klassifizierung (Interesting actinomycete strains) Arch Mikrobiol. 1952;17:361. [Google Scholar]
- 8.Bitzer J, Gesheva V, Zeeck A. Actinomycins with altered threonine units in the β-peptidolactone. J Nat Prod. 2006;69(8):1153–1157. doi: 10.1021/np060063g. [DOI] [PubMed] [Google Scholar]
- 9.Bitzer J, Streibel M, Langer H-J, Grond S. First Y-type actinomycins from Streptomyces with divergent structure-activity relationships for antibacterial and cytotoxic properties. Org Biomol Chem. 2009;7(3):444–450. doi: 10.1039/b815689a. [DOI] [PubMed] [Google Scholar]
- 10.Waksman SA, Gregor FJ. Actinomycin II. Classification of organisms producing different forms of actinomycin. Antibiot Chemother. 1954;4(10):1050–1056. [PubMed] [Google Scholar]
- 11.Goss WA, Katz E. Actinomycin formation by Streptomyces cultures. Appl Microbiol. 1957;5(2):95–102. doi: 10.1128/am.5.2.95-102.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katz E, Pienta P, Sivak A. The role of nutrition in the synthesis of actinomycin. Appl Microbiol. 1958;6(4):236–241. doi: 10.1128/am.6.4.236-241.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallo M, Katz E. Regulation of secondary metabolite biosynthesis: catabolite repression of phenoxazinone synthase and actinomycin formation by glucose. J Bacteriol. 1972;109(2):659–667. doi: 10.1128/jb.109.2.659-667.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones GH. Regulation of actinomycin synthesis in Streptomyces antibioticus. J Nat Prod. 1985;49(6):981–987. doi: 10.1021/np50048a003. [DOI] [PubMed] [Google Scholar]
- 15.Jones GH. Regulation of phenoxazinone synthase expression in Streptomyces antibioticus. J Bacteriol. 1986;163(3):1215–1221. doi: 10.1128/jb.163.3.1215-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller U. Actinomycin synthetases. Multifunctional enzymes responsible for the synthesis of the peptide chains of actinomycin. J Biol Chem. 1987;262(12):5852–5856. [PubMed] [Google Scholar]
- 17.Katz E, Weissbach H. Biosynthesis of the actinomycin chromophore; Enzymatic conversion of 4-methyl-3-hydroxyanthranilic acid to actinocin. J Biol Chem. 1962;237:882–886. [PubMed] [Google Scholar]
- 18.Weissbach H, Redfield B, Beaven V, Katz E. 4-methyl-3-hydroxyanthranilic acid, an intermediate in actinomycin biosynthesis. Biochem Biophys Res Commun. 1965;19:524–530. doi: 10.1016/0006-291x(65)90157-9. [DOI] [PubMed] [Google Scholar]
- 19.Troost T, Hitchcock MJ, Katz E. Distinct kynureninase and hydroxykynureninase enzymes in an actinomycin-producing strain of Streptomyces parvulus. Biochem Biophys Acta. 1980;612(1):97–106. doi: 10.1016/0005-2744(80)90282-x. [DOI] [PubMed] [Google Scholar]
- 20.Brown DD, Hitchcock MJ, Katz E. Evidence for a constitutive and inducible form of kynurenine formamidase in an actinomycin-producing strain of Streptomyces parvulus. Arch Biochem Biophys. 1980;202(1):18–22. doi: 10.1016/0003-9861(80)90400-2. [DOI] [PubMed] [Google Scholar]
- 21.Hitchcock MJ, Katz E. Purification and characterization of tryptophan dioxygenase from Streptomyces parvulus. Arch Biochem Biophys. 1988;261(1):148–160. doi: 10.1016/0003-9861(88)90113-0. [DOI] [PubMed] [Google Scholar]
- 22.Haese A, Keller U. Genetics of actinomycin C production in Streptomyces chrysomallus. J Bacteriol. 1988;170(3):1360–1368. doi: 10.1128/jb.170.3.1360-1368.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller U, Kleinkauf H, Zocher R. 4-methyl-3-hydroxyanthranilic acid activating enzyme from actinomycin-producing Streptomyces chrysomallus. Biochemistry. 1984;23(7):1479–1484. doi: 10.1021/bi00302a022. [DOI] [PubMed] [Google Scholar]
