Abstract
NAC transcription factor (TF) family proteins are expressed in various developmental stages and following various stresses. NAC TFs are involved in mediating various physiological functions of plants and participate in various signaling pathways under biotic or abiotic stress. The present study provided a comprehensive functional analysis of members of the MtNAC TF family. Via screening of Medicago truncatula genome information, we identified 97 MtNAC TFs in M. truncatula and compared the phylogenetic analysis of 14 conserved groups with their Arabidopsis and rice counterparts. The NAC TFs were categorized into 14 groups based on their conserved motifs and gene structure. The predicted M. truncatula NAC genes were distributed among eight chromosomes, and in addition, we found that these genes showed mass gene duplication. Through expression profiling of RNA-seq data analysis, we determined that NAC family members were expressed significantly under different abiotic stresses. This indicates that the NAC TF shows different functions in M. truncatula. Together, this genome-wide analysis of the NAC gene family in M. truncatula, could be applied to improving stress tolerance in plants.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-017-0421-3) contains supplementary material, which is available to authorized users.
Keywords: NAC transcription factors, Phylogenetic analysis, Expression profile, Medicago truncatula
Introduction
Alfalfa is an important perennial forage legume species. It is a high—yielding perennial grass species that has high nutritional value and nitrogen fixation capacity. It is an important germplasm resource in the world. They make a contribution to modern society end economy construction (Samac et al. 2006; Sanderson et al. 2004; Yang et al. 2011).
The NAC domain is a highly conserved amino acid motif in one of the largest groups of plant-specific transcription factors (TFs). No apical meristem (NAM) was the first characterized NAC protein found in petunias, and Arabidopsis transcription activation factor (ATAF) and cup-shaped cotyledon (CUC) were found in Arabidopsis (Tran et al. 2004; Zhong et al. 2006). Several NAC proteins have been identified in other plant species, including rice, wheat, and soybean (Hussey et al. 2015).
The NAC TF family members have a highly conserved NAC domain about 150 amino acids, which include N-terminal ends containing five subdomains (A–E) and highly variable domain at C-terminal ends (Ernst et al. 2004). At their C-terminal end, the basic region also contains α-helical transmembrane motifs (TMs) (Puranik et al. 2012). The special structure is related to specific biotic functions, and the NAC TFs are involved in biotic and abiotic stress processes (Mao et al. 2012). These genes influence plant growth, enhance the absorption of mineral elements, and improve crop nutrition and quality. The NAC TFs are involved in mediating a variety of physiological activities in plants, such as auxin conduction and cell apoptosis (Yoshii et al. 2010). When plants are affected by biotic or abiotic stress, NAC TFs participate in various signaling pathways to cope with these adverse conditions (Pinheiro et al. 2009).
Earlier studies have shown that NAC TFs play an important regulatory role in plants subjected to abiotic stress including salinity, drought, cold, or abscisic acid (ABA) (Olsen et al. 2005). For example, over accumulation of ANAC (019, 055, 072) in Arabidopsis lead to enhanced stress tolerance in transgenic plants (Bu et al. 2008). The gene ANAC062 is involved in the cold stress signal regulation (Yang et al. 2014). OsNAC5 is a senescence-associated ABA-dependent NAC TF. OsNAC045 and OsNAC063 can enhance drought and salt tolerance in rice (Xu et al. 2015). The expression level of OsNAC19 is increased after Magnaporthe grisea infection to regulate the defense responses in rice (Nuruzzaman et al. 2015). NAC proteins are involved in responses to viral infections during plant vegetative development. In soybean, GmNAC11 and GmNAC20 are involved in responses to low temperature, and overexpression of these two genes enhanced low temperature tolerance (Hao et al. 2011).
GmNAC1-6 are differentially expressed during seed development and other physiological processes in Medicago sativa. A novel M. sativa NAC transcription factor has been characterized during drought stress. In M. truncatula, the MtNAC969 is involved in root system architecture by several pathways.
With the rapid development of high-throughput sequencing technologies, several NAC members have been studied in model plants such as A. thaliana (117), Glycine max (152), O. sativa (151), and Vitis vinifera (74) (Le et al. 2011; Nuruzzaman et al. 2010; Ooka et al. 2003; Wang et al. 2013). The theoretical basis for the present study was provided by the completed M. truncatula genome sequence and previous studies on NAC TFs.
NAC proteins are found in most plant species, but their research is poorly understood in M. truncatula. M. truncatula is an excellent model organism for leguminous plants due to small genome and high genetic transformation efficiency (Bell et al. 2001). Legumes are the second most important family of crop plants after Poaceae and significantly contribute to agricultural production (Graham and Vance 2003). For efficient agricultural production, it is necessary to enhance crop resistance to abiotic stress and therefore the study of NAC TF is prerequisite for improving stress tolerance in plants (Chen et al. 2015).
In this study, we used bioinformatic approach to analyse the NAC family in M. truncatula. We have performed comprehensive study of gene structures, motif composition, chromosomal locations, gene duplication, sequence homologies, and expression patterns during different stresses. The transcriptome information was used to characterize the functions of these TFs during abiotic stress. The results of the present study will be helpful for future investigations to enhance the stress tolerance of plants.
Materials and methods
Identification of NAC gene information
The Hidden Markov Model (HMM) profiles of the NAM domain PF02365 were downloaded from the Pfam database (Punta et al. 2011). HMM searched NAM (PF02365) domains from the M. truncatula protein database with values (e-value) cut-off at 1.0 (Johnson et al. 2010). The integrity of the NAM domain was determined using the online program SMART (http://smart.embl-heidelberg.de/) with an e-value < 0.1 (Letunic et al. 2012). In addition, the three fields (length, molecular weight, and isoelectric point) of each NAC protein were predicted by the online ExPasy program (http://www.expasy.org/tools/) (Rueda et al. 2015).
Phylogenetic analysis and motif prediction
To investigate the phylogenetic relationship of the NAC gene families in Arabidopsis, rice, and M. truncatula, NAC protein sequences were downloaded from phytozomes (http://www.phytozome.org) (Goodstein et al. 2012). NAC TFs were aligned using the BioEdit program. A neighbor-joining (NJ) phylogenetic tree was constructed using the MEGA6 program (Tamura et al. 2011). Bootstrapping was performed with 1000 replications. The online MEME analysis used to identify the unknown conserved motifs (http://meme.ebi.edu.au/meme/intro.html) using the following parameters: site distribution: zero or one occurrence (of a contributing motif site) per sequence, maximum number of motifs: 25, and optimum motif width ≥6 and ≤200 (Bailey et al. 2015). Detailed information of M. truncatula NAC proteins can be found in Table A1.
