Abstract
Xp11 translocation renal cell carcinoma (RCC) are defined by chromosome translocations involving the Xp11 breakpoint which results in one of a variety of TFE3 gene fusions. TFE3 break-apart florescence in situ hybridization (FISH) assays are generally preferred to TFE3 immunohistochemistry as a means of confirming the diagnosis in archival material, as FISH is less sensitive to the variable fixation which can result in false positive or false negative immunohistochemistry. Prompted by a case report in the cytogenetics literature, we identify three cases of Xp11 translocation RCC characterized by a subtle chromosomal inversion involving the short arm of the X chromosome, resulting in an RBM10-TFE3 gene fusion. TFE3 rearrangement was not detected by conventional TFE3 break-apart FISH, but was suggested by strong diffuse TFE3 immunoreactivity in a clean background. We then developed novel fosmid probes to detect the RBM10-TFE3 gene fusion in archival material. These cases validate RBM10-TFE3 as a recurrent gene fusion in Xp11 translocation RCC, illustrate a source of false negative TFE3 break-apart FISH, and highlight the complementary role of TFE3 immunohistochemistry and TFE3 FISH.
Keywords: Renal Neoplasm, TFE3, Translocation
Introduction
Xp11 translocation renal cell carcinoma (RCC) encompasses a variety of gene fusions involving TFE3 on Xp11.2 with different gene partners, which triggers oncogenic activation of the TFE3 transcription factor. Known TFE3 fusion partners include ASPSCR1 (ASPL), PRCC, SFPQ1 (PSF), NONO, CLTC, PARP14, LUC7L3, DVL2, and KHSRP 1, 2, 3, 4, 5, 6, 7. Xp11 translocation RCC comprise the majority of pediatric RCC and approximately 1–4% of adult RCC 8–11. While a wide range of morphologic appearances have been reported12, the most common is that of an RCC with papillary architecture, clear cells, and psammoma bodies. By immunohistochemistry, these tumors underexpress cytokeratins, but often express melanocytic markers and the cysteine protease cathepsin k, which distinguishes them from more common RCC subtypes13–15. Overall, outcome is similar to that of clear cell RCC; increased age and advanced stage are poor prognostic factors16, 17. Immunohistochemistry to detect overexpressed TFE3 fusion proteins using an overnight incubation protocol was shown to be a highly sensitive and specific assay to confirm this diagnosis in formalin-fixed, paraffin embedded archival material18; however, variable fixation (particularly when automated immunostaining techniques are used) significantly reduces the specificity of TFE3 immunohistochemistry19. Break-apart fluorescence in situ hybridization (FISH) assays demonstrating TFE3 gene rearrangement are less affected by variable fixation and now are generally considered the preferred method11, 12, 20, 21.
Despite recent advances, some RCC displaying clinical, morphologic, and immunohistochemical profiles typical of Xp11 translocation RCC do not demonstrate TFE3 gene rearrangements by FISH. A subset of these prove to be the related t(6;11) translocation RCC resulting in the MALAT1 (Alpha)-TFEB gene fusion, which are grouped together in the current 2016 World Health Organization Classification of Renal Tumors with Xp11 translocation RCC as MiT family translocation RCC22,23. These cases can be detected by TFEB immunohistochemistry or, preferably, by TFEB break-apart FISH24. Nonetheless, some cases with a translocation RCC phenotype remain negative by conventional TFE3 and TFEB FISH assays. One possibility is that these cases demonstrate subtle cytogenetic alterations involving the TFE3 gene at the Xp11.2 locus which are not detectable by FISH resolution. Along these lines, the NONO-TFE3 gene fusion resulting from the intra-chromosomal X inversion, inv(X) (p11.2;q13.1), can be difficult to detect by TFE3 break-apart FISH, often resulting in small constant gaps between the TFE3 signals7, 12.
Recently, a case report of an Xp11 translocation RCC with fusion between the RBM10 gene at Xp11.23 and TFE3 at Xp11.2 was described in the cytogenetics literature25. As RBM10 is located only 1.8 mb apart from TFE3 on the short arm of the X chromosome, the abnormality was difficult to identify by TFE3 break-apart FISH. The gene fusion was confirmed by RNAseq and reverse transcriptase-polymerase chain reaction demonstrating the fusion of RBM10 exon 17 with TFE3 exon 5. This report suggests that RBM10-TFE3 gene fusions could potentially represent the underlying genetic alteration of at least a subset of the group of RCC with a translocation phenotype but which appear to be negative by conventional TFE3 and TFEB FISH12.
