Abstract
The formation of C-C bonds in an enantioselective fashion to create complex polycyclic scaffolds in the hapalindole/fischerindole type alkaloids from Stigonematales cyanobacteria represents a compelling and urgent challenge in adapting microbial biosynthesis as a catalytic platform in drug development. Here we determine the biochemical basis for tri- and tetracyclic core formation in these secondary metabolites, involving a new class of cyclases that catalyze a complex cyclization cascade.
Carbon-carbon bond construction is the backbone of organic chemistry and encompasses some of the most challenging reactions in synthetic methodology.1 Enzymes that control enantioselectivity during C-C bond formation are of great interest because of their potential to catalyze difficult transformations under mild, environmentally sustainable conditions.2 While biocatalysts have been investigated extensively during the past few decades, those responsible for complex cyclization reactions remain underexplored,3–5 despite some notable progress in terpenoid and polyketide systems,6, 7 and several examples of Diels-Alderases.8–10 Here, we report a new class of Stigonematales (Stig) cyclases that catalyze a highly stereoselective intramolecular ring formation, creating four new chiral centers via a Cope rearrangement and C-C bond forming cascade, the mechanism of which was recently described.11
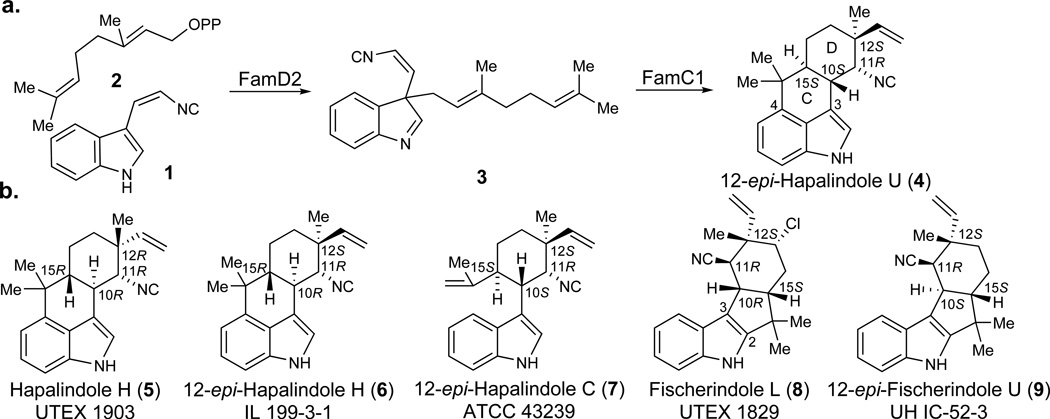
Hapalindoles and the related ambiguines, fischerindoles, and welwitindolinones are bioactive indole alkaloids from cyanobacteria that have attracted substantial attention because of their unique pharmacological profiles and complex chiral structures.12, 13 The most notable structural feature of this family is their stereo- and regiochemically diverse polycyclic ring system (Figure 1). We recently disclosed a novel pathway intermediate (3, Figure 1a), which completely revised the prevailing hypotheses about the formation of this complex heterocyclic core.11 Instead of a cationic catalysis that directly forms hapalindoles (Figure 1b, 4–7) from cis-indole isonitrile (1) and geranylpyrophosphate (GPP, 2), the action of an aromatic prenyltransferase (FamD2) appends 2 onto the C3 position of 1, and the resulting product (3) is subsequently processed into a hapalindole (4) via a Cope rearrangement and intramolecular ring cyclization catalyzed by FamC1.11 However, these initial results prompted two intriguing questions: what explains the regioselective C-ring divergence between hapalindoles and fischerindoles; and what sets the chirality of the C-10, C-11, C-12, and C-15 stereocenters?
Figure 1. Select hapalindole -type alkaloids.
a) Biosynthetic pathway leading to 12-epi- hapalindole U;11 b) representative indole alkaloids from related cyanobacterial metabolic pathways illustrating the variant polycyclic core systems.14–18
We originally identified Stig cyclase FamC1 through cell-free lysate fractionation and demonstrated its direct role in formation of 12-epi-hapalindole U (4).11 However, in vitro assays with purified protein were hampered by the apparent inability to heterologously express FamC1 in Escherichia coli. Through protein sequence analysis, an N-terminal 23 amino acid region was predicted to be a transmembrane segment, which we reasoned led to the failed expression. Mass spectrometry analysis of the isolated functional enzyme showed a mass that exactly matched the protein lacking the predicted transmembrane sequence (Supplementary Results, Supplementary Fig. 1). This result indicated that a post-translational modification is required to form the mature cyclase. Thus, by cloning the corresponding truncated gene, we successfully overexpressed FamC1 in E. coli as a soluble protein and confirmed its ability to convert intermediate 3 into 4 in vitro (Figure 1a).
