Abstract
Background
We assessed whether 234 established dyslipidemia-associated loci modify the effects of metformin treatment and lifestyle intervention (vs. placebo control) on lipid and lipid sub-fraction levels in the Diabetes Prevention Program (DPP) randomized controlled trial.
Methods and Results
We tested gene-treatment interactions in relation to baseline adjusted follow-up blood lipid concentrations (high and low density lipoprotein cholesterol [HDL-C, LDL-C], total cholesterol, triglycerides) and lipoprotein sub-fraction particle concentrations and size in 2,993 participants with pre-diabetes. Of the previously reported SNP associations, 32.5% replicated at P<0.05 with baseline lipid traits. Trait-specific genetic risk scores (GRS) were robustly associated (3×10−4>P>1.1×10−16) with their respective baseline traits for all but two traits. Lifestyle modified the effect of the GRS for large HDL particle numbers, such that each risk allele of the GRSHDL-large was associated with lower concentrations of large HDL particles at follow-up in the lifestyle arm (β=−0.11 μmol/l per GRS risk allele; 95%CI −0.188, −0.033; P=5×10−3; Pinteraction=1×10−3 for lifestyle vs. placebo), but not in the metformin or placebo arms (P>0.05). In the lifestyle arm, participants with high genetic risk had more favorable or similar trait levels at 1-yr compared to participants at lower genetic risk at baseline for 17 of the 20 traits.
Conclusions
Improvements in large HDL particle concentrations conferred by lifestyle may be diminished by genetic factors. Lifestyle intervention, however, was successful in offsetting unfavorable genetic loading for most lipid traits.
Keywords: genetic epidemiology; genetic polymorphism; lifestyle; intervention; genotype; genetics, human
Introduction
Dyslipidemia is a highly prevalent 1 and heritable 2 risk factor for coronary heart disease (CHD) 3, 4. The clinical diagnosis of dyslipidemia includes elevations in total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), and triacylglycerol (TG), and low levels of high-density lipoprotein cholesterol (HDL-C), in addition to other risk factors 5, 6. Not all of the lipid traits used in the diagnosis of dyslipidemia are causally related to CHD 7, 8, and their associations with CHD in observational studies may be attributable to underlying correlations with lipid and lipoprotein subfractions 9.
Although dyslipidemia has a strong heritable basis, in many patients it can be effectively managed through lifestyle modification 10 and/or a range of pharmacotherapies such as statins, bile acid sequestrants, niacin and fibrates 11. Of these treatment options, lifestyle modification, dietary changes, regular moderate intensity exercise, smoking cessation and weight reduction are the frontline therapy for the prevention and treatment of the condition 5, 6.
While lifestyle modification favorably impacts dyslipidemia at a population level, the individual-level response to such interventions is variable 12-14, which to some extent may be governed by a person’s genotype. Recent genome-wide association studies (GWAS) have identified more than 200 single nucleotide polymorphisms (SNPs) for lipids and lipoprotein sub-fraction concentrations 8, 15, 16 that underlie the relatively high heritability estimates (~50%) observed for these traits 17. The evaluation of these genetic variants in the context of lifestyle and drug intervention trials is an important part of the process of clinical translation, as it may identify genetic subgroups of the population that are more or less responsive to the lipid-modulating effects of diet, exercise and weight loss, potentially guiding targeted treatment decisions.
The overarching aim of this study was to examine whether comprehensive sets of lipid- and lipoprotein-associated genetic variants modulate the effects of lifestyle and metformin interventions on lipids and lipoproteins concentrations in pre-diabetic, overweight adults from the Diabetes Prevention Program (DPP). The specific aims of this study were to i) validate established genetic associations with lipid traits at baseline; ii) assess established genetic associations in relation to traits correlated to primary lipid traits; iii) assess genotype × treatment interactions in relation to baseline adjusted 1-yr lipid trait levels; iv) assess whether unfavorable genetic predisposition to dyslipidemia can be overcome by intensive lifestyle intervention.
Methods
Ethics Statement
Each participant provided written informed consent and institutional review board approval was obtained by each of the 27 DPP study centers before the study protocol was initiated.
Participants
The DPP is a multi-center randomized controlled trial of metformin or intensive lifestyle modification for diabetes prevention, as described in detail elsewhere 18, 19. Briefly, persons with elevated, non-diabetic fasting and post-load glucose concentrations and who were overweight or obese were randomized to one of three interventions (placebo, metformin [850 mg twice daily] or intensive lifestyle modification). The lifestyle arm included group-based and individual counseling sessions through which participants were encouraged to engage in ~150 min/wk of physical activity and a fat gram goal of 25% of calories from fat. If necessary, these interventions were followed by further caloric restriction in order to induce a weight loss of ~0.5-1kg/wk. The principal endpoint of the DPP was diabetes incidence, confirmed by a semiannual fasting plasma glucose or annual 75g oral glucose tolerance test (OGTT). Of those participants consented, 2,993 participants (placebo, lifestyle and metformin arms) had DNA available and were not taking lipid lowering medications at baseline. It is this subgroup that constitutes the sample for the current analyses. Due to incomplete measurements and sample exclusions from analyses, interaction analyses were comprised of smaller subgroups for all traits (ApoB [n=2,567]; TC [n=2,584]; TG [n=2,584]; LDL-C [n=2,584]; HDL-C [n=2,584]; IDL-C [n=1,710]; small LDL [n=1,714]; large LDL [n=1,632]; total LDL [n=1,714]; small HDL [n=1,713]; medium HDL [n=1,714]; large HDL [n=1,712]; total HDL [n=1,711]; small VLDL [n=1,708]; medium VLDL [n=1,707]; large VLDL [n=1,714]; total VLDL [n=1,714]; LDL size [n=2,585]; HDL size [n=1,714]; VLDL size [n=1,648]).
Measurements
Blood was drawn from an antecubital vein after an overnight fast (≥12 hrs). Measurements of TG, TC and HDL-C were made at the DPP central biochemistry laboratory using enzymatic methods standardized to the Centers for Disease Control and Prevention reference methods 20. HDL-C concentrations were obtained by precipitation of apolipoprotein B-containing lipoproteins by the dextran sulfate Mg2+ treatment 21. The Friedewald equation was used to calculate LDL-C 22. Where TG levels exceeded 4.5 mmol/l, the lipoprotein fractions were separated using preparative ultracentrifugation of plasma by β quantification 23. Nuclear magnetic resonance (NMR) spectroscopy (LipoScience Inc., Raleigh, NC) was used to quantify IDL-C and ApoB concentration, VLDL particle numbers (total and small, medium and large subfractions), LDL particle numbers (total and small and large subfractions) and HDL particle numbers (total and small, medium and large subfractions) as well as their average total particle sizes 24.