We now report three further cases of RBM10-TFE3 RCC. All 3 occurred in adults and were initially reported to be negative for TFE3 rearrangement by FISH. However, based upon strong morphologic suspicion of Xp11 translocation RCC, strong cathepsin K and TFE3 immunoreactivity, custom fosmid probes were developed to demonstrate the RBM10-TFE3 gene fusion.
MATERIALS AND METHODS
IRB Approval
This study was approved by the Institutional Review Boards at our institutions.
Case Selection and FISH analysis
The cases studied derive from the consultation files of the authors. Immunohistochemistry was performed as previously described7.
FISH on interphase nuclei from paraffin-embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BAC) and fosmids, covering and flanking genes of interest. Fosmids are similar to cosmids but utilize the bacterial F-plasmid to allow cleavage of DNA fragments in the range of 35–40kb. TFE3 break-apart FISH was performed as previously described12. BAC clones and fosmid clones were chosen according to UCSC genome browser (http://genome.ucsc.edu), see Supplementary Table 1. The BAC and fosmid clones were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI)(Oakland, CA)(http://bacpac.chori.org). DNA from individual BACs and fosmids was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described26. The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA). A positive score was interpreted when at least 20% of the nuclei showed a split-apart or come-together signal, depending upon the type of assay applied, either break-apart or fusion assay, respectively. Nuclei with incomplete set of signals were omitted from the score.
Results
Clinical Features and Morphology
All three cases occurred in adults. Case one was a 61 year-old female who had a radical nephrectomy for a 3.8 cm pT3 RCC associated with a 3cm tumor deposit in perirenal adipose tissue, and then developed a peritoneal recurrence 1 year later. Case two was a 54 year-old male with a 2.5 cm pT1bNX organ-confined RCC treated by partial nephrectomy. Case 3 was a 45 year-old female with a 3.8 cm pT1NX organ-confined RCC treated by partial nephrectomy. As the latter two cases are recent, no meaningful clinical follow-up is available. All three cases demonstrated clear cells and papillary architecture. Case 1 also had prominent solid architecture. Case 2 had prominent psamomma bodies, while case 3 demonstrated focally a second population of smaller cells as has been described previously in both t(6;11) RCC and Xp11 translocation RCC12, 23, especially those with the SFPQ-TFE3 gene fusion 27. Cases 1 and 3 demonstrated foci of subnuclear cytoplasmic vacuolization as often seen in the SFPQ-TFE3 and NONO-TFE3 RCCs7. Melanin pigment was not identified in any case. All three cases demonstrated diffuse immunoreactivity for cathepsin k and focal labeling for melan A (Figures 1–3). Cytokeratin 7 was negative in 2 cases in which was tested; in one of these cases, cytokeratin Cam5.2 and EMA were only focally positive. All three cases were initially reported as negative for TFE3 FISH using previously published break-apart probes12, and the two cases tested (cases 1 and 2) were negative for TFEB rearrangements by break-apart FISH24. However, all three cases demonstrated diffuse, strong TFE3 labeling by immunohistochemistry. Therefore, further evaluation was performed to detect the RMB10-TFE3 gene fusion.
