Abstract
Fanconi anemia (FA) is a rare autosomal and X-linked genetic disease characterized by congenital abnormalities, progressive bone marrow failure (BMF), and increased cancer risk during early adulthood. The median lifespan for FA patients is approximately 33 years. The proteins encoded by the FA genes function together in the FA-BRCA pathway to repair DNA damage and to maintain genome stability. Within the past two years, five new FA genes have been identified - RAD51/ FANCR, BRCA1/FANCS, UBE2T/FANCT, XRCC2/FANCU, and REV7/FANCV - bringing the total number of disease-causing genes to 21. This review summarizes the discovery of these new FA genes and describes how these proteins integrate into the FA-BRCA pathway to maintain genome stability and critically prevent early-onset BMF and cancer.
Keywords: Fanconi anemia, Genome instability, DNA repair, homologous recombination, ubiquitin
1. Introduction
In the 1920’s, Swiss pediatrician Guido Fanconi first described the disease later to become formally recognized as Fanconi anemia (FA).1 In a family with five children, three brothers died of a severe condition that resembled pernicious anemia. The three patients’ disease manifested between the ages of five and seven and was associated with congenital microcephaly, café au lait spots, cutaneous hemorrhage, and hypoplasia of the testes, concurrent with a current day diagnosis of FA.2
With an estimated incidence of 1 in 360,000 live births and a carrier frequency of approximately 1 in 181, FA is relatively uncommon and can be difficult to diagnose due to patients presenting with a wide variety of symptoms.3, 4 Symptoms that could raise suspicion of FA among physicians include various congenital anomalies including microcephaly, microphthalmia, abnormal thumbs or radii, and slow growth rate. Hematological signs can include early-onset aplastic anemia, myelodysplastic syndrome (MDS), acute myeloid leukemia (AML) at an atypically young age, and one or more unexplained cytopenias of any cell lineage, including thrombocytopenia, neutropenia, and anemia. Additionally, clinicians should be mindful of the presentation of solid tumors at a particularly young age, specifically, head, neck, esophageal, and gynecological squamous cell carcinomas. Familial cancer predisposition or a history of chemotherapeutic hypersensitivity can also raise the suspicion of FA (see Practice points).3, 5, 6
After many decades of work, the FA genes are rapidly being identified. The FANCC gene was identified in 1992.7, 8 Subsequent discoveries of the FANCA,9, 10 FANCG,11 FANCE,12 FANCF,13 and FANCD214 genes followed. Correlating with rapid advances in genetic and biochemical technologies, the rate of FA gene identification has promptly increased. With the most recent additions, there are now 21 confirmed FA genes; FANCA, FANCB, FANCC, BRCA2/ FANCD1, FANCD2, FANCE, FANCF, FANCG, FANCI, BRIP1/FANCJ, FANCL, FANCM, PALB2/FANCN, RAD51C/FANCO, SLX4/FANCP, ERCC4/FANCQ, and most recently, and as highlighted in this review, RAD51/FANCR, BRCA1/FANCS, UBE2T/FANCT, XRCC2/FANCU, and MAD2L2/REV7/FANCV. The previously designated FA-H complementation group was found to be analogous to FA-A, and as such FANCH was removed.15, 16. The addition of five new FA genes over the past two years is testament to the continued resolve of the international FA research community and the Fanconi Anemia Research Fund.
2. The FA-BRCA pathway
One major function of the FA-BRCA pathway is to orchestrate the repair of DNA interstrand crosslinks (ICLs).17, 18 Examples of ICL-inducing agents include diepoxybutane (DEB) and mitomycin C (MMC). ICLs pose a direct physical block to DNA replication and RNA transcription and result in cellular cytotoxicity and chromosome structural aberrations if not properly repaired. FA patient cells from all complementation groups are characteristically hypersensitive to ICLs, and this phenotype forms the basis of the clinical FA diagnostic test.3, 19 Specifically, FA patient cells exhibit increased radial chromosome formations following ICL induction, a consequence of a molecular roadblock in ICL repair. ICL repair mediated by the FA proteins can be described as a tri-phasic process.
