Abstract
Progranulin (PGRN) is implicated in Alzheimer’s disease (AD) as well as frontotemporal lobar degeneration. Genetic studies demonstrate an association of the common GRN rs5848 variant that results in reduced PGRN levels with increased risk for AD. However, the mechanisms by which PGRN reduction from the GRN AD risk variant or mutation exacerbates AD pathophysiology remain ill defined. Here, we show that the GRN AD risk variant has no significant effects on florbetapir positron emission tomographic amyloid imaging and cerebrospinal fluid (CSF) Aβ levels, whereas it is associated with increased CSF tau levels in human subjects of the Alzheimer’s disease neuroimaging initiative studies. Consistent with the human data, subsequent analyses using the APPswe/PS1ΔE9 (APP/PS1) mouse model of cerebral amyloidosis show that PGRN deficiency has no exacerbating effects on Aβ pathology. In contrast and unexpectedly, PGRN deficiency significantly reduces diffuse Aβ plaque growth in these APP/PS1 mice. This protective effect is due, at least in part, to enhanced microglial Aβ phagocytosis caused by PGRN deficiency-induced expression of TYROBP network genes (TNG) including an AD risk factor Trem2. PGRN-deficient APP/PS1 mice also exhibit less severe axonal dystrophy and partially improved behavior phenotypes. While PGRN deficiency reduces these amyloidosis-related phenotypes, other neuronal injury mechanisms are increased by loss of PGRN, revealing a multidimensional interaction of GRN with AD. For example, C1q complement deposition at synapses is enhanced in APP/PS1 mice lacking PGRN. Moreover, PGRN deficiency increases tau AT8 and AT180 pathologies in human P301L tau-expressing mice. These human and rodent data suggest that global PGRN reduction induces microglial TNG expression and increases AD risk by exacerbating neuronal injury and tau pathology, rather than by accelerating Aβ pathology.
Keywords: progranulin, Alzheimer’s disease, microglia, TREM2, C1q, tau
INTRODUCTION
Progranulin (PGRN) is a secreted glycoprotein widely expressed in several tissues and plays a critical role in a variety of biological processes including development, wound healing, and tumorigenesis. In the central nervous system, PGRN is expressed in neurons and microglial cells and is thought to have neurotrophic and anti-inflammatory properties [11,60].
Mutations in the PGRN gene (GRN) that result in its haploinsufficiency cause frontotemporal lobar degeneration (FTLD) with ubiquitin-positive inclusions containing TAR DNA binding protein 43 (TDP-43) [11,60]. However, genetic studies have also suggested a role of PGRN in other neurodegenerative disorders. Homozygous loss-of-function mutation in GRN was found to cause neuronal ceroid lipofuscinosis (NCL) [71]. In addition, PGRN has been implicated in Alzheimer’s disease (AD). Several studies have reported GRN mutation carriers with an AD clinical presentation [41,59,62,43,17,7]. Common GRN haplotypes were associated with increased risk for AD in a Belgian late-onset AD patient group [8]. The rs5848 T allele of GRN, which causes at least 10–20% reduction of PGRN protein levels in plasma and cerebrospinal fluid (CSF) [57,63], increases risk for AD as well as FTLD [50,81,70,63].
The mechanism(s) by which GRN variation or mutation modifies AD pathophysiology is(are) poorly defined. AD is pathologically characterized by extracellular senile plaques composed of amyloid-β (Aβ) peptides (Aβ plaques) and intracellular neurofibrillary tangles comprised of hyperphosphorylated tau. Synapse loss is widespread and pronounced. In AD, the chronic Aβ accumulation causes cerebral neuroinflammation by activating microglia [35]. PGRN expression has been shown to correlate with dense-core plaques [58], and is increased in activated microglia around Aβ plaques [58,3] and peripheral blood [16]. A previous study has described greater Aβ plaque and worsened behavioral impairment in transgenic AD mice with conditional PGRN deletion using LysM-cre mice [56]. However, a recent study as well as earlier publications raises concerns about the efficient and specific targeting using the LysM-cre mice and cautions against conclusions drawn from the experiments with the mouse line [5,79].
In the present study, we investigate the mechanism by which PGRN reduction from the GRN rs5848 T allele as well as GRN mutations increase AD risk and modify AD pathophysiology. To this end, we first analyze florbetapir positron emission tomographic (PET) amyloid imaging and CSF biomarkers in human subjects with the GRN AD risk variant using Alzheimer’s disease neuroimaging initiative (ADNI) database. In addition, we examine the APPswe/PS1 E9 (APP/PS1) mouse model of cerebral amyloidosis and human P301L tau-expressing mice with global PGRN reduction. Previous studies have suggested that there is a different level of vulnerability in the human versus mouse brain to PGRN reduction. Although PGRN null (Grn−/−) mice recapitulate some of pathological features of FTLD such as microgliosis and retinal degeneration, Grn heterozygous (Grn+/−) mice do not, and both fail to show TDP-43-positive inclusions, a key pathological hallmark of FTLD. Therefore, Grn−/− mice have been studied as the best available mouse model for human PGRN haploinsufficiency-related FTLD [42,1,61,80,27,78,29,47,53]. Given this evidence, although GRN rs5848 T allele causes 10–20% reduction of PGRN in humans and GRN mutation carriers with AD clinical presentation are all heterozygotes, we utilize Grn−/− as well as Grn+/− mice. Our human and rodent data show that PGRN reduction has minimal effect on dense-core amyloid plaques and is protective against diffuse Aβ plaque growth, while it is associated with increased tau pathology. Thus, PGRN reduction may affect AD pathophysiology by causing tau dysfunction, distinct from Aβ pathology.
METHODS
Mice
All animal studies were conducted with approval of the Yale Institutional Animal Care and Use Committee. APP/PS1 mice and Grn−/− mice were described previously [26,38]. All strains were extensively backcrossed onto the C57BL6 background and were maintained on this background.
Brain tissue collection
Six- and sixteen-month-old WT, Grn+/−, Grn−/−, APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice were killed and immediately perfused with ice-cold PBS. The brains were then dissected out. One hemisphere was immediately frozen using liquid nitrogen and stored at −80°C for biochemical or RNA analysis and the other was fixed in freshly prepared 4% paraformaldehyde (PFA) (ACROS ORGANICS 416780030 or Sigma 158127) in PBS (pH 7.4) overnight for immunohistochemistry.
Brain protein extraction
Brain protein extraction was performed as previously reported [26], with slight modifications. The brain hemispheres were homogenized in three-fold volume (w/v) of 50 mM Tris-HCl, 150 mM NaCl, pH 7.6 (TBS) containing a protease inhibitor cocktail (Roche complete Mini EDTA-free 11 836 170 001) and a phosphatase inhibitor cocktail (Roche phosSTOP 04 906 837 001). After ultracentrifugation at 100,000 × g for 20 min at 4°C, the supernatant was collected and frozen as the TBS-soluble fraction. The pellet was resuspended to the same volume as the original homogenate in TBS with 1% Triton X-100 (TBST) containing a protease inhibitor cocktail and a phosphatase inhibitor cocktail. The homogenates were ultracentrifuged at 100,000 × g for 20 min. The supernatant was collected and frozen as the TBST-soluble fraction. The remaining pellet was then resuspended to the same volume in 70% formic acid (FA), homogenized, and ultracentrifuged. The supernatant was frozen as the FA-soluble fraction.
Immunoblot
Immunoblot was performed as previously reported [26], with slight modification. Precast 4–20% tis-glycine or 10–20% tris-tricine gels were used (Bio-Rad). The iBlot™ Gel transfer device (Invitrogen) was used for transfer. The membranes were incubated in blocking buffer (Blocking Buffer for Fluorescent Western Blotting Rockland MB-070) for 1 h at room temperature. The following primary antibodies were used: 6E10 (Covance SIG-39300, 1:1000), APP, C-terminal (Sigma A8717, 1:1000), actin (Sigma A2066, 1:1000), actin (Sigma A3853, 1:1000), phospho-tau Ser404 (Thermo Fisher Scientific #44-758G, 1:2000), phospho-tau pThr181 (Thermo Fisher Scientific #701530, 1:1000), tau (Thermo Fisher Scientific #AHB0042, 1:2000). Antibodies were diluted in the blocking buffer, and membranes were incubated overnight at 4°C. The membranes were washed three times with TBST and secondary antibodies (Odyssey IRDye 680 or 800, all 1:10000) were applied for 1 h at room temperature. After washing four times, proteins were visualized using a Licor Odyssey Infrared imaging system. Blots were analyzed using ImageJ 1.47v software (National Institutes of Health). For the detection of Aβ, the membranes were microwaved in PBS to unmask the epitopes and increase the immunoblot sensitivity before blocking [69].
Immunohistochemistry
Immunohistochemistry was performed as previously reported [26], with slight modification. The fixed brain hemispheres were embedded in 10% gelatin (Sigma G1896) and placed in 4% PFA for 3 days at 4°C. Sagittal sections (30 μm) were then cut using a Leica WT1000S vibratome. The following steps were performed at room temperature. For AT8 and AT180 staining, the free-floating sections were treated with 10% formic acid for 20 min for antigen retrieval before blocking. The sections were blocked with 10% normal goat or donkey serum, 0.2% Triton X-100 in PBS for 1 h, followed by incubation with primary antibody overnight. Primary antibodies were diluted in 1% normal goat or donkey serum, 0.2% Triton X-100 in PBS. The following antibodies were used: β-amyloid (Cell Signaling Technology #2454, 1:500), CD68 (AbD Serotec MCA1957, 1:1000), Iba1 (Wako 019-19741 for immunocytochemistry, 1:600), LAMP-1 (Santa Cruz sc-8334, 1:250), C1q (Abcam ab11861, 1:50), C1q (Abcam ab182451, 1:1000), PSD-95 (Thermo Fisher Scientific 51-6900, 1:200) AT8 (Thermo Fisher Scientific MN1020, 1:500), AT180 (Thermo Fisher Scientific MN1040,1:500), HT7 (Thermo Fisher Scientific MN1000, 1:500), GFP (Santa Cruz sc-8334, 1:500). The sections were washed three times with PBS, incubated in secondary fluorescent antibody (Invitrogen Alexa Fluor, all 1:500) for 2 h. In order to quench autofluorescence, the sections were treated with 10 mM CuSO4 in ammonium acetate for 15 min [66]. The images were taken using the Zeiss AxioImager Z1 microscopy with a 5x objective lens. To obtain high magnification images, we utilized the Zeiss LSM710 confocal microscopy using 63x objective lens. To take images in Supplementary Figure 3, we used 40x objective lens and “tile scan” function in Zen software. The maximum intensity projection function was used in all confocal images.
Thioflavin S (ThioS) staining
The free-floating sagittal sections were stained with 0.01% ThioS (Sigma T1892) in PBS for 15 min, followed by washing three times with PBS. For double and triple staining with antibodies, after secondary antibody incubation, the free-floating sections were stained with 0.01% thioflavin S in PBS, followed by treatment with 10 mM CuSO4.
Image quantification
Quantitative analyses of ThioS-positive plaque, Aβ, CD68, Iba1, C1q, and LAMP-1-positive dystrophic neurite areas and AT8, AT180, and HT7 signals were done using ImageJ 1.47v software (National Institutes of Health). All images taken using the Zeiss AxioImager Z1 microscopy with a 5x objective lens were uniformly thresholded and binarized. For CD68, Iba1, and LAMP-1-positive dystrophic neurite staining, background was subtracted (Rolling ball radius: 200 pixels for CD68 and Iba1, 50 pixels for LAMP-1) before binarization. Aβ, CD68, Iba1, and LAMP-1-positive dystrophic neurite areas were calculated by using the “analyze particles” of ImageJ. For ThioS-positive plaques, plaque area, and plaque number were calculated with the “watershed” algorithm and “include holes” function. Only male was used for Aβ and ThioS analyses with 6-month-old mice in order to avoid a potential gender effect previously reported [45]. For the other analyses, both male and female were used with similar ratio between genotypes and no significant difference between male and female was observed. Three brain sections per mouse, each separated by 150 μm, were used for quantification. The mean of 3 sections was used to represent for each mouse. For the cortical analysis, frontal cortex, the cortex dorsal to the hippocampus, and occipital cortex were assessed and sums of the areas were used to calculate area percentages. In Supplementary Figure 4b, whether microgliosis (> 3 Iba1-positive microglia) occurs around ThioS(+) or ThioS(−) Aβ plaques was manually examined under the Zeiss AxioImager Z1 microscopy. The area occupied by PSD-95-immunoreactive puncta from the layer III of FC was measured as described previously [26]. PSD-95 and C1q colocalization was determined after background subtraction and binarization of C1q-immnoreactive area. For AT8, AT180, and HT7 analyses, the images were taken using Zeiss AxioImager Z1 microscopy and “MosaiX acquisition” function in AxioVision Rel. 4.8 software as shown in Supplementary Figure 8a and uniformly thresholded GFP-positive area was used for quantification. The mean signal of 5 sections from lateral ~2.76 to ~3.25 mm (~0.12 mm interval) were used to represent for each mouse. All analyses were performed in a blinded manner.
Aβ42 ELISA
For mouse samples, Aβ42 ELISA (Invitrogen KHB3441) was performed according to the manufacturer’s manual. The optimal dilution for the FA fraction was empirically determined to be 1:20,000. The absorbance at 450 nm was measured using Victor 3V plate reader (Perkin Elmer). GraphPad Prism (version 5.0d) was used to generate the standard curve (four-parameter dose-response curve) and to interpolate the concentrations of unknown samples.
Primary microglial cell culture
Primary cultured microglia were prepared from the brains of postnatal day 3 WT or Grn−/− mice. Meninges were removed mechanically, and the cells were dissociated and cultured in DMEM supplemented with 10% heat-inactivated (65°C, 30 min) FBS (hiFBS) and penicillin/streptomycin. After 14 days (DIV14), the culture flasks were shaken (190 rpm) for 3 h to collect microglial cells.