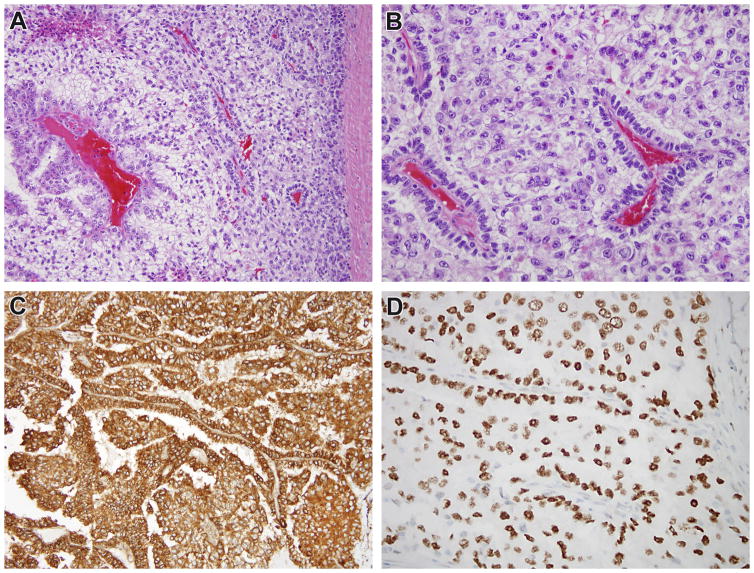
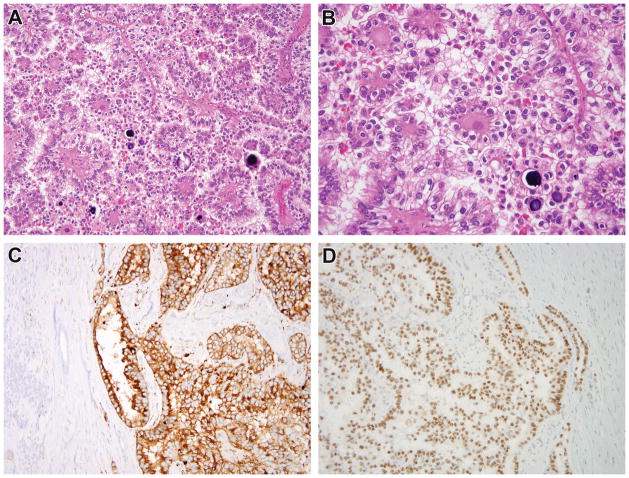
Figure 1.
Case 1. This recurrent renal cell carcinoma demonstrates solid papillary architecture and epithelioid cells with clear cytoplasm (A, B). The neoplastic cells are diffusely immunoreactive for cathepsin k (C). While conventional TFE3 FISH did not demonstrate rearrangement, the neoplasm demonstrated diffuse, strong nuclear labeling for TFE3 protein with a clean background (D), prompting additional FISH fusion studies which demonstrated the RBM10-TFE3 gene fusion.
Figure 3.
Case 3. This renal cell carcinoma demonstrates nested and papillary architecture and epithelioid cells with clear to eosinophilic cytoplasm (A, B). A subpopulation of smaller cells is present within the lumens of acini. The neoplastic cells are diffusely immunoreactive for cathepsin k (C). While conventional TFE3 FISH did not demonstrate rearrangement, the neoplasm demonstrated diffuse, strong nuclear labeling for TFE3 protein with a clean background (D), prompting additional FISH fusion studies which demonstrated the RBM10-TFE3 gene fusion.
RBM10-TFE3 Fusion FISH
As a 1.8 mb chromosomal inversion that would result in an RBM10-TFE3 fusion remains cryptic for the standard FISH resolution using the TFE3 BAC flanking probes, we applied a custom design using two fosmid clones covering the most likely breakpoint for each of the RBM10 and TFE3 genes. A positive inversion event was interpreted when each color signal (green for RBM10 and red for TFE3) splits into half-sized pairs, while a negative result revealed non-split signals (Figure 4). Despite the smaller-sized signals detected using fosmids compared to the more common BACs, all three RCC demonstrated a RBM10-TFE3 fusion using this novel assay (Figure 5). Adjacent normal cells did not show this alteration (Supplementary Figure 1).
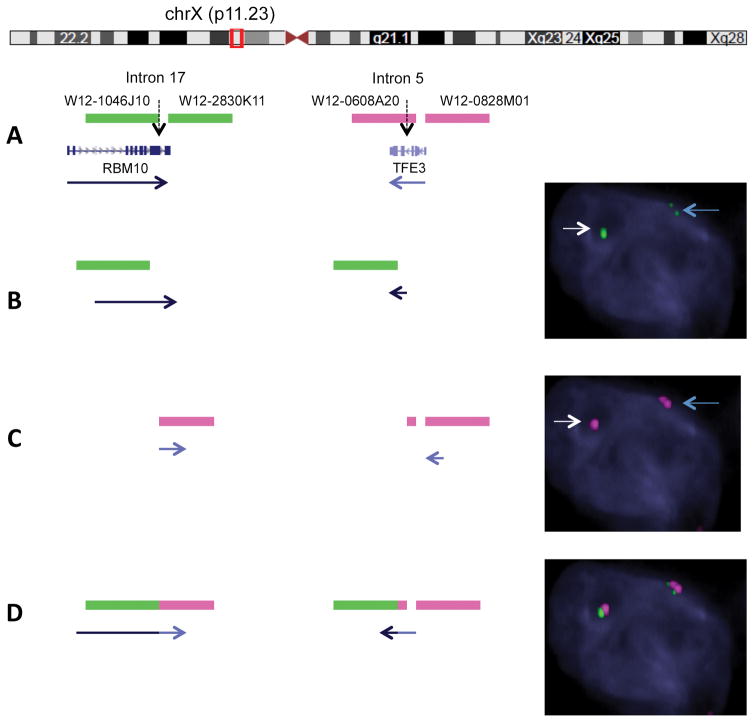
Figure 4.