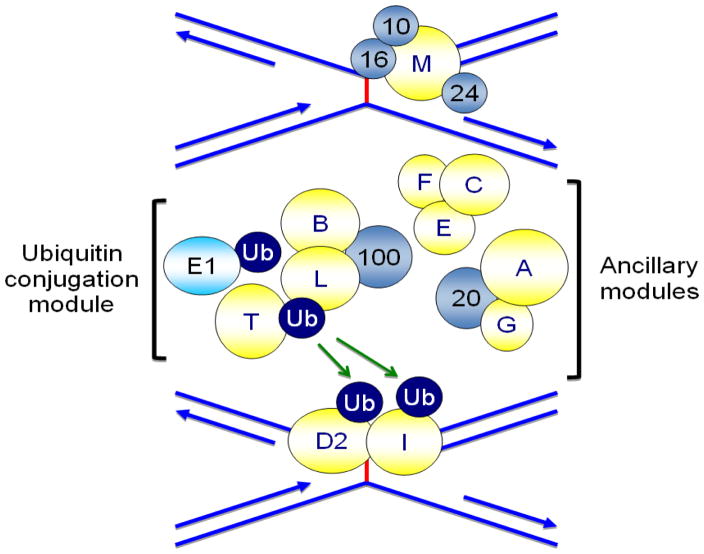
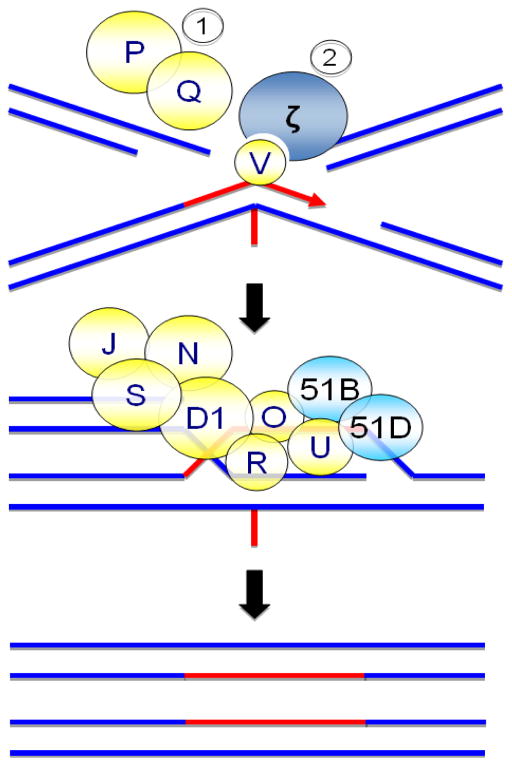
In the first phase, the upstream FA proteins FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, and FANCL, as well as the Fanconi anemia-associated proteins FAAP20 and FAAP100, assemble to form the FA core complex (Fig. 1). Upon DNA damage, the FA core complex is recruited to chromatin where it interacts with UBE2T/FANCT, which is constitutively chromatin localized.20, 21 Recent studies have identified three distinct modules within the FA core complex: the FANCB-FANCL-FAAP100 module, which provides the essential monoubiquitination catalytic activity, and the FANCA-FANCG-FAAP20 and FANCC-FANCE-FANCF modules, the exact functions of which have yet to be determined.22, 23 The FANCM anchor complex, comprising FANCM, FAAP24, FAAP16/MHF1 and FAAP10/MHF2, promotes the chromatin recruitment of the FA core complex.24–27 Together, the FA core complex and UBE2T/FANCT constitute an active multi-subunit E2/E3 ubiquitination enzyme complex. The RING domain-containing FANCL subunit functions as the E3 ubiquitin ligase while UBE2T/FANCT functions as the E2 ubiquitin-conjugating enzyme.28–30 This complex catalyzes the second phase of the pathway, the conjugation of a single ubiquitin moiety (monoubiquitin) to K561 of FANCD2 and K523 of FANCI.31–33 FANCD2 and FANCI form a heterodimer known as ID2.34 Ubiquitin is a 76-amino acid protein that is covalently posttranslationally attached to target proteins. Monoubiquitin functions as a molecular signal that regulates diverse cellular processes including the targeting of proteins to distinct subcellular locations and the promotion of protein-protein interactions.35 As evidence of the critical role the FA core complex plays in this process, FA patient cells harboring deleterious mutations in FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, or FANCT lack the capacity to monoubiquitinate the ID2 heterodimer.31–33 Monoubiquitinated ID2 localizes to chromatin where it associates with several established DNA repair proteins, including BRCA1/FANCS, BRCA2/FANCD1, and RAD51/ FANCR31, 36, 37 Monoubiquitinated ID2 is thought to function in the recruitment of several DNA repair proteins, including CtIP, Fanconi anemia associated nuclease 1 (FAN1), SLX4/FANCP, and ERCC4/FANCQ.38–44 SLX4/FANCP and the ERCC4/FANCQ endonuclease catalyze the unhooking of the ICL,39 enabling translesion DNA synthesis (TLS) beyond the ICL by the multi-subunit TLS polymerase Polζ, one subunit of which is REV7/FANCV (Fig. 2).45, 46
Figure 1.
Schematic of FANCD2 and FANCI monoubiquitination. Following exposure to DNA damaging agents and during S-phase of the cell cycle, the FANCM anchor complex, comprising FANCM, FAAP24, FAAP16/MHF1 and FAAP10/MHF2, recognizes the damage, remodels the fork, and promotes the recruitment of the FA core complex. The FA core complex, which is comprised of three sub-complexes - FANCB/FANCL/FAAP100, FANCC/FANCE/FANCF, and FANCA/FANCG/FAAP100 - together with the E2 ubiquitin-conjugating enzyme UBE2T/FANCT, constitutes an active multisubunit E2/E3 ubiquitination enzyme complex. This E2/E3 enzyme complex catalyzes the site-specific monoubiquitination of FANCD2 K561 and FANCI K523.
Figure 2.
Schematic of downstream steps of the FA pathway. During the later stages of ICL repair, SLX4/FANCP and ERCC4/FANCQ catalyze the unhooking of the ICL (Step 1). The unhooked ICL is then bypassed by Polζ (Step 2). REV7/FANCV is a structural component of Polζ that enhances the polymerase activity of the catalytic subunit, REV3. Downstream of FANCD2 and FANCI monoubiquitination, BRCA2/FANCD1, BRIP1/FANCJ, PALB2/FANCN, RAD51C/FANCO, RAD51/FANCR, BRCA1/FANCS, and XRCC2/FANCU all function cooperatively to repair the remaining broken duplex by homologous recombination (HR). Following 5′–3′ DNA strand resection, RAD51/FANCR forms nucleoprotein filaments on single-stranded DNA and catalyzes homologous pairing and DNA strand invasion. BRCA2/FANCD1, PALB2/FANCN, RAD51C/FANCO, and XRCC2/FANCU facilitate these RAD51/FANCR-mediated processes.
