SUMMARY
Quorum sensing (QS) is a cell–cell communication process that enables bacteria to track cell population density and orchestrate collective behaviors. QS relies on production, detection, and response to extracellular signal molecules called autoinducers. In Vibrio cholerae, multiple QS circuits control pathogenesis and biofilm formation. Here, we identify and characterize a new QS autoinducer-receptor pair. The autoinducer is 3,5-dimethylpyrazin-2-ol, which we call DPO. DPO is made from threonine and alanine, and its synthesis depends on threonine dehydrogenase (Tdh). DPO binds to and activates a transcription factor, VqmA. The VqmA-DPO complex activates expression of vqmR, which encodes a small regulatory RNA. VqmR represses genes required for biofilm formation and toxin production. We propose that DPO allows V. cholerae to regulate collective behaviors to, among other possible roles, diversify its QS output during colonization of the human host.
Keywords: Quorum sensing, autoinducer, Vibrio cholerae, biofilm, VqmA/R, mucin, DPO
INTRODUCTION
Quorum sensing (QS) is a cell-cell communication process that allows bacteria to synchronize behavior in response to changes in cell population density and local species composition1. QS involves the production, release, and subsequent detection of extracellular signal molecules called autoinducers that accumulate with increasing cell density. Once a threshold autoinducer concentration is achieved, global changes in gene expression are elicited. Processes controlled by QS are ones that require many cells acting in concert to be effective. For example, QS controls bioluminescence, secretion of virulence factors, biofilm formation, and the exchange of DNA. Thus, QS is a mechanism that allows bacteria to function as multi-cellular organisms.
In the major human pathogen Vibrio cholerae, multiple QS pathways converge to control virulence and biofilm formation2–5. The two identified V. cholerae autoinducers are CAI-1 ((S)-3-hyroxytridecan-4-one) produced by the CqsA enzyme and AI-2 (4,5-dihydroxy-2,3-pentanedione) that is synthesized by LuxS2,6,7. CqsA is conserved in all Vibrio species supporting a role for CAI-1 in intra-genus communication8. By contrast, homologs of luxS exist in hundreds of bacterial species, indicating a role for AI-2 in inter-species communication9–11. Using two autoinducers presumably allows V. cholerae to track the number of other Vibrios and the total number of bacteria in the vicinity. We note that not all AI-2 producing bacterial species encode an obvious AI-2 receptor, so it is not clear, in some cases how AI-2 is detected. In. V. cholerae, CAI-1 and AI-2 are detected by the membrane-bound receptors CqsS and LuxPQ, respectively8. At low cell density, when autoinducers are scarce, CqsS and LuxPQ function as kinases channeling phosphate to the phosphotransfer protein LuxU, which passes it to the response regulator LuxO12,13. LuxO~P, in concert with the alternative sigma factor σ54, activates the expression of genes encoding four homologous regulatory small RNAs (sRNAs), called Qrr1–414,15. The Qrr sRNAs activate translation of the low cell density master regulator called AphA16 and destabilize the mRNA encoding the master high cell density regulator called HapR14,17. Thus, at low cell density, AphA is produced while HapR is not. AphA activates expression of genes required for biofilm formation and for pathogenicity. As V. cholerae cells grow to high cell density, autoinducers accumulate. Autoinducer binding to the receptors converts them from kinases to phosphatases, resulting in dephosphorylation and inactivation of LuxO, and cessation of qrr expression. This event terminates Qrr activation of aphA translation. hapR mRNA is stabilized, leading to HapR production. Thus, at high cell density, HapR is made while AphA is not16. HapR controls a large regulon, including genes underpinning collective behaviors18. Important for the present work is that HapR represses vpsT, encoding a transcriptional activator that is crucial for biofilm formation19,20. Simultaneously, HapR activates genes encoding proteases that enable V. cholerae to disperse from the host to return to the environment21.
New studies suggest that additional V. cholerae QS systems exist5,22. Indeed, our recent global transcriptome analyses revealed a V. cholerae QS system that depends on the transcription factor called VqmA, a so-called “LuxR-solo”23,24. These orphan LuxR-type proteins are thought to play roles in QS by binding self-produced autoinducers that remain unidentified or heterologous autoinducers made by other organisms in the environment. In the latter case, LuxR-solos could expand the range of chemicals to which a bacterium can ‘tune in‘ and use in QS-census taking24–28. At high cell density, VqmA activates expression of vqmR, which encodes the VqmR sRNA. In concert with the chaperone Hfq, VqmR post-transcriptionally controls target genes, including vpsT and rtx, a key regulator of biofilm formation and a major V. cholerae toxin, respectively24. VqmA has recently been implicated in the control of V. cholerae pathogenicity via modulation of interactions with particular members of the human microbiota29. No cognate ligand for VqmA has been identified.
Here, we discover that VqmA is activated by an extracellular small molecule that accumulates at high cell density, i.e., an autoinducer. The autoinducer controls VqmA activity, not VqmA levels and, furthermore, the molecule acts directly by binding VqmA. Isolation of VqmA bound to the ligand enabled us to capture, purify, and identify the autoinducer as 3,5-dimethylpyrazin-2-ol (DPO, designated compound 1); a new type of autoinducer and a new molecule to biology. We show that DPO is produced from threonine and alanine and requires the ubiquitous threonine dehydrogenase (Tdh) enzyme for its synthesis. VqmA, in complex with DPO, activates expression of the vqmR gene encoding the VqmR small RNA, which represses genes required for V. cholerae biofilm formation. We find that this new QS autoinducer-receptor pathway operates in parallel to the canonical V. cholerae QS pathways. Notably, the canonical QS circuits and the DPO-VqmA circuit control some of the same genes. This network arrangement provides V. cholerae with a mechanism to, when necessary, uncouple expression of a particular subset of QS-regulated genes from expression of the remainder of the QS-controlled regulon.
RESULTS
VqmA is activated by an extracellular factor
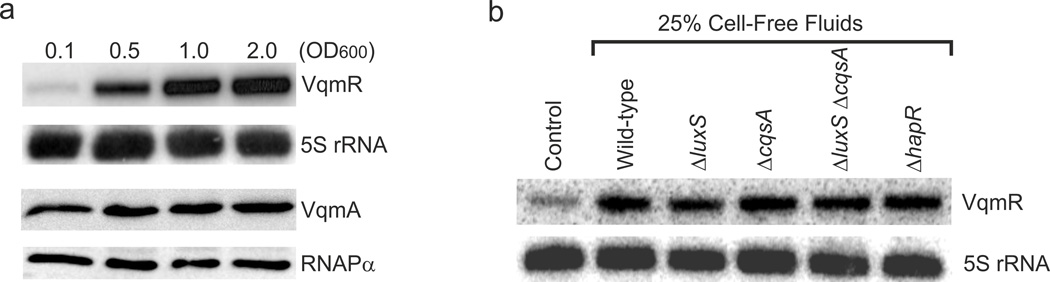
We recently discovered the VqmR sRNA and showed that expression of vqmR is activated by the transcription factor VqmA. We demonstrated that VqmA/R repress biofilm formation in V. cholerae24. To characterize the mechanism of VqmA-controlled activation of vqmR, here, we analyzed production of both factors throughout growth using Western and Northern blotting, respectively (Fig. 1A). To detect VqmA, we used a C-terminal 3X-FLAG fusion expressed from the native vqmA chromosomal locus. VqmA levels were constant at all stages of growth. By contrast, and consistent with our previous data24, minimal VqmR production occurred at low cell densities, and VqmR production increased with cell growth. These results indicate that VqmA activity, not VqmA levels, must change over growth (Fig. 1A), which, in turn, promotes cell-density-dependent changes in VqmR production.
Fig. 1. VqmR production is regulated by an extracellular activity.
(A) RNA and protein samples were collected from wild-type V. cholerae at the indicated ODs. VqmR and VqmA levels were assessed by Northern and Western Blots, respectively, with 5S rRNA and RNAPα as loading controls. (B) Northern blot of VqmR in V. cholerae wild-type cells supplied with cell-free culture fluids (25% final conc.) from the indicated strains. The control sample remained untreated. Total RNA samples were collected from cells at OD600 = 0.1. 5S rRNA is the loading control. Source files with blots showing the full data for panels A and B can be found in Supplementary Fig. 8A.