Fosmid FISH design and novel application to detect RBM10-TFE3 fusion. A. Diagrammatic representation of custom design of fosmid probes covering the likely breakpoints of RBM10 (2 green probes flanking intron 17) and TFE3 (2 red proves flanking intron 5) genes. Red box illustrates the near proximity (1.8 mb) of the 2 genes on chromosome X (Xp11.23 locus). B. Single color channel assay using the above design for RBM10 in a tumor cell reveals one intact green signal (wild type RBM10 allele, white arrow) and one split green signal into two half-sized fragments, in keeping with a RMB10 break and inversion event (small gap corresponds to the 1.8 mb distance, blue arrow). C. Similar single color assay for TFE3 showing one intact red probe (wild type TFE3 allele, white arrow) and one split signal into two smaller red fragments pieces (TFE3 break/inversion, blue arrow). D. Two-color fusion assay FISH showing one normal sized fused green-red signal due to close proximity of wild type RBM10 and TFE3 genes, and two pairs of split-fused smaller signals RBM10 (green) and TFE3 (red) in keeping with the RBM10-TFE3 fusion/inversion.
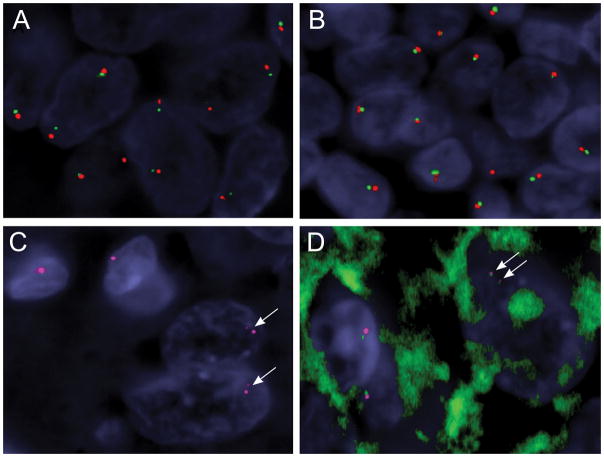
Figure 5.
Identification of RMB10-TFE3 fusion/inversion by the custom fosmid FISH assay, which is not detected by the standard BAC FISH assay resolution. (Case 2)
A,B. Conventional break-apart assay using typical flanking TFE3 probes (red, centromeric; green, telomeric) show small gaps in the tumor cells (A), compared to the normal lymphocytes adjacent to the tumor (B), but interpreted as a negative result in the clinical setting. However the small gaps may suggest the presence of a TFE3 cryptic inversion in the setting of strong TFE3 immunoreactivity and should trigger additional custom FISH assays to investigate a potential RMB10-TFE3 fusion (C, D).
C. Single color channel FISH using 2 fosmids flanking the TFE3 breakpoint (red) shows two split smaller red signals in the 2 tumor cells (lower right, arrows), in contrast to 2 normal cells showing one intact red signal (upper left).
D. Two-color fusion assay using custom fosmids showing 2 pairs of smaller red-green signals in keeping with a RBM10-TFE3 inversion in a tumor cell (upper right, arrows), compared to two normal cells showing only one pair of intact size red-green signals (lower left).
Discussion
We report 3 cases of RBM10-TFE3 RCC. Previously, a single case of an RCC with this gene fusion was reported in the cytogenetics literature25. The patient was a 32 year-old female with a 4.5cm pT1b RCC with solid, papillary, and trabecular architecture, and epithelioid cells with clear to eosinophilic cytoplasm. The neoplasm was strongly immunoreactive for TFE3 protein by immunohistochemistry, and the gene fusion was confirmed by RNAseq and reverse transcriptase-polymerase chain reaction demonstrating the fusion of RBM10 exon 17 with TFE3 exon 5. The RMB10-TFE3 gene fusion appears to be a recurrent cytogenetic alteration, as two additional RCC cases harboring the RMB10-TFE3 fusion transcript were identified in two large sequencing studies of cases characterized as clear cell28 or papillary29 RCC, though the details of the clinical, morphologic and immunohistochemical features of these cases were not provided. Our results confirm the recurrent nature of this cytogenetic alteration, and document that the RBM10-TFE3 RCC have malignant potential, as one of our cases recurred.