During the final phase of ICL repair, the downstream FA proteins - BRCA2/FANCD1, BRIP1/FANCJ, PALB2/FANCN, RAD51C/FANCO, BRCA1/FANCS, RAD51/ FANCR, and XRCC2/FANCU - function cooperatively to repair the remaining broken duplex via homologous recombination (HR) (Fig. 2). HR is predominantly a conservative and error-free process whereby DNA damage is repaired using a homologous DNA template, typically the sister chromatid.47, 48 RAD51/FANCR is the major eukaryotic HR repair protein. RAD51/FANCR forms nucleoprotein filaments on 5′–3′ resected single-stranded DNA and catalyzes homologous pairing and DNA strand invasion and exchange. Many of the downstream FA proteins, e.g. BRCA2/FANCD1, PALB2/FANCN, and RAD51C/FANCO, are known to facilitate RAD51 function.49–51. Disruption of the FA-BRCA pathway leads to defective HR and an increased dependence on the typically error-prone nonhomologous DNA end joining (NHEJ) repair pathway.52–54
3. UBE2T/FANCT is responsible for a new FA subtype
The covalent attachment of ubiquitin to proteins regulates a variety of cellular pathways. This transfer is completed through a cascade of ubiquitin-related enzymes: an ubiquitin-activating enzyme (E1), an ubiquitin-conjugating enzyme (E2), and an ubiquitin ligase (E3) (Fig. 1).55 Monoubiquitination of FANCD2/FANCI is the central step of the FA-BRCA pathway, and it is estimated that this step is defective in >90% FA patients, and intricately tied to patients’ increased BMF and cancer risk. UBE2T, newly termed FANCT, is the E2 ubiquitin-conjugating enzyme for FANCD2 and FANCI.28, 29, 56 In 2000, Zhang et al. cataloged three hundred cDNAs of previously undefined genes expressed in CD34 positive hematopoietic stem cells, one of which, HSPC150, contained a ubiquitin-conjugating motif.57 HSPC150 was mapped to 1q31, a region known to be amplified in a range of cancers, including breast cancers, hepatomas, and cervical carcinomas.58 This protein, later renamed UBE2T, was demonstrated in biochemical assays to interact with FANCL, the E3 ubiquitin ligase of the FA pathway.29 UBE2T was subsequently shown to be required for FANCD2 monoubiquitination. Consequently, similar to other FA patient cells, loss of UBE2T leads to increased sensitivity to ICL-induced chromosome damage.29 These data clearly implicated UBE2T as the ubiquitin-conjugating enzyme for the FA-BRCA pathway, yet stopped short of classification of a new subtype.
In 2015, Hira et al. from the DNA damage signaling laboratory of Kyoto University reported two unrelated FA patients who, through WES and Sanger sequencing, were found to have biallelic UBE2T mutations.59 Both patients presented with congenital malformations, hematological abnormalities, and early onset BMF at ages 8 and 13. A heterozygous missense mutation leading to p.Q2E was uncovered in both patients.59 This highly conserved glutamine residue is found in an amino-terminal helix, which is part of the E2–E3 interacting interface.60 Disruption of this interface results in less efficient FANCD2 ubiquitination.59 Additionally, both patients harbored unique mutations in their second alleles, a 23 kilobase genomic deletion in one patient and skipped exon resulting in a frameshift and premature stop codon in the other. Complementation of patient cells with wild-type UBE2T restored efficient FANCD2 monoubiquitination and nuclear foci formation, and rescued the increased sensitivity to MMC-induced chromosome breakage.59 Hira et al. suggested that this new FA complementation group be named FA-T.
Coinciding with this study, the Laboratory of Genome Maintenance at Rockefeller University described an individual with biallelic mutations in UBE2T, born with microcephaly and bilateral thumb malformations, who tested positive in the DEB chromosome breakage test.61 However, this patient had normal bone marrow cellularity, normal leukocyte and thrombocyte counts, and only mild anemia, preventing a firm FA diagnosis for the first 16 years of life. Proband cells lacked the ability to monoubiquitinate FANCD2 and FANCI, failed to form FANCD2 nuclear foci, and were hypersensitive to ICL-inducing agents. Just months later, Virts and Jankowska et al. at Indiana University School of Medicine reported on the same 16 year old FA patient with biallelic mutations in UBE2T.62 In both cases, Sanger sequencing of genomic DNA identified two germline mutations, a paternal deletion and a maternal duplication of exons 2–6, both caused by aluY-mediated recombination events. Just as with the patients reported by Hira et al., retroviral complementation with wild-type UBE2T rescued the ICL-induced cell cycle arrest and chromosome breakage phenotypes.59, 61, 62 With these three announcements of biallelic UBE2T mutations leading to FA, the new FA-T complementation group has been clearly established and demonstrates that the FANCT alias is warranted.
4. Mutations in MAD2L2/REV7 underlie FA complementation group V
The most recent addition to the family of FA-BRCA genes is MAD2L2/REV7, a subunit of DNA polymerase ζ (Polζ). Involved in translesion DNA synthesis (TLS), Polζ is capable of synthesizing directly across template DNA lesions; specifically inserting nucleotides opposite DNA adducts, abasic sites, and, of particular interest in FA, DNA crosslinks. Through this process, the cell can tolerate the effects of DNA lesions during replication, effectively guarding against genomic instability. Structurally, Polζ is comprised of REV1, REV3, REV7, POLD2, and POLD3.63 REV7 has been shown to stabilize and enhance the polymerase activity of the catalytic subunit, REV3 by a factor of 20–30 demonstrating its role as a critical subunit.63 Additionally, the interaction between REV7 and REV3 has been shown to be necessary to confer DNA damage resistance.64 In 2005, Okada et al. demonstrated that Rev1, Rev3, and Rev7 depletion in chicken lymphocytes lead to hypersensitivity to various DNA damaging agents, including crosslink-inducing UV irradiation.65 These data suggested a possible phenotypic link to the FA-BRCA pathway.