Gene structure and chromosomal localization
The whole-genomic sequence of M. truncatula and the summary of gene localization information were downloaded from the phytozome database (http://phytozome.jgi.doe.gov/medicago.php). The genomic sequence for each NAC gene was extracted from the whole-genomic sequence according to gene localization information using a programmed Perl script. A gene structure display server program (http://gsds.cbi.pku.edu.cn/index.php) was used to display the M. truncatula NAC gene structures (Guo et al. 2007). Duplications between the NAC genes were identified and complemented using the PGDD database (http://chibba.agtec.uga.edu/duplication/), and were identified as tandem duplications (TD). Ideograms were created using Circos (Krzywinski et al. 2009).
Transcriptome analysis of the NAC gene in different tissues and under five abiotic stress
M. truncatula transcriptome data in different tissues during development were downloaded from the NCBISRA database (http://www.ncbi.nlm.nih.gov, Accession numbers SRX099057–SRX099062). The transcriptome data were derived from six tissues, including roots, nodules, blades, buds, seedpods, and flowers. M. truncatula transcriptome data under different abiotic stresses were downloaded from the NCBISRA database (http://www.ncbi.nlm.nih.gov, Accession numbers SRX1056987–92) (Li et al. 2009). The transcriptome data were derived under six stress factors, including cold, freezing, drought, salt, and high levels of ABA. We performed RNA-seq to detect the expression levels of NAC TF genes under different stresses, including cold, freezing, drought, salt and ABA. Clean reads from six samples were mapped to the M. truncatula genome sequencing using Samtool (Li et al. 2009). Tophat and Cufflinks were used to analyze RPKM (Trapnell et al. 2012). The RPKM values for NAC genes were utilized for generating the heatmap and k-means clustering using R (software) (Gentleman et al. 2004).
Plant material and treatments
M. truncatula (Jemalong) A17 was used in this study. Seeds were planted in a soil and sand mixture (3:1), germinated, and irrigated with half-strength Hoagland solution once every 2 d. The seedlings were grown in the following environmental conditions: temperature of 18 °C (night) and 24 °C (day), and relative humidity of 60–80%. The seedlings that germinated after 8 weeks were subjected to the following environmental conditions: temperatures of 4 (cold) or −8 °C (freezing), treated with 300 mM mannitol (drought) or 200 mM NaCl solution (salt), and the seedling leaves were sprayed with 100 μM ABA solution (ABA). Control and treated seedlings were harvested 3 h after treatment. All samples were frozen in liquid nitrogen and stored at −80 °C until use.
RNA extraction and quantitative reverse transcription PCR (qRT-PCR)
The transcriptome sequencing analysis was validated and quantified by qRT-PCR. Primers were designed according to NAC cds with Primer Express 3.0 software (Untergasser et al. 2012), the primer pairs are listed in Table A1. Total RNA were extracted with an RNA prep pure Plant Kit (Tiangen, Beijing, China), and cDNA was synthesized from total RNA using a Rever Tra Ace (Toyobo, Shanghai, China), and qRT-PCR was performed in an ABI 7300 Real-time Detection System (Applied Biosystems, Foster City, CA, USA). The thermal profile for SYBR green RT-PCR consisted of initial denaturation at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s, and a final dissociation at 95 °C for 15 s, followed by one cycle at 60 °C for 20 s and one cycle at 95 °C for 15 s. To confirm that a single PCR product was amplified and detected, a dissociation curve analysis of amplification products was performed at the end of each PCR reaction. After amplification, data were analyzed with ABI 7300 SDS software (Applied Biosystems, USA). The comparative CT method (2-△△Ct method) was used to analyze the expression level of different genes. All of the samples were tested in triplicate, and the experiments were performed on three biological replicates.
Results
Genome-wide identification of NAC family genes in M. truncatula
Searching for NAC genes in the M. truncatula genome, all proteins of the M. truncatula genome from phytozomes were annotated (Town, 2006). Finally, 97 non-redundant and complete NAC-domain-containing protein sequences were selected for further analysis—the amino acid sequence length was between 54 and 672 (average length 341.3)—and named from MtNAC1 to MtNAC97 based on the coordinate order on M. truncatula chromosomes information, including protein properties in Table 1 (Committee, 1999). To further understand NAC TF functions, these target genes were downloaded from database of M. truncatula.
