Fission yeast have adapted to retrotransposon invasion by RNAi-mediated silencing, which has coevolved into a mechanism involving CENPB-mediated heterochromatinization together with ablation of RNAi components via accumulation of recombinogenic repeats in recently diverged species of Schizosaccharomyces. Similar trends are seen in the metazoans.
Abstract
Inactivation of retrotransposons is accompanied by the emergence of centromere-binding protein-B (CENPB) in Schizosaccharomyces, as well as in metazoans. The RNA interference (RNAi)-induced transcriptional silencing (RITS) complex, comprising chromodomain protein-1 (Chp1), Tas3 (protein with unknown function), and Argonaute (Ago1), plays an important role in RNAi-mediated heterochromatinization. We find that whereas the Ago1 subunit of the RITS complex is highly conserved, Tas3 is lost and Chp1 is truncated in Schizosaccharomyces cryophilus and Schizosaccharomyces octosporus. We show that truncated Chp1 loses the property of heterochromatin localization and silencing when transformed in Schizosaccharomyces pombe. Furthermore, multiple copies of CENPB, related to Tc1/mariner and Tc5 transposons, occur in all Schizosaccharomyces species, as well as in humans, but with loss of transposase function (except Schizosaccharomyces japonicus). We propose that acquisition of Tc1/mariner and Tc5 elements by horizontal transfer in S. pombe (and humans) is accompanied by alteration of their function from a transposase/endonuclease to a heterochromatin protein, designed to suppress transposon expression and recombination. The resulting redundancy of RITS may have eased the selection pressure, resulting in progressive loss or truncation of tas3 and chp1 genes in S. octosporus and S. cryophilus and triggered similar evolutionary dynamics in the metazoan orthologues.
INTRODUCTION
Transposons are mobile elements that are widespread in eukaryotes and have the potential to disrupt coding sequences by introducing repeats. Also called junk DNA, they arise abnormally under conditions of stress (Flavell et al., 1980). Eukaryotes have developed strategies to defend their genome by silencing transposons through mechanisms such as methylation of DNA and RNA inhibition (Huda et al., 2010). RNA interference (RNAi) machinery provides cells with fine modulation of gene expression via heterochromatin formation at these transposable elements. Recent whole-genome studies show that Schizosaccharomyces japonicus, the earliest-diverging species belonging to the genus Schizosaccharomyces (Vavouri et al., 2008; Burns et al., 2010; Rhind et al., 2011), harbors the highest number of retrotransposable elements in addition to heterochromatin loci (Rhind et al., 2011), whereas Schizosaccharomyces pombe, which diverged from Saccharomyces cerevisiae 200 million to 1 billion years ago (Vavouri et al., 2008; Burns et al., 2010), contains an active Tf2 (multicopy integration) and an inactive Tf1 retrotransposon element (in the form of solo long terminal repeats [LTRs]). The relatively recently diverged species Schizosaccharomyces cryophilus and Schizosaccharomyces octosporus (which diverged from S. cerevisiae only 10–100 million years ago) have undergone a drastic reduction of transposable elements (Levin et al., 1990). This change in structural organization has been accompanied by further specialization by compartmentalization of the respective functions of RNAi and heterochromatin machineries: although RNAi plays a role in silencing of both transposable elements and heterochromatic loci in S. japonicus, it targets only the heterochromatin for silencing and not the retrotransposable elements in S. pombe. Furthermore, heterochromatin persists in S. cryophilus and S. octosporus, and there is a further egress of retrotransposons accompanied by the replacement of retrotransposons with solo LTRs in S. octosporus and LTRs with a recently reported transposon (Tcry) in S. cryophilus (Rhind et al., 2011).
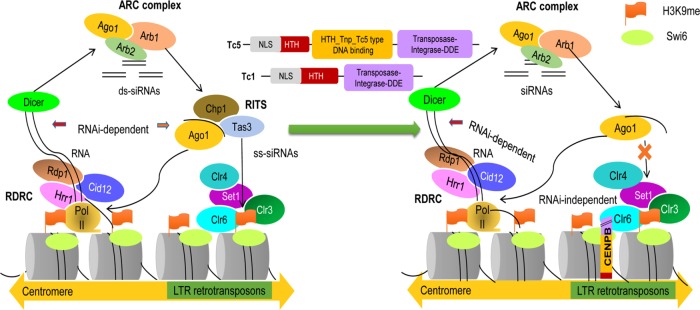
In both metazoans and fission yeast, the RNAi pathway is initiated by Dicer, which cleaves the nascent double-stranded RNA precursors to 21- to 22-nucleotide-long small interfering RNAs (siRNAs), which function as effectors in silencing (Provost et al., 2002). Gene silencing by the heterochromatin pathway is facilitated by the methylation of histone H3 at the Lys-9 position, followed by binding of Swi6/HP1 to H3K9me2 through its chromodomain, resulting in the establishment of heterochromatin and leading to gene silencing (Grewal and Jia, 2007). In S. pombe, deletion of the genes involved in RNAi, namely ago1, dcr1, and rdp1, leads to loss of heterochromatin formation at cen, mat, and telomere loci, accompanied by loss of H3-K9 methylation (which is mediated by the methyltransferase Clr4) and Swi6. One of the effector complexes, RNAi-induced transcriptional silencing (RITS) complex, contains Argonaute (Ago1), the chromodomain protein Chp1, and Tas3, which is of unknown function. RITS and the RNA-directed RNA polymerase complex (RDRC) work in a concerted manner by which chromatin-bound RITS cooperates with Rdp1 and Dcr1 to process nascent transcripts in to siRNAs, whereas the RITS complex and Rdp1 work together to localize the siRNAs at centromeres, which results in methylation at H3K9, with the help of Clr4 (Zhang et al., 2008; Sugiyama et al., 2007a). A schematic representation of the current model linking heterochromatin with RNAi machinery will be discussed later in relation to Figure 9 (left).
FIGURE 9:
Model depicting reciprocal coevolution involving the loss of the RITS and the emergence of CENPB (abp1/cbh1) in Schizosaccharomyces. (Left) Chp1/Tas3 in RITS complex in S. pombe and S. japonicus help to link the RNAi pathway with retrotransposon silencing (adapted from Moazed, 2011). (Right) With Chp1 truncated and Tas3 absent, the retrotransposon-silencing role is taken over by CENPB proteins, which engage in heterochromatin formation. This is brought about by acquisition of Tc1/mariner and Tc5 elements. The conserved domains acquired from Tc1 and Tc5 are shown in three different colors for CENPB: red depicts the partial helix-turn-helix domain present in both Tc1 and Tc5 transposons; this domain binds to DNA but shows nonspecificity. This domain is present in human CENPB, but the S. pombe does not show its complete presence. The yellow CENP DNA–binding domain shows its acquisition from the Tc5- transposase family. The domain is again a helix-turn-helix domain but with specificity for binding to centromeric DNA. The purple part shows the partial domain of DDE endonuclease. The NLS region is depicted with black lines as a short linker.
It was shown that Chp1 binds to Tas3 independently of Ago1, whereas Tas3 binds with Ago1 independently of Chp1 (Debeauchamp et al., 2008; Schalch et al., 2011). Of importance, Tas3 interacts through its N-terminal region with the C-terminal domain of Chp1, and this interaction is necessary for Tas3 stability (Debeauchamp et al., 2008). In cells lacking ago1, Chp1 and Tas3 still bind to noncentromeric regions, although their association with centromeres is lost (Petrie et al., 2005; Schalch et al., 2011). In addition, intrinsic nucleic acid binding by Chp1 plays an important role in silencing (Ishida et al., 2012).
In fission yeast, CENPB homologues (also known as cbp1, abp1, cbh1, or cbh2) are important for regulation of retrotransposons, particularly Tf1 and Tf2 (Levin et al., 1990; Casola et al., 2008; O’Donnell and Boeke, 2008). Earlier reports suggested that inception of CENPB gene family might have provided the switch from RNAi-mediated transposon silencing in S. japonicus to a cbp1 (CENPB) heterochromatin formation–based mechanism in S. pombe, S. octosporus, and S. cryophilus (Rhind et al., 2011). CENPBs are suggested to have descended from Tc1/ mariner-Pogo–like transposases, and in many species, including humans, Caenorhabditis elegans, and Candida albicans, CENPB and CENPB-like proteins play a pivotal role in heterochromatin assembly at centromeres (Casola et al., 2008; O’Donnell and Boeke, 2008).
During coevolution, sequences of different components within and across genes often manifest coordinated changes in order to maintain the structure or function of specific genes (Pazos and Valencia, 2008; Brandman et al., 2012). In this study, we provide evidence in support of reciprocal evolution among the RITS and heterochromatin components in the Schizosaccharomyces genus and in metazoans. Of interest, analyses of the distribution of key components of RNAi pathway genes across Schizosaccharomyces species revealed that whereas tas3 is present in S. japonicus and S. pombe, it is absent in S. cryophilus and S. octosporus. Evidence shows that besides the loss of Tas3, Chp1 is retained only in a truncated form in S. octosporus and S. cryophilus. Cross-species experiments show the inability of chp1 from S. cryophilus and S. octosporus to complement the chp1∆ silencing defect in S. pombe. Our results suggest that concerted reciprocal evolution, involving loss of genes encoding some of the RITS components in conjunction with the loss of transposons, may have been causally occasioned by the acquisition and evolution of CENPB structure and function exclusively for the silencing of residual retrotransposons/transposons in the heterochromatin context, independently of RNAi, during the evolution of species of the Schizosaccharomyces genus. Similar coevolutionary trends seem to be at play in metazoans.
