The yeast Polo kinase Cdc5 changes its localization at centrosomes during the cell cycle. Cdc5 localizes to the nuclear centrosome surface in early mitosis and relocalizes to the cytoplasmic centrosome side in late anaphase. Cdc14 and Bfa1 play important roles in regulating Cdc5 centrosome localization.
Abstract
The budding yeast Polo-like kinase Cdc5 is a key regulator of many mitotic events. Cdc5 coordinates its functions spatially and temporally by changing its localization during the cell cycle: Cdc5 is imported into the nucleus in G2 phase and released to the cytoplasm in anaphase, where it accumulates at the bud neck. Cdc5 also localizes to the spindle pole bodies (SPBs) from S phase until the end of mitosis. Whether Cdc5 changes its SPB population during the cell cycle is not known. We find that Cdc5 localizes to distinct SPB subpopulations, depending on the mitotic stage. Cdc5 localizes to the nuclear side of the SPBs during metaphase and early anaphase and to the cytoplasmic surface of the SPBs during late anaphase. Cdc14 is necessary to relocalize Cdc5 from the nuclear SPB plaque. Accumulation of Cdc5 at the daughter SPB in late anaphase is controlled by Bfa1. We also show that Cdc5 and Bfa1 are found in spatially distinct locations at the SPBs during G2/M arrest after DNA damage. Collectively our data reveal that Cdc5 is a dynamic component of the SPBs during mitosis and provide new insight into its regulation during both late mitotic events and DNA damage–induced G2/M arrest.
INTRODUCTION
Polo-like kinases are highly conserved from yeast to humans and serve essential functions in mitosis (Lee et al., 2005). The budding yeast Polo-like kinase Cdc5 regulates multiple mitotic events, including mitotic entry, chromosome segregation, mitotic exit, and cytokinesis (Archambault and Glover, 2009). Cdc5 phosphorylates its many substrates in a coordinated manner by changing its localization during the cell cycle. Cdc5 acting in the cytoplasm promotes mitotic entry (Park et al., 2003, 2004b; Sakchaisri et al., 2004; Asano et al., 2005). Subsequently Cdc5 is imported into the nucleus in G2/M, where it further promotes mitotic entry (Nakashima et al., 2008) and phosphorylates the cohesin ring subunit Scc1 (Alexandru et al., 2001; Hornig and Uhlmann, 2004). In anaphase, Cdc5 helps release the Cdc14 phosphatase (Shou et al., 2002; Yoshida and Toh-e, 2002; Yoshida et al., 2002; Visintin et al., 2003), a key driver of mitotic exit (Visintin et al., 1998), from the nucleolus as part of the Cdc fourteen early anaphase release (FEAR) mitotic exit pathway (Rock and Amon, 2009). Cdc5 also phosphorylates the nuclear condensin subunits (St-Pierre et al., 2009) and, together with Cdc14, promotes the elongation of the mitotic spindle (Roccuzzo et al., 2015). In late anaphase, Cdc5 is released from the nucleus to the cytoplasm in a Cdc14-dependent manner (Botchkarev et al., 2014), which allows Cdc5 to phosphorylate its cytoplasmic late anaphase substrate Bfa1 (Hu et al., 2001; Geymonat et al., 2003; Botchkarev et al., 2014) and activate the second mitotic exit pathway, the mitotic exit network (MEN; Bardin and Amon, 2001). Cdc5 is also required to recruit the MEN kinase Cdc15 to spindle pole bodies (SPBs; Rock and Amon, 2011). Cdc5 accumulates at the bud neck in late anaphase (Botchkarev et al., 2014), where it promotes cytokinesis by activating Rho1 and phosphorylating Hof1 (Yoshida et al., 2006; Meitinger et al., 2011). Aside from its cell cycle stage–dependent nuclear-cytoplasmic relocalization, Cdc5 also localizes to the SPBs from S phase until the end of mitosis (Song et al., 2000; Park et al., 2004a, b; Maekawa et al., 2007; Botchkarev et al., 2014). However, it is unknown whether the SPB localization represents a single population of Cdc5 or whether Cdc5 is targeted to SPBs using functionally distinct interactions during different cell cycle stages.
The SPBs in budding yeast are functionally analogous to mammalian centrosomes and act as the microtubule-organizing centers in the cell (Jaspersen and Winey, 2004). The SPBs are embedded in the nuclear envelope and consist of a central plaque that connects the outer plaque, which is exposed to the cytoplasm, to the inner plaque, which faces the nuclear interior. Many MEN components, including Bfa1, localize to the cytoplasmic outer plaque of the SPBs (Bardin and Amon, 2001). Bfa1 localizes to both mother and daughter SPBs (mSPBs and dSPBs, respectively) during early mitosis but preferentially localizes to the dSPB in anaphase (Li, 1999; Gruneberg et al., 2000; Pereira et al., 2000; Kim et al., 2012). This Bfa1 localization pattern at the SPBs is very similar to the SPB localization of Cdc5, which localizes to both SPBs in early mitosis and preferentially localizes to the dSPB in anaphase (Song et al., 2000; Park et al., 2004a, b; Maekawa et al., 2007; Botchkarev et al., 2014). Because Cdc5 phosphorylates Bfa1 only in late anaphase to promote mitotic exit (Hu et al., 2001; Maekawa et al., 2007), it is not clear whether Cdc5 is found in the same population as Bfa1 at the SPBs in earlier cell cycle stages before the timing of Bfa1 phosphorylation by Cdc5. The recent finding that impairing nuclear release of Cdc5 in anaphase prevents Bfa1 inactivation (Botchkarev et al., 2014) suggests that even though Cdc5 and Bfa1 are found at the SPBs during most of the cell cycle, the interactions of these proteins at the SPBs may vary in a cell cycle–dependent manner.
In this study, we investigated the regulation of Cdc5 localization at the SPBs during mitosis. We found that Cdc5 is found in two distinct populations at the SPBs during the cell cycle. Cdc5 localizes to the nuclear SPB surface in metaphase and early anaphase. In late anaphase, Cdc5 localization to the dSPB largely depends on Bfa1. We find that Cdc14 is necessary and sufficient to remove Cdc5 from its Bfa1-independent population at the SPBs. The FEAR network plays a role in removing Cdc5 from its Bfa1-independent population in anaphase. Collectively our data reveal a new molecular mechanism of Cdc5 regulation at the SPBs and provides clues to the regulation of late mitotic events.