Table 1.
List of all MtNAC genes information identified in the Medicago truncatula genome
| Gene name | Gene ID | Chromosome location | Length (aa) | Family group | PI | Molecular weight (kDa) |
|---|---|---|---|---|---|---|
| MtNAC1 | Medtr0036s0150 | scaffold0036:60708-55826 | 539 | XIII | 4.51 | 60,016.90 |
| MtNAC2 | Medtr2157s0010 | scaffold2157:1006-209 | 266 | VII | 9.01 | 30,458.60 |
| MtNAC3 | Medtr1g008740 | chr1:1057858-1061279 | 362 | I | 9 | 40,725.70 |
| MtNAC4 | Medtr1g045470 | chr1:17058321-17056452 | 325 | XI | 5.91 | 37,435.10 |
| MtNAC5 | Medtr1g053575 | chr1:22598992-22597045 | 323 | XI | 4.74 | 37,448.70 |
| MtNAC6 | Medtr1g069805 | chr1:30480393-30484636 | 275 | I | 5.88 | 31,799.80 |
| MtNAC7 | Medtr1g087190 | chr1:39063064-39062900 | 54 | XIII | 6.01 | 6,453.50 |
| MtNAC8 | Medtr1g090720 | chr1:40728357-40734069 | 482 | VI | 6.42 | 53,997.70 |
| MtNAC9 | Medtr1g090723 | chr1:40735770-40738910 | 630 | XIII | 4.42 | 70,012.50 |
| MtNAC10 | Medtr1g093670 | chr1:42006623-42004062 | 345 | X | 5.1 | 39,407.20 |
| MtNAC11 | Medtr1g093680 | chr1:42018089-42012426 | 481 | X | 4.81 | 54,345.30 |
| MtNAC12 | Medtr1g096430 | chr1:43438375-43439601 | 339 | II | 6.53 | 39,409.80 |
| MtNAC13 | Medtr1g097300 | chr1:43891021-43886555 | 571 | XIII | 4.58 | 64,727.80 |
| MtNAC14 | Medtr2g014680 | chr2:4232801-4236401 | 308 | X | 6.76 | 34,805.90 |
| MtNAC15 | Medtr2g062730 | chr2:26494431-26498275 | 344 | II | 6.85 | 39,737.40 |
| MtNAC16 | Medtr2g064090 | chr2:27141403-27145406 | 346 | III | 5.13 | 39,985.10 |
| MtNAC17 | Medtr2g064470 | chr2:29175407-29181087 | 306 | I | 8.07 | 34,667.20 |
| MtNAC18 | Medtr2g068880 | chr2:28613602-28616289 | 312 | XII | 6.56 | 35,958.10 |
| MtNAC19 | Medtr2g068920 | chr2:28628371-28630420 | 291 | XII | 6 | 33,721.50 |
| MtNAC20 | Medtr2g078700 | chr2:32897238-32899105 | 381 | I | 8.17 | 42,221.90 |
| MtNAC21 | Medtr2g079990 | chr2:33727407-33729223 | 354 | IX | 7.05 | 40,053.90 |
| MtNAC22 | Medtr2g080010 | chr2:33761666-33764021 | 354 | VII | 8.85 | 39,359.90 |
| MtNAC23 | Medtr2g086690 | chr2:36452124-36453846 | 249 | XIII | 6.61 | 28,467.60 |
| MtNAC24 | Medtr2g086880 | chr2:36530061-36538039 | 589 | III | 8.82 | 66,653.20 |
| MtNAC25 | Medtr2g090735 | chr2:38885938-38887846 | 285 | X | 8.72 | 32,439.60 |
| MtNAC26 | Medtr2g093810 | chr2:40013370-40012105 | 323 | VII | 8.79 | 37,217.80 |
| MtNAC27 | Medtr3g064580 | chr3:29101701-29105835 | 672 | III | 5.74 | 76,914.70 |
| MtNAC28 | Medtr3g070030 | chr3:31349143-31346724 | 352 | I | 6.68 | 40,119.70 |
| MtNAC29 | Medtr3g070040 | chr3:31359786-31357396 | 335 | I | 5.73 | 37,705.10 |
| MtNAC30 | Medtr3g088110 | chr3:39954022-39955471 | 288 | VIII | 7.65 | 33,062.30 |
| MtNAC31 | Medtr3g093040 | chr3:42540067-42535957 | 312 | VI | 5.86 | 35,072.70 |
| MtNAC32 | Medtr3g093050 | chr3:42546323-42542510 | 292 | VIII | 5.94 | 33,796 |
| MtNAC33 | Medtr3g096140 | chr3:43934316-43939062 | 336 | XIII | 4.8 | 37,484.70 |
| MtNAC34 | Medtr3g096400 | chr3:44063892-44061980 | 208 | IX | 4.3 | 23,721.30 |
| MtNAC35 | Medtr3g096920 | chr3:44370629-44372461 | 292 | VIII | 5.94 | 33,796 |
| MtNAC36 | Medtr3g098810 | chr3:45276406-45275099 | 241 | VI | 8.84 | 27,405.90 |
| MtNAC37 | Medtr3g109340 | chr3:50581341-50579226 | 358 | I | 8.14 | 40,490.20 |
| MtNAC38 | Medtr3g116070 | chr3:54270247-54268085 | 353 | I | 6.64 | 40,385.60 |
| MtNAC39 | Medtr3g435150 | chr3:11464191-11461627 | 303 | I | 9.22 | 34,581.40 |
| MtNAC40 | Medtr4g035590 | chr4:12274680-12278280 | 354 | II | 6.9 | 41,163.80 |
| MtNAC41 | Medtr4g036030 | chr4:12849080-12853325 | 346 | II | 5.41 | 40,428.90 |
| MtNAC42 | Medtr4g052620 | chr4:19088028-19085634 | 261 | XIII | 5.69 | 29,816 |
| MtNAC43 | Medtr4g075980 | chr4:29048799-29051425 | 299 | XII | 7.05 | 34,465.30 |
| MtNAC44 | Medtr4g078875 | chr4:30504183-30506651 | 284 | X | 9.02 | 31,985.90 |
| MtNAC45 | Medtr4g081870 | chr4:31873646-31871462 | 271 | VII | 6.51 | 31,265.30 |
| MtNAC46 | Medtr4g088245 | chr4:34833163-34829374 | 318 | X | 5.66 | 35,933.20 |
| MtNAC47 | Medtr4g089135 | chr4:35772138-35774193 | 344 | VII | 7.23 | 38,660.70 |
| MtNAC48 | Medtr4g094302 | chr4:37671153-37672443 | 191 | IX | 4.82 | 22,243.60 |
| MtNAC49 | Medtr4g098630 | chr4:40652597-40651083 | 318 | III | 5.52 | 36,214.40 |
| MtNAC50 | Medtr4g101680 | chr4:42049132-42045198 | 364 | II | 6.25 | 42,406.90 |
| MtNAC51 | Medtr4g108760 | chr4:45058271-45060202 | 359 | I | 8.56 | 40,556.60 |
| MtNAC52 | Medtr4g134460 | chr4:56311697-56314294 | 434 | XII | 6.15 | 48,628 |
| MtNAC53 | Medtr5g012080 | chr5:3572221-3576785 | 349 | II | 6.27 | 40,487 |
| MtNAC54 | Medtr5g014300 | chr5:4775644-4779821 | 335 | III | 4.98 | 37,963.20 |