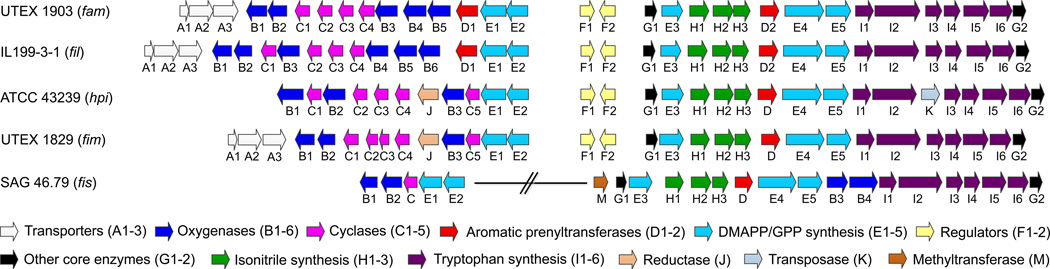
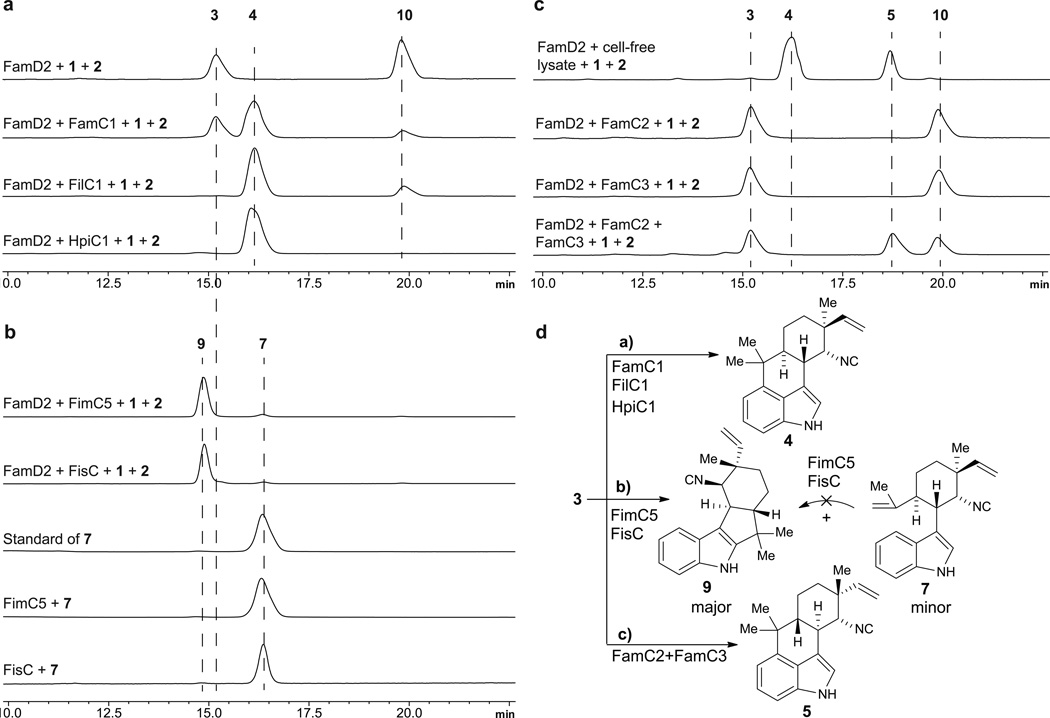
12-epi-Hapalindole U is an unexpected new metabolite bearing a pattern of stereocenters (C-10S/11R/12S/15S) that had not been reported from UTEX 1903 cyanobacterium, suggesting that our knowledge of the full spectrum of natural products generated by this strain is likely incomplete. This discovery inspired us to search for this metabolite in other biosynthetic pathways based on previously characterized compounds and the presence of FamC1 homologs. Thus, genome sequencing and bioinformatic analysis of hapalindole- producing cyanobacteria led us to identify several gene clusters (Figure 2), including the fil gene cluster from Fischerella sp. IL-199-3-1 (ref. 15) and the hpi gene cluster from Fischerella sp. ATCC 4323919. Protein sequence analysis of the nine annotated Stig cyclases (FilC1–FilC4 and HpiC1–HpiC5) against FamC1 resulted in identification of two FamC1 homologs, FilC1 (94% identity) from IL 199-3-1, and HpiC1 (85% identity) from ATCC 43239 (Supplementary Fig. 2). To verify our hypothesis that FilC1 and HpiC1 catalyze formation of the same product as FamC1, we overexpressed and purified these two proteins from E. coli using the strategy of transmembrane-segment truncation to achieve in vitro activity. Both of these FamC1 homologs exhibited the ability to cyclize 3 to 4 (Figure 3a; see Supplementary Fig. 3 for the structure of 10), indicating that proteins of this subclass are responsible for catalyzing formation of a hapalindole-type tetracyclic core containing the C-10S/11R/12S/15S stereocenter motif.