RESULTS
Cdc5 localizes to distinct SPB populations during the cell cycle
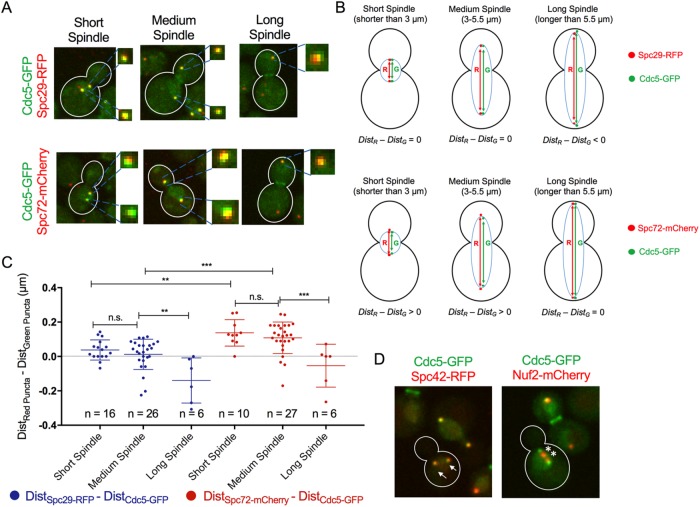
Cdc5 and Bfa1 both localize to the SPBs throughout mitosis (Li, 1999; Gruneberg et al., 2000; Pereira et al., 2000; Song et al., 2000; Park et al., 2004a, b; Maekawa et al., 2007; Kim et al., 2012; Botchkarev et al., 2014). Because Cdc5 is known to phosphorylate Bfa1 only in late anaphase (Hu et al., 2001; Maekawa et al., 2007), we wanted to test whether Cdc5 is spatially separated from Bfa1 at the SPBs in early mitosis. Bfa1 localizes to the cytoplasmic SPB surface during the cell cycle (Gruneberg et al., 2000; Pereira et al., 2000; Gryaznova et al., 2016). Cdc5 accumulates in the nucleus in metaphase and is released to the cytoplasm in anaphase (Botchkarev et al., 2014). We investigated whether, similarly, Cdc5 localizes to the nuclear SPB surface in early mitosis and to the cytoplasmic SPB surface in anaphase. We monitored the localization of endogenously expressed Cdc5–green fluorescent protein (GFP) in mitotic cells in an asynchronous cell culture. We analyzed the colocalization of Cdc5-GFP at SPBs and either Spc29–red fluorescent protein (RFP; SPB inner plaque component) or Spc72-mCherry (SPB outer plaque component; Figure 1A). Cell cycle stage was determined by measuring the distance between opposite SPBs as an indication of the length of the mitotic spindle in each cell. We found that Cdc5-GFP and Spc29-RFP puncta at SPBs were overlapping in cells with short (<3 µm) and medium-length (3–5.5 µm) spindles. In cells with long mitotic spindles (>5.5 µm), Cdc5-GFP SPB puncta were more widely separated in relation to Spc29-RFP. We quantified these observations by measuring the distances between the centroids of Cdc5-GFP (DistCdc5-GFP) and Spc29-RFP (DistSpc29-RFP) puncta at opposite SPBs (Figure 1B). In most cells with short and medium-length spindles as defined here, the difference between DistSpc29-RFP and DistCdc5-GFP was approximately zero, indicating that the Cdc5-GFP and Spc29-RFP puncta were equidistant at opposite SPBs (Figure 1C). The distance between Cdc5-GFP SPB centroids in anaphase cells with long mitotic spindles was significantly greater than the distance between Spc29-RFP SPB centroids in the same cells (Figure 1C). We also quantified these observations by monitoring the red and green fluorescence intensities across a line scan in representative cells. We found that Cdc5-GFP and Spc29-RFP fluorescence signals peaked as single points at SPBs of cells with short and medium-length mitotic spindles (Supplemental Figure S1A). However, in cells with long spindles, the Cdc5-GFP SPB fluorescence signal peak at the dSPB appeared to face the cytoplasm relative to Spc29-RFP (Supplemental Figure S1A). This indicates that Cdc5 and Spc29 are likely found in the same SPB population in metaphase and early anaphase and in distinct SPB populations in late anaphase.
FIGURE 1:
Cdc5 colocalizes at SPBs with Spc29 in early mitosis but with Spc72 only in cells with a long mitotic spindle. (A) Representative images of Cdc5-GFP localization relative to Spc29-RFP and Spc72-mCherry during mitosis in cells cultured asynchronously. (B) Calculation used to determine whether Cdc5 localizes to the nuclear or cytoplasmic SPB surface. (C) Quantified results of experiment in A using calculation in B. (D) Cdc5-GFP colocalizes with SPB component Spc42-RFP (arrows) but not with kinetochore component Nuf2-mCherry (asterisks) during early mitosis.
In contrast, Cdc5-GFP SPB puncta appeared to be closer together relative to Spc72-mCherry SPB puncta in most cells with short and medium-length spindles (Figure 1A). In late anaphase, Cdc5-GFP and Spc72-mCherry SPB puncta appeared to be overlapping (Figure 1A). Comparing the distances between Cdc5-GFP SPB centroids at opposite SPBs, and Spc72-mCherry SPB centroids at opposite SPBs confirmed these observations (Figure 1, B and C, and Supplemental Figure S1B).
It was recently reported that Cdc5 localizes to centromeres during mitosis (Mishra et al., 2016). Because centromeres localize adjacent to SPBs during majority of the cell cycle (Guacci et al., 1997; Pearson et al., 2001; Winey and O’Toole, 2001), it is a formal possibility that the fluorescence signal emitted by the Cdc5-GFP puncta in mitotic cells stems from both centromeres and SPBs. To test this possibility, we compared the localization of Cdc5-GFP and Nuf2-mCherry, a component of the Ndc80 protein complex at kinetochores (Osborne et al., 1994; Ciferri et al., 2008; Wang et al., 2008; Suzuki et al., 2016). We found that in G2/M cells, in which the centromeres have not yet fully separated, the Nuf2-mCherry puncta are distinct from Cdc5-GFP puncta (Figure 1D). In contrast, Cdc5-GFP puncta colocalized with Spc42-RFP in G2/M cells (Figure 1D). Therefore, although it is formally possible that Cdc5 localizes to centromeres as it accumulates in the nucleus in G2 and metaphase, it is highly unlikely that the Cdc5-GFP puncta we are analyzing stem from centromeres but that instead they are components of the SPBs. Collectively our analysis of Cdc5-GFP SPB localization in relation to inner and outer plaque markers strongly suggests that Cdc5 is found in distinct mitotic stage–dependent populations. Cdc5 accumulates at the nuclear SPB surface in metaphase and early anaphase and relocalizes to the cytoplasmic SPB surface in late anaphase.
Cdc5 localization to the dSPB depends on Bfa1 in late anaphase
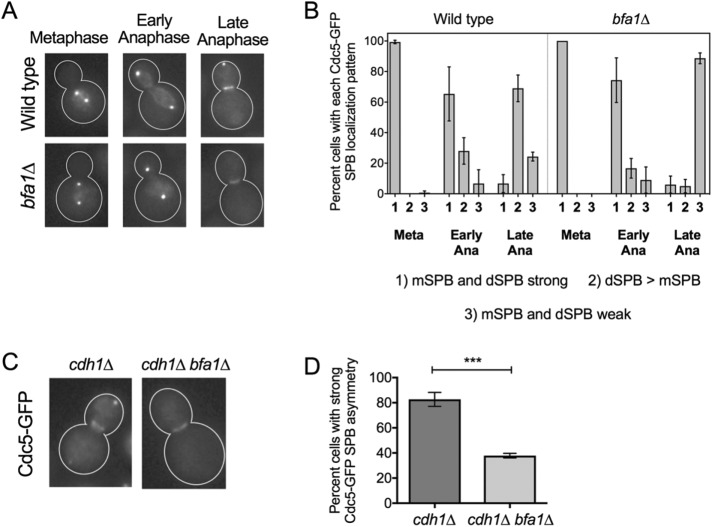
A key Cdc5 substrate in anaphase is the mitotic exit network inhibitor Bfa1 (Hu et al., 2001; Geymonat et al., 2003). Bfa1 localizes to the outer plaque of the SPBs throughout the cell cycle (Gruneberg et al., 2000; Pereira et al., 2000; Gryaznova et al., 2016). Because Bfa1 is a Cdc5 substrate at the dSPB in anaphase, we asked whether Bfa1 played a role in Cdc5 outer plaque localization to the dSPB in anaphase. We monitored the SPB localization of Cdc5-GFP in a wild-type and a bfa1∆ strain (Figure 2, A and B). Cdc5-GFP strongly decorates both the mSPB and dSPB in metaphase and in early anaphase (cell cycle stage was judged by the distance between the two fluorescent foci). In late anaphase cells, we found that strong Cdc5-GFP signal is retained at the dSPB, albeit dimmer than earlier in mitosis, but a strong Cdc5-GFP signal from the mSPB could no longer be detected. This observation is in line with previous reports on Cdc5 SPB localization (Maekawa et al., 2007; Botchkarev et al., 2014). In bfa1∆ cells, Cdc5-GFP localized strongly to both the mSPBs and dSPBs in metaphase and early anaphase cells, similar to wild-type cells (Figure 2A); Cdc5-GFP colocalizes with Spc29-RFP in these cells (Figure 1, A and C). However, in late anaphase cells, when Cdc5-GFP appears to colocalize with Spc72-mCherry (Figure 1, A and C), we were not able to detect a strong Cdc5-GFP signal at the dSPB in bfa1∆ cells (Figure 2, A and B). Instead, Cdc5-GFP showed very weak mSPB and dSPB localization. The absence of Cdc5-GFP from the SPBs without Bfa1 in late anaphase did not impair a strong Cdc5-GFP localization to the bud neck (Figure 2A), suggesting that the loss of SPB Cdc5-GFP signal at the SPB was not due to degradation. Cdc5 is targeted for degradation by the anaphase-promoting complex (APC) adaptor protein Cdh1 (Charles et al., 1998). Indeed, even when we blocked Cdh1-dependent Cdc5 degradation, Cdc5-GFP was still defective in localizing to the dSPB in anaphase in the absence of Bfa1 (Figure 2, C and D). These results further confirm that Cdc5 is a dynamic component of the SPBs and that Bfa1 is important for the strong Cdc5 localization to the dSPB outer plaque in late anaphase.