In August 2016, Bluteau et al. reported a patient presenting with clinical and cellular phenotypes of FA.66 The patient exhibited various physical anomalies including short stature, microcephaly, and renal abnormalities, and presented with severe multi-lineage BMF. Patient-derived cells exhibited increased susceptibility to ICL-induced chromosome breakage and cell cycle arrest, concurrent with an FA diagnosis. Sequencing of known FA genes failed to identify pathogenic mutations. Whole-exome sequencing analysis revealed a homozygous mutation in the MAD2L2/REV7 gene, c.354T>A, resulting in a nonsynonymous p.V85E change in the translated protein. This substitution was predicted to be pathogenic, as it lies in a highly conserved domain of the protein that is known to facilitate interaction between catalytic subunits REV1 and REV3.64 No REV7 protein could be detected in patient cells despite the presence of normal transcript levels.66 Retroviral complementation with wild type REV7 rescued the increased susceptibility of patient cells to ICL-induced chromosome breakage and cell cycle arrest, establishing REV7 as FANCV. Confirming a role for REV7 in the FA-BRCA pathway and ICL repair, homozygous mutation of MAD2L2/REV7 using CRISPR/Cas9 recapitulated the hallmark FA cellular phenotypes.66 Furthermore, in support of an important role for REV7 in hematopoiesis, murine hematopoietic stems cells (HSC) deficient in Rev7 exhibited impaired colony forming ability and increased differentiation, similar to Fancg-deficient HSCs.66 Taken together, these findings implicate REV7/FANCV as a necessary downstream effector of the FA-BRCA pathway, most likely functioning in the translesion DNA synthesis step of ICL repair (Fig. 2). It remains to be seen if mutations in other Polζ subunit genes are causative for FA.
5. Discovery of BRCA1 as an FA gene
BRCA1 is one of the most well known breast and ovarian cancer susceptibility genes, along with BRCA2/FANCD1. In 1990, Mary-Claire King’s group at University of California, Berkeley mapped a gene responsible for inherited breast cancer to chromosome 17q21 and, four years later, BRCA1 was identified by positional cloning.67, 68 In 2001, a biochemical link between the FA pathway and BRCA1 was uncovered, leading to the coining of the phrase the FA-BRCA pathway. Monoubiquitinated FANCD2 was shown to co-localize with BRCA1 in discrete sub-nuclear foci following exposure to DNA damage, as well as in synaptonemal complexes of meiotic chromosomes.31 In late 2012, at the Perelman School of Medicine at the University of Pennsylvania, the laboratory of Roger A. Greenberg identified a patient with one BRCA1 allele with a deleterious mutation and a second allelic variant of unknown clinical significance (VUS): the first report of a patient with biallelic BRCA1 mutations.69 While never formally diagnosed with FA, the patient presented with classical FA-like phenotypes, including physical anomalies, microcephaly, developmental delay, and cellular sensitivity to chemotherapeutic agents, which is not typically displayed with single allelic mutations. The proband also exhibited an increased susceptibility to cancer, evident in the development of stage IV papillary serous ovarian carcinoma by 28 years of age (Table 1). The patient’s VUS was predicted to result in a valine to alanine change at amino acid 1736 (p.V1736A) in the carboxy-terminal BRCT domain.69 This domain plays a critical role in BRCA1 function by mediating interaction with phosphorylated proteins.70 The V1736A change led to a diminished interaction with the RAP80 protein and reduced localization of BRCA1 to DSBs. The second allele contained a truncating mutation, 2576delC, leading to a premature stop codon in exon 11.69
Table 1.
Summary of the clinical phenotypes of FA-R, FA-S, FA-T, FA-U, and FA-V patients described in this review article.
| Patient | Microcephaly | Growth retardation | Skin pigmentation | Radial- ray defects | Microphthalmia | Renal anomalies | Brain anomalies | Genital anomalies | Hipdysplasia | Cancer | BMF (y) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FA- R57 | + | + | + | + | + | + | + | NR | NR | NR | NR |
| FA- R56 | + | + | NR | + | NR | NR | + | + | NR | − | − |
| FA-S63 | + | + | NR | NR | NR | NR | NR | NR | NR | +a | − |
| FA-S65 | + | + | + | + | NR | + | NR | NR | + | +b | − |
| FA- T71 (PNG S-252) | NR | + | NR | + | NR | NR | NR | + | NR | NR | 13 |
| FA- T71 (PNG S-255) | NR | + | NR | + | NR | NR | NR | NR | NR | + | 8 |
| FA- T73,74 (10016 6/1) | + | + | + | + | NR | + | NR | NR | NR | NR | − |
| FA- U75 | + | + | + | + | NR | + | NR | + | NR | NR | − |
| FA- V66 (EGF- 123) | + | + | NR | NR | NR | + | NR | NR | NR | NR | 8 |
Stage IV papillary serous ovarian cancer
Ductal breast carcinoma
NR, Not reported
Early in 2015, the Greenberg laboratory reported the presence of biallelic BRCA1 mutations in another female patient exhibiting phenotypes concordant with FA and presenting with ductal breast carcinoma at the age of 23.71 Patient cells were deficient in localization of BRCA1 to DNA damage sites, demonstrated increased sensitivity to ICL-inducing agents, and had their disease state phenotypes ameliorated by expression of wild-type BRCA1. WES of the proband revealed biallelic compound heterozygous variants in BRCA1. One allele contained a 4 base pair deletion at exon 10 leading to a frame shift in exon 11 (p.S198Rfs*35) and the second harbored a point mutation in exon18 (p.A1699T), predicted to result in misfolding of the BRCT domain and protein instability.71, 72 After chromosome breakage results were found to be in the range for a FA diagnosis, the patient was formally diagnosed with FA subtype S (FA-S), finally establishing BRCA1 as an FA gene.71
Numerous biochemical connections between BRCA1/FANCS and the FA-BRCA pathway have been established. For example, BRCA1/FANCS associates with BRCA2/FANCD1 and RAD51/FANCR (see below) and phosphorylated BRIP1/FANCJ interacts with the carboxy-terminal BRCT domains of BRCA1/FANCS.73–75 Nevertheless, the exact role(s) of BRCA1/FANCS in ICL repair remains to be clearly determined. Recent studies have shown that BRCA1/FANCS is necessary for the removal of the CMG replicative helicase during ICL repair.76 And previous studies indicate that BRCA1 competes with 53BP1 early in the repair process to promote HR and restrict error-prone NHEJ.77, 78 Therefore, it seems likely that BRCA1/FANCS may play several distinct roles during the process of ICL repair.