VqmA harbors a conserved PAS-domain (Per-ARNT-Sim30) at its N-terminus and a DNA-binding domain at the C-terminus23. PAS-domains are ubiquitous sensors of intra- and extracellular ligands and are frequently involved in signal transduction processes in prokaryotes and eukaryotes30. The presence of a PAS-domain, coupled with our finding of increased VqmA activity at high cell density (Fig. 1A), inspired us to investigate the possibility that an extracellular molecule functions together with VqmA to activate VqmR production. To test this idea, we added cell-free spent culture fluids prepared from high-cell density V. cholerae cultures to low cell density V. cholerae cells and assessed VqmR production. Indeed, addition of 25% (final conc.) conditioned medium caused VqmR production to increase ~3-fold (Fig. 1B, compare the first and second lanes). To exclude the possibility that the known V. cholerae autoinducers CAI-1 and AI-2 were responsible, we likewise tested culture fluids from ΔcqsA (i.e., CAI-1−), ΔluxS (i.e., AI-2−), and ΔcqsA ΔluxS (i.e., CAI-1− and AI-2−) strains for VqmR induction. In all three cases, VqmR levels increased exactly as they did following supplementation with wild-type culture fluids (Fig. 1B). We also examined culture fluids from a V. cholerae ΔhapR mutant that is completely deficient in all known QS capabilities. Full induction of VqmR production occurred following administration of the ΔhapR fluids (Fig. 1B). Thus, VqmA requires an extracellular factor to induce vqmR expression and neither CAI-1, AI-2, nor any function controlled by the known QS cascade is involved.
Genome-wide screen for factors affecting vqmR expression
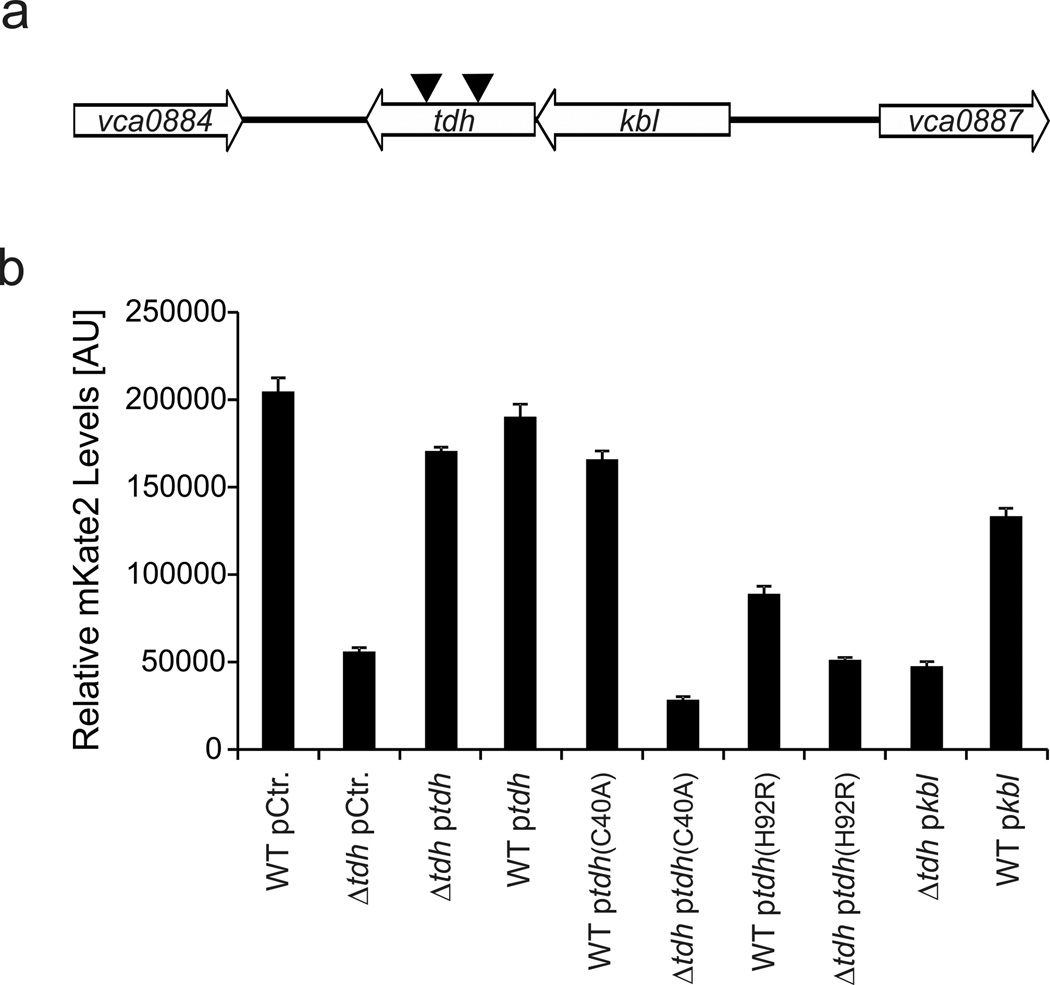
To discover the extracellular factor impinging on the VqmA/R circuit, we fused the vqmR promoter to the mkate2 gene and integrated this construct onto the V. cholerae chromosome at the lacZ locus. We randomly mutagenized this strain using Tn5. We arrayed ~40,000 mutant colonies in 96-well plates containing selective growth medium and screened them for mKate2 production following 12 h of incubation. Our rationale for separating colonies was to avoid cross-feeding from diffusion of the putative autoinducer between mutant colonies. We identified two transposon insertion mutants that displayed significantly (~4-fold) reduced fluorescence compared to the wild-type. Both insertions resided in the tdh (vca0885) gene (Fig. 2A). To verify this result, we generated an in-frame deletion of tdh in V. cholerae carrying the chromosomal PvqmR::mkate2 reporter. The Δtdh mutant also produced ~4-fold less mKate2 than the parent (Fig. 2B, compare bars 1 and 2). Introduction of a plasmid carrying tdh fully rescued the mutant phenotype, whereas introduction of the tdh-containing plasmid into the tdh+ wild-type strain had no effect on PvqmR::mkate2 expression (Fig. 2B).
Fig. 2. Threonine dehydrogenase is required for VqmA activation.
(A) Schematic of the V. cholerae kbl-tdh genomic locus. Black triangles indicate transposon insertion sites from a genome-wide screen for regulators of vqmR expression. (B) vqmR promoter activity was measured using PvqmR::mkate2 in wild-type and Δtdh V. cholerae strains carrying the indicated plasmids following growth for 10 h in LB and normalization of OD600. The pCtr. plasmid is the empty parent vector. Plasmids ptdh and pkbl produce Tdh and Kbl, respectively. Plasmids ptdh(C40A) and ptdh(H92R) produce catalytically inactive Tdh variants. Errors bars denote the standard deviation of three independent biological replicates.
tdh encodes threonine dehydrogenase which is ubiquitous among bacteria, archaea, and eukarya31. Tdh converts L-threonine to 2-amino-3-ketobutyric acid (AKB), which undergoes one of two fates. AKB either spontaneously decomposes to aminoacetone and CO2 or the enzyme Kbl acts on AKB to generate glycine and acetyl-CoA32. Consistent with the predicted function of Tdh, mass spectrometry analysis showed that purified V. cholerae Tdh protein produces aminoacetone (Supplementary Results, Supplementary Fig. 1). Previous Tdh studies in Escherichia coli revealed two critical amino acid residues required for catalytic activity: Cys-38 and His-9033. To test if Tdh catalytic activity is necessary for production of the molecule required for VqmA-directed activation of PvqmR::mkate2 expression, we mutated the corresponding residues in the V. cholerae Tdh enzyme to alanine (C40A) and arginine (H92R), respectively. In both cases, the plasmids expressing the mutant tdh alleles failed to complement mKate2 production in the Δtdh strain (Fig. 2B). Production of the Tdh C40A variant had no effect in wild-type cells (Fig. 2B), whereas Tdh H92R caused a ~2-fold reduction in PvqmR::mkate2 production, indicating potential interference with endogenously produced wild-type Tdh (Fig. 2B). Tdh is a tetramer in E. coli34, so one possible explanation is formation of partially functional or inactive hetero-tetramers composed of wild-type and Tdh H92R protomers. tdh is the second gene in the kbl-tdh operon24,35. Consistent with the results from our in-frame Δtdh mutant analysis, deletion of kbl from wild-type V. cholerae did not alter VqmR production (Supplementary Fig. 2A), and transformation of the Δtdh mutant with a plasmid expressing kbl did not restore PvqmR::mkate2 expression (Fig. 2B). Overproduction of kbl in wild-type V. cholerae modestly repressed PvqmR::mkate2 expression (Fig. 2B) suggesting that increased Kbl activity drains a substrate required to produce the molecule necessary for VqmA activation.
