Apical junctions in epithelial cells are important in the regulation of the epithelial barrier. Phosphorylation of Rab11-FIP2 by MARK2 regulates Rab1l-FIP2’s interaction with Eps15, the composition of these junctions, and the establishment of polarity.
Abstract
MARK2 regulates the establishment of polarity in Madin–Darby canine kidney (MDCK) cells in part through phosphorylation of serine 227 of Rab11-FIP2. We identified Eps15 as an interacting partner of phospho-S227-Rab11-FIP2 (pS227-FIP2). During recovery from low calcium, Eps15 localized to the lateral membrane before pS227-FIP2 arrival. Later in recovery, Eps15 and pS227-FIP2 colocalized at the lateral membrane. In MDCK cells expressing the pseudophosphorylated FIP2 mutant FIP2(S227E), during recovery from low calcium, Eps15 was trapped and never localized to the lateral membrane. Mutation of any of the three NPF domains within GFP-FIP2(S227E) rescued Eps15 localization at the lateral membrane and reestablished single-lumen cyst formation in GFP-FIP2(S227E)–expressing cells in three-dimensional (3D) culture. Whereas expression of GFP-FIP2(S227E) induced the loss of E-cadherin and occludin, mutation of any of the NPF domains of GFP-FIP2(S227E) reestablished both proteins at the apical junctions. Knockdown of Eps15 altered the spatial and temporal localization of pS227-FIP2 and also elicited formation of multiple lumens in MDCK 3D cysts. Thus an interaction of Eps15 and pS227-FIP2 at the appropriate time and location in polarizing cells is necessary for proper establishment of epithelial polarity.
INTRODUCTION
Rab11-FIP2, a member of the Rab11 family of interacting proteins (Rab11-FIPs), plays an important role in apical recycling in epithelial cells (Cullis et al., 2002; Lindsay and McCaffrey, 2002; Fan et al., 2004; Naslavsky et al., 2006; Ducharme et al., 2007, 2011; Nedvetsky et al., 2007; Schwenk et al., 2007; Chu et al., 2009). Previous studies identified multiple proteins that directly interact with Rab11-FIP2 and regulate recycling. Our lab identified the ternary trafficking complex of Rab11-FIP2, members of the Rab11 family of small GTPases (Rab11a, Rab11b, and Rab25), and the motor protein myosin Vb (MYO5B) in the regulation of the plasma membrane recycling system (Hales et al., 2001, 2002). Other labs identified members of the Eps15 homology (EH)–domain family of endocytic proteins EHD1–3 (Naslavsky et al., 2006) and Reps1 (Cullis et al., 2002) as direct interactors with Rab11-FIP2. We previously showed that Rab11-FIP2 has a second role, independent of Rab11a and MYO5B, in the establishment of cellular polarity and apical junction composition (Ducharme et al., 2006; Lapierre et al., 2012). These polarity-associated functions of Rab11-FIP2 are regulated via phosphorylation on serine-227 by the polarity-regulating kinase MARK2/Par1b. In this work, we identify an interaction of phospho-S227-Rab11-FIP2 (pS227-Rab11-FIP2) with Eps15 that is also involved in the establishment and maintenance of epithelial cell polarity.
Epidermal growth factor receptor (EGFR) pathway substrate clone 15 (Eps15; Fazioli et al., 1993) is an adaptor protein that is involved in endocytosis through its interaction with the clathrin adaptor AP-2 at the plasma membrane (Chi et al., 2008) and trafficking from the trans-Golgi network through its interaction with another clathrin adaptor, AP-1 (Benmerah et al., 1995). At the plasma membrane, Eps15 localizes to the clathrin-coated pits, forming a ring around the edge of a pit and recruiting AP-2 (Tebar et al., 1996; Benmerah et al., 2000), which in turn recruits clathrin into the pit, thus forming the clathrin basket that will mature and bud off as a clathrin-coated vesicle. Eps15 is not a component of the clathrin-coated vesicle; instead, clathrin competes Eps15 away from AP-2 (Cupers et al., 1998). Eps15 is also involved in the exit of secretory proteins from the trans-Golgi network through its interaction with AP-1 (Chi et al., 2008). AP-1 and AP-2 bind at two separate but adjacent domains in the carboxyl terminus of Eps15. There are also two ubiquitin-interacting motifs in the carboxyl terminus of Eps15 (van Bergen en Henegouwen, 2009).
The amino terminus of Eps15 contains three EH domains predicted to bind to NPF (asparagine-proline-phenylalanine) domains. Previously four other EH domain–containing proteins—EHD1, EHD2 and EHD3 (Naslavsky et al., 2006), and Reps (Cullis et al., 2002)—were shown to interact with Rab11-FIP2 and control transferrin and EGFR trafficking, respectively. Here we identify a fifth EH domain–containing protein that interacts with Rab11-FIP2, Eps15, which exhibits an in situ preference for the phosphorylated version of Rab11-FIP2. This interaction of Eps15 with pS227-Rab11-FIP2 occurs at a discrete time and location at the lateral membrane to properly regulate apical junction composition.
RESULTS
Identification of phospho-S227-Rab11-FIP2 interaction with Eps15
To investigate proteins that interact with phospho-S227-Rab11-FIP2 (pS227-FIP2), we used an anti–pS227-FIP2 antibody (Lapierre et al., 2012) to immunoprecipitate pS227-FIP2 either from Madin–Darby canine kidney (MDCK) cells lysed with 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) detergent or from a vesicle preparation of mechanically disrupted MDCK cells. The immunoprecipitated material was processed, and tryptic digests were analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) proteomics (Supplemental Table S1). Within our proteomic results, we identified Eps15 tyrosine kinase substrate as interacting with pS227-FIP2 (Supplemental Table S1). Although Eps15 did not give the highest fold difference, and other members of the EH domain family were shown to interact with FIP2, we chose to evaluate the Eps15/pS227-FIP2 interaction. Nonconfluent MDCK cells stained for Eps15 and pS227-FIP2 exhibited colocalization at corners of the leading edge (indicated by the arrowhead in Supplemental Figure S1A). Little colocalization was observed between Eps15 and either of two known interactors of FIP2 involved in receptor trafficking, myosin Vb or Rab11a (Supplemental Figure S1B). These results are consistent with our previous findings that neither myosin Vb nor Rab11a is involved with pS227-FIP2 (Lapierre et al., 2012).