Phagocytosis assay
Primary microglia (DIV16 or 17) from WT and Grn−/− mice were plated on PDL-coated 8-well slides (Lab-Tek, 154941) (8 × 107 cells/well) using DMEM with 8% hiFBS 3 days before experiments. The cells were incubated with FluoSpheres carboxylate, 1.0 μm, red (580/605) (Invitrogen F8821) diluted in DMEM with hiFBS at a final dilution 1:400 or 1:800 for 90 min at 37°C, washed once with PBS, and fixed with 4% formaldehyde in PBS. The cells were then stained with anti-Iba1 antibody (abcam ab107159, 1:500) as described above. Three images per wells were taken in one independent experiment using the Zeiss AxioImager Z1 microscopy with a 10× objective lens and the total area from the 3 images was used for quantification. Quantitative analyses of fluorescent beads and Iba1 areas were performed using ImageJ 1.47v software (National Institutes of Health).
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was purified from snap-frozen frontal cortex using TRIzol reagent (Ambion 15596026) and the PureLink RNA Mini kit (Ambion 12183018A) and reverse transcribed using the SuperScript III first-strand synthesis system (Invitrogen 18080-051) according to the manufacturer’s instructions. Real-time quantitative PCR was performed using the C1000 Thermal Cycler and quantified using CFX96 Real-Time System (Bio-Rad). The TaqMan gene expression assay and iQ supermix (Bio-rad 170-8862) was used for PCR amplification and real time detection of PCR products. mRNA expression values were normalized to the level of GAPDH expression. The following probes from Invitrogen were used: GAPDH (Mm99999915_g1), TNFα (Mm00443258_m1), IL-1β (Mm00434228_m1), iNOS (Mm00440485_m1), IL-6 (Mm00446190_m1), Arg-1 (Mm00475988_m1), TGFβ (Mm00498255_m1), Fizz-1 (Mm00445109_m1), Ym1 (Mm00657889_m1), TREM2 (Mm00451744_m1), TYROBP (Mm00449152_m1), MS4A7 (Mm01197655_m1), CD68 (Mm03047343_m1), CD22 (Mm00515432_m1), Lyz2 (Mm01612741_m1), CatS (Mm01255859_m1), C1qA (Mm00432142_m1), EMR1 (Mm00802529_m1), CCL8 (Mm01297183_m1).
Behavior test
Morris Water Maze testing was performed over a six-day period, consisted of a three-day learning trial and a three-day reversal trial, as previously reported [26]. Both trials were performed in a ~1 m diameter open-water pool and used a submerged, nonvisible escape platform located in the center of one of the pool’s four quadrants. This location remained constant for each three-day trial; for the reversal trial, the platform was placed, for the duration of the trial, in the quadrant diagonally across from its original location in the learning trial. Over the course of each day, the mice swam in the pool a total of eight times which were divided into two blocks of four attempts. One block took place during the morning while the second block took place during the afternoon. Over the course of a block, each mouse would begin its swim in each of four distinct locations around the wall of the pool, and was timed for its latency to reach the escape platform for a maximum time of 60 seconds. In the event that a mouse did not find the submerged platform within the time limit, the mouse would be gently guided to the platform and allowed ~10 seconds on the platform before being taken from the pool.
The water maze probe trial was performed 24 hours after the last day of the forward and reversal trials, and in the same pool described above. During the probe trial, the platform was removed from the pool. All mice were started from a location along the pool wall diagonally opposed to the location of the platform in the reversal trial and permitted to swim for 60 seconds. The probe trials were recorded on a JVC Everio, G-series camcorder and analyzed using Panlab’s Smart tracking and analysis program, v2.5. The observer was blinded to genotype for the duration of behavioral testing.
Florbetapir-PET scan and CSF biomarker analysis
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5- year public-private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI-GO and ADNI-2. To date these three protocols have recruited over 1500 adults, ages 55 to 90, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow up duration of each group is specified in the protocols for ADNI-1, ADNI-2 and ADNI-GO. Subjects originally recruited for ADNI-1 and ADNI-GO had the option to be followed in ADNI-2. For up-to-date information, see www.adni-info.org.
For Figure 1, Table 1, and Supplementary Table 1, Data from ADNI-GO and ADNI-2 participants were analyzed, as GRN rs5848 genotype was available in these groups (ADNIGO2_GWAS_Set_1-15 files). APOE ε4 genotypes and diagnostic categories were examined using APOERES and ARM files, respectively. ADNI florbetapir summary data (UCBERKELEYAV45_7_30_14 file) including standardized uptake value ratio (SUVr) results normalized by whole cerebellum region were downloaded and analyzed. CSF biomarker data in ADNI-GO and ADNI-2 (UPENNBIOMK5, UPENNBIOMK6, and UPPENNBIOMK7 data files) were used for this study. This cross-sectional analysis across the three files was performed by combining all baseline results, as recommended (ADNI_Methods_UPENN_Third_Batch_Analysis_ of CSF_Biomarker_20140611).
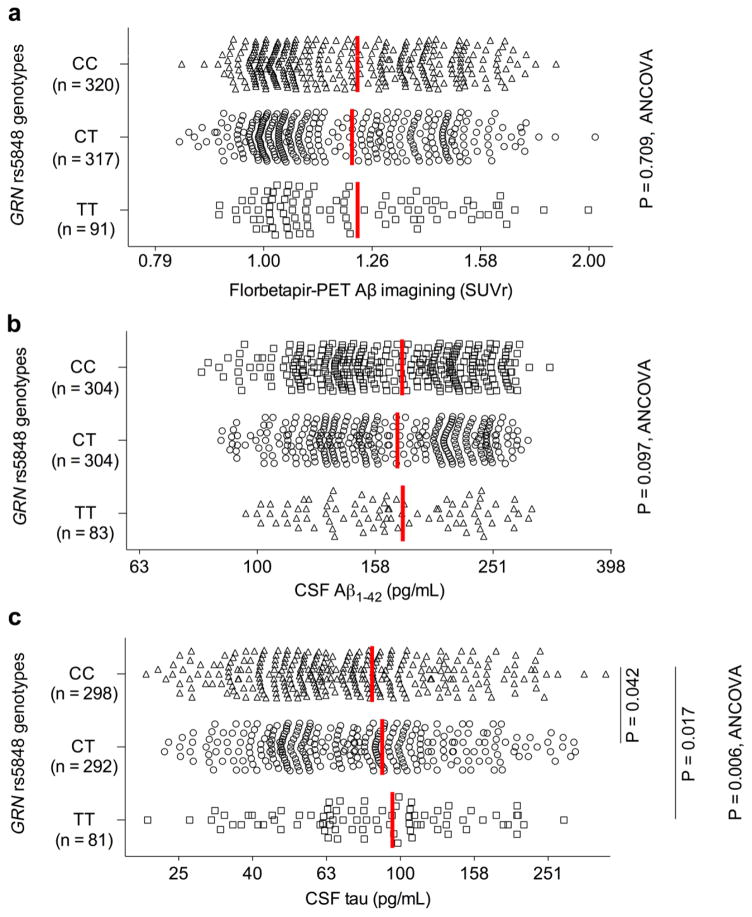
Figure 1. GRN rs5848 AD risk variant has no effects on florbetapir-PET amyloid imaging while it increases CSF tau levels in humans.
a. Mean cortical Aβ levels (SUVr) as quantified by florbetapir-PET in the ADNI participants are segregated by GRN rs5848 genotypes. Log scale is used for X-axis. Red vertical bars indicate mean. See Table 1 for the statistical analyses used in this figure.
b. CSF Aβ levels in the ADNI participants are segregated by GRN rs5848 genotypes. Log scale is used for X-axis. Red vertical bars indicate mean. See Table 1 for the statistical analyses used in this figure.
c. CSF tau levels in the ADNI participants are segregated by GRN rs5848 genotypes. Log scale is used for X-axis. Red vertical bars indicate mean. See Table 1 for the statistical analyses used in this figure.
Table 1.
A summary of florbetapir-PET and CSF Aβ42, tau, and p-tau181 in the ADNI participants segregated by sex, disease stage, APOE ε4 allele number, or GRN rs5848 genotypes
| N* | Florbetapir- PET(SUVr), median (IQR) | CSF Aβ42 (pg/mL), median (IQR) | CSF tau (pg/mL), median (IQR) | CSF p-tau181 (pg/mL), median (IQR) |
|---|---|---|---|---|
| Sex | ||||
| F (n = 411, 352–362) | 1.16 (1.03–1.41) | 163 (134–217) | 77.5 (47.8–122.2) | 36.0 (23.7–54.6) |
| M (n = 455, 395–410) | 1.13 (1.01–1.38) | 165 (129–220) | 67.6 (49.1–98.6) | 36.2 (23.3–51.9) |
| P** | 0.0712 | 0.4064 | 0.0348 | 0.6578 |
| Disease stage | ||||
| NL (n = 163, 141–144) | 1.06 (1.00–1.16) | 207 (158–235) | 56.9 (45.1–84.6) | 28.9 (21.9–42.9) |
| SMC (n = 81, 96) | 1.06 (1.00–1.19) | 207 (156–240) | 57.7 (42.3–79.3) | 31.2 (23.5–44.7) |
| EMCI (n = 293, 251–262) | 1.10 (1.01–1.32) | 187 (140–229) | 64.1 (45.9–90.9) | 30.4 (21.1–46.6) |
| MCI (n = 149, 137–142) | 1.28 (1.05–1.48) | 140 (125–183) | 86.9 (55.0–134.6) | 42.1 (31.4–61.3) |
| AD (n = 139, 122–128) | 1.43 (1.28–1.53) | 131 (114–149) | 117.5 (84.1–163.6) | 51.3 (36.9–66.9) |
| P*** | < 0.0001a | < 0.0001b | < 0.0001c | < 0.0001d |
| APOE ε4 allele copies | ||||
| ε4 (0) (n = 449, 415–423) | 1.05 (1.00–1.19) | 207(153–238) | 57.8 (43.3–86.0) | 29.3 (21.0–43.0) |
| ε4 (1) (n = 290, 263–273) | 1.30 (1.12–1.49) | 142 (126–179) | 87.2 (60.0–131.4) | 42.0 (30.5–61.7) |
| ε4 (2) (n = 79, 68–75) | 1.39 (1.27–1.50) | 116 (99–135) | 105.0 (70.6–163.1) | 48.6 (36.9–72.3) |
| P*** | < 0.0001e | < 0.0001f | < 0.0001g | < 0.0001g |
| GRN rs5848 genotype | ||||
| CC (n = 320,298–304) | 1.16 (1.02–1.40) | 166 (134–218) | 68.4 (47.6–100.9) | 36.3 (22.5–54.6) |
| CT (n = 317, 292–304) | 1.12 (1.02–1.39) | 160 (130–219) | 74.4 (48.5–111.0) | 34.6 (24.2–50.2) |
| TT (n = 91,81–83) | 1.13 (1.03–1.39) | 166 (133–224) | 80.0 (61.2–119.8) | 38.1 (23.8–55.5) |
| P **** | 0.733 | 0.097 | 0.006h | 0.391 |
NL, individuals with no neurological disease; SMC, significant memory concern; EMCI, early mild cognitive impairment; MCI, mild cognitive impairment; AD, Alzheimer’s disease
The first and second n indicate the number of participants available for SUVr and CSF biomarker data, respectively.
Mann Whitney test,
Kruskal-Wallis test,
ANCOVA
Post hoc Dunn’s test: NL vs EMCI, p < 0.05; NL vs MCI, p < 0.001; NL vs AD, p < 0.001; SMC vs EMCI, p < 0.001; SMC vs MCI, p < 0.001; SMC vs AD, p < 0.001; EMCI vs MCI, p < 0.001; EMCI vs AD, p < 0.001; MCI vs AD, p < 0.001
Post hoc Dunn’s test: NL vs MCI, p < 0.001; NL vs AD, p < 0.001; SMC vs MCI, p < 0.001; SMC vs AD, p < 0.001; EMCI vs MCI, p < 0.001; EMCI vs AD, p < 0.001; MCI vs AD, p < 0.05
Post hoc Dunn’s test: NL vs MCI, p < 0.001; NL vs AD, p < 0.001; SMC vs MCI, p < 0.001; SMC vs AD, p < 0.001; EMCI vs MCI, p < 0.001; EMCI vs AD, p < 0.001; MCI vs AD, p < 0.001
Post hoc Dunn’s test: NL vs MCI, p < 0.001; NL vs AD, p < 0.001; SMC vs MCI, p < 0.01; SMC vs AD, p < 0.001; EMCI vs MCI, p < 0.001; EMCI vs AD, p < 0.001
Post hoc Dunn’s test: ε4(0) vs ε4(1), p < 0.001; ε4(0) vs ε4(2), p < 0.001; ε4(1) vs ε4(2), p < 0.05
Post hoc Dunn’s test: ε4(0) vs ε4(1), p < 0.001; ε4(0) vs ε4(2), p < 0.001; ε4(1) vs ε4(2), p < 0.001
Post hoc Dunn’s test: ε4(0) vs ε4(1), p < 0.001; ε4(0) vs ε4(2), p < 0.001
Post hoc Sidak test: CC vs CT, p = 0.042; CC vs TT, p = 0.017
Adeno-associated virus (AAV) and surgery
Stereotaxic AAV injection was performed as previously described [2,4], with modification. An AAV vector that expresses human P301L tau 1–441 or GFP under the control of the neuron-specific synapsin-1 promoter was prepared as previously described [2]. Both male and female WT and Grn−/− mice at 6 to 8 months of age were used for this analysis. Briefly, mice were anesthetized with an intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg). After mice were head-fixed on a stereotaxic frame, AAV was unilaterally injected into entorhinal cortex via the following coordinates: anteroposterior, −4.7 mm; lateral, 3.0 mm; dorsoventral, −4.6 mm. Twenty-eight days post infection, mice were perfused with ice-cold PBS followed by 4% PFA in PBS. All surgery was performed in a blinded manner.