| MtNAC55 | Medtr5g021710 | chr5:8449579-8452636 | 352 | II | 5.12 | 40565.4 |
| MtNAC56 | Medtr5g040420 | chr5:17777239-17779842 | 391 | XII | 6.27 | 44699.2 |
| MtNAC57 | Medtr5g041940 | chr5:18429509-18427695 | 260 | VII | 6.91 | 30,003.7 |
| MtNAC58 | Medtr5g053430 | chr5:22012794-22017536 | 442 | X | 4.84 | 49,671 |
| MtNAC59 | Medtr5g069030 | chr5:29216428-29220286 | 630 | VI | 4.82 | 70,313.2 |
| MtNAC60 | Medtr5g076850 | chr5:32791723-32786597 | 644 | III | 5.37 | 74,164.4 |
| MtNAC61 | Medtr5g090970 | chr5:39632529-39636258 | 311 | XII | 8.51 | 36,029.5 |
| MtNAC62 | Medtr6g011860 | chr6:3555895-3560091 | 391 | XII | 6.94 | 44,591.2 |
| MtNAC63 | Medtr6g012670 | chr6:3892753-3894308 | 321 | I | 5.01 | 37,140.5 |
| MtNAC64 | Medtr6g032770 | chr6:10318538-10315903 | 379 | I | 5.35 | 43,087 |
| MtNAC65 | Medtr6g084430 | chr6:31607391-31604165 | 325 | I | 7.18 | 36,958.40 |
| MtNAC66 | Medtr6g477900 | chr6:28664418-28659313 | 239 | IV | 4.9 | 27,812.20 |
| MtNAC67 | Medtr7g005280 | chr7:47539-49564 | 256 | VI | 9.15 | 27,812.20 |
| MtNAC68 | Medtr7g011120 | chr7:2918496-2920585 | 310 | I | 5.84 | 35,229.10 |
| MtNAC69 | Medtr7g011130 | chr7:2923772-2925970 | 352 | I | 5.66 | 40,054.40 |
| MtNAC70 | Medtr7g033320 | chr7:11902390-11906760 | 501 | X | 4.84 | 56,139.70 |
| MtNAC71 | Medtr7g070140 | chr7:25851047-25851665 | 137 | VI | 8.52 | 15,558.40 |
| MtNAC72 | Medtr7g070150 | chr7:25852151-25851784 | 92 | VI | 10.07 | 10,692.10 |
| MtNAC73 | Medtr7g083330 | chr7:32051182-32049757 | 194 | V | 9.12 | 22,900.10 |
| MtNAC74 | Medtr7g083360 | chr7:32063997-32063118 | 155 | V | 9.14 | 18,494 |
| MtNAC75 | Medtr7g083370 | chr7:32071800-32070846 | 143 | V | 6.08 | 17,385.60 |
| MtNAC76 | Medtr7g085220 | chr7:32967009-32968969 | 340 | VII | 8.87 | 38,464.50 |
| MtNAC77 | Medtr7g085260 | chr7:32990543-32992707 | 383 | I | 6.52 | 43,927.10 |
| MtNAC78 | Medtr7g097090 | chr7:39019290-39014420 | 291 | I | 5.43 | 33,546.60 |
| MtNAC79 | Medtr7g100990 | chr7:40734227-40732629 | 328 | VII | 7.69 | 37,535.30 |
| MtNAC80 | Medtr7g105170 | chr7:42639031-42640463 | 257 | XIII | 5.75 | 29,660.10 |
| MtNAC81 | Medtr7g116460 | chr7:48060498-48059630 | 206 | XI | 8.97 | 23,610.70 |
| MtNAC82 | Medtr8g023840 | chr8:8690689-8692417 | 400 | IV | 5.47 | 45,950.20 |
| MtNAC83 | Medtr8g023860 | chr8:8704958-8707169 | 419 | IV | 5.28 | 47,510 |
| MtNAC84 | Medtr8g023880 | chr8:8712893-8717845 | 269 | IV | 5.56 | 30,475.30 |
| MtNAC85 | Medtr8g023900 | chr8:8722035-8725267 | 470 | IV | 5.65 | 55,139.10 |
| MtNAC86 | Medtr8g023930 | chr8:8738431-8747309 | 473 | IV | 6.74 | 55,551.40 |
| MtNAC87 | Medtr8g024480 | chr8:9002671-9004590 | 434 | II | 6.06 | 48,952.90 |
| MtNAC88 | Medtr8g059170 | chr8:20701024-20703544 | 329 | IX | 5.93 | 36,960.50 |
| MtNAC89 | Medtr8g063550 | chr8:26582418-26578891 | 444 | XIII | 4.31 | 48,649.20 |
| MtNAC90 | Medtr8g069160 | chr8:28951511-28948614 | 259 | X | 6.32 | 29,439.30 |
| MtNAC91 | Medtr8g076110 | chr8:32226602-32223015 | 311 | II | 6.39 | 36,440.20 |
| MtNAC92 | Medtr8g093580 | chr8:39133468-39136666 | 489 | XIII | 4.16 | 53,787.50 |
| MtNAC93 | Medtr8g093790 | chr8:39242842-39241213 | 185 | IX | 4.52 | 21,408.70 |
| MtNAC94 | Medtr8g094580 | chr8:39492258-39494179 | 285 | VIII | 6.08 | 32,727 |
| MtNAC95 | Medtr8g099750 | chr8:40364682-40363158 | 227 | VI | 8.32 | 25,747.80 |
| MtNAC96 | Medtr8g102240 | chr8:43013735-43018918 | 485 | X | 6.62 | 55,002.80 |
| MtNAC97 | Medtr8g467490 | chr8:24258825-24255680 | 395 | XII | 8.43 | 44,571.40 |
Phylogenetic analysis and identification of additional motifs
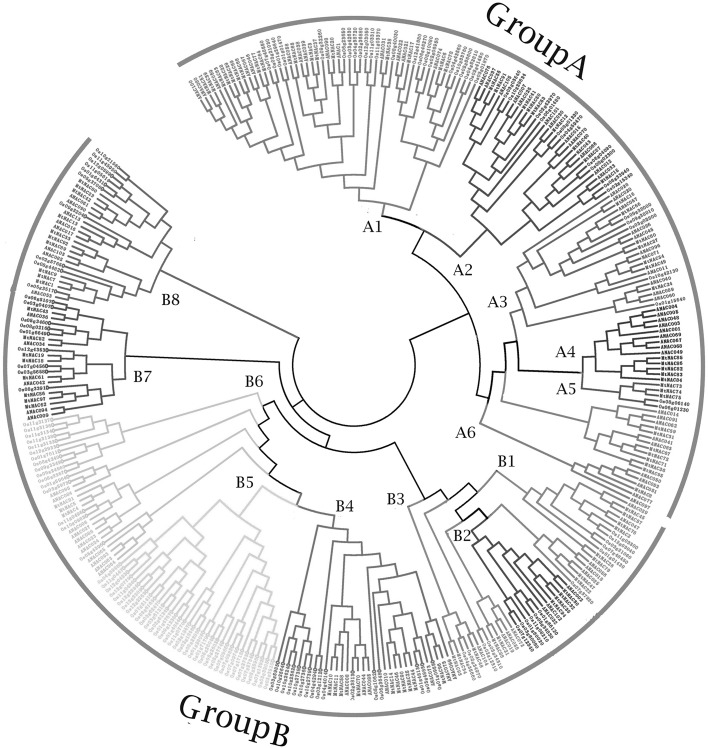
To understand the evolutionary history among MtNAC genes, we constructed a phylogenetic tree for M. truncatula, A. thaliana (dicot), and O. sativa (monocot) (Zhu et al. 2012). From the results, NAC proteins can be divided into 14 subfamilies, characterized by highly conserved motifs with the exception of the NAC domain motif A. The motif pattern was clustered in the same way as the subgroup pattern, and therefore the results demonstrated our phylogenetic clustering results were accurate. The MtNAC proteins were divided into o two groups (A and B). The tree was divided into six groups in Group A (A1–6). Group B possessed eight phylogenetic subgroups (B1–8) (Fig. 1). Most of the subgroups had highly conserved motifs excluding the NAC domain motifs, such as Group (I–VI) belonging to (A1–6) Group (VII–XIII) belonging to (B1–8, except B5). The motifs in the NAC family proteins in M. truncatula were investigated using MEME software, revealing 25 conserved motifs (motifs 1–25), (Fig. 2). Thus, the results show that gene sequences belong to 14 groups.
