Abstract
Plant development requires coordination among complex signaling networks to enhance plant’s adaptation to changing environments. The transcription regulators DELLAs, originally identified as repressors of phytohormone gibberellin (GA) signaling, play a central role in integrating multiple signaling activities via direct protein interactions with key transcription factors. Here, we showed that DELLA was mono-O-fucosylated by a novel O-fucosyltransferase SPINDLY (SPY) in Arabidopsis thaliana. O-fucosylation activates DELLA by promoting its interaction with key regulators in brassinosteroid (BR)- and light-signaling pathways, including BRASSINAZOLE-RESISTANT1 (BZR1), PHYTOCHROME-INTERACTING-FACTOR3 (PIF3), and PIF4. Consistently, spy mutants displayed elevated responses to GA and BR, and increased expression of common target genes of DELLAs, BZR1 and PIFs. Our study revealed that SPY-dependent protein O-fucosylation plays a key role in regulating plant development. This finding has broader importance as SPY orthologs are conserved from prokaryotes to eukaryotes, suggesting that intracellular O-fucosylation may regulate a wide range of biological processes in diverse organisms.
Plant adaptation and survival require strict coordination of internal signaling pathways in response to external biotic and abiotic cues1. A class of important central integrators of multiple signaling pathways in flowering plants is the DELLA proteins. DELLAs were initially identified as repressors of phytohormone gibberellin (GA) signaling2,3. Recent studies further revealed that DELLAs are master growth repressors4,5, which restrict plant growth by causing transcriptional reprogramming of genes that function in cell division, expansion and differentiation, through direct protein-protein interactions with key transcription factors6–8. DELLAs contain a conserved N-terminal DELLA domain required for GA-receptor GID1 complex-induced degradation9–12, followed by a divergent and disordered sequence rich in Ser and Thr residues (PolyS/T), and a conserved C-terminal GRAS domain with transcription regulator function2,8,13 (Fig. 1a). Internal deletions of the DELLA motif (e.g., gai-1 and rga-Δ17 in Arabidopsis) result in a constitutively active repressor that does not respond to GA-induced degradation3,14. The dwarf-phenotype of these dominant DELLA mutants can be partially rescued by hypomorphic spy alleles15,16. Both SPY and its paralog SECRET AGENT (SEC) in Arabidopsis were predicted to encode O-linked β-N-acetylglucosamine (O-GlcNAc) transferases (OGTs), each of which contains an N-terminal protein-interaction domain [tetratricopeptide repeats (TPRs)] and a C-terminal putative OGT catalytic domain (Supplementary Results, Supplementary Fig. 1a)17–19. Because of the presence of multiple putative OGT target sites in the PolyS/T region of the DELLA proteins, it has been long hypothesized that SPY may O-GlcNAcylate DELLAs5,15. We recently showed that the Arabidopsis DELLA protein RGA is indeed O-GlcNAcylated by MS analysis20. However, SEC is the OGT for this modification, whereas SPY is not required. Intriguingly, mutant analysis indicates that SEC and SPY play opposite roles in GA signaling, with SEC being a positive regulator and SPY being a negative regulator20,21, although the biochemical function of SPY remained unsolved.
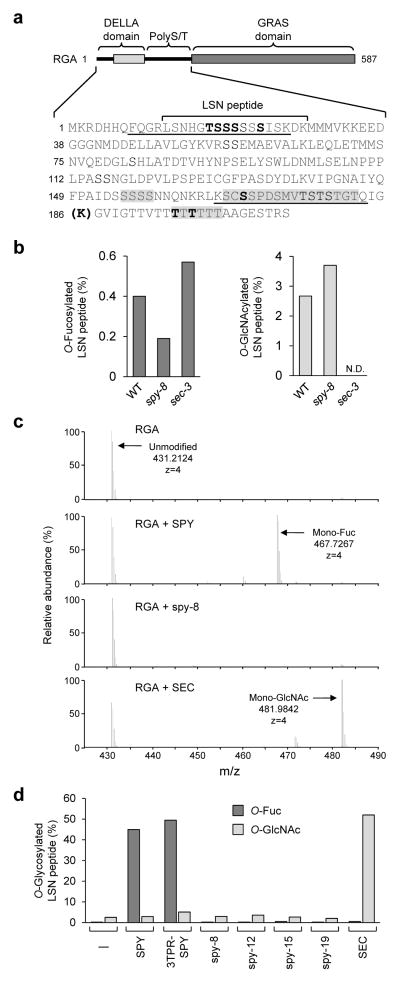
Figure 1. RGA shows SPY-dependent O-fucosylation in planta.
(a) O-fucosylation sites in RGA. In RGA schematic (top), solid lines indicate two structurally disordered regions. The sequences of the DELLA domain and PolyS/T region are listed below the schematic. The LSN peptide was mono-O-fucosylated in FLAG-RGA from transgenic Arabidopsis. Additional O-fucosylation sites were identified using FLAG-RGAGKG from tobacco that was co-expressed with SPY. Boldface S/T indicates modification sites confirmed by MS/MS. Sequences shaded in gray contain one or more additional unmapped sites (see Supplementary Table 1 for detailed information). The boldface K in parenthesis indicates the extra Lys residue in RGAGKG. RGApep1 and RGApep2 are underlined. (b) LC-ETD-MS/MS analysis showed that O-fucosylation levels in the RGA LSN peptide were reduced in spy-8 compared to those in WT and sec-3. (c) MS1 spectra of the RGA LSN peptide from tobacco that expressed FLAG-RGAGKG alone (RGA), or co-expressed with SPY, spy-8 or SEC. Unmodified peptide, predicted m/z = 431.2130. Mono-O-fucosylated RGA peptide (predicted m/z = 467.7195) was only detected in RGA+SPY, whereas mono-O-GlcNAcylated peptide (predicted m/z = 481.9823) was only detected in RGA+SEC. (d) MS analysis showed that O-fucosylation levels in the RGA LSN peptide were dramatically increased only when co-expressed with SPY or 3TPR-SPY. Four biological replicates for WT SPY and SEC, and two biological replicates for mutant spy proteins were analyzed with similar results. Immunoblots showed similar ratios of SPY or mutant spy vs. FLAG-RGAGKG in tobacco protein extracts (Supplementary Fig. 2).
In this report, through sensitive MS analysis and in vitro enzyme assays, we uncovered the biochemical function of SPY as a novel protein O-fucosyltransferase, which modifies the Arabidopsis DELLA protein RGA by attaching mono-fucose to specific Ser and Thr residues. Importantly, this unusual O-fucosylation enhances DELLA activity by promoting DELLA binding to key transcription factors in BR and light-signaling pathways. Our study revealed that SPY-dependent O-fucosylation plays an important role in modulating plant development by regulating activities of nuclear transcription factors. Our finding has broader implication because SPY orthologs are conserved in diverse organisms17, although their biochemical function has not been characterized.
RESULTS
RGA shows SPY-dependent O-fucosylation in planta
A major advance in uncovering the SPY function came from our close interrogation of the LC (liquid chromatography)-ETD (electron transfer dissociation)-MS/MS spectrum of FLAG-tagged RGA isolated from transgenic Arabidopsis carrying PRGA:FLAG-RGA. In this experiment, FLAG-RGA was affinity-purified from Arabidopsis and digested with endoproteinase trypsin, followed by LC-ETD-MS/MS analyses. We discovered a novel post-translational modification, mono O-fucose, attached to specific Ser/Thr residues, in RGA (Fig. 1a–1b, Supplementary Note 1); O-fucosylation of Ser or Thr residues has not been identified in nuclear proteins. Surprisingly, we found that O-fucosylation levels in RGA were reduced in spy-8, but were unaffected by sec-3 (Fig. 1b), suggesting that SPY directly or indirectly promotes O-fucosylation of RGA. However, we could only detect very low levels of O-fucosylation in RGA from purified Arabidopsis samples, possibly because this type of modification is easily lost during purification and MS analysis.