FIGURE 2:
Bfa1 is required for proper localization of Cdc5 to the dSPB in late anaphase. (A) Representative images of asynchronously cycling wild-type and bfa1∆ cells expressing Cdc5-GFP from the endogenous locus. Mitotic stage was judged by the distance between SPBs and appearance of Cdc5-GFP localization at the bud neck in late anaphase. Strong Cdc5-GFP signal is not retained at the dSPB of late anaphase bfa1∆ cells. (B) Quantification of A (n = 489 wild-type; n = 330 bfa1∆). (C) Representative images of Cdc5-GFP localization in late anaphase cdh1∆ and cdh1∆ bfa1∆ cells. (D) Quantification of C. Cdc5-GFP did not show strong accumulation at the dSPB of most late anaphase cdh1∆ bfa1∆ cells. All experiments were performed in triplicate.
Cdc14 is required to liberate Cdc5 from its Bfa1-independent population in anaphase
Our results reveal that Cdc5 is found in distinct populations at the SPBs, depending on mitotic stage, and that Bfa1 controls Cdc5 localization to the dSPB only in late anaphase. Our data further suggest that Cdc5 appears to localize to the nuclear SPB surface before late anaphase. We reported that Cdc14 phosphatase, a key driver of mitotic exit (Visintin et al., 1998), is required for Cdc5 release from the nucleus to the cytoplasm in anaphase (Botchkarev et al., 2014). We wanted to test whether Cdc14 promotes the relocalization of Cdc5 from its Bfa1-independent to a Bfa1-dependent SPB population in anaphase. First, we tested whether Cdc14 is required to liberate Cdc5 from its Bfa1-independent SPB location. If Cdc14 were required to release Cdc5 from its Bfa1-independent SPB location, then Cdc5 localization to the SPBs would be restored in bfa1∆ lacking adequate Cdc14 activity in anaphase. The cdc14-1 mutant arrests in telophase at its restrictive temperature of 37°C, but even at its permissive room temperature (25°C), this mutant is still hypomorphic because it shows synthetic lethal genetic interactions with various mutants, unlike CDC14 (Jaspersen et al., 1998; Yuste-Rojas and Cross, 2000; Rossio et al., 2010; Zhai et al., 2010; Yellman and Roeder, 2015). We found that Cdc5 was retained at the dSPB in bfa1∆ cdc14-1 cells at 25°C (Figure 3, A and B). Of interest, compared with wild-type cells, in which Cdc5 localizes preferentially to the dSPB in anaphase, Cdc5 heavily decorated both the mSPB and dSPB of bfa1∆ cdc14-1 cells (Figures 3, A and B). This result reveals that the Bfa1-independent SPB binding partner of Cdc5 is found at both the mSPB and dSPB in late anaphase, consistent with previous observations of Cdc5 localizing symmetrically to the mSPB and dSPB in metaphase and early anaphase (Figure 2A).
FIGURE 3:
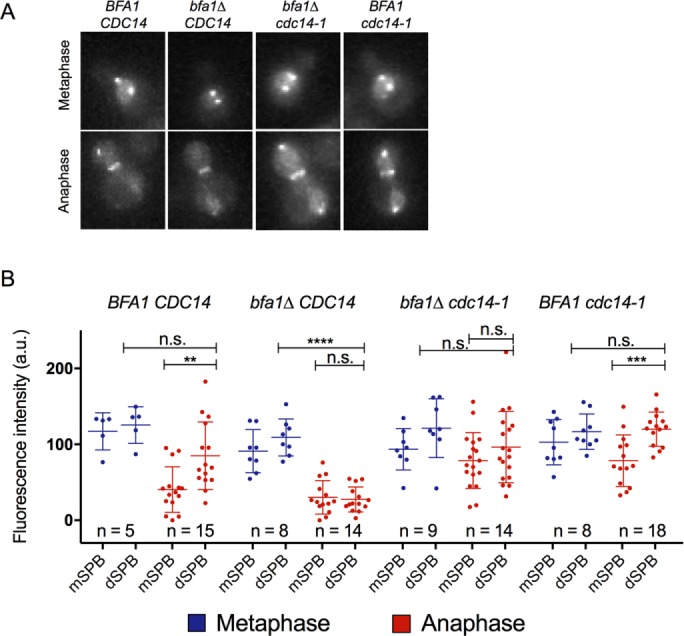
Cdc14 is required to release Cdc5 from its Bfa1-independent SPB population. (A) Representative images of Cdc5-GFP localization in metaphase and late anaphase wild type, bfa1∆, bfa1∆ cdc14-1, and cdc14-1 cells cultured at room temperature. Cdc5-GFP signal was retained at both the mSPB and dSPB of cdc14-1 and cdc14-1 bfa1∆ late anaphase cells. (B) Quantification of A.
Although cdc14-1 cells at their permissive temperature showed strong Cdc5 localization to both SPBs in late anaphase, there was still a preference for the dSPB (Figure 3, A and B). At 25°C, the cdc14-1 strain maintains adequate phosphatase activity and can divide normally to form colonies. Therefore it is likely that at the dSPB of anaphase cdc14-1 cells at permissive temperature, there is both a Bfa1-independent and a Bfa1-dependent Cdc5 signal. Of importance, in cdc14-1 anaphase cells, Bfa1 maintains asymmetry to the dSPB (Geymonat et al., 2009). Our results reveal that Cdc14 regulates Cdc5 dynamics at the SPBs during mitosis and is required to remove Cdc5 from its Bfa1-independent SPB population in anaphase.