Genotyping
Standard methods were used to extract DNA from peripheral blood leukocytes. The DPP was genotyped using the MetaboChip genotyping array (Illumina Inc.) 25. From the MetaboChip array, we selected 71 TC associated, 37 TG associated, 68 HDL-C associated and 54 LDL-C associated SNPs (with overlaps, 150 individual SNPs for the four main lipid traits) 8, 16 and 91 lipoprotein subfraction associated SNPs 15 that had been identified through recent GWAS meta-analyses. All together, we extracted 234 SNPs from the MetaboChip array. To ensure quality control, study participants with failed genotyping (n=1), gender inconsistency (n=14), or cryptic familial relatedness (n=47) were excluded. From the 234 SNPs, none deviated from Hardy-Weinberg equilibrium (P<10−7) in any ethnic groups. The SNPs associated with the various lipoprotein traits are listed in S1 Table. Where the index SNPs were not available on the MetaboChip array (e.g. they had dropped out during the quality control stage) suitable HapMap proxies (r2>0.80) were identified and these variants were used in place of the index SNPs. The genotyping success rate for the 234 SNPs was 99.6%.
Statistical Analysis
Analyses were performed using STATA (version 13.1, StataCorp LP, TX, USA) and PLINK (v1.07) 26. We conducted two parallel sets of analyses. First, dependent variables were analyzed in their native distribution. Second, all analyses were performed with inverse normalized (mean=0, standard deviation=1) variables as outcomes. In the first case, effect sizes and SEs are reported in the outcome traits’ native unit. In the second case, effect sizes are reported in standard deviation units in order to facilitate comparisons across traits.
Pairwise Pearson correlations between traits were determined (S2 Table). As the four primary lipid traits strongly correlate with multiple subfractions, we hypothesized that some genetic variants identified for the primary lipid traits might also associate with lipoprotein subfractions. Thus, SNPs from the Global Lipids Genetics Consortium meta-analysis 8 were evaluated (for marginal and treatment interaction effects) for their respective standard lipid traits and any sub-fraction that was correlated |r|≥0.5 with the associated traits. Thus, guided by the results in S2 Table, TG-associated SNPs were also evaluated for association with LDL particle size, large VLDL, medium VLDL, VLDL particle size and total VLDL; TC- and LDL-C-associated SNPs were also evaluated for association with ApoB and total LDL; HDL-C-associated SNPs were also evaluated for association with large HDL, large LDL, small LDL, LDL particle size and total HDL. In addition, SNPs associated with lipoprotein subfractions 15 were evaluated for associations and treatment interactions with those respective traits. In analyses seeking to replicate the previously reported genetic association results 8, 15, 16 we used the baseline DPP data.
Additive genetic effects were assumed for each SNP, with a value of 0, 1 or 2 being assigned based on the number of minor allele copies. In these analyses, baseline traits were adjusted for age, age2, sex, and principal components for genetic markers of ancestry (to minimize confounding by population stratification). Individual SNP analyses that focused on that SNP’s primary lipid trait(s) at baseline (i.e., the trait for which it was established at a genome-wide level of significance to be associated with in published literature) were not corrected for multiple comparisons, as the prior probability for association is high in these cases given existing replication data. Bonferroni correction, however, was applied in cases where we investigated associations between correlated lipid traits, as described above.
To test whether the SNPs modified response to the DPP interventions, multiple linear regression was used to model the product of the SNP and the treatment condition (lifestyle vs. placebo and metformin vs. placebo) against the value of the lipid or lipoprotein trait measured 1 year after baseline (dependent variables). In the regression models, we fitted the 1-yr (follow-up) trait levels as dependent variables, the SNP × treatment interaction term as the independent variable, and SNP, treatment condition, the corresponding baseline trait, baseline age, baseline age2, sex and genetic principal components as covariates. As there was no difference in lipid medication use (P>0.05) by treatment arm at baseline or 1-yr follow-up, we did not adjust for lipid-lowering medication use. In total, we ran 1,101 interaction tests. As these gene × treatment interaction tests aim to test different biological associations than the regressions testing baseline associations, we corrected for multiple testing in this set of results. The Bonferroni corrected α type 1 error rate was set to 0.05/1,101=4.5×10−5.
Aggregated genetic risk was assessed by constructing trait-specific genetic risk scores (GRS). All SNPs previously associated in published GWAS for a given trait (Table S1) were used to create the respective trait’s GRS. GRSs were calculated in two ways; in the first instance, we assumed an equal magnitude of effect for each risk allele (unweighted GRS) by adding the number of risk alleles (0, 1 or 2) that a participant carried for each SNP associated with the trait of interest. In the second instance, we followed the same principle, but assigned weights to the allele counts based on published effect sizes reported by large-scale GWAS 8, 15 for each SNP and constructed a weighted GRS (wGRS). Regardless of the GRS approach used, and with the exception of HDL-associated SNPs, alleles at each SNP locus were designated ‘risk alleles’ if, within published meta-analyses, they were related with elevated concentrations of the respective lipid or lipoprotein subfractions. Risk alleles for HDL-associated SNPs were those associated with lower HDL-related trait concentrations in published meta-analyses 8, 15. In the event that, for a given participant, SNP data was missing (up to four SNPs of those required to construct a given GRS) and we were unable to replace it with an appropriate proxy variant, genotypes were imputed within each of the five DPP ethnic groups, as previously described 27. GRS and wGRS descriptives are shown in S3 Table. The GRSs were modeled as continuous independent variables in multiple regression analyses; dependent variables were the lipid or lipoprotein traits (at baseline or follow-up, depending on the model), and they were adjusted in the same way as the individual SNP analyses outlined above. In interaction analyses, the Bonferroni corrected α type 1 error rate was set to 0.05/34=0.0015. For figurative purposes, we dichotomized the GRSs based on their median values.
To assess the public health impact of lifestyle and metformin interventions across participants with low and high risk genotypes, we stratified the cohort by above and below the median GRS value and compared the groups’ phenotype levels for each trait at baseline and follow-up in the metformin and lifestyle arms separately. For these analyses, we used independent samples t-tests to determine the statistical significance of any differences between groups over time. Our purpose with these analyses was to determine whether the relevant genetic effects can be offset by metformin or lifestyle interventions.
