Ste23 is a novel mitochondrial protease located in the matrix. Ste23 degrades presequence peptides derived from imported preproteins. Impaired peptide degradation leads to inhibited presequence-processing machinery and accumulation of immature preproteins. Thus efficient peptide turnover is crucial for mitochondrial proteostasis.
Abstract
Approximately 70% of mitochondrial precursor proteins are imported from the cytosol via N-terminal presequences, which are cleaved upon exposure to the mitochondrial processing protease MPP in the matrix. Cleaved presequence peptides then need to be efficiently degraded, and impairment of this clearance step, for example, by amyloid β peptides, causes feedback inhibition of MPP, leading ultimately to accumulation of immature precursor proteins within mitochondria. Degradation of mitochondrial peptides is performed by Cym1 in yeast and its homologue, PreP, in humans. Here we identify the novel mitochondrial matrix protease Ste23 in yeast, a homologue of human insulin-degrading enzyme, which is required for efficient peptide degradation. Ste23 and Cym1 tightly cooperate to ensure the correct functioning of the essential presequence processing machinery.
INTRODUCTION
Mitochondria harbor between 1000 (yeast) and 1500 (human) different proteins. With a few exceptions of the proteins encoded by the mitochondrial genome, almost the entire organellar proteome is built up via import of nuclear-encoded precursor proteins (Habib et al., 2007; Neupert and Herrmann, 2007; Meisinger et al., 2008; Endo et al., 2011; Sokol et al., 2014; Schulz et al., 2015). Approximately 70% of these precursors possess N-terminal presequences, which direct them to the protein import machinery and promote their translocation across the two mitochondrial membranes (Gakh et al., 2002; Millar et al., 2006; Habib et al., 2007; Vögtle et al., 2009). Mitochondrial presequences are typically positively charged amphipathic helices and require the membrane potential Δψ as a major driving force for preprotein translocation across the inner membrane (Gakh et al., 2002; Habib et al., 2007; Neupert and Herrmann, 2007; Vögtle et al., 2009; Endo et al., 2011; Harbauer et al., 2014; Sokol et al., 2014; Schulz et al., 2015). On emerging on the matrix side, the presequence is usually cleaved by the mitochondrial processing protease (MPP), a heterodimer consisting of the two subunits Mas1 and Mas2. Presequence processing by MPP is essential for cell viability, and presequence peptides resulting from MPP processing have to be efficiently degraded because they are toxic for the cell (Hawlitschek et al., 1988; Yang et al., 1991; Gakh et al., 2002; Mossmann et al., 2012; Teixeira and Glaser, 2013; Kmiec et al., 2014; Quiros et al., 2015; van Wijk, 2015). We recently sowed that this toxicity is due to an impairment of the MPP activity by feedback inhibition, which in turn causes accumulation of immature precursor proteins and eventually an imbalanced organellar proteome (Mossmann et al., 2014; Burkhart et al., 2015). Of note, amyloid β peptides, which accumulate in mitochondria of Alzheimer’s disease patients (Hansson Petersen et al., 2008; Alikhani et al., 2011a), also limit peptide degradation capacity and lead to accumulation of immature precursor proteins (Mossmann et al., 2014). Presequence peptide degradation is mediated by metalloproteases—for example, PreP in humans (also known as PITRM1) and plants or their homologue, Cym1, in yeast mitochondria (Kambacheld et al., 2005; Hansson Petersen et al., 2008; Alikhani et al., 2011a,b; Mossmann et al., 2012, 2014; Kmiec et al., 2013; Teixeira and Glaser, 2013).
Here we identify Ste23, a homologue of human insulin-degrading enzyme (IDE), as a novel protease in the mitochondrial matrix of yeast. Mitochondrial Ste23 cooperates with Cym1 in efficient peptide clearance and can also degrade amyloid β peptides such as human IDE and PreP. Moreover, we show by in vitro assays as well as in vivo that Ste23 is required for functional presequence processing machinery.