Threonine dehydrogenase is required for VqmA activation
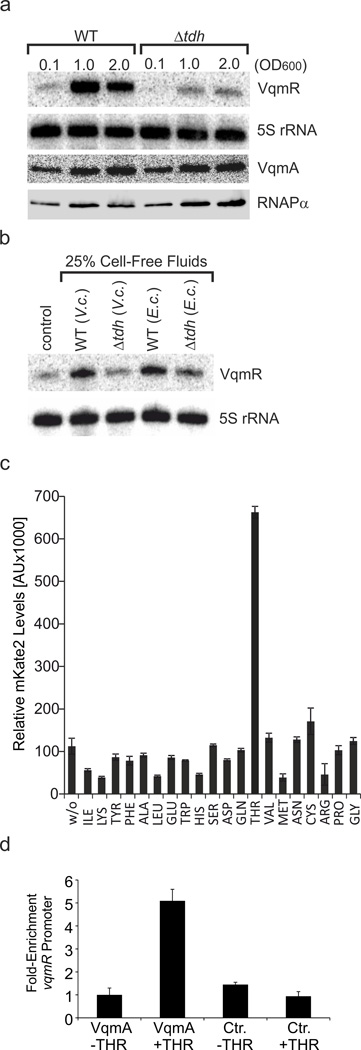
We considered three possible mechanisms that could account for reduced VqmR production in the V. cholerae Δtdh mutant. First, Tdh is required for VqmA production. Second, Tdh is required to detect the extracellular autoinducer that stimulates VqmA activity. Third, Tdh is required for production of the extracellular autoinducer that activates VqmA. To test the first possibility, we compared VqmA and VqmR production in the wild-type to that in the Δtdh strain. While reduced VqmR production occurred in the Δtdh mutant, VqmA levels remained constant (Fig. 3A). These data indicate that Tdh does not influence VqmA production, eliminating the first possibility.
Fig. 3. The autoinducer that controls VqmA activity is produced from threonine.
(A) RNA and protein were collected from the wild-type and Δtdh V. cholerae strains at the indicated ODs. VqmR and VqmA levels were assessed by Northern and Western Blots, respectively, with 5S rRNA and RNAPα as loading controls. (B) Northern blot analysis of VqmR levels in Δtdh V. cholerae following growth in the presence of cell-free culture fluids (25% final conc.) obtained from V. cholerae and E. coli wild-type and Δtdh strains. The control sample remained untreated. Total RNA samples were collected when V. cholerae reached an OD600 of 0.1. 5S rRNA served as loading control. Source files with blots showing the full data for panels A and B can be found in Supplementary Fig. 8B. (C) Wild-type V. cholerae carrying the PvqmR::mkate2 reporter was cultivated in M9 minimal medium supplemented with the indicated amino acid (final conc. according to48) and mKate2 production was measured after 8 h of incubation at 37°C. No additional amino acids were added to the ‘w/o’ sample. (D) Quantitative real-time PCR analysis of the vqmR promoter DNA sequence following co-immunoprecipitation of DNA with VqmA::3XFLAG. V. cholerae was cultivated in M9 medium in the absence or presence of 0.4 mM threonine. Wild-type V. cholerae lacking the FLAG epitope on VqmA (Ctr.) served as the control. In panels C and D, error bars denote the standard deviation of three independent replicates.
To determine whether Δtdh cells are unable to detect or unable to produce the autoinducer that stimulates VqmA, we grew the Δtdh V. cholerae strain with cell-free spent culture fluids prepared from wild-type and from Δtdh cells. Subsequently, we measured VqmR production by Northern blotting. Our prediction was that, if Tdh was required for autoinducer production, Δtdh mutants would be complemented for VqmR production by cell-free culture fluids. By contrast, if Tdh was required for detection of the autoinducer, no complementation would occur. Fig. 3B shows that conditioned medium from wild-type cells induced VqmR production in the Δtdh mutant, while Δtdh-derived culture fluids failed to induce VqmR production under the same conditions. We also added cell-free spent culture fluids from each of 3,156 mutants in an ordered V. cholerae library36 to the Δtdh strain and scored PvqmR::mkate2 expression. In every case except with culture fluids from the tdh::Tn5 mutant, mKate2 production was induced (Supplementary Fig. 2B). The tdh::Tn5 mutant culture fluids caused ~3-fold less fluorescence. Together, these results demonstrate that Tdh is required to make, not to detect, the VqmA-activating autoinducer. As noted, the tdh gene is highly conserved, and indeed, conditioned medium from E. coli was also capable of inducing vqmR expression in V. cholerae to a level comparable to that from fluids prepared from V. cholerae. Moreover, E. coli cells lacking tdh failed to activate expression of vqmR (Fig. 3B). This result suggests that Tdh synthesizes an autoinducer that is released by V. cholerae and E. coli, and likely, other prokaryotes.
Threonine catabolism permits autoinducer production
In enteric bacteria, threonine catabolism provides one route to glycine and serine biosynthesis. Threonine can also be used to synthesize isoleucine via 2-oxobutanoate37. The connections between these amino acid biosynthetic pathways prompted us to investigate which amino acids are capable of yielding the autoinducer that activates VqmA-dependent vqmR transcription. We grew wild-type V. cholerae in minimal medium supplemented with each of the twenty amino acids and measured PvqmR::mkate2 activity. Only L-threonine induced mKate2 production (~6-fold). All other amino acids either had no effect or reduced reporter activity (isoleucine, lysine, leucine, histidine, methionine, and arginine) (Fig. 3C). Therefore, the only relevant pathway for autoinducer production is through threonine and Tdh, and autoinducer synthesis is independent of other downstream reactions mediated by enzymes such as Kbl (see also Supplementary Fig. 2A).
To investigate the effect of threonine on VqmA activity, we performed chromatin-co-immunoprecipitation using VqmA::3XFLAG protein with V. cholerae grown in the presence and absence of threonine. While threonine did not change VqmA production (Supplementary Fig. 3A), quantitative real-time PCR experiments showed that ~5-times more VqmA occupied the vqmR promoter when L-threonine was added to the medium than when the medium lacked threonine (Fig. 3D). Comparison of these results to a control V. cholerae strain containing VqmA but lacking the FLAG epitope furthermore showed that, in the absence of threonine, little to no VqmA is present at the vqmR promoter. This result suggests that not only is the threonine-derived autoinducer required for VqmA activity, it is required to promote protein-DNA interaction.
To examine whether L-threonine is the VqmA autoinducer, we studied the kinetics of VqmR induction (Supplementary Fig. 3B). Specifically, we grew Δtdh V. cholerae in minimal medium to early stationary phase (OD600 = 1.0) and divided the culture into three aliquots. The first aliquot served as a control and was supplemented with fresh minimal medium. To the second aliquot, we added cell-free spent culture fluids from wild-type V. cholerae (25% final vol.) that had been grown in minimal medium containing all 20 amino acids. The third aliquot was supplemented with fresh minimal medium containing excess L-threonine (0.4 mM final conc.). Total RNA samples were collected 1, 3, 5 and 10 min after treatment and assessed for VqmR production by Northern blot. Addition of fresh minimal medium or medium containing L-threonine did not affect VqmR production (Supplementary Fig. 3B, lanes 2–5 and 10–13). However, the cells that had been treated with wild-type culture fluids displayed rapid VqmR production that could be detected 1 min after treatment (lanes 6–9). These data suggest that the autoinducer is not L-threonine, but rather, a molecule derived from L-threonine, and Tdh-mediated processing is required to convert threonine into the active autoinducer.
Capture and identification of DPO
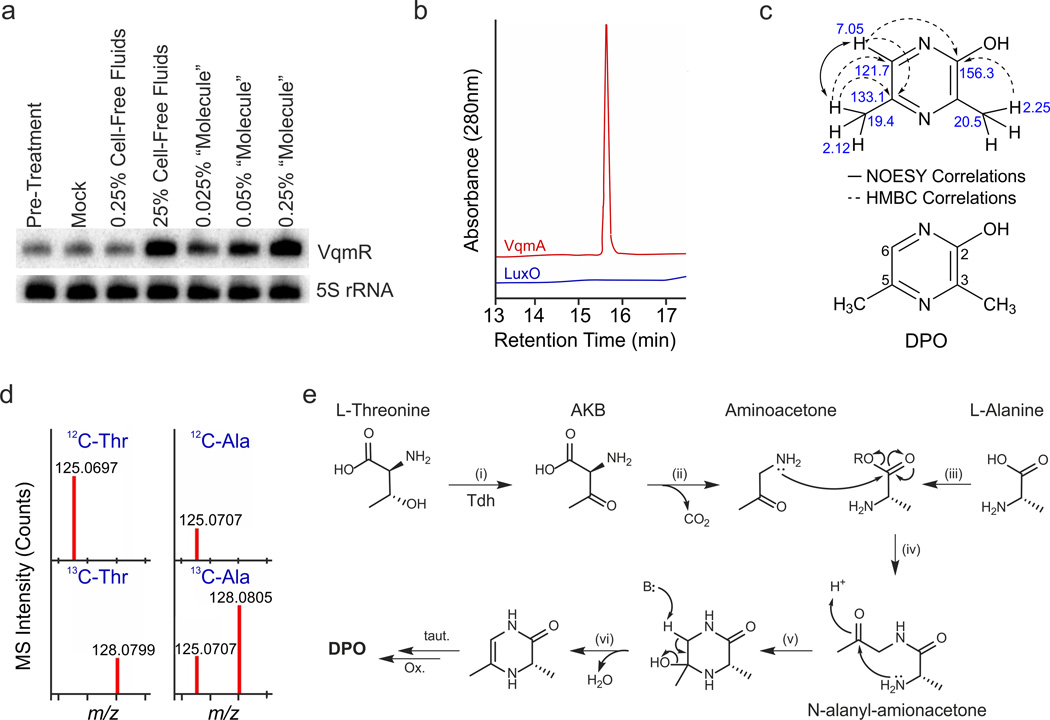
To identify the autoinducer activating VqmA, we built a recombinant system in which we exploited the fact that E. coli also produces the autoinducer (Fig. 3B). We over-expressed 6XHIS::VqmA in E. coli from a plasmid and co-transformed that strain with a PvqmR::gfp reporter. Strong (~20-fold) activation of GFP production occurred (Supplementary Fig. 4A) showing that 6XHIS::VqmA is capable of being activated by the E. coli-produced autoinducer. Next, we purified the 6XHIS::VqmA protein from E. coli (Supplementary Fig. 4B) and released the bound autoinducer through thermal denaturation. We tested the isolated autoinducer for induction of VqmR production and found that we could enrich the specific activity by ~100–1,000-fold (Fig. 4A and Supplementary Fig. 4C). To confirm that our isolated compound was specific for VqmA, we carried out the identical procedure with 6XHIS::LuxO. Preparations from thermally denatured 6XHIS::LuxO were incapable of PvqmR::mkate2 activation (Supplementary Fig. 4C). We used 6XHIS::LuxO as the control rather than 6XHIS::VqmA produced from a Δtdh strain because only low levels of soluble 6XHIS::VqmA protein were made in the Δtdh E. coli strain presumably because, in the absence of ligand, overproduced VqmA is unstable; a common feature of LuxR-type autoinducer binding proteins.