RESULTS
Phylogenetic analysis of heterochromatin and RNAi-regulating genes
RNAi-mediated heterochromatin formation in S. pombe involves RITS, RDRC, and ARB complexes, whereas the Snf2/HDAC-containing repressor complex (SHREC) regulates RNAi-independent heterochromatin assembly (Sugiyama et al., 2007b; Supplemental Table S1). The maximum-likelihood phylogenetic analysis of the components of RDRC, RITS, and SHREC, as well as proteins forming heterochromatin at centromeres, telomeres, and mating-type loci (unpublished data), corroborated a report that S. japonicus is the earliest-branching species to contain the genes regulating heterochromatin formation and RNAi pathways, followed by S. pombe, S. octosporus, and S. cryophilus (Rhind et al., 2011). Specifically, genes like ago1, dcr1, and rdp1 also follow this trend (unpublished data).
Loss of tas3 during Schizosaccharomyces evolution
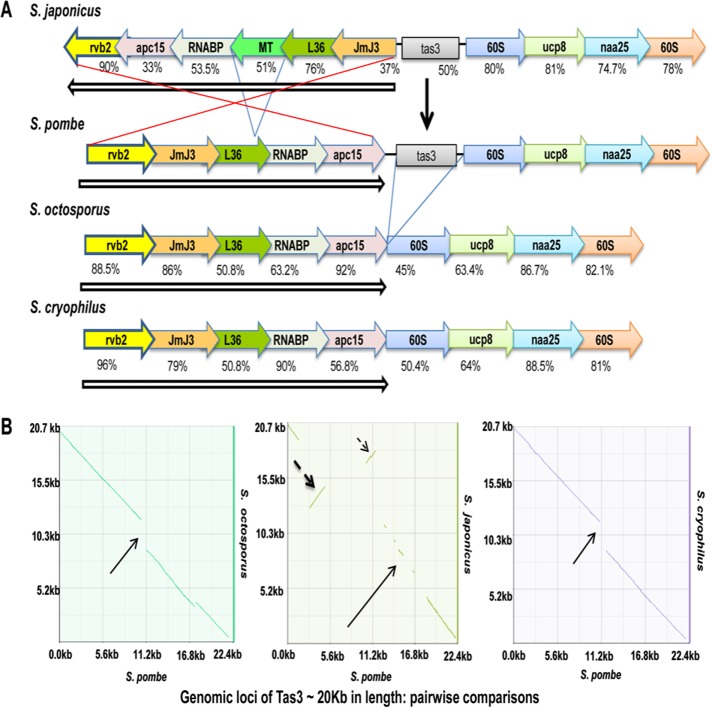
Recent comparative genomics studies indicate a progressive reduction of transposable elements during evolution from S. japonicus to S. pombe, S. octosporus, and S. cryophilus (Rhind et al., 2011). To understand the association of the retrotransposon loss with RNAi machinery, we compared sequences of the genes involved in heterochromatin formation among the species in the genus Schizosaccharomyces (Supplemental Table S1). Sequence comparison of the genomic flanking regions encoding tas3 gene in S. pombe with other Schizosaccharomyces species revealed that there is a complete loss of tas3 in S. octosporus and S. cryophilus. The loss of tas3, a vital component of RITS, did not leave any relics of the original sequences behind (Figure 1A). Furthermore, dot-plot analysis showed a higher level of conservation of the gene order in the tas3-flanking regions among S. pombe, S. octosporus, and S. cryophilus, which was caused by the inversion of the corresponding region comprising the genes apc15, RNABP, L36, and jmj3 (excluding MT, encoding a hypothetical membrane transporter gene) in S. japonicus (Figure 1, A and B). A comparison of the entire region using z-Picture (base positions 1509451–1525726 on chromosome II) showed a dynamic picture of evolutionarily conserved regions (ECRs) above the bottom cut-off of 50%, indicating maximum similarity between S. pombe and S. octosporus and much less similarity of S. pombe with S. cryophilus and S. japonicus (Supplemental Figure S1). Lack of similarity between S. pombe and S. japonicus is understandable, considering the large inversion (Figure 1, A and B).
FIGURE 1:
Precise deletion of tas3 and inversion of tas3-flanking regions in recently diverged species of Schizosaccharomyces. (A) Synteny plot of tas3-containing region in all species of Schizosaccharomyces. The vertical arrow indicates the location of tas3. Open arrows and red lines indicate set of tas3-flanking genes undergoing inversion. (B) Synteny dot plot for tas3-flanking regions from S. japonicus (298738–321148), S. pombe (1505055–1525726), S. octosporus (1361532–1379260), and S. cryophilus (66720–84496).
Truncation of chp1 in the more recently diverged Schizosaccharomyces species
Comparison between the primary sequences of other heterochromatin proteins showed that Chp1 had apparently undergone truncation, losing its C-terminal region in S. octosporus and S. cryophilus in comparison with its homologues in S. japonicus and S. pombe (Figure 2 and Table 1). Domain analysis showed that whereas Chp1S. cryophilus and Chp1S. octosporus have lost the C-terminal region (containing the SPOC and DOMII domains), they retain the N-terminal region comprising the chromodomain and RRM domain (Figure 2, Figure 5A later in this article, Table 1, and Supplemental Figure S2).
FIGURE 2:
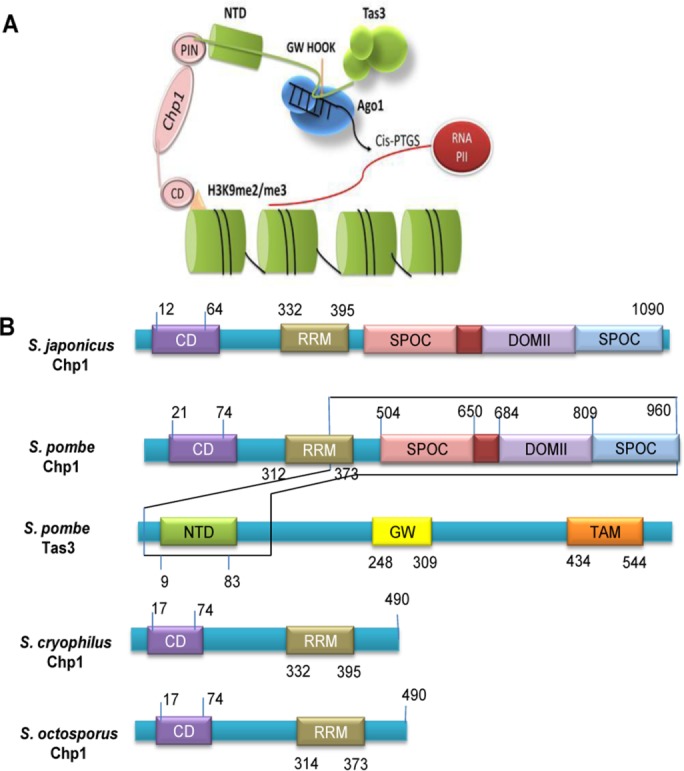
Truncation of Chp1 in recently branched species of Schizosaccharomyces. (A) Schematic representation of the structure of complex of Chp1 with Tas3 (adapted from Debeauchamp et al., 2008). The N-terminal (residues 9–83) of Tas3 attaches to the C-terminal of Chp1 (residues 504–960). Model shows interaction of Chp1 (C-terminal SPOC, DOMII, and PIN domains) with Tas3 (N-terminal domain), which is crucial for centromeric silencing by H3K9 methylation. (B) Comparison of domain structure of Chp1 of S. japonicus, S. pombe, S. octosporus, and S. cryophilus, showing loss of its C-terminal (which interacts with the N-terminal of Tas3) in S. octosporus and S. cryophilus. Also indicated is the domain structure of Tas3 of S. pombe.
TABLE 1:
Level of sequence conservation (percentage identity) of amino acid sequence of Chp1 from different species of Schizosaccharomyces.
| Chp1 | N-terminal (%) | CD domain (%) | C-terminal (%) |
|---|---|---|---|
| S. pombe/S. japonicus | 52.0 | 54.16 | 68.27 |
| S. pombe/S. octosporus | 0 | 91.5 | 0 |
| S. pombe/S. cryophilus | 34.44 | 51 | 0 |
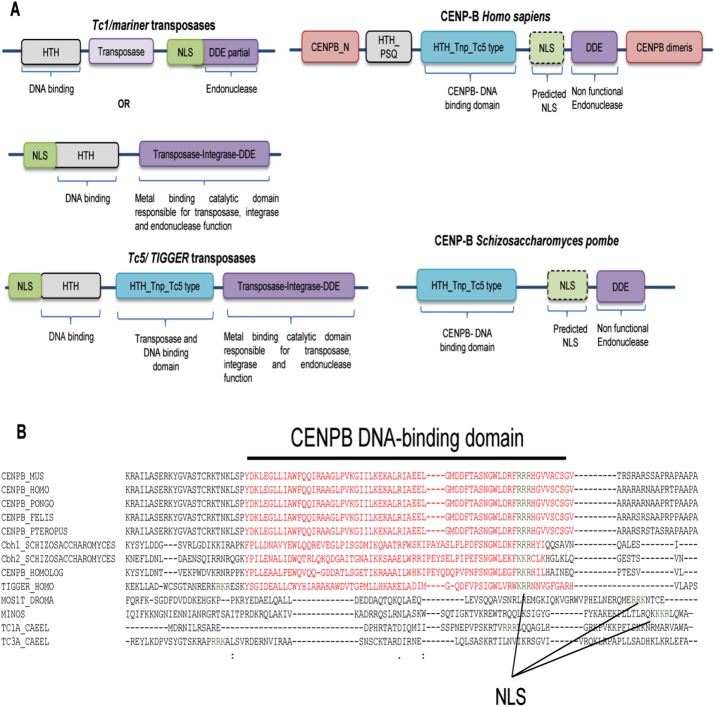
FIGURE 5:
Relocalization of NLS and mutation in endonuclease in CENPB in S. pombe and H. sapiens. (A) Comparison of domains present in Tc1/mariner transposases, Tc5/Tigger transposases, human CENPB, and fission yeast CENPB. (B) Sequence alignment of Tc1/mariner, Tc5/Tigger, and CENPB proteins from different species were studied for the localization of NLS. Data collected from the CDD. The residues represented by red and spanned by the black line represent the HTH-Tn-Tc5 DNA–binding domain (DBD), and green indicates predicted NLS sequences (NLS overlaps with DBD).