Cdc14 is sufficient to remove Cdc5 from its metaphase SPB population
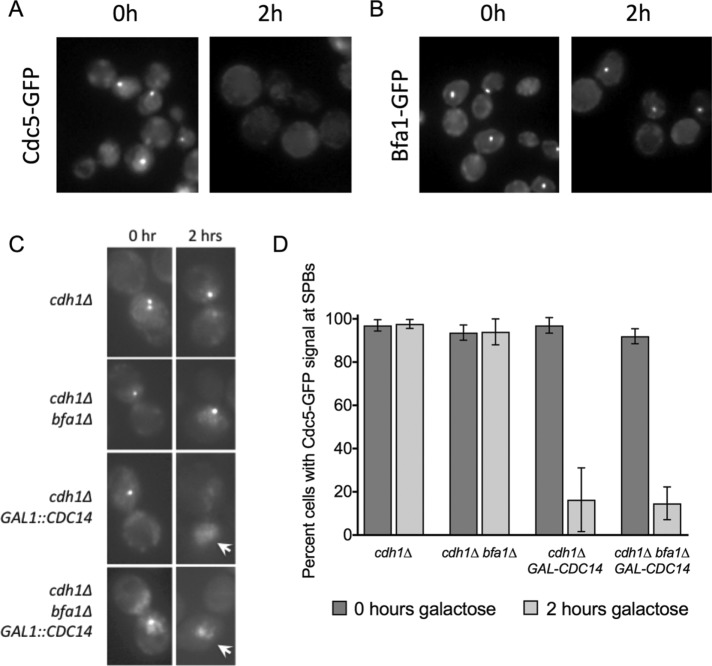
Having established that Cdc14 is required to remove Cdc5 from its Bfa1-independent SPB population in anaphase, we wanted to test whether Cdc14 was sufficient to perform this function. We tested whether Cdc14 overexpression was sufficient to remove Cdc5 from its metaphase SPB population during nocodazole arrest. Indeed, Cdc14 overexpression in nocodazole arrest resulted in complete loss of cellular Cdc5-GFP signal (Figure 4A), whereas Bfa1-GFP localization at the SPBs was not affected (Figure 4B). The loss of Cdc5-GFP signal in this condition was likely due to degradation because Cdc14 activates APC-Cdh1 (Visintin et al., 1998; Jaspersen et al., 1999), the E3 ligase that targets Cdc5 for degradation (Charles et al., 1998). We found that inducing Cdc14 overexpression during nocodazole arrest removed Cdc5-GFP from the SPBs of cdh1∆ bfa1∆ cells (Figure 4, C and D). This result reveals that Cdc14 is sufficient to remove Cdc5 from its Bfa1-independent metaphase SPB population. Despite Cdc5-GFP signal loss from SPBs after Cdc14 overexpression, a strong general Cdc5-GFP signal was still present in the nucleus (Figure 4C). This suggests that although Cdc14 is sufficient to release Cdc5 from the SPB inner plaque, most likely into the general nuclear Cdc5-GFP population, an additional anaphase-specific event aside from Cdc14 activation is required to release Cdc5 from the nucleus to the cytoplasm. Of interest, cdh1∆ cells did not retain Cdc5 at the SPBs after Cdc14 overexpression (Figure 4C). It is possible that Bfa1 is kept protected from binding Cdc5 during nocodazole arrest. Alternatively, it is possible that Cdc14 is also responsible for removal of Cdc5 from Bfa1 in late telophase, and this effect is observed prematurely as a consequence of Cdc14 overexpression. Our results indicate that Cdc14 is sufficient to relocalize Cdc5 from its metaphase SPB population.
FIGURE 4:
Cdc14 is sufficient to release Cdc5 from its metaphase SPB population. (A) Cdc5-GFP fluorescence signal is lost in nocodazole-arrested wild-type cells after 2 h of Gal-Cdc14 overexpression. (B) Bfa1-GFP signal is retained in nocodazole-arrested cells after 2 h of Gal-Cdc14 overexpression. (C) Cdc5-GFP signal is lost from SPBs but not from the nucleus in both cdh1∆ and cdh1∆ bfa1∆ nocodazole-arrested cells after 2 h of Gal-Cdc14 overexpression. Arrows depict nuclei with Cdc5-GFP in the nucleus but not at SPBs. (D) Quantification of C (n > 100 of each strain at time zero; n > 50 of each strain at 2 h; repeated in triplicate).
FEAR contributes to Cdc5 asymmetry at the SPBs in anaphase
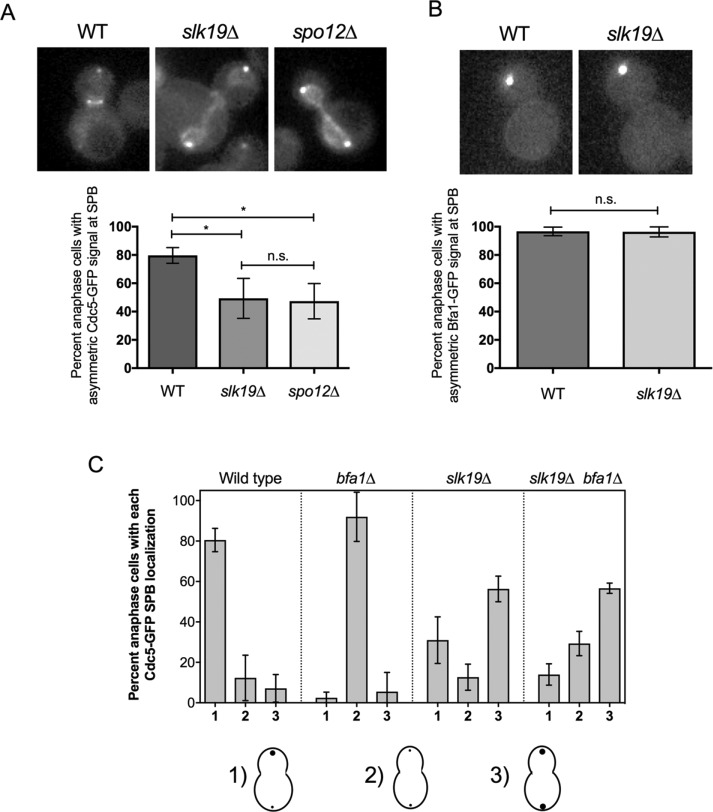
Having determined that Cdc14 is required and sufficient to remove Cdc5 from its Bfa1-independent SPB population in anaphase, we next wanted to understand what controls Cdc14 to remove Cdc5 from its Bfa1-independent population. Cdc14 is activated through two pathways—first, in early anaphase via FEAR, and subsequently in late anaphase via the MEN (Stegmeier and Amon, 2004). Because FEAR but not MEN mutant cells are defective in Cdc5 release from the nucleus in anaphase (Botchkarev et al 2014), we predicted that Cdc14 promotes the release of Cdc5 from its Bfa1-independent SPB population via the FEAR pathway. Indeed, we found that Cdc5-GFP strongly decorates both the mSPB and dSPB in anaphase in the FEAR mutants slk19∆ (on average, 51% of cells) and spo12∆ (on average, 53% of cells; Stegmeier et al., 2002), whereas in wild-type anaphase cells, Cdc5-GFP localized asymmetrically to the dSPB (Figure 5A). Cdc5-GFP SPB puncta faced the nucleus relative to Spc72-mCherry puncta in spo12∆ in anaphase (Supplemental Figure S2). These results suggest that FEAR is important to remove Cdc5 from the mSPB in anaphase. We found that the Bfa1-GFP signal was not restored to the mSPB of anaphase slk19∆ cells (Figure 5B), consistent with findings that Cdc14 is not important to regulation of Bfa1 asymmetry to the dSPB in anaphase (Geymonat et al., 2009). This result strongly suggests that Cdc5 localization to the mSPB in anaphase in cdc14-1 and in FEAR mutants is independent of Bfa1.
FIGURE 5:
Cdc5 is retained in its Bfa1-independent population in FEAR mutants slk19∆ and spo12∆. (A) Cdc5-GFP can localize to both the mSPB and dSPB of asynchronously cycling anaphase slk19∆ and spo12∆ cells (wild-type n = 130; slk19∆ n = 164; spo12∆ n = 97). (B) Bfa1-GFP localizes asymmetrically to the dSPB in late anaphase slk19∆ cells (wild-type n = 123; slk19∆ n = 85). (C) Quantified analysis of the Cdc5-GFP SPB localization pattern at SPBs in wild-type (n = 96), bfa1∆ (n = 81), slk19∆ (n = 198), and slk19∆ bfa1∆ (n = 159) late anaphase cells. All experiments were performed in triplicate.