As DPP is a multiethnic study, we further assessed potential confounding by population stratification by repeating all GRS analyses in the subgroup of self-reported white participants only (N=1,408, the largest ethnic group in the DPP) and compared effect estimates with the overall DPP results.
Detailed a priori power calculations and graphical illustrations are shown in S2 Text.
Functional annotation and pathway analysis
We assessed whether SNPs demonstrate liver-specific expression quantitative trait loci (eQTL) evidence using the The Genotype-Tissue Expression (GTEx) project database 28, as many of the lipids and lipoprotein subfractions studied here are synthesized in the liver. These SNPs were incorporated in eQTL GRSs in a trait specific fashion and the analyses described above were repeated using these GRSs.
We conducted detailed functional annotation of the 234 SNPs analyzed in this study using the ANNOVAR software tool 29. Pathway enrichment analysis for the 20 GRSs were performed using the REACTOME platform 30, 31. As these analyses are not the main scope of this project, we present these results in supplementary material.
Results
Thirty-two of the 234 SNPs included in the current analyses have been studied previously in the DPP 32. Participant characteristics for the DPP study population used in the current analyses are described elsewhere 32, as are the effects of the DPP interventions on 1-yr changes in the lipid and lipoprotein traits studied here 10.
Phenotypic variation explained by genetic factors
Table S4 reports the phenotypic variance explained by the GRSs and wGRSs (adjusted models). The average variance explained by the trait-specific GRSs was 1.7%. The trait-specific wGRSs explained on average 2.4% of the phenotypic variance of the traits. In further analyses, all GRSs (for 20 traits) cumulatively explained 5% of the phenotypic variance on average. All wGRSs explained 6% of the phenotypic variance on average (ranging from 2.7% for IDL-C to 10% for large VLDL particles).
Associations of SNPs with baseline lipid traits
Of the 150 SNPs tested for individual SNP associations with standard lipid traits, 71 were previously associated with TC, 37 with TG, 68 with HDL-C and 54 with LDL-C. As some SNPs were associated with multiple traits, a total of 230 replication analyses of these standard traits were performed. Fifty-nine (25.7%) of these associations replicated at the nominal α=0.05 level. Collectively, 113 SNPs have been previously associated 8, 15, 16 with the lipoprotein sub-fractions that are available in the DPP. For these lipoprotein sub-fractions, of 207 trait-specific associations and 673 associations based on highly correlated traits (in total, 880 association tests), 180 (20.5%) replicated at the nominal α=0.05 level, while 24 (2.7%) replicated at the Bonferroni adjusted level of P<5.7×10−5. S5 Table reports the association of each SNP with each of the baseline lipid and lipoprotein traits. In all, 227/1,110 (20.5%) of these association tests were statistically significant at a critical α=0.05, with 28 (2.5%) replicating at a Bonferroni adjusted level of P<4.5×10−5. Three SNPs previously only associated with the main lipid traits (TC, LDC-C or HDL-C), survived Bonferroni correction for a lipoprotein particle measure or ApoB. These are rs629301 for ApoB (β=0.05 g/l per copy of the risk allele; SE=0.008; P=4.3×10−12), rs3764261 for LDL particle size (β=−0.4 nm per copy of the risk allele; SE=0.08; P=1.8×10−6) and rs1532085 for large HDL (β=−0.43 μmol/l per copy of the risk allele; SE=0.08; P=3.7×10−7).
Associations of GRSs with baseline lipid traits
Table 1 reports all GRS/wGRS trait associations. In the majority of cases (32/34), these tests of association were statistically significant at baseline (P values ranging from 1.3×10−4 for total LDL to 2.4×10−16 for TC), with P>0.05 for tests of association for medium HDL and IDL-C with their respective GRSs. Repeating these models using the inverse normalized traits did not change the results (P values ranging from 1.1×10−4 for total LDL to 1.1×10−16 for TG, with associations for medium HDL and IDL-C P>0.05). Analyses conducted only in self-reported white DPP participants (to help reassure the absence of confounding by population stratification) yielded results that were largely consistent with those observed in the full DPP cohort. Using the wGRS strengthened the results for the majority of the traits (28/34 associations). The GRS was positively correlated with baseline concentrations of TG, TC, LDL-C, small, large and total LDL particle numbers, small, medium, large and total VLDL particle numbers, ApoB; LDL and VLDL particle sizes. The GRS was negatively correlated with IDL-C, HDL-C, HDL particle size, and small, medium, large and total HDL particle numbers.
Table 1.
Unweighted and weighted genetic risk scores - trait associations at baseline (Nmax=2,585)
| Trait (units) | GRS | N | β | SE | P | 95% CI LL |
95% CI UL |
PwGRS | β inv | SEinv | Pinv | PwGRSinv |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ApoB (g/l) | ApoB GRS | 2,567 | 0.014 | 0.002 | 1.9E-10 | 0.01 | 0.018 | 2.2E-25 | 0.058 | 0.009 | 8.0E-11 | 4.3E-26 |