Truncated chp1 is defective in centromeric silencing
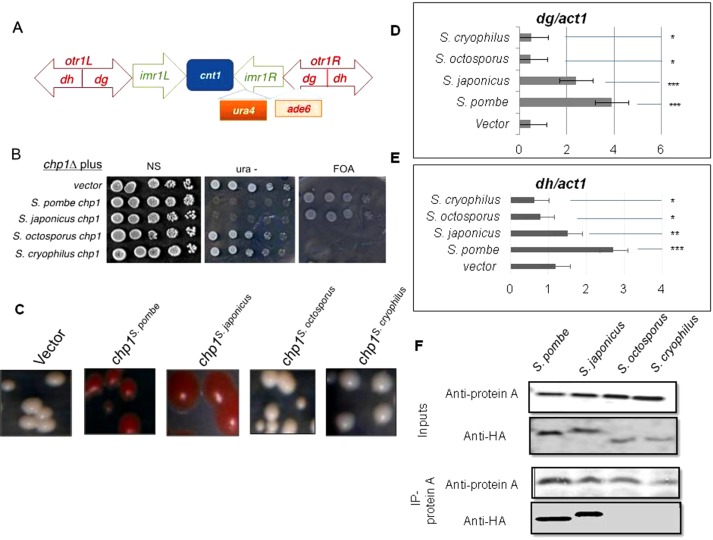
It has been reported that the C-terminal of Chp1, which contains the SPOC and DOMII domains, is indispensable for centromeric and telomeric silencing (Petrie et al., 2005; Debeauchamp et al., 2008; Schalch et al., 2011). Therefore we evaluated whether Chp1 of S. octosporus and S. cryophilus could function in centromeric silencing. Our results show that whereas chp1S. pombe and chp1S. japonicus restored silencing at imr::ura4 (Figure 3, A and B) and otr1R::ade6 (Figure 3, A–C) in chp1∆ strains of S. pombe, the genes chp1S. octosporus and chp1S.cryophilus, whose protein products contain only the CD and RRM domains but lack the Tas3 interacting domains (DOMII and SPOC; Figure 2B), did not restore silencing at either locus (Figure 3, A and C).
FIGURE 3:
Lack of complementation of centromeric silencing defect in chp1∆ strain of S. pombe by chp1 homologues. (A) Schematic diagram of the centromeric locus cen1, showing insertion of ura4 at imr1L and ade6 at otr1R (Allshire et al., 1994). (B) Dilution spotting showing growth levels of the chp1Δ strains carrying the chp1 gene from different species and the imr1::ura4 reporter on 5-FOA, ura–, and complete plates. (C) Transformants shown in B were streaked on complete, leu-, and leu-/low-adenine (15 mg/l) plates. (D, E) Results of real-time PCR for quantitation of localization of Chp1 from different species of Schizosaccharomyces at the dg (D) and dh (E) repeats in S. pombe. The p values were calculated for each of three samples. ***p < 0.001, **p < 0.01, *p < 0.05. (F) Tas3 of S. pombe fails to interact with Chp1 from S. octosporus and S. cryophilus. Coimmunoprecipitation of Tas3 with Chp1 was tested using an S. pombe strain carrying TAP-tagged tas3 gene and chp1Δ, expressing the chp1 gene from S. pombe, S. japonicus, S. octosporus, and S. cryophilus with HA tag. Whole-cell extracts of the indicated strains were immunoprecipitated with anti–protein A antibodies and immunoblotted with anti–protein A and anti-HA antibodies. Because of the barely detectable level of HA-tagged Chp1 in some strains, the reciprocal coimmunoprecipitation was not done.
The centromeric heterochromatin localization of Chp1 from S. pombe, S. japonicus, S. octosporus, and S. cryophilus at the centromeric regions in the S. pombe chp1Δ strain was assessed by chromatin immunoprecipitation (ChIP) assay. We found that Chp1S. pombe could localize to both the dg and dh repeats, and Chp1S. cryophilus and Chp1S. octosporus did not show any enrichment, whereas enrichment of Chp1S.japonicus was relatively more to the dg region (Figure 3, D and E). Taken together, these data demonstrate the ability of full-length Chp1 from S. pombe and S. japonicus to restore silencing, whereas Chp1S. cryophilus and Chp1S. octosporus are unable to do so. The limited enrichment of Chp1S.japonicus at dg versus dh repeats could reflect its ability to restore heterochromatin structure at the inner region of otr1::ade6, which comprises the dg repeats but not the outer region represented by the dh repeats (Figure 3A), possibly due to inefficient functional interactions between heterologous proteins. More pertinently, Chp1S. cryophilus and Chp1S. octosporus, which lack the C-terminal DOMII and SPOC domains required for interactions with Tas3 (Figure 2B; see later discussion), fail to localize to the centromeric dg and dh repeats and to restore centromeric silencing in the chp1Δ strain of S. pombe.
Truncated Chp1 fails to interact with Tas3 of S. pombe
It is pertinent that the N-terminal region of Tas3 interacts with the C-terminal region of Chp1 and is required for maintaining the structural integrity and function of the RITS complex in S. pombe (Petrie et al., 2005; Debeauchamp et al., 2008; Schalch et al., 2011). It is also known that Chp1S. pombe binds to RNA through CD and RRM and that this binding is important for gene silencing (Ishida et al., 2012). However, a truncated derivative of Chp1 of S. pombe lacking the C-terminal region is not functional in establishing heterochromatin, nor does it interact with Tas3 in S. pombe (Debeauchamp et al., 2008). Furthermore, a tas3Δ10-24 mutant that cannot bind Chp1 fails to maintain centromeric heterochromatin, siRNA generation, and recruitment of RITS, RDRC, and Clr4 to heterochromatin (Debeauchamp et al., 2008).
To examine confirm whether the C-terminal region of Chp1 in S. pombe is important for interaction with Tas3, we overexpressed the chp1 gene from S. japonicus, S. pombe, S. cryophilus, and S. octosporus with an N-terminal hemagglutinin (HA) tag in S. pombe strains containing TAP-tagged Tas3. Results of coimmunoprecipitation show that, as expected, Chp1 protein from S. japonicus and S. pombe coimmunoprecipitated with Tas3 of S. pombe. However, Tas3 from S. pombe could not coimmunoprecipitate Chp1 from S. octosporus and S. cryophilus (Figure 3F). Given that Chp1 from S. cryophilus and S. octosporus lacks the C-terminal domain, these results confirm that an intact Chp1 containing the C-terminal region is needed for interaction with Tas3. Thus loss of the tas3 gene is accompanied by loss of the Tas3-interacting, C-terminal region of Chp1 in S. cryophilus and S. octosporus.
Ago1 is highly conserved in all Schizosaccharomyces species
The complete loss of Tas3 and truncation of Chp1 in S. cryophilus and S. octosporus directed our attention to Ago1, the third component of the RITS complex. Surprisingly, Ago1 shows a high level of conservation of both domains PIWI and PAZ, along with the N- and C- terminal sequences, in all four species (Supplemental Figure S3 and Table 2), suggesting that the ago1 gene may be performing some essential functions in all species. We asked whether the ago1Δ mutant showed any phenotype and whether ago1 homologues from other species could complement the defect. We found that whereas dcr1Δ and rdp1Δ strains showed significant a silencing defect at the otr1R::ade6 locus, as indicated by pink colonies compared with red colonies of wild-type strain on adenine-limiting plates, the ago1Δ mutant exhibited only a modest effect: it produced colonies with intermediate phenotype between the wild type and dcr1Δ/ rdp1Δ mutant, suggesting unstable derepression of the ade6 reporter in the ago1Δ mutant (Supplemental Figure S4, A and B). This was confirmed biochemically by the reverse transcriptase PCR reaction, which showed that whereas the otr1R:ade6 locus was derepressed in dcr1Δ and rdp1Δ mutants, it remained repressed in the ago1Δ mutant (Supplemental Figure S4C). This is in contrast to earlier work indicating that ura4 inserted at the otr1R region is derepressed in the dcr1Δ and rdp1Δ mutants, as well as in the ago1Δ mutant (Volpe et al., 2002). Silencing of green fluorescent protein mRNA by hairpin RNA requires that Ago1 and ade6 inserted at mat3 are also derepressed in the ago1Δ mutant (Sigova et al., 2004; Petrie et al., 2005). The discrepancy may be due to different assays used and the fact that in the case of otr1R::ura4, ura4 is a selectable marker, whereas ade6 used in the parent assay is not tested by the selection criterion.
TABLE 2:
Level of sequence conservation (percentage identity) of amino acid sequence of Ago1 from different species of Schizosaccharomyces.
| Ago1 | N-terminal (%) | PIWI domain (%) | PAZ domain (%) | C-terminal (%) |
|---|---|---|---|---|
| S. japonicus/S. pombe | 85.45 | 81.33 | 70.3 | 87 |
| S. japonicus/ S. cryophilus | 79.8 | 74.16 | 91 | 78 |
| S. japonicus/S. octosporus | 66 | 69.8 | 88.6 | 80 |
Conservation of Chp1 and Tas3 domains across the metazoan kingdom
To determine the conservation of domains of Chp1 and Tas3 among other metazoans, we performed BLASTP (40–100% identity; E-value cut-off <10−6), multiple sequence alignment, and Conserved Domain Database (CDD; National Center for Biotechnology Information [NCBI]) searches for putative orthologues. The chromodomain Y–like proteins (CDYLs) in metazoans showed a conserved chromodomain at the N terminal region, along with the accumulation of trinucleotide repeats, but there was a loss of RRM domain (Figure 4A). Similarly, the Ago-hook domain of human protein TNRC6B (having homology with the minimal Tas3 domain, which interacts with SpAgo1) also interacts with human Ago1, indicating a conservation of the binary interaction within the RITS complex. Through conserved domain search, we found that GW182 in humans contains Ago-hook (belonging to the TNR6C family), a part of Tas3, as well as the RRM domain, a part of Chp1 in S. pombe (Figure 4B).