To further confirm that Cdc14 removal of Cdc5 from its Bfa1-independent SPB population occurs through FEAR, we tested whether the Bfa1-independent Cdc5 population was able to persist at the SPBs in anaphase in cells with FEAR defects. We found that bfa1∆ slk19∆ cells were able to retain Cdc5 at the SPBs in anaphase (Figure 5C). We did observe some bfa1∆ slk19∆ anaphase cells without a strong Cdc5-GFP SPB signal. This is likely because Cdc14 maintains some activity in slk19∆ cells (Stegmeier et al., 2002). Because many more bfa1∆ slk19∆ cells maintained strong Cdc5-GFP signal at the dSPB in anaphase than did bfa1∆ cells (Figure 5C), we conclude that FEAR contributes to the relocalization of Cdc5 from its Bfa1-independent population at the SPBs.
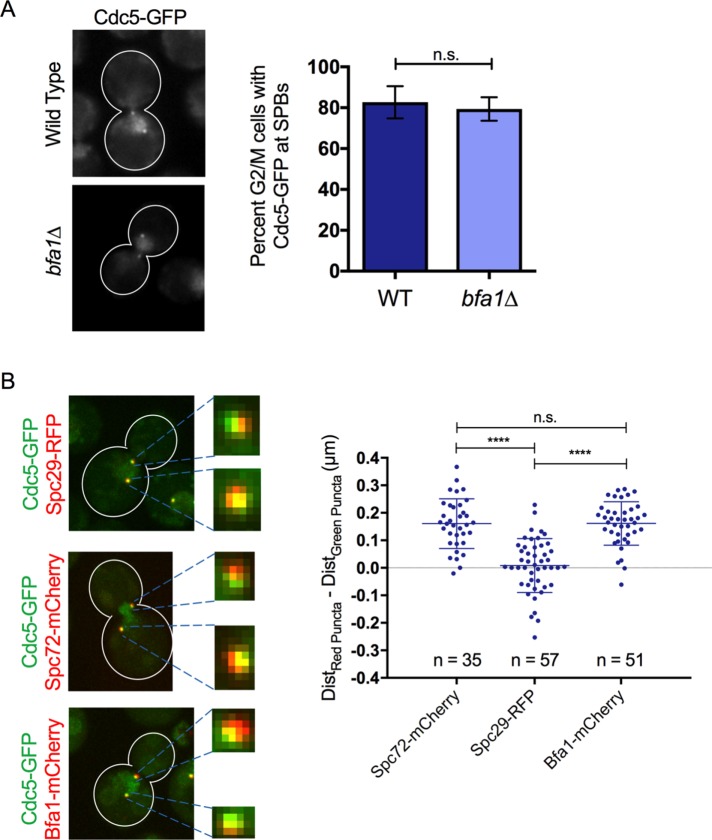
Cdc5 and Bfa1 are found in distinct SPB populations during DNA damage arrest
On DNA damage induction, the DNA damage response arrests cells in the G2/M cell cycle phase. In the presence of irreparable DNA damage, cells maintain a prolonged G2/M arrest but eventually continue dividing through a process called adaptation (Sandell and Zakian, 1993; Toczyski et al., 1997; Lee et al., 1998). Cdc5 is required for adaptation after DNA damage (Toczyski et al., 1997; Donnianni et al., 2010; Vidanes et al., 2010). Considerable evidence indicates that inactivation of Bfa1 is an important mechanism through which Cdc5 promotes adaptation (Hu et al., 2001; Liang and Wang, 2007; Valerio-Santiago et al., 2013; Ratsima et al., 2016; Rawal et al., 2016). Cdc5 localization at the SPBs is required for adaptation to DNA damage (Ratsima et al., 2016; Rawal et al., 2016). We wanted to understand the relationship between Cdc5 and Bfa1 SPB localization during a strong DNA damage response–induced G2/M arrest. We examined adaptation in strain JKM179, where a single, unrepairable double-strand break (DSB) is induced by HO endonuclease (Lee et al., 1998). HO-induced cells arrest at G2/M for 12–15 h before they adapt and resume cell cycle progression. At 6 h after HO induction, Cdc5-GFP was localized in the nucleus and was able to decorate SPBs in the majority of cells (Figure 6A), although ∼20% of these G2/M-arrested cells contained strong nuclear Cdc5-GFP signal but no Cdc5-GFP signal at SPBs. This Cdc5-GFP localization to the SPBs is independent of Bfa1, because no defect in Cdc5-GFP localization at SPBs was observed in a bfa1∆ strain at the same time point (Figure 6A). These results indicate that at 6 h after HO induction, Cdc5-GFP localization to the SPBs does not require Bfa1.
FIGURE 6:
Cdc5 and Bfa1 are found in distinct SPB populations during a DNA damage arrest. (A) Cdc5-GFP localizes to the nucleus and SPBs of JKM179 G2/M-arrested cells 6 h after HO endonuclease induction. Localization of Cdc5-GFP to the SPBs in these cells is not dependent on Bfa1 (experiment performed in triplicate). A small percentage of both wild-type and bfa1∆ G2/M-arrested cells showed strong nuclear Cdc5-GFP accumulation but no obvious localization at SPBs. (B) Cdc5-GFP colocalizes with Spc29-RFP but not Spc72-mCherry or Bfa1-mCherry in most JKM179 G2/M-arrested cells 6 h after HO endonuclease induction. The dot-plot shows the difference in distance between red puncta at opposite SPBs and Cdc5-GFP puncta at opposite SPBs, as calculated according to Figure 1B. Centroids of Spc72-mCherry and Bfa1-mCherry puncta at opposite SPBs were further apart from each other than centroids of Cdc5-GFP SPB puncta in almost every cell. Centroids of Spc29-RFP puncta at opposite SPBs and centroids of Cdc5-GFP puncta at opposite SPBs were nearly indistinguishable in most cells.
We next asked whether Cdc5 localizes to the nuclear SPB surface, as it would in undamaged G2/M cells, during a DNA damage–induced G2/M arrest. We compared the localization of Cdc5-GFP to Spc29-RFP, Spc72-mCherry, and Bfa1-mCherry at SPBs at 6 h after HO induction. We found that Cdc5-GFP puncta at SPBs colocalized with Spc29-RFP (Figure 6B and Supplemental Figure S3A). However, Cdc5-GFP SPB puncta faced toward the nucleus in respect to both Spc72-mCherry and Bfa1-mCherry (Figure 6B and Supplemental Figure S3, B and C). We quantified these observations by comparing the distances between the centroids of Cdc5-GFP (DistCdc5-GFP) and Spc29-RFP (DistSpc29-RFP) to the distances between centroids of Cdc5-GFP (DistCdc5-GFP) and Spc72-mCherry (DistSpc72-mCherry) and Bfa1-mCherry (DistBfa1-mCherry) in the same way as described in Figure 1B. We found that the centroids of both Bfa1-mCherry and Spc72-mCherry puncta were significantly further separated from each other than Cdc5-GFP SPB centroids. However, the mean difference between DistCdc5-GFP and DistSpc29-RFP was almost zero (Figure 6B). Thus our data suggest that during a DSB-induced G2/M arrest, Cdc5 accumulates at the nuclear surface of the SPBs.
