Abstract
Introduction
We describe the rationale and methods of a study designed to compare vaginal and urinary microbiomes in women with mixed urinary incontinence (MUI) and similarly-aged, asymptomatic controls.
Methods
This paper delineates the methodology of a supplementary microbiome study nested in an ongoing randomized controlled trial comparing a standardized perioperative behavioral/pelvic floor exercise intervention plus midurethral sling versus midurethral sling alone for MUI. Women in the parent study had at least ‘moderate bother’ from urgency and stress urinary incontinence symptoms on validated questionnaire and confirmed MUI on bladder diary. Controls had no incontinence symptoms. All participants underwent vaginal and urine collection for DNA analysis and conventional urine culture. Standardized protocols were designed and a central lab received samples for subsequent polymerase chain reaction amplification and sequencing of the bacterial16S rRNA gene. The composition of bacterial communities will be determined by dual amplicon sequencing of variable regions 1-3 and 4-6 from vaginal and urine specimens to compare the microbiome of women with MUI to controls. Sample size estimates determined that 126 MUI and 84 control participants were sufficient to detect a 20% difference in predominant urinary genera with 80% power and 0.05 significance level.
Results
Specimen collection commenced January 2015 and finished April 2016. DNA was extracted and stored for subsequent evaluation.
Conclusions
Methods papers which share information regarding development of genito-urinary microbiome studies, particularly with control populations, are few. We describe the rigorous methodology developed for a novel urogenital microbiome study in women with MUI.
Keywords: Female urinary microbiome, Urinary taxa, Vaginal microbiome, Mixed urinary incontinence, Next-generation sequencing
Introduction
Mixed urinary incontinence (MUI) affects millions of women and imparts an enormous societal and economic burden. Despite the hardships caused by MUI, its optimal treatment and etiology remain unclear. Clinicians have long observed that MUI, defined as a combination of both urgency urinary incontinence (UUI) and stress urinary incontinence (SUI), may improve or resolve following incontinence surgery [1,2,3]. Resolution of SUI following surgery has been related to SUI's anatomic underpinnings [4,5]. It is unclear why UUI may resolve after SUI surgery.
The advent of bacterial genetic sequencing technology enabled researchers to describe specific bacterial communities in women's urine, a female urinary microbiome [6,7,8,9] (Principles of microbiome analyses and terminology are briefly described in Figure 1). The urinary microbiome may play a role in health and disease, contributing to MUI pathophysiology. This microbiome has been reported to differ in women with UUI and its composition has been reported to be associated with UUI treatment response [8,10]. These findings motivated our study of the female urinary microbiome in women with MUI.
Figure 1. NA (embedded in the figure).
This is the description of the methods for a planned, supplementary study of the Effects of Surgical Treatment Enhanced with Exercise for Mixed Urinary Incontinence (ESTEEM) trial. The ESTEEM trial was designed to compare urinary incontinence symptom outcomes in women randomized to a standardized perioperative behavioral/pelvic floor exercise intervention plus midurethral sling versus midurethral sling alone [11]. While ESTEEM interventions are aligned with traditional treatment approaches to SUI and UUI, the ESTEEM population provides a unique opportunity to explore alternative hypotheses regarding the etiology of MUI as well as MUI treatment response.
The primary aim of the Human Microbiome Study in ESTEEM (HMS-ESTEEM) is to evaluate whether the urinary microbiome differs between women with MUI and similarly aged controls. In order to better elucidate the origin of urinary microbiota, the association between the vaginal and urinary microbiome will also be explored. Clinically important study aims include comparison of patient's pre-treatment urinary and vaginal microbiome relative to incontinence symptom severity at baseline and follow-up. We hypothesize that the microbiome of women with MUI differs from asymptomatic controls and that the vaginal and urinary microbiome are inter-related. We will also explore whether the pre-treatment microbiome is associated with clinical outcomes. The purpose of the current report is to i) describe the rationale and ii) illustrate the challenges encountered in the design of the HMS-ESTEEM protocol.
Methods
Study Overview
This manuscript describes the methodology for HMS-ESTEEM, a prospective, multi-institutional, observational study, comparing the microbiota of women with MUI to controls. HMS-ESTEEM received Institutional Review Board approval from each of eight participating sites and the data coordinating center in the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Pelvic Floor Disorders Network (PFDN). All study participants provided written, informed consent.