Fig. 4. Identification and proposed biosynthesis of DPO.
(A) The V. cholerae Δtdh mutant was grown in M9 medium to an OD600 of 1.0 and treated with 0.25% or 25% (final conc.) of wild-type V. cholerae cell-free culture fluids. Alternatively, the cells were treated with the “molecule” released from the 6XHIS::VqmA protein (0.025%, 0.05%, and 0.25% final conc.). Ten min after addition of each preparation, VqmR levels were assessed by Northern Blot. Pre-treatment: collected prior to treatment. Mock: no addition. 5S rRNA served as loading control. Source files with blots showing the full data can be found in Supplementary Fig. 8C. (B) HPLC-Qtof-MS analysis of compounds released from purified VqmA (red trace) and purified LuxO control (blue trace). Shown are elution profiles monitored at 280 nm, with the peak observed in the VqmA sample corresponding to DPO. (C) NOESY and HMBC correlations (top) used to solve the structure of DPO (bottom). The 1H and 13C chemical shifts (top) and the numbering scheme (bottom) for DPO are shown. (D) Mass-spectra of DPO purified from wild-type V. cholerae grown with supplemental L-Thr, uniformly-labeled 13C4-L-Thr, L-Ala, and uniformly labeled 13C3-L-Ala. The presence of a low level of 12C-DPO following supplementation with 13C3-L-Ala indicates that residual 12C-Ala, generated from the carbon-source glucose (via pyruvate), is incorporated into DPO. (E) Proposed biosynthesis of DPO. Tdh catalyzes the oxidation of Thr to AKB (i), which spontaneously decarboxylates to aminoacetone (ii). Ala, activated by an unidentified enzyme (R = phosphoryl, adenylyl, or CoA group) (iii), is condensed with aminoacetone (iv), followed by intramolecular cyclization (v), dehydration (vi), spontaneous tautomerization and oxidation, to yield DPO. We name DPO compound 1 and N-alanyl-aminoacetone compound 2 in the online chemical compound information file.
We subjected the molecules released from 6XHIS::VqmA and 6XHIS::LuxO (control) proteins to differential HPLC-Qtof-MS analysis to characterize the active autoinducer. Absorbance at 280 nm showed a single peak in the VqmA sample, with an [M+H]+obs of 125.0707, and this peak was absent from the LuxO control (Fig. 4B). Furthermore, unlike the wild-type, a V. cholerae Δtdh mutant failed to produce this compound (Supplementary Fig. 4D). 1D and 2D NMR analyses allowed us to elucidate the structure of the autoinducer (Fig. 4C). 1H NMR data showed three distinct peaks, corresponding to two methyl groups and an aromatic proton (Supplementary Fig. 5A–D, Supplementary Table 1). HSQC and HMBC data identified three quaternary carbons arranged in a pyrazine scaffold, consistent with high-resolution mass spectral (HR-MS) data, which suggested the presence of 2 nitrogen atoms (C6H8N2O, [M+H]+calc = 125.0715, Δppm = 6.4). The combination of UV, HR-MS, and NMR data allowed us to propose that the active component is 3,5-dimethylpyrazin-2-ol (DPO, Fig. 4C). The NMR spectra of commercially available DPO were identical to those of our isolated compound, thus confirming our structural assignment (Supplementary Fig. 5E–H, Supplementary Table 2).
Biosynthesis of DPO
DPO is a new bacterial natural product, and thus has no known biosynthetic pathway. To gain insight into its production, we carried out isotope feeding experiments. Inclusion of uniformly 13C-labeled L-Thr (13C4-L-Thr) resulted in a 3 Da shift in DPO mass, suggesting that, upon reaction with Tdh, the AKB product is decarboxylated to aminoacetone, which subsequently serves as the substrate for DPO biosynthesis (Fig. 4D). Taking this into account, a retrobiosynthetic analysis of DPO suggested that alanine could serve as the other DPO precursor. Indeed, supplementation of the growth medium with 13C3-L-Ala caused a 3 Da shift in DPO mass, confirming that L-Ala serves as a precursor for DPO synthesis (Fig. 4D). Our combined results allow us to propose a biosynthetic route for DPO (Fig. 4E). Threonine is converted to AKB by Tdh, and following spontaneous decarboxylation, aminoacetone is formed. Aminoacetone is condensed with L-Ala, which presumably requires ATP and possibly also CoA, as well as the action of an enzyme that remains to be identified. Intramolecular condensation and dehydration yield the penultimate precursor, which upon tautomerization and oxidation would yield DPO. Our transposon screen for mutants defective in DPO production (Fig. 2A), revealed only Tdh. If another enzyme(s) is involved in DPO biosynthesis, we suspect it is either essential or that redundant activities exist.
To acquire further evidence for our suggested biosynthetic route to DPO, we assessed the putative intermediate, N-alanyl-aminoacetone (designated compound 2), made from condensation of alanine and aminoacetone (Fig. 4E, reaction iv) for its ability to activate the PvqmR::mkate2 reporter. Our rationale was that the intermediate should be converted to DPO by V. cholerae and the steps should not rely on Tdh since any spontaneous or enzyme-mediated steps are downstream of the function of Tdh in the putative pathway. Indeed, dose-response analysis demonstrates that administration of N-alanyl-aminoacetone (Supplementary Fig. 5I) to the Δtdh V. cholerae strain induces roughly the same PvqmR::mkate2 output as does an equivalent concentration of DPO (Supplementary Fig. 6). These data establish the involvement of alanine in the DPO biosynthetic pathway, consistent with the mass spectral analyses in Fig. 4D.
DPO inhibits V. cholerae biofilm formation
To verify that DPO is the autoinducer that activates VqmA, we added increasing amounts of synthetic DPO to the Δtdh V. cholerae strain carrying the PvqmR::mkate2 reporter (Supplementary Fig. 6). High levels of DPO (1 mM) triggered strong mKate2 production, significantly above that produced by wild-type cells. 1,000-fold less synthetic DPO (1 µM) promoted mKate2 production to the levels produced by wild-type V. cholerae responding to endogenously produced DPO. Finally, 60 nM synthetic DPO elicited a doubling in PvqmR::mkate2 fluorescence. Comparison of the mKate2 production elicited by cell-free culture fluids from V. cholerae wild-type cells with known amounts of synthetic DPO suggests that DPO accumulates to ~1 µM by stationary phase (Supplementary Fig. 6B). This concentration in line with the concentrations of CAI-1 and AI-2 which are estimated to be ~0.3 µM and ~2 µM, respectively38,39.