| ApoB (g/l) | LDL-C GRS | 2,567 | 0.007 | 0.001 | 2.8E-11 | 0.005 | 0.009 | 1.4E-14 | 0.03 | 0.004 | 1.9E-11 | 4.0E-15 |
| ApoB (g/l) | TC GRS | 2,567 | 0.006 | 0.001 | 4.2E-12 | 0.005 | 0.008 | 1.6E-18 | 0.027 | 0.004 | 2.1E-12 | 6.5E-19 |
| HDL size (nm) |
HDL size GRS |
1,714 | −0.018 | 0.004 | 1.3E-05 | −0.026 | −0.01 | 5.7E-08 | −0.04 | 0.01 | 8.6E-05 | 6.4E-07 |
| HDL-C (mmol/l) |
HDL-C GRS | 2,584 | −0.007 | 0.001 | 1.9E-12 | −0.01 | −0.005 | 4.2E-31 | −0.027 | 0.003 | 1.6E-14 | 1.8E-36 |
| IDL-C (nmol/l) |
IDL-C GRS | 1,710 | −1.568 | 1.409 | 2.7E-01 | −4.332 | 1.197 | 4.6E-01 | −0.011 | 0.013 | 4.1E-01 | 4.4E-01 |
| large HDL (μmol/l) |
HDL-C GRS | 1,712 | −0.052 | 0.01 | 5.6E-07 | −0.073 | −0.032 | 6.6E-15 | −0.023 | 0.004 | 4.8E-08 | 4.4E-16 |
| large HDL (μmol/l) |
large HDL GRS |
1,712 | −0.151 | 0.026 | 5.7E-09 | −0.202 | −0.1 | 4.9E-13 | −0.06 | 0.01 | 6.2E-09 | 1.2E-12 |
| large LDL (nmol/l) |
HDL-C GRS | 1,632 | −4.903 | 1.152 | 2.2E-05 | −7.162 | −2.644 | 4.2E-12 | −0.021 | 0.005 | 7.2E-06 | 4.4E-13 |
| large LDL (nmol/l) |
large LDL GRS |
1,632 | 13.644 | 2.257 | 1.8E-09 | 9.217 | 18.072 | 8.8E-12 | 0.053 | 0.009 | 4.4E-09 | 6.0E-11 |
| large VLDL (nmol/l) |
large VLDL GRS |
1,714 | 0.353 | 0.088 | 6.9E-05 | 0.179 | 0.526 | 1.5E-04 | 0.049 | 0.012 | 2.4E-05 | 1.7E-05 |
| large VLDL (nmol/l) |
TG GRS | 1,714 | 0.261 | 0.045 | 9.3E-09 | 0.172 | 0.349 | 2.0E-17 | 0.035 | 0.006 | 3.5E-09 | 2.4E-18 |
| LDL size (nm) |
HDL-C GRS | 2,585 | −0.073 | 0.011 | 1.7E-11 | −0.094 | −0.052 | 1.7E-16 | −0.023 | 0.004 | 1.1E-10 | 9.6E-16 |
| LDL size (nm) |
LDL size GRS |
2,585 | 0.172 | 0.023 | 5.2E-14 | 0.127 | 0.216 | 2.8E-12 | 0.055 | 0.008 | 6.8E-13 | 1.2E-11 |
| LDL size (nm) |
TG GRS | 2,585 | −0.119 | 0.015 | 2.8E-15 | −0.149 | −0.09 | 5.3E-18 | −0.039 | 0.005 | 5.8E-15 | 7.1E-18 |
| LDL-C (mmol/l) |
LDL-C GRS | 2,584 | 0.027 | 0.004 | 1.4E-12 | 0.02 | 0.034 | 4.2E-18 | 0.032 | 0.004 | 7.7E-13 | 2.6E-18 |
| medium HDL (μmol/l) |
medium HDL GRS |
1,714 | −0.045 | 0.077 | 5.6E-01 | −0.196 | 0.105 | 4.0E-01 | −0.009 | 0.014 | 5.4E-01 | 4.0E-01 |
| medium VLDL (nmol/l) |
medium VLDL GRS |
1,707 | 1.113 | 0.236 | 2.5E-06 | 0.651 | 1.575 | 6.5E-03 | 0.056 | 0.012 | 1.3E-06 | 7.3E-03 |
| medium VLDL (nmol/l) |
TG GRS | 1,707 | 0.483 | 0.123 | 9.1E-05 | 0.241 | 0.725 | 1.1E-11 | 0.024 | 0.006 | 5.6E-05 | 5.0E-12 |
| small HDL (μmol/l) |
small HDL GRS |
1,713 | −0.366 | 0.056 | 6.2E-11 | −0.475 | −0.257 | 6.8E-19 | −0.076 | 0.012 | 7.9E-11 | 9.6E-19 |
| small LDL (nmol/l) |
HDL-C GRS | 1,714 | 8.772 | 1.793 | 1.1E-06 | 5.256 | 12.289 | 1.2E-11 | 0.022 | 0.004 | 6.3E-07 | 6.4E-12 |
| small LDL (nmol/l) |
small LDL GRS |
1,714 | 26.807 | 4.017 | 3.4E-11 | 18.929 | 34.685 | 1.2E-09 | 0.067 | 0.01 | 3.2E-11 | 1.3E-09 |
| small VLDL (nmol/l) |
small VLDL GRS |
1,708 | 0.83 | 0.182 | 5.6E-06 | 0.473 | 1.187 | 6.3E-08 | 0.045 | 0.01 | 1.6E-05 | 2.0E-07 |
| TC (mmol/l) |
TC GRS | 2,584 | 0.029 | 0.004 | 2.4E-16 | 0.022 | 0.036 | 2.2E-24 | 0.031 | 0.004 | 2.1E-16 | 2.2E-24 |
| TG (mmol/l) |
TG GRS | 2,584 | 0.043 | 0.005 | 2.5E-15 | 0.033 | 0.054 | 6.0E-24 | 0.041 | 0.005 | 1.1E-16 | 1.7E-26 |
| total HDL (μmol/l) |
HDL-C GRS | 1,711 | −0.051 | 0.027 | 5.7E-02 | −0.103 | 0.002 | 1.2E-04 | −0.01 | 0.004 | 2.8E-02 | 4.4E-05 |
| total HDL (μmol/l) |
total HDL GRS |
1,711 | −0.517 | 0.094 | 4.7E-08 | −0.702 | −0.332 | 1.6E-10 | −0.087 | 0.015 | 1.9E-08 | 7.0E-11 |
| total LDL (nmol/l) |
LDL-C GRS | 1,714 | 9.782 | 2.064 | 2.3E-06 | 5.734 | 13.829 | 5.0E-08 | 0.027 | 0.005 | 9.9E-07 | 3.0E-08 |
| total LDL (nmol/l) |
TC GRS | 1,714 | 8.442 | 1.786 | 2.5E-06 | 4.94 | 11.945 | 3.9E-09 | 0.023 | 0.005 | 1.1E-06 | 1.7E-09 |
| total LDL (nmol/l) |
total LDL GRS |
1,714 | 14.083 | 3.67 | 1.3E-04 | 6.884 | 21.281 | 3.1E-08 | 0.037 | 0.01 | 1.1E-04 | 2.9E-08 |
| total VLDL (nmol/l) |
TG GRS | 1,714 | 0.885 | 0.205 | 1.6E-05 | 0.483 | 1.286 | 3.8E-11 | 0.026 | 0.006 | 1.7E-05 | 5.4E-11 |
| total VLDL (nmol/l) |
total VLDL GRS |
1,714 | 2.467 | 0.362 | 1.3E-11 | 1.757 | 3.177 | 1.2E-08 | 0.072 | 0.011 | 1.1E-11 | 1.5E-08 |
| VLDL size (nm) |
TG GRS | 1,648 | 0.255 | 0.054 | 2.4E-06 | 0.149 | 0.361 | 1.4E-11 | 0.029 | 0.006 | 8.0E-06 | 5.8E-11 |
| VLDL size (nm) |
VLDL size GRS |
1,648 | 0.579 | 0.131 | 1.0E-05 | 0.323 | 0.835 | 9.5E-08 | 0.066 | 0.015 | 1.9E-05 | 1.6E-07 |
95% CI LL - 95% confidence interval lower limit; 95% CI UL - 95% confidence interval upper limit; ApoB - apolipoprotein B; β - beta effect estimate; βinv – beta effect estimate for inverse normalized traits; DPP - Diabetes Prevention Program; GRS - genetic risk score per GRS risk allele; HDL - high density lipoprotein; IDL - intermediate density lipoprotein; inv – inverse normalized traits; LDL - low density lipoprotein; N - sample size; Pinv – P value for inverse normalized traits; PwGRS – P value for weighted genetic risk score associations; PwGRSinv – P value for weighted genetic risk score associations undertaken with the inverse normalized traits; SE - standard error; SEinv – standard error for inverse normalized traits; TC - total cholesterol; TG - triglyceride; VLDL - very low density lipoprotein; wGRS - weighted genetic risk score.