- 24.Schauwecker F, Pfennig F, Schröder W, Keller U. Molecular cloning of the actinomycin synthetase gene cluster from Streptomyces chrysomallus and functional heterologous expression of the gene encoding actinomycin synthetase II. J Bacteriol. 1998;180(9):2468–2474. doi: 10.1128/jb.180.9.2468-2474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfennig F, Schauwecker F, Keller U. Molecular characterization of the genes of actinomycin synthetase I and of a 4-methyl-3-hydroxyanthranilic acid carrier protein involved in the assembly of the acylpeptide chain of actinomycin in Streptomyces. J Biol Chem. 1999;274(18):12508–12516. doi: 10.1074/jbc.274.18.12508. [DOI] [PubMed] [Google Scholar]
- 26.Schauwecker F, Pfennig F, Grammel N, Keller U. Construction and in vitro analysis of a new bi-modular polypeptide synthetase for synthesis of N-methylated acyl peptides. Chem Biol. 2000;7(4):287–297. doi: 10.1016/s1074-5521(00)00103-4. [DOI] [PubMed] [Google Scholar]
- 27.Keller U, Lang M, Crnovčić I, Pfennig F, Schauwecker F. The Actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: a Genetic hall of mirrors for synthesis of a molecule with mirror symmetry. J Bacteriol. 2010;192(10):2583–2595. doi: 10.1128/JB.01526-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller U. Acyl pentapeptide lactone synthesis in actinomycin-producing streptomycetes by feeding with structural analogs of 4-methyl-3-hydroxyanthranilic acid. J Biol Chem. 1984;259(13):8226–8231. [PubMed] [Google Scholar]
- 29.Keller U, Krengel U, Haese A. Genetic analysis in Streptomyces chrysomallus. J Gen Microbiol. 1985;131(5):1181–1191. doi: 10.1099/00221287-131-5-1181. [DOI] [PubMed] [Google Scholar]
- 30.Hopwood DA, Bibb MJ, Chater KF, et al. Genetic Manipulation of Streptomyces. A Laboratory Manual. Norwich, United Kingdom: John Innes Foundation; 1985. [Google Scholar]
- 31.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholas KB, Nicholas HB, Jr, Deerfield DWII. GeneDoc: analysis and visualization of genetic variation. Embnew News. 1997;4(1):14. [Google Scholar]
- 33.Solovyev V, Salamov A. Automatic annotation of microbial genomes and metagenomic sequences. In: Li RW, editor. Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies. New York: Nova Science Publishers; 2011. pp. 61–78. [Google Scholar]
- 34.de Jong A. Genome2D webserver for analysis and visualization of bacterial genomes and transcriptome data. [Accessd March 21, 2017]. http://pepper.molgenrug.nl/
- 35.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34(Database issue):D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33(18):5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blin K, Medema MH, Kazempour D, et al. AntiSMASH 2.0-a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41(Web Server issue):W204–W12. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crnovčić I. Genetic and Enzymatic Characterization Of Actinomycin Biosynthesis in the Streptomyces [doctoral thesis] Berlin: Technische Universität Berlin, Institut fuer Chemie; 2013. [Google Scholar]
- 40.Wang X, Tabudravu J, Rateb ME, et al. Identification and characterization of the actinomycin G gene cluster in Streptomyces iakyrus. Mol Biosyst. 2013;9(6):1286–1289. doi: 10.1039/c3mb70081j. [DOI] [PubMed] [Google Scholar]
- 41.Crnovčić I, Süssmuth R, Keller U. Aromatic C-methyltransferases with antipodal stereoselectivity for structurally diverse phenolic amino acids catalyze the methylation step in the biosynthesis of the actinomycin chromophore. Biochemistry. 2010;49(45):9698–9705. doi: 10.1021/bi101422r. [DOI] [PubMed] [Google Scholar]
- 42.Fawaz F, Jones GH. Actinomycin synthesis in Streptomyces antibioticus. Purification and properties of a 3-hydroxyanthranilate 4-methyltransferase. J Biol Chem. 1988;263(10):4602–4606. [PubMed] [Google Scholar]