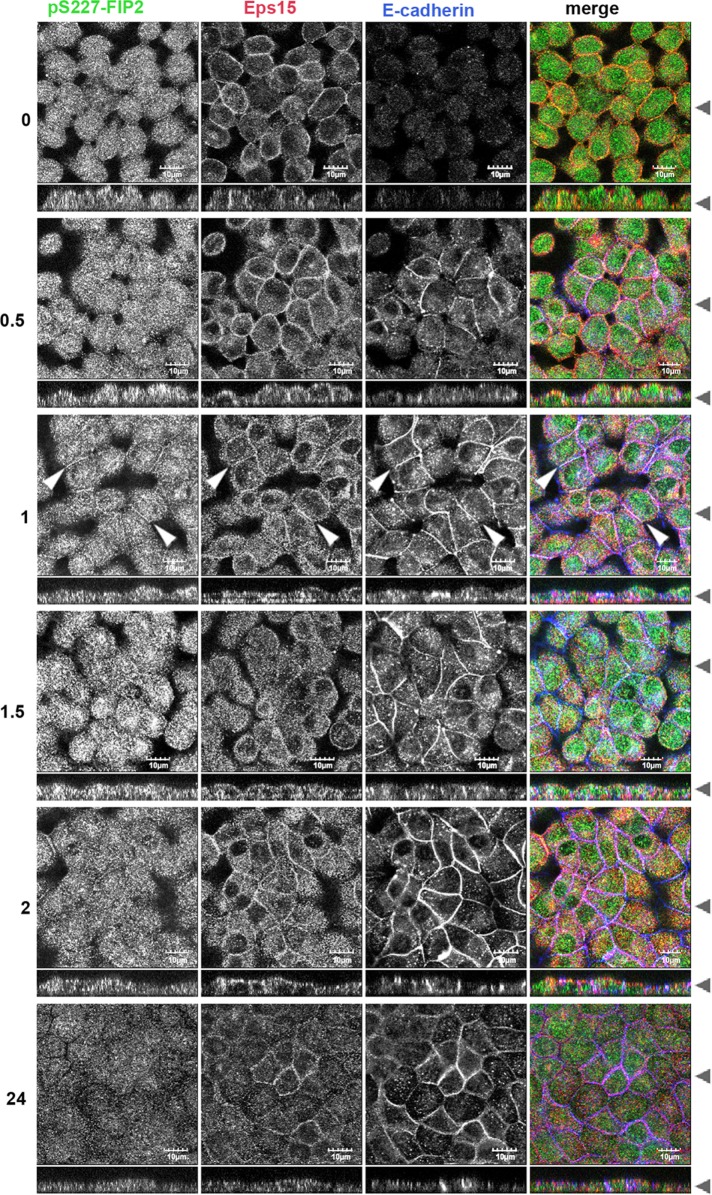
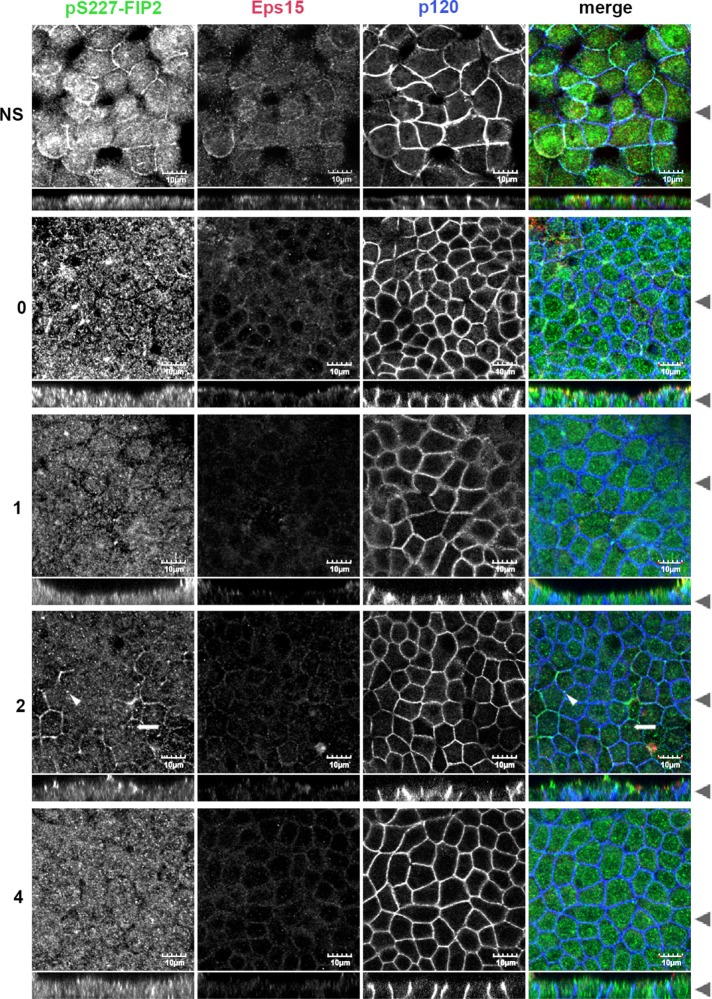
Using a calcium switch assay, we next examined when and where Eps15 and pS227-FIP2 might interact when cells are reestablishing polarity. Five-day-postconfluent monolayers of MDCK cells were exposed to low-calcium medium overnight. The next day, the monolayers were refed with regular-calcium-containing medium, fixed after 0–24 h, and then costained for endogenous pS227-FIP2, Eps15, and E-cadherin (Figure 1). In low-calcium medium (time point 0), endogenous Eps15 was observed near the peripheral membrane, whereas endogenous pS227-FIP2 localized in dispersed vesicles in the cytoplasm. A similar distribution was observed 30 min into the recovery. At 1 h of recovery, both Eps15 and pS2227-FIP2 were observed at the peripheral membrane with E-cadherin (indicated by arrowheads, 1-h time point, Figure 1). Over the next 2 h, pS227-FIP2 staining exhibited a gradual decrease, whereas Eps15 remained at the membrane. At 24 h after the readdition of calcium, little pS227-FIP2 was observed, whereas some Eps15 remained at the membrane (Figure 1). These results suggested that the majority of pS227-FIP2 and Eps15 do not travel together on the same vesicle but instead meet at the periphery of the cell early in the reestablishment of polarity, with Eps15 reaching the lateral membrane before pS227-FIP2 arrival.
FIGURE 1:
During recovery from low calcium, Eps15 arrived at the lateral membrane before pS227-FIP2. MDCK cells were grown on Transwell supports for 5 d postconfluence, incubated overnight in low calcium, and then returned to regular media and allowed to recovery for the hours indicated on the left. Cells were fixed in methanol and stained for pS227-FIP2 (green merge), Eps15 (red in merge), and E-cadherin (blue in merge). White arrowheads indicate areas of colocalization of pS227-FIP2, Eps15, and p120. Black arrowheads indicate where the xy- and xz-slices were taken. Bar, 10 μm.
Eps15 associated preferentially with pseudophosphorylated green fluorescent protein–Rab11-FIP2
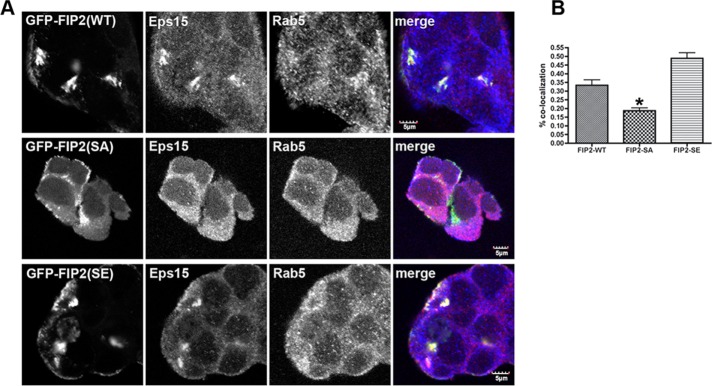
To ascertain whether endogenous Eps15 had a preference for either of the S227 phosphorylation mutants of FIP2 (Ducharme et al., 2006; Lapierre et al., 2012), we stained nonpolarized MDCK cell lines expressing green fluorescent protein (GFP)–FIP2(WT), GFP-FIP2(S227A) (nonphosphorylatable), or GFP-FIP2(S227E) (pseudophosphorylated) for Eps15. As seen in Figure 2, when GFP-FIP2(WT) and GFP-FIP2(S227E), but not GFP-FIP2(S227A), are overexpressed in nonpolar cells, the GFP-FIP2-containing vesicles clustered internally in the cell. Colocalization was observed between Eps15 and GFP-FIP2(WT) and GFP-FIP2(S227E), but not with GFP-FIP2(S227A) (Figure 2A). Although we observed only endogenous Eps15 and endogenous pS227-FIP2 together at the lateral membrane, overexpression of GFP-FIP2(WT) and GFP-FIP2(S227E) caused endogenous Eps15 to redistribute to the internal tubulovesicular structures containing the GFP-FIP2 proteins. Some colocalization was observed between Eps15 and GFP-FIP2(S227A), but the amount was significantly less than that seen with GFP-FIP2(WT) or GFP-FIP2(S227E) (Figure 2B).
FIGURE 2:
Eps15 accumulated in GFP-FIP2(WT) and GFP-FIP2(S227E)–containing compartments but not the GFP-FIP2(S227A)–containing membranes. (A) The GFP-FIP2–expressing MDCK cell lines were grown to subconfluence on coverglass slips, fixed in 4% paraformaldehyde, and stained for Eps15 (red in merge) and Rab5 (blue in merge). Bar, 5 μm. (B) Manders quantification of colocalization in A; a minimum of 60 cells were counted per cell line. Eps15 colocalization with GFP-FIP2(S227A) exhibited a significant decrease compared with Eps15 with GFP-FIP2(WT) or GFP-FIP2(S227E). *p < 0.05 by Dunn’s test.
In a yeast 2-hybrid binary assay, we did observe an interaction between Eps15 and FIP2(S227A) that was not significantly different from that observed between Eps15 and FIP2(WT) or FIP2(S227E) (unpublished data). Yeast 2-hybrid assays are very sensitive and can detect relatively weak interactions. In our previous work (Lapierre et al., 2012), we observed interaction between myosin Vb and FIP2(S227E) in a yeast 2-hybrid assay but not when the GFP-myosin Vb tail–expressing MDCK cell line was stained for endogenous pS227-FIP2. In both cases, we believe that overexpression of the proteins allowed a detectable interaction versus observation of the behavior of the endogenous pool of, in this case, Eps15. These findings suggested that the interaction of Rab11-FIP2 with Eps15 is facilitated by the phosphorylation of serine 227 in Rab11-FIP2.
All three NPF domains of Rab11-FIP2 are necessary for interaction with Eps15
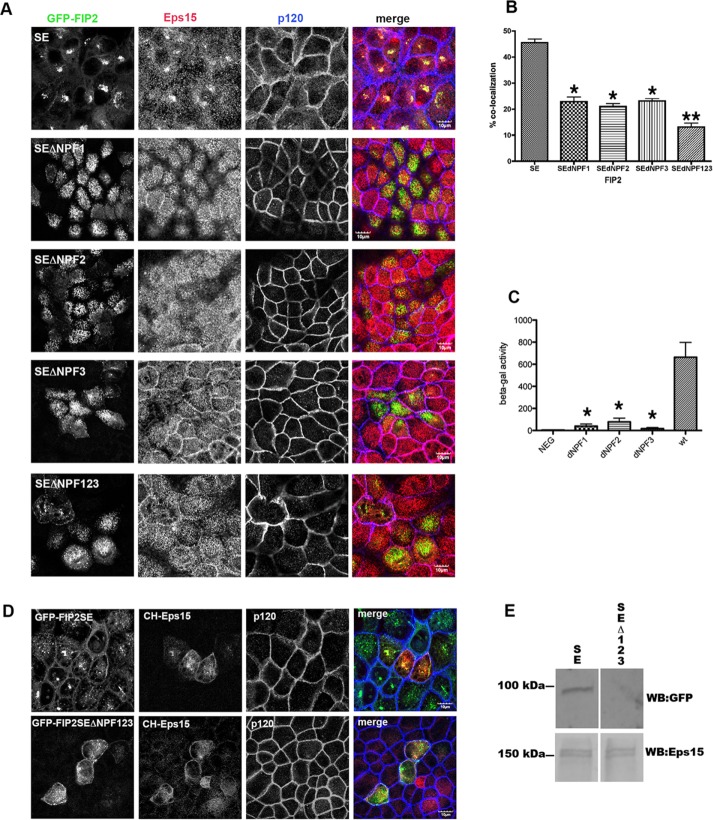
The Caplan lab demonstrated that EHD1 and EHD3 interact with FIP2 mainly though the second NPF domain of FIP2 (Naslavsky et al., 2006). When we overexpressed GFP-FIP2(S227E) (Figure 3) or GFP-FIP2(WT) (unpublished data) in MDCK cells with each individual NPF domain mutated or all three NPF domains mutated, we found loss of colocalization with Eps15 (Figure 3, A and B). These studies were performed in pooled cell lines, and thus some variation in the level of GFP expression was observed, and images were taken only for moderately GFP-expressing cells. This loss of interaction was also observed in a yeast-2-hybrid binary assay (Figure 3C). Our results indicate that unlike EHD1–3 and Reps1, all three of the Rab11-FIP2 NPF domains are necessary for interaction with Eps15. Of interest, when the MDCK cell lines expressingGFP-FIP2(WT) or FIP2(S227E), but not FIP2(S227A), were not fully polarized either by plating cells on glass coverslips (Figure 3A) or by removing calcium from monolayers grown on Transwell filters (the zero time point of Figure 4), a tight nidus of GFP-FIP2 was observed in the cells (Ducharme et al., 2006; Lapierre et al., 2012). However, when any of the NPF domains in GFP-FIP2(S227E) (Figure 3A) or GFP-FIP2(WT) (unpublished data) were mutated, the GFP-FIP2 was observed on dispersed vesicles. Although merged images show some yellow pixel overlap, when the individual grayscale images are compared, the patterns between the GFP-FIP2s and endogenous Esp15 are very different.