DISCUSSION
The mechanism of Cdc5 regulation at the SPBs during the cell cycle
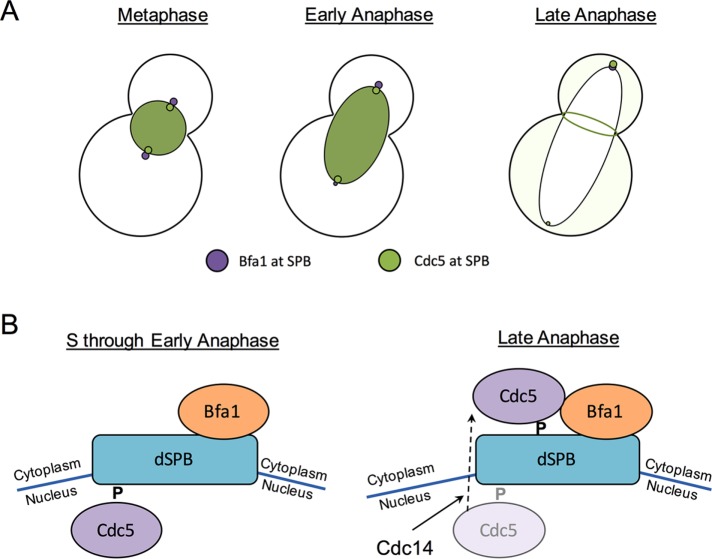
The localization of Cdc5 and Bfa1 at the SPBs has been extensively studied (Li, 1999; Gruneberg et al., 2000; Pereira et al., 2000; Song et al., 2000; Park et al., 2004a, b; Maekawa et al., 2007; Nakashima et al., 2008; Kim et al., 2012; Botchkarev et al., 2014; Gryaznova et al., 2016). Here we provide insight into the molecular mechanism of Cdc5 regulation at the SPBs during the cell cycle. We reveal that Cdc5 changes its localization and binding partners at the SPBs, depending on cell cycle stage, and that multiple factors regulate Cdc5 localization at the SPBs. Consistent with previous reports (Maekawa et al., 2007; Botchkarev et al., 2014), we show here that Cdc5 accumulates at both the mSPB and dSPB in metaphase and early anaphase but has a preference for the dSPB in late anaphase (Figure 7). Cdc5 colocalizes with Spc29 in metaphase and early anaphase. In anaphase, Cdc14 promotes the release of Cdc5 from its early cell cycle SPB population, and Cdc5 relocalizes to the outer plaque of the dSPB in a Bfa1-dependent manner (Figure 7). The FEAR network components Slk19 and Spo12 contribute to Cdc5 relocalization from the inner plaque. Because this Cdc14-dependent switch in Cdc5 SPB populations occurs later in anaphase, our data suggest that there may be other temporally slow steps after Cdc14 activation before we see final Cdc5 localization to the outer plaque.
FIGURE 7:
Model for Cdc5 localization during the cell cycle. (A) In metaphase and early anaphase, Cdc5 localizes to the nucleus and to both SPBs independently of Bfa1. Cdc5 is released from the nucleus in late anaphase, when it localizes strongly to the bud neck and to the daughter SPB. Localization of Cdc5 to the daughter SPB depends largely on Bfa1. (B) Mechanism of Cdc5 localization at the daughter SPB during the cell cycle. In S phase through early anaphase, Cdc5 localizes to the daughter SPB independently of Bfa1, likely to the nuclear daughter SPB surface. A priming phosphorylation event is required for Cdc5 localization to the SPBs. Later in anaphase, Cdc14 releases Cdc5 from its Bfa1-independent population at the SPBs. Cdc5 localizes to the daughter SPB in a Bfa1-dependent manner. The Cdc5 binding partner at the dSPB in late anaphase (likely Bfa1) undergoes a priming phosphorylation event that is required for Cdc5 localization to the dSPB in late anaphase.
The mechanism by which Cdc14 removes Cdc5 from its Bfa1-independent SPB population is unclear. It is possible that Cdc14 directly dephosphorylates Cdc5 in anaphase, because Cdc5 is a Cdk1 substrate (Mortensen et al., 2005); this is unlikely, however, because previous work showed that the cdc5-4A mutant, in which the four Cdk1 phosphorylation sites that are not essential for Cdc5 kinase activity have been mutated to alanine, can localize to the SPBs in metaphase and anaphase (Botchkarev et al., 2014). It was previously shown that the cdc5-16 pincer mutant, which does not bind to substrates that have undergone a priming phosphorylation, fails to accumulate at the SPBs in metaphase and anaphase (Ratsima et al., 2011; Botchkarev et al., 2014). It is therefore likely that Cdc14 dephosphorylates the Cdc5 metaphase SPB-binding partner and thereby releases Cdc5 from the SPB in early anaphase (Figure 7B). Because cdc5-16 does not accumulate at the SPBs in late anaphase (Ratsima et al., 2011; Botchkarev et al., 2014), at the time when Cdc5 localization to the dSPB depends on Bfa1, it is likely that the Cdc5 late anaphase dSPB outer plaque binding partner (most likely Bfa1) undergoes a priming phosphorylation event important for Cdc5 recruitment (Figure 7B). The priming kinase(s) for Cdc5 binding partners at the SPBs remains to be defined.
Regulation of Bfa1-independent Cdc5 localization to the SPBs
In this study, we identified Bfa1 as an important regulator of Cdc5 localization to the dSPB in anaphase. Because Bfa1 is found on the cytoplasmic surface of the SPB during the cell cycle (Gruneberg et al., 2000; Pereira et al., 2000; Gryaznova et al., 2016), our result reveals that Cdc5 localizes mainly to the cytoplasmic surface of the dSPB in anaphase. This is in agreement with past findings that Cdc5 is defective in localizing to SPBs of outer plaque mutants (Park et al., 2004a, b). Our analysis of Cdc5-GFP colocalization with Spc29-RFP and Spc72-mCherry suggests that Cdc5-GFP localizes to the nuclear SPB surface in metaphase and early anaphase. This observation is supported by our finding that the distance between Cdc5-GFP SPB centroids is similar to the distance between Spc29-RFP SPB centroids but shorter than the distance between Spc72-mCherry SPB centroids in almost every metaphase cell. Recent data suggest that Cdc5 is released from the nucleus to the cytoplasm in a Cdc14-dependent manner in anaphase to phosphorylate Bfa1 and promote MEN activity (Botchkarev et al., 2014). Previous work showed that Cdc5 can interact with both nuclear and cytoplasmic SPB components (Park et al., 2004a, b; Snead et al., 2007). Blocking Cdc5 nuclear import in karyopherin mutant strains prevents accumulation of Cdc5 at SPBs in small/medium-budded cells (Nakashima et al., 2008). These studies are consistent with our findings that Cdc5 localizes to the nuclear surface of the SPB in metaphase/early anaphase and relocalizes to the cytoplasmic SPB surface in a Bfa1-dependent manner in late anaphase.
Physiological relevance of Cdc5 retention at the dSPB in anaphase by Bfa1
Our work reveals that Bfa1 is important to retaining Cdc5 at the dSPB in late anaphase. Because Bfa1 is a major substrate of Cdc5 in anaphase for activation of the MEN (Hu et al., 2001; Geymonat et al., 2003), we suggest that Cdc5 and Bfa1 should be found in a complex at the SPB in anaphase for timely MEN activation. In support of this idea, cdc5-16, which cannot localize to the SPBs in anaphase (Ratsima et al., 2011; Botchkarev et al., 2014), is defective in phosphorylation of Bfa1 (Ratsima et al., 2011). It was previously reported that Cdc5 localization to the SPB is important for cytokinesis (Park et al., 2004b). It will be of interest to study whether Bfa1 retains Cdc5 at the dSPB in late anaphase to promote cytokinesis.
In summary, our work revealed that Cdc5 is found in distinct populations at the SPBs, depending on cell cycle stage. We identified Cdc14 to be necessary and sufficient to remove Cdc5 from its early mitotic population at both mother and daughter SPBs and found that Bfa1 is an important regulator of Cdc5 localization to the dSPB in late anaphase. Studying the roles of Cdc5 at each of its SPB populations will be important in explaining the timing of the functions of this very complex protein during the cell cycle.