FIGURE 4:
Structural comparisons among proteins orthologous to Chp1 and Tas3 in different species. Sequence and domainwise structural comparison of the proteins orthologous to (A) Chp1 and (B) Tas3 (based on BLASTp, E-value cutoff <10−6).
Expansion of CENPB copy number during evolution
Because CENPB appears to have taken over the function of transposon silencing, we examined whether there is change in CENPB copy number. Results of BLASTn analysis of the genomes of S. pombe, S. octosporus, and S. cryophilus showed that the loss of tas3 and truncation of Chp1 is accompanied by an increase in the number of CENPB copies during evolution from S. pombe (three copies) to S. octosporus and S. cryophilus (five copies each). Of interest, the different copies of CENPB also share syntenically identical and conserved positions in S. octosporus and S. cryophilus. Moreover, CENPBs in both S. octosporus and S. cryophilus show greater sequence conservation (nearly 90%), whereas the conservation between CENPBs present in S. pombe and S. octosporus/S. cryophilus is <30%. Of interest, no copy of CENPB was found in S. japonicus (Supplemental Figure S5, A–G, and Table 3).
TABLE 3:
Level of conservation of DNA sequence of CENPBs from different species of Schizosaccharomyces.
| ARS-binding protein (CENPB), nucleotide positions | Presence | Corresponding nucleotide positions in S. cryophilus | Percentage identity | |
|---|---|---|---|---|
| S. octosporus | 3184914–3189031 (supercontig 2) | Present in S. octosporus and S. cryophilus | 1360860–1363219 (supercontig 3) | 93 |
| S. octosporus | 1144082–1145599 (supercontig 1) | Present in S. octosporus and S. cryophilus | 2287482–2293026 (supercontig 1) | 96 |
| S. octosporus | 3053103–3054846 (supercontig 2) | Present in S. octosporus and S. cryophilus | 363274–365092 (supercontig 2) | 94.5 |
| S. octosporus | 176616–178456 (supercontig 1) | Present in S. octosporus and S. cryophilus | 186982–190444 (supercontig 4) | 94.17 |
| S. octosporus | 2726353–2728175 (supercontig 1) | Present in S. octosporus and S. cryophilus | 675712–677677 (supercontig 6) | 92.45 |
CENPBs have evolved from the ancient Tc1/mariner and Tc5 family transposons
According to earlier reports, CENPB might have evolved from Tc1/mariner elements (Casola et al., 2008; O’Donnell and Boeke, 2008). We sought to identify the changes CENPB might have acquired during the course of its evolution following horizontal transmission. Through synteny, BLASTn, and the CDD, we found that, like Tc1/ mariner, CENPB genes in mammals and fission yeast have retained the DDE_1 domain. This domain is a member of the DDE superfamily, which contains three carboxylate residues (DDE/ DEK) within 200 residues and is believed to be responsible for coordinating metal ions needed for transposase catalytic activity. The catalytic activity involves DNA cleavage at a specific site, followed by a strand transfer reaction. Of interest, CENP-B shows the presence of the DDE domain at its C-terminal region, but the metal-binding residues of this domain are absent from the CENP-B DDE-endonuclease, implying a loss of endonuclease function (Figure 5A). Moreover, on the basis of the sequence homology, we infer that the CENPB-DNA–binding domain from both mammals and fission yeast probably evolved from the HTH_Tnp_Tc5 transposase superfamily (Figure 5A). The superfamily consists of a helix-loop-helix motif that ensures the centromere DNA–binding capacity of the CENP-B (Figure 5A). The loss of the N-terminal residues of the CENP-B DNA–binding domain disrupts helix formation and leads to the loss of centromeric localization of CENP-B (Pluta et al., 1992). The presence of the functional nuclear localization sequence (NLS) in CENP-B is still questionable. The transposases have a NLS at the N-terminal region of their DNA-binding HTH- motif, which determines their import to the nucleus. The CENP-B does not show the presence of NLS regions within the HTH-DNA–binding regions. However, by performing an analysis of the CENP-B sequence with the cNLS mapper tool (Kosugi et al., 2009), we located a region of monopartite NLS in the CENP-B sequence at a cut-off score of 5.0–6.7, predicting the localization of this protein in both nucleus and cytoplasm (Figure 5A). This NLS region was between the CENP-B DNA–binding and DDE endonuclease domains (Figure 5B). Thus the DDE domain in Tc1, having an endonuclease function, and the transposase-integrase-DDE domain in Tc5 transposases, having integrase and endonuclease functions, have undergone loss of the endonuclease function in the CENPB homologues in humans and S. pombe (Figure 5A), whereas the HTH-Tnp-Tc5 type domain has been retained with the DNA-binding function (Figures 4B and 5A).
Time-scaled evolutionary pattern of CENP-B and Tas3
To understand the divergence and emergence of CENP-B and Tas3 proteins across different phyla, we performed phylogenetic analysis of CENP-B and Tas3 sequences. The CENP-B and Tas3 sequences of S. pombe were subjected independently to PSI-BLAST to retrieve their homologues in yeast, plants, mammals, and other phyla. The matches obtained were further screened to remove redundancy in sequence, as well as in the organism. Further, the maximum-likelihood tree was generated using 174 matches to CENP-B sequences and 56 matches to Tas3 sequences. We found that certain sequences formed a separate clade and diverged, and thus we removed them to further generate maximum-likelihood tree. After this cleaning step, we were left with 87 CENP-B and 64 Tas3 sequences, for which bootstrapped maximum-likelihood and likelihood-dependent time trees were generated.
Using the maximum likelihood and time tree generated for CENP-B protein, we divided species into two large clades, one of which comprised all the fungi along with Schizosaccharomyces, and the other with all other organisms along with a few fungal sequences. The clades were further divided into subclades, which were clearly demarcated based on different organisms. One subclade had plants and fungi with demarcated clades, and the other had Aves, Mammalia, Arthropoda, and Bivalvia in different subclades. The divergence time of the fungal and mammalian clades in the time tree suggested an earlier emergence of CENP-B in the fungal clade than in the mammalian clade (Figure 6).
FIGURE 6:
Maximum likelihood analysis–based phylogenetic relationship of CENPB proteins in various species. The scale indicates relative times from 0 to 1.5 billion years ago. Taxon names are colored based on class: Mammalia, Aves, Schizosaccharomyces, Amphibia, Actinopterygii, alligator, Plantae, and Fungi.
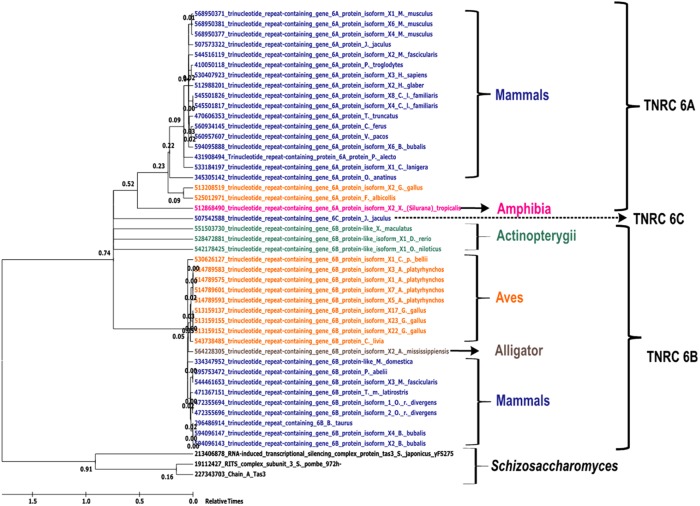
Similarly, the maximum-likelihood and likelihood-dependent time trees for Tas3 sequences were also divided into two large groups. One group contained only the Tas3 from S. pombe and S. japonicus, whereas the other large clade had all of the p branched into demarcated clades comprising Mammalia, Aves, Amphibia, and Actinopterygii. Of interest, similar to CENP-B, the mammalian clade in the Tas3 phylogeny shows the emergence events at approximately the same time, with a time lag in the emergence of the mammalian clade compared with the Schizosaccharomyces clade (Figure 7). In addition, the tas3 gene was found to be ancestral to trinucleotide repeat–containing genes (TNRC6), which had three isoforms, A–C, each of which emerged independently and formed separate clusters regardless of the organism to which they belonged (Eystathioy et al., 2002).
FIGURE 7:
Maximum likelihood analysis–based phylogenetic relationship of TNRC6 proteins in various species. Maximum likelihood–based phylogenetic tree for different proteins containing the Ago-hook domain. The scale indicates relative times from 0 to 1.5 billion years ago. Taxon names are colored based on class: Mammalia, Aves, Schizosaccharomyces, Amphibia, Actinopterygii, alligator, Plantae, and Fungi.
Horizontal gene transfer analysis and prediction of horizontal gene transfer through guanine and cytosine content
In the course of microbial evolution, horizontal transfer appears to be a major force, leading to evolution via “quantum leaps.” Generally, the molecular phylogenies calculated for single gene lead to conflicting results, with alternative branches represented by low bootstrap values. However, comparing conflicting topologies with high bootstrap values in alternative branches with the 16S rRNA phylogenetic tree suggests that the CENP-B genes were horizontally transferred among the Schizosaccharomyces species (Supplemental Figure S6, A and B).