Figure 2. Gene clusters identified from hapalindole producing strains.
fam gene cluster from ambiguine producing strain Fischerella ambigua UTEX 1903;11 fil gene cluster from ambiguine producing strain Fischerella sp. IL-199-3-1 (this study); hpi gene cluster from hapalindole producing strain Fischerella sp. ATCC 4323919 (this nomenclature has been revised for consistency with other newly annotated gene clusters); fim gene cluster from fischerindole producing strain Fischerella muscicola UTEX 1829 (this study); fis gene cluster from fischerindole producing strain Fischerella sp. SAG 46.79 (this study). Full annotation of these gene clusters is in Supplementary Tables 2–5.
Figure 3. In vitro assay involving recombinant cyclases to form hapalindole and fischerindole natural products.
Assays were conducted in a one-pot enzymatic tandem reaction by incubating substrates indole isonitrile (1) and GPP (2) with FamD2 and the selected cyclases. a) Proteins producing 12-epi-hapalindole U (4); b) protein producing 12-epi-fischerindole U (9) and 12-epi-hapalindole C (7); c) proteins producing hapalindole H (5); cell-free lysate is from UTEX 1903 strain11; d) summary of enzymatic hapalindole/fischerindole production determined in this study (see Supplementary Fig. 3 for the structure of 10).
One of the most intriguing questions regarding construction of the broader family of hapalindole core structures is the basis for regiospecific C-ring formation in hapalindoles (4, C-3/4) and fischerindoles (8, C-3/2). Our studies on 12-epi-hapalindole U suggested a novel mechanism involving a concerted FamC1-catalyzed cyclization to afford the tetracyclic hapalindoles. However, it remained unclear whether a similar type of concerted mechanism was employed to generate the fischerindole core, and if tricyclic hapalindoles could serve as active intermediates toward formation of the tetracyclic cores. Thus, we expanded our analysis to include two fischerindole producing strains, Fischerella muscicola UTEX 1829 (ref. 17) and Fischerella sp. SAG 46.79 (Fig. 2)20. The genomic DNA of UTEX 1829 was sequenced and mined for the fischerindole biosynthetic gene cluster (fim), revealing a 39 kb region encoding 31 predicted proteins, including five hypothetical Stig cyclases (FimC1–FimC5). The fis gene cluster from strain SAG 46.79 is split into a 22 kb region encoding 18 proteins and a 5.3 kb region encoding five proteins, with only one open reading frame annotated as a cyclase (FisC).
Our ability to predict the activity of FilC1 and HpiC1 through comparative sequence analysis suggested that the same strategy could be applied to identify the Stig cyclases responsible for fischerindole formation. Thus, all putative cyclase amino acid sequences in our study were compared and classified into subgroups according to their identities. FimC1 to FimC4 showed relatively high identities (>90%) with FamC1 to FamC4, respectively, and were classified into the same subgroups. FimC5 and FisC were 90% identical and displayed relatively low similarity to any FamC subgroup (Supplementary Data Set). These results led us to hypothesize that FimC5 and FisC are responsible for fischerindole formation, and may generate the same stereochemical pattern at the four chiral centers. Thus, the two heterologously expressed and purified proteins were separately incubated with chemoenzymatically generated 3 for activity analysis, and two new compounds were observed from both reactions (Figure 3b). NMR analysis revealed these two proteins catalyzed formation of the same molecules: the major product was 12-epi-fischerindole U (9, C-10S/11R/12S/15S), and the minor one 12-epi-hapalindole C (7), a tricyclic hapalindole possessing the same stereochemistry as 9 (Supplementary Note). 7 also failed to convert to 9 upon incubation with FimC5 or FisC, indicating that it is a shunt metabolite and cannot re-enter the catalytic cycle to form fischerindoles.