Our results also highlight the novel utilization of fosmid probes to detect subtle intrachromosomal inversions that can be missed by standard FISH. Fosmid clones are much smaller (35–40 kb) compared to the typical BAC clones (159–200 kb) applied in the clinical lab setting, and thus interpretation of fosmid results is less robust, due to their weaker and subtle signals. However, despite the smaller-sized signals, all three RCC in our study definitively demonstrated the RBM10-TFE3 gene fusion using this assay. To our knowledge, this technique has not previously been utilized to demonstrate cryptic chromosome inversions and rearrangements in cancer.
Furthermore, our findings illustrate an important pitfall when using TFE3 FISH as the gold standard in diagnosis rather than TFE3 immunohistochemistry. While we generally find TFE3 FISH to be more reliable in variably-fixed consultation material than TFE3 immunohistochemistry12, and typically use it as the first-line test to evaluate for Xp11 translocation RCC, the RBM10-TFE3 fusion may be missed using the conventional TFE3 break-apart FISH assays. Hence, when we encounter an RCC with a phenotype strongly suggestive of Xp11 translocation RCC (clear cells and papillary architecture, cathepsin k and or melanocytic marker immunoreactivity, conventional TFE3 and TFEB FISH negative), we now routinely perform TFE3 immunohistochemistry. Strong immunoreactivity for TFE3 in this setting using a properly calibrated assay suggests the possibility of a cryptic rearrangement involving TFE3, and prompts us to evaluate for the RBM10-TFE3 gene fusion. This highlights the importance of utilizing a properly calibrated TFE3 immunohistochemical assay; strong nuclear TFE3 labeling in a clean background is highly suggestive of the rearrangement, while weak or moderate nuclear staining with high background is likely nonspecific as documented previously19. While young patient age makes the diagnosis of translocation RCC more likely, translocation RCC is numerically more common in adults than children due to the much greater overall incidence of RCC in adults30, so adult age of the patient should not preclude further work-up. Indeed, all cases of RBM10-TFE3 RCC reported to date (including the three in this study) have been in adults.
For pathologists who may not have immunohistochemistry or FISH for TFE3 or TFEB available, utilization of other more common immunohistochemical markers can help suggest the diagnosis of MiT family translocation RCC. Underexpression of cytokeratins is a useful clue to the diagnosis1,2, which can be substantiated by aberrant expression of cathepsin k and /or melanocytic markers. Absence of the latter two markers essentially excludes the t(6;11) RCC, but does not exclude Xp11 translocation RCC which will be cathepsin K negative in approximately 50% of cases depending upon fusion type7,14,15. As our study shows, even a negative conventional TFE3 break-apart FISH study does not exclude the diagnosis of Xp11 translocation RCC, and such cases require more specialized testing for confirmation
Finally, we note that the RBM10-TFE3 gene fusion does not account for all renal neoplasms previously described as having a translocation phenotype (i.e., RCC with papillary architecture and clear cells that label for cathepsin K by immunohistochemistry) but for which TFE3 and TFEB FISH assays are negative12. We tested six additional renal neoplasms with this profile in the course of this study, and none of these six were RBM10-TFE3 positive. Three of these six cases were strongly positive for TFE3 by IHC (Supplementary Table 2). It is possible that some of these cases harbor other cryptic rearrangements resulting in TFE3 gene fusions that remain to be elucidated. Other such cases likely have different but related genes involved in their pathogenesis, and these genes also remain to be determined.
Supplementary Material
Supplementary Figure 1. Case 3 is a female with normal cells (white arrows) showing 2 pairs of TFE3 (red) and RBM10 (green); with a small gap due to the 2 Mb apart between the 2 genes. Right upper corner, shows an abnormal tumor cell (orange arrow) with 2 pairs of split-fused smaller signals RBM10 (green) and TFE3 (red) in keeping with the RBM10-TFE3 fusion/inversion.