Study Protocol Description
Participant Flow
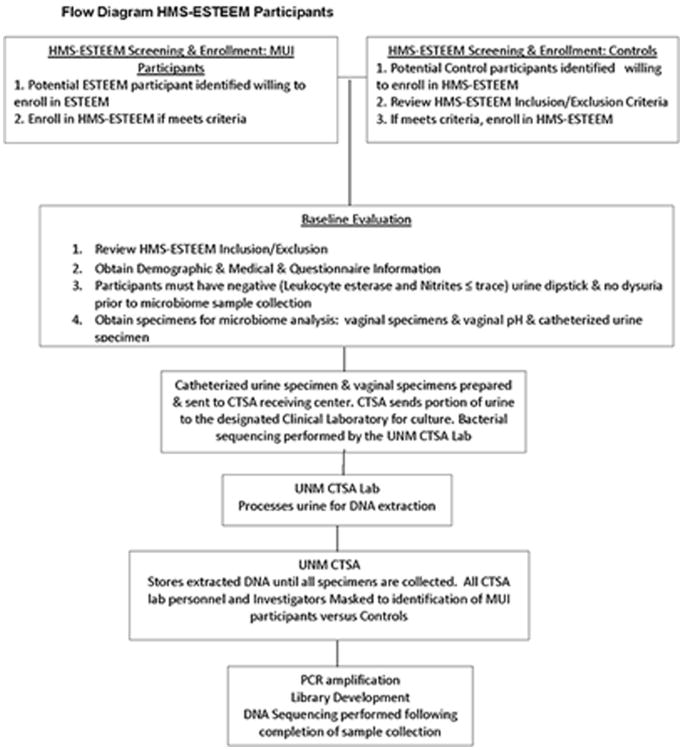
Figure 2 illustrates participant flow from screening/enrollment through sample processing. PFDN sites recruited women with MUI (cases) from the ESTEEM study as well as similarly aged women without urinary symptoms (controls) into HMS-ESTEEM. Controls were recruited from the community and clinics using site-specific recruitment methods, including use of advertisements, online resources and referrals from personnel in primary care and general gynecology clinics. Specimens were sent to University of New Mexico's Clinical and Translational Science Center laboratory which served as this study's biorepository laboratory. This laboratory will perform PCR amplification and 16S rRNA gene sequencing.
Figure 2. Flow Diagram of HMS-ESTEEM Participants.
Inclusion and Exclusion Criteria
The ESTEEM trial methods have been published previously [11]. The ESTEEM parent trial enrolled women with MUI who reported at least “moderate bother” for both UUI and SUI components of the Urinary Distress Inventory (UDI) questionnaire [12]. HMS-ESTEEM inclusion and exclusion criteria for MUI and control participants are outlined in Table 1.
Table 1. Study Inclusion and Exclusion Criteria.
| HMS-ESTEEM MUI Participants |
| Inclusion Criteria |
|
Exclusion Criteria
|
|
|
| HMS-ESTEEM Controls |
Inclusion Criteria
|
Exclusion Criteria
|
Variables Measured and Other Considerations
MUI participants completed bladder diaries and validated questionnaires assessing symptom burden for ESTEEM. All HMS-ESTEEM participants provided demographic and medical information, including whether or not they had symptoms of vaginitis, history of vaginal infection or vaginal medication, oral antibiotic, vaginal douche or tampon use, history of sexual activity, birth control methods, and systemic or local hormone use. To decrease potential transient effects on vaginal microbiota, participants were asked to refrain from vaginal intercourse, douching, or genital spray/wipe use for 48 hours prior to study visits. Specimens were collected at least 48 hours after menstruation ended.
Specimen Collection & Mailing to Central Laboratory & Processing
Study personnel performed urine dipsticks on mid-stream urines to eliminate samples from women with urinary tract infections (UTIs) (Fig.2). Mid-upper vaginal specimens were obtained and placed in commercially available tubes containing DNA protectant (Copan® Swabs, Copan Diagnostics, Murrieta CA). Mid-vaginal pH was recorded. The urethra was then swabbed with antiseptic solution and catheterized urine specimens were obtained for routine culture and DNA analysis; urines were placed in vacutainers for routine culture (BDVacutainer®, Sierra Molecular Corporation, Incline Village, NV) and in commercially available tubes containing DNA protectant (Assay Assure®, Becton, Dickinson and Company Franklin Lakes, NJ) for bacterial DNA analysis. Samples were labeled and shipped on cold-pack to the UNM biorepository laboratory within 1 day of collection. The biorepository laboratory, on the day of sample receipt, forwarded vacutainers to a single clinical laboratory (Tricore Reference Laboratories, Albuquerque, New Mexico) for urine culture.
The biorepository laboratory stored samples at -80° Centigrade until DNA extraction was performed. Following DNA extraction, specimens were stored at -20°Centigrade. After collection of all specimens, PCR amplification and sequencing of variable regions 1-3 and 4-6 of the bacterial 16S rRNA gene will be performed.
DNA Isolation Procedures
DNA was isolated from vaginal swabs using the QIAamp® DNA Investigator Kit (QIAGEN, Hilden, Germany) and the QIAcube liquid handling automation system (QIAGEN) per manufacturer's recommendations and eluted into 100 μl of Tris-EDTA (TE) buffer. DNA isolation from urine was completed manually using the QIAamp® Viral RNA Mini kit (QIAGEN) with the addition of carrier RNA to 280 μl of urine following manufacturer's recommendations. DNA was eluted with 30 μl of TE buffer and DNA concentration was determined by Qubit® fluorometric quantitation (ThermoFisher Scientific, Waltham, MA).