RESULTS AND DISCUSSION
Identification of Ste23 as a peptide-degrading protease in the mitochondrial matrix
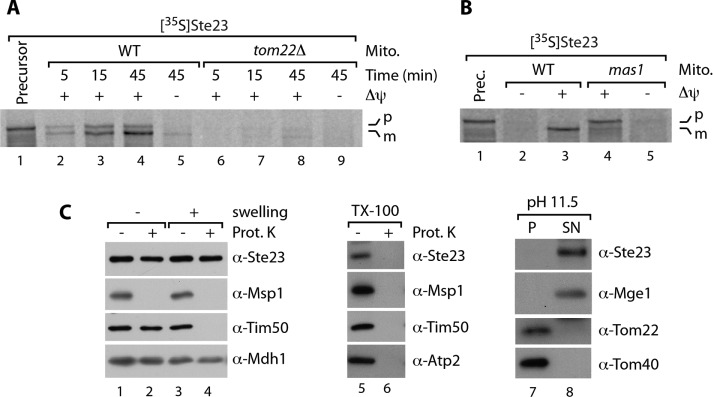
The metalloprotease Cym1 was the only known peptide-degrading enzyme in the yeast mitochondrial matrix. Peptide degradation activity can be monitored in soluble mitochondrial extracts (Alikhani et al., 2011a; Mossmann et al., 2014). Indeed, the degradation kinetics tested thus far revealed an apparently complete loss of peptide degradation capacity in the absence of Cym1 (Alikhani et al., 2011a; Mossmann et al., 2014). Here we analyzed the degradation of presequence peptides in wild-type and cym1Δ soluble mitochondrial extracts for longer time spans and observed residual peptide clearance activity even in the absence of Cym1 (Figure 1A). Of note, we observed such an activity also for the degradation of amyloid β peptides (Supplemental Figure S1, A and B). This led us to speculate about the presence of an additional mitochondrial protease activity involved in peptide degradation. To narrow the class of protease to which this activity could be assigned, we monitored peptide degradation (on prolonged incubation) in cym1Δ soluble mitochondrial extracts in the presence of protease inhibitors. In contrast to inhibition of serine proteases by phenylmethylsulfonyl fluoride (PMSF), the presence of o-phenanthroline/EDTA strongly inhibited peptide degradation capacity in the absence of Cym1, indicating a further, different metalloprotease involved in this process (Figure 1B). To uncover such an enzyme, we searched the yeast genome for a putative metalloprotease with unknown function and a high prediction score for the presence of a mitochondrial presequence at its N-terminus. We found the open reading frame YLR389C encoding Ste23 with a high score prediction of 0.975 (MitoProt II) for a mitochondrial presequence (Claros and Vincens, 1996). This protein has been implicated in pro–a-factor processing and was therefore named STErile23 (Adames et al., 1995). However, this role has been challenged by the fact that Ste23 is expressed in cells that do not produce a-factor, and it was proposed that Ste23 might be involved in other physiological functions (Alper et al., 2009). Of note, Alper et al. (2009) found that proteolytic activity of Ste23 does not require the annotated N-terminal 53 amino acids and speculated about the misassignment of the STE23 gene. We asked whether the N-terminus might be explained by the presence of a cleavable mitochondrial presequence. To test this, we generated [35S]Ste23 precursor protein by in vitro transcription/translation. On incubation with isolated mitochondria, the [35S]Ste23 precursor is imported in a membrane potential–dependent manner and processed into a shorter variant reflecting the mature protein (Figure 2A, lanes 1–5). Import of Ste23 requires the protein import machinery (revealed by the dependence on the central import receptor Tom22; Figure 2A, lanes 6–9), and its presequence is cleaved by MPP (shown by import into mas1 mutant mitochondria [ Witte et al., 1988]; Figure 