- 43.Hu Y, Phelan V, Ntai I, Farnet CM, Zazopoulos E, Bachmann BO. Benzodiazepine biosynthesis in Streptomyces refuineus. Chem Biol. 2007;14(6):691–701. doi: 10.1016/j.chembiol.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Li W, Khullar A, Chou S, Sacramo A, Gerratana B. Biosynthesis of sibiromycin, a potent antitumor antibiotic. Appl Environ Microbiol. 2009;75(9):2869–2878. doi: 10.1128/AEM.02326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthijs S, Baysse C, Koedam N, et al. The Pseudomonas siderophore quinolobactin is synthesized from xanthurenic acid, an intermediate of the kynurenine pathway. Mol Microbiol. 2004;52(2):371–384. doi: 10.1111/j.1365-2958.2004.03999.x. [DOI] [PubMed] [Google Scholar]
- 46.Colabroy KL, Begley TP. Tryptophan catabolism: identification and characterization of a new degradative pathway. J Bacteriol. 2005;187(22):7866–7869. doi: 10.1128/JB.187.22.7866-7869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peschke U, Schmidt H, Zhang HZ, Piepersberg W. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol Microbiol. 1995;16(6):1137–1156. doi: 10.1111/j.1365-2958.1995.tb02338.x. [DOI] [PubMed] [Google Scholar]
- 48.Herbst DA, Boll B, Zocher G, Stehle T, Heide L. Structural basis of the interaction of MbtH-likeproteins, putative regulators of nonribosomal peptide biosynthesis, with adenylating enzymes. J Biol Chem. 2013;288(3):1991–2003. doi: 10.1074/jbc.M112.420182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai X, Teta R, Kohlhaas C, et al. Manipulation of regulatory genes reveals complexity and fidelity in hormaomycin biosynthesis. Chem Biol. 2013;20(6):839–846. doi: 10.1016/j.chembiol.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 50.Dangel V, Eustáquio AS, Gust B, Heide L. NovE and NovG act as positive regulators of novobiocin biosynthesis. Arch Microbiol. 2008;190(5):509–519. doi: 10.1007/s00203-008-0396-0. [DOI] [PubMed] [Google Scholar]
- 51.Eustáquio AS, Li SM, Heide L. NovG, a DNA-binding protein acting as a positive regulator of novobiocin biosynthesis. Microbiology. 2005;151(Pt 6):1949–1961. doi: 10.1099/mic.0.27669-0. [DOI] [PubMed] [Google Scholar]
- 52.Tomono A, Tsai Y, Yamazaki H, Ohnishi Y, Horinouchi S. Transcriptional control by A-factor of strR, the pathway-specific transcriptional activator for streptomycin biosynthesis in Streptomyces griseus. J Bacteriol. 2005;187(16):5595–5604. doi: 10.1128/JB.187.16.5595-5604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dangel V, Härle J, Goerke C, et al. Transcriptional regulation of the novobiocin biosynthetic gene cluster. Microbiology. 2009;155(Pt 12):4025–4035. doi: 10.1099/mic.0.032649-0. [DOI] [PubMed] [Google Scholar]
- 54.Chen L, Wang ZL, Zhao QF, Gao SH, Ye RF. Functional determination of the regulatory gene lmbU in lincomycin biosynthesis gene cluster. Chem Bioengg. 2011;28:37–41. [Google Scholar]
- 55.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, et al. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69(2):326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chandra G, Chater KF. Evolutionary flux of potentially bldA-dependent Streptomyces genes containing the rare leucine codon TTA. Antonie Van Leeuwenhoek. 2008;94(1):111–126. doi: 10.1007/s10482-008-9231-5. [DOI] [PubMed] [Google Scholar]
- 57.Gang L, Chater KF, Chandra G, Niu G, Tan H. Molecular regulation of antibiotic biosynthesis in streptomyces. Microbio Mol Bio Rev. 2013;77(1):112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hutchinson CR. Biosynthetic studies of Daunorubicin and Tetracenomycin C. Chem Rev. 1997;97:2525–2536. doi: 10.1021/cr960022x. [DOI] [PubMed] [Google Scholar]
- 59.Choy HA, Jones GH. Phenoxazinone synthase from Streptomyces antibioticus: purification of the large and small enzyme forms. Arch Biochem Biophys. 1981;211(1):55–65. doi: 10.1016/0003-9861(81)90429-x. [DOI] [PubMed] [Google Scholar]