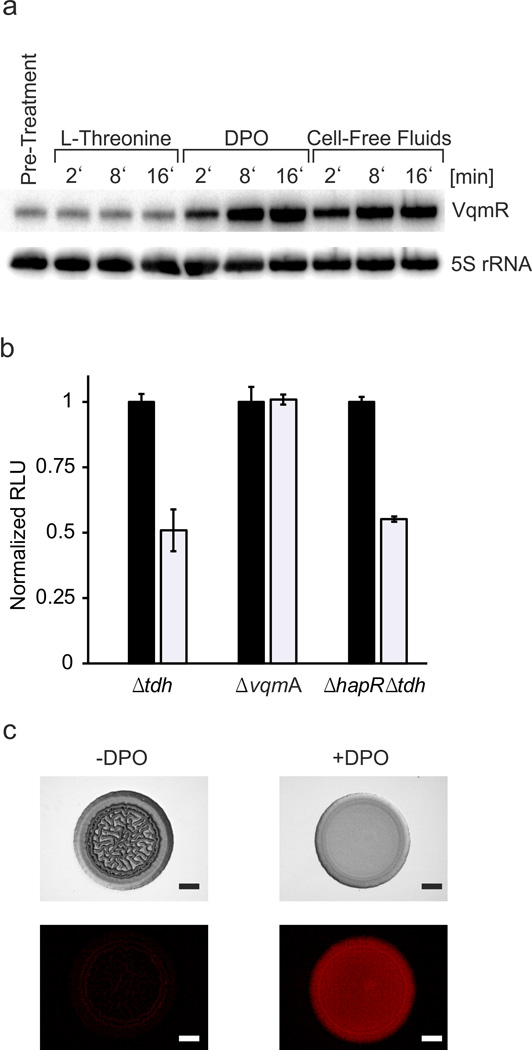
The time-course analysis of VqmA activation shown in Supplementary Fig. 3B indicated that cell-free culture fluids elicit rapid VqmR induction. We tested if synthetic DPO could produce similar response dynamics. Indeed, the V. cholerae Δtdh strain responded to DPO (100 µM final conc.) as rapidly as it responded to cell-free culture fluids (Fig. 5A). In both cases, a significant increase in VqmR could be detected as soon as 2 min after administration, supporting our arguments that DPO is the authentic autoinducer that activates VqmA.
Fig. 5. DPO induces production of VqmR which represses V. cholerae biofilm formation.
(A) Northern blot showing VqmR production in the V. cholerae Δtdh strain grown in M9 medium with casamino acids and collected at the designated time points following addition of 1 mM L-threonine, 100 µM DPO, or cell-free culture fluids prepared from wild-type V. cholerae (25% final conc.). The first lane shows the no addition control. 5S rRNA served as loading control. Source files with blots showing the full data can be found in Supplementary Fig. 8D. (B) The designated V. cholerae mutants carrying a PvpsL::lux reporter fusion were supplied without (black bars) or with (white bars) 100 µM DPO. Bioluminescence emission and OD600 were measured at 5.5 h to obtain relative light units (RLU). In each set, the RLU of the untreated sample was set to 1.0 for normalization. Each experiment was performed in triplicate. Error bars correspond to one standard deviation from the mean. (C) A ΔhapR, tdh::Tn5, pvqmR::mKate2 V. cholerae strain harboring PvqmR on a low copy plasmid was grown in liquid LB medium with chloramphenicol to OD600 = 0.7 prior to being spotted onto an LB agar plate lacking or containing 1 mM DPO. Transmission and fluorescence (mKate2) images were captured after 2 days of growth at 30 °C. The experiment was performed in duplicate 10 times and a representative image is shown for each condition. Scale bar = 1 mm.
To explore the physiological consequences of DPO signal transduction, we focused on biofilms. Our previous work showed that one target of VqmR is vpsT24, a positive regulator of genes required for biofilm formation. VpsT, upon binding the second messenger molecule cyclic-di-GMP, activates expression of multiple genes including vpsL20, the first gene in the vpsII-operon that directs the synthesis of polysaccharides, which are a major component of the V. cholerae biofilm matrix. Disruption of either vpsT or vpsL dramatically impairs V. cholerae biofilm formation40,41. Exogenous DPO repressed expression of a vpsL-lux reporter fusion (Fig. 5B) and DPO also inhibited V. cholerae biofilm formation (Fig. 5C). Notably, we also show that exogenous DPO activates expression of PvqmR::mkate2 in cells inhibited for biofilm formation confirming that the pathway is activated (Fig. 5C). DPO repression is VqmA dependent and HapR independent (Fig. 5B). Thus, DPO and the canonical QS autoinducers signal through parallel pathways to control V. cholerae biofilm formation.
DISCUSSION
In this study, we identify 3,5-dimethylpyrazin-2-ol (DPO), a new type of QS autoinducer and a new molecule to biology. In V. cholerae, DPO is detected by the cytoplasmic transcription factor VqmA, which upon binding DPO, activates expression of vqmR encoding the VqmR regulatory RNA that controls a downstream regulon of 17 genes24. Important for this work is that VqmR represses vpsT, which is required for biofilm formation. Our discovery of DPO and its VqmA-mediated link to VqmR allowed us to demonstrate that synthetic DPO inhibits biofilm formation in V. cholerae (Figs. 5B and C).
DPO could also be relevant for colonization of the small intestine by V. cholerae. A previous study reported that the commensal bacterium Ruminococcus obeum limits the severity of V. cholerae infection by impairing colonization. The resistance conferred by R. obeum was eliminated when animals were infected with a V. cholerae vqmA mutant29. We speculate that R. obeum-mediated resistance to V. cholerae colonization could be a consequence of high level DPO production in the small intestine. Specifically, DPO could be made by V. cholerae and/or commensal bacteria, which activates VqmA. VqmA promotes transcription of vqmR, and VqmR represses biofilm formation causing V. cholerae to disperse back into the environment. The V. cholerae vqmA mutant, because it does not respond to DPO, is not subject to DPO-mediated limitation. Using RNA-seq, the authors of the previous work reported that expression of V. cholerae vqmA was activated in the presence of R. obeum. By contrast, we find that VqmA levels remain constant during growth of V. cholerae, whereas VqmR levels increase significantly (Figs. 1A and 3A). One possible explanation for this discrepancy stems from mis-annotation of vqmA in several V. cholerae genomes. Indeed, vqmR and vqmA are frequently annotated as a single gene. Thus, RNA-sequencing reads that mapped to the vqmR locus would have been incorrectly attributed to the vqmA gene24.
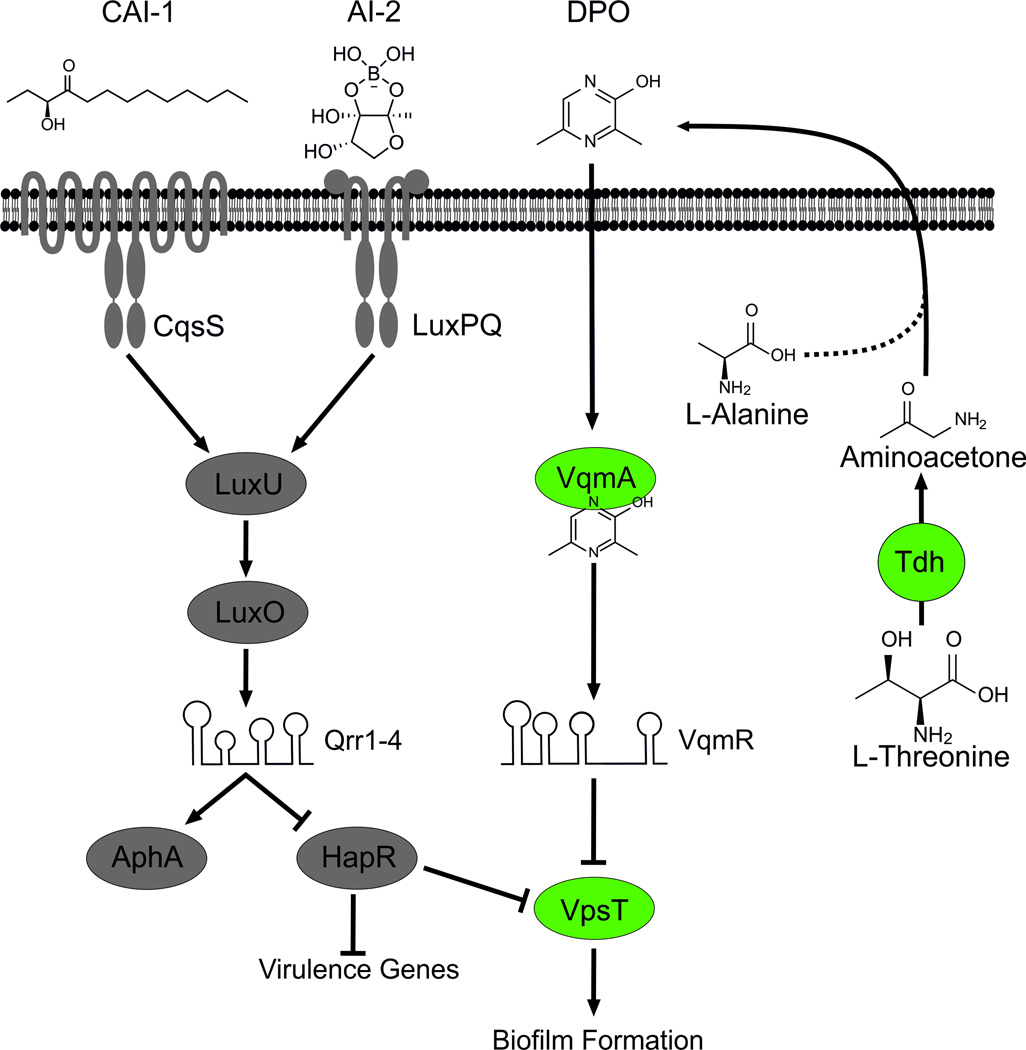
Two QS autoinducer-receptor pathways have previously been characterized in V. cholerae. They involve the autoinducers CAI-12 and AI-26 and their cognate receptors CqsS and LuxPQ, respectively (Fig. 6). Detection of CAI-1 and AI-2 at high cell density results in production of HapR. HapR represses genes required for biofilm formation and virulence factor production3,4,19. DPO signaling could provide a mechanism to uncouple regulation of biofilm formation from regulation of virulence factor production in V. cholerae. Specifically, when HapR is not active, detection of DPO via VqmA/R, and subsequent repression of biofilm formation through post-transcriptional regulation of vpsT could occur while virulence factors continue to be produced. This bypass mechanism could diversify V. cholerae gene regulatory programs to optimize QS functions under particular conditions such as during host colonization.