2B). To confirm mitochondrial matrix localization of endogenous Ste23, we generated specific and affinity-purified antibodies. Ste23 is protected by externally added Proteinase K (PK) under isoosmotic and hypoosmotic conditions (which enable access of PK to the intermembrane space) but is degraded by PK when both membranes are lysed by the detergent Triton X-100 (Figure 2C, lanes 1–6). The protein can also be easily extracted by alkaline pH into the soluble protein fraction (Figure 2C, lanes 7 and 8). In summary, Ste23 is imported into mitochondria via a cleavable presequence and localizes to the matrix.
FIGURE 1:
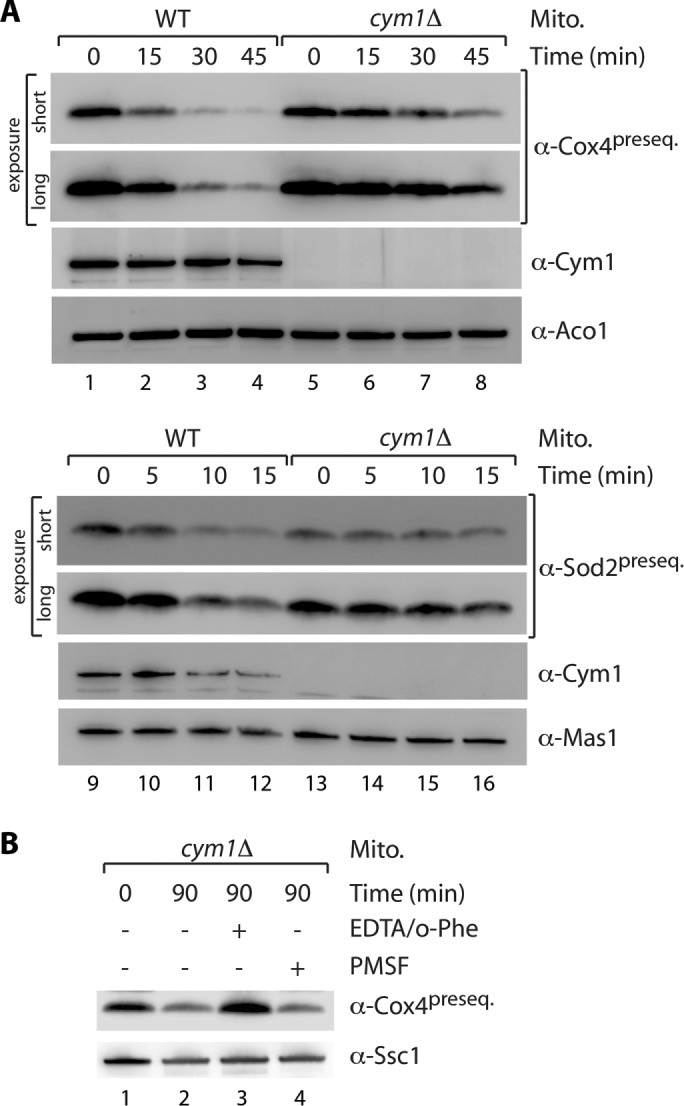
Detection of a peptide-degrading activity in cym1Δ mitochondria. (A) Degradation of Cox41–17 (lanes 1–8) and Sod21–18 (lanes 9–16) presequence peptides in soluble extracts of mitochondria from wild-type (WT) and cym1Δ yeast strains. Samples were analyzed by SDS–PAGE and immunodecoration. Aco1 and Mas1 served as loading controls. (B) Degradation of Cox41–17 presequence peptide in mitochondrial extract from cym1Δ strain in the presence or absence of protease inhibitors (5 mM EDTA and 1 mM o-phenanthroline [o-Phe] or 1 mM PMSF). Ssc1, loading control.
FIGURE 2:
The metalloprotease Ste23 localizes to the mitochondrial matrix. (A) Radiolabeled Ste23 precursor was incubated with WT and tom22Δ mitochondria for various times. Where indicated, the membrane potential (Δψ) was dissipated before the import reaction. Samples were treated with PK and analyzed via SDS–PAGE and autoradiography. m, mature; p, precursor. (B) Import of [35S]Ste23 precursor into mitochondria isolated from WT and mas1 temperature-sensitive mutant strains (Vögtle et al., 2009) after shifting of the culture for 6 h to 37°C. Impaired MPP activity in mas1 allows import but not presequence processing of Ste23 precursor (lane 4 vs. lane 3). Samples were treated as in A. (C) Mitochondria were subjected to hypoosmotic swelling (lanes 1–4), Triton X-100 treatment (lanes 5 and 6), or carbonate extraction at pH 11.5 (lanes 7 and 8). Samples were analyzed by SDS–PAGE and immunodecoration. Prot. K, Proteinase K; P, pellet; SN, supernatant.
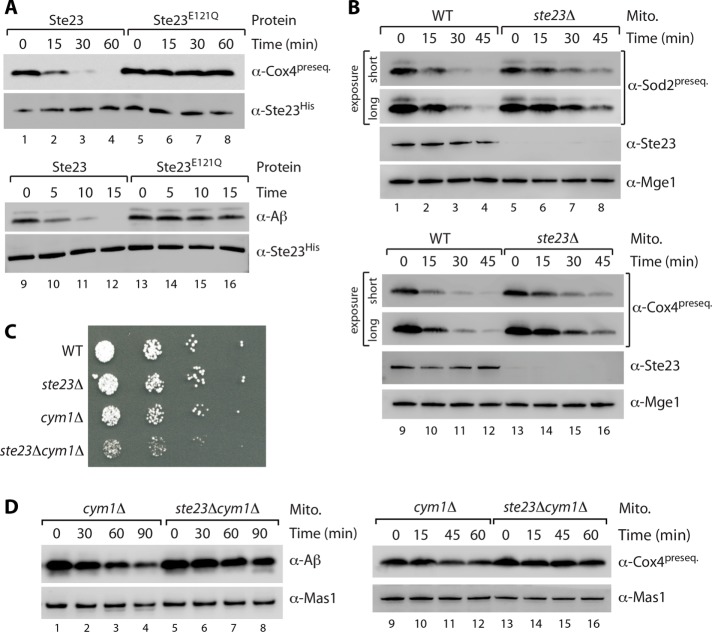
To study the potential role of Ste23 in presequence degradation, we generated wild-type Ste23 and the variant Ste23E121Q with a mutation in the predicted catalytic site by cell-free translation (Alper et al., 2009). Wild-type Ste23 but not the inactive variant can efficiently degrade the presequence peptide of Cox4 and also amyloid β peptide (Figure 3A). Degradation of presequence peptides in soluble mitochondrial fractions of wild type and an ste23Δ mutant revealed a delay in peptide clearance in the absence of Ste23 (Figure 3B; note that Cym1 is still present in these samples). Similarly, we also observed a delay of the degradation of amyloid β peptides in ste23Δ samples compared with wild type (Supplemental Figure S2A). Taken together, the results indicate that Ste23 is a novel mitochondrial matrix protease that appears to play a role in peptide degradation.