To examine further the role of SPY in O-fucosylation of RGA, we transiently expressed FLAG-RGAGKG alone, or co-expressed it with Myc-SPY or Myc-SEC (as a control) in tobacco leaves by agroinfiltration. RGAGKG contains an extra K residue between G185 and G186 in RGA (Fig. 1a), which creates an additional trypsin cleavage site to facilitate the detection of the O-fucosylated peptide that spans the poly-T track within the PolyS/T region. CAD (collisionally activated dissociation)- and ETD-MS/MS analyses showed that the purified FLAG-RGAGKG from the RGA+SPY samples was highly O-fucosylated (Fig. 1c–1d, Supplementary Fig. 2, and Supplementary Notes 2–5). One of the most abundant O-fucosylated sequences was located in the LSN peptide that was also O-fucosylated in Arabidopsis (Fig. 1a, 1c–1d, and Supplementary Table 1). Other major O-fucosylation sites were within the polyT track (Fig. 1a and Supplementary Table 1). In total, a minimum of 9 O-fucosylation sites in 4 RGA peptides were identified, and 8 of these sites were mapped to specific Ser or Thr residues (Fig. 1a, Supplementary Table 1, Supplementary Notes 1–5). In contrast, the RGA alone samples showed very low amounts of O-fucosylation compared to the RGA+SPY samples (Fig. 1c–1d and Supplementary Table 1), indicating that RGA was modified to a lesser extent by endogenous protein O-fucosyltransferase(s) of tobacco. Co-expression of the spy-8 mutant protein or SEC did not increase O-fucosylation in RGA. These results indicate that SPY promotes O-fucosylation of RGA. To investigate whether SPY also promotes O-fucosylation of other AtDELLAs, we examined purified GAI, RGL1 and RGL2 that were co-expressed with SPY in tobacco by MS analysis. We identified an O-fucosylated peptide near the N-terminus (9-ESSAGEGGSSSMTTVIK-25) of RGL1 (Supplementary Fig. 3).
SPY is a novel protein O-fucosyltransferase (POFUT)
Our RGA and SPY co-expression experiments using the tobacco system strongly suggest that SPY is a novel POFUT that modifies RGA. Protein O-fucosyltransferases modify target proteins (the acceptor substrates) by transferring O-fucose from GDP-fucose (the donor substrate) to the hydroxy oxygen of Ser and Thr residues. In fact, protein O-fucosylation is rare, and is mainly known to occur in the ER to promote protein folding and/or ligand binding of a few cell surface transmembrane proteins22: e.g., EGF domain-containing Notch family proteins23, and thrombospondin type 1 repeats, TSRs24. The O-fucosylation of EGF and TSR is catalyzed by ER-localized POFUT1 and POFUT2, respectively22. POFUT1 and POFUT2 have similar 3D structures with 2 β/α/β Rossmann-like folds (GT-B fold)25,26, but are distinct from the structures of OGTs27 (Supplementary Fig. 1b–1c).
To assess whether SPY directly O-fucosylates RGA, we performed in vitro enzyme assays using 2 synthetic RGA peptides that were O-fucosylated in planta (Fig. 1a and Supplementary Table 1) as the acceptor substrates. Purified recombinant 3TPR-SPY protein (Supplementary Fig. 4a), which is truncated by 8 TPRs from the N-terminus, was used in the enzyme assays. This truncated 3TPR-SPY was equally active as the full-length SPY (FL-SPY) in our tobacco assay (Fig. 1d and Supplementary Table 1), consistent with the previous report showing that the truncated human OGT containing only 5 TPRs displayed similar enzymatic activity as the full-length protein28. Also, we could obtain larger amounts of soluble 3TPR-SPY than FL-SPY when expressed in E. coli. In the initial enzyme assays, each RGA peptide was incubated with GDP-fucose and purified 3TPR-SPY in various buffer conditions for 2 hr before it was analyzed by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). As shown in Figure 2a, 88% of RGA peptide 1 (RGApep1) was mono-fucosylated in the presence of both GDP-fucose and 3TPR-SPY, but no modification was detected in the absence of GDP-fucose or SPY, proving that SPY is indeed a POFUT that modifies RGA. Peptide 2 (RGApep2) was also O-fucosylated by SPY, although it was modified at much lower levels compared to those in RGApep1 (Supplementary Fig. 4b). Therefore, RGApep1 was used for further SPY enzyme characterization.
Figure 2. SPY is a novel protein O-fucosyltransferase (POFUT).
(a) MALDI-MS spectra of the RGA peptide 1 (RGApep1) from in vitro enzyme assays ± 3TPR-SPY or GDP-fucose or SEC. Monon-fucosylated RGApep1 (predicted m/z = 2155.17 vs. 2009.11 for unmodified peptide) was only detected in RGA+3TPR-SPY in the presence of GDP-fucose. Peaks labeled A and B (observed m/z =1908.92 for A, 1923.17 for B) are unmodified incomplete peptides (byproducts of RGApep1 synthesis) with a T- or S-deletion, respectively; Peaks labeled A* and B* are mono-fucosylated A and B peptides (observed m/z = 2054.98 for A*, 2069.23 for B*). Three biological replicates were analyzed with similar results. (b) Optimal pH for SPY in vitro enzyme assay determined by malachite green-coupled assay. (c–d) Steady-state kinetics were determined for 3TPR-SPY using malachite green-coupled assays. In (c), GDP-fucose concentration was fixed at 800 μM. In (d), RGApep1 concentration was fixed at 312.5 μM. In (b–d), each reaction was at least triple-duplicated. Means ± SE are shown. Km for each substrate was calculated by the Lineweaver-Burk plot (Supplementary Fig. 6a–6b).
To analyze the donor substrate specificity of SPY, four other nucleotide-sugars were tested. These include UDP-GlcNAc (donor substrate for OGT), GDP-mannose, UDP-galactose, and UDP-glucose. By MALDI-MS analysis, we found that SPY did not exhibit any activity in the presence of these four nucleotide sugars, indicating that SPY displays specific POFUT activity. In addition, our in vitro assay using RGA peptides shows that SEC only displayed OGT activity (Supplementary Fig. 5a–5b), but not POFUT activity (Fig. 2a), consistent with the results of our RGA+SEC coexpression study using the tobacco system (Fig. 1c–1d).
For further POFUT activity characterization of SPY, we used a recently developed method for assaying glycosyltransferase activities, called malachite green-coupled reaction29, because this assay is more efficient than MALDI-MS. In this assay, the glycosyltransferase reaction is coupled with a phosphatase (ectonucleoside triphosphate diphosphohydrolase, ENTPD) that releases the β-phosphate of GDP, which can then be detected by the malachite green reagents29,30. Using this assay, we showed that SPY enzyme activity was not significantly affected by the salt concentration or metal cations (Supplementary Fig. 6a–6b), but was sensitive to pH, with highest activity at pH 8.2 (Fig. 2b). Michaelis-Menten kinetics analysis was then performed under the optimized buffer conditions [50 mM Tris (pH 8.2), 5 mM MgCl2, 50 mM NaCl]. The O-fucosylation reaction has two substrates – RGApep1 and GDP-fucose. To simplify the kinetics analysis, two sets of reactions were performed, one with a fixed GDP-fucose concentration at 800 μM, and the other with a fixed peptide concentration at 312.5 μM (Fig. 2c–2d). For 3TPR-SPY, the Km for RGApep1 was 8.23 ± 0.10 μM, with kcat of 0.50 ± 0.02 sec−1; The Km for GDP-fucose was determined to be 50.48 ± 3.90 μM, with kcat of 0.27 ± 0.01 sec−1 (Supplementary Fig. 7a–7b).