6. Institution of a new alias for RAD51: FANCR
The bacterial recA protein catalyzes strand exchange between homologous regions of two double-stranded DNA molecules in E. coli. The human homolog, RAD51, plays a parallel role in HR during the repair of DNA double-strand breaks (DSBs).79, 80 Multiple biochemical connections between the FA proteins and RAD51 have been established over the past two decades. For example, RAD51 is necessary for resistance to ICL-inducing agents,81, 82 and FANCD2, BRCA1/FANCS, BRCA2/FANCD1 and RAD51 function in a common DNA damage response pathway.31, 36, 37, 75, 83 Nevertheless, a causative genetic link between FA and RAD51 was unexpected given the early embryonic lethality exhibited by rad51 nullizygous mice.84
Within the past year, however, two groups have described heterozygous dominant-negative RAD51 mutations in patients with an FA-like disorder.85, 86 Wang et al. from the Smogorzewska laboratory at Rockefeller University described a de novo heterozygous mutation c.391A>C in RAD51, resulting in a threonine to proline amino acid change (p.T131P).86 Patient cells exhibited ICL hypersensitivity, yet remained proficient for HR, most likely because of a low ratio of mutant to wild-type RAD51.86 These results indicate that RAD51 has additional roles in ICL repair distinct from its recombinase function. For example, several studies have shown that RAD51 plays an important role in replication fork protection prior to DSB formation.87–89 Mutant RAD51-T131P acts antagonistically to wild-type RAD51 causing a disease-state in the context of a single mutant allele, representing the first report of an FA being caused by a de novo dominant-negative heterozygous mutation.86 In the second report, Ameziane et al., described an individual with growth retardation, microcephaly, skeletal abnormalities and other physical phenotypes consistent with FA (Table 1).85 A DEB chromosome breakage test provided grounds for an FA diagnosis; patient cells displayed increased levels of chromosomal aberrations and extensive accrual in late S-G2 phase of the cell cycle. Whole-genome sequencing, whole-exome sequencing (WES), and Sanger sequencing of the patient implicated a de novo g.41022153G>A mutation in RAD51 as the cause of disease. This mutation, leading to alanine 293 being substituted for threonine (p.A293T), is found in a highly conserved region of RAD51 affecting its ability to undergo oligomerization. RAD51 oligomerization is essential for nucleoprotein filament formation, underlying the dominant-negative nature of this mutation. 79, 80 Biochemical studies of mutant RAD51-A293T also revealed impaired D-loop formation and decreased DNA binding.85 Taking into account these two reports, the role of RAD51 in the FA pathway as a downstream FA protein has been clearly established and RAD51 assumes the alias of FANCR.
7. XRCC2/FANCU and establishment of FA-U
The most recent and twentieth FA gene is XRCC2/FANCU (X-ray cross-complementing gene 2).90 XRCC2 is a member of the RAD51 family of proteins, encoding for products integrally involved in HR and critical for the maintenance of genome integrity.51 XRCC2 is a member of the BCDX2 protein complex - comprising RAD51B, RAD51C, RAD51D, and XRCC2 - which is thought to have several functions in HR, including stabilization of RAD51 nucleoprotein filaments and the promotion of RAD51-dependent homologous pairing and strand invasion.51, 91, 92 Underscoring the critical functions of XRCC2, homozygous disruption of murine Xrcc2 results in embryonic lethality occurring from mid-gestation, with Xrcc2−/− embryonic cells exhibiting high levels of chromosome aberrations.93
In 2012, Shamseldin et al. described a 2 year old Saudi Arabian patient presenting with classical FA phenotypes, including microcephaly, bilaterally absent thumbs, and kidney malformations. Positional mapping and WES uncovered a homozygous biallelic stop mutation in XRCC2.94 This mutation, c.643C>T, is predicted to encode p.R215* resulting in the production of a carboxy-terminal truncated protein. However, due to the presence of mutations in other genes, XRCC2 could not be clearly established as the causative gene in the absence of complementation studies.94 In 2016, Park et al. of the Andreassen laboratory at Cincinnati Children’s Hospital Medical Center further examined this patient.90 On a cellular level, the patient exhibited a heightened sensitivity to DEB and accumulation in G2-M phase of cell cycle, in line with FA. Reintroduction of wild-type XRCC2 corrected all pathological cellular phenotypes, establishing that the XRCC2 mutations were responsible for the observed cellular defects. Re-expression of XRCC2 also restored RAD51 nuclear foci to wild-type levels and corrected the increased ionizing radiation and ICL sensitivity. Consistent with XRCC2 functioning at a later stage of the FA-BRCA pathway, FANCD2/FANCI monoubiquitination was not affected in patient cells.90 Heterozygous mutations in XRCC2 have previously been shown to be associated with an increased predisposition to breast cancer.95 This is consistent with other downstream FA genes - BRCA1/FANCS, BRIP1/FANCJ, PALB2/FANCN, and RAD51C/ FANCO - that are also associated with an increased risk of developing breast cancer.96–102
With this new classification of XRCC2 as an FA gene, and in conjunction with the establishment of RAD51C as FANCO, it is clear that the RAD51 paralog complex BCDX2 plays critical functions in the FA-BRCA pathway.90, 103 This leads to speculation that mutations in other RAD51 paralog-encoding genes could lead to FA. Further experiments and genetic screening will be needed to determine the status of other RAD51 paralogs as potential FA genes.