The percentage guanine plus cytosine (%GC) content of a gene is a measure of the horizontal transfer of the gene through the evolutionary cycle. Thus, to examine horizontal transfer in the CENP-B gene, we calculated %GC according to Mann and Chen (2010). We observed a significant difference in the GC content of CENP-B from that of the entire chromosome of the same given species (Rhind et al., 2011; Supplemental Figure S7, A and B, and Supplemental Tables S2–S5), further supporting the possibility of their horizontal transmission from other species. This difference was not apparent, however, in Tas3-related (TNRC6C) genes (unpublished data).
The GC skew and GC profile together show the presence of horizontally transferred genomic regions. In cumulative GC skew plots (with red representing high and blue representing low), an increase in skew refers to the abruptness in the suddenly terminated gene and the presence of a divergent genome having the indication of an inserted gene.
Mobile elements acquired via horizontal transfer have been termed genomic islands. In the GC plots, genomic islands are represented as a negative GC profile. The negative cumulative GC profile for the genomic islands is distinct from that of the rest of the genome, in that the genomic islands have relatively low GC content, accompanied by abrupt drops in the GC profile. The abrupt drop in the negative cumulative GC profile indicates that there are clear boundaries between genomic islands and surrounding regions.
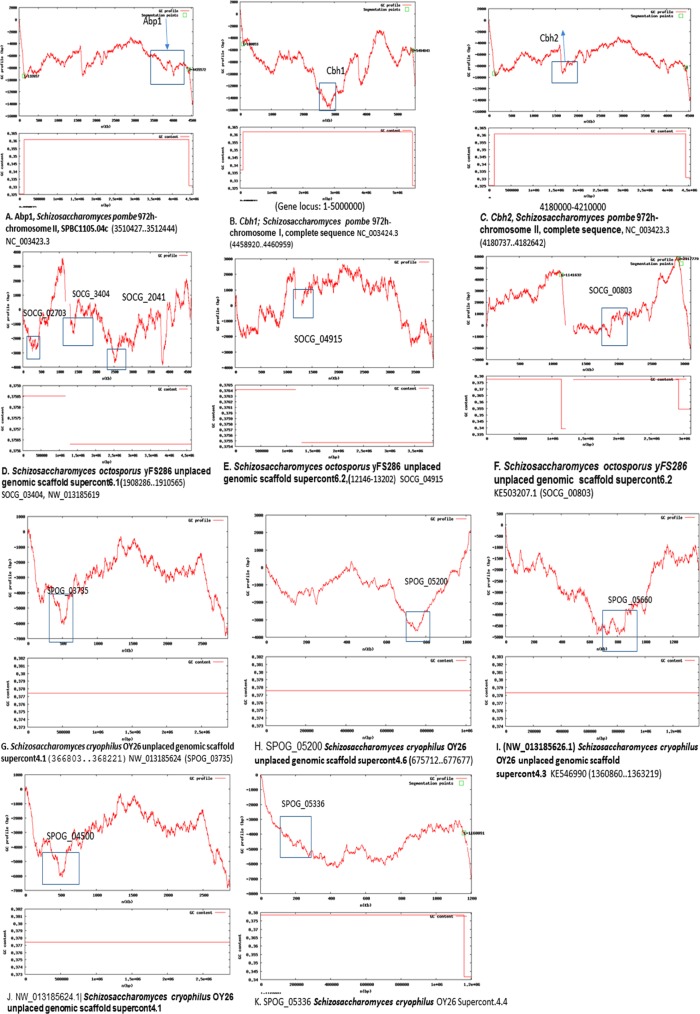
The GC profile for the CENPB regions, along with the whole genome of the Schizosaccharomyces spp., was constructed using the GC Profile tool created by Gao and Zhang (2006; Figure 8 and Supplemental Figure S7C). The negative cumulative GC profile for the genomic islands differs from the rest of the genome due to their relatively low GC content and is represented by abrupt crests in the profile. In Figure 8A, there are two strong segmentation points, and the Abp1 (CENPB protein) of S. pombe, SPBC1105.04c, 3510427–3512444 (base pairs), clearly shows a drop compared with neighboring regions, corroborating the low-GC genomic islands present at regions that are horizontally transferred. A cumulative GC skew plot (Supplemental Figure S8A) for the 3510427–3512444 region containing the abp1 gene shows a sudden elevation, with a plateau in the coding region of abp1 as well, indicating the origin and termination at regions that may have been horizontally transferred. Similarly, the region covering cbh1 (4458920–4460959; Figure 8B) shows a drop compared with regions surrounding chromosome I, for complete sequence NC_003424.3. The drop in the GC skew (Supplemental Figure S8B) is unexpectedly positive, but there is an unexpected convergence around the coding region nonetheless.
FIGURE 8:
GC profile analysis of CENPB homologues in different Schizosaccharomyces species. (A–C) S. pombe abp1, cbh1, and cbh2. (D–H) S. octosporus CENPB genes. (I–K) S. cryophilus CENPB genes.
The region covering cbh2 (Figure 8C) also shows a crest in both the GC profile and multiple small peaks and a skewed, converging GC skew plot (Supplemental Figure S8C).
In both S. octosporus and S. cryophilus, all five CENPB genes follow a similar trend, suggesting these genes too have been horizontally transferred. SOCG_02041 SOCG_02703, SOCG_3404, present in S. octosporus (supercontig 6.1), shows three major drops in the GC and skew profile for the entire supercontig compared with the regions surrounding these genes (Figure 8D and Supplemental Figure S, D–F).
SOCG_04915 (Figure 8E) and SOCG_00803 (Figure 8F), contained in supercontig 6.2, show a sudden drop in GC profile and a GC skew showing multiple bends (Supplemental Figure S8, G and H), sometimes affecting the break in the GC profile.
In S. cryophilus, of the five genes coding for CENPB, SPOG_03735 (Figure 8G) and SPOG_05200 (Figure 8H; S. cryophilus supercontigs 4.1 and 4.6) display abrupt drops, as opposed to the large increase in the neighboring region’s GC profile. SPOG_04500 (supercontig 4.3; Figure 8I), SPOG _05660 (supercontig 4.1; Figure 8J), and SPOG_05336 (Supercontig 4.4; Figure 8K and Supplemental Figure S8, I–L) follow the trend of having sharp bends at these regions, suggesting their existence as genomic islands. The only exceptions are the GC skew plots that seem to be normal for the regions covering SPOG_05200 and SPOG_05336 (Supplemental Figure S8, J and M).
DISCUSSION
Invasion of eukaryotic genomes by retrotransposons seems to have occurred as an early evolutionary process, dating to ∼500 million to 1 billion years ago. In response, eukaryotic cells have evolved RNAi and heterochromatin pathways to silence the invasive DNA elements. RNAi machinery operates at the centromere (heterochromatin) and retrotransposon regions in S. japonicus (Rhind et al., 2011), the earliest branching species of the Schizosaccharomyces. However, its role is limited to heterochromatin silencing and not to retrotransposon silencing in S. pombe (Rhind et al., 2011). In the context of constant coevolutionary arms race between RNAi machinery and exogenous parasitic transcripts exemplified by retrotransposons, there is a dynamic balance between reinforced adaptation and counteradaptation (Dawkins and Krebs, 1979; Brandman et al., 2012). Under evolutionary constraints, this is accompanied by the gradual loss of retrotransposons during evolution from S. japonicus to S. pombe, S. octosporus, and S. cryophilus and the parallel emergence of CENPB proteins in S. pombe, S. octosporus, and S. cryophilus (Rhind et al., 2011). Similar widespread retrotransposon silencing and CENP-B acquisition occurred in metazoans as well, including humans. The reciprocal trend between retrotransposon diminution and CENPB acquisition suggests the possibility of coevolutionary modulation of heterochromatin formation from RNAi-dependent to -independent pathways.
Reduction of RITS components in Schizosaccharomyces
The foregoing considerations prompted us to investigate evolutionary changes in all of the component proteins of the RNAi machinery, including RITS and RDRC complexes and the RNAi- independent SHREC complex, that may affect retrotransposon function. In addition to its role in heterochromatin formation, studies have speculated on the role of the RITS complex in retrotransposon silencing as well. Phylogenetic analysis revealed that all of the components of the RDRC and SHREC complexes are highly conserved. In all Schizosaccharomyces species, however, among the RITS components, only Ago1 is highly conserved; tas3 is completely missing, and Chp1, an interacting partner of Tas3, lacks the C-terminal region in S. octosporus and S. cryophilus. Of interest, the C-terminal of Chp1 binds to the N-terminal of Tas3, and this interaction is important for functional RITS-associated, RNAi-dependent heterochromatin formation in S. pombe (Petrie et al., 2005; Debeauchamp et al., 2008; Schalch et al., 2011; Ishida et al., 2012). Consistent with this, our study shows that whereas full-length Chp1 of S. pombe and S. japonicus interacts efficiently with Tas3 of S. pombe, is able to restore the silencing defect in the chp1Δ mutant, and localizes to the centromere in S. pombe, the truncated Chp1, retaining only the CD and RRM domains in S. octosporus and S. cryophilus, has lost all of these properties. These results can be viewed in light of the H3-Lys9-Me2/3–binding chromodomain and the RRM motif, which are required for nucleic acid binding (Petrie et al., 2005; Debeauchamp et al., 2008; Schalch et al., 2011; Ishida et al., 2012) but are not adequate for heterochromatin formation; the Tas3-interacting region is obligatory for heterochromatin formation (Debeauchamp et al., 2008). Thus the heterochromatin silencing function of RITS is compromised in the more recently diverged species of Schizosaccharomyces.