FIGURE 3:
Cooperation of Ste23 and Cym1 in mitochondrial peptide degradation. (A) In vitro–synthesized Ste23 protein with a C-terminal hexahistidine tag (WT or E121Q mutation in the catalytic center) was incubated with Cox4 presequence peptide and amyloid β peptide (Aβ1–28) for indicated periods of time. Samples were analyzed by SDS–PAGE, followed by immunodecoration with Cox4 presequence specific antibodies and anti-histidine to detect Ste23. (B) Soluble extracts from WT and ste23Δ mitochondria were supplemented with Cox41–17 (lanes 1–8) and Sod21–18 (lanes 9–16) presequence peptides and incubated for indicated time. Peptide degradation was monitored by SDS–PAGE and immunodecoration. Mge1, loading control. (C) Double deletion of STE23 and CYM1 results in severe growth defect. Strains were grown under respiratory growth conditions (YPG) at 19°C. (D) Soluble mitochondrial fractions from cym1Δ and ste23Δcym1Δ yeast cells were incubated with Cox4 presequence peptides or amyloid β peptides for indicated time. Samples were analyzed by SDS–PAGE and immunodecoration. Mas1 served as loading control.
Ste23 and Cym1 form a cooperative system of presequence peptide degradation
Owing to relatively mild impairment of peptide degradation in ste23Δ mitochondria (Figure 3B and Supplemental Figure S2A), in which Cym1 is still present (and conversely; Figure 1A and Supplemental Figure S1, A and B), we wondered whether Ste23 and Cym1 tightly cooperate to ensure efficient peptide clearance. We asked whether the absence of both proteases, Ste23 and Cym1, might more severely affect mitochondrial peptide degradation capacity and eventually cell viability. Indeed, whereas single-deletion mutants of Cym1 and Ste23 grow like wild type at low temperature, their simultaneous deletion leads to a severe growth defect (Figure 3C). This points toward cooperative action of the enzymes that is important for cell viability. This cooperative system of Ste23 and Cym1 is also reflected by the strongly impaired peptide degradation capacity (tested for Cox4 presequence and amyloid β peptides) in mitochondrial extracts from the double mutant ste23Δcym1Δ compared with the (already impaired) activity of cym1Δ samples (Figure 3D).
Ste23 is required for efficient preprotein maturation
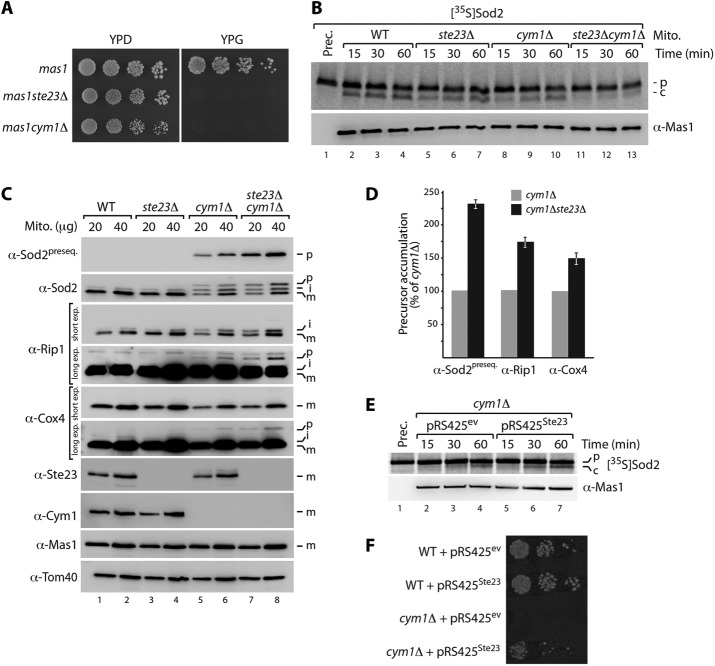
Impaired peptide degradation in cym1Δ mitochondria affects the activity of the essential protease MPP, and this is likely caused by competition of accumulating presequence peptides with the incoming precursor proteins for binding to MPP (Yang et al., 1991; Taylor et al., 2001; Mukhopadhyay et al., 2007; Mossmann et al., 2014). As a consequence, immature precursor proteins accumulate within the organelle, finally leading to impaired mitochondrial proteostasis (Mossmann et al., 2014). We wondered whether loss of Ste23 also affects MPP activity and therefore tested first a potential genetic interaction of STE23 and MPP. Indeed, when we deleted STE23 in the MPP mutant strain mas1, the cells were not viable on respiratory growth medium even at low temperature, indicating a functional link between STE23 and MPP (Figure 4A). A very similar phenotype is observed for the mas1cym1Δ double mutant (Figure 4A; Mossmann et al., 2014). We then sought to analyze MPP activity in soluble mitochondrial extracts from ste23Δcym1Δ double- and single-mutant strains. Presequence processing of [35S]Sod2 precursors was severely impaired in the double-mutant compared with the already decreased activity in the cym1Δ and ste23Δ single-mutant samples (Figure 4B). To test whether the impaired MPP activity might also be detectable in vivo, we inspected typical mitochondrial precursor proteins with classical N-terminal presequences by immunoblotting mitochondria isolated from wild-type, single-deletion, and ste23Δcym1Δ double-deletion strains (Figure 4C). Immature precursors can be identified either by their slower migration during electrophoresis or presequence-specific antibodies that recognize only the precursor and not the mature protein (Mossmann et al., 2014). Whereas lack of Ste23 alone did not reveal detectable precursors (unlike in cym1Δ mitochondria), deletion of both proteases led to a severe increase of nonprocessed immature mitochondrial proteins (Figure 4, C and D). These findings support our view that efficient peptide clearance is an important requirement for proper functioning of the presequence-processing protease MPP.