8. Summary and final remarks
The discovery of these five new bona fide FA genes further strengthens the molecular ties between FA and DNA repair. These discoveries not only further elucidate key facets of the FA-BRCA repair pathway, but also have important implications for FA genetic testing. As there are many FA patients who currently cannot be assigned to any of the existing complementation groups, it is certain that more FA genes will be discovered. The identification of new genes using the powerful combination of next-generation sequencing and classical biochemistry approaches represents one critical aspect of the urgent quest in improve therapeutic options for this devastating disease, and to provide much needed hope for FA patients and their families.
Practice points.
Any of the following clinical symptoms should raise suspicion for FA. If FA is suspected, physicians are advised to refer the patient to a hematologist/oncologist and/or geneticist for FA genetic testing, preferably at a center experienced in FA diagnostic testing (see fanconi.org).
Atypically early-onset myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML), in addition to any of the following
Familial cancer predisposition and/or a history of excessive toxicity to radiation or chemotherapy
Unexplained cytopenias of any cell lineage due to bone marrow failure, e.g. thrombocytopenia, neutropenia, or anemia
Idiopathic macrocytsosis in the absence of identifiable cause, e.g. vitamin B12 or folate deficiency
Congenital anomalies including short stature, microcephaly, microphthalmia, hypo- or hyper-pigmentation of the skin, radial ray defects, male hypogonadism, and structural renal defects
Atypically early-onset tumors, particularly head and neck squamous cell carcinomas in the absence of a history of risk factors such as alcohol or tobacco use
Research agenda.
Continue to combine next-generation sequencing and classical biochemistry approaches to identify new FA genes
While a role for the FA proteins in the repair of ICLs generated by exogenous agents in vitro has been clearly established, the endogenous source(s) of genomic instability in the physiological setting still remains to be determined
Determine the mechanisms underlying the tissue-specificity of FA cancers. For example, why are FA patients at increased risk for head and neck squamous cell carcinomas?
Discover novel strategies to effectively prevent and treat FA BMF and cancers
Acknowledgments
We thank members of the Howlett laboratory for critical reading of this manuscript. This work was supported by National Institutes of Health/National Heart, Lung and Blood Institute grant R01HL101977 (NGH) and Rhode Island IDeA Network of Biomedical Research Excellence (RI-INBRE) grant P20GM103430 from the National Institute of General Medical Sciences.
Footnotes
Conflict of interest statement
There are no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lobitz S, Velleuer E. Guido Fanconi (1892–1979): a jack of all trades. Nat Rev Cancer. 2006;6:893–8. doi: 10.1038/nrc2009. [DOI] [PubMed] [Google Scholar]
- 2.Fanconi G. Familiare, infantile, perniziosaartige Anamie (pernizioses Blutbild und Konstitution) Jahrbuch fur Kinderheilkunde. 1927;117:257–80. [Google Scholar]
- 3.Fanconi Anemia Research Fund, Inc. Fanconi Anemia: Guidelines for Diagnosis and Management. 2014:1–429. [Google Scholar]
- 4.Rosenberg PS, Tamary H, Alter BP. How high are carrier frequencies of rare recessive syndromes? Contemporary estimates for Fanconi Anemia in the United States and Israel. Am J Med Genet A. 2011;155a:1877–83. doi: 10.1002/ajmg.a.34087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alter BP. Fanconi anemia and the development of leukemia. Best Pract Res Cl Ha. 2014;27:214–21. doi: 10.1016/j.beha.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alter BP, Greene MH, Velazquez I, Rosenberg PS. Cancer in Fanconi anemia. Blood. 2003;101:2072. doi: 10.1182/blood-2002-11-3597. [DOI] [PubMed] [Google Scholar]
- 7.Strathdee CA, Duncan AM, Buchwald M. Evidence for at least four Fanconi anaemia genes including FACC on chromosome 9. Nat Genet. 1992;1:196–8. doi: 10.1038/ng0692-196. [DOI] [PubMed] [Google Scholar]
- 8.Strathdee CA, Gavish H, Shannon WR, Buchwald M. Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature. 1992;356:763–7. doi: 10.1038/356763a0. [DOI] [PubMed] [Google Scholar]
- 9.Positional cloning of the Fanconi anaemia group A gene. Nat Genet. 1996;14:324–8. doi: 10.1038/ng1196-324. [DOI] [PubMed] [Google Scholar]
- 10.Lo Ten Foe JR, Rooimans MA, Bosnoyan-Collins L, et al. Expression cloning of a cDNA for the major Fanconi anemia gene, FAA. Nature Genet. 1996;14:320–3. doi: 10.1038/ng1196-320. [DOI] [PubMed] [Google Scholar]
- 11.de Winter JP, Waisfisz Q, Rooimans MA, van Berkel CGM, Bosnoyan-Collins L, Alon N, et al. The Fanconi anaemia group G gene is identical with human XRCC9. Nature Genet. 1998;20:281–3. doi: 10.1038/3093. [DOI] [PubMed] [Google Scholar]
- 12.de Winter JP, Leveille F, van Berkel CGM, Rooimans MA, van der Weel L, Steltenpool J, et al. Isolation of a cDNA representing the Fanconi Anemia Complementation Group E gene. Am J Hum Genet. 2000;67:1306–8. doi: 10.1016/s0002-9297(07)62959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Winter JP, Rooimans MA, van der Weel L, Van Berkel CM, Alon N, Bosnoyan-Collins L, et al. The Fanconi Anemia Complementation Gene FANCF encodes a novel protein with homology to ROM. Nature Genet. 2000;24:15–6. doi: 10.1038/71626. [DOI] [PubMed] [Google Scholar]
- 14.Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, et al. Positional cloning of a novel Fanconi Anemia gene, FANCD2. Mol Cell. 2001;7:241–8. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- 15.Joenje H, Levitus M, Waisfisz Q, D’Andrea A, Garcia-Higuera I, Pearson T, et al. Complementation analysis in Fanconi anemia: assignment of the reference FA-H patient to group A. Am J Hum Genet. 2000;67:759–62. doi: 10.1086/303067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joenje H, Oostra AB, Wijker M, di Summa FM, van Berkel CG, Rooimans MA, et al. Evidence for at least eight Fanconi anemia genes. Am J Hum Genet. 1997;61:940–4. doi: 10.1086/514881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–63. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walden H, Deans AJ. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys. 2014;43:257–78. doi: 10.1146/annurev-biophys-051013-022737. [DOI] [PubMed] [Google Scholar]