16S PCR Planned Procedures & Rationale for Choice of Variable Regions
Rationale
Vaginal microbiome studies have typically sequenced variable regions 1-2 (V1-V2) or variable regions 1-3 (V1-V3) [13,14,15]. Recent work specific to female urinary microbiota and UUI used V-4 to evaluate urine [8,10,16]. To limit bacterial identification discrepancies that could be introduced by using different variable regions for comparing vaginal and urinary microbiomes, HMS-ESTEEM will perform 16S rRNA gene sequencing following PCR amplification of both V1-V3 and V4-V6 regions in urine and vaginal specimens. Resultant sequences derived from urine specimens using V1-V3 amplicons will be compared to sequences derived from vaginal specimens using V1-V3 amplicons. The same will be done for vaginal and urine specimens using V4-V6 amplicons. In doing so we will determine if over or under representation of particular bacteria by the two different amplicons could cause spurious differences between the apparent microbiota of the vagina or urine.
Planned Procedures: Planned Procedures
Amplification of 16S rRNA regions V1-V3 and regions V4-V6 will be conducted on DNA isolated from both vaginal and urine samples by PCR using primers designed to adhere to conserved regions of the gene. Importantly, unique Nextera®XT (Illumina®, San Diego, CA) indexes will be added to V1-V3 and V4-V6 PCR amplicons, respectively, enabling separate analysis of each 16S rRNA region for each sample.
A three-step PCR will be used to amplify the V1-V3 16S rRNA region. The first PCR for the V1-V3 region will utilize regular A17F and 515R primers (Table 2) and 25 cycles, followed by a second 5 cycle PCR using longer primers (A17F-Nextera® & 515R-Nextera®; Table 2) that contain the Illumina Nextera® linker adapter sequence on the ends [17]. A final 8 cycle PCR using Illumina Nextera® XT primers will be conducted to complete the Illumina® adapter sequence and add unique indexes to each sample. For the V4-V6 region, a two-step PCR will be used consisting of 30 cycles of PCR amplification with the longer 515F-Nextera® and 1114R-Nextera® primers (Table 2) followed by a second 8 cycle PCR step to add one of 96 unique Nextera® XT indexes from a different set of Nextera® indexes.
Table 2. 16S Primer Sequences to be used in the HMS-ESTEEM Study.
| Primers Variable Regions 1-3 (v1-v3) | Primers Variable Regions 4-6 (v4-v6) |
|---|---|
| A17F 5′- GTT TGA TCC TGG CTC AG -3′ | 515F-Nextera 5′- TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG GTG CCA GCT GCC GCG GTA ATA -3′ |
| 515R 5′- TTA CCG CGG CMG CSG GCA -3′ | 1114R-Nextera 5′- GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGG GTT GCG CTC GTT GC -3′ |
| A17F-Nextera 5′- TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG GTT TGA TCC TGG CTC AG-3′ | |
| 515R-Nextera 5′- GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GTT ACC GCG GCM GCS GGC A -3′ |
Primer sequences that will be used in the HMS-ESTEEM Study: Sequences underlined indicate the 16S rRNA locus specific priming sequences. Sequences not underlined indicate the Illumina linker adapter sequences to be used for priming during the NexteraXT® PCR step, which will add the unique indexes and complete the Illumina adapter sequence
Prior investigation in our laboratory revealed suboptimal amplification using the longer A17F-Nextera® and 515R-Nextera® (Table 2) for the initial amplification of the 16S rRNA gene. Initial examination of the V1-V3 PCR reaction indicated reduced amplification of the region of the 16S rRNA gene could be ameliorated by using gene specific primers first (A17F + 515R, Table 2) followed by a second PCR using the longer primers with Nextera® linker sequences (A17F-Nextera® and 515R-Nextera®, Table 2). The V4-V6 PCR using the longer Nextera® linker primers for the gene specific PCR did not suffer from reduced amplification; thus, a two-step PCR for the V4-V6 16S rRNA region will be use.
After each of the PCR steps described above, PCR products will be purified with 0.8 volume of AMPure® XP beads (Beckman Coulter, Pasadena, CA) to remove unused primers. For each sample, the successful amplification of the desired 16S rRNA region will be confirmed by electrophoresis using an Agilent Technologies 2100 Bioanalyzer or 4200 TapeStation instrument (Agilent, Santa Clara, CA). Concentration will be determined using Qubit® fluorometric measurement. Equimolar pools of PCR amplicons will be created and resultant pools containing the desired ∼790bp PCR amplicons will be purified using a 1.5% gel and the BluePippin system (Sage Science). Samples will be processed in batches of 96, including two negative (water) controls without addition of any DNA to assess contamination and 7% of samples will be processed in duplicate to assess the reproducibility of the process.
16S rRNA Sequencing Plan
Sequencing will be conducted on the Illumina® MiSeq platform (Illumina®, San Diego, CA) using version 3 sequencing chemistry and 2×300bp read length. Both variable regions (V1-V3 & V4-V6) from each batch of 96 samples will be pooled and sequenced together resulting in a total of 192 normalized and pooled libraries per batch, each with a unique index sequence, per sequencing run (Figure 3). To ensure proper clustering and basecalling, pools of 16S rRNA amplicon libraries will be spiked with 5-15% of a PhiX library (Illumina®, San Diego, CA) and loaded on the MiSeq flowcell at 7-12.5pM to create an optimal cluster density of ∼800-1,200k/mm2.
Figure 3. Overview of the Dual 16S rRNA Amplicon Sequencing that will be Utilized for Urine and Vaginal Swab Samples in the HMS-ESTEEM Study.