FIGURE 4:
Peptide clearance by Ste23 and Cym1 is required for functional presequence processing machinery. (A) Synthetic lethality of mas1ste23Δ and mas1cym1Δ double mutants. Strains were grown under fermentative (YPD) or respiratory (YPG) conditions at 23°C. (B) Presequence-processing activity in soluble mitochondrial extracts from indicated yeast strains was analyzed by incubation with radiolabeled Sod2 precursor, followed by SDS–PAGE and autoradiography. c, cleaved Sod2 protein; p, precursor. (C) Immunoblot analysis of mitochondria isolated from WT, ste23Δ, cym1Δ, and ste23Δcym1Δ yeast cells. i, processing intermediate; m, processed, mature protein. (D) Levels of nonprocessed preproteins (p) analyzed in C were quantified. Quantifications represent mean ± SEM (n = 3). (E) Overexpression of Ste23 in cym1Δ cells stimulates presequence processing of radiolabelled Sod2 precursor in soluble mitochondrial extracts. c, cleaved Sod2 protein; ev, empty vector; p, precursor. (F) Ste23 rescues loss of Cym1 function in vivo. Overexpression of Ste23 from pRS425 plasmid restores viability of cym1Δ cells at 37°C.
To gain further independent evidence of functional cooperation of Cym1 and Ste23 in this process, we overexpressed Ste23 in the cym1Δ mutant strain. We found that the presequence-processing activity of MPP can be stimulated by overexpression of Ste23 in the absence of Cym1 (Figure 4E). Similarly, Cym1 overexpression facilitated MPP activity in ste23Δ samples (Supplemental Figure S2B). Moreover, overexpression of Ste23 can rescue the lethal growth phenotype of the cym1Δ strain at higher temperature, strongly supporting a cooperative role of both enzymes in mitochondrial peptide clearance, which is an important requirement for a functional presequence-processing machinery (Figure 4F).
Our results shed new light on a possible involvement of human IDE in mitochondrial peptide clearance and preprotein maturation. Human IDE is mainly localized to the cytosol; however, a novel variant generated from an initiation codon upstream of the canonical start site that encodes a putative presequence has been identified, and IDE was proposed to cleave a targeting peptide in vitro (Leissring et al., 2004). Of note, although high-throughput proteomic studies detected IDE in mitochondrial fractions (Calvo et al., 2015), the in vivo role of mitochondrial IDE and the submitochondrial localization of the authentic protein had not been studied. Of interest, we find that a significant fraction of the IDE homologue Ste23 also localizes to the cytosol, like its human counterpart (Supplemental Figure S2C). Besides insulin and several further peptides, IDE can also degrade amyloid β peptides, and Leal et al. (2013) found that inhibition of IDE leads to increased levels of mitochondrial amyloid β peptides in vitro. It is therefore tempting to speculate that human IDE, like the yeast homologue Ste23, also localizes to the mitochondrial matrix, where it may contribute to the clearance of presequence peptides (and amyloid β peptides) and consequently ensure proper functioning of the presequence processing machinery. In plants, a novel protease (organellar oligopeptidase [OOP]) has been identified in chloroplasts and mitochondria of Arabidopsis thaliana that can degrade targeting peptides in cooperation with AtPreP (Kmiec et al., 2013). Unlike Ste23 and human IDE, which belong to the M16 clan of proteases (Alper et al., 2009), OOP is an M3 family protease. It will be interesting to see whether the plant OOP/PreP system might also be important for efficient preprotein maturation.