Our studies conclusively demonstrated that 3 is the common intermediate to all hapalindole and fischerindole type core ring systems. However, the functional basis of stereodiversification at positions C-10/11/12/15 by the suite of homologous Stig cyclases remained unclear. We hypothesized that each cyclase subgroup is responsible for a specific stereochemistry observed in the core ring system. In support of this reasoning, we were able to isolate hapalindole H (5, C-10R/11R/12R/15R) as an additional product from the cell-free lysate reaction of 1 with 2 (Figure 3c), a compound with a different stereochemical configuration from previously isolated 4.11, 21 Thus, we sought to use the FamC cyclases as a model to explore the basis for stereochemical control in these indole alkaloids.
In order to probe the hypothesis that the alternative cyclases control the relative stereochemical configurations observed amongst the variant hapalindole metabolites, the three additional FamC proteins (FamC2–FamC4) were overexpressed in E. coli and purified. Surprisingly, in vitro assays with FamC2 and FamC3 individually failed to generate any product (Figure 3c). However, testing of the proteins in pairs revealed that co-incubating FamC2 and FamC3 in a 1:1 ratio resulted in efficient formation of hapalindole H. We also observed that mixing the two proteins helped solubilize FamC2, which precipitated partially when purified as a homogeneous protein. Because size exclusion chromatography showed that the other FamC homologs were active as homodimers, it is reasonable to suggest that the association between FamC2 and FamC3 may involve heterodimer formation (Supplementary Fig. 4).22–24 This hypothesis was tested using a Ni-NTA pull-down assay in which untagged FamC2 co-purified with His- tagged FamC3. After further purification using size-exclusion chromatography, this complex was shown to generate 5. Thus, we propose that the hapalindole biosynthetic machinery may engage various homo- and heterodimeric forms of the cyclase monomers to control the stereo- and regiochemical outcomes of the indole alkaloid metabolites. This hypothesis could address the apparent conflict that different stereochemical patterns in the ambiguine/fischerindole metabolites are produced between UTEX 1903 and UTEX 1829 despite the high sequence identity between the FamC1–FamC4 and FimC1–FimC4 Stig cyclases (Supplementary Figs. 2 and 5), a factor we now attribute to expanded cyclase scope through differential dimer formation.
In summary, in vitro reconstitution of the functional activities of this new class of indole alkaloid cyclases provides conclusive evidence that they are responsible for ring formation and stereochemical control in the biosynthesis of hapalindoles and fischerindoles. We identified and characterized the function of several Stig cyclases from five hapalindole-producing cyanobacterial strains, including three novel gene clusters. This work demonstrates that the FamC1/FilC1/HpiC1 class catalyzes formation of 12-epi-hapalindole U (4) and FimC1/FisC directly produces 12-epi-fischerindole U (9), while hapalindole H (5) is assembled by a heterodimeric combination of FamC2 and FamC3. Moreover, tricyclic hapalindole 7 was not transformed into fischerindole 9, suggesting that tricyclic compounds are shunt products and not intermediates on the pathway to tetracycle formation. These data provide a plausible basis for enzymatic regio- and stereoselective ring formation, which is controlled by the related Stig-type cyclases acting on central bicyclic precursor 3. Importantly, the protein-protein interactions between FamC2 and FamC3 suggest that some functional cyclases are comprised of more than one protein in order to create the variant stereo- and regiochemical motifs observed within this large class of indole alkaloids. Further efforts involving protein structural studies to probe directly the mechanism of these remarkable biocatalysts are underway.