Statistical analysis
For animal studies, two-tailed T test (to compare two groups) and one-way ANOVA followed by Tukey’s multiple comparison test (to compare three and more groups) were prepared using GraphPad Prism (version 5.0d). For human data that did not follow a normal distribution, nonparametric Mann-Whitney (to compare two groups) and Kruskal-Wallis (to compare three and more groups) were performed using GraphPad Prism (version 5.0d). Chi-square test was used for comparison of differences in categorical variables. For GRN rs5848 genotypes analysis, the CSF and PET variables were log-transformed and evaluated by ANCOVA in order to control for effects of potential confounders (age, gender, diagnosis, and APOE ε4 genotypes), using the Statistical Package for the Social Science 21.0 (SPSS Inc.).
RESULTS
GRN rs5848 T allele does not alter florbetapir-PET signal and CSF Aβ42 levels
We first sought to assess how GRN rs5848 T allele might alter AD pathophysiology in the clinical setting. The ADNI database includes SNP genotypes, PET measurement of florbetapir binding to amyloid deposits in the brain [13], and CSF biomarker analyses of individuals with no neurological disease (NL), with significant memory concern (SMC), with early mild cognitive impairment (EMCI) and with mild cognitive impairment (MCI) and with AD. The combined sample size is more than 700 cases. We first analyzed the florbetapir-PET amyloid imaging data. There is progressive increase of the PET radioactive amyloid ligand florbetapir uptake (SUVr) across the clinical spectrum (Table 1). As expected, the APOE ε4 allele modifies the extent of florbetapir-PET signal, as previously described (Table 1) [64]. We segregated the florbetapir-PET results by the GRN rs5848 genotype, which is reportedly linked with decreased PGRN level and increased AD/FTLD risk [50,8,70,22,57,63]. Characteristics of the ADNI participants with different GRN rs5848 genotypes are summarized in Supplementary Table 1. With or without correction for covariates (age, sex, diagnosis, and APOE ε4 copy number), there was no detectable difference in florbetapir-PET signal for those individuals with different GRN rs5848 genotypes (Fig. 1a and Table 1).
CSF Aβ42 levels are lower in AD patients compared to age-matched cognitively healthy controls, and the inverse correlation of CSF levels with plaque deposition is likely due to Aβ42 sequestration in plaques [73,20,35]. We observed the expected progressive decrease of CSF Aβ42 levels across the clinical spectrum and APOE ε4 gene dosage (Table 1). However, consistent with the florbetapir-PET data analysis, the GRN rs5848 T allele had no effect on CSF Aβ42 levels with or without correction for covariates (Fig. 1b and Table 1). Thus, our analyses of ADNI florbetapir-PET and CSF Aβ42 dataset suggest that the GRN rs5848 T allele has no detectable influence on amyloid pathology in AD.
GRN rs5848 T allele increases CSF tau levels
CSF tau and tau phosphorylated at T181 (p-tau181) levels are increased in AD patients [74,31,21]. The CSF increases are thought to reflect neuronal damage accompanied by neurofibrillary tangles in AD [35]. As we did not observe an effect of GRN rs5848 T allele on Aβ pathology, we analyzed ADNI dataset to examine whether the GRN risk allele might affect CSF tau and p-tau181 levels. As previously described [18,32,46], there are progressive increases of CSF tau and p-tau181 levels across the clinical spectrum and the APOE ε4 gene dosage (Table 1). In addition, we observed a difference in CSF tau levels between male and female in this ADNI dataset (Table 1). Strikingly, there is a GRN rs5848 T allele gene dosage-dependent increase of CSF tau levels (Fig. 1c and Table 1). In addition, there is a statistically significant difference between GRN rs5848 genotypes after correction for covariates (age, sex, diagnosis, and APOE ε4 copy number) (Fig. 1c and Table 1). Importantly, the difference in CSF tau between GRN rs5848 genotypes (CC vs. TT) is greater than the one between NL and EMCI in the ADNI dataset (Table 1). GRN variation does not have a significant effect on CSF p-tau181 in this cohort (Table 1). Thus, GRN variation has detectable effects on CSF tau levels in this ADNI group without alterations in amyloid pathology.
PGRN deficiency prevents diffuse Aβ plaque growth in APP/PS1 mice
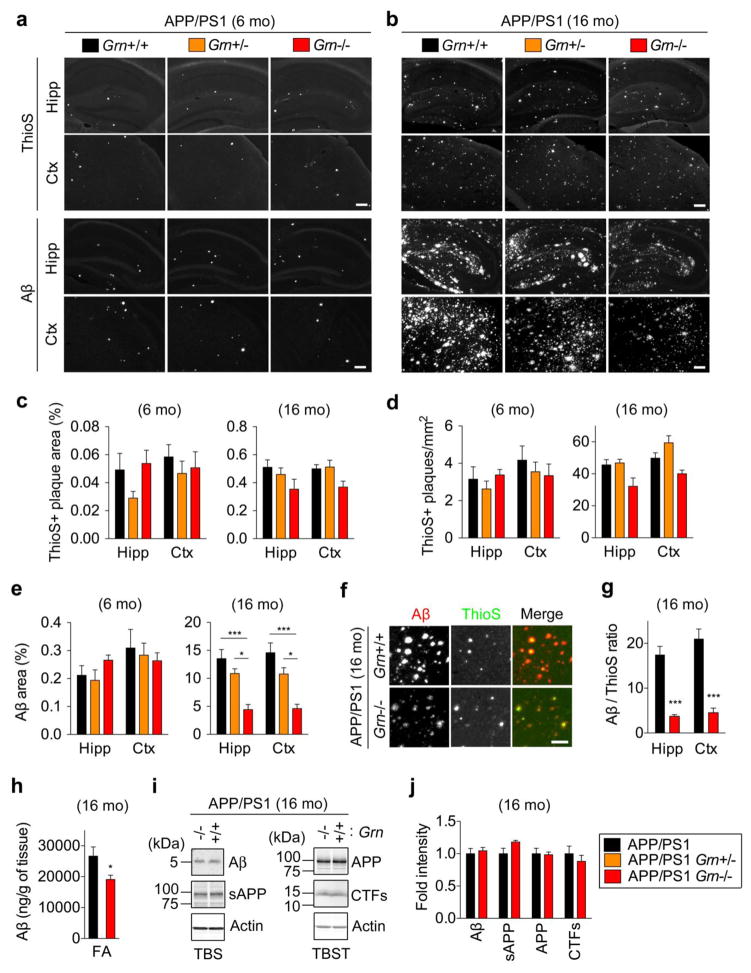
While the human data show no effect of GRN variation, which causes at least 10–20% reduction of PGRN protein, on amyloid pathology, a previous study reported greater Aβ accumulation using 7-month-old APP (J20) mice with conditional PGRN deletion [56]. In addition, potential effects of further global PGRN reduction, which may occur by GRN mutations in humans, on Aβ accumulation are unclear. We therefore examined whether PGRN haploinsufficiency or complete absence affects Aβ pathology using the APP/PS1 mouse model of cerebral amyloidosis. As the APP/PS1 mice begin to develop Aβ deposition from 4–6 months of age with progressive increases up to 12 months [39,24], we performed thioflavin S (ThioS) staining of dense-core amyloid plaques and anti-Aβ (2454) antibody staining of diffuse Aβ plaques using 6- and 16-month old animals. Only male mice were used for Aβ and ThioS analyses at 6 months of age, in order to avoid a potential gender effect previously reported in the APP/PS1 strain [45]. The ThioS staining of dense-core amyloid plaques shows either no change, or a slight decrease, in different regions and with different plaque metrics in the 6-month-old and 16-month-old APP/PS1 mice (Fig. 2a, 2b, 2c, and 2d, Supplementary Fig. 1). Anti-Aβ antibody staining shows no significant difference between Grn genotypes in 6-month-old mice (Fig. 2a and 2e), but reveals a significant reduction in the area of diffuse Aβ plaque pathology in 16-month-old APP/PS1 Grn−/− mice (Fig. 2b and 2e, Supplementary Fig. 1). Co-staining with ThioS and anti-Aβ antibody shows that PGRN deficiency significantly decreases the ratio of diffuse Aβ plaques to dense-core amyloid plaques in 16-month-old APP/PS1 mice (Fig. 2f and 2g, Supplementary Fig. 1). The histological analysis was confirmed by ELISA analysis of tissue from these mice. The insoluble fractions from APP/PS1 Grn−/− mice yield significantly less Aβ42 after extraction with formic acid (Fig. 2h). Immunoblot analysis shows that TBS-soluble Aβ or APP fragments are equal with and without PGRN expression (Fig. 2i and 2j), suggesting that the reduction of diffuse Aβ plaques in 16-month-old APP/PS1 Grn−/− mice is not due to changes of APP metabolism. Thus, neither PGRN haploinsufficiency nor its absence accelerates the onset of Aβ plaque in 6-month-old mice. In contrast, PGRN absence reduces diffuse Aβ plaque significantly, and selectively, compared to dense-core amyloid plaques in 16-month-old APP/PS1 mice.
Figure 2. PGRN deficiency prevents diffuse Aβ plaque growth to a greater extent than dense-core amyloid plaques in APP/PS1 mice.
a. Representative images of Thioflavin S (ThioS) staining and Aβ immunostaining of hippocampus (Hipp) and frontal cortex (Ctx) of 6-month-old male APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice. Bar, 200 μm.
b. Representative images of Thioflavin S (ThioS) staining and Aβ immunostaining of hippocampus (Hipp) and frontal cortex (Ctx) of 16-month-old APP/PS1, APP/PS1 Grn+/− and APP/PS1 Grn−/− mice. Bar, 200 μm.
c. Quantification of ThioS-positive area (%) in hippocampus (Hipp) and cortex (Ctx) of 6-month old male and 16-month-old APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice. Mean ± SEM, n = 4 (APP/PS1 and APP/PS1 Grn+/−) or 3 (APP/PS1 Grn−/−) in 6-month-old animals, n = 5 (APP/PS1 and APP/PS1 Grn−/−) or 4 (APP/PS1 Grn+/−) in 16-month-old animals.
d. Quantification of ThioS-positive plaque number in hippocampus (Hipp) and cortex (Ctx) of 6-month-old male and 16-month-old APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice. Mean ± SEM, n = 4 (APP/PS1 and APP/PS1 Grn+/−) or 3 (APP/PS1 Grn−/−) in 6-month-old animals, n = 5 (APP/PS1 and APP/PS1 Grn−/−) or 4 (APP/PS1 Grn+/−) in 16-month-old animals.
e. Quantification of Aβ immunoreactive area (%) in hippocampus (Hipp) and cortex (Ctx) of 6-month-old male and 16 month-old APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice. Mean ± SEM, n = 6 (APP/PS1, APP/PS1 Grn+/−) or 3 (APP/PS1 Grn−/−) in 6-month-old animals, n = 6 (APP/PS1), n = 4 (APP/PS1 Grn+/−), or n = 5 (APP/PS1 Grn+/−) in 16-month-old animals.
f. Representative images of ThioS and anti-Aβ antibody co-staining of frontal cortex of 16-month-old APP/PS1 and APP/PS1 Grn−/− mice. Bar, 200 μm.
g. Quantification of ratio of diffuse Aβ plaque area to ThioS-positive dense-core amyloid plaque area in hippocampus (Hipp) and cortex (Ctx) of 16-month-old APP/PS1 and APP/PS1 Grn−/− mice. Mean ± SEM, n = 4 (APP/PS1) or 5 (APP/PS1 Grn−/−). **p < 0.01, p < 0.001; T test.
h. Aβ42 ELISA using the FA-soluble fraction from 16-month-old APP/PS1 and APP/PS1, Grn−/− mice. Mean ± SEM, n = 4 (APP/PS1) or 5 (APP/PS1 Grn−/−), *p < 0.05; T test.
i. Representative blots using anti-Aβ, APP, actin antibodies of the TBS-soluble fraction, using anti-APP, CTF, and actin antibodies of the TBST-soluble fraction.