MATERIALS AND METHODS
Yeast strains and growth conditions
The Saccharomyces cerevisiae strains used in this study are listed in Supplemental Table S1. Deletion of the open reading frame was performed using the deletion cassettes pFa6a-kanMX6 and pFa6a-HIS3MX6 by homologous recombination (Sikorski and Hieter, 1989; Longtine et al., 1998). The integration of the deletion cassette was confirmed by PCR and Western blotting. Yeast cells were grown on YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] Bacto Peptone, 2% [wt/vol] glucose), YPS medium (containing 2% [wt/vol] sucrose instead of glucose), or YPG medium (containing 3% [wt/vol] glycerol instead of glucose) at 19, 23, 30, or 37°C. For overexpression of Ste23, wild-type and cym1∆ cells were transformed with pRS425Ste23 or the empty plasmid (pRS425ev). For overexpression of Cym1, the ste23∆ yeast strain was transformed with pRS425Cym1 or the empty plasmid (pRS425ev). Cells were grown on minimal medium (2% [wt/vol] glucose, 0.67% [wt/vol] yeast nitrogen base without amino acids, 0.07% [wt/vol] CSM-LEU dropout medium). For growth tests, yeast cells were grown on YPD medium at 30°C, spotted on YPD or YPG agar plates in serial dilutions, and incubated at various temperatures.
Isolation of mitochondria
Isolation of mitochondria from S. cerevisiae was performed according to standard protocols (Meisinger et al., 2006). Yeast cells grown on YPG, YPD, or YPS were harvested by centrifugation. To obtain spheroplasts, the yeast pellet was treated with dithiothreitol (DTT) buffer (10 mM DTT, 100 mM Tris-H2SO4, pH 9.4) and subsequently with Zymolyase buffer (1.2 M sorbitol, 20 mM K2HPO4-HCl, pH 7.4) containing 3 mg Zymolyase/g wet weight yeast cells. Spheroplasts were homogenized by 20 strokes with a glass-Teflon potter. After homogenization, cell debris and nuclei were removed by centrifugation at 1500 × g at 4°C. Mitochondria were isolated by differential centrifugation. The mitochondrial pellet was resuspended in SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS)–KOH, pH 7.2), and the concentration was determined by Bradford assay and adjusted to a concentration of 10 mg/ml. Isolated mitochondria were aliquoted, snap-frozen in liquid nitrogen, and stored at −80°C.
Cellular and submitochondrial localization of Ste23
For cellular fractionation, yeast cells were homogenized and centrifuged at 20,000 × g at 4°C. The supernatant was subjected to two additional centrifugation steps, followed by centrifugation at 100,000 × g for 1 h. The resulting supernatant (S100) contains mainly cytosolic proteins and the pellet (P100) mainly the microsomal fraction. The pellet was resuspended in EM buffer (1 mM EDTA, 10 mM MOPS-KOH, pH 7.2). Alkaline treatment of mitochondria was performed in 100 mM Na2CO3, pH 11.5. Swelling of mitochondria was performed in EM buffer. Lysis of mitochondria was performed in SEM buffer containing 1% (vol/vol) Triton X-100. All fractions were analyzed via SDS–PAGE and immunoblotting.
Generation of radiolabeled preproteins and in organello import
For generation of radiolabeled Sod2 precursors, the gene was amplified from yeast genomic DNA by PCR using forward primers with SP6 promoter sequences. For generation of [35S] Ste23, the open reading frame was cloned into the pRS416 plasmid. The change of A157G in the coding sequence was generated by site-directed mutagenesis, yielding the plasmid pRS416_Ste23M53A, which was used as template for PCR. Sod2 and Ste23 radiolabeled preproteins were synthesized by in vitro transcription/translation using the rabbit reticulate lysate system (Promega) in the presence of 35S-methionine. Import of [35S]Ste23 precursor into isolated mitochondria was performed in import buffer (3% [wt/vol] bovine serum albumin, 10 mM MOPS-KOH, pH 7.2, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 5 mM KH2PO4, 2 mM ATP, 2 mM NADH) and terminated by addition of 1% (vol/vol) AVO mix (8 μM antimycin A, 1 μM valinomycin, 20 μM oligomycin). Nonimported [35S]Ste23 was removed by PK treatment (50 µg/ml PK for 10 min on ice, followed by inhibition with 1 mM PMSF), and mitochondria were reisolated by centrifugation at 20,000 × g at 4°C. Mitochondrial samples were analyzed by SDS–PAGE and visualized by digital autoradiography.
Preprotein processing and peptide degradation assays in soluble mitochondrial extracts
For preprotein processing, isolated mitochondria were solubilized in reaction buffer (250 mM sucrose, 10 mM MOPS-KOH, pH 7.2, 80 mM KCl, 5 mM MgCl2, 5 mM KH2PO4) or 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (20 mM HEPES-KOH, pH 8.0, 10 mM MgCl2) containing 1% (vol/vol) digitonin for 15 min on ice and then centrifuged at 16,000 × g for 10 min at 4°C. For peptide degradation assays, mitochondria were subjected to sonication (five times 30 s with 30-s breaks on ice; Sonifier250, Branson) in reaction buffer, followed by centrifugation at 100,000 × g for 45 min at 4°C. The obtained supernatants after sonication or digitonin solubilization were incubated with different peptides (presequence amino acids 1–18 of yeast Sod2preseq MFAKTAAANLTKKGGLSL and presequence amino acids 1–17 of yeast Cox4preseq MLSLRQSIRFFKPATRT, with an additional cysteine residue at the C-terminus for generation of antiserum) or radiolabeled Sod2 precursor in the presence of 10 μM Cox4preseq peptides. Reactions were terminated by addition of 4× Laemmli buffer (8% [wt/vol] SDS, 0.08% [wt/vol] bromophenol blue, 40% [vol/vol] glycerol, 240 mM Tris-HCl, pH 6.8) containing 5% (vol/vol) β-mercaptoethanol and analyzed by SDS–PAGE, followed by autoradiography and immunodecoration. Degradation kinetics was established for each different metabolic condition and every precursor individually. For cell-free translation of Ste23WT and Ste23E121Q with a C-terminal hexahistidine tag (mutation generated by site-directed mutagenesis in the pRS416 vector), the RTS100 wheat germ system (5PRIME) was used.