FIGURE 3:
Mutation of all three NPF domains or any individual NPF domain of FIP2 released Eps15 from the GFP-FIP2(S227E) compartment. (A) The GFP-FIP2(S227E) MDCK cell lines with or without the NPF domain mutations were plated onto glass coverslips, fixed with 4% paraformaldehyde, and stained for Eps15 (red in merge) and F-actin (phalloidin; blue in merge). Bar, 5 μm. (B) Manders quantification of the colocalization shown in A; minimum of 30 cells per cell line were counted. The GFP-FIP2(S227E) containing any of the NPF domain mutations (SEdNPF1, SEdNPF2, or SEdNPF3) exhibited a significant loss of colocalization compared with GFP-FIP2(S227E). Mutation of all three NPF domains (SEdNPF123) elicited a further significant decrease in colocalization. *p < 0.05 by Dunn’s test vs. SE. **p < 0.05 vs. all other groups. (C) Results of yeast two-hybrid assay. The amount of β-galactosidase activity was calculated by compassion to a standard curve of known β-galactosidase concentrations. The assay was performed three separate times. NEG, negative control. The GFP-FIP2(S227E) containing any of the NPF domain mutations exhibited a significant loss of colocalization compared with GFP-FIP2(S227E). *p < 0.05 by Dunn’s test. (D) GFP-FIP2(S227E) and GFP-FIP2(S227EΔNFP123) MDCK cells were transfected mCherry-Eps15, fixed, and stained for p120 (blue in merge). mCherry-Eps15 was localized with the GFP-FIP2(S227E) but not with coexpressed GFP-FIP2(S227ΔNPF123). (E) Western blot of mCherry-Eps15 precipitated from GFP-FIP2(S227) or GFP-FIP2(S227ΔNPF123)–expressing cells. The blot was probed simultaneously for GFP (top) and Eps15 (bottom) and imaged on a LiCor Odyssey FC imager. Size makers are shown on the left.
FIGURE 4:
Eps15 localized to the central GFP-FIP2(S2227E) compartment and away from the lateral membrane in low calcium. The MDCK cell line expressing GFP-FIP2(S227E) was grown on Transwells, switched into low-calcium medium, and allowed to recover for the hours listed on the left. Cells were fixed in 4% paraformaldehyde and stained for Eps15 (red in merge) and p120 (blue in merge). Black arrowheads indicate where xy- and xz-slices were taken. Bar, 10 μm.
To validate further the interaction between GFP-FIP2(SE) and Eps15, we expressed mCherry-Eps15 in the MDCK GFP-FIP2(SE)– and GFP-FIP2(SEΔNPF123)–expressing cell lines. As shown in Figure 3D, mCherry-Eps15 localized with GFP-FIP2(SE) but not with GFP-FIP2(SEΔNPF123). When mCherry-Eps15 was precipitated from the two GFP-FIP2–expressing MDCK cell lines, GFP-FIP2(S227E) was coisolated, whereas very little GFP-FIP2(SEΔNPF123) precipitated with the mCherry-Eps15 (Figure 3E). An equal amount of mCherry-Eps15 was precipitated from the mCherry-Eps15 transient GFP-FIP2–expressing cell lines (Figure 3E).
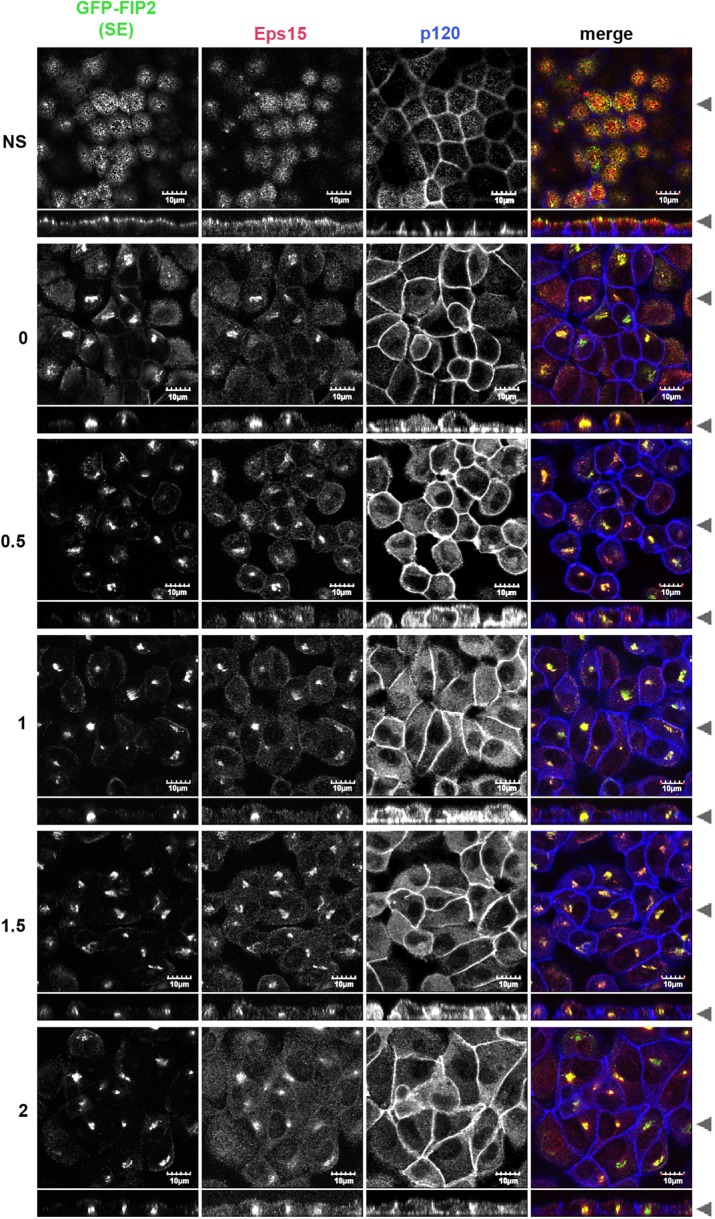
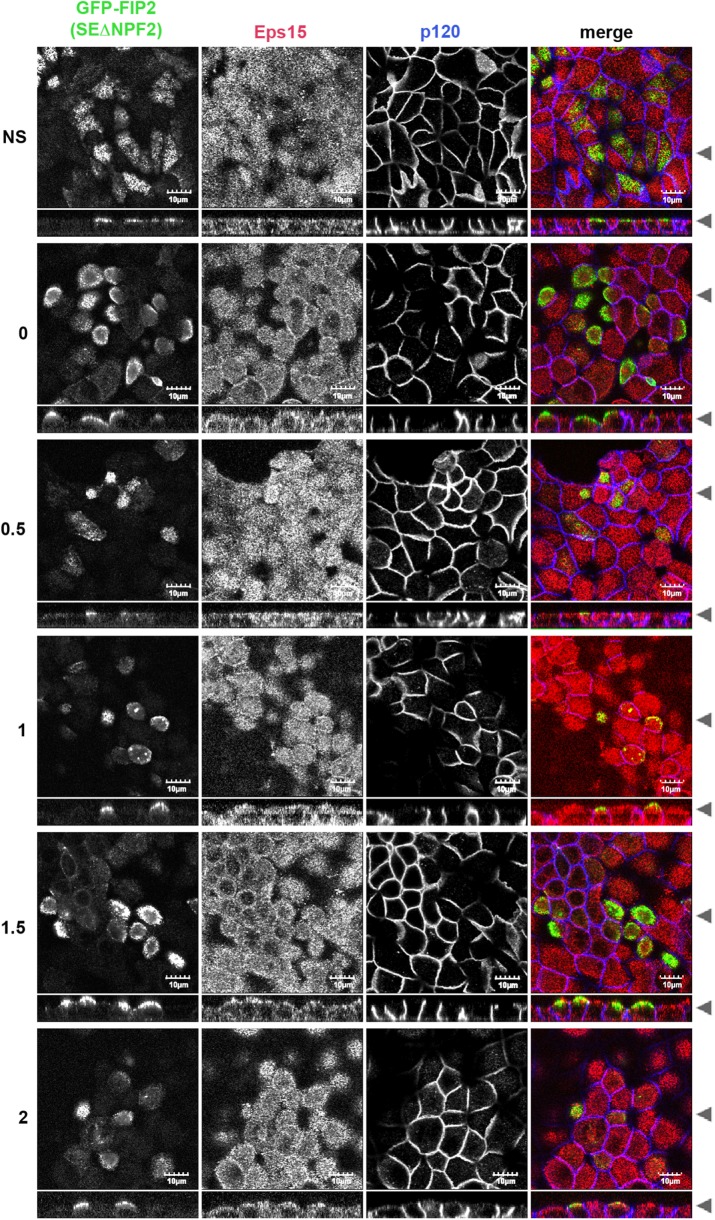
We next tested whether GFP-FIP2(S227E) could alter Eps15 localization during a calcium switch assay. In the nonswitched GFP-FIP2(S227E)–expressing MDCK cell line, GFP-FIP2(S227E) and Eps15 were observed on separate vesicles, and Eps15 could also be observed at the peripheral membrane (Figure 4). In the low-calcium depolarized cells, GFP-FIP2(S227E) collapsed into a central membranous cisternum, and Eps15 localized into this structure. When the GFP-FIP2(S227E)–expressing MDCK cell line recovered from low calcium, endogenous Eps15 remained trapped in the collapsed structure containing GFP-FIP2(S227E) (Figure 4). Thus Eps15 never relocated to the cell membrane. In cells expressing GFP-FIP2(S227E) with mutated versions of the first NPF domain (Supplemental Figure S2), the second NPF domain (Figure 5), or the third NPF domain (Supplemental Figure S3), Eps15 did relocate to the peripheral membrane, although the localization was not as complete as in the parental MDCK cell line (Figure 1).