j. Quantification of immunoblots (i). Mean ± SEM, n = 4 (APP/PS1) or 5 (APP/PS1 Grn−/−)
PGRN deficiency increases CD68-positive microglia near Aβ plaques in APP/PS1 mice
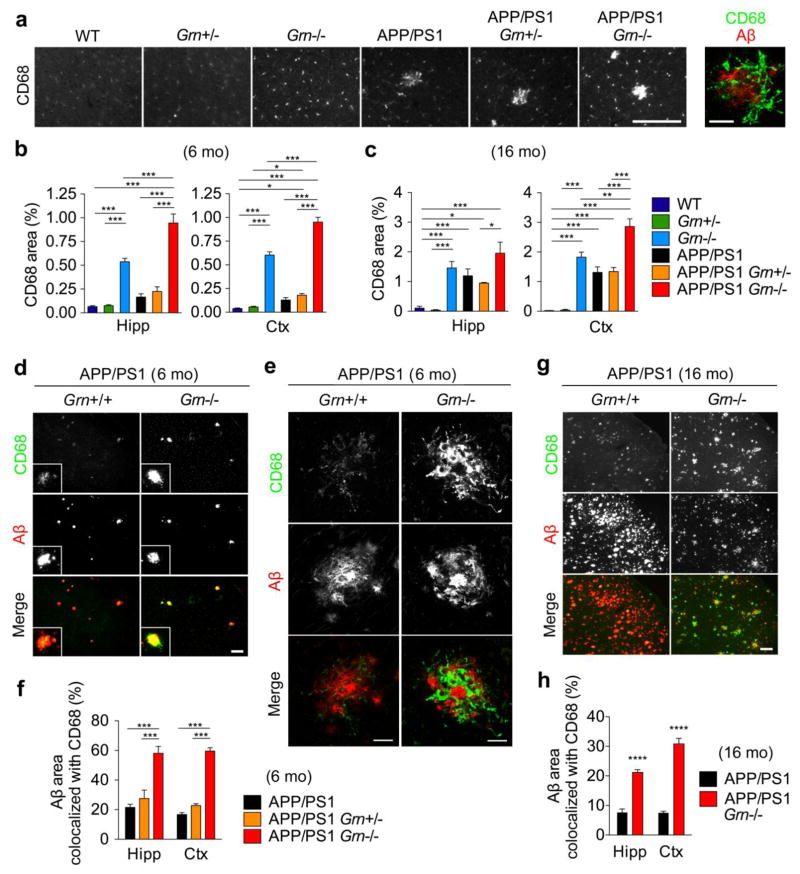
As a reduction in diffuse Aβ plaque in APP/PS1 Grn−/− mice is unexpected and inconsistent with a previous study [56], we sought to examine how PGRN deficiency prevents diffuse Aβ plaque growth. PGRN is involved in neuroinflammation in the brain and an increase in CD68, a common marker for activated microglia, has been reported in several aged Grn−/− strains as well as human PGRN-deficient FTLD cases [82,55,56,75,48]. In addition, accumulating evidence suggests that microglia paly an important role in Aβ clearance in AD [49]. We thus first examined whether PGRN deficiency affects microglial phenotypes by using anti-CD68 antibody in 6- and 16-month-old WT, Grn+/−, Grn−/−, APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice (6 genotypes). CD68 immunohistochemistry shows a significant increase in CD68-immunoreactive area for Grn−/− mice, and an even greater increase for APP/PS1 Grn−/− mice in both hippocampus and cortex of 6- and 16-month-old animals although the increase in hippocampus at 16 months of age does not reach statistical significance (Fig. 3a, 3b, and 3c). At 6 months, the Grn−/− phenotype is substantially more prominent than the APP/PS1 single gene increase of 3-fold magnitude (Fig. 3a and 3b). The CD68 increase is present at 6 months of age, when Aβ plaques are first forming, and at 16 months, when Aβ plaque density is maximal, the increase reaches 30-fold WT levels in APP/PS1 and 50-fold WT levels in APP/PS1 Grn−/− mice (Fig. 3c and 3g). For APP/PS1 hippocampus and cerebral cortex, the increase of CD68-positive microglia is most prominent near Aβ plaques. High-resolution confocal microscopy confirms that the enhanced CD68-immnoreactive microglial processes interdigitate with Aβ plaques in APP/PS1 Grn−/− mice (Fig. 3a and 3e). Quantitatively, the CD68-positive area co-localized with the vicinity of Aβ immunoreactivity is significantly greater for both 6-month and 16-month-old APP/PS1 Grn−/− mice, compared with APP/PS1 mice (Fig. 3f, 3g, and 3h).
Figure 3. PGRN deficiency increases CD68-positive microglia near Aβ plaques in APP/PS1 mice.
a. Representative images of CD68 immunostaining of frontal cortex of 6-month-old WT, Grn+/−, Grn−/−, APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice. Bar, 200 μm. The right panel shows a high-resolution image of CD68 and Aβ double immunostaining in the massive CD68-immunoreactive area in the sample of APP/PS1 Grn−/− mice. Bar, 20 μm.
b. Quantification of CD68-positive area (%) in 6-month-old WT, Grn+/−, Grn−/−, APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice. Mean ± SEM, n = 5/genotype, *p < 0.05, ***p < 0.001; one-way ANOVA, with Tukey post hoc correction.
c. Quantification of CD68-positive area (%) in 16-month-old WT, Grn+/−, Grn−/−, APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice. Mean ± SEM, n = 9 (WT), 4 (Grn+/−, Grn−/−, APP/PS1 Grn+/−), 6 (APP/PS1), or 5 (APP/PS1 Grn−/−), *p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA, with Tukey post hoc correction.
d. Representative images of Aβ and CD68 double immunostaining of frontal cortex of 6-month-old APP/PS1 and APP/PS1 Grn−/− mice. Bar, 200 μm.
e. Representative confocal high-resolution images of CD68 and Aβ co-staining of frontal cortex (corresponding to Figure 3d inset) of 6-month-old APP/PS1 and APP/PS1 Grn−/− mice. The images were taken using the samples identical to the ones used in Figure 3D. Bar, 20 μm.
f. Quantification of CD68-positive Aβ area (%) in 6-month-old APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice. Mean ± SEM, n = 5/genotype, ***p < 0.001; one-way ANOVA, with Tukey post hoc correction.
g. Representative images of CD68 and Aβ co-staining of frontal cortex of 16-month-old APP/PS1 and APP/PS1 Grn−/− mice. Bar, 200 μm
h. Quantification of CD68-positive Aβ area (%) in 16-month-old APP/PS1 and APP/PS1 Grn−/− mice. Mean ± SEM, n = 5/genotype, ****p < 0.0001; T test.
We also examined whether PGRN deficiency affects microgliosis and microglial morphology in 6-month-old mice by using anti-Iba1 antibody, a general marker for microglia. Although PGRN absence slightly increases Iba1-immunocreactive area and Iba1-positive microglia in both hippocampus and cortex, regardless of APP/PS1 transgene, the increases fail to reach a statistical significance (Supplementary Fig 2a, 2b, and 2c). High-resolution confocal microscopy shows no significant morphological change by PGRN deficiency in Iba1-positive microglia despite a significant increase in CD68-immunoreactive area (Supplementary Fig. 3). These results using anti-Iba1 antibody are consistent with previous studies using different Grn−/− strains [75,1]. In addition, triple staining with ThioS, anti-Aβ antibody, and anti-Iba1 antibody shows that microgliosis rarely occurs near small ThioS-negative anti-Aβ-antibody-immunoreactive diffuse Aβ plaques while microglia accumulate robustly around ThioS-positive Aβ plaques in 6-month-old APP/PS1 mice, regardless of the presence of PGRN (Supplementary Fig. 4a and 4b). Together, these results indicate that PGRN deficiency significantly increases CD68-positive microglia in the brain globally, and does so near Aβ plaques in particular.
PGRN deficiency induces expression of TYROBP network genes
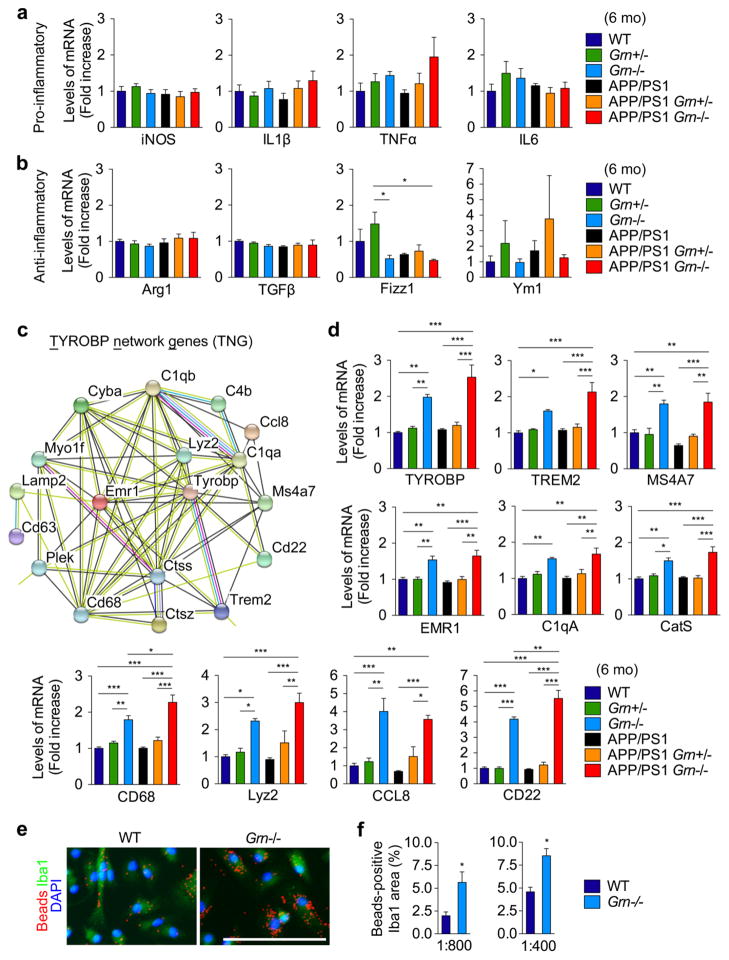
To further characterize microglial phenotypes induced by PGRN deficiency, we analyzed expression of typical pro-inflammatory and anti-inflammatory genes in frontal cortex of 6-month-old mice of the 6 genotypes by qRT-PCR. Despite an increase in CD68-positive microglia in Grn−/− background, there is no significant difference in typical pro-inflammatory (iNOS, IL1β, TNFα, and IL6) and anti-inflammatory genes (Arg-1, TGFβ, Fizz1, and Ym1) between WT and Grn−/− mice or APP/PS1 and APP/PS1 Grn−/− mice (Fig. 4a and 4b). In order to explore changes of any other microglial genes by PGRN deficiency, we performed genome-wide RNA sequencing (RNA-seq) using 2-month-old WT and Grn−/− brains (Z. Klein et al., unpublished observations) and analyzed up-regulated genes in Grn−/− brains (FDR < 0.05, logFC > 0.2) using the database STRING website [76]. In a functional protein-protein interaction module of differentially expressed genes detected by the STRING, there is a cluster with TYROBP forming a major hub that consists of many microglia-related genes including an AD risk factor Trem2 [14], as well as Ms4a7, one member of the AD risk MS4A gene cluster [41]. Details of the 36 genes in the cluster are summarized in Supplementary Table 2. We also sought to validate our data using the microarray dataset in a previous study with 6- or 9-month-old different Grn−/− strain [65]. Interestingly, 18 of 36 genes in the cluster from our study were also up-regulated in the microarray dataset. We further analyzed them by the STRING and named the genes in the cluster TYROBP network gene (TNG) (Fig. 4c, Supplementary Fig. 5). Consistent with the immunohistochemical data, Cd68 is also found in the TNG. Several lysosomal cysteine proteases including cathepsin S and cathepsin Z, which are highly expressed in immune/inflammatory cells, complement system proteins (C1qA, C1qB, and C4B), and Lyz2 (also known as Lysozyme M (LysM)) are also in the TNG. By using qRT-PCR of 6 genotypes, up-regulation of at least 10 of 18 genes in the TNG was confirmed in frontal cortex of 6-month-old Grn−/− mice, regardless of APP/PS1 transgenes (Fig. 4d). Since previous studies have reported that TREM2 and TYROBP are involved in microglial phagocytosis, which in turn is implicated in Aβ accumulation in AD [14], we next sought to test the effect of increased TNG expression by PGRN deficiency on microglial phagocytic function. Grn−/− microglia engulf a significantly greater amount of fluorescent beads compared to WT cells at 2 different concentrations (Fig. 4e and 4f). Together, these results show that PGRN deficiency causes TNG expression and facilitates microglial phagocytosis. Thus, increased phagocytosis by TNG-positive microglia surrounding ThioS-positive Aβ plaques is the likely proximal cause for decreased diffuse Aβ plaque growth in PGRN deficient APP/PS1 mice.
Figure 4. PGRN deficiency induces expression of TYROBP network genes.
a. Quantitative real-time PCR analysis of transcription of pro-inflammatory genes (iNOS, IL1β, TNFα, and IL6) in 6-month-old WT, Grn+/−, Grn−/−, APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice. Mean ± SEM, n = 6 (IL1β and TNFα) or 4 (iNOS and IL6)/genotype.
b. Quantitative real-time PCR analysis of transcription of anti-inflammatory genes (Arg, IL1β, TNFα, and IL6) in 6-month-old WT, Grn+/−, Grn−/−, APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice. Mean ± SEM, n = 6 (Fizz1 and Ym1) or 4 (Arg1 and TGFβ)/genotype.
c. TYROBP network genes (TNG) identified by STRING database analysis of up-regulated genes (FDR < 0.05, logFC > 0.2) in our genome-wide RNA-seq analysis and a previous microarray study. For more detail information, see Supplementary Figure 5 and Supplementary Table 2.
d. Quantitative real-time PCR analysis of transcription of TYROBP network genes (TYROBP, TREM2, MS4A7, EMR1, C1qA, CD68, cathepsin S (CatS), Lyz2, CD22, and CCL8) in 6-month-old WT, Grn+/−, Grn−/−, APP/PS1, APP/PS1 Grn+/−, and APP/PS1 Grn−/− mice. Mean ± SEM, n = 4/genotype. *p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA, with Tukey post hoc correction.
e. Representative images of fluorescent beads (1:400 dilution) phagocytosis assay using WT and Grn−/− primary microglia. Bar, 100 μm.
f. Quantification of Beads-positive Iba1 area (%) in fluorescent beads phagocytosis assay using two different dilutions 1:400 and 1:800. Mean ± SEM, n = 3. *p < 0.05; T test.