Generation and affinity purification of antisera
For generation of antisera against the Ste23 protein and the presequences of Sod2 and Cox4, the following peptides were coupled to keyhole limpet hemocyanin via N- or C-terminal cysteine residues and rabbits immunized: Ste23 Cys-KDFEISAPPKLNNSSESE, amino acids 1–16 of Sod2 presequence (Sod2preseq), and amino acids 1–17 of Cox4 presequence (Cox4preseq). For affinity purification of antisera, corresponding peptides were reduced using Tris (2-carboxyethyl) phosphine and covalently immobilized to agarose beads (Sulfolink Coupling Resin). Peptide-coupled resin was washed, equilibrated, and incubated with protein-specific antiserum. The resin-bound antibodies were eluted with elution buffer containing 0.2 M glycine-HCl (pH 2.2), and eluted fractions were neutralized by 1 M Tris-HCl, pH 8.8. A complete list of antibodies used in this study is given in Supplemental Table S2.
Statistical analysis
Western blots were developed with the LAS 4000 system (Fujifilm). Quantified data are shown as mean ± SEM and were obtained from three independent experiments.
Supplementary Material
Acknowledgments
We thank E. Glaser, P. Teixeira, and B. Kmiec for discussion and B. Schönfisch for technical assistance. Work included in this study was performed in partial fulfillment of the requirements for the doctoral theses of A.A.T., C.K., and D.M. This work was supported by the Emmy Noether Programm of the Deutsche Forschungsgemeinschaft (F.N.V.), the Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS; GSC-4 Spemann Graduate School; C.M. and A.A.T.), the Baden-Württemberg-Stiftung (F.N.V.), and the Deutsche Forschungsgemeinschaft (Me1921 and GRK2202; C.M.).
Abbreviations used:
- IDE
insulin-degrading enzyme
- MPP
mitochondrial presequence protease
- PK
proteinase K
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-10-0732) on February 22, 2017.
REFERENCES
- Adames N, Blundell K, Ashby MN, Boone C. Role of yeast insulin-degrading homologs in pheromone processing and bud site selection. Science. 1995;270:464–467. doi: 10.1126/science.270.5235.464. [DOI] [PubMed] [Google Scholar]
- Alikhani N, Berglund AK, Engmann T, Spanning E, Vögtle FN, Pavlov P, Meisinger C, Langer T, Glaser E. Targeting capacity and conservation of PreP homologues localization in mitochondria of different species. J Mol Biol. 2011b;410:400–410. doi: 10.1016/j.jmb.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Alikhani N, Guo L, Yan S, Du H, Pinho CM, Chen JX, Glaser E, Yan SS. Decreased proteolytic activity of the mitochondrial amyloid-β degrading enzyme, PreP peptidasome, in Alzheimer’s disease brain mitochondria. J Alzheimers Dis. 2011a;27:75–87. doi: 10.3233/JAD-2011-101716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper BJ, Rowse JW, Schmidt WK. Yeast Ste23p shares functional similarities with mammalian insulin-degrading enzymes. Yeast. 2009;26:595–610. doi: 10.1002/yea.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart JM, Taskin AA, Zahedi RP, Vögtle FN. Quantitative profiling for substrates of the mitochondrial presequence processing protease reveals a set of nonsubstrate proteins increased upon proteotoxic stress. J Proteome Res. 2015;14:4550–4563. doi: 10.1021/acs.jproteome.5b00327. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2015;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K, Kawano S. Structural insight into the mitochondrial protein import system. Biochim Biophys Acta. 2011;1808:955–970. doi: 10.1016/j.bbamem.2010.07.018. [DOI] [PubMed] [Google Scholar]
- Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- Habib SJ, Neupert W, Rapaport D. Analysis and prediction of mitochondrial targeting signals. Methods Cell Biol. 2007;80:761–781. doi: 10.1016/S0091-679X(06)80035-X. [DOI] [PubMed] [Google Scholar]
- Hansson Petersen CA, Alikhani N, Behbahani H, Wiehager B, Pavlov PF, Alafuzoff I, Leinonen V, Ito A, Winblad B, Glaser E. The amyloid β-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc Natl Acad Sci USA. 2008;105:13145–13150. doi: 10.1073/pnas.0806192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C. The protein import machinery of mitochondria: a regulatory hub in metabolism, stress and disease. Cell Metab. 2014;19:357–372. doi: 10.1016/j.cmet.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Hawlitschek G, Schneider H, Schmidt B, Tropschug M, Hartl FU, Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988;53:795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- Kambacheld M, Augustin S, Tatsuta T, Müller S, Langer T. Role of the novel metallopeptidase MoP112 and Saccharolysin for the complete degradation of proteins residing in different subcompartments of mitochondria. J Biol Chem. 2005;280:20132–20139. doi: 10.1074/jbc.M500398200. [DOI] [PubMed] [Google Scholar]