Fig. 6. DPO-dependent and independent QS pathways in V. cholerae.
Left (grey): The two previously characterized canonical QS pathways in V. cholerae. CAI-1 and AI-2 are detected by the CqsS and LuxPQ receptors, respectively. At low cell density, CqsS and LuxPQ function as kinases to phosphorylate LuxU. LuxU~P shuttles the phosphate to LuxO, and LuxO~P activates transcription of genes encoding the Qrr1–4 sRNAs. The Qrr sRNAs post-transcriptionally repress hapR and activate aphA promoting virulence gene expression and biofilm formation. At high cell density, binding of the CAI-1 and AI-2 autoinducers to CqsS and LuxPQ, respectively, converts them from kinases to phosphatases, reversing the phosphorylation cascade. Consequently, production of Qrr1–4 ceases. Under this condition, aphA translation is not activated and hapR translation is de-repressed. HapR represses genes required for virulence. HapR also represses biofilm formation via negative regulation of vpsT. Right (green): The DPO-dependent QS pathway in V. cholerae. DPO is produced from threonine catabolism, and DPO production requires the Tdh (threonine dehydrogenase) enzyme. We propose that the product, aminoacetone, is condensed with alanine to form DPO. Additional enzymes may be involved but they are not yet identified. DPO or a DPO precursor is released into the environment. Extracellular DPO, acting as an autoinducer, and possibly intracellularly produced DPO, binds to the VqmA receptor, which activates it. The VqmA-DPO complex activates transcription of the vqmR gene encoding the VqmR sRNA. VqmR inhibits biofilm formation by repressing translation of vpsT.
CAI-1 is an α-hydroxyketone produced by the CqsA enzyme. AI-2 is a furanosyl borate diester synthesized by LuxS (Fig. 6). DPO, the new autoinducer identified here, is a pyrazine unrelated to CAI-1 and AI-2. Although pyrazines are common in nature, few biological roles have been ascribed to them42. Consistent with our findings, pyrazines are known to be efficiently produced by bacteria when L-threonine is provided as the main carbon source43, suggesting that the Tdh-pathway could be a general source for pyrazines in organisms beyond V. cholerae. Indeed, threonine dehydrogenases are conserved among bacteria, archaea, and eukarya31, and cell-free culture fluids from E. coli (Fig. 3B) significantly induce vqmR expression. Together, these findings indicate that DPO production, and possibly its role in cell-cell communication, could be relevant in other bacterial organisms including pathogens.
In contrast to the universality of Tdh, homologs of the VqmA receptor are limited to species in the Vibrio genus. In all cases, VqmA proteins possess both PAS- and DNA-binding domains (Supplementary Fig. 7). PAS domains facilitate protein-protein and protein-small molecule interactions30. Three other QS receptors harbor PAS domains: TraR of Agrobacterium tumefaciens that binds the autoinducer N-3-oxo-octanoyl-homoserine lactone44, Vibrio LuxQ that detects AI-2 via interaction with the periplasmic binding protein LuxP45,46, and RpfR from Xanthomonas campestris that responds to BDSF (cis-2-dodecenoic acid)47. Given that the capacity to make DPO extends to genera distant from Vibrios, it will be interesting to explore whether non-VqmA-type DPO receptors exist, and if so, if they possess PAS domains and if they function in QS circuits.
Online Methods
Bacterial strains, plasmids and growth
Strains, plasmids, and oligonucleotides used in this study are listed in Supplementary Tables 4, 5 and 6, respectively. Strain and plasmid construction methods are provided in Supplementary Methods. V. cholerae and E. coli were grown aerobically in LB or M9 medium (0.4% glucose, final conc.) at 37°C. Growth at 30°C was used for biofilm assays. When indicated, M9 medium was supplemented with casamino acids (0.4% final conc.). Antibiotics were used at the following concentrations: 50 U mL−1 polymyxin B, 200 µg mL−1 ampicillin, 100 µg mL−1 kanamycin, streptomycin 5,000 µg mL−1, and 20 µg mL−1 chloramphenicol.
Construction of V. cholerae strains
A list of all strains used in this study is provided in Supplementary Table 4. V. cholerae C6706 was the wild-type strain. All V. cholerae mutants were generated using established cloning strategies2,24. Plasmids were introduced into V. cholerae by mating with the E. coli S17-1λpir strain carrying the plasmid of interest on LB agar plates for 6–8 h. Subsequently, the mating mixtures of V. cholerae and E. coli were collected and streaked on LB agar plates containing the antibiotic that selects for the plasmid and polymyxin B to eliminate E. coli.
Plasmid construction
A complete list of all plasmids used in this study is in Supplementary Table 5. E. coli S17-1λpir was used for cloning purposes. Restriction enzymes, T4 DNA ligase, and antarctic phosphatase were purchased from New England Biolabs. iProof DNA polymerase (Bio-Rad) was used for all PCR reactions. For plasmids pKP-426 and pKP-427, the vca0885 and vc0886 genes were PCR-amplified using V. cholerae genomic DNA with oligonucleotide pairs KPO-886x887 and KPO-0900x0901, respectively. The PCR products were ligated into plasmid pKP-41824 using KpnI restriction sites. Plasmid pKP-426 served as the template to generate plasmids pKP-429 and pKP-430 using oligonucleotides KPO-906/907 and KPO-910/911, respectively. To generate plasmid pKP-484, the flanking regions of vca0885 were amplified with KPO-888/889 and KPO-890/891 and the fused PCR product was amplified using KPO-892/893. The PCR products were ligated into pKAS32 using AvrII/KpnI restriction sites. The plasmid was conjugated into V. cholerae and exconjugants were selected with ampicillin. Single colonies were transferred to new plates with streptomycin. Lastly, cells were tested for possessing the correct mutation using PCR. For plasmid pKP-418, the 3XFLAG epitope was introduced downstream of vqmA (vca1078) by PCR amplification using oligonucleotides KPO-618/619 and KPO-620/621. Oligonucleotides KPO-622 and KPO-623 were used to amplify the fused PCR product, which was subsequently cloned into pKAS32. For plasmid pKP-483, gene synthesis was used to fuse the V. cholerae micX promoter24 to the superfolder gfp gene49 and the promoter of vqmR to the mKate2 (Evrogen) open reading frame. Both sequences were integrated into the lacZ locus of V. cholerae in opposite genomic orientations. For plasmids pKP-443 and pKP-445, the V. cholerae vca0885 and vqmA genes were PCR-amplified using oligonucleotides KPO-953/954 and KPO-935/972, respectively. Both fragments were ligated into pET15b (Novagen) using NcoI/BamHI and NdeI/BamHI restriction sites, respectively. For plasmid pJES24, full-length vqmR and an origin of transfer were PCR-amplified from pKP-363 (a vector harboring vqmR and the p15a origin of replication) using primer pair JSO-103/104 generating a KpnI site at the oriT end of the PCR product. The insert was subsequently digested with KpnI and ligated to pKP-33724 that was PCR amplified with JSO-109/110 and digested with KpnI.
Northern blot analysis
Total RNA was prepared and blotted as described24. Membranes were hybridized in ULTRAhyb Ultrasensitive Hybridization Buffer (Thermo Fisher) at 42°C with [32P] end-labeled DNA oligonucleotides. Signals were visualized using a Typhoon phosphorimager (GE healthcare) and band intensities were quantified using the GelQuant software (biochemlabsolutions). Oligonucleotides for Northern Blot analyses are provided in Supplementary Table 6.
Western blot analysis
Culture samples were collected over growth and subjected to centrifugation at 16,100 g for 2 min at 4°C. Cell pellets were resuspended in sample loading buffer to a final concentration of 0.01 OD/µL. Following denaturation for 5 min at 95°C, 0.1 OD600 equivalents were separated on SDS gels. Western blot analyses probing for FLAG fusion proteins followed previously published protocols24. Signals were visualized on a ImageQuant LAS 4000 imager (GE healthcare), and band intensities were quantified using the GelQuant software (biochemlabsolutions). The FLAG antibody is available at Sigma (F3165), the antibody that recognizes the RNAPα loading control is available at Neoclone (WP003).
