Abstract
Transcriptional and post-transcriptional regulators including transcription regulator, transcription factor and miRNA are the main endogenous molecular elements which control complex cellular mechanisms such as development, growth and response to biotic and abiotic stresses in a coordinated manner in plants. Utilizing the most recent information on such relationships in a plant species, obtained from high-throughput experimental technologies and advanced computational tools, we can reconstruct its co-regulatory network which consequently sheds light on key regulators involved in its important biological processes. In this article, combined systems biology approaches such as mining the literatures, various databases and network reconstruction, analysis, and visualization tools were employed to infer and visualize the coregulatory relationships between miRNAs and transcriptional regulators in Citrus sinensis. Using computationally and experimentally verified miRNA-target interactions and constructed co-expression networks on array-based data, 10 coregulatory networks and 10 corresponding subgraphs include FFL motifs were obtained for 10 distinct tissues/conditions. Then PPI subnetworks were extracted for transcripts/genes included in mentioned subgraphs in order to the functional analysis of extracted coregulatory circuits. These proposed coregulatory connections shed light on precisely identifying C. sinensis metabolic pathways key switches, which are demanded for ultimate goals such as genome editing.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-017-0416-0) contains supplementary material, which is available to authorized users.
Keywords: Citrus sinensis, miRNA, Transcriptional regulator, Coregulatory network, Co-expression networks
Introduction
Deciphering complicated coregulatory network is a crucial issue in the post-genomic era for understanding the intracellular mechanism of complex biological processes such as development, growth and responses to biotic and abiotic stresses in all plants (Berri et al. 2009; Dodds and Rathjen 2010; Leyva-González et al. 2012; Megraw et al. 2016). Coregulatory network includes transcription regulators and post transcription regulators that can regulate each other as well as themselves.
Major elements in the transcriptional regulatory network include transcription regulators (TRs) and transcription factors (TFs). Identification and classification of these regulatory proteins will facilitate studies on gene function, evolution, and coregulatory network. Moreover, these downstream main regulators themselves are regulated by a class of enzymes called protein kinases (PKs) which operate as an intermediate signal transducer in response to a diverse upstream stimulus. Protein kinases activate TFs and TRs by chemically adding phosphate groups to them and are known to regulate the majority of cellular mechanisms in this way (Treisman 1996; Endicott et al. 2012). After activation by kinases (phosphorylation), TFs and TRs can bind to specific DNA motifs in the target gene promoter and coordinately regulate the amount of RNA transcription (Kadonaga 1998; Venters and Pugh 2010).
On the other hand, translational elements which play a role in post-transcription level include endogenous small non-coding RNAs composed of 19–24 nucleotides, called miRNAs that degrade or inhibit translation procedure of mRNA targets (Voinnet 2009). Increasing evidence suggests that miRNAs play pivotal roles in multiple biological processes such as plant growth, development, and defense response against both biotic and abiotic stress including nutrient stresses (Wu et al. 2014a, b, 2016; Sunkar et al. 2012; Mendoza-Soto et al. 2012; Zhang et al. 2011; Xu et al. 2013a, b; Sunkar et al. 2007; Chiou 2007). Plant miRNAs usually have a near-perfect pairing with their mRNA targets, which induces gene repression through binding to target transcripts (Brodersen et al. 2008). As well as transcriptional regulators, miRNAs are well conserved in plants and are thought to be a vital and evolutionary ancient component of gene regulation (Axtell and Bowman 2008; Song et al. 2010).
Transcriptional and post-transcriptional regulators form network subgraphs and motifs to co-regulate gene transcription and translation in a connective manner (Tam and Lim 2009; Guzzi et al. 2015; Le et al. 2014; Zhang et al. 2013). Utilizing the information on these coregulatory network connections could shed light on key elements involved in various biological processes. Therefore, extracting miRNA-TR coregulatory network subgraphs and motifs could be so useful for many important applications such as biomarker identifying, drug designing, genome editing and so on.
In this article, we aim to reconstruct transcriptional Transcription Factor/Transcription Regulator/Protein Kinase (TF/TR/PK)-translational (miRNA) coregulatory network for Citrus sinensis. Citrus sinensis is an important agricultural product throughout the world. Its fruit is among the world’s most economically important fruit crops consumed as both fresh products and processed juices. Its aromatic peels (rind) are used in several different ways, as are the blossoms (the flowers), leaves and wood of the tree (Kalim et al. 2015; Aruoma et al. 2012; Wang et al. 2014). The draft genome sequencing and annotation project of C. sinensis were finished in recent years providing beneficial information to the researcher of this species (Wang et al. 2014; Xu et al. 2013a, b). For our purpose, we reconstructed several coregulatory networks for different tissues and biological conditions and then we extracted important subgraphs including feed forward loop (FFL) motifs from these networks. Finally, we investigated protein–protein interaction (PPI) network to identify functional role of transcriptional and translational regulators in C. sinensis biological processes.
Materials and methods
miRNAs data collection and pre-processing
Well known databases include miRBASE (Kozomara and Griffiths-Jones 2014), PMRD (Zhang et al. 2009) and PMTED (Sun et al. 2013) were searched for C. sinensis miRNAs, but there was no comprehensive, consistent and homogenous dataset. For example, there were similar mature miRNA with a different sequence in different databases [e.g. csi-MIR393 in (Zhang et al. 2009) and (Xu et al. 2010) or csi-miR172e in (Wu et al. 2015) and (Xu et al. 2010)]. Therefore, referenced C. sinensis mature miRNAs datasets from various articles and databases were searched and collected (Xu et al. 2010; Lu et al. 2015; Wu et al. 2015). After removing duplicated miRNAs with the same name and same sequence and also merging miRNA with the same sequence in a unique name, 614 miRNAs with unique sequence remained. In order to merge miRNAs with the same sequences but different names, we simply concatenated their names in a unique string. So, that readers can distinguish between mature miRNAs which have the same sequence but are located in different genome positions. Furthermore, some of these 614 unique sequences had the same name. For overcoming this issue, unique published date of related dataset added to the name of the sequence as a postfix. All of these efforts were done to simplify further stages of analysis and results presentation.