- 19.Auerbach AD. Fanconi anemia diagnosis and the diepoxybutane (DEB) test. Exp Hematol. 1993;21:731–3. [PubMed] [Google Scholar]
- 20.Alpi A, Langevin F, Mosedale G, Machida YJ, Dutta A, Patel KJ. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol. 2007;27:8421–30. doi: 10.1128/MCB.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurtan AM, D’Andrea AD. Dedicated to the core: understanding the Fanconi anemia complex. DNA Repair (Amst) 2006;5:1119–25. doi: 10.1016/j.dnarep.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Leung JW, Lowery M, Matsushita N, Wang Y, Shen X, et al. Modularized functions of the fanconi anemia core complex. Cell Rep. 2014;7:1849–57. doi: 10.1016/j.celrep.2014.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajendra E, Oestergaard VH, Langevin F, Wang M, Dornan GL, Patel KJ, et al. The genetic and biochemical basis of FANCD2 monoubiquitination. Mol Cell. 2014;54:858–69. doi: 10.1016/j.molcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25:331–43. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Coulthard R, Deans AJ, Swuec P, Bowles M, Costa A, West SC, et al. Architecture and DNA recognition elements of the Fanconi anemia FANCM-FAAP24 complex. Structure. 2013;21:1648–58. doi: 10.1016/j.str.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JM, Kee Y, Gurtan A, D’Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–22. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37:879–86. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32:767–77. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D’Andrea AD, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–96. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 30.Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–70. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–62. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 32.Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–7. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 33.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joo W, Xu G, Persky NS, Smogorzewska A, Rudge DG, Buzovetsky O, et al. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science. 2011;333:312–6. doi: 10.1126/science.1205805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 36.Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D’Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–20. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Andreassen PR, D’Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24:5850–62. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood. 2013;121:54–63. doi: 10.1182/blood-2012-07-441212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, et al. XPF-ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell. 2014;54:460–71. doi: 10.1016/j.molcel.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 41.MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murina O, von Aesch C, Karakus U, Ferretti LP, Bolck HA, Hanggi K, et al. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Rep. 2014;7:1030–8. doi: 10.1016/j.celrep.2014.03.069. [DOI] [PubMed] [Google Scholar]
- 43.Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Unno J, Itaya A, Taoka M, Sato K, Tomida J, Sakai W, et al. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Rep. 2014;7:1039–47. doi: 10.1016/j.celrep.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 45.Budzowska M, Graham TG, Sobeck A, Waga S, Walter JC. Regulation of the Rev1-pol zeta complex during bypass of a DNA interstrand cross-link. EMBO J. 2015;34:1971–85. doi: 10.15252/embj.201490878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Walter JC. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) 2014;19:135–42. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haber JE. Partners and pathways: repairing a double-strand break. Trends Genet. 2000;16:259–64. doi: 10.1016/s0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- 48.Mazon G, Mimitou EP, Symington LS. SnapShot: Homologous recombination in DNA double-strand break repair. Cell. 2011;142:646, e1. doi: 10.1016/j.cell.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 49.Boulton SJ. Cellular functions of the BRCA tumour-suppressor proteins. Biochem Soc Trans. 2006;34:633–45. doi: 10.1042/BST0340633. [DOI] [PubMed] [Google Scholar]
- 50.Park JY, Zhang F, Andreassen PR. PALB2: the hub of a network of tumor suppressors involved in DNA damage responses. Biochim Biophys Acta. 2014;1846:263–75. doi: 10.1016/j.bbcan.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suwaki N, Klare K, Tarsounas M. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Semin Cell Dev Biol. 2011;22:898–905. doi: 10.1016/j.semcdb.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 52.Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 53.Lieber MR, Wilson TE. SnapShot: Nonhomologous DNA end joining (NHEJ) Cell. 2010;142:496– e1. doi: 10.1016/j.cell.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 54.Pace P, Mosedale G, Hodskinson MR, Rosado IV, Sivasubramaniam M, Patel KJ. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–23. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 55.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 56.Longerich S, San Filippo J, Liu D, Sung P. FANCI binds branched DNA and is mono-ubiquitinated by UBE2T-FANCL. J Biol Chem. 2009;284:23182–86. doi: 10.1074/jbc.C109.038075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang QH, Ye M, Wu XY, Ren SX, Zhao M, Zhao CJ, et al. Cloning and functional analysis of cDNAs with open reading frames for 300 previously undefined genes expressed in CD34+ hematopoietic stem/progenitor cells. Genome Res. 2000;10:1546–60. doi: 10.1101/gr.140200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corson TW, Huang A, Tsao MS, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24:4741–53. doi: 10.1038/sj.onc.1208641. [DOI] [PubMed] [Google Scholar]