FIGURE 5:
Mutation of the second NPF domains in GFP-FIP2(S227E) released Eps15 from the GFP-FIP2(S227E) compartment, allowing Eps15 to localize at the lateral membrane. The MDCK cell line expressing GFP-FIP2(S227EΔNPF2) was grown on Transwells, switched into low-calcium medium, and then allowed to recover for the hours listed on the left. Cells were fixed in 4% paraformaldehyde and stained for Eps15 (red in merge) and p120 (blue in merge). Black arrowheads indicate where the xy- and xz-slices were taken. Bar, 10 μm.
The pool of Eps15 that interacts with pS227-Rab11-FIP2 is separate from the pool of Eps15 that interacts with AP-2 or Numb
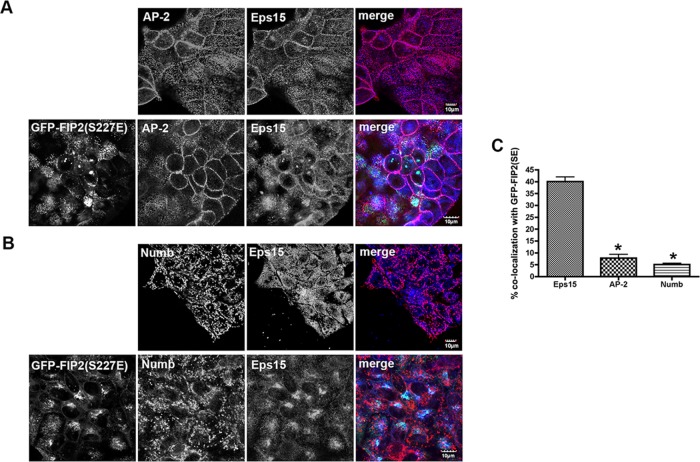
Because AP-2 and Numb are known binding partners of Eps15 (van Bergen en Henegouwen, 2009) and are involved in the formation of clathrin-coated pits during endocytosis, we investigated whether the Eps15 that we observed with pS227-FIP2 could also associate with AP-2 or Numb. In MDCK cells grown on coverslips, AP-2 and Numb were located at the membrane and on internal structures, respectively. In GFP-FIP2(S227E)–expressing MDCK cells grown on coverslips, neither AP-2 nor Numb was observed in the GFP-FIP2(S227E)/Eps15–containing complex (Figure 6). Thus, unlike Eps15, the localization of AP-2 and Numb were not affected by the overexpression of GFP-FIP2(SE). The findings suggest that the pool of Eps15 that is involved in cell polarity with pS227-Rab11-FIP2 is separate from that involved with both AP-2 in clathrin-coated pit establishment (Tebar et al., 1996) and that involved with Numb in endocytosis (Santolini et al., 2000).
FIGURE 6:
pS227-FIP2 did not colocalize with two known Eps15-binding proteins. The parental MDCK cells (top) or the GFP-FIP2(S227E)–expressing MDCK cells (bottom) were grown on coverslips, fixed with paraformaldehyde, and stained for Eps15 (blue in merge) and either AP-2 (A; red in merge) or Numb (B; red in merge). Bar, 10 μm. (C) Quantitation of Eps15, AP-2, and Numb colocalization with GFP-FIP2(S227E). At least 30 cells were counted for each staining pattern. *p < 0.05 by Dunn’s test compared with Eps15 colocalization.
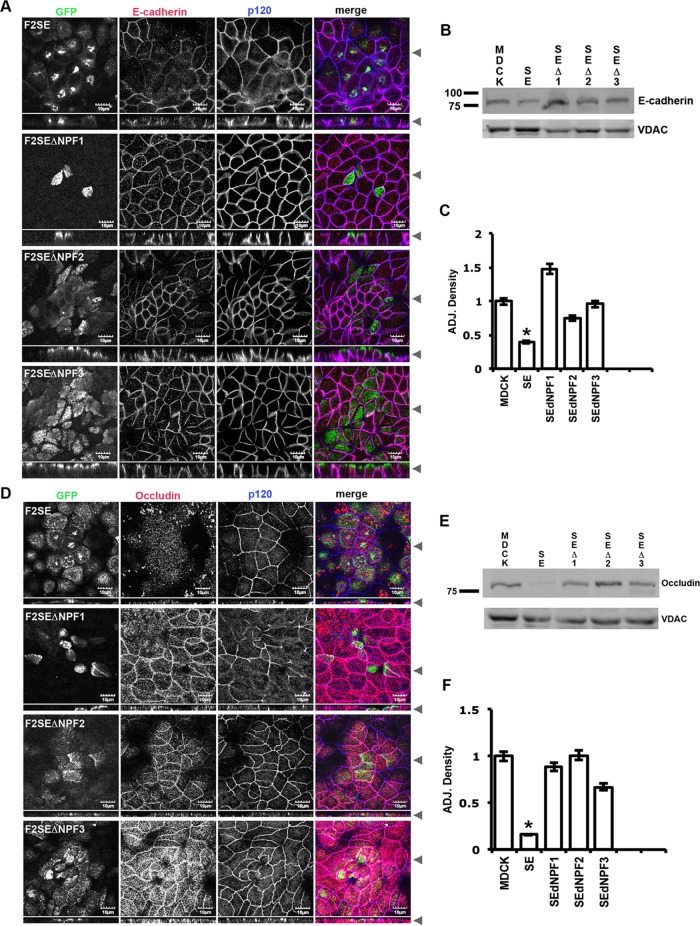
Mutation of any of the NPF domains of GFP-FIP2(S227E) restored expression of E-cadherin and occludin at the apical junctions
Previously we observed that E-cadherin and occludin were lost from their respective junctions in an MDCK cell line expressing GFP-FIP2(S227E), whereas p120 and K-cadherin and ZO-1 remained at the adherens junction and tight junction, respectively (Lapierre et al., 2012). Because we observed that Eps15 was sequestered away from the lateral membrane when GFP-FIP2(S227E) was expressed, we examined whether release of Eps15 from GFP-FIP2(S227E) in the NPF domain mutants would allow recovery of E-cadherin and occludin at the apical junctions. In MDCK cells expressing any of the NPF mutants of GFP-FIP2(S227E), E-cadherin (Figure 7, A–C) and occludin (Figure 7, D–F) were expressed at normal levels, and both proteins were observed at their respective junctions. These results indicate that Rab11-FIP2 association with Eps15 regulates proper localization of E-cadherin and occludin at their respective junctions.
FIGURE 7:
Release of Eps15 from the GFP-FIP2(S227E) compartment by mutation of any of the NPF domains rescued the expression and localization of both E-cadherin and occludin at the junctions. The GFP-FIP2(S227E)–expressing MDCK cell lines were grown on Transwells, fixed with methanol, and stained for (A) E-cadherin (red in merge) and p120 (blue in merge) or (D) occludin (red in merge) and ZO-1 (blue in merge). Black arrowheads indicate where the xy- and xz-slices were taken. Bars, 10 μm. The cell lines were also grown in 10-cm tissue culture dishes for Western blots (B, E). (B) Representative blot probed for E-cadherin (top) and VDAC (bottom). (C) The adjusted density to VDAC of three similar blots for E-cadherin. (E) A representative blot probed for occludin (top) and VDAC (bottom). (F) The adjusted density to VDAC of three similar blots for occludin. *p < 0.05 by Dunn’s test vs. parental MDCK cells.