MATERIALS AND METHODS
Yeast genetics and plasmid construction
The strains and plasmids are listed in Supplemental Tables S1 and S2. All strains in this study were isogenic or congenic to BY4741 (MATa leu2∆0 his3∆1 met15∆0 ura3∆0) and JKM179, which is a derivative of S288C. Standard yeast genetics was used to generate the strains. SY strains were gifts from D. Pellman (Dana-Farber Cancer Institute, Boston, MA) and S. Yoshida (Gunma University, Japan). p100 was a gift of A. Amon (Massachusetts Institute of Technology, Cambridge, MA). GAL1-CDC14 was integrated at the CDC14 locus after plasmid linearization with AflII. Gene deletions and modifications were performed using pFA6a vectors provided by J. Pringle (Stanford University, Stanford, CA) as previously described (Longtine et al., 1998).
Cell culture
Cells in Figures 1 and 3 and Supplemental Figures S1 and S2 were cultured in yeast extract/peptone/dextrose (YPD) medium at room temperature, fixed with 4% paraformaldehyde (Boston Bio Products), and washed at least three times in 1× phosphate-buffered saline (PBS) before imaging. For live-cell imaging in Figures 2 and 5, cells were cultured in synthetic complete plus 2% dextrose medium at room temperature. For fluorescence microscopy of Figure 4, cells were cultured in YP plus 2% raffinose at room temperature and arrested in 10 μg/ml nocodazole for 4 h, and 2% galactose was subsequently added for an additional 2 h. Cells were fixed in 4% paraformaldehyde (Boston Bio Products) and washed at least three times in 1× PBS before imaging. Some cells of the cdh1∆ strains appeared abnormally large and were excluded from the analysis. Cells in Figure 6A were cultured in YP plus 2% raffinose overnight at 30°C overnight and released at low density into YP plus 2% galactose medium at 30°C the next day for 6 h. For the fluorescence microscopy of Figure 6B and Supplemental Figure S3, cells were cultured in YP plus lactate medium overnight at 30°C and released at low density into YP + 2% galactose medium at 30° the next day for 6 h. Cells were fixed in 4% paraformaldehyde (Boston Bio Products) and washed at least three times in 1× PBS before imaging.
Fluorescence microscopy
Fluorescence images in Figures 1 and 6 and Supplemental Figures S1–S3 were acquired as z-stacks with the Nikon Ni-E fluorescence microscope equipped with a charge-coupled device camera (DU-897; Andor) and a numerical aperture (NA) 1.45/100× oil objective. Fluorescence images in Figures 2– 5 were acquired as z-stacks with the Eclipse E600 Nikon fluorescence microscope equipped with a charge-coupled device camera (DC350F; Andor) and NA 1.4/60× oil objective. All images were captured using NIS-Elements software (Nikon), and figures were assembled using GraphPad Prism 7.
Fluorescence intensity quantification and statistics
Image analysis of Figures 1 and 6 and Supplemental Figures S1–S3 was performed using the Fiji image processing package of ImageJ. ND2 image files acquired on the Nikon Ni-E fluorescence microscope were converted to TIFFs, and maximum-intensity projections were generated for each channel. Centroid X and Y-coordinates were determined from the maximum-intensity projection of an entire red or green SPB fluorescent punctum. The X- and Y-coordinates of fluorescent puncta at opposite SPBs were used to determine the distance between centroids of a given protein at opposite SPBs. A line scan in Fiji was used to quantify the relative location and fluorescence intensity of red and green pixels passing through a straight line of maximum-intensity projected cells to determine whether the red and green SPB signal peaks are distinct or overlapping. Fluorescence intensity in Figure 3B was quantified in NIS-Elements software and normalized for background signal. All statistical analyses were performed using GraphPad Prism software, using an unpaired t test. Statistical significance of is represented as follows: n.s., p > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; and ****p ≤ 0.00001. Error bars represent SD.
Supplementary Material
Acknowledgments
We are very grateful to Satoshi Yoshida for significant input on the experimental design and analysis of results, as well as for initial guidance of this work. We are grateful to Angelika Amon, Steve Elledge, Bruce Goode, Sue Lovett, David Pellman, John Pringle, Kiwon Song, and especially Satoshi Yoshida for gifts of reagents. We thank Valentina Rossio, Qiuqin Wu, Baris Avsaroglu, David Engel, and Valerie Onyeukwu for technical assistance. We are grateful to Iain Cheeseman, Valentina Rossio, and Yuko Nakajima for critical reading of versions of the manuscript. We thank Angelika Amon, Stan Burgess, Iain Cheeseman, Bruce Goode, Avi Rodal, and members of the Amon, Haber, Goode, Rodal, and Yoshida labs for helpful discussions. This work was supported by a Massachusetts Life Sciences Center Young Investigator Award to S.Y. and National Institutes of Health Grant GM61766 to J.E.H.
Abbreviations used:
- APC
anaphase-promoting complex
- dSPB
daughter spindle pole body
- FEAR
Cdc fourteen early anaphase release
- MEN
mitotic exit network
- mSPB
mother spindle pole body
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-05-0324) on February 22, 2017.
REFERENCES
- Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- Asano S, Park JE, Sakchaisri K, Yu LR, Song S, Supavilai P, Veenstra TD, Lee KS. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J. 2005;24:2194–2204. doi: 10.1038/sj.emboj.7600683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin AJ, Amon A. Men and sin: what’s the difference? Nat Rev Mol Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Botchkarev VV, Jr, Rossio V, Yoshida S. The budding yeast Polo-like kinase Cdc5 is released from the nucleus during anaphase for timely mitotic exit. Cell Cycle. 2014;13:3260–3270. doi: 10.4161/15384101.2014.953882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnianni RA, Ferrari M, Lazzaro F, Clerici M, Tamilselvan Nachimuthu B, Plevani P, Muzi-Falconi M, Pellicioli A. Elevated levels of the polo kinase Cdc5 override the Mec1/ATR checkpoint in budding yeast by acting at different steps of the signaling pathway. PLoS Genet. 2010;6:e1000763. doi: 10.1371/journal.pgen.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, de Bettignies G, Sedgwick SG. Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J Cell Biol. 2009;187:497–511. doi: 10.1083/jcb.200905114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Walker PA, Johnston LH, Sedgwick SG. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J Biol Chem. 2003;278:14591–14594. doi: 10.1074/jbc.C300059200. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryaznova Y, Caydasi AK, Malengo G, Sourjik V, Pereira G. A FRET-based study reveals site-specific regulation of spindle position checkpoint proteins at yeast centrosomes. Elife. 2016:5, e14029. doi: 10.7554/eLife.14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig NC, Uhlmann F. Preferential cleavage of chromatin-bound cohesin after targeted phosphorylation by Polo-like kinase. EMBO J. 2004;23:3144–3153. doi: 10.1038/sj.emboj.7600303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F, Wang Y, Liu D, Li Y, Qin J, Elledge SJ. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Kim J, Luo G, Bahk YY, Song K. Cdc5-dependent asymmetric localization of bfa1 fine-tunes timely mitotic exit. PLoS Genet. 2012;8:e1002450. doi: 10.1371/journal.pgen.1002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Park JE, Asano S, Park CJ. Yeast polo-like kinases: functionally conserved multitask mitotic regulators. Oncogene. 2005;24:217–229. doi: 10.1038/sj.onc.1208271. [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Li R. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc Natl Acad Sci USA. 1999;96:4989–4994. doi: 10.1073/pnas.96.9.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Wang Y. DNA damage checkpoints inhibit mitotic exit by two different mechanisms. Mol Cell Biol. 2007;27:5067–5078. doi: 10.1128/MCB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maekawa H, Priest C, Lechner J, Pereira G, Schiebel E. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J Cell Biol. 2007;179:423–436. doi: 10.1083/jcb.200705197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev. 2011;25:875–888. doi: 10.1101/gad.622411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK, Ciftci-Yilmaz S, Reynolds D, Au WC, Boeckmann L, Dittman LE, Jowhar Z, Pachpor T, Yeh E, Baker RE, et al. Polo kinase Cdc5 associates with centromeres to facilitate the removal of centromeric cohesin during mitosis. Mol Biol Cell. 2016;27:2286–2300. doi: 10.1091/mbc.E16-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen EM, Haas W, Gygi M, Gygi SP, Kellogg DR. Cdc28-dependent regulation of the Cdc5/Polo kinase. Curr Biol. 2005;15:2033–2037. doi: 10.1016/j.cub.2005.10.046. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Maruki Y, Imamura Y, Kondo C, Kawamata T, Kawanishi I, Takata H, Matsuura A, Lee KS, Kikkawa U, et al. The yeast Tor signaling pathway is involved in G2/M transition via polo-kinase. PLoS One. 2008;3:e2223. doi: 10.1371/journal.pone.0002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne MA, Schlenstedt G, Jinks T, Silver PA. Nuf2, a spindle pole body-associated protein required for nuclear division in yeast. J Cell Biol. 1994;125:853–866. doi: 10.1083/jcb.125.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Song S, Giddings TH, Jr, Ro HS, Sakchaisri K, Park JE, Seong YS, Winey M, Lee KS. Requirement for Bbp1p in the proper mitotic functions of Cdc5p in Saccharomyces cerevisiae. Mol Biol Cell. 2004a;15:1711–1723. doi: 10.1091/mbc.E03-07-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Song S, Lee PR, Shou W, Deshaies RJ, Lee KS. Loss of CDC5 function in Saccharomyces cerevisiae leads to defects in Swe1p regulation and Bfa1p/Bub2p-independent cytokinesis. Genetics. 2003;163:21–33. doi: 10.1093/genetics/163.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Park CJ, Sakchaisri K, Karpova T, Asano S, McNally J, Sunwoo Y, Leem SH, Lee KS. Novel functional dissection of the localization-specific roles of budding yeast polo kinase Cdc5p. Mol Cell Biol. 2004b;24:9873–9886. doi: 10.1128/MCB.24.22.9873-9886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Salmon ED, Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- Ratsima H, Ladouceur AM, Pascariu M, Sauve V, Salloum Z, Maddox PS, D’Amours D. Independent modulation of the kinase and polo-box activities of Cdc5 protein unravels unique roles in the maintenance of genome stability. Proc Natl Acad Sci USA. 2011;108:E914–E923. doi: 10.1073/pnas.1106448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratsima H, Serrano D, Pascariu M, D’Amours D. Centrosome-dependent bypass of the DNA damage checkpoint by the polo kinase Cdc5. Cell Rep. 2016;14:1422–1434. doi: 10.1016/j.celrep.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Rawal CC, Riccardo S, Pesenti C, Ferrari M, Marini F, Pellicioli A. Reduced kinase activity of polo kinase Cdc5 affects chromosome stability and DNA damage response in S. cerevisiae. Cell Cycle. 2016;15:2906–2919. doi: 10.1080/15384101.2016.1222338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccuzzo M, Visintin C, Tili F, Visintin R. FEAR-mediated activation of Cdc14 is the limiting step for spindle elongation and anaphase progression. Nat Cell Biol. 2015;17:251–261. doi: 10.1038/ncb3105. [DOI] [PubMed] [Google Scholar]
- Rock JM, Amon A. The FEAR network. Curr Biol. 2009;19:R1063–R1068. doi: 10.1016/j.cub.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JM, Amon A. Cdc15 integrates Tem1 GTPase-mediated spatial signals with Polo kinase-mediated temporal cues to activate mitotic exit. Genes Dev. 2011;25:1943–1954. doi: 10.1101/gad.17257711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossio V, Galati E, Ferrari M, Pellicioli A, Sutani T, Shirahige K, Lucchini G, Piatti S. The RSC chromatin-remodeling complex influences mitotic exit and adaptation to the spindle assembly checkpoint by controlling the Cdc14 phosphatase. J Cell Biol. 2010;191:981–997. doi: 10.1083/jcb.201007025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakchaisri K, Asano S, Yu LR, Shulewitz MJ, Park CJ, Park JE, Cho YW, Veenstra TD, Thorner J, Lee KS. Coupling morphogenesis to mitotic entry. Proc Natl Acad Sci USA. 2004;101:4124–4129. doi: 10.1073/pnas.0400641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Shou W, Azzam R, Chen SL, Huddleston MJ, Baskerville C, Charbonneau H, Annan RS, Carr SA, Deshaies RJ. Cdc5 influences phosphorylation of Net1 and disassembly of the RENT complex. BMC Mol Biol. 2002;3:3. doi: 10.1186/1471-2199-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead JL, Sullivan M, Lowery DM, Cohen MS, Zhang C, Randle DH, Taunton J, Yaffe MB, Morgan DO, Shokat KM. A coupled chemical-genetic and bioinformatic approach to Polo-like kinase pathway exploration. Chem Biol. 2007;14:1261–1272. doi: 10.1016/j.chembiol.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Grenfell TZ, Garfield S, Erikson RL, Lee KS. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol Cell Biol. 2000;20:286–298. doi: 10.1128/mcb.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Douziech M, Bazile F, Pascariu M, Bonneil E, Sauve V, Ratsima H, D’Amours D. Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol Cell. 2009;34:416–426. doi: 10.1016/j.molcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Badger BL, Haase J, Ohashi T, Erickson HP, Salmon ED, Bloom K. How the kinetochore couples microtubule force and centromere stretch to move chromosomes. Nat Cell Biol. 2016;18:382–392. doi: 10.1038/ncb3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toczyski DP, Galgoczy DJ, Hartwell LH. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- Valerio-Santiago M, de Los Santos-Velazquez AI, Monje-Casas F. Inhibition of the mitotic exit network in response to damaged telomeres. PLoS Genet. 2013;9:e1003859. doi: 10.1371/journal.pgen.1003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidanes GM, Sweeney FD, Galicia S, Cheung S, Doyle JP, Durocher D, Toczyski DP. CDC5 inhibits the hyperphosphorylation of the checkpoint kinase Rad53, leading to checkpoint adaptation. PLoS Biol. 2010;8:e1000286. doi: 10.1371/journal.pbio.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Visintin R, Stegmeier F, Amon A. The role of the polo kinase Cdc5 in controlling Cdc14 localization. Mol Biol Cell. 2003;14:4486–4498. doi: 10.1091/mbc.E03-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol. 2008;383:894–903. doi: 10.1016/j.jmb.2008.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, O’Toole ET. The spindle cycle in budding yeast. Nat Cell Biol. 2001;3:E23–E27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]
- Yellman CM, Roeder GS. Cdc14 early anaphase release, FEAR, is limited to the nucleus and dispensable for efficient mitotic exit. PLoS One. 2015;10:e0128604. doi: 10.1371/journal.pone.0128604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Asakawa K, Toh-e A. Mitotic exit network controls the localization of Cdc14 to the spindle pole body in Saccharomyces cerevisiae. Curr Biol. 2002;12:944–950. doi: 10.1016/s0960-9822(02)00870-9. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kono K, Lowery DM, Bartolini S, Yaffe MB, Ohya Y, Pellman D. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science. 2006;313:108–111. doi: 10.1126/science.1126747. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Toh-e A. Budding yeast Cdc5 phosphorylates Net1 and assists Cdc14 release from the nucleolus. Biochem Biophys Res Commun. 2002;294:687–691. doi: 10.1016/S0006-291X(02)00544-2. [DOI] [PubMed] [Google Scholar]
- Yuste-Rojas M, Cross FR. Mutations in CDC14 result in high sensitivity to cyclin gene dosage in Saccharomyces cerevisiae. Mol Gen Genet. 2000;263:60–72. doi: 10.1007/pl00008676. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Yung PY, Huo L, Liang C. Cdc14p resets the competency of replication licensing by dephosphorylating multiple initiation proteins during mitotic exit in budding yeast. J Cell Sci. 2010;123:3933–3943. doi: 10.1242/jcs.075366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.