P values are based on linear regression models. SNP associations were tested by fitting the genetic risk scores as the independent variables with the different lipoprotein subfractions at baseline as dependent variables. β coefficients reflect the association of one genetic risk score unit (effect allele) with the trait (expressed in native units and inverse normalized units).
Age, age2, sex and genomic principal components were used as covariates in all models.
Interactions between interventions and SNPs on 1-yr lipid traits
Results for all SNPs are shown in S6 and S7 Tables, for lifestyle and metformin interactions, respectively. One interaction test passed the Bonferroni corrected critical α level (α=0.05/1101=4.5×10−5). The rs581080 variant in tetratricopeptide repeat domain 39B (TTC39B) showed evidence for lifestyle treatment modification with large HDL particle numbers (Pinteraction=2.8×10−6 for lifestyle vs. placebo). The treatment interaction effect for this SNP was less statistically significant when assessed using the inverse normalized large HDL particle numbers variable (Pinteraction=1.7×10−4 for lifestyle vs. placebo). The interaction for rs581080 was no longer statistically significant when assessed only in European ancestry participants (Pinteraction=0.12 for lifestyle vs. placebo), which may reflect lower statistical power owing to the smaller sample size of this subcohort.
Interactions between interventions and GRSs on 1-yr lipid traits
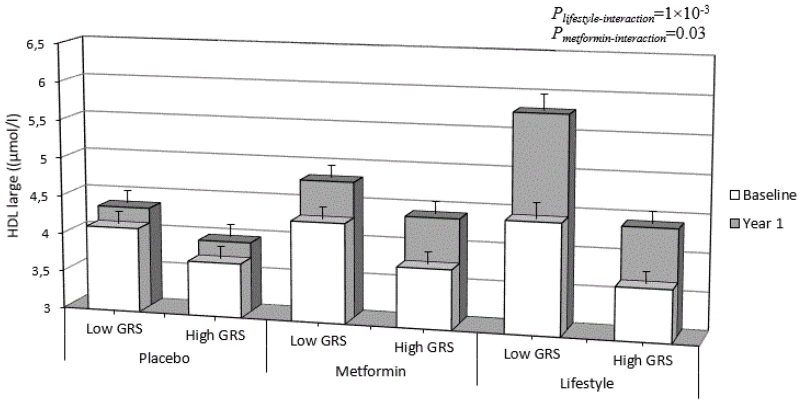
The lifestyle intervention modified the effect of the GRS for large HDL particle numbers, such that a higher GRSHDL large was associated with lower 1-year baseline-adjusted large HDL particle numbers in the lifestyle group (β=−0.11 μmol/l per GRS risk allele; 95%CI −0.188, −0.033; P=5×10−3; Pinteraction=1×10−3 for lifestyle vs. placebo), but not in the metformin group (β=−0.08 μmol/l per GRS risk allele; 95%CI −0.141, −0.008; P=0.027; Pinteraction=0.07 for metformin vs. placebo) or the placebo group (β=−0.02 μmol/l per GRS risk allele; 95%CI −0.086, 0.042; P=0.50) (Fig. 1.). Using the wGRS attenuated this result, such that the interaction between lifestyle intervention and GRSHDL large on large HDL particle number (Pinteraction=6×10−3 for lifestyle vs. placebo) became nominally statistically significant. Repeating the analyses with inverse normalized large HDL particle number did not materially change the results (Pinteraction=5×10−3). The exclusion of those individuals initiated on lipid lowering medication (n=226) between baseline and follow-up did not materially impact the results (Pinteraction=6×10−3). GRS results for large HDL particle numbers per treatment arm are shown in Table 2, while all GRS and wGRS × lifestyle and metformin interactions are shown in S8 Table and S9 Table, respectively. Repeating analyses only in European ancestry DPP participants (Nmax=1,408) attenuated the statistical significance of the interactions observed in all participants, although the pattern of the interaction effects remained the same for large HDL (β=−0.16 μmol/l per GRS risk allele; 95%CI −0.283, −0.047; P=6×10−3; Pinteraction=0.054 for lifestyle vs. placebo).
Figure 1.
Large HDL particle numbers at baseline and 1-year later stratified by treatment group and high and low levels of the trait-specific genetic risk score (GRS). GRS by treatment interactions are shown for each active treatment group compared with the placebo group. Error bars represent standard deviations of the means.
Table 2.
Statistically significant GRS interactions with baseline adjusted 1-yr traits (Nmax=1,377)
| Trait | GRS | Intervention arm |
N | β | SE | P | 95% CI LL | 95% CI UL | Pinteraction |
|---|---|---|---|---|---|---|---|---|---|
| large HDL (μmol/l) |
large HDL GRS | Placebo | 463 | −0.022 | 0.032 | 0.499 | −0.086 | 0.042 | - |
| large HDL (μmol/l) |
large HDL GRS | Metformin | 461 | −0.075 | 0.034 | 0.027 | −0.141 | −0.008 | 0.07 |
| large HDL (μmol/l) |
large HDL GRS | Lifestyle | 450 | −0.111 | 0.039 | 5×10−3 | −0.188 | −0.033 | 1×10−3 |
95% CI LL - 95% confidence interval lower limit; 95% CI UL - 95% confidence interval upper limit; β - beta effect estimate per GRS risk allele; DPP - Diabetes Prevention Program; HDL - high density lipoprotein; GRS - genetic risk score; N - sample size; SE - standard error.
P values are based on linear regression models. GRS associations were modeled by fitting the GRSs as the independent variables with the different lipoprotein subfractions as dependent variables.