A genetic screen for factors controlling PvqmR::mkate2 expression
Wild-type V. cholerae carrying a chromosomal copy of the PvqmR::mkate2 reporter (strain KPS-661) was mutagenized with Tn5 as described4. Briefly, a library of ~100,000 mutants was assembled of which ~40,000 were transferred into 96-well plates containing LB supplemented with kanamycin using a colony-picking robot (Molecular Devices). The plates were incubated for ~12 h at 37°C without shaking followed by sub-culturing into fresh 96-well plates. After an additional 8 h of growth, the cells in each well were tested for mKate2 production using a plate reader (Perkin Elmer). mKate2 levels were corrected for optical density (OD600) to obtain the final fluorescence levels.
Tdh activity assay
Tdh activity assays were carried out using the previously reported Fmoc-chloride conjugation method50. Reactions contained 10 µM Tdh, 1 mM L-Thr, 0.5 mM NAD in a final volume of 0.5 mL in elution buffer (50 mM HEPES, 50 mM KCl, 250 mM imidazole, 5 mM 2-mercaptoethanol). After 1 h of incubation at room temperature, reactions were quenched by addition of 0.25 mL of a 0.8 M K2B4O7 solution (pH 10.0), followed by vigorous vortex for 1 min. Reactions were worked-up and analyzed by HPLC-MS as described50.
Fluorescence and bioluminescence assays
V. cholerae reporter strains were grown overnight and back-diluted 1:1,000 in M9 medium supplemented with casamino acids. At OD600 = 0.1, 150 µL of culture was transferred to the wells of 96 well microtiter plates. Subsequently, 50 µL of cell-free culture fluids or medium was added to the wells. Mineral oil (50 µL) was layered on top of each well to prevent evaporation and the plates were incubated overnight in a 37 °C plate-reader (BioTek SynergyMx). Fluorescence intensity from PvqmR::mkate2 was measured every 15 min along with OD600. PvpsL::lux assays were performed using the identical protocol except at 30 °C. Bioluminescence, PvqmR::mkate2 fluorescence, and OD600 were measured at 15 min intervals.
VqmA::3XFLAG co-immunoprecipitation
The co-immunoprecipitation protocol is based on previously published methods51. Overnight cultures of V. cholerae wild-type cells with and without the chromosomal VqmA::3XFLAG were diluted 1:1,000 and cultivated in M9 medium (with or without 0.4 mM L-threonine final conc.) until cells reached OD600=1.0. Fifty OD600 units of cells were cross-linked and washed as previously described51, and cells were lysed in 1 mL of lysis buffer (1X protease inhibitors, (Sigma), 50 µg/mL lysozyme, 1X Bugbuster, 1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride (PMSF)) for 20 min at room temperature on a rotator. Following lysis, the DNA was sheared by sonication to an average size of 100 to 1,000 bp. The supernatant was clarified at 13,000 rpm for 10 min at 4°C. Immunoprecipitation reaction mixtures contained a 200 µL aliquot of input sample, 800 µL of IP buffer (50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 1 mM PMSF), and 40 µL EZ-view anti-FLAG agarose beads (Sigma) (equilibrated in Tris-buffered saline). Immunoprecipitations were carried out for 2 h at room temperature on a rotator. Subsequently, beads were collected and washed. Immunoprecipitated complexes were eluted, and cross-links were reversed as described51. Samples were analyzed by qRT-PCR using oligonucleotides specific to the vqmR promoter sequence. Identical analyses using oligonucleotides specific to the control hfq gene were used for normalization. Data analyses followed previously established protocols24.
DPO purification from recombinant 6XHIS::VqmA in E. coli
The active molecule was produced and purified from a total of 72 L of E. coli cultures and used in NMR and MS studies for DPO identification. Production (12 L scale): The pET15b vector carrying 6XHIS::VqmA was transformed into BL21-Gold (DE3) E. coli cells (Agilent) and plated on LB plates supplemented with 200 µg/mL ampicillin (Amp). A 250 mL overnight culture was prepared from a freshly transformed colony, back-diluted 1:50 (240 mL) into 12 L LB-Amp and divided into 16 × 2 L baffled flasks, each filled to 750 mL. Cultures were grown at 37 °C with shaking (260 rpm) to OD600=0.5–0.7. IPTG was added to a final concentration of 1 mM and the cultures grown for an additional 4 h. Pellets were collected and stored at −80 °C prior to DPO molecule purification. 6XHIS::LuxO was produced and purified identically to 6XHIS::VqmA except that Kan was used instead of Amp. 24 L of 6XHIS::LuxO culture was purified side-by-side with the first 24 L of 6XHIS::VqmA culture. In both cases, 5 mg of protein per 1 L culture was obtained. The total DPO collected is estimated to be 1 mg, corresponding to ~8 µmol DPO per 10 µmol VqmA protein.
Purification of DPO by capture in the VqmA protein
Thawed 6XHIS::VqmA-expressing pellets were resuspended in lysis buffer (20 mM imidazole and 10 mM BME in PBS supplemented with 1× Halt Protease Inhibitor Cocktail (Thermo)) using ~ 20 mL buffer per 10 g bacterial cell pellet, and lysed using French press. The lysate was centrifuged at 17,000 rpm for 30 min at 4 °C, and the supernatant recovered. Ni-NTA Superflow resin (Qiagen) was washed once in Lysis buffer and applied to the clarified lysates at a ratio of 2 mL resin per 1 L cell culture. The resin-lysate mixture was divided equally into 50 mL conical tubes and rotated end-over-end for 80 min at 4 °C. Resin was washed 5X with Wash buffer (40 mM imidazole in PBS) by centrifugation at 1,500 rpm for 3 min at 4 °C, fresh Wash buffer was added, and this step was repeated. The resin was subsequently washed twice, in the same manner, with ice cold PBS to remove residual imidazole and, twice more, in ice cold Milli-Q water to remove salts. The DPO molecule was isolated by resuspending the final volume of resin in 40% ethanol and heating the slurry to 70 °C in a water bath for 20 min with occasional vortex. The heated slurry was pelleted by centrifugation at 4,000 rpm for 10 min and the molecule was recovered in the supernatant. These supernatants were concentrated by rotovap to approximately 5 mL and stored at −80 °C prior to further purification. The batches were pooled, lyophilized to dryness, and resuspended in a small volume of water (~0.5 mL). DPO was subsequently purified on an Agilent 1260 Infinity Series analytical HPLC system equipped with a photodiode array detector and an automated fraction collector. Approximately 100 µL of sample was injected iteratively onto a Thermo Scientific Hypercarb column (4.6 × 100 mm, 3 µm) operating at a flow-rate of 0.7 mL/min. Elution was monitored at 280 nm and initially carried out isocratically using 8% acetonitrile (MeCN) in water for 6 min followed by a gradient of 8–50% MeCN (in water) over 15 min. DPO activity was monitored in the fractions using the PvqmR::mkate2 reporter assay. DPO typically eluted at 42% MeCN (17.3 min) under these conditions. Fractions that contained DPO were combined, dried in vacuum, and dissolved in D2O (for NMR analysis) or H2O (for further bioactivity assays). DPO was also purchased from Oxchem Corporation and the commercial analysis calculates >95% purity. Our in-house NMR estimate is ~80% purity.
NMR data acquisition
NMR spectra were acquired at the Princeton University Department of Chemistry Facility. NMR spectra were collected in D2O using a triple resonance cryoprobe in a Bruker A8 Avance III HD 800 MHz NMR spectrometer. 1D and 2D NMR data were analyzed with MestReNova software.
Production of cell-free culture fluids
Cell-free culture fluids were prepared by back-diluting V. cholerae overnight cultures 1:1,000 in the relevant media and incubating at 37 °C with shaking for 7–12 h. Cultures were subjected to centrifugation at 4,000 rpm for 10 min at 4 °C and culture fluids were filtered through 0.22 µm vacuum driven syringe filters (Millipore).
Isotopic labeling of culture fluids
Isotopically labeled culture fluids were prepared in the same fashion as unlabeled culture fluids but standard amino acids were replaced with uniformly 13C labeled amino acids (Cambridge Isotope Laboratories). Additions to the standard M9 medium recipe were as follows:
Unlabeled Thr: M9 + 0.4% glucose + 0.4 mM 12C L-Thr
Unlabeled Ala: M9 + 0.4% glucose + 0.4 mM 12C L-Thr + 0.4 mM 12C L-Ala
Labeled Thr: M9 + 0.4% glucose + 0.4 mM 13C L-Thr
Labeled Ala: M9 + 0.4% glucose + 0.4 mM 12C L-Thr + 0.4 mM 13C L-Ala
Overnight cultures of wild-type V. cholerae were divided into aliquots and back-diluted into unlabeled medium or medium containing a labeled amino acid(s). Cell-free culture fluids were prepared as above and loaded onto a Thermo Scientific Hypersep Hypercarb column (100–700 mg resin), which had been washed with 5 column volumes (CV) of MeOH, 5 CV of MeCN, and equilibrated with 10 CV of water. After loading, the column was washed with 5 CV of water and eluted with 5 CV of the following: 15% MeCN, 50% MeCN, and 100% MeCN. DPO and its isotopomers typically eluted in the 15% MeCN fraction, which was subsequently dried in vacuo and resuspended in 100 µL of water. 35 µL of this solution was analyzed by HPLC-Qtof-MS as described below. Note that Strata-C18, Strata-C8, and Strata-CN Seppak resins (Phenomenex) all failed to retain DPO. Optimal retention of DPO by Hypercarb chromatography was achieved by increasing the resin size to 200–1,000 mg.