Also, other available data include tissue and condition expression (four tissues include fruit, leaf, flower and callus and three conditions include normal, red-flesh mutant and boron deficiency) and experimentally confirmed target genes (1071 targets) were obtained from these 614 sequence referenced sources. Supplementary file 1 contains the list of all miRNAs and their corresponding sequences.
miRNAs target prediction and/or collection
Then PSRNAT (Dai and Zhao 2011) and TAPIRYBRID (Bonnet et al. 2010) servers were used to predict gene targets of these 614 sequences computationally. As recommended by Srivastava et al. (2014), a combination of intersection for these tools could bring about very particular predictions in non-Arabidopsis species. For this purpose, the list of miRNAs was uploaded on PSRNAT server in FASTA format and cDNA library of C. sinensis was selected from the embedded list. All the input parameters were considered as default, excluding the number of the top target gene for each miRNA that changed to 20 which consisted of Srivastava et al. observations and previous hypothesis (Srivastava et al. 2014; German et al. 2008). Next, the sequence of PSRNAT results was extracted from CAP dataset and the sequence of each miRNA and its targets uploaded on TAPIRHYBRID server one by one due to its executive limitation. Input parameters for TAPIRHYBRID server were left default too. The results were 1375 highly specific computationally predicted target genes.
In addition to computationally predicted targets, some experimentally confirmed targets had been obtained from various datasets, as mentioned before. About 198 of these targets were in GJI gene ID format (Goodstein et al. 2012). The gene ID of targets with GJI format converted for CAP format using the Arabidopsis homologue equivalents. For these targets, we add “h-” to their gene IDs as a prefix, because of the inaccuracy of format conversion. Finally, 2446 miRNA-target interactions were obtained. It should be noted that targets in these interactions are genes or transcripts due to data collection from various datasets. These interactions formed five miRNAs-target genes network based on miRNAs tissue expression including fruit, leaf, flower, callus and overall (all resulted in interactions). These five networks are accessible through supplementary file 2. Tissue expression for some of the targets was obtained from Ding et al. (2014).
Gene co-expression network reconstruction
Citrus sinensis gene expression data were collected from NICCE project and constructed network using random matrix theory by Dongliang Du et al. in order to discover the co-expression relationships between transcriptional regulators and genes (Wong et al. 2014; Du et al. 2015). NICCE C. sinensis specific co-expression network data was downloaded (Wong et al. 2014). The co-expression network was constructed using Aracne2 algorithm (Margolin et al. 2006) with a cutoff p value of 1e-7 and a cutoff MI (mutual information) of 0.3 for all PKs, TRs and TFs included in downloaded data. The C. sinensis transcriptional regulatory element lists were obtained from iTAK (Zheng and Fei 2014). Another co-expression network dataset in Citrus clementina gene ID format was also downloaded (Wong et al. 2014). Then format conversion was done using mapping file included in dataset folder and co-expression networks were obtained for 7 tissues or conditions including flesh, leaf, albedo, flavedo, HLB (Da Graca et al. 2004), Canker (Das 2003) and condition independent (overall) for C. sinensis genes. In this paper, the co-expression network derived from the NICCE dataset are transcripts and called Coex1, and nodes in other 7 networks are genes and called Coex2.
Transcriptional and post-transcriptional network integration