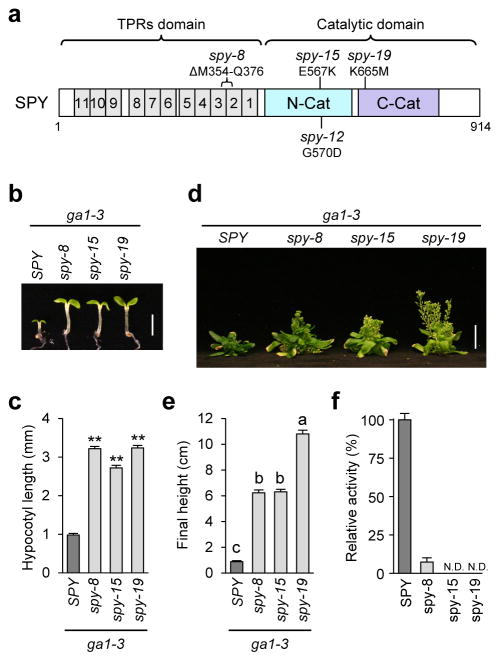
spy alleles define key residues for POFUT activity
Previous studies have identified a number of hypomorphic spy alleles, some of which are located in the TPR domain and others are in the C-terminal catalytic domain15 (Fig. 3a). For example, the spy-8 protein contains an in-frame 23 amino-acid deletion (M354-Q37615) in TPR3-TPR2, while spy-12 (G570D15), spy-15 (E567K15) and spy-19 (K665M, identified in this study) each contains a single amino acid substitution in the catalytic domain. Previous phenotype characterization of the single spy mutants showed that spy-12, spy-15 and spy-19 display more severe fertility defects and earlier flowering time than spy-8, indicating that these catalytic domain mutations are stronger alleles than spy-815. To investigate the specific effects of spy alleles on GA signaling, we analyzed phenotypes of spy mutants in the GA-deficient mutant ga1-3 background. At the seedling stage, spy-8 rescued the hypocotyl growth of ga1-3 to a similar extent as spy-15 and spy-19 (Fig. 3b–3c). However, at the adult stage, spy-19 rescued the stem growth defect of ga1-3 more efficiently than spy-8 and spy-15 (Fig. 3d–3e). To test whether spy-8 protein (with mutations in the TPR region) still retains some catalytic activity, we expressed and purified spy-8, spy-15 and spy-19 mutant proteins (3TPR-SPY truncated version) for in vitro enzyme assays. Malachite green-coupled assay showed that the POFUT activity of spy-8 was 7.3% of WT, whereas no activity was detected for spy-15 or spy-19 (Fig. 3f, Supplementary Fig. 8a). In an attempt to detect residual enzymatic activities of spy-15 and spy-19, we performed in vitro enzyme assays with a 16 hr incubation time, followed by MALDI-MS. The results of this assay are consistent with the malachite green assay with spy-8 showing a low POFUT activity (8.5% of WT), whereas spy-15 and spy-19 did not exhibit any POFUT activity (Supplementary Fig. 9). Similar results were also observed when these spy mutant proteins (plus spy-12) were co-expressed with FLAG-RGAGKG in tobacco, although no residual POFUT activity was detected for spy-8 (Fig. 1d). These results indicate that E567, G570 and K665 are essential for the POFUT activity of SPY, and TPR2 and TPR3 (partially deleted in spy-8) also play an important role in SPY function. These results confirm that SPY is a novel POFUT, although its paralog SEC is an OGT.
Figure 3. Phenotype and enzyme activity analyses of the spy mutants.
(a) Schematic of SPY protein structure. Mutation in spy-8 is located in TPR2-3, whereas mutations in spy-12, spy-15 and spy-19 are located in the C-terminal catalytic domain. (b–c) All spy alleles rescued the hypocotyl growth defect of the GA-deficient mutant ga1-3. Hypocotyl lengths of 6d-old seedlings grown in short-day conditions were measured. In (b), bar = 2.5 mm. In (c), average hypocotyl lengths are means ± SE. n=20. ** p<0.01, significantly different from WT. Statistical analyses were performed using Student’s t-tests. Three biological replicates were analyzed with similar results. (d) Photo of 67-day-old plants. bar = 2 cm. (e) Average final heights of ga1-3 and ga1-3 spy mutants. The data are means ± SE. n=13. Different letters above the bars indicate significant differences, p<0.01. Two biological replicates were analyzed with similar results. In (d–e), plants were grown under 16 hr-light conditions. (f) Malachite green-coupled assay shows that spy-8 retains a low POFUT activity, but no activity was detected for spy-15 or spy-19. Truncated 3TPR-SPY and -spy proteins were expressed and purified as His10-MBP fusion proteins for this assay using RGApep1 and GDP-fucose as substrates. Means ± SE are shown. Three biological replicates were analyzed with similar results. The immunoblot detected by anti-SPY antibodies indicates that similar amounts of WT and mutant enzymes were used in the assays (Supplementary Fig. 8a).
Three dimensional (3D)-structure modeling using the HsOGT crystal structure31 predicts that SPY and SEC fold similarly to HsOGT (Supplementary Fig. 1b, 1d–1e). K665 in SPY (mutated in spy-19) is absolutely conserved in all OGT and SPY-like sequences (Supplementary Fig. 10b), and is equivalent to K842 in HsOGT, which binds to the β-phosphate of UDP-GlcNAc and is critical for OGT activity27,32,33. It is possible that K665 in SPY serves a similar function as K842 in HsOGT by interacting with the β-phosphate of GDP-fucose to help position this nucleotide sugar in its substrate-binding pocket. E567 and G570 in SPY (mutated in spy-15 and spy-12, respectively) are also highly conserved in almost all OGT and SPY-like sequences (Supplementary Fig. 10a). However, the transition helix (H3, between the TPRs and catalytic domain) and the first two helices (H1 and H2) of N-Cat, which are important for the catalytic activity of HsOGT, are more divergent in SPY (Supplementary Fig. 1b, 1e, and Supplementary Fig. 10a). Most notably, two key residues (H498–H499 in HsOGT, and F540–H541 in AtSEC), which are important for OGT activity20,27,32,33, are missing in SPY. These differences may contribute to the distinct enzymatic activity of SPY.