- Kmiec B, Teixeira PF, Berntsson RP, Murcha MW, Branca RM, Radomiljac JD, Regberg J, Svensson LM, Bakali A, Langel U, et al. Organellar oligopeptidase (OOP) provides a complimentary pathway for targeting peptide degradation in mitochondria and chloroplasts. Proc Natl Acad Sci USA. 2013;110:E3761–E3769. doi: 10.1073/pnas.1307637110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec B, Teixeira PF, Glaser E. Shredding the signal: targeting peptide degradation in mitochondria and chloroplasts. Trends Plant Sci. 2014;19:771–778. doi: 10.1016/j.tplants.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Leal MC, Magnani N, Villordo S, Buslje CM, Evelson P, Castano EM, Morelli L. Transcriptional regulation of insulin-degrading enzyme modulates mitochondrial amyloid β (Aβ) peptide catabolism and functionality. J Biol Chem. 2013;288:12920–12931. doi: 10.1074/jbc.M112.424820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leissring MA, Farris W, Wu X, Christodoulou DC, Haigis MC, Guarente L, Selkoe DJ. Alternative translation initiation generates a novel isoform of insulin-degrading enzyme targeted to mitochondria. Biochem J. 2004;383:439–446. doi: 10.1042/BJ20041081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M, McKenzie A, Demarini D, Shah N, Wach A, Brachat A, Philippsen P, Pringle J. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Sickmann A, Pfanner N. The mitochondrial proteome: from inventory to function. Cell. 2008;134:22–24. doi: 10.1016/j.cell.2008.06.043. [DOI] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Small I. Recent surprises in protein targeting to mitochondria and plastids. Curr Opin Plant Biol. 2006;9:610–615. doi: 10.1016/j.pbi.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Mossmann D, Meisinger C, Vögtle FN. Processing of mitochondrial presequences. Biochim Biophys Acta. 2012;1819:1098–1106. doi: 10.1016/j.bbagrm.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Mossmann D, Vögtle FN, Taskin AA, Teixeira PF, Ring J, Burkhart JM, Burger N, Pinho CM, Tadic J, Loreth D, et al. Amyloid-β peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metab. 2014;20:662–669. doi: 10.1016/j.cmet.2014.07.024. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Yang CS, Wei B, Weiner H. Precursor protein is readily degraded in mitochondrial matrix space if the leader is not processed by mitochondrial processing peptidase. J Biol Chem. 2007;282:37266–37275. doi: 10.1074/jbc.M706594200. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Quiros PM, Langer T, Lopez-Otin C. New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- Schulz C, Schendzielorz A, Rehling P. Unlocking the presequence import pathway. Trends Cell Biol. 2015;25:265–275. doi: 10.1016/j.tcb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol AM, Sztolsztener ME, Wasilewski M, Heinz E, Chacinska A. Mitochondrial protein translocases for survival and wellbeing. FEBS Lett. 2014;588:2484–2495. doi: 10.1016/j.febslet.2014.05.028. [DOI] [PubMed] [Google Scholar]
- Taylor AB, Smith BS, Kitada S, Kojima K, Miyaura H, Otwinowski Z, Ito A, Deisenhofer J. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure. 2001;9:615–625. doi: 10.1016/s0969-2126(01)00621-9. [DOI] [PubMed] [Google Scholar]
- Teixeira PF, Glaser E. Processing peptidases in mitochondria and chloroplasts. Biochim Biophys Acta. 2013;1833:360–370. doi: 10.1016/j.bbamcr.2012.03.012. [DOI] [PubMed] [Google Scholar]
- van Wijk KJ. Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu Rev Plant Biol. 2015;66:75–111. doi: 10.1146/annurev-arplant-043014-115547. [DOI] [PubMed] [Google Scholar]
- Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- Witte C, Jensen RE, Yaffe MP, Schatz G. MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 1988;7:1439–1447. doi: 10.1002/j.1460-2075.1988.tb02961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MJ, Géli V, Oppliger W, Suda K, James P, Schatz G. The MAS-encoded processing protease of yeast mitochondria. Interaction of the purified enzyme with signal peptides and a purified precursor protein. J Biol Chem. 1991;266:6416–6423. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.