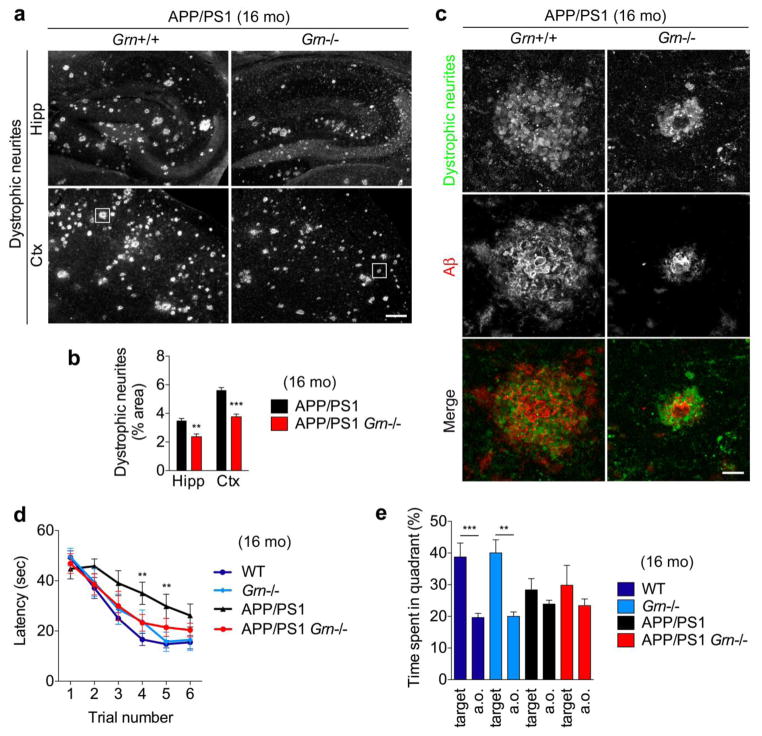
PGRN deficiency reduces dystrophic neurites and partially restores behavior deficits of APP/PS1 mice
Given the detectable reduction of diffuse Aβ plaques in 16-month-old APP/PS1 Grn−/− mice, we examined whether PGRN deficiency affects axonal dystrophy and a memory deficit in APP/PS1 mice. LAMP-1-positive dystrophic neurites are associated with all sizes and ages of Aβ plaques and are present from the earliest detectable stages of Aβ deposition in AD mouse models [28]. We performed triple staining using ThioS, anti-Aβ (2454) antibody, and anti-LAMP-1 antibody and detected the dystrophic neurites even in ThioS-negative and anti-Aβ-antibody-immunoreactive diffuse Aβ plaques in both 6- and 16-month-old APP/PS1 mice (Supplementary Fig. 6a and 6b). This demonstrates an association of the LAMP-1-positive dystrophic neurites with diffuse Aβ plaques, regardless of the presence of ThioS-positive amyloid filaments. Consistent with diffuse Aβ plaque results, the dystrophic neurites are significantly reduced in both hippocampus and cortex in APP/PS1 Grn−/− mice (Fig. 5a, 5b and 5c). To examine spatial memory, we tested 16-month-old APP/PS1 Grn−/− mice in the Morris water maze. The APP/PS1 mice require a longer latency time to locate a hidden platform across forward learning trials and spend less time in the target quadrant during the probe trial 24 hours after learning trials compared to WT mice, as previously reported [26] (Fig. 5d and 5e). PGRN deficiency reduces the spatial learning acquisition deficit in APP/PS1 mice at certain time points, although APP/PS1 Grn−/− mice are indistinguishable from APP/PS1 mice in the probe trial (Fig. 5d and 5e). For APP/PS1 mice, the PGRN null state reduces axonal dystrophy and improves memory impairment moderately, in contradistinction to enhanced AD risk in humans with PGRN reduction.
Figure 5. PGRN deficiency reduces axonal dystrophy and a memory deficit in APP/PS1 mice.
a. Representative images of LAMP-1-positive dystrophic neurites of hippocampus (Hipp) and cortex (Ctx) of 16-month-old APP/PS1 and APP/PS1 Grn−/− mice. Bar, 200 μm.
b. Quantification of Lamp-1-positive dystrophic neurite area (%) in hippocampus (Hipp) and cortex (Ctx) of 16-month-old APP/PS1 and APP/PS1 Grn−/− mice. Mean ± SEM, n = 5. **p < 0.01, ***p < 0.001; T test.
c. Representative confocal high-resolution images of LAMP-1-positive dystrophic neurites and Aβ co-staining of frontal cortex (corresponding to Figure 6a white square) of APP/PS1 and APP/PS1 Grn−/− mice. Bar, 20 μm.
d. MWM forward learning trial of Aged WT, Grn−/−, APP/PS1, and APP/PS1 Grn−/− mice. Spatial learning is plotted as latency to find hidden platform. Mean ± SEM, WT, n = 14; Grn−/−, n=8; APP/PS1, n=12; APP/PS1 Grn−/−, n=7. Performance differed across the last 4 trials by genotypes. There was an interaction between APP/PS1 transgenes and Grn genotype. **p < 0.01; two-way RM ANOVA. By Bonferroni post hoc comparisons, the APP/PS1 group differed from the WT group (***p < 0.001). In the trial number 3 and 4, the APP/PS1 group differed from the WT group, whereas none of the other groups differed from each other, as shown in Figure. **p < 0.01; one-way ANOVA, with Tukey post hoc correction.
e. MWM probe trial after reverse trial of Aged WT, Grn−/−, APP/PS1, and APP/PS1 Grn−/− mice. Percentage of time spent in the target quadrant and averaged time spent in all other (a.o.) quadrants. Mean ± SEM, WT, n = 14; Grn−/−, n=8; APP/PS1, n=12; APP/PS1 Grn−/−, n=7. **p < 0.01, ***p < 0.001; T-test.
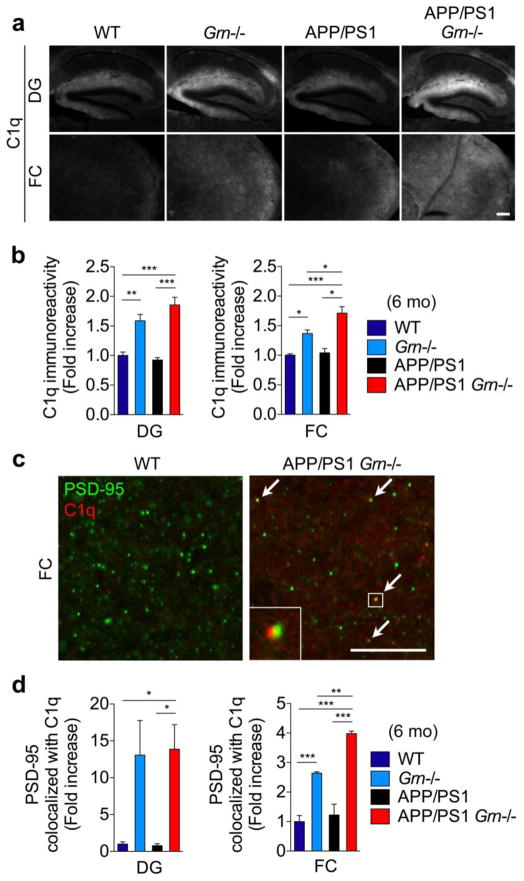
PGRN deficiency increases accumulation of C1q on neuronal synapses
To explain the contradiction described above as well as moderate effects of PGRN deficiency on behavior deficits despite a significant reduction in diffuse Aβ plaques in APP/PS1 mice, we considered whether other phenotypes and molecular pathways might be modulated differentially. Our human data with the GRN AD risk variant have shown an increase in CSF tau levels, which reflect neuronal damage accompanied by neurofibrillary tangles. Recent studies indicate that complement activation, especially an increase in C1q, mediates neuronal synaptic damage in AD and PGRN null mice [53,36]. As the TNG also includes C1qA and C1qB, we examined whether PGRN deficiency affects synaptic accumulation of C1q in 6-month-old APP/PS1 mice. Consistent with the RNA analyses, C1q immunoreactivity is increased in dentate gyrus and frontal cortex in Grn−/− mice (Fig. 6a and 6b). Although there is no significant increase in the C1q immunoreactivity in APP/PS1 mice, the immunoreactivity is further increased in frontal cortex of APP/PS1 Grn−/− mice, compared to Grn−/− mice (Fig. 6a and 6b). Importantly, the staining pattern of the anti-C1q antibody is consistent with several previous studies [72,53,12]. We also tested another anti-C1q antibody that is validated by C1q null mice [72,36], and confirmed a similar distribution pattern of C1q in both dentate gyrus and frontal cortex and an increase in the immunoreactivity in APP/PS1 Grn−/− mice (Supplementary Fig. 7). High-resolution imaging analyses of PSD-95 and C1q co-localization show an increase in the co-localization in Grn−/− and APP/PS1 Grn−/− mice (Fig. 6c and 6d). The co-localization pattern of PSD-95 and C1q is also similar to that in a previous study [36]. These results suggest that PGRN deficiency enhances synaptic accumulation of C1q that has been reported to cause neuronal damage [53,36] in APP/PS1 mice. Thus, while PGRN deficiency-induced microglial TNG expression protects against diffuse Aβ plaques, it simultaneously has a detrimental effect on neurons, a downstream target of Aβ accumulation, which can also, at least in part, explain the moderate effects on behavior deficits in PGRN-deficient APP/PS1 mice.
Figure 6. PGRN deficiency increases C1q deposition on PSD-95 positive synapses.
a. Representative images of C1q staining of dentate gyrus (DG) and frontal cortex (FC) of 6-month-old WT, Grn−/−, APP/PS1 and APP/PS1 Grn−/− mice. Bar, 200 μm.
b. Quantification of C1q-immunoreactivity in DG and FC of 6-month-old WT, Grn−/−, APP/PS1 and APP/PS1 Grn−/− mice. Mean ± SEM, n = 4/genotype, *p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA, with Tukey post hoc correction.
c. Representative confocal high-resolution images of PSD-95 and C1q co-staining of FC of 6-month-old WT and APP/PS1 Grn−/− mice. Arrows indicate colocalization of PSD-95 and C1q puncta. Bar, 10 μm.
d. Quantification of PSD-95 and C1q colocalization in FC of 6-month-old WT, Grn−/−, APP/PS1 and APP/PS1 Grn−/− mice. Mean ± SEM, n = 4/genotype, **p < 0.01, ***p < 0.001; one-way ANOVA, with Tukey post hoc correction.
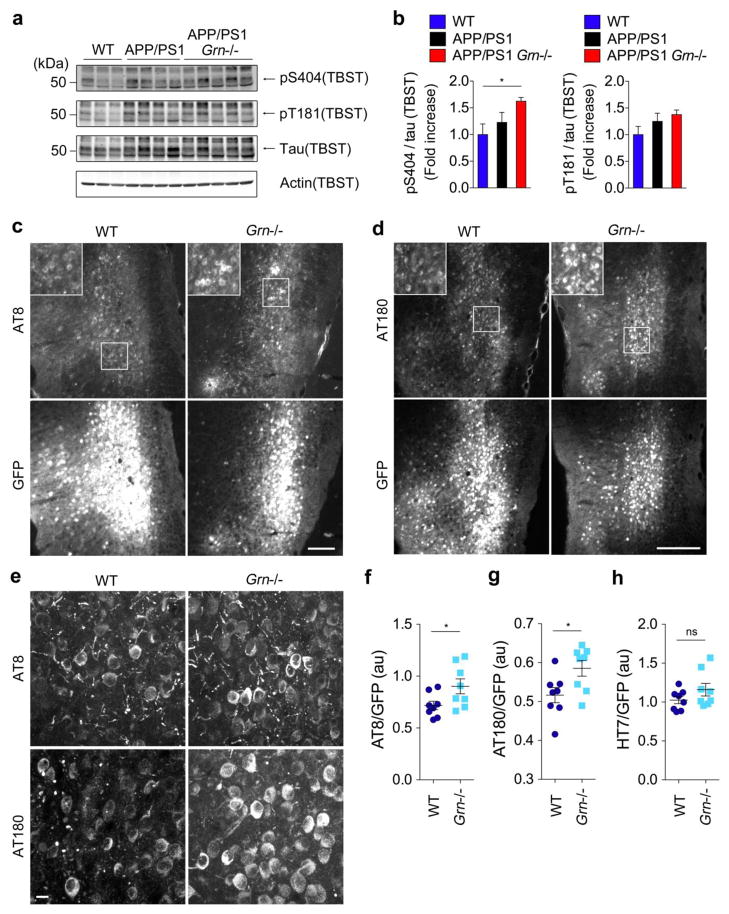
Tau pathology is exacerbated in human P301L tau-expressing Grn−/− mice
The CSF tau data and the results of enhanced synaptic C1q accumulation in Grn−/− mice prompted us to further examine the effects of PGRN deficiency on neuronal tau dysfunction. We first tested whether tau phosphorylation is increased in APP/PS Grn−/− mice by immunoblot analysis using phospho-specific antibodies. Although no difference in tau phosphorylation between 16-month-old WT and APP/PS1 mice is observed, there is a significant increase in phosphorylation of tau at S404 in 16-month-old APP/PS1 Grn−/− mice, compared with WT mice (Fig. 7a and 7b). A trend toward an increased tau phosphorylation at T181 is also observed but does not reach statistical significance. It should be noted that APP/PS1 mice do not exhibit tau pathologies including neurofibrillary tangles. Therefore, to further investigate human tau pathology in the absence of Aβ accumulation, we took advantage of adeno-associated virus (AAV) vector that expresses human P301L tau 1–441 under the control of the neuron-specific synapsin-1 promoter. This AAV vector has been used to accelerate tau pathologies in mice and prominent AT8 pathology has been detected at least at 28 days post injection [2]. We injected the AAV expressing human P301L tau (and GFP) into the entorhinal cortex of WT and Grn−/− mice (Supplementary Fig. 8a) and analyzed tau pathologies at 28 days post injection. In order to detect tau pathology, we used AT8 (PHF phospho-S202 and phospho-T205) and AT180 (PHF phospho-T231) antibodies because they have been most commonly used for this purpose and are commercially available. Immunohistochemical analysis shows that both AT8 and AT180 signals are significantly increased in Grn−/− mice compared to WT (Fig. 7c, 7d, 7f, and 7g). High-resolution confocal microscopy confirms an increase in neuronal AT8 and AT180 pathologies in Grn−/− mice (Fig. 7e). In contrast, HT7 staining for human tau shows no difference in total viral-mediated expression of human mutant tau in entorhinal cortex between WT and Grn−/− mice (Fig. 7h, Supplementary Fig. 8b). It should be noted that the human CSF (Fig. 1) and mouse brain tissue (Fig. 7) showed differential effects of GRN on phospho-tau and total tau. Nonetheless, the human data from GRN AD risk variants together with mouse data from human P301L tau-expression in knockouts show that PGRN reduction exacerbates tau pathology in AD irrespective of Aβ pathology.