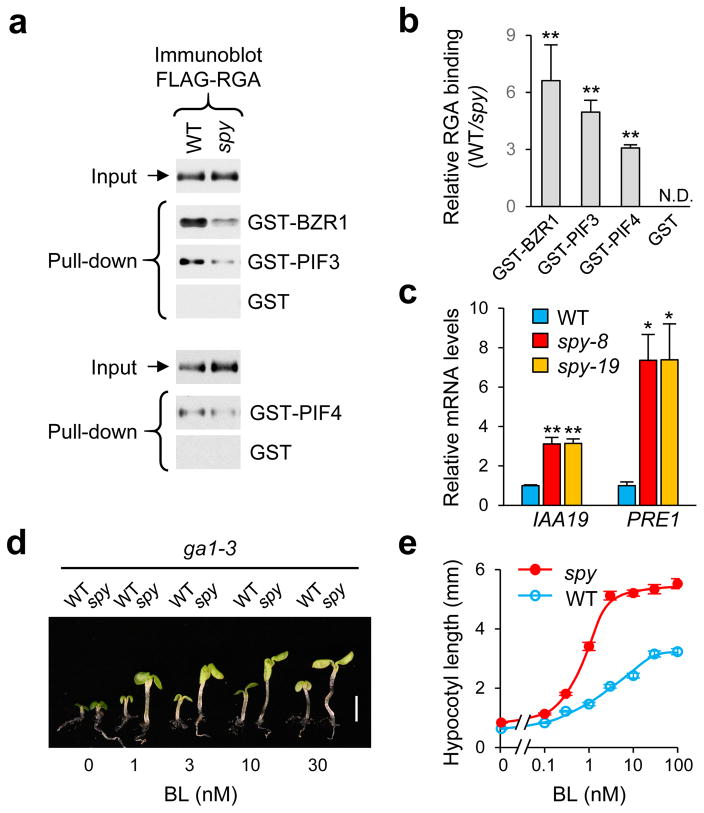
O-Fucosylation promotes RGA binding to its interactors
Previously, we showed that the spy mutation does not promote RGA degradation or affect RGA nuclear localization15. One possible role of RGA O-fucosylation by SPY is to regulate binding affinity between RGA and its interacting transcription factors, as this is a major regulatory mechanism for DELLA-modulated plant development6–8. Recent studies showed that BRZ1 (a BR-signaling activator) and PIFs (light-signaling regulators) are key transcription factors for promoting hypocotyl elongation in response to BR and light conditions, and that DELLAs inhibit hypocotyl growth by interacting with BZR1 and PIFs to repress expression of BZR1- and PIFs-induced genes6,7,34. To test the effect of spy on DELLA interaction with BZR1 and PIFs, an in vitro pull-down assay was performed using Arabidopsis protein extracts of PRGA:FLAG-RGA transgenic lines in the WT SPY or spy-8 background, and three recombinant RGA-interacting proteins, BZR1, PIF3 and PIF4, which were expressed individually in E. coli as glutathione S-transferase (GST) fusion proteins. GST was also included as a negative control. We found that FLAG-RGA (O-fucosylated) from WT displayed stronger binding to GST-BZR1, GST-PIF3 and GST-PIF4 than FLAG-RGA (less fucosylated) from the spy-8 mutant (Fig. 4a–4b, Supplementary Fig. 8b). These results suggest that O-fucosylation of RGA by SPY increases RGA activity by enhancing RGA interactions with BZR1 and PIFs. This model would predict that spy mutations should cause up-regulated expression of DELLA-repressed and BZR1/PIF-induced genes, which will lead to increased BR- and PIF-dependent responses. INDOLE 3-ACETIC ACID INDUCIBLE19 (IAA19) and PACLOBUTRAZOL RESISTANCE1 (PRE1) are 2 common target genes of BZR1, PIFs and DELLAs34–36. RT-qPCR analysis showed that transcript levels of IAA19 and PRE1 were up-regulated by spy-8 and spy-19 (Fig. 4c). We also found that ga1-3 spy seedlings had longer hypocotyls than ga1-3 (Fig. 3b–3c), which is known to be promoted by PIF3, PIF4 and PIF537,38. Moreover, in the presence of BR biosynthesis inhibitor, spy-8 enhanced BR response in hypocotyl elongation (Fig. 4d–4e). These results indicate that SPY negatively regulates GA-, BR- and PIF-dependent pathways by O-fucosylating RGA and enhancing its activity.
Figure 4. O-fucosylation enhances RGA activity by promoting RGA binding to its interactors.
(a) In vitro pull-down assay. Recombinant GST, GST-BZR1, GST-PIF3 and GST-PIF4 bound to glutathione sepharose beads were used separately to pull-down FLAG-RGA from protein extracts from Arabidopsis in WT SPY or spy-8 background. Immunoblots containing input Arabidopsis extracts and pulled-down samples, and FLAG-RGA was detected using an anti-FLAG antibody. Full blot images are shown in Supplemetary Fig. 12. Ponceau S-stained blots showed similar amounts of the GST/GST-fusion proteins were used in each pair of the pull-down assays (Supplementary Fig. 8b). (b) Relative binding of FLAG-RGA (from WT vs spy background) to GST-fusion proteins. The data are means ± SE (3 biological replicates). ** p<0.01. Statistical analyses were performed using Student’s t-tests. (c) RT-qPCR analysis shows that transcript levels of IAA19 and PRE1 (common target genes of BZR1, PIF and DELLA) were increased in spy-8 and spy-19 compared to those in WT (all in the ga1-3 background). PP2A was used to normalize samples. Data are means ± SE of 3 biological replicas and 2 technical repeats. * p<0.05, ** p<0.01. Statistical analyses were performed using Student’s t-tests. (d–e) spy-8 conferred an elevated BR response in hypocotyl growth comparing to WT SPY. The ga1-3 and spy-8 ga1-3 seedlings (labeled as WT and spy-8) were grown in media containing 2 μM propiconazole (a BR biosynthesis inhibitor), and varying concentrations of brassinolide (BL). Hypocotyl lengths were measured at day 6 (n=20). Bar in (d): 2.5 mm. Average hypocotyl lengths in (e) are means ± SE.
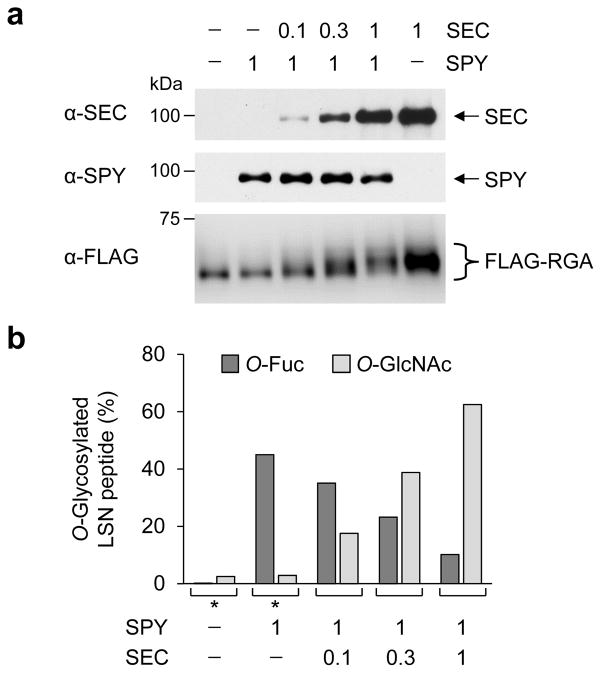
In contrast to the role of SPY in activating RGA, we showed previously that O-GlcNAcylation of RGA by SEC reduces RGA activity through interfering with RGA and BZR1/PIFs interactions20. Interestingly, the sites of O-fucosylation in RGA largely overlap with the sites of O-GlcNAcylation within the two structurally disordered regions: the LSN peptide near the N-terminus, and the PolyS/T region (Fig. 1a and Supplementary Fig. 11a)20. To test whether SPY and SEC compete with each other in modifying RGA, we transiently expressed FLAG-RGA together with a fixed amount of SPY and varying amounts of SEC in tobacco (Fig. 5). FLAG-RGA was then purified and analyzed by immunoblot and LC-ETD-MS/MS analyses. Increasing ratios of SEC-to-SPY correlated with slower gel mobility of FLAG-RGA (Fig. 5a), due to elevated O-GlcNAcylation20. MS analysis further showed that increasing levels of SEC led to reduced O-fucosylation and elevated O-GlcNAcylation in the RGA LSN peptide (Fig. 5b), supporting the notion that SPY and SEC compete with each other in regulating RGA activity by reciprocal modifications.