Pinteraction values are based on linear regression models. GRS associations were tested by fitting the GRS × lifestyle vs. placebo intervention and GRS × metformin vs. placebo intervention interaction terms as the independent variables with the different lipoprotein subfractions at follow-up as dependent variables. β coefficients reflect the association of one genetic risk score unit (effect allele) with the trait (expressed in native units and inverse normalized units).
Age, age2, sex, baseline lipoprotein subfraction values and genomic principal components were used as covariates in all models.
Lipid profile change from baseline to 1-yr
In the lifestyle arm, participants at higher genetic risk (GRS above median) had more favorable (P<0.05) or similar (P>0.05) trait levels at 1-yr than participants with lower genetic risk (GRS below median) at baseline (see Fig. 1) for all traits, except for large LDL, small VLDL particle numbers and LDL size (3 out of 20 traits). In the metformin arm, participants at higher genetic risk had more favorable trait levels at 1-yr than participants at lower genetic risk at baseline for TG, LDL-C, HDL-C, IDL-C, ApoB, small, medium, large and total HDL, small and total LDL, medium and large VLDL particle numbers and HDL size. No difference was observed for TC, large LDL, small and total VLDL particle number, nor LDL or VLDL size.
Functional annotation and pathway analysis
Two SNPs for TC (rs10893499, rs4530754), LDL-C (rs10893499, rs4530754) and total VLDL (rs10889353, rs646776) demonstrated liver-specific eQTL evidence in GTEx, therefore we repeated our interaction analyses with three trait specific eQTL GRSs comprised of these SNPs for TC, LDL-C and total VLDL, respectively. Although all three GRSs demonstrated nominal statistical significance (β=−0.268 mmol/l per GRS risk allele; 95%CI −0.530, −0.005; Pinteraction=0.050 for lifestyle vs. placebo for LDL-C; β=−0.341 mmol/l per GRS risk allele; 95%CI −0.644, −0.037; Pinteraction=0.028 for lifestyle vs. placebo for TC; β=4.120 nmol/l per GRS risk allele; 95%CI 0.539, 7.700; Pinteraction=0.024 for lifestyle vs. placebo for total VLDL), none of these associations remained significant after correction for multiple testing.
Detailed functional annotation of all SNPs is shown in Table S10, while results from trait-specific pathway enrichment analyses are shown in Table S11.
Discussion
This is to our knowledge the most comprehensive assessment to date of established lipid- and lipoprotein-associated loci in the context of human diabetes prevention interventions. It is also the first study to our knowledge to examine the effects of these loci on changes in lipid and lipoprotein concentrations over time.
The major finding of this is study is that genetic predisposition to a higher large HDL particle number modifies the response to lifestyle intervention. Although in GRS analyses, lifestyle intervention robustly increased the number of large HDL particles at 1 year DPP participants, the intervention was less effective in participants at higher genetic risk. The participants at higher genetic risk also had fewer large HDL particles at baseline than those at lower genetic risk. Nevertheless, lifestyle intervention generally improved lipoprotein values in people at higher genetic risk to a level that was similar or more favorable than observed in participants with lower genetic burden assigned to the control arm (17 out of the 20 traits), suggesting that lifestyle intervention can overcome genetic risk for dyslipidemia. Of note, analyzing treatment interactions with GRSs constructed exclusively from SNPs demonstrating eQTL evidence in the liver did not yield clinically relevant results.
In SNP analyses, one SNP (TTC39B rs581080) × lifestyle interaction passed the predefined conservative threshold for multiple test-corrected statistical significance for large HDL particle numbers; no such interaction with metformin was observed. However we believe this interaction with lifestyle to be spurious, as the interaction is driven by differences in the genetic effect on large HDL particle numbers by treatment arm prior to randomization, and not by the joint effect of the interventions and genotypes (which was apparent when the data were visualized).
The minor ‘C’ allele of the TTC39B rs581080 variant was originally associated with lower HDL-C and TC concentrations, and in vivo knockdown of its mouse homolog correlates with higher HDL-C concentrations 16, 33, 34. The function of the TTC39B gene in humans is presently unknown. No human studies of gene-lifestyle interaction for this locus have been reported to our knowledge.
Lifestyle modification is the frontline therapy to combat dyslipidemia; our data help understand better why some people are more responsive than others to lifestyle interventions. In addition, lifestyle and other therapies that target specific lipoprotein subfractions might be clinically more relevant than only modifying the major fractions, such as LDL-C, HDL-C or TG levels 35. This is support by data showing that particle numbers, lipoprotein associated protein levels (such as ApoA1 or ApoB) and their relative amounts predict cardiovascular risk and other hard clinical outcomes with higher accuracy than the major lipids 36, 37.
Metformin treatment, unlike lifestyle intervention, appears to act independently on changes in VLDL, LDL and HDL, suggesting that the two interventions influence these traits through different mechanisms. In support of this we found that the GRS-intervention interactions were only apparent for lifestyle and not for metformin. All of these changes are thought to favorably impact CVD risk. For example, pharmacologically increased small HDL particle numbers (with fibrates) reduces CVD risk in some studies 38.
In previous analyses within the DPP, we observed interactions between a GRS and lifestyle intervention for LDL-C and small LDL particle numbers 32. A key distinction between those analyses and the ones reported here is that the GRS used in the former analysis was not trait-specific, but included a set of 32 SNPs with heterogeneous roles in lipid biology, whereas the GRSs studied here were fitted to the specific lipid traits. Elsewhere in the DPP, Goldberg et al examined the lipid and lipoprotein traits examined here for their relationships with various cardiometabolic outcomes 10. Compared to placebo intervention, the DPP lifestyle intervention lowered VLDL particle numbers, especially large VLDL particles, which are prominent in diabetic dyslipidemia, and VLDL particle size. Possibly as a consequence of the DPP lifestyle intervention’s effects on VLDL, the intervention also lowered LDL particle numbers, especially for small LDL particles, increased average LDL particle size (which associate with fasting insulin, hepatic lipase and CETP concentrations), and increased large HDL particle numbers by ~1 μmol/l and size by ~1.5 nm. By contrast, metformin did not affect VLDL particle numbers or size in the DPP. Metformin did however lower LDL sub-fraction concentrations and increased small and total HDL particle numbers. Despite the robust and wide-ranging effects of the DPP lifestyle and metformin interventions on lipoprotein subfractions reported by Goldberg et al, only one of these traits (large HDL particle numbers) appears to be influenced by gene × treatment interactions in the current analyses. Although recent evidence suggests that HDL-C is not causal in the development of cardiovascular disease 39, 40, the findings of this analysis might represent underlying causal effects of HDL-C or its correlates through gene × environment interactions.