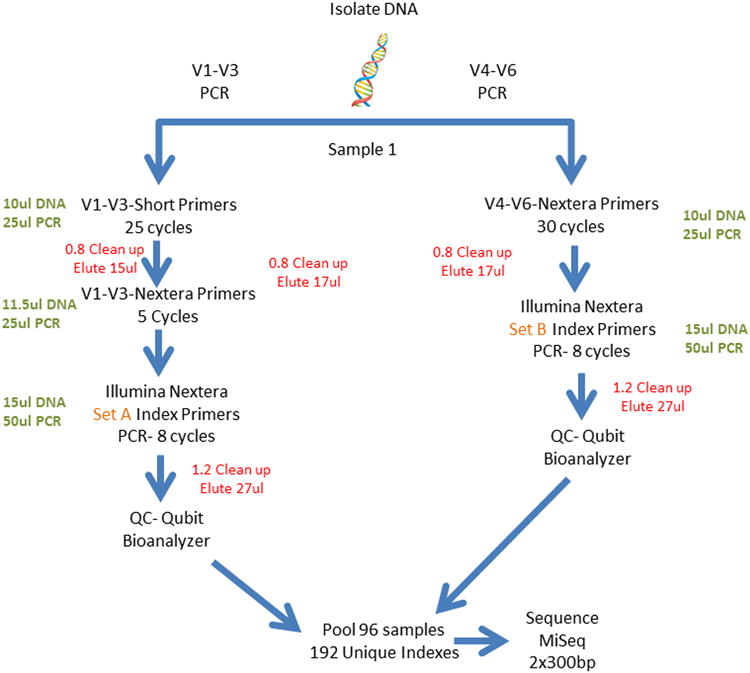
The samples are processed in batches of 96 that include 88 clinical samples, 2 no template controls (NTC), and 6 repeats of clinical samples to test for reproducibility and variability of the method.
Bioinformatics
Sequences will be analyzed using the Illumina BaseSpace 16S Metagenomics App version 1.0.1. The 16S Metagenomics App classifies sequencing reads against the Illumina-curated version of the GreenGenes taxonomic database using ClassifyReads. ClassifyReads is a high-performance implementation of the Ribosomal Database Project Classifier [18]. The rationale for using the BaseSpace 16S Metagenomics App is that it is a free software package with version controls and an easy to use graphical user interface, and is available to everyone. By using a standardized bioinformatic pipeline the data generated in this study can be more easily compared to sequence results generated from other studies also using the same bioinformatic pipeline.
Study Outcomes
Study outcomes will be the composition of bacterial communities identified by DNA sequencing using the described 16S rRNA techniques. The primary outcome will compare urinary microbiota in women with MUI compared to controls (See Statistical Approach). An important secondary outcome will be to determine the vaginal microbiome profile compared to that of the urinary microbiome. An additional outcome, enabled by our unique dual 16S rRNA amplicon study design, will be to evaluate whether these profiles as determined by sequencing of V1-V3 and V4-V6 regions are similar.
Other clinically relevant outcomes include evaluation of bacterial community characteristics and incontinence severity at baseline and follow-up and urinary tract infections. Specifically, HMS-ESTEEM will describe whether pre-treatment bacterial community characteristics are associated with baseline symptom severity. Consistent with the ESTEEM parent trial, HMS-ESTEEM incontinence symptom severity will be primarily based on Urogenital Distress Inventory (UDI) questionnaire scores; UUI will be measured by the UDI-irritative subscale scores, SUI will be measured by the UDI-stress subscale scores, and MUI will be measured by UDI total scores. Additional analyses will be based on incontinence episodes on 3-day bladder diary, including UUI, SUI and overall incontinence episodes. HMS-ESTEEM will also describe whether pre-treatment bacterial community characteristics are associated with change in MUI, UUI and SUI symptom severity at 1 year follow-up. Last, investigators will assess whether baseline bacterial community characteristics are associated with development of post-operative urinary tract infection (UTI), and will describe the congruence or difference between 16S urine sequencing and urine culture results.
Power Calculations & Statistical Approach
Unlike previous studies [7-10], HMS-ESTEEM investigators performed sample size calculations in the study design. These calculations were based on data from prior microbiome studies.
Sample Size Calculation
Analyses of the vaginal microbiome indicate that Lactobacillus is the predominant vaginal genus and that there are age-dependent differences in Lactobacillus contributions to the vaginal microbiome [13,19]. Lactobacillus is dominant vaginally in 73%-83% of reproductive aged women [13,19]. A recent study reported predominant vaginal genera based on menopausal status; Lactobacillus was dominant in 83% of pre and peri-menopausal women compared to 54% of postmenopausal women (P=.004) [19].
In the Pearce UUI microbiome study, predominance was defined as any genus comprising >45-50% of sequences in a sample [10]. The patients' mean age in that study placed them in the menopausal range (56.7-59.3 years), and 25% of patients had Lactobacillus predominant urine. For the current study, we assumed Lactobacillus would be the predominant urinary genus in 50% of controls and 30% of ESTEEM participants. With that assumption, 200 participants (120 ESTEEM participants and 80 controls) would be required to find a 20% difference in predominant urinary genera between groups with 80% power and a two-sided test with a significance level of 0.05. Based on the Pearce microbiome study [10], we assumed that 5% of participants would have urine which could not be sequenced and increased enrollment by 5%. The study aimed to enroll 210 women (126 cases and 84 controls).