Horizontal transmission of Tc1/mariner and Tc5 elements and their evolution into CENPBs
The CENPB homologues in fission yeast display extensive sequence and domain-wise similarity to the Tc1-mariner class of transposases (Casola et al., 2008; O’Donnell and Boeke, 2008). It has been proposed that regardless of the concurrent time of evolution, these CENPBs arose independently in humans and fission yeast: whereas human CENPBs arose and were domesticated from pogo-like transposases, Cbh1/Abp1/CENPB in S. pombe are believed to have arisen from Tc1/mariner-like transposases (Casola et al., 2008; O’Donnell and Boeke, 2008). Our comparison of sequences of conserved domains and the NLS for Tc1/mariner and Tc5 transposase elements showed a shift in placement of the NLS motif in CENPBs from humans and S. pombe. The NLS overlaps with the DNA-binding domain and is required for integration and receptor-mediated nuclear uptake of transposases (Dingwall and Laskey, 1991; Plasterk et al., 1999); mutations in this motif are detrimental to overall transposase function (Plasterk et al., 1999). In contrast, in the CENPB of both humans and S. pombe, a monopartite NLS is found toward the C-terminal of the sequence. This region does not overlap with either the DNA-binding or the nonfunctional DDE_1 endonuclease domain. A unique WVPHEL stretch upstream of this NLS required for Tc1/transposase integration also seems to be lacking in humans, mice, and S. pombe/S. octosporus/S. cryophilus. However, the HTH-Tc5 DNA–binding domain retains its DNA-binding function in CENPB. This strongly suggests that CENPB lost its transposition activity and acquired the function of folding retrotransposons into heterochromatin, leading to suppression of transposition.
Thus the loss of transposons and the shift in their mode of silencing from RNA to heterochromatin during evolution from S. japonicus to S. pombe, S. octosporus, and S. cryophilus has been associated with the emergence of CENPB. This association may also be true in other organisms, including humans and C. elegans. Although S. japonicus completely lacks the CENP-B homologue, phylogenetic and synteny analyses show that CENPB homologues are more abundant in other species—three copies in S. pombe and five each in S. octosporus and S. cryophilus—and share syntenically conserved locations in S. cryophilus and S. octosporus (Table 3). Of interest, the origin of CENPB in S. pombe coincides with the timing of its inception in mammals, lending further support to the coevolution hypothesis (Figure 6).
Putative Tas3 homologues in other eukaryotes
The presence of tas3 in only S. japonicus and S. pombe prompted us to screen for their homologues in higher eukaryotes. Of interest, a conserved domain in Tas3 named Argonaute hook (Ago-hook) is present in many species, including humans and mice (Till et al., 2007). TNRC6B (having homology with the minimal Tas3 domain, which interacts with SpAgo1) interacts with human Ago1, indicating a conservation of the binary interaction within the RITS complex. GW182 (human TNRC6B) contains both Ago-hook (belonging to the TNR6C family)—a part of Tas3—and the RRM domain, a part of Chp1 in S. pombe (Petrie et al., 2005; Debeauchamp et al., 2008; Schalch et al., 2011; Ishida et al., 2012). Thus GW182 could integrate the RNAi and heterochromatin pathways. Indeed, GW182 has been shown to be responsible for the generation of PIWI RNAs through P-bodies (Eystathioy et al., 2002). Similarly, in Drosophila, Ago-hook protein SDE3 is involved in transposon silencing via piRNAs (Garcia et al., 2012).
A phylogenetic analysis of Ago-hook proteins also indicates that the emergence of the Ago-hook protein Tas3 in S. pombe and S. japonicus took place around the same time as the emergence of TNRC6 proteins in mammals (Supplemental Figure S7D and Table 3), supporting the possibility that coevolutionary pressure ensuing from the acquisition of CENPB and evolution of its role in transposon silencing via heterochromatin formation may have led to redundancy of RITS. We also speculate that, as in GW182, both Chp1 (RRM domain) in fission yeast and Tas3 (Ago-hook domain) might have once been parts of the same protein and hence may have coevolved. Of interest, analogous to GW182 protein P-bodies, CENP-B helps to organize the 13 copies of Tf2 as a part of one or two foci, referred to as Tf bodies, that perform a retrotransposon surveillance function in S. pombe (Cam et al., 2008; see later discussion). Subsequent expansion of CENPB copy number in S. octosporus and S. cryophilus may have been instrumental in further reduction of the retrotransposons in these species.
These results may exemplify the phenomenon of coevolution of proteins involved in interaction between retrotransposons and the host Schizosaccharomyces species. The loss of retrotransposons, which began with S. pombe, may have caused accumulation of mutations in Tas3 and Chp1,or, alternatively, caused the loss of Tas3, together with drastic reduction of retrotransposons in S. octosporus and S. cryophilus. Loss of retrotransposons leading to the mutants of Tas3 and Chp1 must have overlapped with the time scale when S. octosporus and S. cryophilus diverged. Thus the functions performed by Tas3 and the C-terminal domain of Chp1 may have been rendered redundant by the loss of retrotransposons, leading to their loss. However, the retention of the chromodomain that binds to H3-K9-me2/me3 and RNA in Chp1 may imply that the residual Chp1 in S. cryophilus and S. octosporus may continue to perform some function related to the role of RNAi in heterochromatin assembly. Similar coevolution may have occurred in higher eukaryotes, as a few Ago-hook proteins like Tas3, such as SDE3 in Drosophila and GW182 (TNRC6) protein in humans, perform a specialized function of controlling retrotransposons through their Ago-hook domain, suggesting that Tas3 might also perform such a function in Schizosaccharomyces.
In light of these observations, the retention and high level of conservation of Ago1 in all of the species is quite surprising, suggesting that Ago1 may still be performing some essential function in S. octosporus and S. cryophilus. In agreement, Ago1 shows a high level of conservation among the species: sequence identity is 63–69%; sequence similarity is 78–81%. Of importance, all four species contain the PIWI domain, which is responsible for binding to the GW-hook in Tas3 (Table 2). In addition to RITS (Verdel et al., 2002), Ago1 also exists in a complex called ARC in association with proteins Arb1 and Arb2 in S. pombe (Buker et al., 2007). The ARC complex associates primarily with double-stranded (ds) siRNAs (Buker et al., 2007). It was proposed that the ds-siRNAs are first associated with ARC; later, Ago1 associates with the proteins Chp1 and Tas3 and single-stranded (ss) siRNA in the RITS complex (Verdel et al., 2002). RITS, in turn, channels the ss siRNA to the RDRC complex for the generation of longer dsRNAs (Halic and Moazed, 2010). Because of the high degree of conservation of Ago1, we checked for the presence and conservation of ARC components Arb1 and Arb2. Of interest, both Arb1 and Arb2 are present and are well conserved in all of the Schizosaccharomyces species (unpublished data). Thus the ARC complex present in S. octosporus and S. cryophilus may retain the function of linking with RDRC to generate dsRNAs, albeit less effectively than the RITS because of lower levels of ss siRNAs. Furthermore, Ago1 can also work independently of the RNAi pathway, as it can also associate with primal RNAs, which are degradation products of abundant transcripts and trigger siRNA amplification and heterochromatin formation (Halic and Moazed, 2010).
Ablation of retrotransposons in S. pombe
There are 10 types of Gypsy retrotransposons in S. japonicus (Rhind et al., 2011). There are only two types of retrotransposons in S. pombe: Tf1, an extinct retrotransposon present in the form of solo-LTRs, and Tf2. Tcry1 is the only transposon in S. cryophilus. The difference between Tf1 and Tf2 that might contribute to their independent occurrence in S. pombe is that the 5’ untranslated region (UTR) and 3’UTR regions differ drastically between the two retrotransposon families—in particular, in the central portions of the LTRs and a region extending from just downstream of the 5’ LTR to the middle of the putative capsid protein domain. Tf1 is also closer to retroviruses and retrotransposable elements in the 5’ regions of 3’UTR that contain a polypurine stretch (Levin et al., 1990). It is these regions that are involved in dsRNA generation and transposition and comprise the least conserved regions between Tf1 and Tf2 (Cam et al., 2008). However, retrotransposon Tf1 apparently underwent drastic truncation, and it is no longer able to generate dsRNAs (Atwood-Moore et al., 2006); this truncation may also have contributed to the redundancy of the RITS components, leading to their elimination in newly emerging/branching species.
Role of CENPB in transposon silencing in fission yeast
The parallel coevolutionary emergence of CENPB from S. pombe to S. octosporus and S. cryophilus and the fact that it suppresses Tf2 retrotransposon recombination may signify that CENPB acquired the role of controlling retrotransposition through an RNAi-independent process (Figure 9). CENPB evolved as a protein-coding transposon that controls propagation of transposon Tf2 in S. pombe (Levin et al., 1990; Cam et al., 2008). Of interest, CENPB negatively regulates the expansion of Tf2 with the help of heterochromatin components, Set1 and histone deacetylases, but not the Tf1, because Tf1 is extinct and present as solo-LTRs only (Atwood-Moore et al., 2006; Cam et al., 2008; Lorenz et al., 2012). A synthetic chimera of Tf1 is expressed at augmented levels compared with Tf2 (Levin et al., 1990; Cam et al., 2008). CENPB performs an additional role of suppressing recombination between Tf1 and Tf2, thus preventing their expansion. Of note, this function is independent of RNAi but is heterochromatin mediated. Note that the dg/dh repeats in the centromeres of S. pombe also represent the remnants of retrotransposons of the Tf1 family that are also lost in S. octosporus and S. cryophilus. Thus CENPB acquisition and transposition of retrotransposons show a reciprocal association during evolution.
Our results suggest an ongoing adaptation between RNAi and transposons during evolution of the species in the genus Schizosaccharomyces. In the first stage of this adaptation, the RNAi-RITS may have evolved the role of controlling the transposable elements by domesticating them as part of the centromeres in S. japonicus and S. pombe. Through evolution, the widely prevalent retrotransposon may have undergone inactivation and mutation, and hence the components of RITS that silenced them might have become superfluous, leading to their elimination. This continuous adaptation and evolution causing reduction of RNAi components may have been facilitated in turn by acquisition of the Tc1/Tc5 transposase elements their its evolution from transposase/endonuclease into a structural protein, CENPB, with a role in regulating the transposon function as a heterochromatin protein, independent of RNAi (Figure 9). It is interesting to note recent genome studies on the Japanese pufferfish, Fugu rubripes, which has a genome size 7.5 times smaller than the human genome (Elgar et al., 1996). Remarkably, the fugu genome contains a low level of retrotransposons (2.7%) and also lacks CENPB (unpublished data).