Figure 5. SPY and SEC compete with each other in reciprocal modifications of RGA.
(a) Immunoblots showing fixed amounts of SPY and increasing levels of SEC in tobacco extracts that were co-expressed with FLAG-RGA. The blots were probed with anti-SEC, anti-SPY or anti-FLAG antibodies. Tobacco agro-infiltrations used mixed agrobacterium cultures containing FLAG-RGAGKG and/or Myc-SPY, and/or Myc-SEC constructs (0.1× = OD600 0.06; 0.3× = OD600 0.2; 1× = OD600 0.6). The reduction in FLAG-RGA mobility correlated with increasing amounts of co-expressed SEC, due to greater numbers of GlcNAcylated residues in FLAG-RGA. Full blot images are shown in Supplemetary Fig. 12. (b) MS analysis shows that increasing expression of SEC results in elevated O-GlcNAcylation, but is inversely correlated with O-fucosylation levels in the RGA LSN peptide. Peptide abundances were calculated from MS1 ion currents integrated across chromatographic elution of peptide. * MS data from Fig. 1d are included here for comparison.
DISCUSSION
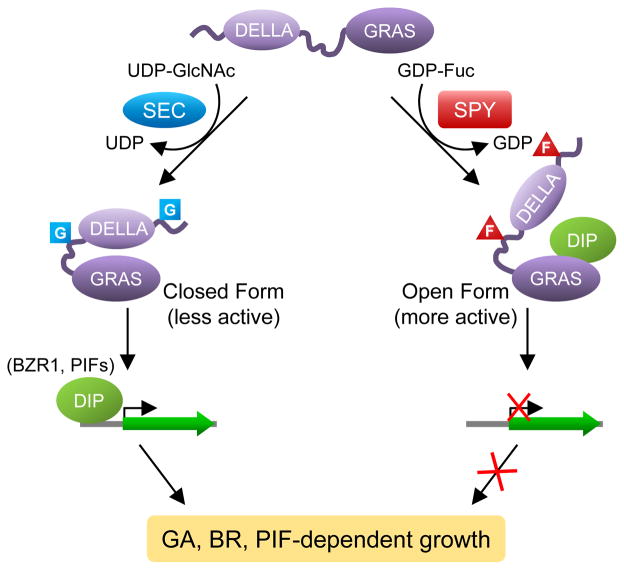
Our study discovered the novel O-fucosylation of nuclear DELLA repressor by SPY in Arabidopsis. Sequence comparison and 3D structure prediction indicate that SPY is highly similar to the TPR-containing OGTs17 (Supplementary Fig. 1 and Supplementary Fig. 10). This similarity to the animal OGTs led to the longstanding hypothesis for SPY’s function as an OGT17. However, results of previous enzymatic assays were inconclusive. We now show that SPY is an unusual POFUT, which is unrelated to the ER-localized POFUTs that modify secreted cell surface proteins in animals22. We further show that SPY-dependent O-fucosylation enhances DELLA activity, which is opposite to the role of SEC as an inhibitor of DELLA by O-GlcNAcylation. In addition, SPY and SEC compete with each other in modifying RGA in the transient co-expression assay in tobacco. We propose that O-GlcNAcylation may stabilize RGA in a “closed form”, whereas O-fucosylation may induce a conformational transition of RGA to an “open form” (Fig. 6). The O-GlcNAcylated closed form of RGA is a less active growth repressor because this conformation prevents interaction with DELLA-interacting proteins (DIPs, such as BZR1 and PIFs), whereas the O-fucosylated open form of RGA is a more active growth repressor as its GRAS domain can bind DIPs efficiently.
Figure 6. Model for the role of O-fucosylation vs. O-GlcNAcylation of RGA.
O-Fucosylation (labeled as F) by SPY may induce RGA to an open conformation that is a more active growth repressor; this open form promotes binding of the RGA GRAS domain to interacting transcription factors (e.g., BZR1 and PIFs), which leads to down-regulated expression of target genes of BZR1 and PIFs to restrict plant growth. In contrast, O-GlcNAcylation (labeled as G) by SEC may cause RGA to fold into a closed conformation that is less active because this form reduces the binding affinity of RGA to BZR1 and PIFs so that growth-related target genes can be activated. DIP, DELLA-interacting protein.
The sites of O-fucosylation and O-GlcNAcylation in RGA are clustered within two regions: the LSN peptide near the N-terminus, and the PolyS/T region (Fig. 1a and Supplementary Fig. 11a)20. These two regions in RGA are structurally disordered, and the sequences surrounding the glycosylation sites are not conserved among the five AtDELLAs (RGA, GAI, RGL1, RGL2 and RGL3; Supplementary Fig. 11a and 11b). Nonetheless, co-expression of SEC with each AtDELLA in the tobacco system resulted in slower gel mobility of these AtDELLAs, supporting that SEC O-GlcNAcylates all AtDELLAs20. To investigate whether SPY also modifies the other AtDELLAs, MS analysis was performed because O-fucosylation of RGA did not cause visible gel mobility shift (Supplementary Fig. 2)20. Our initial analysis showed that RGL1 is O-fucosylated by SPY (Supplementary Fig. 3). These results indicate that, in addition to RGA, SEC and SPY also glycosylate and regulate the activities of at least some of the other AtDELLAs. This idea is supported by the dramatically longer hypocotyl and taller final height of the ga1 rga spy triple mutant in comparison to the ga1 rga double mutant39. Our results also suggest that SEC and SPY may modify the flexible regions of target proteins by interacting with the substrate amide backbone, similar to HsOGT27.
The truncated 3TPR-SPY, which lacks TPR4-11, was as active as FL-SPY to modify RGA when they were overexpressed in tobacco. However, TPR4-11 of SPY may be important for SPY’s function in vivo, for example, in recruiting target proteins and/or forming protein complexes with its interactors. This idea could be tested by expression of 3TPR-SPY under the control of the SPY promoter in the spy mutant background. Among several spy alleles with mutations either in TPR2-3 (spy-8) or catalytic domain (spy-12, -15, -19), we found that spy-19 (K665M) displayed a more severe phenotype at the adult stage, although all spy alleles tested showed similar hypocotyl phenotype at the seedling stage (Fig. 3b–3e). The elevated GA signaling caused by the leaky spy-8 might already saturate the GA response for hypocotyl growth. Therefore, no difference was observed for this phenotype and expression of corresponding growth-related genes (IAA19 and PRE1) among the different spy mutants. In contrast, inflorescence stem growth may require higher GA signaling activities to reach saturated GA response.
OGT-mediated protein O-GlcNAcylation has been studied extensively in animals, and is known to play a key role in regulating a plethora of intracellular signaling events in response to nutrient and stress status40,41. In plants, O-fucosylation and O-GlcNAcylation are likely to play important roles in regulating multiple intracellular processes. The spy mutants display pleiotropic phenotypes, some of which (e.g., fused cotyledons and altered phyllotaxy) are unrelated to GA responses15. Previous genetic studies further showed that SPY promotes phytohormone cytokinin signaling42, and regulates circadian rhythms43. Our studies indicate that SPY and SEC play opposite roles in regulating DELLA activity. Intriguingly, the spy sec double mutations confer embryo-lethal phenotype18. The impaired embryogenesis of the spy sec double mutant is likely due to defect(s) in other pathways that are not directly related to GA signaling. O-GlcNAcylation in animals affects many fundamentally important cellular functions, including epigenetic regulation of chromatin structure, transcription, translation and protein degradation40,41. If O-GlcNAcylation and O-fucosylation also modulate these processes in plants, disruption of multiple cellular functions/pathways in the spy sec double mutant would be deleterious to the developing embryo. Interestingly, SPY-like genes are absent in animals, but are present in diverse organisms, ranging from prokaryotes, protists, algae to all plants17, although their biochemical functions have not been well characterized. These conserved SPY orthologs in other organisms may have POFUT or other novel glycosyltransferase activities that are distinct from OGT. Future studies will reveal the role of intracellular protein O-fucosylation in regulating developmental processes in diverse organisms.