Online Methods
General Materials and Methods
All NMR spectra were acquired on Varian 400, 600 and 700 MHz spectrometers. Proton and carbon signals are reported in parts per million (δ) using residual solvent signals as an internal standard. The LCMS analysis was performed on a Shimadzu 2010 EV APCI spectrometer equipped with an Agilent Extend-C18 5 µm 4.6 × 150 mm column, using a mobile phase gradient of 70–90% acetonitrile in water over 22 min and was monitored by UV absorption at 220 nm. Preparative-scale HPLC was performed using an Agilent Extend-C18 10 µm 10 × 250 mm column, using a mobile phase gradient of 70–90% acetonitrile in water over 28 min. High-resolution APCIMS spectra and protein mass spectrometry sequence analysis were obtained from an Agilent 6520 Q-TOF mass spectrometer equipped with an Agilent 1290 HPLC system at the University of Michigan core facility in the Department of Chemistry, with MS grade solvents. Optical rotations were measured in CH2Cl2 at 25 °C at the sodium D line.
Escherichia coli strain DH5α (Invitrogen) was used for plasmid manipulation, BL21(DE3/pRARE) was used for protein expression. KOD Xtreme Hot Start DNA polymerase (EMD Millipore) was used for polymerase chain reactions. Restriction endonucleases (NheI, XhoI, NdeI and HindIII) and T4 DNA ligase were purchased from New England BioLabs. Primers were purchased from Integrated DNA Technologies. PureLink Quick Plasmid Miniprep Kit (Invitrogen) was used to prepare plasmid DNA. All cloned plasmids were confirmed by Sanger sequencing at the University of Michigan DNA Sequencing Core. Isopropyl-D-thiogalactopyranoside (IPTG, GoldBio) was used to induce expression; benzonase and lysozyme used in purification were purchased from Sigma- Aldrich; phenylmethane sulfonyl fluoride (PMSF) was dissolved in isopropanol and used as serine protease inhibitor during protein purification. Ni-NTA agarose from Invitrogen was used to purify 6×His-tag proteins. LB broth and agar (EMD Millipore) were used for all E. coli culturing.
Cyanobacterial culturing, Genomic DNA extraction and sequencing
Cyanobacteria strains Fischerella ambigua UTEX 1903 and Fischerella muscicola UTEX 1829 were purchased from the UTEX Culture Collection of Algae at the University of Texas at Austin. Fischerella sp. IL-199-3-1 was obtained from Shmuel Carmeli and Avi Raveh (Tel Aviv University), Fischerella sp. ATCC 43239 was purchased from the ATCC microbial collection, and Fischerella sp. SAG 46.79 was obtained from Jimmy Orjala (University of Illinois at Chicago). The cyanobacterial culturing, genomic DNA isolation, and whole genome sequencing of these strains followed the established protocols.11
Protein constructs
Vector pET28a (NdeI and HindIII sites) was used to construct the FamD2 expression system. Vector pET28a (NheI and XhoI sites) was used for the cloning of all seven cyclases. The famD2 gene was amplified from UTEX 1903 genomic DNA, filC1 from IL-199-3-1 genomic DNA, and fisC from SAG 46.79 genomic DNA. For the other five cyclase genes, famC1/famC2/famC3 (UTEX 1903), hpiC5 (ATCC 43239), and fimC5 (UTEX 1829), the DNA sequences used for cloning were purchased from Integrated DNA Technologies (IDT) after rare codon optimized because of low expression levels in E. coli using the native DNA sequence. All primers used are listed in Supplementary Table 1.