Nearly 44% of the human genome is constituted of diverse transposon elements (Mills et al., 2003), which are maintained in the heterochromatic state by proteins like CENPB, which, in turn, was derived and evolved from transposases. Significantly, similar to S. pombe, human cells and other metazoans may display similar coevolutionary dynamics between the RNAi- and CENPB-mediated control of the widely prevalent transposons. We propose that retention of the transposable element (Tc1/Tc5) and its evolution from transposase to the heterochromatin-bound, centromeric protein CENPB may also have occurred in parallel with or been caused by the inactivation of retrotransposons in a coevolutionary manner in humans.
MATERIALS AND METHODS
Data acquisition
We obtained sequences of proteins involved in heterochromatin in S. pombe, S. japonicus, S. octosporus, and S. cryophilus from the databases of the Broad Institute (Rhind et al., 2011) and Pombase (Wood et al., 2002) to study their conservation among these species. We acquired sequences of homologues of Chp1 from other fission yeasts and metazoans using BLASTn (Altschul et al., 1990). All sequences were obtained from NCBI’s genetic sequence database (GenBank) and compared using BLASTn to confirm their identity and similarity to the annotated chp1 genes. Sequence IDs for various genes as retrieved from NCBI databases for these species are as follows:
CDYL (Homo sapiens) NC_000006.12, Cdyl (Mus musculus) NC_000079.6, Cdyl (Bos taurus) AC_000180.1, Cdyl (Takifugu rubripes) NW_004075257.1, CDYL (Canis lupus familiaris) NC_006617.3, chp1 (S. pombe) NC_003424.3, chp1 (S. japonicus) NW_011627861.1, chp1 (S. octosporus) NW_013185621.1, and chp1 (S. cryophilus; SPOG_00205) NW_013185626.1.
TNRC6A (H. sapiens) NC_000016.10, TNRC6A (M. musculus) NC_000073.6, TNRC6A (B. taurus) AC_000182.1, TNRC6A (Pteropus alecto) NW_006436286.1, TNRC6A (T. rubripes) NC_018894.1, TNRC6A (C. l. familiaris) NC_006588.3, tas3 (S. japonicus) NW_011627861.1, and tas3 (S. pombe) NC_003423.3.
TUBB (H. sapiens) NC_000006.12, tubb (P. alecto) NW_006436809.1, tubb1 (M. musculus) NC_000068.7, tubb (T. rubripes) NC_018896.1, tubb (B. taurus) AC_000170.1, tubb1 (C. l. familiaris) NC_006594.3, nda3 tubulin (S. pombe) SPBC26H8.07c, NC_003423.3, nda3 (S. japonicus), SJAG_04003, NW_011627861.1, nda3 (S. octosporus) SOCG_01442, NW_013185621.1, and nda3 (SPOG_03182; S. cryophilus), NW_013185624.1.
Synteny analysis
Synteny can be described as a comparison of genes within the loci of the same chromosome or two chromosomes among two or more species to assess the positions of genes. We accessed the Broad Institute Schizosaccharomyces database for sequence information of loci containing tas3, ago1, chp1, and CENPB homologues in S. pombe, S. japonicus, S. octosporus, and S. cryophilus. The GeVO (COGE) and DOTPLOT (dcode; NCBI) software were used for constructing the synteny maps of these genes.
Comparative phylogenetic analysis of genes involved in heterochromatinization across the Schizosaccharomyces genus
Phylogenetic analysis was performed using MEGA version 6.0 software (Tamura et al., 2013). All inferences were drawn using the maximum likelihood method, with the initial tree drawn using the maximum parsimony method. The WAG + F model was used, in which each model was applied in the +F form, with one set of amino acid frequencies estimated from the entire database and applied to the analysis of all protein families. This model was used, and the subtree-pruning-regrafting (SPR) algorithm at level 5 was implemented along with bootstrapping at a value of 100. The WAG + F model was specifically chosen because it gives a better likelihood and better fit to the data, and p values are very small.
The time tree was also calculated using MEGA version 6.0 (Tamura et al., 2013), which calculates the times of divergence based on the distances between the branching points (RelTime method). The constraints were set for the most recent common ancestor (MRCA), and we provided no constraints during the study to provide for the absolute divergence times. The sequence-based analysis performed by maximum likelihood analysis was independently allowed to calculate the relative time differences between the branches. The relative times were from 0.0 to 1.5, where 1.5 represents the earliest divergence time and 0.0 represents the protein sequences of the species of most recent origin. Ago-hook proteins were retrieved from the literature (Buker et al., 2007).
Ago-hook proteins from S. pombe were subjected to PSI-BLAST to retrieve Ago-hook–like proteins from other species. CENP-B homologues were retrieved from the NR database by using the S. pombe CENP-B sequence as query. Many sequences obtained from PSI-BLAST were further filtered based on their functional annotation and redundancy in the same species. Sequences that were too diverged and misleading and were present in a single clade were removed from the final tree-making process. To analyze conserved domains and conserved residues, the sequences for CENPB, Tas3, and Chp1 were subjected to CDD to identify conserved domains, and the alignment was performed using Clustal omega (EBI) followed by computation of the completely conserved residues as 100% alignment. The synteny maps were created using COGE (GeVo) software. Codon usage bias analysis was carried out by CODONW (Peden, 2013) and YASS (Noe and Kucherov, 2005).
GC-skew plots
We used the software GenSkew, which computes the normal and cumulative skew of two given nucleotides for a given sequence. The results are displayed in red for irregular skew (with unusual GC content) and blue for regular GC skew. For the pair of nucleotides G and C, the skew is given by
Skew = (G − C) / (G + C)
For the computation, the genome is separated into parts called windows. For every window, the value of the skew is calculated with the foregoing formula. The normal graph (blue) displays every value related to the particular position in the sequence. The cumulative graph adds the values for all previous windows up to the given position. The windows are shifted along the sequence by a certain step size (in kilobases). We computed portions of the genomic locations covering CENPB proteins in S. pombe, S. octosporus, and S. cryophilus.
GC skew plots describe a minimum (origin of replication) and maximum (termination). Positive GC skew represents G > C, and negative GC skew represents C > G.
Cloning of chp1 of S. octosporus, S cryophilus, S. japonicus, and S. pombe
The chp1 genes of S. pombe, S. japonicus, S. octosporus, and S. cryophilus chp1 were PCR amplified and cloned into the vector pREP41HAN, designed to express Chp1 with an HA tag. These plasmids were transformed into strains having an epitope-tagged tas3 gene and the chp1Δ mutation.
Serial dilution assay
Expression of imr::ura4 reporter was checked by dilution assay by spotting 5 μl of 10-fold serial dilutions of overnight cultures of the required strains on nonselective plates, plates lacking uracil, and those containing 5-fluoroorotic acid (5-FOA; Koski et al., 2001). Expression of heterochromatic ade6 (otr1R::ade6) was assessed by streaking the cells for single colonies on plates containing limiting adenine (15 mg/l). Cells expressing ade6 form light-pink or white colonies on these plates, and cells failing to express ade6 gene produce dark-red colonies. Determining the percentage of such colonies provides a semiquantitative indication of the silencing defect. The strains used in this study are listed in Table 4.
TABLE 4:
Strains used in this study.
| Strain name | Genotype |
|---|---|
| USP 21 | h+ leu1-32 ade6-216 ura4DS/E imr1R (NcoI)::ura4+ oriI tas3-TAP-Kanr kanMX6 pREP41HAN::chp1S. japonicus |
| USP22 | h+ leu1-32 ade6-216 ura4DS/E imr1R (NcoI)::ura4+ oriI tas3-TAP- Kanr kanMX6 pREP41HAN::chp1S. pombe |
| USP23 | h+ leu1-32 ade6-216 ura4DS/E imr1R (NcoI)::ura4+ oriI tas3-TAP- Kanr kanMX6 pREP41HAN::chp1S. octosporus |
| USP24 | h+ leu1-32 ade6-216 ura4DS/E imr1R (NcoI)::ura4+ oriI tas3-TAP- Kanr kanMX6 pREP41HAN::chp1S. cryophilus |
| USP25 | h+ leu1-32 ade6-216 ura4DS/E imr1R (NcoI)::ura4+ oriI chp1∆-kanMX6 pREP41HAN |
| USP26 | h+ leu1-32 ade6-216 ura4DS/E imr1R (NcoI)::ura4+ oriI chp1-∆-kanMX6 pREP41HAN::chp1S. pombe |
| USP27 | h+ leu1-32 ade6-216 ura4DS/E imr1R (NcoI)::ura4+ oriI chp1-∆-kanMX6 pREP41HAN::chp1S. japonicus |
| USP28 | h+ leu1-32 ade6-216 ura4DS/E imr1R (NcoI)::ura4+ oriI chp1-∆-kanMX6 pREP41HAN::chp1S. octosporus |
| USP29 | h+ leu1-32 ade6-216 ura4DS/E imr1R (NcoI)::ura4+ oriI chp1-∆-kanMX6 pREP41HAN::chp1S. cryophilus |
| USP30 | h+ ura4DS/E ade6-210/ ade6 DN/N, imr1L(NcoI)::ura4, Otr1 R:: ade6 chp1-∆-kanMX6 pREP41HAN::chp1S. pombe |
| USP31 | h+ ura4DS/E ade6-210/ ade6 DN/N, imr1L(NcoI)::ura4, Otr1 R:: ade6 chp1-∆-kanMX6 pREP41HAN::chp1S. japonicus |
| USP32 | h+ ura4DS/E ade6-210/ ade6 DN/N, imr1L(NcoI)::ura4, Otr1 R:: ade6 chp1-∆-kanMX6 pREP41HAN::chp1S. octosporus |
| USP33 | h+ ura4DS/E ade6-210/ ade6 DN/N, imr1L(NcoI)::ura4, Otr1 R:: ade6 chp1-∆-kanMX6 pREP41HAN::chp1S. cryophilus |
Coimmunoprecipitation of Chp1 and Tas3
Coimmunoprecipitation was carried out to assess the interaction between Chp1 of S. japonicus, S. pombe, S. octosporus, and S. cryophilus and Tas3 of S. pombe by following the protocol of Lee (2007) using protein A–Sepharose beads (GE healthcare) and anti–protein A antibody specific for TAP tag (Sigma-Aldrich) and anti-HA antibody for HA tag (Santa Cruz Biotechnology).