ONLINE METHODS
Plant materials, plant growth conditions, and plant transformation
The spy-8, spy-12, spy-15 and spy-19 mutants were described previously15,39. The DNA mutation (1994A-to-T) in spy-19 was identified by sequencing of PCR-amplified spy-19 genomic DNA. For in vitro pull-down assays and mass spectrometry analyses, a PRGA:FLAG-RGA transgenic Arabidopsis line in the sly1-10 rga-24 background20 was crossed with spy-8 to generate PRGA:FLAG-RGA sly1-10 spy-8 rga-24. The F-box sly1-10 mutation promotes accumulation of RGA11.
Plant growth conditions were as previously described44, with the following modifications: for affinity purification of FLAG-RGA protein for MS analysis, Arabidopsis seedling cultures were not supplemented with PuGNAc or glucosamine.
For BR response, 2 μM propiconazole was included in the media in addition to increasing concentrations of BL; for RT-qPCR analyses of DELLA, BZR1 and PIF target genes, seedlings were grown under constant light for 9 days. For tobacco agro-infiltrations, 3-week old plants of Nicotiana benthamiana were used.
Statistical analysis
Statistical analyses were performed with the statistical package JMP Pro 10.0.2 (SAS Institute Inc.), using Student’s t-tests.
Plasmid construction
Primers for plasmid constructions are listed in Supplementary Table 2. All DNA constructs generated from PCR amplification were sequenced to ensure that no mutations were introduced.
For transient co-expression in tobacco, 35S:His-FLAG-RGAGKG, 35S:Myc-SPY and 35S:Myc-SEC constructs have been described previously20. To generate additional His-FLAG-AtDELLA constructs, the coding region of RGA in 35S:His-FLAG-RGA was substituted by the coding sequences of GAI, RGL1, RGL2 and RGL3 that had been cloned into pCR8/GW/TOPO20. For 35S:His-FLAG-GAI, -RGL1 and -RGL3 constructs, the 35S:His-FLAG-RGA construct was cut with BamHI/XbaI to remove RGA sequence, and then ligated with DNA fragments of GAI (BamHI/XbaI), RGL1 (BamHI/XbaI) and RGL3 (BglII/SpeI), respectively. For 35S:His-FLAG-RGL2, 35S:His-FLAG-RGA was cut with BamHI/XmaI and ligated with the BglII/XmaI fragment from pCR8-RGL2 containing the RGL2 coding sequence. For constructs 35S:Myc-spy-8 and 35S:Myc-spy-12, cDNA from mutant plants was PCR-amplified with primers AtSPY-B1F/AtSPY-B2R and the product cloned into pDONR221 by BP-recombination and moved to pEarlyGate-20345 by LR-recombination. To generate 35S:Myc-spy-15, site-directed mutagenesis of pDONR221-SPY was performed using primers spy15+/spy15− and the QuickChange Lightning strategy (Agilent Technologies). The resulting plasmid pDONR221-spy-15 was then LR recombined with pEarlyGate-203. To generate 35S:Myc-spy-19, the template pDONR221-SPY was PCR-amplified with primers attB1-site/spy-19-R and attB2-site/spy-19-F. The two PCR products were mixed for a second round of amplification with primers attB1-site/attB2-site. The resulting product was cloned into pDONR207 by BP-recombination and moved to pEarlyGate-203 by LR-recombination. 35S:3TPR-SPY construct was obtained by PCR amplification with primers 3TPR-SPY-ATG-F/SPY-STOP-R and cloning into pCR8/GW/TOPO (Thermo Fisher Scientific) and moved to pEarlyGate-203 by LR-recombination.
For in vitro enzyme assays, pTrc-His-MBP-3TPR-SPY and pTrc-His-MBP-5TPR-SEC constructs were generated by PCR-amplification of 3TPR-SPY and 5TPR-SEC from pDONR221-SPY and pENTR1A-SEC, respectively, using primers SPY-TPR3/SPY- and SEC-TPR5/SEC-; the products were digested with BamHI and KpnI and cloned into the pTrc-His10-MBP-TEV vector derived from pTrcHisB (Thermo Fisher Scientific), a gift from Jae Cho in Pei Zhou’s lab. A similar procedure was used to create pTrc-His-MBP-3TPR-spy-8, -15 and -19, except that the templates used were pDONR221-spy-8, -spy-15 and pDONR207-spy-19, respectively.
GST-fusion constructs for pull-down assays have been previously described20,46.
Tobacco agro-infiltrations
Tobacco leaf co-infiltration procedures were performed as described20,47. For most co-expression experiments, equal amounts of agrobacterium cultures harboring different expression constructs were mixed prior to infiltration. For the SPY and SEC competition experiments, cultures containing FLAG-RGA-GKG and Myc-SPY constructs were used at full-strength (1× = OD600 0.6), whereas Myc-SEC was included at different dilutions (0.1× = OD600 0.06; 0.3× = OD600 0.2; 1× = OD600 0.6).
Protein purification for MS analysis
His-FLAG-RGA was purified from PRGA:FLAG-RGA transgenic Arabidopsis lines in rga-24 ga1-3 (WT for both SPY and SEC), spy-8 rga-24 ga1-3 or sec-3 rga-24 ga1-3 backgrounds. The tandem affinity purification procedures were as described previously20 with the following modifications: 5 g of tissue of lines PRGA:FLAG-RGA sly1-10 rga-24 with or without spy-8 or sec-3 mutations were homogenized in 3 volumes (m/v) of Buffer B. The cleared extract was incubated with 0.4 mL of His-Bind resin for 1.5 h at 4°C and loaded onto a disposable plastic column. The resin was washed with 10 bed vol. of Buffer B followed by 10 bed vol. Buffer B supplemented with 20 mM imidazole prior to elution. The second purification step was carried out with 5 μL of anti-FLAG-agarose beads (Sigma-Aldrich).
The purification procedure of His-FLAG-RGAGKG from tobacco samples agro-infiltrated with 35S:His-FLAG-RGAGKG alone or co-infiltrated with 35S:Myc-SPY or -spy, -3TPR-SPY or 35S:Myc-SEC was as described previously20. However, only 5 g of tissue per sample were used and an additional wash with 20 mM imidazole in Buffer B was included prior to elution from the His-Bind column.