- 60.Jones GH. Actinomycin production persists in a strain of Streptomyces antibioticus lacking phenoxazinone synthase. Antimicrob Agents Chemother. 2000;44(5):1322–1327. doi: 10.1128/aac.44.5.1322-1327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eggert C, LaFayette PR, Temp U, Eriksson KE, Dean JF. Molecular analysis of a laccase gene from the white rot fungus Pycnoporus cinnabarinus. Appl Environ Microbiol. 1998;64(5):1766–1772. doi: 10.1128/aem.64.5.1766-1772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toussaint O, Lerch K. Catalytic oxidation of 2-aminophenols and ortho hydroxylation of aromatic amines by tyrosinase. Biochemistry. 1987;26(26):8567–8571. doi: 10.1021/bi00400a011. [DOI] [PubMed] [Google Scholar]
- 63.Lombo F, Brana AF, Mendez C, Salas JA. The mithramycin gene cluster of Streptomyces argillaceus contains a positive regulatory gene and two repeated DNA sequences that are located at both ends of the cluster. J Bacteriol. 1999;181(2):642–647. doi: 10.1128/jb.181.2.642-647.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ikeda H, Kazuo SY, Omura S. Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J Ind Microbiol Biotechnol. 2014;41(2):233–250. doi: 10.1007/s10295-013-1327-x. [DOI] [PubMed] [Google Scholar]
- 65.Mei X, Xu K, Yang L, Yuan Z, Mahillon J, Hu X. The genetic diversity of cereulide biosynthesis gene cluster indicates a composite transposon Tnces in emetic Bacillus weihenstephanensis. BMC Microbiology. 2014;14:149. doi: 10.1186/1471-2180-14-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mauger AB, Lackner H. The actinomycins. In: Kingston DGI, Cragg GM, Newman DJ, editors. Anticancer Agents from Natural Products. Boca Raton, FL: CRC Press; 2005. pp. 281–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignments and nearest neighbor tree of different flavine monooxygenase protein sequences from Streptomyces antibioticus (AFM16) and Streptomyces chrysomallus (JI76) with similarity to kynurenine-3-monooxygenase. Included are bacterial KMO sequences Orf23 from Streptomyces refuineus (ABW71854.1), SibC from Streptosporangium sibiricum (ACN39726.1), QbsG from Stenotrophomonas maltophilia (CRX68935.1). The tree consists of two main clades, of which the upper clade has a subclade (shaded) containing conserved KMO sequences. Sequences in the other subclade are less similar to KMOs having 30% identity and less to SibC or Orf23. Red numbers denote branch values representing a measure of support for the node. The bar at the bottom denotes phylogenetic distance.
Abbreviation: KMO, kynurenine-3-monooxygenase.
Alignments of AcmT and AcmE sequences from Streptomyces chrysomallus (sc), Streptomyces antibioticus (sa) and Streptomyces mutabilis (WP_043377479.1 and WP_052412530.1, respectively).
Nearest neighbor tree of AcmE and AcmT sequences from different streptomycetes.
Notes: From the listed strains sa (Streptomyces antibioticus), sc (Streptomyces chrysomallus), Streptomyces mutabilis and Streptomyces iakyrus are known Acm producers. The tree shows two main clades each for AcmE or AcmT sequences. Accession numbers are: Streptomyces flavovariabilis (WP_031142912.1); Streptomyces variegatus (WP_052686596.1); S. iakyrus E (CCO61882.1); S. mutabilis E (WP_052412530.1); Streptomyces sp. CNS654 (WP_032769128.1); Streptomyces sp. NRRL F-5008 (WP_051781089.1); S. mutabilis T (WP_043377479.1); Streptomyces sp.1 (WP_030987180.1); Streptomyces sp.2 (WP_030653030.1); S. iakyrus T (WP_033306586.1); Streptomyces sp. NRRL WC-3742 (WP_051838397.1). Red numbers denote branch values. The bar at the bottom indicates phylogenetic distance.