Eventually, five miRNAs-targets and six co-expression networks were merged. Albedo and flavedo co-expression networks were ignored due to unavailability of miRNAs-targets network data for these tissues. Canker and HLB co-expression networks were merged with overall miRNAs-target networks (because of the effect of these conditions on more than one tissue of the C. sinensis), and also callus and flower miRNA-target networks were merged with overall co-expression network. Obtained co-expression network for C. sinensis from NICCE was merged with four tissue-specific miRNAs-target networks too. All the merging processes was done using Cytoscape software default network merger tool (Shannon et al. 2003). At the end, ten merged networks were resulted shown in Table 1 and their visualized formats are accessible through supplementary file 3.
Table 1.
Merged networks and their constituent parts
| miRNA-target network | Co-expression network | Merged network |
|---|---|---|
| Fruit | Coex1 | Fruit-Coex1 |
| Leaf | Coex1 | Leaf-Coex1 |
| Flower | Coex1 | Flower-Coex1 |
| Callus | Coex1 | Callus-Coex1 |
| Leaf | Coex2 leaf | Leaf-C2 leaf |
| Fruit | Coex2 flesh | Fruit-C2 flesh |
| Overall | Coex2 HLB | Overall-C2 HLB |
| Overall | Coex2 Canker | Overall-C2 Canker |
| Flower | Coex2 overall | Flower-C2 overall |
| Callus | Coex2 overall | Callus-C2 overall |
Identification of transcriptional regulator-target connections
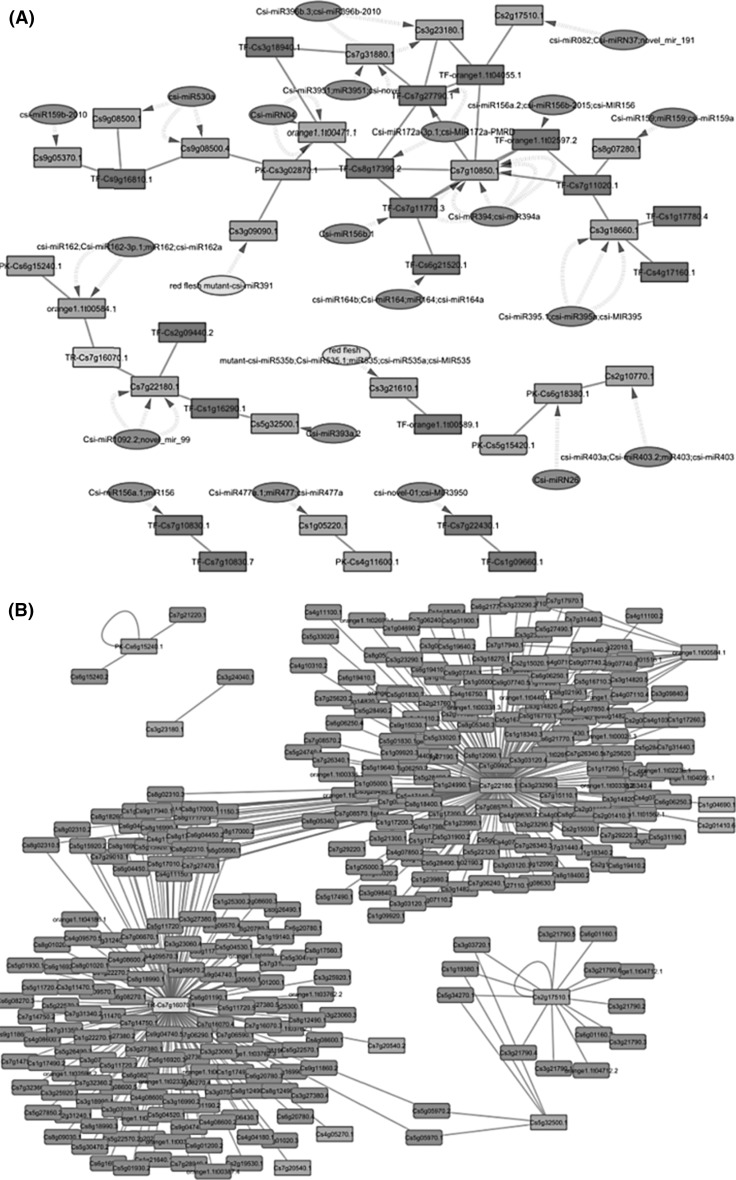
All gene nodes in above-merged networks were searched in iTak datasets to identify PKs, TRs, and TFs existing in these networks. Those genes which were identified by gene ID conversion based on homology with Arabidopsis were searched in PlantTFDB 3.0 dataset (Jin et al. 2013) with GJI format too. Interestingly all TFs identified genes with GJI format in PlantTFDB were available in iTAK TF dataset after gene format conversion. Then for each network, a subgraph includes only miRNA-gene-(PK or TR or TF) or miRNA-(PK or TR or TF)-gene interactions were extracted. Visualization of Fruit-Coex1 in Fig. 3 and Leaf-C2 leaf in Fig. 4 was done using Cytoscape and all subgraphs are accessible through supplementary file 4 (compressed file). It is obvious that these ten subnetworks include all possible FFL motifs as well.
Fig. 3.
a Fruit-Coex1 subgraph. All miRNA-target interactions are predicted computationally. Blue nodes miRNAs; cyan nodes condition (red-flesh mutant Citrus sinensis) dependent miRNAs; orange nodes genes; purple nodes protein kinases; red nodes: transcription factors. b Its corresponding PPI subnetworks. Blue nodes genes which don’t exist in overall miRNA-target; orange nodes genes which exist in overall miRNA-target; purple nodes protein kinase; beige nodes transcription regulators
Fig. 4.
a Leaf-C2 leaf subgraph. Dashed edges miRNA-target computationally predicted interactions; solid edges experimentally verified interactions; Green edges positive co-expression relationship; red color negative co-expression relationship; nodes which their labels begin with star don’t exist in overall miRNA-target network, whereas other nodes exist in miRNA-target network; dark blue node condition (Boron deficiency) dependent miRNA. b Its corresponding PPI subnetworks. Green nodes don’t exist in miRNA-target and co-expression networks
PPI investigation for targets included in reconstructed networks
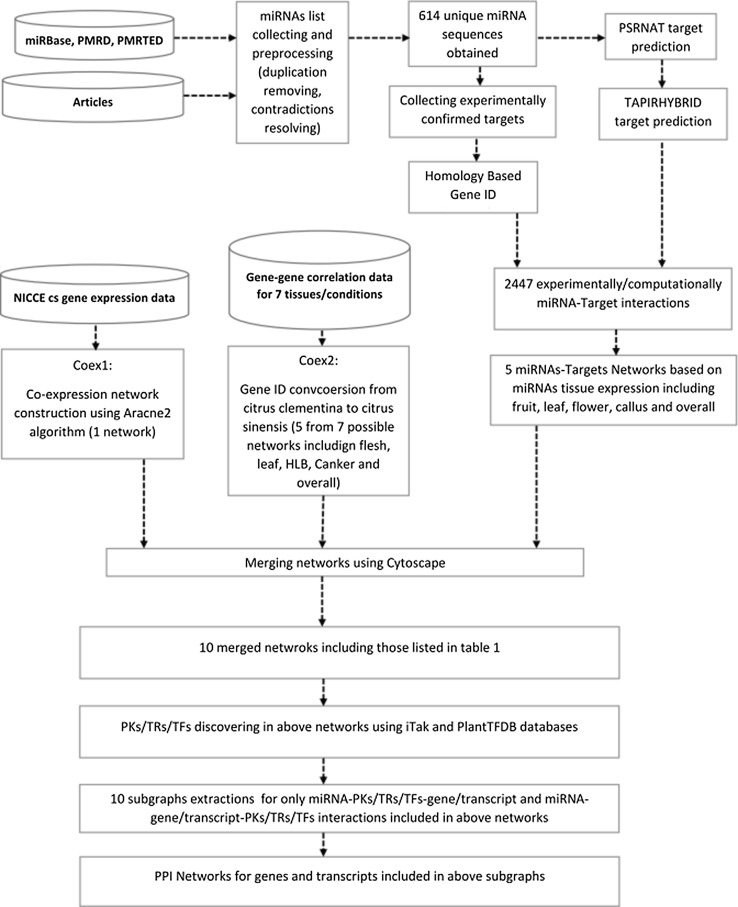
For functional analysis and interpretation of mentioned subgraphs, C. sinensis PPI network (CitrusNet) were searched for all of their genes and/or transcripts (Ding et al. 2014). Then PPI subnetwork for genes and/or transcripts in each of the 10 subgraphs were extracted and visualized by Cytoscape (Shannon et al. 2003). All the subgraphs represented protein–protein interactions, their target genes/transcripts annotations were given in details in the supplementary files 5 and 6, respectively. The flowchart of C. sinensis coregulatory network reconstruction steps has been shown in Fig. 1.