Protein purification for in vitro enzyme assays
Recombinant 3TPR-SPY (spanning residues 326–914) protein was expressed in E.coli BL21 with an N-terminal decahistidine-maltose binding protein (His10-MBP) followed by a TEV protease cleavage site (ENLYFQG/S). Cells were grown in LB media at 37°C until A600 0.5, at which point the culture was cooled on ice before induced with 0.5mM IPTG and incubated at 25°C for 4 h. Cells were pelleted, resuspended in lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, 10 mM 2-mercaptoethanol), and disrupted using French pressure cell, followed by centrifugation at 40,000g for 60min. The supernatant was incubated with Ni-NTA agarose superflow resin (Qiagen), washed with 10-column volumes of wash buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 50 mM imidazole, 10 mM 2-mercaptoethanol), and eluted with 4-column volumes of elution buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 250 mM imidazole, 10 mM 2-mercaptoethanol). The elute was concentrated with centrifugal concentrators (Millipore) and buffer exchanged into TEV digestion buffer (50 mM NaH2PO4, pH 7.5, 200 mM NaCl, 0.5 mM EDTA) before treated with TEV protease at 4°C for 16 h. A second Ni-NTA column was used to remove His10-MBP tag as well as the TEV protease. The target protein was further purified with size-exclusion chromatography (HiPrep Sephacryl S-300 HR, GE Healthcare). The final protein was concentrated to 10mg/ml in storage buffer (20 mM Tris, pH 8.0, 50 mM NaCl, 1 mM DTT) and stored at −80°C.
3TPR-spy mutants and 5TPR-SEC (spanning residues 356–977) were purified using similar method, except that the His10-MBP tag was not removed to enhance the solubility of the purified protein.
Immunoblot analysis
To determine FLAG-RGA, Myc-SPY and Myc-SEC protein levels in Arabidopsis or tobacco tissues, total proteins were extracted and analyzed by SDS-PAGE and immunoblotting as described48. An anti-cMyc-HRP antibody (Sigma-Aldrich, Cat. A5598; 8,000× dilution) was used to detect Myc-SPY. A monoclonal anti-FLAG-HRP antibody (Sigma-Aldrich, Cat. A8592; 10,000× dilution) was used to detect FLAG-RGA. An anti-SEC antisera20 (2,000× dilution) and anti-rabbit IgG-HRP (Thermo-Fisher, Cat. 31462; 20,000× dilution) were used to detect Myc-SEC. Affinity-purified polyclonal anti-SPY antibodies (anti-LQKE49; 6,000× dilution), and anti-rabbit IgG-HRP (same as above) were used to detect recombinant WT and mutant SPY proteins for the in vitro enzyme assays, and for Myc-SPY in the tobacco samples.
Identification of O-fucosylation and -GlcNAc sites by liquid chromatography (LC)-Electron Transfer Dissociation (ETD) and Collisionally Activated Dissociation (CAD)-tandem MS (MS/MS) analyses
His-FLAG-RGA proteins, purified from Arabidopsis and tobacco respectively, were digested with Trypsin and then separated by HPLC and analyzed by tandem MS using an Orbitrap Fusion Tribrid Mass Spectrometer equipped with ETD (Thermo Scientific)50,51. Sample processing and HPLC were as described previously20. MS1 spectra were acquired in the Orbitrap at a resolution of 120,000 followed by a data-dependent, 2-second Top-N, MS/MS experiment alternating between CAD and ETD. Precursors were isolated by resolving quadrupole with a 2 m/z window. MS2 spectra were acquired in the linear ion trap at a normal scan rate, and ETD reaction times were varied according to precursor charge state. Dynamic exclusion was turned on with a repeat count of 1 and exclusion duration of 10 sec.
In vitro SPY and SEC activity assays by MALDI-MS
Two RGA peptides were synthesized (ChinaPeptides) for the in vitro enzyme assays: peptide 1, FQGRLSNHGTSSSSSSISK (amino acid residues 8–26, RGApep1); peptide 2, KSCSSPDSMVTSTSTGTQGGW (amino acid residues 166–183 +GGW, RGApep2). Each 20 μL reaction includes 125 μM RGA peptide, 200 μM GDP-fucose (or UDP-GlcNAc, GDP-mannose, UDP-galactose and UDP-glucose in substrate specificity test) and 10 μg 3TPR-SPY or 5TPR-SEC. For SPY, the reaction buffer contains 50 mM Tris-HCl, pH 8.2, 50 mM NaCl, 5 mM MgCl2 (unless otherwise stated for buffer condition optimization). For SEC, the reaction buffer contains 20 mM Tris-HCl, pH 7.2, 12.5 mM MgCl2. After incubating for 2 h (unless specified to be 16 h) at 25°C, the peptides were purified using C18 Ziptips (MilliporeSigma) according to the manufacturer’s protocol, and analyzed by MALDI-MS analysis using .an Ultraflex II TOF/TOF mass spectrometer (Bruker Daltonics). Sample processing was performed as described previously52. For each reaction, 4 μL sample was mixed with 6 μL α-cyano-4-hydroxycinnamic acid (HCCA) matrix, and 1.2 μL of the mixture was spotted onto the plate and dried before detected by MALDI-MS.
In vitro SPY activity assay by malachite green-coupled reaction
Kinetics
Steady-state kinetics of 3TPR-SPY were determined using malachite green-coupled reaction as described before29. Briefly, a 130 μL reaction containing 50 mM Tris-HCl (pH 8.2), 50 mM NaCl, 5 mM MgCl2, 50 μg of 3TPR-SPY, 0.5 μg of CD39L3 (NTPDase 3, R&D Systems) with different concentrations of substrates was incubated at 25°C. A 25 μL aliquot was collected at 2, 4, 6, 8 and 10 min, separately, and inactivated at 95°C for 30 sec before assaying free phosphate release using malachite-green reagent. Kinetics parameters for GDP-fucose was measured with a fixed RGApep1 concentration of 312.5 μM and varying GDP-fucose concentrations of 800, 400, 200, 100, 50, 25 and 12.5 μM. Kinetics parameters for the peptide was measured with a fixed GDP-fucose concentration of 800 μM and varying RGApep1 concentrations of 100, 50, 37.5, 25, 12.5, 6.25, 3.125 and 1.56 μM. Data were fitted to Lineweaver-Burk plot to determine the kinetics parameters.
Activity assay of spy mutants
25 μL reactions containing 50 mM Tris-HCl, pH 8.2, 50 mM NaCl, 5 mM MgCl2, 125 μM RGApep1, 200 μM GDP-fucose, 0.1 μg of NPTDase-3 and 7 μg of 3TPR-SPY or mutant spy protein (His-MBP fusion proteins) were incubated for 10 min at 25°C before quenched and detected by malachite-green reagent. Each reaction was at least triple-duplicated.
In vitro pull-down assays and RT-qPCR analysis
Pull-down assays using Arabidopsis extracts and GST-fusion proteins produced in E. coli were carried out as described previously20. Recombinant GST, GST-BZR1, GST-PIF3 and GST-PIF4 bound to glutathione sepharose beads were used separately to pull-down FLAG-RGA from protein extracts from PRGA:FLAG-RGA transgenic Arabidopsis in SPY sly1-10 rga-24 or spy-8 sly1-10 rga-24 background.
For RT-qPCR analysis, total RNA was isolated from 9-day old seedlings of ga1-3, ga1-3 spy-8 and ga1-3 spy-19 using a Quick-RNA MiniPrep kit (Zymo Reseaarch). First strand cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis kit (Roche Applied Science). For qPCR, the FastStart Essential DNA Green Master mix was used on a LigthCycler 96 Instrument (Roche Applied Science).
In silico predictions of 3D protein structures
SPY and SEC 3D structure models based on the human OGT structure (4N3C)31 were generated using the online SWISS-MODEL Workspace (http://swissmodel.expasy.org/)53,54. 3D structure alignment, visualization and manipulations were performed with PyMOL (www.pymol.org).
Sequence alignment
Protein sequences were downloaded from the National Center for Biotechnology Information website, and sequence alignment was carried out with the DNAStar Lasergene Core Suite Ver. 12.0.0.
Data availability
Authors can confirm that all relevant data are included in the paper and in its Supplementary Information file and Supplementary Notes.