Fig. 1.
Phylogenetic tree analysis of the NAC transcription factor amily in Medicago truncatula, Arabidopsis thaliana (dicot) and Oryza sativa. The phylogenetic tree was constructed using MEGA 6.0 by the neighbor-joining method. The Bootstrap value was 1000 replicates. The three plant-specific clusters were designated as A (A1–A6), B (B1–B8) and indicated in a specific color (color figure online)
Fig. 2.
Distribution of conserved motifs within MtNAC transcription factor family in Medicago truncatula. Summary for the distribution of conserved motifs identified from 93 MtNAC proteins by each group given separately. Each motif is represented by a number in colored box (color figure online)
In general, the clusters of NAC proteins had similar motif compositions and most of the conserved motifs were found in N-terminal. Motifs 1–20 were conserved in the NAC protein family (Fig. S1). Most groups possessed an N-terminal NAC domain that included Motif1-5 (Hu et al. 2010). Most of these proteins had a special motif in the C-terminal; Group I (28, 29, 51, 63, 68, and l69) possessed an NAC domain Motif 8, Group II possessed an NAC domain Motif 14, and Group VIII possessed a composite motif (19.20) in the C-terminal. Detailed information is listed in Table S2.
Gene structure, gene chromosomal location, and gene duplication events of MtNAC
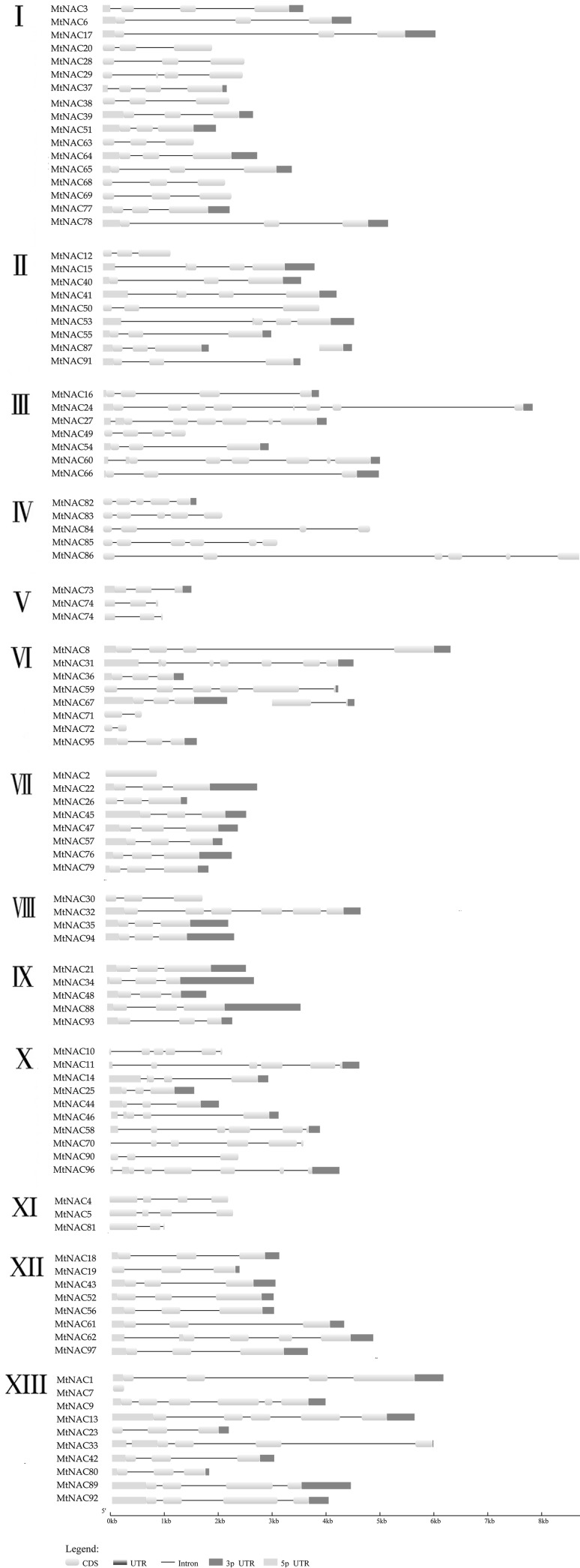
We found the same groups members had the same or similar gene structures (Fig. 3). Groups I and II had two introns and the same CDS distribution in genes, and the last coding region was longer than the others. Most members had three introns and up/downstream in Group III, except MtNAC7. Groups IV and XI had large quantities of introns, Groups V and VI also showed obvious genetic structure characteristics with longer intron and shorter exon positions. Group VIII had numerous members, most members having three exon positions besides every area being longer. The other groups had shorter gene lengths and all groups had similar gene structures.
Fig. 3.