Statistical Approach
For the primary outcome, urinary genera (including the definition of bacterial predominance as >45-50% of sequences in a sample), will be described using methods similar to those used by Pearce [10]. We anticipate that, similar to the previously mentioned findings, we will describe approximately 4-5 “Urotypes” or groups. The proportion of ESTEEM participants whose urine samples are Lactobacillus predominant will be compared between cases and controls using chi-square or Fisher's exact tests.
A Dirichlet multinomial mixture (DMM)-based approach will cluster urine and vaginal samples based on the overall distribution of genera found in the samples [20]. The relative abundance of various genera within the resulting clusters, or “community types,” will be described. Community type membership will be correlated between the urine and vagina to explore whether individuals belonging to certain urinary community types are more likely to belong to specific vaginal community types.
General linear modeling will be used to assess whether the predominant genera and/or community types in the urine and/or vagina are associated with pre-treatment MUI, SUI, or UUI symptom severity as measured by UDI total and subscale scores, and UI, SUI, and UUI episodes on the voiding diary, and whether bacterial community characteristics predict post-treatment outcomes. Similarly, generalized linear modeling will explore whether pre-treatment bacterial community characteristics are predictive of post-operative UTI development.
Urine cultures results will be characterized using methods similar to those for the urinary microbiome (identification of predominant species and clustering using DMM methods). Chi-square or Fischer's exact tests will assess whether there is an association between the predominant genera in the urine culture and sequencing results. Additional comparisons between the urinary genera in the urine culture and sequencing results will be performed using DMM methods.
Discussion
There were several challenges encountered while designing this study. The following discussion addresses our approach to these challenges and highlights the importance and clinical relevance of the study.
Challenges Encountered in Designing HMS-ESTEEM
Design challenges encountered in this multi-center microbiome study included coordination and standardization of specimen collection/processing, communication over multiple sites, selection of age-appropriate controls, masking of case/control status, and the issues encountered in comparing urine and vaginal microbiota.
Coordination between Sites, Standardization of Procedures & Masking
This study's multi-site design complicated its coordination. To ameliorate the problem, investigators developed specimen collection kits, a procedure manual and identified a central biorepository laboratory to integrate research activities with the data coordinating center (DCC). Biorepository personnel forwarded urine vacutainers to a clinical lab for standard culture and communicated specimen receipt information to the DCC. The DCC tracked recruitment, managed specimen accrual information and ensured inappropriately collected specimens were excluded from analysis. To protect the integrity of sequencing results, the DCC was aware of a specimen's case or control status, whereas the biorepository personnel performing genetic analyses were masked.
Enrollment of age appropriate controls
Given the effect of age on the vaginal microbiome [14], HMS-ESTEEM aimed to decrease age discrepancies between cases and controls. Prior to recruitment for HMS-ESTEEM, the age distribution of women already enrolled in ESTEEM was reviewed, and age cohorts for controls were determined for recruitment. Initially, only 63 of the total 84 control slots (75%) were allocated to the age cohorts and opened to enrollment. Toward the end of recruitment, the age distribution of HMS-ESTEEM participants was assessed, and the remaining 21 control slots were allocated to the age cohorts to achieve optimal age matching. Similar numbers of controls were recruited from each PFDN site.
Urine cultures performed by a single clinical laboratory
Catheterized urine samples were collected for DNA analysis, as recommended by Wolfe [21]. Urines were sent for routine culture to a single clinical laboratory which minimized variability in culture results. Results will be used to estimate the prevalence of bacteriuria in this cohort. Investigators will also compare results from bacterial profiles obtained from routine culture to 16S rRNA gene sequencing. Hilt reported that expanded urine culture techniques (increasing sample volumes, incubation times, culturing under aerobic and anaerobic conditions) detected similar bacteria compared to sequencing [16]. Few studies have compared conventional urine culture to sequencing results.
Rationale for sequencing both vaginal and urine specimens
This study will compare the vaginal and urinary microbiomes as there is a paucity of literature regarding their inter-relationship. This planned comparison resulted in an additional challenge; choosing comparable variable regions for vaginal and urine PCR amplification and sequencing as described in the Methods.
Significance of the HMS-ESTEEM study
Advancing knowledge regarding the pathophysiology of MUI is important. Affected women commonly have more symptoms and may be more refractory to treatment than those with UUI or SUI alone [3,22,23,24]. Whereas the parent trial for HMS-ESTEEM addresses treatment outcomes for women with MUI, HMS-ESTEEM addresses MUI's potential microbiologic underpinnings. Though urine is traditionally considered sterile, studies using genomic technology have not only reported the existence of a distinct female urinary microbiome, but also that this urinary microbiome differs in women with UUI [6-9,8,10]. Researchers have previously proposed that UUI pathophysiology was associated with inflammation and activation of afferent neural pathways and tissue remodeling [29,30]. The co-occurrence of inflammatory urinary biomarkers and alteration in the urinary biome in women with UUI raises the possibility that these disturbances in the bladder environment are inter-related and may affect the genesis of UUI.