Supplementary Material
Acknowledgments
We thank Cliff Toleman for his help with in vitro enzyme assay, and George Dubay at Duke Chemistry Instrument Facilities for help with the MALDI-MS analysis. We also thank Zhen-Ming Pei and Jim Siedow for helpful comments on the manuscript. This work was supported by the National Institutes of Health (R01 GM100051 to TPS, R01 GM037537 to DFH), the National Science Foundation (MCB-0923723 to TPS, MCB-0516690, -0820666 and -1158089 to NEO), Department of Energy (DE-SC0014077 to TPS), and US Department of Agriculture (2014-67013-21548 to TPS).
Footnotes
Author contributions
R.Z., N.S., and T.S. conceived and designed the research project. R.Z., N.S., W.H., J.H. performed molecular biology and biochemical analysis; R.Z., N.S., P.Z. and T.S. analyzed data. B.B. performed LC-ETD-MS/MS analysis, and B.B., J.S. and D.H. analyzed MS data. P.Z. also provided advice on protein purification and structure modeling. M.B. provided reagents and advice for in vitro enzyme assay using MALDI-MS; N.E.O provided experimental materials and shared unpublished results. R.Z., N.S. and T.S. wrote the manuscript.
Competing financial interests statements
The authors declare no competing financial interests.
References
- 1.Vert G, Chory J. Crosstalk in cellular signaling: background noise or the real thing? Dev Cell. 2011;21:985–991. doi: 10.1016/j.devcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silverstone AL, Ciampaglio CN, Sun T-p. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng J, et al. The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21:R338–345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 5.Daviere JM, Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- 6.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 7.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H, Liu Q, Yao T, Fu X. Shedding light on integrative GA signaling. Curr Opin Plant Biol. 2014;21:89–95. doi: 10.1016/j.pbi.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Ueguchi-Tanaka M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 10.Murase K, Hirano Y, Sun T-p, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- 11.McGinnis KM, et al. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell. 2003;15:1120–1130. doi: 10.1105/tpc.010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki A, et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299:1896–1898. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths J, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dill A, Jung HS, Sun T-p. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci SA. 2001;98:14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverstone AL, et al. Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 2007;143:987–1000. doi: 10.1104/pp.106.091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson RN, Somerville CR. Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 1995;108:495–502. doi: 10.1104/pp.108.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olszewski NE, West CM, Sassi SO, Hartweck LM. O-GlcNAc protein modification in plants: Evolution and function. Biochimi Biophys Acta. 2010;1800:49–56. doi: 10.1016/j.bbagen.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartweck LM, Scott CL, Olszewski NE. Two O-Linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics. 2002;161:1279–1291. doi: 10.1093/genetics/161.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zentella R, et al. O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 2016;30:164–176. doi: 10.1101/gad.270587.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luther KB, Haltiwanger RS. Role of unusual O-glycans in intercellular signaling. Int J Biochem Cell Biol. 2009;41:1011–1024. doi: 10.1016/j.biocel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 24.Hofsteenge J, et al. C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J Biol Chem. 2001;276:6485–6498. doi: 10.1074/jbc.M008073200. [DOI] [PubMed] [Google Scholar]
- 25.Lira-Navarrete E, et al. Structural insights into the mechanism of protein O-fucosylation. PloS One. 2011;6:e25365. doi: 10.1371/journal.pone.0025365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CI, et al. Structure of human POFUT2: insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J. 2012;31:3183–3197. doi: 10.1038/emboj.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase: role of the tetratricopeptide repeats. J Biol Chem. 1999;274:32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- 29.Wu ZL, Ethen CM, Prather B, Machacek M, Jiang W. Universal phosphatase-coupled glycosyltransferase assay. Glycobiology. 2011;21:727–733. doi: 10.1093/glycob/cwq187. [DOI] [PubMed] [Google Scholar]
- 30.Baykov AA, Evtushenko OA, Avaeva SM. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 31.Lazarus MB, et al. HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science. 2013;342:1235–1239. doi: 10.1126/science.1243990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schimpl M, et al. O-GlcNAc transferase invokes nucleotide sugar pyrophosphate participation in catalysis. Nat Chem Biol. 2012;8:969–974. doi: 10.1038/nchembio.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Fleites C, et al. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat Struct Mol Biol. 2008;15:764–765. doi: 10.1038/nsmb.1443. [DOI] [PubMed] [Google Scholar]
- 34.Bai MY, et al. Brassinosteroid, gibberellin, and phytochrome signalling pathways impinge on a common transcription module in Arabidopsis. Nat Cell Biol. 2012;14:810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh E, et al. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife. 2014;3:e03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallego-Bartolome J, et al. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 2012;109:13446–13451. doi: 10.1073/pnas.1119992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 38.Soy J, et al. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 2012;71:390–401. doi: 10.1111/j.1365-313X.2012.04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silverstone AL, Mak PYA, Casamitjana Martínez E, Sun T-p. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics. 1997;146:1087–1099. doi: 10.1093/genetics/146.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol. 2015;208:869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenboim-Wainberg Y, et al. Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell. 2005;17:92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tseng TS, Salome PA, McClung CR, Olszewski NE. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell. 2004;16:1550–1563. doi: 10.1105/tpc.019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zentella R, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 46.Dill A, Thomas SG, Hu J, Steber CM, Sun T-p. The Arabidopsis F-box protein SLEEPY1 targets GA signaling repressors for GA-induced degradation. Plant Cell. 2004;16:1392–1405. doi: 10.1105/tpc.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang ZL, et al. SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing DELLA in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:2160–2165. doi: 10.1073/pnas.1012232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silverstone AL, et al. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13:1555–1566. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swain SM, Tseng T-s, Thornton TM, Gopalraj M, Olszewski N. SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiol. 2002;129:605–615. doi: 10.1104/pp.020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Udeshi ND, Compton PD, Shabanowitz J, Hunt DF, Rose KL. Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry. Nat Protoc. 2008;3:1709–1717. doi: 10.1038/nprot.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berk JM, et al. O-GlcNAc regulates emerin binding to BAF in a chromatin- and lamin B-enriched ‘niche’. J Biol Chem. 2013;288:30192–30209. doi: 10.1074/jbc.M113.503060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu Y, Strickland EC, Fitzgerald MC. Thermodynamic analysis of protein folding and stability using a tryptophan modification protocol. Anal Chem. 2014;86:7041–7048. doi: 10.1021/ac501278j. [DOI] [PubMed] [Google Scholar]
- 53.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 54.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sokol KA, Olszewski NE. The putative eukaryote-like O-GlcNAc transferase of the cyanobacterium Synechococcus elongatus PCC 7942 hydrolyzes UDP-GlcNAc and is involved in multiple cellular processes. J Bacteriol. 2015;197:354–361. doi: 10.1128/JB.01948-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Banerjee S, Robbins PW, Samuelson J. Molecular characterization of nucleocytosolic O-GlcNAc transferases of Giardia lamblia and Cryptosporidium parvum. Glycobiology. 2009;19:331–336. doi: 10.1093/glycob/cwn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ostrowski A, Gundogdu M, Ferenbach AT, Lebedev AA, van Aalten DM. Evidence for a Functional O-Linked N-Acetylglucosamine (O-GlcNAc) System in the Thermophilic Bacterium Thermobaculum terrenum. J Biol Chem. 2015;290:30291–30305. doi: 10.1074/jbc.M115.689596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Authors can confirm that all relevant data are included in the paper and in its Supplementary Information file and Supplementary Notes.