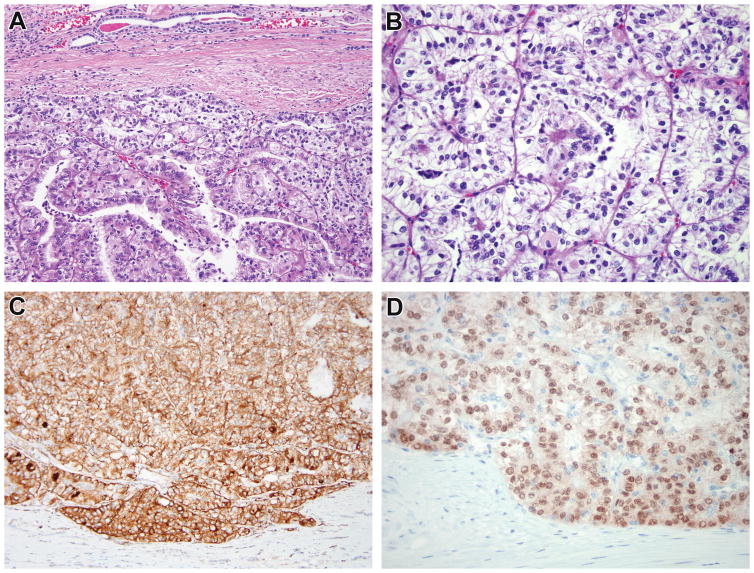
Figure 2.
Case 2. The renal cell carcinoma demonstrates papillary architecture, clear cells, and abundant psammoma bodies, highly suggestive of Xp11 translocation RCC (A, B). The neoplasm was diffusely immunoreactive for cathepsin k (C), with surrounding kidney being appropriately negative. While conventional TFE3 FISH was negative for rearrangement, immunostain for TFE3 demonstrated diffuse strong nuclear staining in the neoplastic cells with absence of staining in the normal kidney (D), leading to additional FISH fusion studies which demonstrated the RBM10-TFE3 gene fusion.
Acknowledgments
We thank Norman Barker MA, MS, RBP for expert photographic assistance.
Footnotes
Disclosures: Supported in part by: P01CA47179 (CRA), P50 CA 140146-01 (CRA), Cycle for Survival (CRA), Kristin Ann Carr Foundation (CRA), Dahan Translocation Carcinoma Fund (PA)
References
- 1.Argani P, Antonescu CR, Illei PB, Lui MY, Timmons CF, Newbury R, Reuter VE, Garvin AJ, Perez-Atayde AR, Fletcher JA, Beckwith JB, Bridge JA, Ladanyi M. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: A distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol. 2001;159:179–92. doi: 10.1016/S0002-9440(10)61684-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argani P, Antonescu CR, Couturier J, Fournet J, Sciot R, Debiec-Rychter M, Hutchinson B, Reuter VE, Boccon-Gibod L, Timmons C, Hafez N, Ladanyi M. PRCC-TFE3 renal carcinomas: Morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11. 2;q21) Am J Surg Pathol. 2002;26:1553–66. doi: 10.1097/00000478-200212000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, Hamoudi R, Linehan WM, Shipley J, Cooper CS. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–9. doi: 10.1038/sj.onc.1201394. [DOI] [PubMed] [Google Scholar]
- 4.Argani P, Lui MY, Couturier J, Bouvier R, Fournet J, Ladanyi M. A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11. 2;q23) Oncogene. 2003;22:5374–8. doi: 10.1038/sj.onc.1206686. [DOI] [PubMed] [Google Scholar]
- 5.Huang W, Goldfischer M, Babyeva S, Mao Y, Volyanskyy K, Dimitrova N, Fallon JT, Zhong M. Identification of a novel PARP14-TFE3 gene fusion from 10-year-old FFPE tissue by RNA-seq. Genes Chromosomes Cancer. 2015 May 29; doi: 10.1002/gcc.22261. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Malouf GG, Monzon FA, Couturier J, et al. Genomic heterogeneity of translocation renal cell carcinoma. Clin Cancer Res. 2013;19:4673–84. doi: 10.1158/1078-0432.CCR-12-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Argani P, Zhong M, Reuter VE, Fallon JT, Epstein JI, Netto GJ, Antonescu CR. TFE3-Fusion Variant Analysis Defines Specific Clinicopathologic Associations Among Xp11 Translocation Cancers. Am J Surg Pathol. 2016;40:723–37. doi: 10.1097/PAS.0000000000000631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altinok G, Kattar MM, Mohamed A, Poulik J, Grignon D, Rabah R. Pediatric renal carcinoma associated with Xp11. 2 translocations/TFE3 gene fusions and clinicopathologic associations. Pediatr Dev Pathol. 2005;8:168–80. doi: 10.1007/s10024-004-9106-3. [DOI] [PubMed] [Google Scholar]
- 9.Ramphal R, Pappo A, Zielenska M, Grant R, Ngan BY. Pediatric renal cell carcinoma: Clinical, pathologic, and molecular abnormalities associated with the members of the mit transcription factor family. Am J Clin Pathol. 2006;126:349–64. doi: 10.1309/98YE9E442AR7LX2X. [DOI] [PubMed] [Google Scholar]