Fig. 1.
The flowchart of Citrus sinensis coregulatory network reconstruction steps
Results
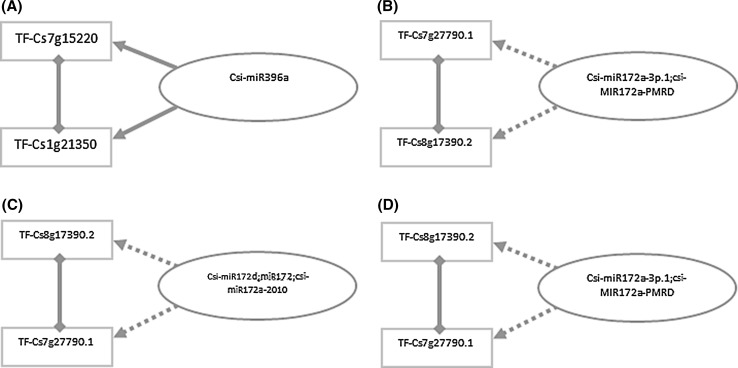
In the present work, we gathered 614 unique sequences of mature miRNA and reconstructed 10 miRNAs-Targets-Transcriptional containing coregulatory networks for C. sinensis. We extracted 10 FFL motif included subgraphs from these networks. These subnetworks suggest important nodes and connections which may play a key role in biological processes involved in this plant species. We discovered 4 miRNA-FFL motifs as introduced by Zhang et al. (2013) and Hamed et al. (2015) in previous coregulatory network studies. One of them has experimentally verified miRNA-target edge and other 3 miRNA-FFL motifs have computationally predicted miRNA-target interactions as shown in Fig. 2. FFL motifs are important loops in gene coregulatory networks and play critical roles in various biological processes such as cell differentiation, development, signal transduction, and diseases (Guzzi et al. 2015; Zhang et al. 2013; Hsieh et al. 2015; Osella et al. 2011).
Fig. 2.
Four extracted miRNA-FFL motifs from a Flower-C2 overall, b Fruit-Coex1, c Callus-Coex1, d Flower-Coex1. Dashed lines represent computational interactions, whereas normal lines indicate an experimental relationship
Furthermore, 9 PPI subnetworks were extracted from C. sinensis PPI network for genes and transcripts included in subgraphs (no PPI were found for genes included in Callus-C2 overall subgraph). Using protein–protein interaction information, key miRNAs that target components of important protein complexes and their regulatory functions can be identified. For example, as shown in Figs. 3 and 4, two subgraphs and their corresponding PPI networks suggest the important role of interactions listed in Table 2 in the regulation of C. sinensis fruit and leaf biological processes. Target annotation has been done using MapMan annotation tool (Thimm et al. 2004) by Ding et al. (2014) in CitrusNet project. Nearly all of these interactions directly effect an essential molecular functions. Using further information of target involved protein complexes, more aspects of these interactional roles may be revealed.
Table 2.
Important regulatory connections in Fruit-Coex1 and Leaf-C2 leaf subgraphs, their target annotations, and their target homologues in Arabidopsis thaliana
| Important interactions | Tissue | Target annotation | Homologue |
|---|---|---|---|
| Csi-miR2111 → Cs3g07490 | Leaf | DNA.synthesis/chromatin structure | at1g08260 |
| Csi-miR403.1 → Cs3g01650 | Leaf | Protein.degradation.AAA type | at3g01610 |
| B-deficiency-novel_mir_35 → TR-Cs7g16070 → Cs1g15850 | Leaf | RNA.processing | at4g32850 |
| Csi-miR5054.2 → Cs7g07710 | Leaf | DNA.synthesis/chromatin structure | at4g31210 |
| Csi-miR477a-3p → TF-Cs6g19680 → Cs6g16540 | Leaf | RNA.regulation of transcription.Global transcription factor group | at1g65440 |
| Csi-miR1092.2;novel_mir_99 → Cs7g22180.1 | Fruit | RNA.processing.RNA helicase | at5g47010 |
| Csi-miR162;Csi-miR162-3p.1;miR162;Csi-miR162a → orange1.1t00584.1 | Fruit | RNA.processing.degradation dicer | at1g01040 |
| Csi-miR082;Csi-miRN37;novel_mir_191 → Cs2g17510.1 | Fruit | Protein.degradation.ubiquitin.E3.SCF.FBOX | at1g68050 |
| Csi-miR393a.2 → Cs5g32500.1 | Fruit | Hormone metabolism.auxin.signal transduction | at3g62980 |
| Csi-miR162;Csi-miR162-3p.1;miR162;csi-miR162a → orange1.1t00584.1 ← PK-Cs6g15240.1, TR-Cs7g16070.1 | Fruit | RNA.processing.degradation dicer | at1g01040 |
| Csi-miR1092.2;novel_mir_99 → Cs7g22180.1 ← TF-Cs2g09440.2, TF-Cs1g16290.1, TR-Cs7g16070.1 | Fruit | RNA.processing.RNA helicase | at5g47010 |
Discussion and conclusion
In this study, reviewing literature, open access databases beside network reconstruction, analysis, and visualization tools were employed to infer and visualize the coregulatory relationships between miRNAs and transcriptional regulators in C. sinensis. Using computationally and experimentally verified miRNA-target interactions and constructed co-expression networks on array-based data, 10 coregulatory networks and 10 corresponding subgraphs include FFL motifs were obtained for 10 distinct tissues/conditions. Then PPI subnetworks were extracted for transcripts/genes included in above-mentioned subgraphs in order to determine the functional importance of extracted coregulatory circuits.