Work comparing bacterial profiles in women with UUI and without UUI reported that the UUI microbiome was composed of increased Gardnerella and decreased Lactobacilli, with cohorts differing in predominance of Lactobacillus species [8]. Although these findings are important, the study's cases and controls differed in characteristics which may have influenced the results including age, estrogen status and body mass index. We have addressed these issues in HMS-ESTEEM, carefully characterizing cases and controls.
Little has been published regarding SUI and the female urinary microbiome. HMS-ESTEEM will investigate whether SUI symptoms, as measured by UDI-stress subscale scores, are associated with a characteristic microbiome in women in this study. It will explore the genito-urinary microbiome's relationship with MUI, including both SUI and UUI components. Further investigation of bacterial characteristics possibly associated with UUI and SUI symptoms and investigation of characteristics that may be predictive of treatment success is essential.
Relative to the urinary microbiome, more data exist regarding the vaginal microbiome, an initial habitat studied by the Human Microbiome Project [25]. Investigators have found that specific vaginal communities, particularly those that are Lactobacillus deficient, are associated with disease (bacterial vaginosis, sexually transmitted infections, HIV) [13]. Lactobacilli produce hydrogen peroxide which maintains a normal vaginal pH, a mechanism thought to confer disease resistance [26,27]. Evidence suggests that in women with vaginal atrophy and recurrent UTIs, vaginal estrogen normalizes pH and decreases recurrence of these infections, suggesting a relationship between the urinary and vaginal microbiological niches [28].
No studies currently compare a woman's urinary and vaginal microbiome [6]. HMS-ESTEEM will lead to a better understanding of a previously undescribed genito-urinary microbiome, characterizing the vaginal and urinary microbiomes in women undergoing treatment for MUI versus controls. It will assess whether urinary and/or vaginal microbiomes are associated with symptom severity and treatment success.
This study does have limitations. These include the possibility that yet unrecognized covariates affecting urinary or vaginal microbiota were unaccounted for. It is also possible that current gene sequencing techniques reliably characterize microbiota to the genus level but less reliably to the species level. If MUI differences in microbiota exist solely at the species level, this study may be unable to demonstrate these differences. Despite potential limitations, this study has unique features; these include its well characterized cases and controls in the setting of a multi-center clinical trial, the standardization of vaginal and urine specimen collection and processing, and utilization of similar bacterial variable regions to compare urinary and vaginal microbiomes. Results from HMS-ESTEEM could ultimately improve understanding of MUI with important implications for MUI treatment outcomes.
In conclusion, we have described the rationale, methods and challenges encountered by HMS-ESTEEM study investigators. This methods paper serves to disseminate information regarding challenges in the design of this multi-center genito-urinary microbiome study.
Synopsis.
This report describes the design of a prospective comparison of the urogenital microbiome in women with mixed urinary incontinence and similarly aged asymptomatic controls.
Acknowledgments
We thank Drs. Amy Overby, the UNM CTSA T-Laboratory and Karissa Culbreath (of Tricore Laboratories) for their invaluable help with this project
Supported by grants from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (1-U10-HD069025-01, 2-U10-HD041261-11, 2-U10-HD041267-12, 1-U10-HD069013-01, 2-U10-HD054214-06, 2-U10-HD054215-06, 1-U10-HD069010-01, 1-U10-HD069006-01, 1-U01HD069031-01) and the National Institutes of Health Office of Research on Women's Health, and the National Center for Research Resources and the National Center for Advancing Translational Sciences of the National Institutes of Health (Grant Number ULTR001449). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH
Footnotes
All the above for the Pelvic Floor Disorders Network
Conflicts of Interest: Y. Komesu: NIH 5R-01AT007171-03 (Primary Institute: NCCIH)
H. Richter: Symposia Medicus, Pelvalon, Astellas, Ferring, Statking, Society of Gynecologic Surgeons Board of Directors, UpToDate
D. Dinwiddie: None
N. Siddiqui: Medtronic
E. Lukacz: AMS/Astora, Axonics, Boston Scientific, Pfizer, Uroplasty, UpToDate
V. Sung: Society of Gynecologic Surgeons Executive Committee
B. Ridgeway: Coloplast
L. Arya: None
H. Zyczynski: American Urogynecologic Society Board of Directors, NICHD U10HD069006
R. Rogers: UpToDate, DSMB Chair TRANSFORM trial (sponsored by AMS/Astora), Mc Graw Hill, American Board of Obstetrics and Gynecology, International Urogynecology Journal
M. Gantz: None
Contributions: Y. Komesu: Protocol Development, Data Collection, Manuscript writing/editing
H. Richter: Protocol Development, Data Collection, Manuscript writing/editing
D. Dinwiddie: Protocol Development, Data Collection, Manuscript writing/editing. Oversight of laboratory procedures
N. Siddiqui: Protocol Development, Data Collection, Manuscript writing/editing
E. Lukacz: Protocol Development, Data Collection, Manuscript writing/editing
V. Sung: Protocol Development, Data Collection, Manuscript writing/editing
B. Ridgeway: Protocol Development, Data Collection, Manuscript writing/editing
L. Arya: Protocol Development, Data Collection, Manuscript writing/editing
H. Zyczynski: Protocol Development, Data Collection, Manuscript writing/editing
R. Rogers: Protocol Development, Data Collection, Manuscript writing/editing
M. Gantz: Protocol Development, Data Collection, Manuscript writing/editing
Contributor Information
Yuko M. Komesu, Division of Female Pelvic Medicine and Reconstructive Surgery, Department of Obstetrics & Gynecology, University of New Mexico Health Sciences Center, Albuquerque, NM.