Figure 7. PGRN deficiency exacerbates tau pathology in human P301L tau-expressing Grn−/− mice.
a. Immunoblot analysis using anti-phospho-S404, anti-phospho-T181, anti-tau, and anti-actin antibodies of the TBST-soluble fractions in 16-month-old WT, APP/PS1, and APP/PS1 Grn−/− mice.
b. Quantification of immunoblot (a). Mean ± SEM, n = 3–5/genotype. *p < 0.05; one-way ANOVA, with Tukey post hoc correction.
c. Representative images of AT8 and GFP double immunostaining of entorhinal cortex of human P301L tau and GFP-expressing WT and Grn−/− mice. Bar, 200 μm.
d. Representative images of AT180 and GFP double immunostaining of entorhinal cortex of human P301L tau and GFP-expressing WT and Grn−/− mice. Bar, 200 μm.
e. Representative confocal high-resolution images of AT8 and A180 immunostaining of entorhinal cortex (corresponding to Figure 7c and 7d white squares) of human P301L tau and GFP-expressing WT and Grn−/− mice. Bar, 10 μm.
f. Quantification of AT8 signals normalized by GFP signals in human P301L tau and GFP-expressing WT and Grn−/− mice. Mean ± SEM, n = 8/genotype. *p < 0.05; T test.
g. Quantification of AT180 signals normalized by GFP signals in human P301L tau and GFP-expressing WT and Grn−/− mice. Mean ± SEM, n = 8/genotype. *p < 0.05; T test.
h. Quantification of HT7 signals normalized by GFP signals in human P301L tau and GFP-expressing WT and Grn−/− mice. Mean ± SEM, n = 8/genotype.
DISCUSSION
The major conclusions of the present study are that GRN AD risk variant increases CSF tau levels without affecting Aβ pathology in humans and that PGRN deficiency exacerbates tau pathology while it protects against diffuse Aβ plaques in mice. The opposing effects of PGRN deficiency observed in mice are, at least in part, associated with induction of microglial TNG expression, including TYROBP, an AD risk factor TREM2, and a complement protein C1q. Importantly, PGRN deficiency can modify microglial phenotype and enhance tau pathology independent of Aβ pathology.
The purpose of this study was to investigate how PGRN reduction by GRN rs5848 variant or GRN mutation could modify AD pathophysiology and increase AD risk and we unexpectedly found that GRN rs5848 T allele and PGRN deficiency have no exacerbating effects on Aβ pathology in humans and rodents respectively. Moreover, complete absence of PGRN prevents diffuse Aβ plaque growth leading to less severe axonal dystrophy and a partial rescue of behavior phenotypes in an APP/PS1 amyloidosis mouse model. This protective effect could be explained by an induction of the TNG expression by PGRN deficiency. Interestingly, the enhanced TNG expression is not due to an increase in microgliosis because PGRN absence has no significant effects on the number of Iba1-positive microglia or typical pro-inflammatory and anti-inflammatory genes at 6 months of age. There is also a further increase in expression of several TNG components in APP/PS1 Grn−/− mice, although we detect no significant changes in the TNG or pro-inflammatory or anti-inflammatory genes in APP/PS1 mice, in distinction to previous studies investigating neuroinfllammation in AD mouse models [68]. This discrepancy is likely explained by a difference in mouse age [33]. Many previous studies have used aged AD mouse models while we performed qRT-PCR analysis using 6-month-old animals. In fact, we observed a dramatic increase in CD68-immunoreactive area in 16-month-old APP/PS1 mice while there is no such change at 6 months of age. We also showed that PGRN deficiency facilitates microglial phagocytosis in vitro, consistent with a previous study using PGRN-deficient peripheral macrophages [40]. It is thus possible that increased TNG expression cooperatively facilitates microglial phagocytosis to reduce diffuse Aβ plaque pathology. Importantly, a recent study has shown up-regulation of very similar microglial gene network including TREM2, TYROBP, MS4A7, CD68, C1qA, C1qB, and Lyz2 by PGRN deficiency [53], supporting this conclusion. The mechanisms by which PGRN deficiency induces microglial TNG expression will require further investigation.
CD68 has been commonly used as a marker for “activated” microglia. However, we detected no significant changes in microglial morphology nor in expression of typical pro- and anti-inflammatory genes by 6-month-old Grn−/− mice despite the dramatic increase in CD68-immunoreactivity and TNG mRNA levels. It appears likely that CD68 is up-regulated as a part of the TNG involved in protein degradation following phagocytosis together with cathepsins CatS and CatZ in Grn−/− mice, rather than as a marker of typical microglial “activation”, although the precise role of CD68 in lysosome remains unclear. Our data thus caution against simple interpretation of CD68 immunohistochemistry as microglial activation. In future studies, careful characterization of microglia by gene expression analyses, in addition to CD68 immunostaining, will be necessary in order to determine specific microglial phenotypes.
A reduction in diffuse Aβ plaques is detected only in 16-month-old APP/PS1 Grn−/− mice despite increased TNG expression by PGRN deficiency at 6 months of age in our study. We found that microglia are rarely present around small ThioS-negative and anti-Aβ (2454) antibody-positive diffuse Aβ plaques whereas pronounced microglial accumulation is observed around ThioS-positive amyloid plaques even in 6-month-old APP/PS1 mice. Consistent with this result, we also found that PGRN deficiency has no significant effect on Iba-1-immunoreactive microgliosis on APP/PS1 background. Importantly, recent studies have shown that microglia and TREM2/TYROBP play a critical role in reducing the diffuse Aβ deposition associated with already-formed Aβ plaques and in reducing axonal dystrophy [83,77,15]. Our results thus suggest that Grn−/− microglia may prevent diffuse Aβ plaque growth from ThioS-positive dense-core amyloid plaques most efficiently, rather than delaying the initial formation (onset) of diffuse Aβ plaques. Furthermore, this is mediated, at least in part, by enhanced phagocytosis. In human florbetapir-PET analysis with the ADNI dataset, we have not seen a reduction of Aβ pathology by GRN AD risk variant. This result might be due to the PET tracer possessing a higher affinity for dense-core amyloid plaques, although the full in vivo specificity of the tracer remains to be determined [44].
Our Aβ plaque results are, however, inconsistent with a previous study using a different AD mouse model with conditional PGRN deletion [56]. One potential explanation for this contradiction is that the previous study utilized the LysM-cre mice lacking endogenous Lyz2, a gene significantly up-regulated in Grn−/− and APP/PS1 Grn−/− mice in previous [53,65] and our studies. Humans have one lysozyme gene, while mice have two encoding LyzM and LyzP [23], and the gene product lysozyme is reportedly increased in CSF of AD patients and has a protective role in Aβ aggregation in vitro and in fly [34]. Lyz2 deficiency alone might affect Aβ pathology directly or indirectly by compromising TNG-related microglial phenotypes by PGRN deficiency. Alternatively, artificial PGRN reduction in a subset of neurons and microglia by LysM-cre mice [5] and/or AD mouse models used may yield complicated outcomes reflecting the varied and multifactorial and indirect actions of PGRN on Aβ pathology. However, regardless of difference in mouse models, our analyses of human florbetapir-PET and CSF Aβ42 dataset clearly suggest that GRN rs5848 T allele increases AD risk not by altering Aβ accumulation.
In contrast to Aβ pathology, our analysis of ADNI database shows that GRN rs5848 T allele increases CSF tau levels in a T allele dose-dependent manner. The difference between GRN rs5848 genotype CC and TT is greater than the one between NL and EMCI in the ADNI cohort, suggesting an importance of this change for increasing AD risk. We also found that PGRN deficiency increases tau AT8 and AT180 pathologies in human P301L tau-expressing mice. PGRN haploinsufficiency causes FTLD with tau-negative and TDP-43-positive inclusions [3,60,19]. However, in several GRN mutations, tau pathology is observed in addition to TDP-43 inclusion, showing an association of GRN mutation with tau pathology [52,59,62]. Although we utilized the AAV vector to examine tau pathologies in mice, AAV injection can be associated with impairment of the blood-brain barrier and inflammation during the period in which abnormal tau aggregates are formed. In addition, as expression from AAV vector may be transient, germline-modified tauopathy models may be closer to chronic human diseases. Of note, the recent report that PGRN haploinsufficiency increases tau phosphorylation in P301L tau transgenic mice [37], strongly supports our conclusion from AAV studies. The mechanism by which PGRN reduction increases CSF tau levels and tau pathology will be a fruitful site for further investigation. Both cell-autonomous and non-cell-autonomous mechanisms are possible, since neuroinflammation has been implicated in tau pathology as described previously [54]. The TNG-related microglial phenotype by PGRN deficiency might exacerbate tau phosphorylation despite a reduction in diffuse Aβ plaque pathology in a non-cell-autonomous manner, similar to previous studies using IL-1β-overexpressing mice and CX3CL1-deficient mice [25,51]. Our results of C1q immunostaining suggest that greater neuronal injury by enhanced C1q accumulation may exacerbate tau phosphorylation and release into the CSF in GRN AD risk variants. It will be interesting to analyze C1q levels in CSF or brain sections of humans with GRN AD risk variant.
It also remains unclear how the GRN AD risk variant affects CSF tau but not CSF p-tau181 in our human study and how it is related to our mouse studies showing an increase in AT8 and AT180 pathologies by PGRN deficiency. CSF phospho-tau is thought to reflect phosphorylation states of tau in the brain [6,30,67]. While CSF p-tau181 has been widely used in CSF biomarker studies, other p-tau epitopes such as p-tau199, p-tau231, p-tau404 are also increased in AD patients [6,30,67]. A study comparing p-tau181, p-tau199, and p-tau231 using the same samples shows similar discrimination between AD and control [6,31]. Previous CSF biomarker studies as well as the ADNI dataset in this study suggest that p-tau181-negative p-tau species are also present in the brain and released into the CSF of AD patients. Interestingly, although CSF p-tau181 shows a discrimination between AD and control, the relationship between CSF p-tau181 and AT8 (p-tau202+205) and AT180 (p-tau231) pathologies is still controversial and CSF p-tau231 showed a better correlation in AD in previous studies [9,10]. In the present study of APP/PS1 mice lacking PGRN, we detected no significant difference in phospho-T181 tau while there was an increase in phospho-S404. Therefore, one potential hypothesis to connect between our human and rodent results is that the GRN AD risk variant or PGRN reduction might affect p-tau181-negative p-tau species to cause AT8 and AT180 tau pathologies. Again, further investigations using the postmortem brain with GRN AD risk variant as well as genetically modified tauopathy mouse model will be needed to confirm this hypothesis.
Despite evidence that PGRN haploinsufficiency causes FTLD in human, most of previous studies have failed to see significant effects with mechanistic insight in Grn+/− mice [42,1,61,80,27,53]. In the present study, we also observed no obvious phenotypic changes in Grn+/− mice with or without APP/PS1 transgene, compared to WT. The mechanism by which PGRN reduction differentially affects human and mouse brain functions is currently unclear and further investigation will be required.
In summary, studies with an amyloidosis mouse model lacking PGRN and genetic associations in the ADNI database argue against global PGRN reduction having an exacerbating effect on Aβ accumulation. Critically, as an alternative hypothesis for an increased AD risk by PGRN reduction, our studies of human CSF tau levels and human P301L tau-expressing mice show a role of PGRN in tau pathology. Furthermore, identification and characterization of TNG and a TNG-related microglial phenotype in this study provide novel insights into understanding of neurodegenerative disorders including AD and FTLD.
Supplementary Material
Acknowledgments
We thank Stefano Sodi and Yiguang Fu for assistance with mouse husbandry. We thank Janghoo Lim for help with the C1000 Thermal Cycler. This work was supported by grants from NIH, BrightFocus Foundation, Alzheimer’s Association and Falk Medical Research Trust to S.M.S. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare;; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Cellular Neuroscience, Neurodegeneration and Repair Program, Departments of Neurology and Neurobiology, Yale University School of Medicine, New Haven, CT 06536 USA.
Disclosure: S.M.S. is a co-founder of Axerion Therapeutics, seeking to develop NgR- and PrP-based therapeutics.
AUTHOR CONTRIBUTIONS
H.T. and S.M.S. designed various aspects of the research. Z.A.K. performed the RNA-seq and the behavior test. S.M.B. and A.C.K. performed the surgeries. T.I. prepared the AAV. H.T. performed the other experiments. H.T., Z.A.K., M.A.K. and S.M.S. analyzed various parts of the data. H.T., S.M.B., and S.M.S. contributed to the writing of the paper.