Exon-intron structure analyses of MtNAC genes were performed by using the online tool GSDS. Lengths of exons and introns of each MtNAC gene were exhibited proportionally. Exon/intron organization of 97 MtNAC genes was depicted for each group. The exons and introns are represented by box and lines, respectively
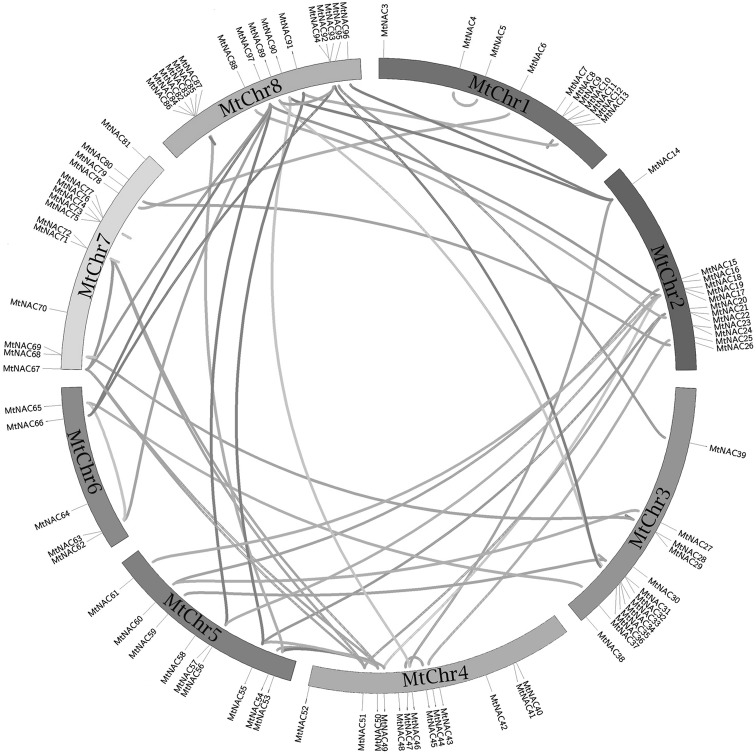
A total of 97MtNACs were located on eight chromosomes of M. truncatula, with 63 pairs of genes in tandem duplication. Different color links were used to distinguish gene indices of similarity. A total of 95 MtNAC genes could be located in eight chromosomes (1–8), and MtNAC1 and 2 could not be conclusively mapped on any chromosome. There were only 5 NAC genes in MtChr6, whereas others possessed at least 10 MtNAC genes. Multi-member groups were widely distributed among chromosomes, for example Group I was distributed among 6 chromosomes, excluding MtChr8 and MtChr5. Most chromosomes had more than six different groups. The result showed gene clusters and hot regions is produced by tandem duplications in MtNAC, for instance the MtNAC73–77 and MtNAC82–87 clusters on Chr1. Segmental duplication produced homologous NAC genes, which expanded the numbers of MtNAC genes in genome. For example, MtNAC6 and MtNAC78 from Group I were distributed on different chromosomes (MtNAC6, MtNAC78, MtNAC56 and 57), which were segmental duplication in M. truncatula (Fig. 4).
Fig. 4.
Duplicated genes between different chromosomes or loci were linked with colored lines in the diagrams using Circos as described previously. Genes were identified using the BLASTP using parameters; e-value ≤ 1e − 10 and minimum percent identity = 70%
Expression profiles of MtNAC genes among different tissues
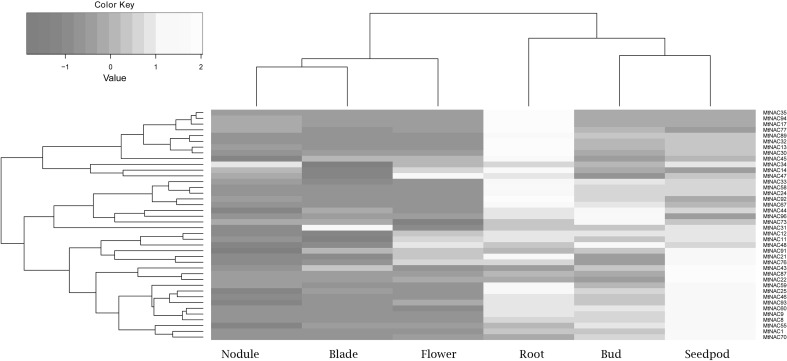
The heatmap showed that 40 NAC were expressed in all six tissues: Mt (35, 94, 17, 77, 89, 32, 13, 30, 45) were highly expressed in the root; Mt (34,14,47) were specifically expressed in buds; Mt (43, 87, 22, 59, 25, 49, 93, 60, 9, 8, 55, 1, 70) were highly expressed in seedpods; Mt (12, 11, 48, 91, 21, 76) were highly expressed in seedpods and flowers, and Mt (33, 58, 24, 92, 67, 44, 96, 73, 31) were highly expressed in roots and seedpods (Fig. 5).
Fig. 5.
Number of differentially expressed MtNAC genes involved in tissue development. Clustering of legume MtNAC genes according to their expression profiles in tissues including roots, nodules, blades, buds, seedpods and flowers
Expression responses of MtNAC genes among abiotic stress
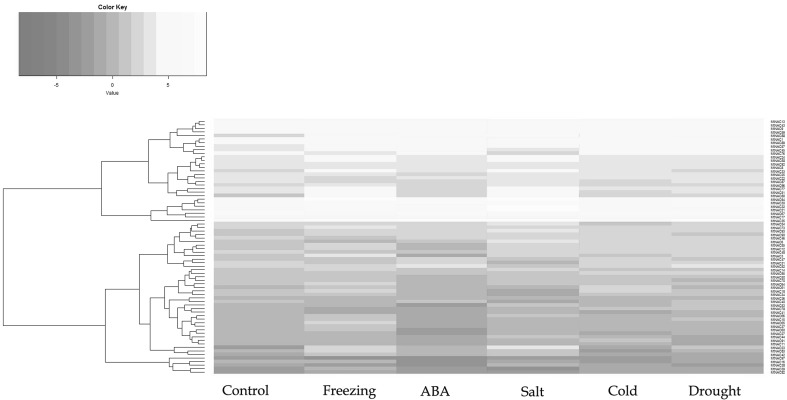
We used RNA-seq to analyse the expression of MtNAC genes under different stresses, such as cold-stress, freezing-stress, drought-stress, salt-stress and ABA-stress. We attempted to evaluate the 44 genes detected in at least one library (Fig. 6). Most genes were exclusively induced and partial genes were exclusively repressed. There were 17 genes up regulated under all five stresses (Fig. S2). Only MtNAC1 was downregulated under all five stresses. During cold stress, 6 genes showed no obvious change and 5 genes were repressed, whereas 33 genes were exclusively induced. In the freezing stress treatments, 8 genes were repressed and 36 were induced. In the drought conditions, 6 genes showed no obvious change, 5 genes were repressed, and 33 genes were exclusively induced. MtNAC (17, 21, 30, 32, 35, 67, and 94) were highly expressed and showed the greatest variation in most abiotic stresses. MtNAC (24, 58, 92, 8, 33, 25, 22, 87, 96, 77, 51, and 80) were specifically expressed in response to freezing and salt stresses. MtNAC (97, 16, 38, 39, and 82) showed very low expression in various stress conditions. MtNAC (13, 43, 9, 59, 88, 1, 89, 57, 45, and 76) were not expressed in response to abiotic stresses.
Fig. 6.