ChIP assay
ChIP assay was performed according to a protocol modified from Ekwall and Partridge (1999).
Real-time PCR analysis of ChIP samples
Real-time PCR was done with ChIP samples to check the localization of Chp1 from different species of Schizosaccharomyces. A chp1∆ strain from S. pombe was transformed with HA-tagged chp1 genes from S. pombe, S. japonicus, S. octosporus, and S. cryophilus cloned in the pREP41HAN plasmid, and fold enrichment of Chp1 at the dh region was quantitated using act1 as a control visualized by SYBR green dye (Realplex Real Time PCR machine; Thermo Scientific). Results were analyzed by calculating the DCt value (normalized to the input samples) for each sample: Ct(sample) − Ct(input) (Mukhopadhyay et al., 2008).
Supplementary Material
Acknowledgments
We thank Jun Ichi Nakayama for the chp1∆ and tas3 TAP-tagged strains and S. Haldar for critically reading the manuscript. This work received intramural support from the Institute of Microbial Technology (Council of Scientific and Industrial Research) and an Indian Council of Medical Research and University Grants Commission Fellowship to U.U. and I.K., respectively.
Abbreviations used:
- Abp
ARS-binding protein
- Ago
Argonaute
- ARC
Argonaute siRNA chaperone
- chp1
chromodomain protein
- clr
cryptic loci regulator
- DBD
DNA-binding domain
- dcr
Dicer
- HTH
helix turn helix
- NLS
nuclear localization sequence
- otr
outer repeat
- rdp
RNA-dependent RNA polymerase
- RDRC
RNA-directed RNA polymerase complex
- RITS
RNA-induced transcriptional silencing
- RRM
RNA recognition motif
- swi
switch
- Tas3
targeting complex subunit
- TNR
trinucleotide repeat.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-07-0485) on February 22, 2017.
REFERENCES
- Allshire RC, Javerzat JP, Redhead NJ, Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994;76:157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Atwood-Moore A, Yan K, Judson RL, Levin HL. The self primer of the long terminal repeat retrotransposon Tf1 is not removed during reverse transcription. J Virol. 2006;80:8267–8271. doi: 10.1128/JVI.01915-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman R, Brandman Y, Pande VS. Sequence co-evolution between RNA and protein characterized by mutual information between residue triplets published. PLoS One. 2012;10:1371. doi: 10.1371/journal.pone.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buker SM, Iida T, Buhler J, Villen J, Gygi SP, Nakayama J, Moazed D. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat Struct Mol Biol. 2007;14:200–207. doi: 10.1038/nsmb1211. [DOI] [PubMed] [Google Scholar]
- Burns C, Stajich JE, Rechtsteiner A, Casselton L, Hanlon SE, Wilke SK, Savytskyy OP, Gathman AC, Lilly WW, Lieb JD, et al. Analysis of the basidiomycete Coprinopsis cinerea reveals conservation of the core meiotic expression program over half a billion years of evolution. PLoS Genet. 2010;10:1001135. doi: 10.1371/journal.pgen.1001135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Noma K, Ebina H, Levin HL, Grewal SI. Host genome surveillance for retrotransposon by transposon-derived proteins. Nature. 2008;451:431–436. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- Casola C, Hucks D, Feschotte C. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol Biol Evol. 2008;25:29–41. doi: 10.1093/molbev/msm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc Lond Biol Sci. 1979;205:489–551. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- Debeauchamp JL, Moses A, Noffsinger VJP, Ulrich DL, Job G, Kosinski AM, Partridge JF. Chp1-Tas3 interaction is required to recruit RITS to fission yeast centromeres and for maintenance of centromeric heterochromatin. Mol Cell Biol. 2008;28:2154–2166. doi: 10.1128/MCB.01637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Partridge JF. In: Chromosome Structural Analysis. A Practical Approach. Oxford, UK: Oxford University Press; 1999. Fission yeast chromosome analysis: fluorescence in situ hybridisation (FISH) and chromatin immunoprecipitation (ChIP) pp. 39–57. [Google Scholar]
- Elgar G, Sandford R, Aparicio S, Macrae A, Venkatesh B, Brenner S. Small is beautiful: comparative genomics with the pufferfish (Fugu rubripes) Trends Genet. 1996;12:145–150. doi: 10.1016/0168-9525(96)10018-4. [DOI] [PubMed] [Google Scholar]
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell AJ, Ruby SW, Toole JJ, Roberts BE, Rubin GM. Translation and developmental regulation of RNA encoded by the eukaryotic transposable element copia. Proc Natl Acad Sci USA. 1980;77:7107–7111. doi: 10.1073/pnas.77.12.7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Zhang CT. GC-Profile: A web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 2006;1(34(Web Server issue)): W686–W691. doi: 10.1093/nar/gkl040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Garcia S, Pontier D, Marchais A, Renou JP, Lagrange T, Voinnet O. Ago hook and RNA helicase motifs underpin dual roles of SDE3 in antiviral defense and silencing of non-conserved intergenic regions. Mol Cell. 2012;48:109–120. doi: 10.1016/j.molcel.2012.07.028. [DOI] [PubMed] [Google Scholar]
- Grewal SIS, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huda A, Mariño-Ramírez L, Jordan IK. Epigenetic histone modifications of human transposable elements: genome defense versus exaptation. Mobile DNA. 2010;1:2. doi: 10.1186/1759-8753-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida M, Shimojo H, Hayashi A, Kawaguchi R, Ohtani Y, Uegaki K, Nishimura Y, Nakayama JI. Intrinsic nucleic acid-binding activity of Chp1 chromodomain is required for heterochromatic gene silencing. Mol Cell. 2012;47:228–241. doi: 10.1016/j.molcel.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Koski LB, Morton RA, Golding GB. Codon bias and base composition are poor indicators of horizontally transferred genes. Mol Biol Evol. 2001;18:404–412. doi: 10.1093/oxfordjournals.molbev.a003816. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of yeast cell cycle-dependent nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Coimmunoprecipitation assay. Methods Mol Biol. 2007;362:401–406. doi: 10.1007/978-1-59745-257-1_31. [DOI] [PubMed] [Google Scholar]
- Levin HL, Weaver DC, Boeke JD. Two related families of retrotransposon from Schizosaccharomyces pombe. Mol Cell Biol. 1990;10:6791–6798. doi: 10.1128/mcb.10.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz D, Mikheyeva IV, Johansen P, Meyer L, Berg A, Grewal SI, Cam HP. CENP-B cooperates with Set1 in bidirectional transcriptional silencing and genome organization of retrotransposon. Mol Cell Biol. 2012;32:4215–4225. doi: 10.1128/MCB.00395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann S, Chen YP. Bacterial genomic G+C composition-eliciting environmental adaptation. Genomics. 2010;95:7–15. doi: 10.1016/j.ygeno.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2003;23:183–187. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011;146:510–518. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Deplancke B, Walhout AJM, Tissenbaum HA. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real- time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protocols. 2008;3:698–709. doi: 10.1038/nprot.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noe L, Kucherov G. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 2005;33:W540–W543. doi: 10.1093/nar/gki478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Boeke JD. Domesticated DNA transposon proteins mediate retrotransposon control. Cell Res. 2008;18:331–333. doi: 10.1038/cr.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F, Valencia A. Protein co-evolution, co-adaptation and interactions. Plasmid. 2008;69:1–15. doi: 10.1038/emboj.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden J. Correspondence analysis of codon usage. Available at. 2013. http://codonw.sourceforge.net/ (accessed 22 February 2017)
- Petrie VJ, Wuitschick JD, Givens CD, Kosinski M, Partridge JF. RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast. Mol Cell Biol. 2005;25:2331–2346. doi: 10.1128/MCB.25.6.2331-2346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk RH, Izsvák Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- Pluta AR, Saitoh N, Goldberg I, Earnshaw WC. Identification of a subdomain of CENPB that is necessary and sufficient for localization to the human centromere. J Cell Biol. 1992;116:1081–1093. doi: 10.1083/jcb.116.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Silverstein RA, Dishart D, Walfridsson J, Djupedal I, Kniola B, Wright A, Samuelsson B, Radmark O, Ekwall K. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc Natl Acad Sci USA. 2002;99:16648–16653. doi: 10.1073/pnas.212633199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Job G, Shanker S, Partridge JF, Joshua-Tor L. The Chp1- Tas3 core is a multifunctional platform critical for gene silencing by RITS. Nat Struct Mol Biol. 2011;18:1351–1357. doi: 10.1038/nsmb.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova A, Rhind N, Zamore PD. A single argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev. 2004;18:2359–2367. doi: 10.1101/gad.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of self-reinforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2007a;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam HP, Sugiyama R, Noma K, Zofall M, Kobayashi R, Grewal SIS. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007b;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Lehner B. Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet. 2008;24:485–488. doi: 10.1016/j.tig.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi mediated targeting of heterochromatin by the RITS complex. Science. 2002;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Grace T, Grewal SIS, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SIS. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.