References
- 1.Ahmed Z, Sheng H, Xu YF, Lin WL, Innes AE, Gass J, Yu X, Wuertzer CA, Hou H, Chiba S, Yamanouchi K, Leissring M, Petrucelli L, Nishihara M, Hutton ML, McGowan E, Dickson DW, Lewis J. Accelerated lipofuscinosis and ubiquitination in granulin knockout mice suggest a role for progranulin in successful aging. Am J Pathol. 2010;177:311–324. doi: 10.2353/ajpath.2010.090915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai H, Ikezu S, Tsunoda S, Medalla M, Luebke J, Haydar T, Wolozin B, Butovsky O, Kugler S, Ikezu T. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci. 2015;18:1584–1593. doi: 10.1038/nn.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, Cannon A, Dwosh E, Neary D, Melquist S, Richardson A, Dickson D, Berger Z, Eriksen J, Robinson T, Zehr C, Dickey CA, Crook R, McGowan E, Mann D, Boeve B, Feldman H, Hutton M. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 4.Bhagat SM, Butler SS, Taylor JR, McEwen BS, Strittmatter SM. Erasure of fear memories is prevented by Nogo Receptor 1 in adulthood. Molecular psychiatry. 2016;21:1281–1289. doi: 10.1038/mp.2015.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blank T, Prinz M. CatacLysMic specificity when targeting myeloid cells? Eur J Immunol. 2016;46:1340–1342. doi: 10.1002/eji.201646437. [DOI] [PubMed] [Google Scholar]
- 6.Blennow K, Hampel H. CSF markers for incipient Alzheimer’s disease. The Lancet Neurology. 2003;2:605–613. doi: 10.1016/s1474-4422(03)00530-1. [DOI] [PubMed] [Google Scholar]
- 7.Brouwers N, Nuytemans K, van der Zee J, Gijselinck I, Engelborghs S, Theuns J, Kumar-Singh S, Pickut BA, Pals P, Dermaut B, Bogaerts V, De Pooter T, Serneels S, Van den Broeck M, Cuijt I, Mattheijssens M, Peeters K, Sciot R, Martin JJ, Cras P, Santens P, Vandenberghe R, De Deyn PP, Cruts M, Van Broeckhoven C, Sleegers K. Alzheimer and Parkinson diagnoses in progranulin null mutation carriers in an extended founder family. Arch Neurol. 2007;64:1436–1446. doi: 10.1001/archneur.64.10.1436. [DOI] [PubMed] [Google Scholar]
- 8.Brouwers N, Sleegers K, Engelborghs S, Maurer-Stroh S, Gijselinck I, van der Zee J, Pickut BA, Van den Broeck M, Mattheijssens M, Peeters K, Schymkowitz J, Rousseau F, Martin JJ, Cruts M, De Deyn PP, Van Broeckhoven C. Genetic variability in progranulin contributes to risk for clinically diagnosed Alzheimer disease. Neurology. 2008;71:656–664. doi: 10.1212/01.wnl.0000319688.89790.7a. [DOI] [PubMed] [Google Scholar]
- 9.Buerger K, Alafuzoff I, Ewers M, Pirttila T, Zinkowski R, Hampel H. No correlation between CSF tau protein phosphorylated at threonine 181 with neocortical neurofibrillary pathology in Alzheimer’s disease. Brain: a journal of neurology. 2007;130:e82. doi: 10.1093/brain/awm140. [DOI] [PubMed] [Google Scholar]
- 10.Buerger K, Ewers M, Pirttila T, Zinkowski R, Alafuzoff I, Teipel SJ, DeBernardis J, Kerkman D, McCulloch C, Soininen H, Hampel H. CSF phosphorylated tau protein correlates with neocortical neurofibrillary pathology in Alzheimer’s disease. Brain: a journal of neurology. 2006;129:3035–3041. doi: 10.1093/brain/awl269. [DOI] [PubMed] [Google Scholar]
- 11.Cenik B, Sephton CF, Kutluk Cenik B, Herz J, Yu G. Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J Biol Chem. 2012;287:32298–32306. doi: 10.1074/jbc.R112.399170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung WS, Verghese PB, Chakraborty C, Joung J, Hyman BT, Ulrich JD, Holtzman DM, Barres BA. Novel allele-dependent role for APOE in controlling the rate of synapse pruning by astrocytes. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:10186–10191. doi: 10.1073/pnas.1609896113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark CM, Pontecorvo MJ, Beach TG, Bedell BJ, Coleman RE, Doraiswamy PM, Fleisher AS, Reiman EM, Sabbagh MN, Sadowsky CH, Schneider JA, Arora A, Carpenter AP, Flitter ML, Joshi AD, Krautkramer MJ, Lu M, Mintun MA, Skovronsky DM, Group A-AS. Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-beta plaques: a prospective cohort study. The Lancet Neurology. 2012;11:669–678. doi: 10.1016/S1474-4422(12)70142-4. [DOI] [PubMed] [Google Scholar]
- 14.Colonna M, Wang Y. TREM2 variants: new keys to decipher Alzheimer disease pathogenesis. Nat Rev Neurosci. 2016;17:201–207. doi: 10.1038/nrn.2016.7. [DOI] [PubMed] [Google Scholar]
- 15.Condello C, Yuan P, Schain A, Grutzendler J. Microglia constitute a barrier that prevents neurotoxic protofibrillar A beta 42 hotspots around plaques. Nature communications. 2015;6 doi: 10.1038/Ncomms7176. Artn 6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coppola G, Karydas A, Rademakers R, Wang Q, Baker M, Hutton M, Miller BL, Geschwind DH. Gene expression study on peripheral blood identifies progranulin mutations. Ann Neurol. 2008;64:92–96. doi: 10.1002/ana.21397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortini F, Fenoglio C, Guidi I, Venturelli E, Pomati S, Marcone A, Scalabrini D, Villa C, Clerici F, Dalla Valle E, Mariani C, Cappa S, Bresolin N, Scarpini E, Galimberti D. Novel exon 1 progranulin gene variant in Alzheimer’s disease. European journal of neurology. 2008;15:1111–1117. doi: 10.1111/j.1468-1331.2008.02266.x. [DOI] [PubMed] [Google Scholar]
- 18.Cruchaga C, Kauwe JS, Harari O, Jin SC, Cai Y, Karch CM, Benitez BA, Jeng AT, Skorupa T, Carrell D, Bertelsen S, Bailey M, McKean D, Shulman JM, De Jager PL, Chibnik L, Bennett DA, Arnold SE, Harold D, Sims R, Gerrish A, Williams J, Van Deerlin VM, Lee VM, Shaw LM, Trojanowski JQ, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD, Peskind ER, Galasko D, Fagan AM, Holtzman DM, Morris JC, Goate AM Consortium G, Alzheimer’s Disease Neuroimaging I, Alzheimer Disease Genetic C. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;78:256–268. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, van Duijn C, Peeters K, Sciot R, Santens P, De Pooter T, Mattheijssens M, Van den Broeck M, Cuijt I, Vennekens K, De Deyn PP, Kumar-Singh S, Van Broeckhoven C. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 20.Fagan AM, Mintun MA, Mach RH, Lee SY, Dence CS, Shah AR, LaRossa GN, Spinner ML, Klunk WE, Mathis CA, DeKosky ST, Morris JC, Holtzman DM. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Annals of neurology. 2006;59:512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- 21.Fagan AM, Mintun MA, Shah AR, Aldea P, Roe CM, Mach RH, Marcus D, Morris JC, Holtzman DM. Cerebrospinal fluid tau and ptau(181) increase with cortical amyloid deposition in cognitively normal individuals: implications for future clinical trials of Alzheimer’s disease. EMBO molecular medicine. 2009;1:371–380. doi: 10.1002/emmm.200900048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenoglio C, Galimberti D, Cortini F, Kauwe JS, Cruchaga C, Venturelli E, Villa C, Serpente M, Scalabrini D, Mayo K, Piccio LM, Clerici F, Albani D, Mariani C, Forloni G, Bresolin N, Goate AM, Scarpini E. Rs5848 variant influences GRN mRNA levels in brain and peripheral mononuclear cells in patients with Alzheimer’s disease. J Alzheimers Dis. 2009;18:603–612. doi: 10.3233/JAD-2009-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganz T, Gabayan V, Liao HI, Liu L, Oren A, Graf T, Cole AM. Increased inflammation in lysozyme M-deficient mice in response to Micrococcus luteus and its peptidoglycan. Blood. 2003;101:2388–2392. doi: 10.1182/blood-2002-07-2319. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Alloza M, Robbins EM, Zhang-Nunes SX, Purcell SM, Betensky RA, Raju S, Prada C, Greenberg SM, Bacskai BJ, Frosch MP. Characterization of amyloid deposition in the APPswe/PS1dE9 mouse model of Alzheimer disease. Neurobiology of disease. 2006;24:516–524. doi: 10.1016/j.nbd.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh S, Wu MD, Shaftel SS, Kyrkanides S, LaFerla FM, Olschowka JA, O’Banion MK. Sustained interleukin-1beta overexpression exacerbates tau pathology despite reduced amyloid burden in an Alzheimer’s mouse model. J Neurosci. 2013;33:5053–5064. doi: 10.1523/JNEUROSCI.4361-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gimbel DA, Nygaard HB, Coffey EE, Gunther EC, Lauren J, Gimbel ZA, Strittmatter SM. Memory impairment in transgenic Alzheimer mice requires cellular prion protein. J Neurosci. 2010;30:6367–6374. doi: 10.1523/JNEUROSCI.0395-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gotzl JK, Mori K, Damme M, Fellerer K, Tahirovic S, Kleinberger G, Janssens J, van der Zee J, Lang CM, Kremmer E, Martin JJ, Engelborghs S, Kretzschmar HA, Arzberger T, Van Broeckhoven C, Haass C, Capell A. Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol. 2014;127:845–860. doi: 10.1007/s00401-014-1262-6. [DOI] [PubMed] [Google Scholar]
- 28.Gowrishankar S, Yuan P, Wu Y, Schrag M, Paradise S, Grutzendler J, De Camilli P, Ferguson SM. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc Natl Acad Sci U S A. 2015;112:E3699–3708. doi: 10.1073/pnas.1510329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hafler BP, Klein ZA, Jimmy Zhou Z, Strittmatter SM. Progressive retinal degeneration and accumulation of autofluorescent lipopigments in Progranulin deficient mice. Brain Res. 2014;1588:168–174. doi: 10.1016/j.brainres.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hampel H, Blennow K, Shaw LM, Hoessler YC, Zetterberg H, Trojanowski JQ. Total and phosphorylated tau protein as biological markers of Alzheimer’s disease. Exp Gerontol. 2010;45:30–40. doi: 10.1016/j.exger.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hampel H, Buerger K, Zinkowski R, Teipel SJ, Goernitz A, Andreasen N, Sjoegren M, DeBernardis J, Kerkman D, Ishiguro K, Ohno H, Vanmechelen E, Vanderstichele H, McCulloch C, Moller HJ, Davies P, Blennow K. Measurement of phosphorylated tau epitopes in the differential diagnosis of Alzheimer disease: a comparative cerebrospinal fluid study. Archives of general psychiatry. 2004;61:95–102. doi: 10.1001/archpsyc.61.1.95. [DOI] [PubMed] [Google Scholar]
- 32.Han MR, Schellenberg GD, Wang LS Alzheimer’s Disease Neuroimaging I. Genome-wide association reveals genetic effects on human Abeta42 and tau protein levels in cerebrospinal fluids: a case control study. BMC Neurol. 2010;10:90. doi: 10.1186/1471-2377-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heiss JK, Barrett J, Yu Z, Haas LT, Kostylev MA, Strittmatter SM. Early Activation of Experience-Independent Dendritic Spine Turnover in a Mouse Model of Alzheimer’s Disease. Cerebral cortex. 2016 doi: 10.1093/cercor/bhw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helmfors L, Boman A, Civitelli L, Nath S, Sandin L, Janefjord C, McCann H, Zetterberg H, Blennow K, Halliday G, Brorsson AC, Kagedal K. Protective properties of lysozyme on beta-amyloid pathology: implications for Alzheimer disease. Neurobiology of disease. 2015;83:122–133. doi: 10.1016/j.nbd.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 35.Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: the challenge of the second century. Science translational medicine. 2011;3:77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li SM, Ramakrishnan S, Merry KM, Shi QQ, Rosenthal A, Barres BA, Lemere CA, Selkoe DJ, Stevens B. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosokawa M, Arai T, Masuda-Suzukake M, Kondo H, Matsuwaki T, Nishihara M, Hasegawa M, Akiyama H. Progranulin reduction is associated with increased tau phosphorylation in P301L tau transgenic mice. Journal of neuropathology and experimental neurology. 2015;74:158–165. doi: 10.1097/NEN.0000000000000158. [DOI] [PubMed] [Google Scholar]