Differential expression analysis of MtNAC genes involved in the response to abiotic stress. Heatmap of MtNAC genes expressed among five stresses. The relative expression values were log2 transformed using the R soft
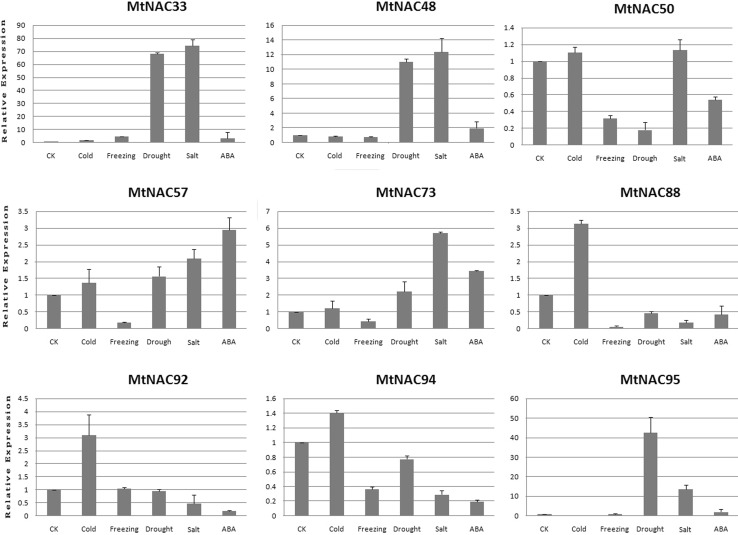
qRT-PCR of MtNAC genes in abiotic stress
To verify the authenticity of transcriptome data, we selected 12 genes to detect their expression profiles under 6 stress conditions using qTR-PCR (Fig. 7). The results of qRT-PCR were consistent with the transcriptome data. The results showed that MtNAC33 and 48 shared the same expression pattern under five different stress conditions. In addition, the expression of the two genes increased significantly in salt and drought conditions. MtNAC50 showed high expression levels only in cold- and salt-stress conditions. MtNAC57 and 73 were upregulated under all stresses, except freezing. MtNAC88, MtNAC92, and MtNAC94 expression only increased in cold stress. MtNAC95 expression increased in salt-, drought- and ABA-stress conditions. These results corroborate the findings of transcriptome data.
Fig. 7.
qRT-PCR analysis reveals NAC genes under five stress conditions: cold, freezing, salt, drought, and ABA treatments compared to the controls. Stress treatments and time course are described in ‘‘Materials and methods’’
Discussion
We performed a comprehensive in silico study and characterized 97 MtNAC genes in the Medicago genome (Table 1). The NAC TF gene family were surveyed in Arabidopsis (117), rice (151), soybean (152), grape (Vitis vinifera) (163), and tobacco (152) (Le et al. 2011; Nuruzzaman et al. 2010; Ooka et al. 2003; Wang et al. 2013).
The phylogenetic analysis classified the MtNACs into 13 groups with their AtNAC and OsNAC (Fig. 1). These members were widely distributed in different groups, but only B5 contained OsNACs (Cenci et al. 2014). Soybean showed similar results, which confirmed that differences appeared between monocots and dicots in the course of their evolutionary history (Le et al. 2011).
All NAC TFs have important conserved motifs for their function. We identified 25 motifs in MtNAC, motifs 1–5 are related to the NAC domain, and motifs (3, 4, 1, 2, 5) represent the A–E subdomain (De Zélicourt et al. 2012). Subdomain A is component of functional dimmer, subdomains C and D can bind to DNA terminal, and the divergent subdomains B and E play an important role in gene function. Subdomain E is NAC DB domain that has five motifs. Interestingly, the function of NAC TFs can regulate downstream gene expression level through the complex interaction between the DB domain, NARD, and the activation domain. Additionally, plant physiology could also mediate transcriptional regulation by the NAC protein domain (Hao et al. 2010). NAC gene family promoter regions (NACBS) showed positive response to stress (JENSEN et al. 1997). The TR domain represents transcription activation repression and protein binding in the C-terminal. A single or few motifs constitute TR domains such as motifs (8, 14, 19, and 20).
The plants gene duplication phenomenon plays an important role in dealing with environmental change. However information about functionality of duplicated genes is still limited (Bowers et al. 2003).
This phenomenon increase the diversity of the gene family through changing gene expression (Friedman and Hughes 2003). The differential expresion pattern of genes may result in functional diversity (Soskine and Tawfik 2010).
However, the high number of TCP genes in M. truncatula was possibly caused by gene duplication. In the paralogous pairs, most TCPs shared conserved exons/introns and organization such as MtNAC4 and 5, and most paralogous pairs of MtTCPs exhibited conserved motif composition such as MtNAC14 and MtNAC46, with several motifs disappearing in some MtTCP members, such as motif 16 for MtTCP6-78. Several motifs were observed, such as motifs 9 and 12 for MtTCP14-96. These specific motifs may contribute to which paralogous member obtains a new function after gene duplication, and duplicated genes are changed by a series of synonymous or nonsynonymous mutations during evolution.
NAC family genes regulate different tissue development such as shoot, meristem and organ, therefore they have different expression patterns in plant (Kim et al. 2007). CUC1, CUC2, NAM, NACL, AT5G07680, and AT5G61430 regulate plant and meristem development through miR164 (Kim et al. 2009). NST1 and NST3 play an important role in woody secondary walls regeneration. Most types of leaf vein are regulated by the VND7 in roots and shoots (Mitsuda et al. 2007). Mt (35, 94, 17, 77, 89, 32, 13, 30, and 45) were only highly expressed in root tissue (Fig. 5). MtNAC47 was only expressed in nodule tissue, and its homologous gene, RhNAC100 participates in flower petals cell expansion through ethylene regulatory pathway (Mitsuda et al. 2007). The closest homolog of MtNAC969 in Arabidopsis is AtNAP/ANAC029 (De Zélicourt et al. 2012). This gene takes part in floral and stamen formation through APETALA3 PISTILLATA regulating. This phenomenon demonstrate that NAC family genes may have significant role in root and flower formation in M. truncatula.
This study shows that NAC TFs are involved in various environmental stresses. The transcript of MtNAC30 had higher expressions in drought than normal condition and the homolog of the ANAC002 was previously upregulated during drought in Arabidopsis (Balazadeh et al. 2010). In our study MtNAC35 was upregulated during ABA treatment. The homolog of the MtNAC35 291 OsNAC19 has a role in ABA and ethylene (ETH) induction (Lin et al. 2007). Based on recent studies and abiotic stress expression data from genes, we found that MtNAC genes take part in diverse signaling pathways and stress responses. The SNAC1 transgenic lines possess higher drought and salt tolerance than wild plants in dry fields, and the expression of several MtNAC genes improved by drought and salt stress, such as MtNAC35 (You et al. 2014).