Protein expression and purification
Positive plasmids from gene cloning were transformed into electro-competent BL21 (DE3/pRARE) E. coli cells. A single colony was picked and inoculated into LB medium (10 mL, 50 µg/mL kanamycin), shaken overnight at 37 °C (200 rpm), and used to inoculate a 2.8 L Fernbach flask containing pre-warmed LB medium (1 L, 50 µg/mL kanamycin). After incubation (37 °C, 200 rpm) to an optical density of 0.6, the culture was cooled to 16 °C. IPTG was added to a final concentration of 0.2 mM. After overnight incubation (16 °C, 200 rpm), the cells were centrifuged (6,000 × g, 4 °C, 15 min). The following procedures were performed at 4 °C in a temperature controlled room. The cell pellets were re-suspended in 20 mL of lysis buffer (10 mM HEPES, 50 mM NaCl, 20 mM imidazole, 0.2 mM TCEP, 10% glycerol), containing 1 mM PMSF, 0.5 mg/mL of lysozyme, and 1 µL of benzonase. The mixture was incubated for 30 min and sonicated on ice for 2 min using 10 s pulses followed by a 50 s pause. The sample was centrifuged (60,000 × g, 4 °C, 35 min) to remove cellular debris, and the supernatant was loaded onto Ni-NTA agarose prewashed with lysis buffer and loaded by gravity. The column was washed with buffer (10 mM HEPES, 300 mM NaCl, 0.2 mM TCEP, 10% glycerol, 20 mM imidazole) to remove unbound proteins, followed by 10 mL of elution buffer (10 mM HEPES, 50 mM NaCl, 0.2 mM TCEP, 10% glycerol, 300 mM imidazole) to elute the desired protein. The protein-containing fractions were combined and desalted using a PD10-desalting column (GE Healthcare) pre-equilibrated with storage buffer (10 mM HEPES, 50 mM NaCl, 0.2 mM TCEP, 10% glycerol). The purified protein was analyzed by SDS-PAGE gel for homogeneity, assessed by Nanodrop (Company) for concentration using calculated molar extinction coefficients, flash-frozen in liquid nitrogen, and stored at -80 °C.
In vitro enzymatic assays
The synthesis of indole isonitrile (1) was described previously11, GPP (2) was purchased from supplier Isoprenoids.com (purity > 95%). The activity assays for cyclases (FamC1/FilC1/HpiC1/FimC5/FisC/FamC2/FamC3) were initially conducted by incubating them with pure intermediate 3, which was produced by incubating FamD2 with substrates 1 and 2 at pH 10.5.11 However, compound 3 was not stable in extended storage. Thus, the reactions were set up in one-pot by incubating substrates 1 and 2 with enzyme cascades (FamD2 and the selected cyclase(s)), which also resulted in the generation of byproduct 10 (Supplementary Fig. 1)11. The general protocol for enzymatic reactions is as follows. A 50 µL reaction containing 5 µM of FamD2, 10 µM of cyclase(s) or 1 mg/mL of cell-free lysate,11 1 mM of 1, 1 mM of 2, 5 mM of MgCl2, and 50 mM of Tris buffer (pH 7.8) was incubated at 37 °C. The reaction was quenched after 5 h and extracted three times with an equal volume of ethyl acetate. The organic layers were combined, dried, and re- dissolved in 100 µL of acetonitrile for LCMS analysis. The same conditions were used to test the activity of FamC2 and FamC3 in combination by mixing 10 µM of each protein with FamD2. Fresh FamC2 was used for in vitro assay because of precipitation during the freeze-thawing process. For the conversion assay between 12-epi-hapalindole C (7) and 12-epi-fischerindole U (9), compound 7 was purified by HPLC from the reaction with FimC5, and dissolved in DMSO. 0.5 mM of 7 was incubated at 37 °C with 15 µM FimC5 or FisC in 50 mM Tris buffer (pH 7.8). The reactions were quenched and analyzed as described above.
For structure analysis of enzymatic products (4/5/7/9), the reactions were scaled up to 5 mL and incubated under identical conditions. The extracted products were purified by HPLC (See section General Materials and Methods), then concentrated, dissolved in C6D6, CDCl3, or CD2Cl2, and analyzed by NMR (Supplementary Fig. 6, Supplementary Tables 6,7, and Supplementary Note). The spectra of known compounds 5, 7, and 9 were identical to those previously reported.18, 25
Data availability
The nucleotide sequence of the gene clusters were deposited to NCBI GenBank under the following accession numbers: KX451322 for UTEX 1903 (fam), KY026488 for IL-199-3-1 (fil), KY026487 for UTEX 1829 (fim), KY026489 for SAG 46.79 (fis). All other data generated or analyzed during this study are included in this published article (and its supplementary information files) or are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
The authors thank the National Science Foundation under the CCI Center for Selective C-H Functionalization (CHE-1205646), the National Institutes of Health (CA70375 to RMW and DHS), and the Hans W. Vahlteich Professorship (to DHS) for financial support. We are grateful to Shmuel Carmeli and Avi Raveh (Tel Aviv University) for Fischerella sp. IL-199-3-1, and Jimmy Orjala (University of Illinois at Chicago) for Fischerella sp. SAG 46.79.