Holly E. Richter, Division of Urogynecology and Pelvic Reconstructive Surgery, Department of Obstetrics & Gynecology, University of Alabama at Birmingham, Birmingham, AL.
Darrell L. Dinwiddie, Clinical Translational Sciences Center & Department of Pediatrics, University of New Mexico of New Mexico Health Sciences Center, Albuquerque, NM.
Nazema Y. Siddiqui, Division of Urogynecology & Reconstructive Pelvic Surgery, Department of Obstetrics & Gynecology, Duke University Medical Center, Durham, NC.
Vivian W. Sung, Division of Urogynecology and Reconstructive Pelvic Surgery, Department of Obstetrics & Gynecology, Alpert Medical School of Brown University, Providence, RI.
Emily S. Lukacz, Division of Female Pelvic Medicine & Reconstructive Surgery, Department of Reproductive Medicine, UC San Diego Health System, San Diego, CA.
Beri Ridgeway, Center for Urogynecology and Reconstructive Pelvic Surgery, Obstetrics & Gynecology and Women's Health Institute, Cleveland Clinic, Cleveland, OH.
Lily A. Arya, Division of Urogynecology and Reconstructive Pelvic Surgery, Hospital of University of Pennsylvania, Philadelphia, PA.
Halina M. Zyczynski, Division of Urogynecology and Reconstructive Pelvic Surgery, Department of Obstetrics, Gynecology and Reproductive Sciences, University of Pittsburgh School of Medicine, Pittsburgh, PA.
Rebecca G. Rogers, Division of Female Pelvic Medicine and Reconstructive Surgery, Department of Obstetrics & Gynecology, University of New Mexico Health Sciences Center, Albuquerque, NM.
Marie Gantz, Social, Statistical & Environmental Sciences, RTI International, Research Triangle Park, NC.
References
- 1.Richter HE, Litman HJ, Lukacz ES, et al. Demographic and clinical predictors of treatment failure one year after midurethral sling surgery. Obstet Gynecol. 117(4):913–21. doi: 10.1097/AOG.0b013e31820f3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palva K, Nilsson CG. Prevalence of urinary urgency symptoms decreases by mid-urethral sling procedures for treatment of stress incontinence. Int Urogynecol J. 22(10):1241–7. doi: 10.1007/s00192-011-1511-3. [DOI] [PubMed] [Google Scholar]
- 3.Abdel-Fattah M, Ramsay I, Pringle S, et al. Randomised prospective single-blinded study comparing ‘inside-out’ versus ‘outside-in’ transobturator tapes in the management of urodynamic stress incontinence: 1-year outcomes from the E-TOT study. BJOG. 117(7):870–8. doi: 10.1111/j.1471-0528.2010.02544.x. [DOI] [PubMed] [Google Scholar]
- 4.DeLancey JO. Structural support of the urethra as it relates to stress urinary incontinence: the hammock hypothesis. Am J Obstet Gynecol. 1994;170:1713–20. doi: 10.1016/s0002-9378(94)70346-9. [DOI] [PubMed] [Google Scholar]
- 5.Petros PE, Ulmsten U. An integral theory of female urinary incontinence. Acta Scand. 1990;69(27)(153):1–79. doi: 10.1111/j.1600-0412.1990.tb08027.x. [DOI] [PubMed] [Google Scholar]
- 6.Schneeweiss J, Koch M, Umek W. The human urinary microbiome and how it relates to urogynecology. Int Urogynecol J. 2016 doi: 10.1007/s00192-016-2944-5. [DOI] [PubMed] [Google Scholar]
- 7.Brubaker L, Nager CW, Richter HE, Visco A, Nygaard I, Barber MD, et al. Urinary bacteria in adult women with urgency urinary incontinence. Int Urogynecol J. 2014;25(9):1179–84. doi: 10.1007/s00192-013-2325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearce MM, Hilt EE, Rosenfeld AB, Zilliox MJ, Thomas-White K, Fok C, et al. The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. MBio. 2014;5(4):e01283–14. doi: 10.1128/mBio.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siddiqui H, Nederbragt AJ, Lagesen K, et al. Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol. 2011;11:244. doi: 10.1186/1471-2180-111-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pearce MM, Zilliox MJ, Rosenfeld AB, Thomas-White KJ, Richter HE, Nager CW, et al. The female urinary microbiome in urgency urinary incontinence. Am J Obstet Gynecol. 2015;213(3):347 e1–47 e11. doi: 10.1016/j.ajog.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung V, Borello-France D, Dunivan G, Gantz M, Lukacz E, Moalli P, et al. for the Pelvic Floor Disorders Network Methods for a multicenter randomized trial for mixed urinary incontinence: Rationale and patient-centeredness of the ESTEEM trial. International Urogynecology Journal. doi: 10.1007/s00192-016-3031-7. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shumaker SA, Wyman JF, Uebersax JS, McClish D, Fantl JA. Health-related quality of life measures for women with urinary incontinence: the Incontinence Impact Questionnaire and the Urogenital Distress Inventory. Continence Program in Women (CPW) Research Group. Qual Life Res. 1994;3:291–306. doi: 10.1007/BF00451721. [DOI] [PubMed] [Google Scholar]