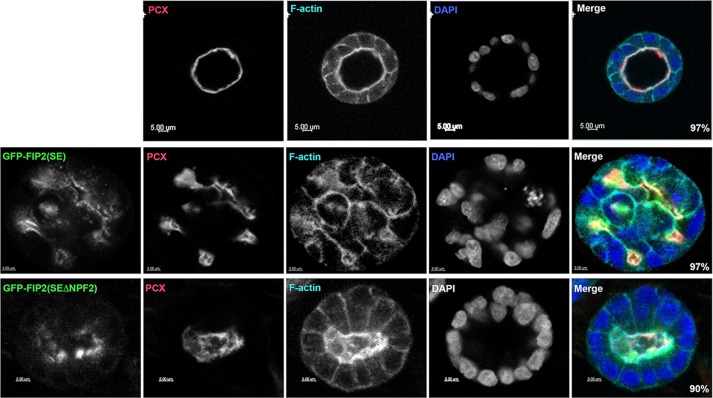
Mutation of the second NPF domain returned cysts to a single-lumen morphology
We previously noted that the MDCK cells expressing Rab11-FIP2(S227E) developed multilumen cysts when grown in Matrigel (Lapierre et al., 2012). We grew the GFP-FIP2(S227EΔNPF2) MDCK cell line in Matrigel to determine whether the disconnection between GFP-FIP2(S227EΔNPF2) and Eps15 could restore a single lumen to the double-mutated GFP-FIP2(S227EΔNPF2) cysts. Parental MDCK cells formed cysts with a single lumen (Figure 8, top row, and Supplemental Video S1), whereas the GFP-FIP2(S227E)–expressing MDCK cells formed multiple luminal cysts (Figure 8, middle row, and Supplemental Video S2), as we previously reported (Lapierre et al., 2012). When the GFP-FIP2(S227EΔNPF2)–expressing MDCK line was grown in Matrigel (Figure 8, bottom, and Supplemental Video S3), the cysts formed a single lumen. The single lumens formed by the GFP-FIP2(S227EΔNPF2) cell line tended to be more oval than those formed by the parental MDCK cell line. These results indicate that release of Eps15 from the tubulovesicular GFP-FIP2(S227E)–containing structure by mutation of the NPF domains allowed the return of E-cadherin and occludin to their respective junctions and the return of the cellular coordination necessary for the formation of a single-lumen cyst.
FIGURE 8:
Release of Eps15 from the GFP-FIP2(S227E) compartment by mutation of the second NPF domain resolved the multilumen GFP-FIP2(S227E) morphology back to a single- lumen morphology. Parental MDCK cells (top), GFP-FIP2(S227)–expressing MDCK cells (middle, green in merge), and GFP-FIP2(S227ΔNPF2)–expressing MDCK cells (bottom, green in merge) were grown in Matrigel for 7 d, fixed, and stained for podocalyxin (red in merge), F-actin (cyan in merge), and DAPI (blue in merge). Bars, 5 μm. Numbers in the lower right corner of the merged images indicate the percentage of cysts exhibiting the illustrated morphology; 15 cysts from three separate experiments were counted.
Knockdown of Eps15 alters the localization of pS227-FIP2 but not the ability of Rab11-FIP2 to be phosphorylated
To explore whether Eps15 is necessary for proper phosphorylation or localization of pS227-FIP2, we knocked down Eps15 expression in MDCK cells (Figure 9) and examined the localization of endogenous pS227-FIP2 during recovery from low calcium. We achieved 63% knockdown of Eps15 in MDCK cells (Supplemental Figure S4A). During recovery from calcium switch, Rab11-FIP2 was phosphorylated on serine 227 in the Eps15-knockdown MDCK cell line. Although we observed pS227-FIP2 early (0–2 h) in the recovery from low calcium in the control knockdown MDCK cell line (Supplemental Figure S4B) and parental MDCK cells (Figure 1), the timing of arrival of pS227-FIP2 at the lateral membrane was altered in the Eps15-knockdown MDCK cell line (Figure 9). In the Eps15-knockdown cell line, pS227-FIP2 was observed at the lateral membrane in the nonswitch cells. At the zero time point, there was still some pS227-FIP2 at the lateral membrane, but at the 1-h time point, very little pS227-FIP2 was observed at the lateral membrane. After 2 h of recovery, the pS227-FIP2 was observed at the lateral membrane in “corners” where more then two cells intersected (indicated by the arrowhead in Figure 9) and in small, tight structures in the center of the cells (indicated by the arrow in Figure 9). This timing of pS227-FIP2 localization at the lateral membrane is different from the pattern observed in either the control knockdown MDCK cells (Supplemental Figure S4) or the MDCK parent cells (Figure 1). In these control cell lines, the greatest amount of pS227-FIP2 in the lateral membranes was observed at the 1-h time point, with little at the 2-h or nonswitched time points. These results, along with our previous work, indicate that Eps15 is necessary for the proper spatial and temporal localization of pS227-FIP2 and this localization is necessary for proper junctional composition and cellular polarity.
FIGURE 9:
The temporal and spatial timing of pS227-FIP2 was altered when Eps15 protein expression was knocked down. An MDCK cell line expressing shRNA to Eps15 was grown on Transwells, switched into low-calcium medium, allowed to recover from low calcium for the indicted hours, and then fixed in methanol and stained for pS227-FIP2 (green in merge), Eps15 (red in merge), and p120 (blue in merge). pS227-FIP2 was observed in almost all of the time points, with the least seen at 1 h. White arrowheads indicate lateral “corners” of cells where pS227-FIP2 was observed. White arrows indicate central areas where pS227-FIP2 was observed. Black arrowheads indicate where the xy- and xz-slices were taken. Bar, 10 μm.
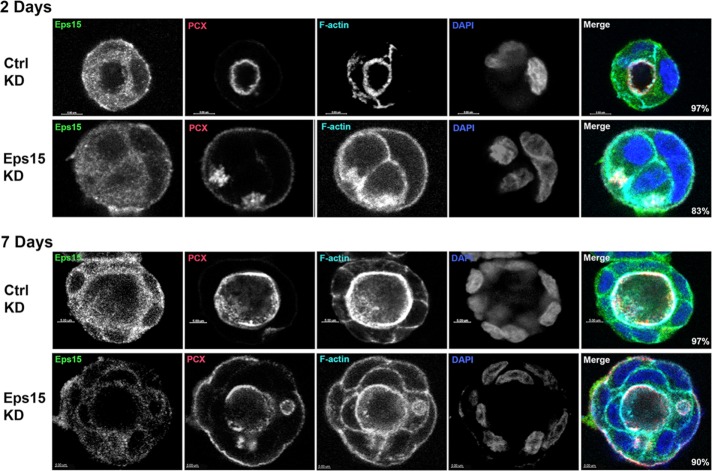
Knockdown of Eps15 caused multilumen cysts
We previously showed that GFP-FIP2(S227E)–expressing MDCK cells exhibited abnormal polarity when grown as cysts in Matrigel, with the formation of multiple lumens (Lapierre et al., 2012). MDCK cells expressing a control nonsilencing short hairpin RNA (shRNA; Figure 10, top, and Supplemental Videos S4 and S6) or an MDCK cell line expressing shRNA against Eps15 (Figure 10, bottom, and Supplemental Videos S5 and S7) were grown in Matrigel for 2 d (Figure 10, top) or 7 d (Figure 10, bottom), fixed, and stained for Eps15, podocalyxin, F-actin, and 4′,6-diamidino-2-phenylindole (DAPI). In the control knockdown cells (Figure 10, top), endogenous Eps15 was observed on vesicles just under the apical membrane and along the lateral membrane. At both the early cyst stage and the later cyst stage, the Eps15-knockdown MDCK cells formed multilumen cysts similar to the multilumen cysts formed by the MDCK GFP-FIP2(S227E)–expressing cells (Lapierre et al., 2012; Figure 8, middle, and Supplemental Video S2). These results indicate that loss of Eps15 alters the ability of the cells to form single-lumen polarized cysts, thus implicating Eps15 in the formation and maintenance of cellular polarity.
FIGURE 10:
Growth of the Eps15-knockdown MDCK cell line in Matrigel resulted in multilumen cysts. The MDCK control knockdown cell line (top) and the MDCK Eps15 knockdown cell line (bottom) were grown in Matrigel for 2 d (top) or 7 d (bottom), fixed, and stained for Eps15 (green in merge), podocalyxin (red in merge), F-actin (cyan in merge), and DAPI (blue in merge). Bars, 5 μm. Numbers in the lower right corner of the merged images indicate the percentage of cysts exhibiting the illustrated morphology; 15 cysts from three separate experiments were counted.
DISCUSSION
We first identified Rab11-FIP2 through its interaction with the Rab11 family of small GTPases (Hales et al., 2001). Subsequently we described its role in apical trafficking not only through its interaction with Rab11a, but also through its interaction with the motor protein MYO5b (Hales et al., 2002). Previous work showed the importance of phosphorylation of Rab11-FIP2 on serine 227 in regulating both tight and adherens junction composition (Ducharme et al., 2006; Lapierre et al., 2012). Of interest, unlike the role of Rab11-FIP2 in apical recycling, the function of pS227-Rab11-FIP2 in epithelial cell polarity does not require Rab11a or MYO5B (Lapierre et al., 2012). In the present work, we provide evidence that the action of pS227-Rab11-FIP2 in establishing cell polarity involves interaction with Eps15.