- 38.Hu F, Padukkavidana T, Vaegter CB, Brady OA, Zheng Y, Mackenzie IR, Feldman HH, Nykjaer A, Strittmatter SM. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68:654–667. doi: 10.1016/j.neuron.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, Copeland NG, Lee MK, Younkin LH, Wagner SL, Younkin SG, Borchelt DR. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 40.Kao AW, Eisenhut RJ, Martens LH, Nakamura A, Huang A, Bagley JA, Zhou P, de Luis A, Neukomm LJ, Cabello J, Farese RV, Jr, Kenyon C. A neurodegenerative disease mutation that accelerates the clearance of apoptotic cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4441–4446. doi: 10.1073/pnas.1100650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karch CM, Cruchaga C, Goate AM. Alzheimer’s Disease Genetics: From the Bench to the Clinic. Neuron. 2014;83:11–26. doi: 10.1016/j.neuron.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kayasuga Y, Chiba S, Suzuki M, Kikusui T, Matsuwaki T, Yamanouchi K, Kotaki H, Horai R, Iwakura Y, Nishihara M. Alteration of behavioural phenotype in mice by targeted disruption of the progranulin gene. Behav Brain Res. 2007;185:110–118. doi: 10.1016/j.bbr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 43.Kelley BJ, Haidar W, Boeve BF, Baker M, Shiung M, Knopman DS, Rademakers R, Hutton M, Adamson J, Kuntz KM, Dickson DW, Parisi JE, Smith GE, Petersen RC. Alzheimer disease-like phenotype associated with the c.154delA mutation in progranulin. Arch Neurol. 2010;67:171–177. doi: 10.1001/archneurol.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kepe V, Moghbel MC, Langstrom B, Zaidi H, Vinters HV, Huang SC, Satyamurthy N, Doudet D, Mishani E, Cohen RM, Hoilund-Carlsen PF, Alavi A, Barrio JR. Amyloid-beta positron emission tomography imaging probes: a critical review. J Alzheimers Dis. 2013;36:613–631. doi: 10.3233/JAD-130485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J, Castellano JM, Jiang H, Basak JM, Parsadanian M, Pham V, Mason SM, Paul SM, Holtzman DM. Overexpression of low-density lipoprotein receptor in the brain markedly inhibits amyloid deposition and increases extracellular A beta clearance. Neuron. 2009;64:632–644. doi: 10.1016/j.neuron.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S, Swaminathan S, Shen L, Risacher SL, Nho K, Foroud T, Shaw LM, Trojanowski JQ, Potkin SG, Huentelman MJ, Craig DW, DeChairo BM, Aisen PS, Petersen RC, Weiner MW, Saykin AJ Alzheimer’s Disease Neuroimaging I. Genome-wide association study of CSF biomarkers Abeta1–42, t-tau, and p-tau181p in the ADNI cohort. Neurology. 2011;76:69–79. doi: 10.1212/WNL.0b013e318204a397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kleinberger G, Capell A, Haass C, Van Broeckhoven C. Mechanisms of granulin deficiency: lessons from cellular and animal models. Mol Neurobiol. 2013;47:337–360. doi: 10.1007/s12035-012-8380-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lant SB, Robinson AC, Thompson JC, Rollinson S, Pickering-Brown S, Snowden JS, Davidson YS, Gerhard A, Mann DM. Patterns of microglial cell activation in frontotemporal lobar degeneration. Neuropathol Appl Neurobiol. 2014;40:686–696. doi: 10.1111/nan.12092. [DOI] [PubMed] [Google Scholar]
- 49.Lee CY, Landreth GE. The role of microglia in amyloid clearance from the AD brain. J Neural Transm (Vienna) 2010;117:949–960. doi: 10.1007/s00702-010-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee MJ, Chen TF, Cheng TW, Chiu MJ. rs5848 variant of progranulin gene is a risk of Alzheimer’s disease in the Taiwanese population. Neurodegener Dis. 2011;8:216–220. doi: 10.1159/000322538. [DOI] [PubMed] [Google Scholar]
- 51.Lee S, Xu G, Jay TR, Bhatta S, Kim KW, Jung S, Landreth GE, Ransohoff RM, Lamb BT. Opposing effects of membrane-anchored CX3CL1 on amyloid and tau pathologies via the p38 MAPK pathway. J Neurosci. 2014;34:12538–12546. doi: 10.1523/JNEUROSCI.0853-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leverenz JB, Yu CE, Montine TJ, Steinbart E, Bekris LM, Zabetian C, Kwong LK, Lee VM, Schellenberg GD, Bird TD. A novel progranulin mutation associated with variable clinical presentation and tau, TDP43 and alpha-synuclein pathology. Brain. 2007;130:1360–1374. doi: 10.1093/brain/awm069. [DOI] [PubMed] [Google Scholar]
- 53.Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, Shang Y, Oldham MC, Martens LH, Gao F, Coppola G, Sloan SA, Hsieh CL, Kim CC, Bigio EH, Weintraub S, Mesulam MM, Rademakers R, Mackenzie IR, Seeley WW, Karydas A, Miller BL, Borroni B, Ghidoni R, Farese RV, Jr, Paz JT, Barres BA, Huang EJ. Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell. 2016;165:921–935. doi: 10.1016/j.cell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maphis N, Xu G, Kokiko-Cochran ON, Jiang S, Cardona A, Ransohoff RM, Lamb BT, Bhaskar K. Reactive microglia drive tau pathology and contribute to the spreading of pathological tau in the brain. Brain. 2015;138:1738–1755. doi: 10.1093/brain/awv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martens LH, Zhang J, Barmada SJ, Zhou P, Kamiya S, Sun B, Min SW, Gan L, Finkbeiner S, Huang EJ, Farese RV., Jr Progranulin deficiency promotes neuroinflammation and neuron loss following toxin-induced injury. J Clin Invest. 2012;122:3955–3959. doi: 10.1172/JCI63113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minami SS, Min SW, Krabbe G, Wang C, Zhou Y, Asgarov R, Li Y, Martens LH, Elia LP, Ward ME, Mucke L, Farese RV, Jr, Gan L. Progranulin protects against amyloid beta deposition and toxicity in Alzheimer’s disease mouse models. Nat Med. 2014;20:1157–1164. doi: 10.1038/nm.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicholson AM, Finch NA, Thomas CS, Wojtas A, Rutherford NJ, Mielke MM, Roberts RO, Boeve BF, Knopman DS, Petersen RC, Rademakers R. Progranulin protein levels are differently regulated in plasma and CSF. Neurology. 2014;82:1871–1878. doi: 10.1212/WNL.0000000000000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pereson S, Wils H, Kleinberger G, McGowan E, Vandewoestyne M, Van Broeck B, Joris G, Cuijt I, Deforce D, Hutton M, Van Broeckhoven C, Kumar-Singh S. Progranulin expression correlates with dense-core amyloid plaque burden in Alzheimer disease mouse models. J Pathol. 2009;219:173–181. doi: 10.1002/path.2580. [DOI] [PubMed] [Google Scholar]
- 59.Perry DC, Lehmann M, Yokoyama JS, Karydas A, Lee JJ, Coppola G, Grinberg LT, Geschwind D, Seeley WW, Miller BL, Rosen H, Rabinovici G. Progranulin mutations as risk factors for Alzheimer disease. JAMA Neurol. 2013;70:774–778. doi: 10.1001/2013.jamaneurol.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petkau TL, Leavitt BR. Progranulin in neurodegenerative disease. Trends Neurosci. 2014;37:388–398. doi: 10.1016/j.tins.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Petkau TL, Neal SJ, Milnerwood A, Mew A, Hill AM, Orban P, Gregg J, Lu G, Feldman HH, Mackenzie IR, Raymond LA, Leavitt BR. Synaptic dysfunction in progranulin-deficient mice. Neurobiology of disease. 2012;45:711–722. doi: 10.1016/j.nbd.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 62.Rademakers R, Baker M, Gass J, Adamson J, Huey ED, Momeni P, Spina S, Coppola G, Karydas AM, Stewart H, Johnson N, Hsiung GY, Kelley B, Kuntz K, Steinbart E, Wood EM, Yu CE, Josephs K, Sorenson E, Womack KB, Weintraub S, Pickering-Brown SM, Schofield PR, Brooks WS, Van Deerlin VM, Snowden J, Clark CM, Kertesz A, Boylan K, Ghetti B, Neary D, Schellenberg GD, Beach TG, Mesulam M, Mann D, Grafman J, Mackenzie IR, Feldman H, Bird T, Petersen R, Knopman D, Boeve B, Geschwind DH, Miller B, Wszolek Z, Lippa C, Bigio EH, Dickson D, Graff-Radford N, Hutton M. Phenotypic variability associated with progranulin haploinsufficiency in patients with the common 1477C-->T (Arg493X) mutation: an international initiative. The Lancet Neurology. 2007;6:857–868. doi: 10.1016/S1474-4422(07)70221-1. [DOI] [PubMed] [Google Scholar]
- 63.Rademakers R, Eriksen JL, Baker M, Robinson T, Ahmed Z, Lincoln SJ, Finch N, Rutherford NJ, Crook RJ, Josephs KA, Boeve BF, Knopman DS, Petersen RC, Parisi JE, Caselli RJ, Wszolek ZK, Uitti RJ, Feldman H, Hutton ML, Mackenzie IR, Graff-Radford NR, Dickson DW. Common variation in the miR-659 binding-site of GRN is a major risk factor for TDP43-positive frontotemporal dementia. Hum Mol Genet. 2008;17:3631–3642. doi: 10.1093/hmg/ddn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, Shen L, Foroud TM, Hakonarson H, Huentelman MJ, Aisen PS, Petersen RC, Green RC, Jack CR, Koeppe RA, Jagust WJ, Weiner MW, Saykin AJ Alzheimer’s Disease Neuroimaging I. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol Psychiatry. 2014;19:351–357. doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosen EY, Wexler EM, Versano R, Coppola G, Gao F, Winden KD, Oldham MC, Martens LH, Zhou P, Farese RV, Jr, Geschwind DH. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron. 2011;71:1030–1042. doi: 10.1016/j.neuron.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schnell SA, Staines WA, Wessendorf MW. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J Histochem Cytochem. 1999;47:719–730. doi: 10.1177/002215549904700601. [DOI] [PubMed] [Google Scholar]
- 67.Schraen-Maschke S, Sergeant N, Dhaenens CM, Bombois S, Deramecourt V, Caillet-Boudin ML, Pasquier F, Maurage CA, Sablonniere B, Vanmechelen E, Buee L. Tau as a biomarker of neurodegenerative diseases. Biomark Med. 2008;2:363–384. doi: 10.2217/17520363.2.4.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwab C, Klegeris A, McGeer PL. Inflammation in transgenic mouse models of neurodegenerative disorders. Biochim Biophys Acta. 2010;1802:889–902. doi: 10.1016/j.bbadis.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 69.Shankar GM, Welzel AT, McDonald JM, Selkoe DJ, Walsh DM. Isolation of low-n amyloid beta-protein oligomers from cultured cells, CSF, and brain. Methods in molecular biology. 2011;670:33–44. doi: 10.1007/978-1-60761-744-0_3. [DOI] [PubMed] [Google Scholar]
- 70.Sheng J, Su L, Xu Z, Chen G. Progranulin polymorphism rs5848 is associated with increased risk of Alzheimer’s disease. Gene. 2014;542:141–145. doi: 10.1016/j.gene.2014.03.041. [DOI] [PubMed] [Google Scholar]
- 71.Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, Morbin M, Rossi G, Pareyson D, Mole SE, Staropoli JF, Sims KB, Lewis J, Lin WL, Dickson DW, Dahl HH, Bahlo M, Berkovic SF. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet. 2012;90:1102–1107. doi: 10.1016/j.ajhg.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stephan AH, Madison DV, Mateos JM, Fraser DA, Lovelett EA, Coutellier L, Kim L, Tsai HH, Huang EJ, Rowitch DH, Berns DS, Tenner AJ, Shamloo M, Barres BA. A dramatic increase of C1q protein in the CNS during normal aging. J Neurosci. 2013;33:13460–13474. doi: 10.1523/JNEUROSCI.1333-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Strozyk D, Blennow K, White LR, Launer LJ. CSF Abeta 42 levels correlate with amyloid-neuropathology in a population-based autopsy study. Neurology. 2003;60:652–656. doi: 10.1212/01.wnl.0000046581.81650.d0. [DOI] [PubMed] [Google Scholar]
- 74.Sunderland T, Linker G, Mirza N, Putnam KT, Friedman DL, Kimmel LH, Bergeson J, Manetti GJ, Zimmermann M, Tang B, Bartko JJ, Cohen RM. Decreased beta-amyloid1–42 and increased tau levels in cerebrospinal fluid of patients with Alzheimer disease. Jama. 2003;289:2094–2103. doi: 10.1001/jama.289.16.2094. [DOI] [PubMed] [Google Scholar]
- 75.Tanaka Y, Chambers JK, Matsuwaki T, Yamanouchi K, Nishihara M. Possible involvement of lysosomal dysfunction in pathological changes of the brain in aged progranulin-deficient mice. Acta neuropathologica communications. 2014;2:78. doi: 10.1186/s40478-014-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y, Ulland TK, Ulrich JD, Song W, Tzaferis JA, Hole JT, Yuan P, Mahan TE, Shi Y, Gilfillan S, Cella M, Grutzendler J, DeMattos RB, Cirrito JR, Holtzman DM, Colonna M. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J Exp Med. 2016;213:667–675. doi: 10.1084/jem.20151948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ward ME, Taubes A, Chen R, Miller BL, Sephton CF, Gelfand JM, Minami S, Boscardin J, Martens LH, Seeley WW, Yu G, Herz J, Filiano AJ, Arrant AE, Roberson ED, Kraft TW, Farese RV, Jr, Green A, Gan L. Early retinal neurodegeneration and impaired Ran-mediated nuclear import of TDP-43 in progranulin-deficient FTLD. The Journal of experimental medicine. 2014;211:1937–1945. doi: 10.1084/jem.20140214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wieghofer P, Knobeloch KP, Prinz M. Genetic targeting of microglia. Glia. 2015;63:1–22. doi: 10.1002/glia.22727. [DOI] [PubMed] [Google Scholar]
- 80.Wils H, Kleinberger G, Pereson S, Janssens J, Capell A, Van Dam D, Cuijt I, Joris G, De Deyn PP, Haass C, Van Broeckhoven C, Kumar-Singh S. Cellular ageing, increased mortality and FTLD-TDP-associated neuropathology in progranulin knockout mice. J Pathol. 2012;228:67–76. doi: 10.1002/path.4043. [DOI] [PubMed] [Google Scholar]
- 81.Xu HM, Tan L, Wan Y, Tan MS, Zhang W, Zheng ZJ, Kong LL, Wang ZX, Jiang T, Tan L, Yu JT. PGRN Is Associated with Late-Onset Alzheimer’s Disease: a Case-Control Replication Study and Meta-analysis. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-9698-4. [DOI] [PubMed] [Google Scholar]
- 82.Yin F, Banerjee R, Thomas B, Zhou P, Qian L, Jia T, Ma X, Ma Y, Iadecola C, Beal MF, Nathan C, Ding A. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J Exp Med. 2010;207:117–128. doi: 10.1084/jem.20091568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yuan P, Condello C, Keene CD, Wang Y, Bird TD, Paul SM, Luo W, Colonna M, Baddeley D, Grutzendler J. TREM2 Haplodeficiency in Mice and Humans Impairs the Microglia Barrier Function Leading to Decreased Amyloid Compaction and Severe Axonal Dystrophy. Neuron. 2016;90:724–739. doi: 10.1016/j.neuron.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.