- 13.Ravel J, Gajer P, Abdo Z, Schneider GM, Doenig SS, McCulle SL, Karlebach S, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(S1):4680–7. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gajer P, Brotman RM, Bai G, Sakamoto J, Schütte UM, Zhong X, et al. Temporal Dynamics of the Human Vaginal Microbiota. Sci Transl Med. 2012;4(132):132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fettweis JM, Serrano MG, Sheth HU, Mayer CM, Glascock AL, Jefferson KK. Vaginal Microbiome Consortium, Buck GA. Species-level classification of the vaginal microbiome. BMC Genomics. 2012;13(8):S17. doi: 10.1186/1471-2164-13-S8-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilt E, McKinley K, Pearce MM, Rosenfeld AB, Zilliox JM, Mueller ER, et al. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flor in the adult female bladder. J Clin Microbiol. 2014;52(3):871–6. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar PS, Brooker MR, Dowd SE, Camerlengo T. Target region selection is a critical determinant of community fingerprints generated by 16S pyrosequencing. PloS one. 2011;6(6):e20956. doi: 10.1371/journal.pone.0020956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Applied and Environmental Microbiology. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brotman RM, Shardell MD, Gajer P, Fadrosh D, Chang K, Silver MI, et al. Association between the vaginal microbiota, menopause status and signs of vulvovaginal atrophy. Menopause. 2013;21(5):1–9. doi: 10.1097/GME.0b013e3182a4690b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7(2):e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, FitzGerald MP, et al. Evidence of Uncultivated Bacteria in the Adult Female Bladder. J Clin Microbiol. 2012;50(4):1376–83. doi: 10.1128/JCM.05852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dooley Y, Lowenstein L, Kenton K, FitzGerald M, Brubaker L. Mixed incontinence is more bothersome than pure incontinence subtypes. Int Urogynecol J Pelvic Floor Dysfunct. 2008;19:1359–62. doi: 10.1007/s00192-008-0637-4. [DOI] [PubMed] [Google Scholar]
- 23.Monz B, Chartier-Kastler E, Hampel C, Samsioe G, Hunskaar S, Espuna-Pons M, Wagg A, Quail D, Castro R, Chinn C. Patient characteristics associated with quality of life in European women seeking treatment for urinary incontinence: results from PURE. Eur Urol. 2007;51:1073–81. doi: 10.1016/j.eururo.2006.09.022. discussion 81-82. [DOI] [PubMed] [Google Scholar]
- 24.Dmochowski R, Staskin D. Mixed incontinence: definitions, outcomes, and interventions. Curr Opin Urol. 20015;15:374–9. doi: 10.1097/01.mou.0000183946.96411.76. [DOI] [PubMed] [Google Scholar]
- 25.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, et al. The human microbiome project. Nature. 2007;449:804. doi: 10.3389/fcimb.2013.00041.eCollection02013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borges S, Silva J, Teixeira P. The role of lactobacilli and probiotics in maintaining vaginal health. Arch Gynecol Obstet. 2014;289:479–89. doi: 10.1007/s00404-013-3064-9. [DOI] [PubMed] [Google Scholar]
- 27.Gupta K, Stapleton AE, Hooten TM, et al. Inverse association of H2O2 –producing Lactobacilli and vaginal Escherichia coli colonization in women with recurrent urinary tract infections. J Infect Dis. 1998;178:446–50. doi: 10.1086/515635. [DOI] [PubMed] [Google Scholar]
- 28.Rahn DD, Carberry C, Sanses TV, Mamik MM, Ward RM, Meriwether KV, Olivera CK, Abed H, Balk EM, Murphy M Society of Gynecologic Surgeons Systematic Review Group. Vaginal estrogen for genitourinary syndrome of menopause: a systematic review. Obstet Gynecol. 2014;124(6):1147–56. doi: 10.1097/AOG.0000000000000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antunes-Lopes T, Pinto R, Barros SC, Btelho F, Silva CM, Cruz CD, Cruz F. Urinary Neurotrophic Factors in Health Individuals and Patients with Overactive Bladder. J Urol. 2013;189:359–365. doi: 10.1016/j.juro.2012.08.187. [DOI] [PubMed] [Google Scholar]
- 30.Tiyagi P, Tiyagi V, Qu X, Lina HT, Kuo HC, Chuan YC, Chancellor M. Association of Inflammaging (inflammation + aging) with higher prevalence of OAB in elderly population. Int Urol Nephrol. 2014;46(5):871–7. doi: 10.1007/s11255-013-0621-x. [DOI] [PubMed] [Google Scholar]