Other investigators showed that interactions between Rab11-FIP2 and members of the EH domain–containing family can regulate receptor trafficking. The interaction of Rab11-FIP2 with Reps regulates trafficking of the EGFR (Cullis et al., 2002), whereas the interaction of Rab11-FIP2 with EHD1 and EHD3 regulates transferrin trafficking (Cullis et al., 2002; Naslavsky et al., 2006). Both studies involved regulation of cargo transport and not regulation of cellular polarity. Reps1, EHD1, and EHD3 all contain only one EH domain, which preferentially binds the second NPF domain in Rab11-FIP2. On the other hand, Eps15 contains three EH domains, and all three of Rab11-FIP2’s NPF domains were necessary for the Eps15/Rab11-FIP2 interaction. Although Rab11-FIP2 colocalizes with Reps, EHD1, or EHD3 on vesicles within cells (Cullis et al., 2002; Naslavsky et al., 2006), we did not observe Eps15 and pS227-Rab11-FIP2 together on vesicles. Eps15 localized at the lateral membrane before the arrival of the pS227-FIP2–containing vesicles, suggesting that at least in the early stages of recovery from low calcium, Eps15 and pS227-Rab11-FIP2 are on separate membranes. This observation could indicate that Eps15 and Rab11-FIP2 interact in-trans, forming a bridge from Eps15 at the lateral membrane to Rab11-FIP2 on a vesicle. This interaction would require a high degree of avidity between the two proteins, consistent with the requirement of all three NPF domains in Rab11-FIP2 for the interaction with the three EH domains in Eps15. Alternatively, Eps15 and pS227-Rab11-FIP2 might not interact until after the pS227-Rab11-FIP2–containing vesicles fuse with the lateral membrane. Further investigation of the orientation of Eps15 and pS227-Rab11-FIP2 at the time of binding will be needed to address this issue. Nevertheless, this investigation supports the interaction of Eps15 and pS227-Rab11-FIP2 at the lateral membrane in the early stages of cell polarization as important for proper junctional composition.
During recovery from loss of polarity, Eps15 located to the lateral membrane within the first hour, followed soon after by pS227-Rab11-FIP2–containing vesicles. After calcium switch in the GFP-Rab11-FIP2(S227E)–expressing cell line, Eps15 became trapped in the central nidus formed by GFP-FIP2(S227E). If any of the three NPF domains of GFP-FIP2(S227E) were mutated, Eps15 was released and localized to the lateral membrane. In previous work, we showed that mutating Rab11-FIP2 at serine 227 to a glutamic acid (Rab11-FIP2(S227E)) caused loss of E-cadherin and occludin from their respective junctions (Lapierre et al., 2012). Here we show that mutating individually any of the three NPF domains of Rab11-FIP2(S227E) released Eps15 from the GFP-FIP2(S227E) compartment and rescued E-cadherin and occludin expression at their proper junctions. Thus dissociation of Eps15 from the mutated Rab11-FIP2(S227E) allowed proper localization of the junctional proteins. Furthermore, in previous work we showed that, when the GFP-FIP2(S227E)–expressing MDCK cell line was grown in three-dimensional (3D) culture, multiple lumens were observed (Lapierre et al., 2012). In the present studies, when we grew the MDCK cell line expressing GFP-FIP2(S227EΔNPF2), which contains both the S227E and the NPF2 mutations, in 3D culture, it restored the single-lumen morphology. Thus dissociation of the Eps15 from the mutated GFP-FIP2(S227E) allowed the coordination of cells to form a single lumen. When we knocked down Eps15 expression, the timing of pS227-FIP2 appearance at the lateral membrane was altered, such that little pS227-FIP2 was observed at the lateral membrane at the 1-h time point but instead was observed at later time points. When the Eps15-knockdown MDCK cell line was grown in 3D culture, the resulting cysts contained multiple lumens, similar to the pattern we observed with the GFP-FIP2(S227E)–expressing MDCK cell line grown in 3D culture (Lapierre et al., 2012). Thus Eps15, in concert with pS227-Rab11-FIP2, contributes to the coordination of cells to form a single-lumen cyst when grown in three dimensions.
Previous studies established a role for Eps15 in the formation of clathrin-coated pits and regulation of endocytosis through its interaction with AP-2 (Benmerah et al., 1998). Eps15 recruits AP-2 to the clathrin-coated pit (Tebar et al., 1996) and is dissociated from AP-2 as clathrin is recruited into the maturing clathrin-coated vesicle (Cupers et al., 1998). Potentially, during repolarization, a pool of Eps15 may mark a region of the membrane where junctional components are recruited during cell polarization. The Nelson lab proposed the existence of a Triton X-100–insoluble plaque on the lateral membrane of repolarizing epithelial cells where junctional components are first recruited (Adams et al., 1996, 1998). Further studies will be required to define whether Eps15 and pS227-Rab11–FIP2 define such an area on the lateral membrane during epithelial cell polarization.
MATERIALS AND METHODS
Site-directed mutagenesis and stable line production
Site-directed mutagenesis of the NPF sites to APA of Rab11-FIP2WT or S227E in the pEGFP vectors was performed using the QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) with 4-min extension time. Primers were synthesized (Sigma Aldrich, St. Louis, MO) with multiple nucleotide changes per oligonucleotide sequence using the Agilent on-line primer design site. T23 MDCK cells were transfected with Effectene (Qiagen, Waltham, MA) and plated in normal medium. The next day, the cells were refed with medium containing 200 μg/ml G418 (Cellgro, Holly Hill, FL). G418-resistent cells were then flow sorted for midrange GFP expression (Vanderbilt Flow Cytometry Shared Resource) and kept as pools.
For the Eps15-knockdown cell line, a lentiviral pLKO.1 shRNA vector targeting human Eps15 (TRCN0000007976; Sigma Aldrich) that shares 100% homology to the canine sequence was used for transduction of MDCK cells. HEK 293 cells were plated on 10-cm dishes and grown to 30% confluence. To transfect the HEK 293 cells, polyethylenimine (1 mg/ml stock) was used at a 3:1 ratio with total micrograms of DNA consisting of the Eps15 targeting vector, the packaging plasmid pR8.2, and the ENV plasmid pMDG.2. The HEK 293 cells were washed with phosphate-buffered saline (PBS) after 24 h and incubated for an additional 48 h. The medium from the 293 HEK cells was then filtered and used with Polybrene at 5 μg/ml to transduce plated MDCK cells at 30% confluence. The MDCK cells were then selected in medium containing 2 μg/ml puromycin (Cellgro).
The GFP and pBD FIP2(WT) and FIP2(SE) vectors were previously described (Ducharme et al., 2006). The FIP2 constructs were cloned into the EcoI-SalI sites in the pBD-Gal vector (Stratagene). Eps15 was amplified from the pAD vector and cloned into the XhoI-SacI sites in mCherry-C2 (Clontech). All vectors were submitted for sequencing (GenHunter, Nashville, TN), and the sequences were confirmed.
Cell culture
The GFP-Rab11-FIP2 and mutant tet-off MDCK cell lines (kind gift of Keth Mostov, University California, San Francisco, San Francisco, CA) were grown as previously described (Ducharme et al., 2006) in DMEM supplemented with l-glutamine, nonessential amino acids, and G418 (all from Cellgro) and 10% fetal bovine serum (FBS; GE Heathcare Life Sciences-Hyclone, Logan, UT). The cell lines were also grown in the presence of 20 μg/ml doxycycline to keep then in the off position. At the time of plating for experiments, the doxycycline was removed to turn on the expression of the GFP-tagged Rab11-FIP2s constructs. The GFP-Rab11a cell line was as previously described (Casanova et al., 1999). The cells were plated onto glass coverslips or Transwells (CoStar Corning, Corning, NY) at confluence and then grown for 5 d before treatment. All cell lines were tested monthly for mycoplasma.
For the calcium switch assay, the cells were washed once in PBS and then refed in S-MEM (Joklik-Modified; Lonza, Walkersville, MD) supplemented with l-glutamine, nonessential amino acids, and 10% dialyzed FBS. The cells were then incubated 18–24 h, refed with regular-calcium medium, and allowed to recover for the indicated times.
For growth of cysts in Matrigel, 150 μl of cell stock (1.5 × 106 cells/ml) was mixed with 75 μl of ice-cold Matrigel (Sigma-Aldrich) per well and plated into a Lab-Tek eight-chambered coverglass slides (Thermo Scientific/Nunc). The mixture was allowed to gel and then overlain with medium, and new medium was added every 2 d.
Antibodies
The rabbit anti–phospho-serine 227-Rab11-FIP2 antibody and the chicken anti–myosin Vb antibody were previously described (Lapierre et al., 2012). Commercial antibodies were, from BD Biosciences (San Jose, CA), mouse anti–E-cadherin (610181, immunofluorescence [IF] 1:200), mouse anti-p120 (610133, IF 1:200), and rabbit anti-occludin (71–1500, IF 1:200); from the Developmental Studies Hybridoma Bank (Iowa City, IA), rat anti–ZO-1 (R26.4C, IF 1:400); from Santa Cruz Biotechnology (Dallas, TX), goat anti-Eps15 (SC-11716, IF 1:300) and mouse anti-Numb (SC-136554, IF 1:200); and from Abcam (Cambridge, MA), rabbit anti–voltage-dependent anion channel 1 (VDAC1)/porin (ab15895, Western blot 1:5000). All secondary antibodies were made in donkey and absorbed for multistaining use (Jackson ImmunoResearch, West Grove, PA).