For example, Cs7g22180.1 and Cs7g16070.1 transcripts are the components of C. sinensis important protein complexes, as shown in Fig. 3b. Similarly, as shown in Fig. 4b, Cs3g07490 and Cs7g16070 genes have very important role in C. sinensis protein–protein interaction network. These genes and transcripts, their annotations and their regulatory connections are listed in Table 2. Csi-miR2111, Csi-miR403.1, and Csi-miR5054.2 regulate vital biological processes such as DNA synthesis/chromatin structure and protein degradation in the C. sinensis leaf. Also, according to homologue annotation in TAIR (Lamesch et al. 2012), these miRNAs are key players in growth, protein synthesis, and cell division. In addition, novel_mir_35 and Csi-miR477a-3p indirectly affect biological processes including RNA processing, transcription initiation, and chromatin structure/assembly according to MapMan and TAIR annotations (Thimm et al. 2004; Lamesch et al. 2012). Notice that novel_mir_35 is expressed only in a boron-deficiency condition; therefore, a closer experimental inspection could reveal its target annotation (Cs1g15850) more accurately.
On the other hand, Csi-miR1092.2, novel_mir_99, csi-miR162, Csi-miR162-3p.1, miR162, csi-miR162a, Csi-miR082, Csi-miRN37, novel_mir_191 and Csi-miR393a.2 seem to be post-transcriptional key regulators in C. sinensis fruit. These miRNAs control biological processes including RNA processing, protein degradation, hormone metabolism and signal transduction. Furthermore, Csi-miR162, Csi-miR162-3p.1, miR162, Csi-miR162a, PK-Cs6g15240.1, TR-Cs7g16070.1 and Csi-miR1092.2, novel_mir_99, TF-Cs2g09440.2, TF-Cs1g16290.1, TR-Cs7g16070.1 respectively regulate orange1.1t00584.1 and Cs7g22180.1 in a cooperative manner. These transcriptional and translational key regulators play important roles in the control of RNA processing by increasing (via transcriptional regulators) or decreasing (via miRNAs) the synthesis of degradation dicer and RNA helicase enzymes.
On the other hand, to date there is not sufficient precise and experimentally validated information about the role of all known genes and their regulators in C. sinensis relevant biological processes. Therefore, our proposed co-regulatory relationships could be so useful in order to identifying the role of some genes and/or regulators based on the known function of other connected genes and/or regulators in well studied biological processes. For example, Fig. 2a represent 2 transcription factors (TF-Cs7g15220 and TF-Cs1g21350) which are involved in leaf and fruit development (Liu et al. 2016). Therefore, Csi-miR396a could be a very important citrus growth inhibitor, because it decreases the translation of both Cs7g15220 and Cs1g21350 genes simultaneously in a synergistic manner. Interestingly, the importance of this post transcription regulator homologue in response to drought stress in Solanum tuberosum has been proven before (Hwang et al. 2011).
Moreover, as shown in Fig. 3a, Cs3g23180.1 which is known as a C. sinensis fruit ripening-associated transcript (Wu et al. 2014a, b) is regulated by Csi-miR396b.3, Csi-miR396b-2010, TF-orange1.1t04055.1 and TF-Cs7g27790.1. Therefore, these transcriptional and post-transcriptional regulators are also involved in fruit ripening pathways and even may be the key regulators of this biological process. TF-Cs3g18940.1 encodes ARF17 (auxin response factor 17) protein and ARFs have been reported that are involved in the regulation of various aspects of fruit development in recent plant studies (Liu et al. 2015). So its regulated transcripts and their associated regulators including Cs7g31880.1, orange1.1t00471.1, Csi-miRN04, Csi-miR3951, miR3951 and csi-novel-03 may also be involved in the C. sinensis fruit development process.
TF-Cs7g11020.1 was reported to be a WRKY gene and possibly involved in the response of C. sinensis to drought and salinity (5). Moreover, this regulator expression is always affected by pathogen infections which make it a key regulator of defense against plant diseases (Ayadi et al. 2016). Cs7g10850.1, Cs8g07280.1 and Cs3g18660 are connected to TF-Cs7g11020.1 and may have the similar bilogical roles. Thus their inhibitors including Csi-miR159, miR159, csi-miR159a, Csi-miR394, csi-miR394a, Csi-miR395.1, csi-miR395a and csi-MIR395 also could be important post transcription regulator of these biotic and abiotic stress mediators. Cs3g01650 gene encodes a cell division control protein which regulates the citrus flowering time and floweral development by controlling the transcription of APETALA1-like homologue gene in C. sinensis (Sun et al. 2014). So, based on our proposed Leaf-C2 leaf subgraph (Fig. 3b), Csi-miR403.1 and TF-Cs4g01030 regulators have negative and positive roles in the citrus flowering transition process. Exploring other subgraphs and their corresponded PPI subnetworks, researchers can find most likely Citrus sinensis biological pathways considerable transcriptional and post-transcriptional regulators for further experiments and investigations.
According to our studies, there are only a few published research about miRNA-transcriptional regulator coregulatory network in plants. Nevertheless, some outstanding researches were done on mammals, particularly human and mouse. AtmiRNET is a web service resource that employed machine learning and statistical approaches to infer transcription factor-miRNA connections in Arabidopsis thaliana. It also used experimentally and computationally verified miRNA-target interactions to reconstruct Arabidopsis coregulatory network (Chien et al. 2015). Zho et al. (n.d.) obtained and compared statistically significant TF-miRNA motifs for the drought and salt stresses, and different development stages in Arabidopsis.