Sequences of directed repeats (DRs) flanking the acm biosynthetic gene cluster of Streptomyces chrysomallus.
Table S1.
Comparisons of G+C mole% contents of genes in the acm biosynthetic gene clusters of Streptomyces antibioticus and Streptomyces chrysomallus
| Gene in S. antibioticus | % G+C | 3rd-letter % G+C | Gene in S. chrysomallus | Gene in S. chrysomallus
|
|
|---|---|---|---|---|---|
| % G+C left arm/right arm | 3rd-letter % G+C left arm/right arm | ||||
| saacmrC | 68.9 | 94 | scacmrC/scacmY | 67.1/70.1 | 91.1/95.8 |
| saacmrB | 69.4 | 95.3 | scacmrB/scacmX | 68.2/71.5 | 91.4/97.3 |
| saacmrA | 72.1 | 90.8 | scacmrA/scacmW | 71.6/73.5 | 89.3/95.4 |
| saacmQ | 73 | 95.6 | scacmQ/scacmV | 72.2/73.7 | 92.5/98.7 |
| saacmP | 73.8 | 90.7 | scacmP/scacmU | 70.3/74 | 85.1/91.5 |
| saacmO | 71.6 | 92.6 | scacmO/scacmJ | 69.4/72.5 | 83.3/90.5 |
| saacmN | 70.5 | 81.2 | scacmN | 72.5 | 92.8 |
| saacmM | 71.7 | 91 | scacmM | 69.6 | 87 |
| saacmL | 68.1 | 91 | scacmL/scacmI | 69.1/70.5 | 93.4/97.4 |
| saacmK | 72.5 | 89 | scacmH/scacmK | 74.1/72.8 | 95/90.6 |
| saacmG | 70.8 | 92.3 | scacmG | 70.8 | 94.7 |
| saacmF | 74.4 | 90.6 | scacmF | 75.7 | 94.8 |
| saacmE | 70.3 | 92.9 | scacmE (acmT) | 70.1 (68.2) | 90.3 (93.4) |
| saacmC | 70.5 | 89.8 | scacmC | 73.2 | 95.6 |
| saacmB | 71.2 | 88.6 | scacmB | 73.4 | 94.3 |
| saacmA | 70.4 | 88 | scacmA | 71.5 | 90.7 |
| saacmD | 62.4 | 88.6 | scacmD | 66.7 | 97.4 |
| saacmR | 68.2 | 92.5 | scacmR | 69.7 | 97 |
| saacmS | 66 | 88.8 | scacmS | 68.4 | 95.7 |
| saacmT | 69.2 | 93.4 | scacmT (scacmE) | 68.2 (70.1) | 93.4 (90.3) |
Table S2.
IS elements flanking the acm biosynthetic gene cluster of Streptomyces chrysomallus
| Name | Length | Family | Highest similarity to | Length |
|---|---|---|---|---|
| ISSc1.1 | 421 (fragment) | IS5 | ISStu1 (Streptomyces turgidiscabies) AY707081 | 899 bp (4e−05) |
| ISSc1.2 | 189 (fragment) | IS481 | ISsco1 (Streptomyces coelicolor) AF099014 | 1057 bp (3e−20) |
| ISSc1.3 | 882 | IS5 | IS1650 (S. coelicolor) AL079356 | 898 bp (9e−11) |
| ISSc1.4 | 1300 | IS3 | ISTesp3 (Terrabacter sp.) AP008980 | 1373 bp (1e−16) |
| ISSc1.5 | 1071 | IS5 | IS1648 (S. coelicolor) AL023861 | 969 bp (4e−07) |
| ISSc1.6 | 1622 | IS3 | IS1372 (Streptomyces lividans) U50076 | 1304 bp (0) |
| ISSc1.7 | 1397 | IS701 | ISSav3 (Streptomyces avermitilis) NC_003155 | 1400 bp (0) |
| ISSc1.8 | 779 | IS5 | IS1647 (S. coelicolor) AL023702 | 878 bp (8e−48) |