HPLC-Qtof-MS analysis
High-resolution HPLC-MS was carried out on an Agilent UHD Accurate Mass Q-tof LC-MS system, equipped with a 1260 Infinity Series HPLC, an automated liquid sampler, a photodiode array detector, a JetStream ESI source, and the 6540 Series Qtof. Samples were subjected to separation on a Thermo Scientific Hypercarb column (4.6 × 100 mm, 3 µm) operating at a flow rate of 0.7 mL/min using water and MeCN. Elution was initially carried out isocratically with 8% MeCN in water for 6 min followed by a gradient of 8–50% MeCN over 15 min.
Synthesis of the ligation product of aminoacetone and L-Ala
The ligation product of aminoacetone and L-Ala (N-alanyl-aminoacetone, the product of step (iv) in Figure 4E) was synthesized by WuXi AppTec. The manufacturer estimates >95% purity which was verified by in-house NMR analysis (Supplementary Fig. 5I, Supplementary Table 3). A 100 mM stock solution of the compound was prepared in water and tested in the PvqmR::mkate2 reporter assay as described above for DPO.
Supplementary Material
Acknowledgments
This work was supported by the Howard Hughes Medical Institute, NIH Grant 2R37GM065859, and National Science Foundation Grant MCB-0948112 (to B.L.B.). K.P. was supported by a career development award from the Human Frontiers Science Program (CDA00024/2016-C) and DFG Grant PA2820/1. J.E.S. was supported by the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG) Program. M.R.S gratefully acknowledges support from the Pew Biomedical Scholars Program. K.R.S was supported by the Eli Lilly-Edward C. Taylor Fellowship in Chemistry. We thank the members of the Bassler lab for helpful discussions. We especially thank Mohamed Donia, Tharan Srikumar, István Pelczer, and Saw Kyin for help in initial analyses of DPO.
Footnotes
AUTHOR CONTRIBUTIONS
KP, JES, MRS and BLB designed the experiments; KP, JES, KRS, JPC and MRS performed the experiments. Specifically, KP, JES and JPS performed Northern and Western Blot experiments. KP, JES, KRS and MRS were involved in the autoinducer-capture experiments. KP, JES, KRS, JPS, MRS and BLB analyzed the data; and KP, JES, MRS and BLB wrote the paper.
Data availability statement: No large datasets were generated or analyzed during this study.
Competing financial interests
The authors declare no competing financial interests.
REFERENCES
- 1.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Higgins DA, et al. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 5.Jung SA, Chapman CA, Ng WL. Quadruple quorum-sensing inputs control Vibrio cholerae virulence and maintain system robustness. PLoS Pathog. 2015;11:e1004837. doi: 10.1371/journal.ppat.1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 7.Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci U S A. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2 doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013;37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- 10.Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 11.Miller ST, et al. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004;15:677–687. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Bassler BL, Wright M, Silverman MR. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol Microbiol. 1994;12:403–412. doi: 10.1111/j.1365-2958.1994.tb01029.x. [DOI] [PubMed] [Google Scholar]
- 13.Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 14.Lenz DH, et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Lilley BN, Bassler BL. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol Microbiol. 2000;36:940–954. doi: 10.1046/j.1365-2958.2000.01913.x. [DOI] [PubMed] [Google Scholar]
- 16.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev. 2011;25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng L, et al. A qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell. 2015;160:228–240. doi: 10.1016/j.cell.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsou AM, Cai T, Liu Z, Zhu J, Kulkarni RV. Regulatory targets of quorum sensing in Vibrio cholerae: evidence for two distinct HapR-binding motifs. Nucleic Acids Res. 2009;37:2747–2756. doi: 10.1093/nar/gkp121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krasteva PV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 22.Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat Rev Micro. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Hsiao A, Joelsson A, Zhu J. The transcriptional regulator VqmA increases expression of the quorum-sensing activator HapR in Vibrio cholerae. J Bacteriol. 2006;188:2446–2453. doi: 10.1128/JB.188.7.2446-2453.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papenfort K, Forstner KU, Cong JP, Sharma CM, Bassler BL. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc Natl Acad Sci U S A. 2015;112:E766–E775. doi: 10.1073/pnas.1500203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez JF, Venturi V. A novel widespread interkingdom signaling circuit. Trends Plant Sci. 2013;18:167–174. doi: 10.1016/j.tplants.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Brameyer S, Kresovic D, Bode HB, Heermann R. LuxR solos in Photorhabdus species. Front Cell Infect Microbiol. 2014;4:166. doi: 10.3389/fcimb.2014.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venturi V, Ahmer BM. Editorial: LuxR Solos are Becoming Major Players in Cell-Cell Communication in Bacteria. Front Cell Infect Microbiol. 2015;5:89. doi: 10.3389/fcimb.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramoni S, Florez Salcedo DV, Suarez-Moreno ZR. A bioinformatic survey of distribution, conservation, and probable functions of LuxR solo regulators in bacteria. Front Cell Infect Microbiol. 2015;5:16. doi: 10.3389/fcimb.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsiao A, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henry JT, Crosson S. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjee JJ, Dekker EE. Purification, properties, and N-terminal amino acid sequence of homogeneous Escherichia coli 2-amino-3-ketobutyrate CoA ligase, a pyridoxal phosphate-dependent enzyme. J Biol Chem. 1987;262:14441–14447. [PubMed] [Google Scholar]
- 33.Johnson AR, Dekker EE. Site-directed mutagenesis of histidine-90 in Escherichia coli L-threonine dehydrogenase alters its substrate specificity. Arch Biochem Biophys. 1998;351:8–16. doi: 10.1006/abbi.1997.0501. [DOI] [PubMed] [Google Scholar]
- 34.Boylan SA, Dekker EE. L-threonine dehydrogenase. Purification and properties of the homogeneous enzyme from Escherichia coli K-12. J Biol Chem. 1981;256:1809–1815. [PubMed] [Google Scholar]
- 35.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cameron DE, Urbach JM, Mekalanos JJ. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci U S A. 2008;105:8736–8741. doi: 10.1073/pnas.0803281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reitzer L. Catabolism of Amino Acids and Related Compounds. EcoSal Plus. 2005;1 doi: 10.1128/ecosalplus.3.4.7. [DOI] [PubMed] [Google Scholar]
- 38.Kelly RC, et al. The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA. Nat Chem Biol. 2009;5:891–895. doi: 10.1038/nchembio.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thiel V, Kunze B, Verma P, Wagner-Dobler I, Schulz S. New structural variants of homoserine lactones in bacteria. Chembiochem. 2009;10:1861–1868. doi: 10.1002/cbic.200900126. [DOI] [PubMed] [Google Scholar]
- 40.Fong JC, Syed KA, Klose KE, Yildiz FH. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology. 2010;156:2757–2769. doi: 10.1099/mic.0.040196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casper-Lindley C, Yildiz FH. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J Bacteriol. 2004;186:1574–1578. doi: 10.1128/JB.186.5.1574-1578.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller R, Rappert S. Pyrazines: occurrence, formation and biodegradation. Appl Microbiol Biotechnol. 2010;85:1315–1320. doi: 10.1007/s00253-009-2362-4. [DOI] [PubMed] [Google Scholar]
- 43.Besson I, Creuly C, Gros BJ, Larroche C. Pyrazine production by Bacillus subtilis in solid-state fermentation on soybeans. Applied Microbiology and Biotechnology. 1997;47:489–495. [Google Scholar]
- 44.Vannini A, et al. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 2002;21:4393–4401. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neiditch MB, et al. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 2005;18:507–518. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 47.Deng Y, et al. Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc Natl Acad Sci U S A. 2012;109:15479–15484. doi: 10.1073/pnas.1205037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 49.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 50.Kazachkov M, Yu PH. A novel HPLC procedure for detection and quantification of aminoacetone, a precursor of methylglyoxal, in biological samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;824:116–122. doi: 10.1016/j.jchromb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 51.van Kessel JC, Ulrich LE, Zhulin IB, Bassler BL. Analysis of activator and repressor functions reveals the requirements for transcriptional control by LuxR, the master regulator of quorum sensing in Vibrio harveyi. MBio. 2013;4 doi: 10.1128/mBio.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.