The major strength of this analysis is that it was conducted in a tightly controlled randomized clinical trial, which limits the extent to which confounding, reverse causality and some other sources of bias are likely to underlie our findings. As the DPP is a multiethnic trial, we dealt with potential confounding by population stratification using genomic control and ethnic-specific quality control. We also conducted subgroup analyses in self-reported white participants, but we did not observe major differences between these set of results and the ones we obtained from analyzing the whole study. Although DPP is one of the largest clinical trials investigating the effects of metformin and lifestyle, our a priori power calculations indicate that some of our apparently negative findings are likely to be false negatives owing to insufficient statistical power to detect small interaction effects. However, the objective of this study was to determine if established dyslipidemia-associated loci are likely to be of clinical relevance, and the small effects that this study is unpowered to detect are unlikely to be clinically useful.
We have replicated the effects of genetic variants previously associated with lipid and lipoprotein sub-fraction traits. We provide evidence that the deleterious effects of some established lipid- and lipid sub-fraction-associated loci modify the effects of intensive lifestyle interventions. Specifically, individuals genetically predisposed to low large HDL particle concentrations are less responsive to the ability of these interventions to increase these levels. Nonetheless, participants at higher genetic risk assigned to lifestyle intervention had comparable lipid profiles at 1-year post-randomization to those at lower genetic risk at baseline, indicating that these interventions are of value to individuals with high-risk genetic profiles. While this study provides some evidence of gene-lifestyle interactions at a few loci and for specific lipid traits, most tests yielded no compelling evidence of gene-lifestyle interactions, indicating that most GWAS-derived loci do not affect response to lifestyle interventions to a clinically relevant degree.
Supplementary Material
Clinical Perspective.
In our study in the Diabetes Prevention Program randomized clinical trial we aimed to detect gene environment interactions of known lipid and lipoprotein subfraction loci (individually and amalgamated in genetic risk scores) and metformin/lifestyle intervention vs. the placebo arm. We detected statistically significant interactions between the genetic risk score of large HDL particle concentrations and the lifestyle arm (vs. placebo) in relation to large HDL particle concentrations. Those at higher genetic risk fewer large HDL particles at baseline than those at lower genetic risk and lifestyle intervention elevated the number of large HDL particles at 1 year, but the intervention was less effective in people at higher genetic risk. The clinical relevance of our study is that participants at higher genetic risk assigned to lifestyle intervention had comparable lipid profiles for most traits, (including large HDL particle concentrations and HDL size) at 1-year post-randomization to those at lower genetic risk who had been assigned to the placebo-control intervention, indicating that these interventions are of value to individuals with high-risk genetic profiles.
Acknowledgments
PWF, TVV and TIP conceived and designed the research, ESH, JCF, KEW, KAJ, PWF, RG, TIP and WCK acquired the data, AHW and TVV performed the statistical analysis, ESH, FR, JCF, KEW, KAJ, PWF, RG, TIP and WCK handled funding and supervision, PWF, TVV and TIP drafted the manuscript. All authors made critical revision of the manuscript for key intellectual content. The guarantors of this article are PWF and TIP. We thank Angela Estampador for undertaking the functional annotation for this project. We thank Kathy Ryan and Tushar Dave for their help with additional analyses.
Sources of Funding: The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health provided funding to the clinical centers and the Coordinating Center for the design and conduct of the study, and collection, management, analysis, and interpretation of the data. The Southwestern American Indian Centers were supported directly by the NIDDK and the Indian Health Service. The General Clinical Research Center Program, National Center for Research Resources, supported data collection at many of the clinical centers. Funding for data collection and participant support was also provided by the Office of Research on Minority Health, the National Institute of Child Health and Human Development, the National Institute on Aging, the Centers for Disease Control and Prevention, the Office of Research on Women’s Health, and the American Diabetes Association. Bristol-Myers Squibb and Parke-Davis provided medication. This research was also supported, in part, by the intramural research program of the NIDDK. LifeScan, Health O Meter, Hoechst Marion Roussel, Merck-Medco Managed Care, Merck, Nike Sports Marketing, Slim Fast Foods, and Quaker Oats donated materials, equipment, or medicines for concomitant conditions. McKesson BioServices, Matthews Media Group, and the Henry M. Jackson Foundation provided support services under subcontract with the Coordinating Center. The opinions expressed are those of the investigators and do not necessarily reflect the views of the Indian Health Service or other funding agencies. A complete list of centers, investigators, and staff is shown in the Acknowledgments. The DPP Genetics Project was supported by NIH/NIDDK grant R01DK072041; additional funding from the Swedish Heart-Lung Foundation supported the costs of data analysis (for TVV). Part of the work described in this manuscript was undertaken as part of the 2013-2014 BLUE ScY educational exchange program, which was supported by the Faculty of Medicine at Umeå University and EPiHealth (Lund University) (for TVV).
Footnotes
Clinical Trial Registration - https://clinicaltrials.gov/; Unique Identifier: NCT00004992.
Disclosures: PWF has been a paid consultant for Eli Lilly and Sanofi Aventis and has received research support from several pharmaceutical companies as part of a European Union Innovative Medicines Initiative (IMI) project.