Guo et al. (2010) for the first time constructed a miRNA-TF regulatory network for a complex disease, schizophrenia, based on 32 identified feed-forward loops (FFLs) among their compiled disease-related miRNAs, TFs and genes. Wang et al. (2010) extracted 243 out of 5000 reports in the literature on TF-miRNA relationships in human and made a database called TransmiR. Nazarov et al. (2013) investigated simultaneously the transcriptional changes of miRNA and mRNA expression levels in microarray time-series data derived from Jak/STAT activated melanoma cells and observed the delayed response of miRNAs with respect to mRNAs. They also extracted network FFL motifs involving transcriptional regulators, mRNAs and miRNAs. Cantini et al. (2015) proposed a multi-network-based strategy to integrate different layers of genomic information including transcription factor co-targeting, microRNA co-targeting, protein–protein interaction and gene co-expression networks in order to find new candidate driver cancer genes in four cancer types. Liu et al. (2013) defined the significant triple relations among miRNAs, TFs and mRNAs from mouse lung development time course data. This study reveals that circuits vary in different stages of development and play different roles in biological pathways. Utilizing functional annotation information, Chen et al. (2011) suggested that cross-layer co-regulation (e.g. transcriptional-translational co-regulation) may show higher specificity than intra-layer co-regulation in various biological conditions.
Shao et al. (2015) developed a pipeline capable of inferring regulatory relationships between miRNAs and transcription factors from Chip-Seq data, and used it to derive coregulatory network in mouse embryonic stem cells. Afshar et al. (2014) proposed a method that collectively utilizes various data, algorithms, and statistical techniques to infer miRNA-mediated regulatory circuits at the transcriptional, post-transcriptional, and signaling levels. Naeem (2012) developed a database of miRNA/Transcription factor-target relationship by means of literature text mining, applied statistical tests on regulator perturbation datasets to predict activity state of miRNA/Transcription factor, and used gene set enrichment methods to predict regulation cascades.
In order to identify the regulation mechanism of miRNAs in Homo sapiens and Mus musculus (Georgakilas et al. 2015), the DIANA-miRGen v3.0 using accurate cell-line-specific miRNA gene transcription start sites (TSSs) and genome-wide maps of transcription factor binding sites were combined. GeneNetworkBuilder facilitates identification of the complex coregulatory network including TFs, miRNAs and mRNAs by combining binding site identifying and gene expression technologies such as chip-seq, chip–chip, microarray, and RNA-Seq (Ou and Zhu 2016).
The availability of valid data is fundamental for Network reconstruction tasks. Using only computational data often leads to many false positive connections in a reconstructed network. Therefore, it is essential to use the maximum number of experimentally unveiled relationships and applying an approved pipeline for computational relation discovery. Although present work has tried to follow these constraints, further experiments on C. sinensis are needed to validate computationally predicted relationships in this study. Additionally, as many other non-model species, since the C. sinensis genome sequencing project is short lived, insufficient information about miRNA–miRNA and TF-miRNA interactions in its co-regulatory network is provided here.
Furthermore, gene regulatory mechanism is not fully known yet and it is expected that more components are involved in it. For example, Zhao et al. (2016) recently found that miRNA-mediated motifs require the cooperation of epigenetic marks to effect on gene regulatory mechanism. Thus, co-regulatory network reconstruction for some species such as C. sinensis will still remain an open area for research, requiring more precise experimental results and more enhanced computational tools for improving the inferences of this complex network.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Prof. Qiang and his database team at Huazhong agricultural university for providing us the whole PPI network data of Citrus sinensis. We would also like to thank two reviewers, who’s comments make the manuscript improved and better to understand.
Contributor Information
Ali Ashraf Mehrabi, Email: a.mehrabi@ilam.ac.ir.
Ali Masoudi-Nejad, Phone: +98-21-6695-9256, Email: amasoudin@ibb.ut.ac.ir, http://LBB.ut.ac.ir.
References
- Afshar AS, Xu J, Goutsias J. Integrative identification of deregulated miRNA/TF-mediated gene regulatory loops and networks in prostate cancer. PloS One. 2014;9(6):e100806. doi: 10.1371/journal.pone.0100806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruoma OI, Landes B, Ramful-Baboolall D, Bourdon E, Neergheen-Bhujun V, Wagner K-H, Bahorun T. Functional benefits of citrus fruits in the management of diabetes. Prev Med. 2012;54:S12–S16. doi: 10.1016/j.ypmed.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008;13(7):343–349. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Ayadi M, Hanana M, Kharrat N, Merchaoui H, Marzoug RB, Lauvergeat V, Rebaï A, Mzid R. The WRKY transcription factor family in citrus: valuable and useful candidate genes for citrus breeding. Appl Biochem Biotechnol. 2016 doi: 10.1007/s12010-016-2114-8. [DOI] [PubMed] [Google Scholar]
- Berri S, Abbruscato P, Faivre-Rampant O, Brasileiro ACM, Fumasoni I, Satoh K, Kikuchi S, Mizzi L, Morandini P, Pè ME, Piffanelli P. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 2009;9(120):1–22. doi: 10.1186/1471-2229-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, He Y, Billiau K, van de Peer Y. TAPIR, a web server for the prediction of plant microRNA targets, including target mimics. Bioinformatics. 2010;26(12):1566–1568. doi: 10.1093/bioinformatics/btq233. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320(5880):1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Cantini L, Medico E, Fortunato S, Caselle M. Detection of gene communities in multi-networks reveals cancer drivers. Sci Rep. 2015;5:17386. doi: 10.1038/srep17386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-Y, Chen S-T, Fuh C-S, Juan H-F, Huang H-C. Coregulation of transcription factors and microRNAs in human transcriptional regulatory network. BMC Bioinformatics. 2011;12(Suppl 1):S41. doi: 10.1186/1471-2105-12-S1-S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CH, Chiang-Hsieh YF, Chen YA, Chow CN, Wu NY, Hou PF, Chang WC. AtmiRNET: a web-based resource for reconstructing regulatory networks of Arabidopsis microRNAs. Database. 2015 doi: 10.1093/database/bav042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T-J. The role of microRNAs in sensing nutrient stress. Plant Cell Environ. 2007;30(3):323–332. doi: 10.1111/j.1365-3040.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- Da Graca JV, Korsten L, Graca JVD, Korsten L. Citrus huanglongbing: review, present status and future strategies. Dis Fruits Veg. 2004;1:229–245. [Google Scholar]
- Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39(suppl 2):W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das AK. Citrus canker—a review. J Appl Hortic. 2003;5(1):52–60. [Google Scholar]
- Ding YD, Chang JW, Guo J, Chen D, Li S, Xu Q, Deng XX, Cheng YJ, Chen LL. Prediction and functional analysis of the sweet orange protein-protein interaction network. BMC Plant Biol. 2014;14:213. doi: 10.1186/s12870-014-0213-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet. 2010;11(8):539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Du D, Rawat N, Deng Z, Gmitter FG. Construction of citrus gene coexpression networks from microarray data using random matrix theory. Hortic Res. 2015;2:15026. doi: 10.1038/hortres.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott JA, Noble MEM, Johnson LN. The structural basis for control of eukaryotic protein kinases. Annu Rev Biochem. 2012;81:587–613. doi: 10.1146/annurev-biochem-052410-090317. [DOI] [PubMed] [Google Scholar]
- Georgakilas G, Vlachos IS, Zagganas K, Vergoulis T, Paraskevopoulou MD, Kanellos I, Tsanakas P, Dellis D, Fevgas A, Dalamagas T, Hatzigeorgiou AG. DIANA-miRGen v3.0: accurate characterization of microRNA promoters and their regulators. Nucleic Acids Res. 2015;44(D1):D190–D195. doi: 10.1093/nar/gkv1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong D-H, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, De Paoli E, Lu C, Schroth G, Meyers BC, Green PJ. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26(8):941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A-Y, Sun J, Jia P, Zhao Z. A novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC Syst Biol. 2010;4:10. doi: 10.1186/1752-0509-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi PH, Di Martino MT, Tagliaferri P, Tassone P. Cannataro M (2015) Analysis of miRNA, mRNA, and TF interactions through network-based methods. EURASIP J Bioinf Syst Biol. 2015;1:1–11. doi: 10.1186/s13637-015-0023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamed M, Spaniol C, Nazarieh M, Helms V. TFmiR: a web server for constructing and analyzing disease-specific transcription factor and miRNA co-regulatory networks. Nucleic Acids Res. 2015;43(W1):W283–W288. doi: 10.1093/nar/gkv418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh W-T, Tzeng K-R, Ciou J-S, Tsai JJ, Kurubanjerdjit N, Huang C-H, Ng K-L. Transcription factor and microRNA-regulated network motifs for cancer and signal transduction networks. BMC Syst Biol. 2015;9(Suppl 1):S5. doi: 10.1186/1752-0509-9-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E-W, Shin S-J, Kwon H-B. Identification of microRNAs and their putative targets that respond to drought stress in Solanum tuberosum. J Korean Soc Appl Biol Chem. 2011;54(3):317–324. doi: 10.3839/jksabc.2011.051. [DOI] [Google Scholar]
- Jin J, Zhang H, Kong L, Gao G, Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92(3):307–313. doi: 10.1016/S0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- Kalim B, Mughal MS, Ali NM, Rehman S, Faiz R, Mazhar B, Sarwar MS. Estimation of organic and inorganic components of Citrus sinensis linn (musambi) and its effects on early growth of broiler chicks. Merit Res J Agric Sci Soil Sci. 2015;3(7):106–112. [Google Scholar]
- Kozomara A, Griffiths-Jones S. MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014 doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Singh S, Wensel A, Huala E. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012 doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TD, Liu L, Zhang J, Liu B, Li J. From miRNA regulation to miRNA-TF co-regulation: computational approaches and challenges. Brief Bioinform. 2014;16(3):475–496. doi: 10.1093/bib/bbu023. [DOI] [PubMed] [Google Scholar]
- Leyva-González MA, Ibarra-Laclette E, Cruz-Ramírez A, Herrera-Estrella L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PloS One. 2012 doi: 10.1371/journal.pone.0048138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ye X, Wu F-X. Characterizing dynamic regulatory programs in mouse lung development and their potential association with tumourigenesis via miRNA-TF-mRNA circuits. BMC Syst Biol. 2013;7(Suppl 2):S11. doi: 10.1186/1752-0509-7-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li J, Huang M, Chen J. Mechanisms for the influence of citrus rootstocks on fruit size. J Agric Food Chem. 2015;63(10):2618–2627. doi: 10.1021/jf505843n. [DOI] [PubMed] [Google Scholar]
- Liu X, Guo L-X, Jin L-F, Liu Y-Z, Liu T, Fan Y-H, Peng S-A. Identification and transcript profiles of citrus growth-regulating factor genes involved in the regulation of leaf and fruit development. Mol Biol Rep. 2016;43(10):1059–1067. doi: 10.1007/s11033-016-4048-1. [DOI] [PubMed] [Google Scholar]
- Lu Y-B, Qi Y-P, Yang L-T, Guo P, Li Y, Chen L-S. Boron-deficiency-responsive microRNAs and their targets in Citrus sinensis leaves. BMC Plant Biol. 2015;15(1):271. doi: 10.1186/s12870-015-0642-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M, Cumbie JS, Ivanchenko MG, Filchkin SA. Small genetic circuits and microRNAs: big players in Pol-II transcriptional control in plants. Plant Cell. 2016 doi: 10.1105/tpc.15.00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Soto AB, Sánchez F, Hernández G. MicroRNAs as regulators in plant metal toxicity response. Front Plant Sci. 2012;3:1–6. doi: 10.3389/fpls.2012.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem H (2012) Activity of microRNAs and transcription factors in gene regulatory networks. Doctoral dissertation, LMU