Immunoprecipitation of the pS227-FIP2 complex
Sheep anti-rabbit immunoglobulin G (IgG) Dynabeads (125 μl; Thermo Fisher Scientific) were washed three times for 5 min with PBS and then resuspended in 500 μl of PBS, and 1.5 μl of rabbit anti–pS227-FIP2 or rabbit IgG (Jackson ImmunoResearch) was added and incubated with rotation overnight at 4°C. The beads were washed three times for 5 min with PBS, then twice for 5 min with 0.2 M triethanolamine, pH 8.2, resuspended in fresh coupling solution (20 mM dimethyl pimelimidate [Pierce-Thermo Fisher Scientific] and 0.2 M triethanolamine, pH 8.2), and incubated at room temperature with rotation for 30 min. The beads were quenched by first incubating with rotation for 15 min at room temperature with 50 mM Tris, pH 7.5, and then by incubating for 5 min at room temperature with rotation in 20 mM ethanolamine. The beads were washed a final three times for 5 min with PBS. For immunoisolation, MDCK cells were grown on 10-cm Transwells (CoStar Corning) for 5 d postconfluence, switched into low-calcium-containing medium for 18–24 h, and then allowed to recover for 1 h. Cells were lysed in 1% CHAPS in PBS, and then 25 μg of protein was diluted into a final volume of 500 μl of PBS with protease and phosphatase inhibitor cocktail (Sigma Aldrich), added to the antibody-conjugated beads, and incubated 2 h at room temperature with rotation. For proteomic analysis, four separate immunoisolations were performed and then combined at the first of the following washes. The beads were washed three times for 20 min in PBS with rotation, and the material was eluted from the beads with SDS–PAGE sample buffer (200 mM Tris, pH 7.5, 0.9% SDS, 12 mM EDTA, 4% sucrose, 10 mM dithiothreitol, and Pyronin Y), heated for 5 min at 70°C, and then applied to a 10% SDS–PAGE gel with no stacker. Electrophoresis continued until the dye front had migrated into the gel ∼1–2 cm. The gel was then stained with Gel Code Blue (Pierce-Thermo Fisher Scientific), and the protein band was excised from the gel, minced, and washed. The proteins were subjected to in-gel trypsin digestion (Ham, 2005), and the resulting peptides were eluted from the gel and submitted for liquid chromatography and tandem mass spectrometry (LC-MS-MS) analysis.
LC-MS-MS analysis and protein identification
LC-MS-MS analysis of the resulting peptides was performed using a Thermo Finnigan LTQ ion trap mass spectrometer equipped with a Thermo MicroAS autosampler and Thermo Surveyor HPLC pump, nanospray source, and Xcalibur 1.4 instrument control. The peptides were separated on a packed capillary tip, 100 μm × 11 cm, with C18 resin (Monitor C18, 5 μm, 100 Å; Column Engineering, Ontario, CA), using an in-line solid-phase extraction column that was 100 μm × 6 cm, packed with the same C18 resin (using a frit generated with liquid silicate Kasil 1; Cortes et al., 1987), similar to the process as previously described (Licklider et al., 2002), except that the flow from the HPLC pump was split before the injection valve. The flow rate during the solid-phase extraction phase of the gradient was 1 ml/min and during the separation phase was 700 nl/min. Mobile phase A was 0.1% formic acid, and mobile phase B was acetronitrile with 0.1% formic acid. A 95-min gradient was performed with a 15-min washing period (100% A for the first 10 min, followed by a gradient to 98% A at 15 min) to allow for solid-phase extraction and removal of any residual salts. After the initial washing period, a 60-min gradient was performed in which the first 35 min was a slow, linear gradient from 98 to 75% A, followed by a faster gradient to 10% A at 65 min and an isocratic phase at 10% A to 75 min. The MS/MS spectra of the peptides were performed using data-dependent scanning in which one full MS spectrum, using a full mass range of 400–200 amu, was followed by three MS/MS spectra. Proteins were identified using the cluster version of the SEQUEST algorithm (Yates et al., 1995), using the canine subset of the Uniref 100 database (www.uniprot.org). The database was concatenated with the reverse sequences of all the proteins in the database to allow for the determination of false-positive rates. Protein matches were preliminarily filtered using the following criteria: if the charge state of the peptide is 1, the xcorr is ≥1, the RSp is ≤5, and the Sp is ≥350. If the charge state is 2, the xcorr is ≥1.8, the RSp is ≤5, and the Sp is ≥350. If the charge state is 3, the xcorr is ≥2.5, the RSp is ≤5, and the Sp is ≥350. Once filtered based on these scores, all protein matches that had fewer than two peptide matches were eliminated. These filtering criteria achieved a false-positive rate of <1% in all data sets.
Immunofluorescence
Cells were washed twice with PBS, fixed with either −20°C methanol for 5 min at −20°C or 4% paraformaldehyde/PBS for 20 min at room temperature. Cells were washed three times with PBS and then block/extracted for 30 min at room temperature in 10% normal donkey serum (Jackson ImmunoResearch) and 0.3% Triton X-100 in PBS. Cells were incubated overnight at 4°C with primary antibodies diluted in 1% normal donkey serum and 0.05% Tween-20 in PBS. Cells were washed three times for 15 min at room temperature with 0.05% Tween-20 in PBS (PBS-T) and then incubated for 1 h at room temperature with secondary antibodies diluted as the primaries, washed twice in PBS-T and once in PBS, and then rinsed in water and mounted with ProLong Gold (Invitrogen-ThermoFisher Scientific).
All images were captured with an Olympus FV1000 confocal microscope (Vanderbilt Cell Imaging Shared Resource) using a 60× Plan-Apochromat oil immersion objective with a numerical aperture (NA) of 1.45 and a 3× optical zoom using the FV1000 software. The individual images were converted into tiff files with the FV1000 software, and then Adobe Photoshop was used to produce the final figures. Mean fluorescence intensity was quantified using the JaCoP plug-in (Bolte and Corpelieres, 2006) of ImageJ (National Institutes of Health, Bethesda, MD). Quantitated intensities were compared by a Kruskal–Wallis test with post hoc analysis of significant means with Dunn’s test.
Staining of the cysts was performed as for the Transwells, with the exception that all incubations, with the exception of the primary overnight incubation, were doubled in time for the fixation and the block and extraction steps and in the number of washes. The cysts were imaged on a Zeiss LSM 710 Meta Inverted confocal microscope using a 40× LD C-Apochromat water immersion lens with NA of 1.10 (Vanderbilt Cell Imaging Shared Resource). The individual images were converted into tiffs using Zeiss Zen software and converted into movies using Imaris software (Bitplane; Vanderbilt Cell Imaging Shared Resource).
Precipitation of mCherry-Eps15
The mCherry-Eps15 plasmid was transfected into 10-cm dishes of GFP-FIP2(SE) or GFP-FIP2(SEΔNPF123) MDCK cells with PolyJet (Signagen). The cells were scrap harvested and lysed in 1% Triton-100/PBS with protease and phosphatase inhibitors (Sigma-Aldrich). Then mCherry was precipitated with rabbit anti-RFP (ab28664; Abcam) covalently attached to sheep anti-rabbit IgG Dynabeads (Life Technologies/Dynal), as described earlier. The beads were washed with PBS, and the mCherry-Eps15 complex was eluted off the beads with sample buffer.
Western blots
For the MDCK Eps15-knockdown Western blots, nonconfluent cells were lysed in CHAPS (1% CHAPS, 0.5 mM EDTA, 20 mM magnesium acetate, 30 mM Tris, pH 7.5, 150 NaCl) supplemented with protease (P8340; Sigma-Aldrich) and phosphatase (P0044, P5726; Sigma-Aldrich) inhibitors for 30 min on ice and then centrifuged for 10 min at 16,000 × g at 4°C to clear the lysates. For the E-cadherin and occludin Western blots, cells were grown 5 d postconfluence on Transwells, lysed in RIPA (1% CHAPS, 0.5 mM EDTA, 20 mM magnesium acetate, 30 mM Tris, pH 7.5, 150 mM NaCl) supplemented with protease (P8340) and phosphatase (P0044, P5726) inhibitors for 10 min on ice, and then centrifuged for 10 min at 100,000 × g at 4°C to clear the lysates. For all samples, protein concentrations were measured by DirectDetect (EMD Millipore, Billerica, MA), and 80 μg of protein was loaded onto a 10% Laemmli polyacrylamide gel (Laemmli, 1970). The proteins were transferred onto Odyssey nitrocellulose membranes (LI-COR, Lincoln, NE). Blots were air-dried for 1 h at room temperature, blocked for 1 h in Odyssey Tris-buffered saline (TBS) Blocking Buffer (LI-COR), and probed with primary antibodies for 18 h in 0.2% Tween-20/Odyssey TBS Blocking Buffer at 4°C. Blots were washed in TBS/0.05% Tween (TBS-T), followed by a 1-h incubation with secondary antibodies labeled with 680- or 800-nm fluorescent dyes (LI-COR) and diluted as the primary antibody. Blots were washed three times in TBS-T, and fluorescence was detected using the Odyssey Fc (LI-COR). The resulting JPEGs were opened in ImageJ, the area under the peak was calculated, and the relative density was normalized to the parental MDCK value. The densities were then adjusted to the control (VDAC), and statistical significance was determined by an unpaired Student’s t test.