- 59.Hira A, Yoshida K, Sato K, Okuno Y, Shiraishi Y, Chiba K, et al. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am J Hum Genet. 2015;96:1001–7. doi: 10.1016/j.ajhg.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hodson C, Purkiss A, Miles JA, Walden H. Structure of the human FANCL RING-Ube2T complex reveals determinants of cognate E3–E2 selection. Structure. 2014;22:337–44. doi: 10.1016/j.str.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rickman KA, Lach FP, Abhyankar A, Donovan FX, Sanborn EM, Kennedy JA, et al. Deficiency of UBE2T, the E2 ubiquitin ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T subtype of Fanconi anemia. Cell Rep. 2015;12:35–41. doi: 10.1016/j.celrep.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Virts EL, Jankowska A, MacKay C, Glaas MF, Wiek C, Kelich SL, et al. AluY-mediated Germline Deletion, Duplication and Somatic Stem Cell Reversion in UBE2T Defines a New Subtype of Fanconi Anemia. Hum Mol Genet. 2015;24:5093–5108. doi: 10.1093/hmg/ddv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma S, Helchowski CM, Canman CE. The roles of DNA polymerase zeta and the Y family DNA polymerases in promoting or preventing genome instability. Mutat Res. 2013;743–744:97–110. doi: 10.1016/j.mrfmmm.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tomida J, Takata K, Lange SS, Schibler AC, Yousefzadeh MJ, Bhetawal S, et al. REV7 is essential for DNA damage tolerance via two REV3L binding sites in mammalian DNA polymerase zeta. Nucleic Acids Res. 2015;43:1000–11. doi: 10.1093/nar/gku1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okada T, Sonoda E, Yoshimura M, Kawano Y, Saya H, Kohzaki M, et al. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol Cell Biol. 2005;25:6103–11. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bluteau D, Masliah-Planchon J, Clairmont C, Rousseau A, Ceccaldi R, Dubois d’Enghien C, et al. Biallelic inactivation of REV7 is associated with Fanconi anemia. J Clin Invest. 2016;126:3580–4. doi: 10.1172/JCI88010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–9. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 68.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 69.Domchek SM, Tang J, Stopfer J, Lilli DR, Hamel N, Tischkowitz M, et al. Biallelic deleterious BRCA1 mutations in a woman with early-onset ovarian cancer. Cancer Discov. 2013;3:399–405. doi: 10.1158/2159-8290.CD-12-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–42. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 71.Sawyer SL, Tian L, Kahkonen M, Schwartzentruber J, Kircher M, et al. University of Washington Centre for Mendelian G. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–42. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coquelle N, Green R, Glover JN. Impact of BRCA1 BRCT domain missense substitutions on phosphopeptide recognition. Biochemistry. 2011;50:4579–89. doi: 10.1021/bi2003795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–60. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 74.Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–28. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 75.Chen JJ, Silver D, Cantor S, Livingston DM, Scully R. BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res. 1999;59:1752s–6s. [PubMed] [Google Scholar]
- 76.Fullbright G, Rycenga HB, Gruber JD, Long DT. p97 promotes a conserved mechanism of helicase unloading during DNA crosslink repair. Mol Cell Biol. 2016 doi: 10.1128/MCB.00434-16. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell. 2012;46:125–35. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–54. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–4. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 80.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 81.Abe H, Wada M, Kohno K, Kuwano M. Altered drug sensitivities to anticancer agents in radiation-sensitive DNA repair deficient yeast mutants. Anticancer Res. 1994;14:1807–10. [PubMed] [Google Scholar]
- 82.Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 83.Dong Y, Hakimi MA, Chen X, Kumaraswamy E, Cooch NS, Godwin AK, et al. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol Cell. 2003;12:1087–99. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 84.Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–43. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ameziane N, May P, Haitjema A, van de Vrugt HJ, van Rossum-Fikkert SE, Ristic D, et al. A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51. Nat Commun. 2015;6:8829. doi: 10.1038/ncomms9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, et al. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol Cell. 2015;59:478–90. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–11. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–7. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–16. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park JY, Virts EL, Jankowska A, Wiek C, Othman M, Chakraborty SC, et al. Complementation of hypersensitivity to DNA interstrand crosslinking agents demonstrates that XRCC2 is a Fanconi anaemia gene. J Med Genet. 2016 doi: 10.1136/jmedgenet-2016-103847. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, et al. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21:2858–66. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000;19:6675–85. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shamseldin HE, Elfaki M, Alkuraya FS. Exome sequencing reveals a novel Fanconi group defined by XRCC2 mutation. J Med Genet. 2012;49:184–6. doi: 10.1136/jmedgenet-2011-100585. [DOI] [PubMed] [Google Scholar]
- 95.Park DJ, Lesueur F, Nguyen-Dumont T, Pertesi M, Odefrey F, Hammet F, et al. Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet. 2012;90:734–9. doi: 10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Erkko H, Xia B, Nikkila J, Schleutker J, Syrjakoski K, Mannermaa A, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–9. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 97.Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–2. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 98.Lux MP, Fasching PA, Beckmann MW. Hereditary breast and ovarian cancer: review and future perspectives. J Mol Med. 2006;84:16–28. doi: 10.1007/s00109-005-0696-7. [DOI] [PubMed] [Google Scholar]
- 99.Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42:410–4. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 100.Melchor L, Benitez J. The complex genetic landscape of familial breast cancer. Hum Genet. 2013;132:845–63. doi: 10.1007/s00439-013-1299-y. [DOI] [PubMed] [Google Scholar]
- 101.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–41. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 103.Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–9. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]