- Nazarov PV, Reinsbach SE, Muller A, Nicot N, Philippidou D, Vallar L, Kreis S. Interplay of microRNAs, transcription factors and target genes: linking dynamic expression changes to function. Nucleic Acids Res. 2013;41(5):2817–2831. doi: 10.1093/nar/gks1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osella M, Bosia C, Corá D, Caselle M. The role of incoherent microRNA-mediated feedforward loops in noise buffering. PLoS Comput Biol. 2011 doi: 10.1371/journal.pcbi.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J, Liu H, Tissenbaum HA, Zhu LJ (2016) GeneNetworkBuilder: build regulatory network from ChIP-chip/ChIP-seq and expression data. R package version 1.16.0
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, Sun Y, Zhou S. Identifying TF-miRNA regulatory relationships using multiple features. PloS One. 2015 doi: 10.1371/journal.pone.0125156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Wang C, Zhang C, Korir NK, Yu H, Ma Z, Fang J. Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata) BMC Genomics. 2010;11(1):1. doi: 10.1186/1471-2164-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava PK, Moturu TR, Pandey P, Baldwin IT, Pandey SP. A comparison of performance of plant miRNA target prediction tools and the characterization of features for genome-wide target prediction. BMC Genomics. 2014;15(1):1. doi: 10.1186/1471-2164-15-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Dong B, Yin L, Zhang R, Du W, Liu D, Shi N, Li A, Liang Y, Mao L. PMTED: a plant microRNA target expression database. BMC Bioinform. 2013;14(1):174. doi: 10.1186/1471-2105-14-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L-M, Zhang J-Z, Mei L, Hu C-G. Molecular cloning, promoter analysis and functional characterization of APETALA 1-like gene from precocious trifoliate orange (Poncirus trifoliata L. Raf.) Sci Hortic. 2014;178:95–105. doi: 10.1016/j.scienta.2014.08.015. [DOI] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu J-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12(7):301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Tam W-L, Lim B (2009) The interface of MicroRNAs and transcription factor networks. Syst Biomed Concepts Perspect 5:109–137. doi:10.1016/B978-0-12-372550-9.00005-5
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37(6):914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/S0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit Rev. 2010;44(Pol II):117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(2009):669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Wang J, Lu M, Qiu C, Cui Q. TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Res. 2010;38(Database issue):D119–D122. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen D, Lei Y, Chang JW, Hao BH, Xing F, Li S, Xu Q, Deng XX, Chen LL. Citrus sinensis annotation project (CAP): a comprehensive database for sweet orange genome. PloS One. 2014 doi: 10.1371/journal.pone.0087723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DCJ, Sweetman C, Ford CM. Annotation of gene function in citrus using gene expression information and co-expression networks. BMC Plant Biol. 2014;14(1):186. doi: 10.1186/1471-2229-14-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang D, Liu Y, Wang L, Qiao X, Zhang S. Identification of miRNAs involved in pear fruit development and quality. BMC Genomics. 2014;15(1):953. doi: 10.1186/1471-2164-15-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Xu Z, Zhang Y, Chai L, Yi H, Deng X. An integrative analysis of the transcriptome and proteome of the pulp of a spontaneous late-ripening sweet orange mutant and its wild type improves our understanding of fruit ripening in citrus. J Exp Bot. 2014 doi: 10.1093/jxb/eru044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X-M, Kou S-J, Liu Y-L, Fang Y-N, Xu Q, Guo W-W. Genomewide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: microRNA-and siRNA-mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol J. 2015;13(3):383–394. doi: 10.1111/pbi.12317. [DOI] [PubMed] [Google Scholar]
- Wu X, Ding D, Shi C, Xue Y, Zhang Z, Tang G, Tang J. microRNA-dependent gene regulatory networks in maize leaf senescence. BMC Plant Biol. 2016;16(1):73. doi: 10.1186/s12870-016-0755-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Chen L-L, Ruan X, Chen D, Zhu A, Chen C, Bertrand D, Jiao W-B, Hao B-H, Lyon MP, Chen J, Gao S, Xing F, Lan H, Chang J-W, Ge X, Lei Y, Hu Q, Miao Y, Wang L, Xiao S, Biswas MK, Zeng W, Guo F, Cao H, Yang X, Xu X-W, Cheng Y-J, Xu J, Liu J-H, Luo OJ, Tang Z, Guo W-W, Kuang H, Zhang H-Y, Roose ML, Nagarajan N, Deng X-X, Ruan Y. The draft genome of sweet orange (Citrus sinensis) Nat Genet. 2013;45(1):59–66. doi: 10.1038/ng.2472. [DOI] [PubMed] [Google Scholar]
- Xu Q, Liu Y, Zhu A, Wu X, Ye J, Yu K, Guo W, Deng X. Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genomics. 2010;11:246. doi: 10.1186/1471-2164-11-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MY, Zhang L, Li WW, Hu XL, Wang M-B, Fan YL, Zhang CY, Wang L. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. J Exp Bot. 2013 doi: 10.1093/jxb/ert353. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yu J, Li D, Zhang Z, Liu F, Zhou X, Wang T, Ling Y, Su Z. PMRD: plant microRNA database. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhou X, Zhang X, Fromuth N, Coutino G, Coffey M, Jin H. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol. 2011;75(1):93–105. doi: 10.1007/s11103-010-9710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Kuang S, Xiong X, Gao T, Liu C, Guo AY. Transcription factor and microRNA co-regulatory loops: important regulatory motifs in biological processes and diseases. Brief Bioinform. 2013;16(1):45–58. doi: 10.1093/bib/bbt085. [DOI] [PubMed] [Google Scholar]
- Zhou X, Zhang W (2007) Regulatory circuits of microRNAs and transcription factors in plant. https://www.semanticscholar.org/paper/Regulatory-Circuits-of-Micrornas-and-Transcription-Zhou-Zhang/63580ab2bb249d5b89b736e0d67776b6dde4ced9. Accessed 3 Jan 2016
- Zhao H, Zhang G, Pang L, Lan Y, Wang L, Yu F, Hu J, Li F, Zhao T, Xiao Y, et al. ‘Traffic light rules’: chromatin states direct miRNA-mediated network motifs running by integrating epigenome and regulatome. Biochim Biophys Acta (BBA) 2016;7:1475–1488. doi: 10.1016/j.bbagen.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Fei Z (2014) iTAK-identification and classification of plant transcription factors and protein kinases. In: Plant and animal genome XXII conference
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.