Yeast two-hybrid assay
Yeast were transfected and tested for β-galactosidase activity using the colony lift assay as previously described (Lapierre et al., 1999). For the plate assay, three colonies from each plate (triplicates) were inoculated into separate 4 ml of YPDA broth (yeast extract/peptone/dextrose [Clontech, Mountain View, CA] supplemented with 0.003% adenine hemisulfate [Sigma-Aldrich]) and grown overnight at 30°C. Yeast cells were spun for 3 min at 3000 × g, resuspended in 300 μl of PBS, transferred to preweighed 1.5-μl tubes, and spun for 3 min at 3000 × g. The supernatant was removed, and the tubes were reweighed. To lyse the yeast, the pellet was suspended in 2.5 μl of Y-PER (ThermoFisher) per milligram of yeast, vortexed for 5 s, and then incubated for 20 min at room temperature with agitation. The yeast lysate was spun for 10 min at 14,000 × g at 4°C, and 100 μl of the supernatant was added to a 96-well round-bottom plate (CoStar Corning), followed by 100 μl of o-nitrophenyl β-d-galactopyranoside (Sigma-Aldrich) at 4 mg/ml in Z-buffer (60 mM Na2HPO, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7.0). The plate was read every minute for 2 h on a BioTek Synergy 4 plate reader at 410 nm. The β-galactosidase activity was calculated by compassion to a standard curve of known β-galactosidase concentrations, and levels were compared by a Kruskal–Wallis test with post hoc analysis of significant means with Dunn’s test.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01 DK48370 and R01 DK70856 to J.R.G. Confocal microscopy and protein mass spectrometry were performed through the VUMC Cell Imaging Shared Resource and the Vanderbilt Mass Spectrometry Resource, respectively, which are supported by National Institutes of Health Grants CA68485, DK20593, DK58404, and HD15052.
Abbreviations used:
- pS227-FIP2
phospho-S227-Rab11-FIP2
- Rab11-FIP
Rab11 family of interacting proteins
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-04-0214) on February 22, 2017.
REFERENCES
- Adams CL, Chen Y-T, Smith SJ, Nelson WJ. Mechanisms of epithelial cell-cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J Cell Biol. 1998;142:1105–1119. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams CL, Nelson WJ, Smith SJ. Quantitative analysis of cadherin-catenin-actin reorganization during development of cell-cell adhesion. J Cell Biol. 1996;135:1899–1911. doi: 10.1083/jcb.135.6.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A, Gagnon J, Begue B, Megarbane B, Dautry-Varsat A, Cerf-Bensussan N. The tyrosine kinase substrate eps15 is constitutively associated with the plasma adaptor AP-2. J Cell Biol. 1995;131:1831–1838. doi: 10.1083/jcb.131.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A, Lamaze C, Begue B, Schmid SL, Dautry-Varsat A, Cerf-Bensussan N. AP-2/Eps15 interaction is required for receptor-mediated endocytosis. J Cell Biol. 1998;140:1055–1062. doi: 10.1083/jcb.140.5.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A, Poupon V, Cerf-Bensussan N, Dautry-Varsat A. Mapping of Eps15 domains involved in its targeting to clathrin-coated pits. J Biol Chem. 2000;275:3288–3295. doi: 10.1074/jbc.275.5.3288. [DOI] [PubMed] [Google Scholar]
- Bolte S, Corpelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Casanova JE, Wang X, Kumar R, Bhartur SG, Navarre J, Woodrum JE, Altschuler Y, Ray GS, Goldenring JR. Association of Rab25 and Rab11a with the apical recycling system of polarized Madin-Darby canine kidney cells. Mol Biol Cell. 1999;10:47–61. doi: 10.1091/mbc.10.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi S, Cao H, Chen J, McNiven M. Eps15 mediates vesicle trafficking from the trans-Golgi network via an interaction with the clatherin adaptor AP-1. Mol Biol Cell. 2008;19:3564–3575. doi: 10.1091/mbc.E07-10-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B-B, Ge L, Xie C, Zhao Y, Miao H-H, Wang J, Li B-L, Song B-L. Requirement of Myosin Vb-Rab11a-Rab11-FIP2 complex in cholesterol-regulated translocation of NPC1L1 to the cell surface. J Biol Chem. 2009;284:22481–22490. doi: 10.1074/jbc.M109.034355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes HJ, Pfeiffer CD, Richter BE, Stevens T. Porous ceramic bed supports for fused silica packed capillary columns used in liquid chromatography. J High Resolut Chromatogr. 1987;10:446–448. [Google Scholar]
- Cullis DN, Philip B, Baleja JD, Feig LA. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J Biol Chem. 2002;277:49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
- Cupers P, Jadhav AP, Kirchhausen T. Assembly of clathrin coats disrupts the association between Eps15 and AP-2 adaptors. J Biol Chem. 1998;273:1847–1850. doi: 10.1074/jbc.273.4.1847. [DOI] [PubMed] [Google Scholar]
- Ducharme NA, Hales CM, Lapierre LA, Ham A-JL, Oztan A, Apodaca G, Goldenring JR. MARK2/EMK1/Par-1Ba phosphorylation of Rab11-Family Interacting Protein 2 is necessary for the timely establishment of polarity in Madin-Darby Canine Kidney cells. Mol Biol Cell. 2006;17:3625–3637. doi: 10.1091/mbc.E05-08-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducharme NA, Ham AJ, Lapierre LA, Goldenring JR. Rab11-FIP2 influences multiple components of the endosomal system in polarized MDCK cells. Cell Logist. 2011;1:57–68. doi: 10.4161/cl.1.2.15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducharme NA, Williams JA, Oztan A, Apodaca G, Lapierre LA, Goldenring JR. Rab11-FIP2 regulates differentiable steps in transcytosis. Am J Physiol Cell Physiol. 2007;293:1059–1072. doi: 10.1152/ajpcell.00078.2007. [DOI] [PubMed] [Google Scholar]
- Fan G-H, Lapierre LA, Goldenring JR, Sai J, Richmond A. Rab11-family interacting protein 2 and myosin Vb are required for CXCR2 recycling and receptor-mediated chemotaxis. Mol Biol Cell. 2004;15:2456–2469. doi: 10.1091/mbc.E03-09-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazioli F, Minichiello L, Matoskova B, Wong WT, DiFlore PP. eps15, a novel tyrosine kinase substrate, exhibits transforming activity. Mol Cell Biol. 1993;13:5814–5828. doi: 10.1128/mcb.13.9.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales CM, Griner R, Dorn MC, Hardy D, Kumar R, Navarre J, Chan EKC, Lapierre LA, Goldenring JR. Identification and characterization of a Family of Rab11 Interacting Proteins. J Biol Chem. 2001;276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- Hales CM, Vaerman J-P, Goldenring JR. Rab11 family interacting protein 2 associates with myosin Vb and regulates plasma membrane recycling. J Biol Chem. 2002;277:50415–50421. doi: 10.1074/jbc.M209270200. [DOI] [PubMed] [Google Scholar]
- Ham A-JL. Proteolytic digestion protocols. In: Caprioli RM, Gross ML, editors. Biological Applications Part A: Peptides and Proteins. Vol. 2. Oxford, UK: Elsevier; 2005. pp. 10–17. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lapierre LA, Avant KM, Caldwell CM, Oztan A, Apodaca G, Knowles BC, Roland JT, Ducharme NA, Goldenring JR. Phosphorylation of Rab11-FIP2 regulates polarity in MDCK cells. Mol Biol Cell. 2012;23:2302–2318. doi: 10.1091/mbc.E11-08-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LA, Tuma PL, Goldenring JR, Navarre J, Anderson JM. VAP-33 localizes with occludin at the intercellular tight junction and also with intracellular vesicles. J Cell Sci. 1999;112:3723–3732. doi: 10.1242/jcs.112.21.3723. [DOI] [PubMed] [Google Scholar]
- Licklider LJ, Thoreen CC, Peng J, Gygi SP. Automation of nanoscale microcapillary liquid chromatography-tandem mass spectrometry with a vented column. Anal Chem. 2002;74:3076–3083. doi: 10.1021/ac025529o. [DOI] [PubMed] [Google Scholar]
- Lindsay AJ, McCaffrey MW. Rab11-FIP2 functions in transferrin recycling and associates with endosomal membranes via its COOH-terminal domain. J Biol Chem. 2002;277:27193–27199. doi: 10.1074/jbc.M200757200. [DOI] [PubMed] [Google Scholar]
- Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Mol Biol Cell. 2006;17:163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedvetsky PI, Stefan E, Frische S, Santamaria K, Burkhard W, Valenti G, Hammer JAI, Nielsen S, Goldenring JR, Rosenthal W, Klussmann E. A role of myosin Vb and Rab11-FIP2 in the Aquaporin-2 shuttle. Traffic. 2007;8:110–123. doi: 10.1111/j.1600-0854.2006.00508.x. [DOI] [PubMed] [Google Scholar]
- Santolini E, Puri C, Salcini AE, Gagliani C, Pelicci PG, Tacchetti C, DiFlore PP. Numb is an endocytic protein. J Cell Biol. 2000;151:1345–1351. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk RW, Luiken JJFP, Eckel J. FIP2 and Rip11 specify Rab11a-mediate cellular distribution of GLUT4 and FAT/CD36 in H9c2-hIR cells. Biochem Biophys Res Commun. 2007;363:119–125. doi: 10.1016/j.bbrc.2007.08.111. [DOI] [PubMed] [Google Scholar]
- Tebar F, Sorkina T, Sorkin A, Ericsson M, Kirchhausen T. Eps15 is a component of clathrin-coated pits and vesicles and is located at the rim of coated pits. J Biol Chem. 1996;271:28727–28730. doi: 10.1074/jbc.271.46.28727. [DOI] [PubMed] [Google Scholar]
- van Bergen en Henegouwen PM. Eps15: a multifunctional adaptor protein regulating intracellular trafficking. Cell Commun Signal. 2009;7:24–35. doi: 10.1186/1478-811X-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;6:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.