- 10.Komai Y, Fujiwara M, Fujii Y, Mukai H, Yonese J, Kawakami S, Yamamoto S, Migita T, Ishikawa Y, Kurata M, Nakamura T, Fukui I. Adult Xp11 translocation renal cell carcinoma diagnosed by cytogenetics and immunohistochemistry. Clin Cancer Res. 2009;15:1170–6. doi: 10.1158/1078-0432.CCR-08-1183. [DOI] [PubMed] [Google Scholar]
- 11.Zhong M, De Angelo P, Osborne L, Keane-Tarchichi M, Goldfischer M, Edelmann L, Yang Y, Linehan WM, Merino MJ, Aisner S, Hameed M. Dual-color, break-apart FISH assay on paraffin-embedded tissues as an adjunct to diagnosis of Xp11 translocation renal cell carcinoma and alveolar soft part sarcoma. Am J Surg Pathol. 2010;34:757–766. doi: 10.1097/PAS.0b013e3181dd577e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green WM, Yonescu R, Morsberger L, Morris K, Netto GJ, Epstein JI, Illei PB, Allaf M, Griffin C, Argani P. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am J Surg Pathol. 2013;37:1150–1163. doi: 10.1097/PAS.0b013e31828a69ae. [DOI] [PubMed] [Google Scholar]
- 13.Argani P, Hicks J, DeMarzo A, Albadine R, Illei P, Ladanyi M, Reuter VE, Netto G. Xp11 Translocation Renal Cell Carcinoma (RCC): Extended Immunohistochemical (IHC) Profile Emphasizing Novel RCC Markers. Am J Surg Pathol. 2010;34:1295–1303. doi: 10.1097/PAS.0b013e3181e8ce5b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martignoni G, Pea M, Gobbo S, Brunelli M, Bonetti F, Segala D, Pan CC, Netto G, Doglioni C, Hes O, Argani P, Chilosi M. Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol. 2009;22:1016–22. doi: 10.1038/modpathol.2009.58. [DOI] [PubMed] [Google Scholar]
- 15.Martignoni G, Gobbo S, Camparo P, Brunelli M, Munari E, Segala D, Pea M, Bonetti F, Illei PB, Netto G, Ladanyi M, Chilosi M, Argani P. Differential expression of cathepsin-K in neoplasms harbouring TFE3 gene fusions. Mod Pathol. 2011;24:1313–1319. doi: 10.1038/modpathol.2011.93. [DOI] [PubMed] [Google Scholar]
- 16.Sukov WR, Hodge JC, Lohse CM, et al. TFE3 rearrangements in adult renal cell carcinoma: clinical and pathologic features with outcome in a large series of consecutively treated patients. Am J Surg Pathol. 2012;36:663–670. doi: 10.1097/PAS.0b013e31824dd972. [DOI] [PubMed] [Google Scholar]
- 17.Ellis CL, Eble JN, Subhawong AP, Martignoni G, Zhong M, Ladanyi M, Epstein JI, Netto GJ, Argani P. Clinical heterogeneity of Xp11 translocation renal cell carcinoma: impact of fusion subtype, age and stage. Mod Pathol. 2014;27:875–86. doi: 10.1038/modpathol.2013.208. [DOI] [PubMed] [Google Scholar]
- 18.Argani P, Lal P, Hutchinson B, Lui MY, Reuter VE, Ladanyi M. Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: A sensitive and specific immunohistochemical assay. Am J Surg Pathol. 2003;27:750–61. doi: 10.1097/00000478-200306000-00005. [DOI] [PubMed] [Google Scholar]
- 19.Argani P, Aulmann S, Illei PB, Netto GJ, Ro J, Cho HY, Dogan S, Ladanyi M, Martignoni G, Goldblum JR, Weiss SW. A distinctive subset of PEComas harbors TFE3 gene fusions. Am J Surg Pathol. 2010;34:1395–406. doi: 10.1097/PAS.0b013e3181f17ac0. [DOI] [PubMed] [Google Scholar]
- 20.Rao Q, Williamson SR, Zhang S, Eble JN, Grignon DJ, Wang M, Zhou XJ, Huang W, Tan PH, Maclennan GT, Cheng L. TFE3 break-apart FISH has a higher sensitivity for Xp11. 2 translocation associated renal cell carcinoma compared with TFE3 or cathepsin K immunohistochemical staining alone: expanding the morphologic spectrum. Am J Surg Pathol. 2013;37:804–15. doi: 10.1097/PAS.0b013e31827e17cb. [DOI] [PubMed] [Google Scholar]