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, et al. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaess B, Fischer M, Baessler A, Stark K, Huber F, Kremer W, et al. The lipoprotein subfraction profile: heritability and identification of quantitative trait loci. J Lipid Res. 2008;49:715–723. doi: 10.1194/jlr.M700338-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Otvos JD, Collins D, Freedman DS, Shalaurova I, Schaefer EJ, McNamara JR, et al. Low-density lipoprotein and high-density lipoprotein particle subclasses predict coronary events and are favorably changed by gemfibrozil therapy in the Veterans Affairs High-Density Lipoprotein Intervention Trial. Circulation. 2006;113:1556–1563. doi: 10.1161/CIRCULATIONAHA.105.565135. [DOI] [PubMed] [Google Scholar]
- 4.Rosenson RS, Otvos JD, Freedman DS. Relations of lipoprotein subclass levels and low-density lipoprotein size to progression of coronary artery disease in the Pravastatin Limitation of Atherosclerosis in the Coronary Arteries (PLAC-I) trial. Am J Cardiol. 2002;90:89–94. doi: 10.1016/s0002-9149(02)02427-x. [DOI] [PubMed] [Google Scholar]
- 5.Genest J, McPherson R, Frohlich J, Anderson T, Campbell N, Carpentier A, et al. 2009 Canadian Cardiovascular Society/Canadian guidelines for the diagnosis and treatment of dyslipidemia and prevention of cardiovascular disease in the adult - 2009 recommendations. Can J Cardiol. 2009;25:567–579. doi: 10.1016/s0828-282x(09)70715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson TJ, Gregoire J, Hegele RA, Couture P, Mancini GB, McPherson R, et al. 2012 update of the Canadian Cardiovascular Society guidelines for the diagnosis and treatment of dyslipidemia for the prevention of cardiovascular disease in the adult. Can J Cardiol. 2013;29:151–167. doi: 10.1016/j.cjca.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 7.Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krauss RM. Lipoprotein subfractions and cardiovascular disease risk. Curr Opin Lipidol. 2010;21:305–311. doi: 10.1097/MOL.0b013e32833b7756. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg R, Temprosa M, Otvos J, Brunzell J, Marcovina S, Mather K, et al. Lifestyle and metformin treatment favorably influence lipoprotein subfraction distribution in the Diabetes Prevention Program. J Endocrinol Metab. 2013;98:3989–3998. doi: 10.1210/jc.2013-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundy SM, Cleeman JI, Merz CN, Brewer HB, Jr., Clark LT, Hunninghake DB, et al. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. Circulation. 2004;110:227–239. doi: 10.1161/01.CIR.0000133317.49796.0E. [DOI] [PubMed] [Google Scholar]
- 12.Leon AS, Gaskill SE, Rice T, Bergeron J, Gagnon J, Rao DC, et al. Variability in the response of HDL cholesterol to exercise training in the HERITAGE Family Study. Int J Sports Med. 2002;23:1–9. doi: 10.1055/s-2002-19270. [DOI] [PubMed] [Google Scholar]
- 13.Cox C, Mann J, Sutherland W, Ball M. Individual variation in plasma cholesterol response to dietary saturated fat. Br Med J. 1995;311:1260–1264. doi: 10.1136/bmj.311.7015.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefevre M, Champagne CM, Tulley RT, Rood JC, Most MM. Individual variability in cardiovascular disease risk factor responses to low-fat and low-saturated-fat diets in men: body mass index, adiposity, and insulin resistance predict changes in LDL cholesterol. Am J Clin Nutr. 2005;82:957–963. doi: 10.1093/ajcn/82.5.957. quiz 1145-1146. [DOI] [PubMed] [Google Scholar]
- 15.Chasman DI, Pare G, Mora S, Hopewell JC, Peloso G, Clarke R, et al. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goode EL, Cherny SS, Christian JC, Jarvik GP, de Andrade M. Heritability of longitudinal measures of body mass index and lipid and lipoprotein levels in aging twins. Twin Res Hum Genet. 2007;10:703–711. doi: 10.1375/twin.10.5.703. [DOI] [PubMed] [Google Scholar]
- 18.The Diabetes Prevention Program Design and methods for a clinical trial in the prevention of type 2 diabetes. Diabetes Care. 1999;22:623–634. doi: 10.2337/diacare.22.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warnick GR. Enzymatic methods for quantification of lipoprotein lipids. Methods Enzymol. 1986;129:101–123. doi: 10.1016/0076-6879(86)29064-3. [DOI] [PubMed] [Google Scholar]
- 21.Warnick GR, Benderson J, Albers JJ. Dextran sulfate-Mg2+ precipitation procedure for quantitation of high-density-lipoprotein cholesterol. Clin Chem. 1982;28:1379–1388. [PubMed] [Google Scholar]
- 22.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 23.Hainline A Jr, Karon J, Lippel K, editors. Manual of laboratory operations: Lipid and lipoprotein analysis. 2nd ed. National Heart, Lung and Blood Institute, Lipid Research Clinics Program; Bethesda, MD: HEW Pub. No. (NIH) 75-628 (rev.), U.S. Government Printing Office Publication No. 1982-361-132:678. [Google Scholar]
- 24.Otvos JD. Measurement of lipoprotein subclass profiles by nuclear magnetic resonance spectroscopy. Clin Lab. 2002;48:171–180. [PubMed] [Google Scholar]
- 25.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fontaine-Bisson B, Renstrom F, Rolandsson O, Magic Payne F, Hallmans G, et al. Evaluating the discriminative power of multi-trait genetic risk scores for type 2 diabetes in a northern Swedish population. Diabetologia. 2010;53:2155–2162. doi: 10.1007/s00125-010-1792-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44:D481–487. doi: 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollin TI, Isakova T, Jablonski KA, de Bakker PI, Taylor A, McAteer J, et al. Genetic modulation of lipid profiles following lifestyle modification or metformin treatment: the Diabetes Prevention Program. PLoS Genet. 2012;8:e1002895. doi: 10.1371/journal.pgen.1002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holmes MV, Harrison S, Talmud PJ, Hingorani AD, Humphries SE. Utility of genetic determinants of lipids and cardiovascular events in assessing risk. Nat Rev Cardiol. 2011;8:207–221. doi: 10.1038/nrcardio.2011.6. [DOI] [PubMed] [Google Scholar]
- 34.Waterworth DM, Ricketts SL, Song K, Chen L, Zhao JH, Ripatti S, et al. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol. 2010;30:2264–2276. doi: 10.1161/ATVBAHA.109.201020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikolic D, Katsiki N, Montalto G, Isenovic ER, Mikhailidis DP, Rizzo M. Lipoprotein subfractions in metabolic syndrome and obesity: clinical significance and therapeutic approaches. Nutrients. 2013;5:928–948. doi: 10.3390/nu5030928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sniderman AD, Kiss RS. The strengths and limitations of the apoB/apoA-I ratio to predict the risk of vascular disease: a Hegelian analysis. Curr Atheroscler Rep. 2007;9:261–265. doi: 10.1007/s11883-007-0031-6. [DOI] [PubMed] [Google Scholar]
- 37.Fizelova M, Miilunpohja M, Kangas AJ, Soininen P, Kuusisto J, Ala-Korpela M, et al. Associations of multiple lipoprotein and apolipoprotein measures with worsening of glycemia and incident type 2 diabetes in 6607 non-diabetic Finnish men. Atherosclerosis. 2015;240:272–277. doi: 10.1016/j.atherosclerosis.2015.03.034. [DOI] [PubMed] [Google Scholar]
- 38.Barter PJ, Rye KA. Cardioprotective properties of fibrates: which fibrate, which patients, what mechanism? Circulation. 2006;113:1553–1555. doi: 10.1161/CIRCULATIONAHA.105.620450. [DOI] [PubMed] [Google Scholar]
- 39.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, et al. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.