Footnotes
Author contributions
S.L. and D.H.S. designed the research. S.L. performed all experiments. A.N.L. synthesized the indole isonitrile. S.L., S.A.N., A.N.L. and D.H.S. conducted data analysis and interpretation. F.Y. performed bioinformatics analyses. S.L., A.N.L., D.H.S., and R.M.W. contributed to manuscript preparation.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Li CJ. Chem. Rev. 2005;105:3095–3165. doi: 10.1021/cr030009u. [DOI] [PubMed] [Google Scholar]
- 2.Koeller KM, Wong CH. Nature. 2001;409:232–240. doi: 10.1038/35051706. [DOI] [PubMed] [Google Scholar]
- 3.Tsunematsu Y, et al. Nat. Chem. Biol. 2013;9:818–825. doi: 10.1038/nchembio.1366. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez C, Mendez C, Salas JA. Nat. Prod. Rep. 2006;23:1007–1045. doi: 10.1039/b601930g. [DOI] [PubMed] [Google Scholar]
- 5.Jakubczyk D, Cheng JZ, O'Connor SE. Nat. Prod. Rep. 2014;31:1328–1338. doi: 10.1039/c4np00062e. [DOI] [PubMed] [Google Scholar]
- 6.Fesko K, Gruber-Khadjawi M. ChemCatChem. 2013;5:1248–1272. [Google Scholar]
- 7.Miao Y, Rahimi M, Geertsema EM, Poelarends GJ. Curr. Opin. Chem. Biol. 2015;25:115–123. doi: 10.1016/j.cbpa.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 8.Klas K, Tsukamoto S, Sherman DH, Williams RM. J. Org. Chem. 2015;80:11672–11685. doi: 10.1021/acs.joc.5b01951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auclair K, et al. J. Am. Chem. Soc. 2000;122:11519–11520. [Google Scholar]
- 10.Fage CD, et al. Nat. Chem. Biol. 2015;11:256–258. doi: 10.1038/nchembio.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, et al. J. Am. Chem. Soc. 2015;137:15366–15369. doi: 10.1021/jacs.5b10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhat V, Dave A, MacKay JA, Rawal VH. The Alkaloids: Chemistry and Biology. San Diego: Academic Press; 2014. Ch. 2. [DOI] [PubMed] [Google Scholar]
- 13.Baran PS, Maimone TJ, Richter JM. Nature. 2007;446:404–408. doi: 10.1038/nature05569. [DOI] [PubMed] [Google Scholar]
- 14.Moore RE, et al. J. Org. Chem. 1987;52:1036–1043. [Google Scholar]
- 15.Raveh A, Carmeli S. J. Nat. Prod. 2007;70:196–201. doi: 10.1021/np060495r. [DOI] [PubMed] [Google Scholar]
- 16.Becher PG, Keller S, Jung G, Sussmuth RD, Juttner F. Phytochem. 2007;68:2493–2497. doi: 10.1016/j.phytochem.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 17.Park A, Moore RE, Patterson GML. Tetrahedron Lett. 1992;33:3257–3260. [Google Scholar]
- 18.Stratmann K, et al. J. Am Chem. Soc. 1994;116:9935–9942. [Google Scholar]
- 19.Micallef ML, et al. BMC Microbiol. 2014;14:213–230. doi: 10.1186/s12866-014-0213-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H, et al. Tetrahedron. 2012;68:3205–3209. doi: 10.1016/j.tet.2012.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smitka TA, et al. J. Org. Chem. 1992;57:857–861. [Google Scholar]
- 22.Pyriochou A, Papapetropoulos A. Cell. Signal. 2005;17:407–413. doi: 10.1016/j.cellsig.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Newmister SA, Chan CH, Escalante-Semerena JC, Rayment I. Biochemistry. 2012;51:8571–8582. doi: 10.1021/bi301142h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, et al. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 25.Lu Z, Yang M, Chen P, Xiong X, Li A. Angew. Chem. Int. Ed. 2014;53:13840–13844. doi: 10.1002/anie.201406626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequence of the gene clusters were deposited to NCBI GenBank under the following accession numbers: KX451322 for UTEX 1903 (fam), KY026488 for IL-199-3-1 (fil), KY026487 for UTEX 1829 (fim), KY026489 for SAG 46.79 (fis). All other data generated or analyzed during this study are included in this published article (and its supplementary information files) or are available from the corresponding author on reasonable request.