- 21.Mosquera JM, Dal Cin P, Mertz KD, Perner S, Davis IJ, Fisher DE, Rubin MA, Hirsch MS. Validation of a TFE3 break-apart FISH assay for Xp11 translocation renal cell carcinomas. Diagn Mol Pathol. 2011;20:129–37. doi: 10.1097/PDM.0b013e31820e9c67. [DOI] [PubMed] [Google Scholar]
- 22.Argani P, Hawkins A, Griffin CA, Goldstein JD, Haas M, Beckwith JB, Mankinen CB, Perlman EJ. A distinctive pediatric renal neoplasm characterized by epithelioid morphology, basement membrane production, focal HMB45 immunoreactivity, and t(6;11)(p21.1;q12) chromosome translocation. Am J Pathol. 2001;158:2089–96. doi: 10.1016/S0002-9440(10)64680-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Argani P, Laé M, Hutchinson B, Reuter VE, Collins MH, Perentesis J, Tomaszewski JE, Brooks JS, Acs G, Bridge JA, Vargas SO, Davis IJ, Fisher DE, Ladanyi M. Renal carcinomas with the t(6;11)(p21;q12): clinicopathologic features and demonstration of the specific alpha-TFEB gene fusion by immunohistochemistry, RT-PCR, and DNA PCR. Am J Surg Pathol. 2005;29:230–40. doi: 10.1097/01.pas.0000146007.54092.37. [DOI] [PubMed] [Google Scholar]
- 24.Argani P, Yonescu R, Morsberger L, Morris K, Netto GJ, Smith N, Gonzalez N, Illei PB, Ladanyi M, Griffin CA. Molecular confirmation of t(6;11)(p21;q12) renal cell carcinoma in archival paraffin-embedded material using a break-apart TFEB FISH assay expands its clinicopathologic spectrum. Am J Surg Pathol. 2012;36:1516–26. doi: 10.1097/PAS.0b013e3182613d8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Just PA, Letourneur F, Pouliquen C, Dome F, Audebourg A, Biquet P, Vidaud M, Terris B, Sibony M, Pasmant E. Identification by FFPE RNA-Seq of a new recurrent inversion leading to RBM10-TFE3 fusion in renal cell carcinoma with subtle TFE3 break-apart FISH pattern. Genes Chromosomes Cancer. 2016;55:541–8. doi: 10.1002/gcc.22356. [DOI] [PubMed] [Google Scholar]
- 26.Antonescu CR, Zhang L, Chang NE, et al. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Argani P, Ladanyi M. Distinctive neoplasms characterised by specific chromosomal translocations comprise a significant proportion of paediatric renal cell carcinomas. Pathology. 2003;35:492–8. doi: 10.1080/00313020310001619901. [DOI] [PubMed] [Google Scholar]
- 28.Gotoh M, Ichikawa H, Arai E, Chiku S, Sakamoto H, Fujimoto H, Hiramoto M, Nammo T, Yasuda K, Yoshida T, Kanai Y. Comprehensive exploration of novel chimeric transcripts in clear cell renal cell carcinomas using whole transcriptome analysis. Genes Chromosomes Cancer. 2014;53:1018–32. doi: 10.1002/gcc.22211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. N Engl J Med. 2016;374:135–45. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Argani P, Olgac S, Tickoo SK, Goldfischer M, Moch H, Chan DY, Eble JN, Bonsib SM, Jimeno M, Lloreta J, Billis A, Hicks J, De Marzo AM, Reuter VE, Ladanyi M. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol. 2007;31:1149–60. doi: 10.1097/PAS.0b013e318031ffff. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Case 3 is a female with normal cells (white arrows) showing 2 pairs of TFE3 (red) and RBM10 (green); with a small gap due to the 2 Mb apart between the 2 genes. Right upper corner, shows an abnormal tumor cell (orange arrow) with 2 pairs of split-fused smaller signals RBM10 (green) and TFE3 (red) in keeping with the RBM10-TFE3 fusion/inversion.