Homologous gene of MtNAC83, SsNAC23 was strongly induced at 4 °C, indicating a positive response to cold stress (Nogueira et al. 2005). MtNAC83 was also upregulated under cold (4 °C) conditions. The SiNAC improve the plant resistance by three different pathway, such as ABA-independent, JA and SA (Puranik et al. 2011). Several NAC genes help the plant to adapt to large environmental changes through promoting plant growth. Their versatility ensures the longevity and multiplication of plants (Puranik et al. 2012). Furthermore, our results suggest that Group VIII were more sensitive to abiotic stress, whereas Group VIII were active in tissue development (Figs. 5, 6). The results of our studies verify those of previous results. We used genome information and transcriptome data along with qRT-PCR validation to determine the function of a many NAC genes in M. truncatula. Understanding the gene function will help to provide useful information for genetic model systems. The information about NAC genes will be helpful for improving plant resistance and crop yield in the future. Furthermore, it can help us to understand the NAC transcription factors regulatory mode during adverse stress condition.
Conclusions
In summary, 97 putative NAC transcription factors were identified from the M. truncatula genome sequence, one of the most important model organisms for leguminous plants. We investigated the structure, phylogeny, and gene duplication of the conserved motifs and gene organization. Furthermore, the differential expression profile of MtNAC genes suggest their responsiveness to different stress. The present study shows that NAC TF family has responsiveness to abiotic stress (cold, freezing, drought, salt, and ABA) in M. truncatula.
The study provides advantageous information for understanding the molecular basis of the NAC TF family in M. truncatula, and the results can be used to engineer the plants with enhanced stress resistance. Further study of the NAC genes’ function will be helpful for transgenic applications.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Conserved motifs identified from members of the NAC proteins in Medicago truncatula (JPEG 45871 kb)
Venn diagram of shared expression MtNAC genes among cold stress freezing drought salt and ABA stresses (JPEG 18518 kb)
List of primers used in qRT-PCR (XLS 19 kb)
Detailed information of 97 Medicago truncatula NAC proteins (XLS 75 kb)
Acknowledgements
This work was supported by the MOST 863 Project (2013AA102607-5) and the Graduate Innovation Fund of Harbin Normal University (HSDBSCX2014-04) Key Scientific and Technological Project of Heilongjiang Province of China (GA15B105-1) the Natural and Science Foundation of China (No. 31302019 and 31470571) and the research was supported by the National Major Project for Cultivation of Transgenic Crops (2011ZX08004-002-003).
Compliance with ethical standards
Conflicts of interest
The authors declare no conflict of interest.
References
- Bailey TL, Johnson J, Grant CE, Noble WS. The MEME Suite. Nucleic Acids Res. 2015;43(W1):W39–W49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S, Wu A, Mueller-Roeber B. Salt-triggered expression of the ANAC092-dependent senescence regulon in Arabidopsis thaliana. Plant Signal Behav. 2010;5:733–735. doi: 10.4161/psb.5.6.11694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Dixon RA, Farmer AD, Flores R, Inman J, Gonzales RA, Harrison MJ, Paiva NL, Scott AD, Weller JW. The Medicago genome initiative: a model legume database. Nucleic Acids Res. 2001;29:114–117. doi: 10.1093/nar/29.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Bu Q, Jiang H, Li C, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008;18:756–767. doi: 10.1038/cr.2008.53. [DOI] [PubMed] [Google Scholar]
- Cenci A, Guignon V, Roux N, Rouard M. Genomic analysis of NAC transcription factors in banana (Musa acuminata) and definition of NAC orthologous groups for monocots and dicots. Plant Mol Biol. 2014;85:63–80. doi: 10.1007/s11103-013-0169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lu S, Wang Y, Zhang X, Lv B, Luo L, Xi D, Shen J, Ma H, Ming F. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J. 2015;82:302–314. doi: 10.1111/tpj.12819. [DOI] [PubMed] [Google Scholar]
- Committee NRN. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/S0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- De Zélicourt A, Diet A, Marion J, Laffont C, Ariel F, Moison M, Zahaf O, Crespi M, Gruber V, Frugier F. Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J. 2012;70:220–230. doi: 10.1111/j.1365-313X.2011.04859.x. [DOI] [PubMed] [Google Scholar]
- Ernst HA, Olsen AN, Skriver K, Larsen S, Leggio LL. Structure of the conserved domain of ANAC a member of the NAC family of transcription factors. EMBO Rep. 2004;5:297–303. doi: 10.1038/sj.embor.7400093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R, Hughes AL. The temporal distribution of gene duplication events in a set of highly conserved human gene families. Mol Biol Evol. 2003;20:154–161. doi: 10.1093/molbev/msg017. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PH, Vance CP. Legumes: importance and constraints to greater use. Plant Physiol. 2003;131:872–877. doi: 10.1104/pp.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display serve. Yi chuan = Hereditas/Zhongguo yi chuan xue hui bian ji. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- Hao Y, Song Q, Chen H, Zou H, Wei W, Kang X, Ma B, Zhang W, Zhang J, Chen S. Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta. 2010;232:1033–1043. doi: 10.1007/s00425-010-1238-2. [DOI] [PubMed] [Google Scholar]
- Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011;68:302–313. doi: 10.1111/j.1365-313X.2011.04687.x. [DOI] [PubMed] [Google Scholar]
- Hu R, Qi G, Kong Y, Kong D, Gao Q, Zhou G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 2010;10:145. doi: 10.1186/1471-2229-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey SG, Sa MN, Hefer CA, Myburg AA, Grima Pettenati J. Structural evolutionary and functional analysis of the NAC domain protein family in Eucalyptus. New Phytol. 2015;206:1337–1350. doi: 10.1111/nph.13139. [DOI] [PubMed] [Google Scholar]
- Jensen PH Højrup, Hager P, Nielsen H, Jacobsen MS, Olesen L, Gliemann OF, Jakes R. Binding of Aβ to α-and β-synucleins: identification of segments in α-synuclein/NAC precursor that bind Aβ and NAC. Biochem J. 1997;323:539–546. doi: 10.1042/bj3230539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Binformat. 2010;11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Park BO, Yoo JH, Jung MS, Lee SM, Han HJ, Kim KE, Kim SH, Lim CO, Yun D. Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J Biol Chem. 2007;282:36292–36302. doi: 10.1074/jbc.M705217200. [DOI] [PubMed] [Google Scholar]
- Kim JH, Woo HR, Kim J, Lim PO, Lee IC, Choi SH, Hwang D, Nam HG. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science. 2009;323:1053–1057. doi: 10.1126/science.1166386. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 2011;18(4):263–276. doi: 10.1093/dnares/dsr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Zhao W, Meng X, Wang M, Peng Y. Rice gene OsNAC19 encodes a novel NAC-domain transcription factor and responds to infection by Magnaporthe grisea. Plant Sci. 2007;172:120–130. doi: 10.1016/j.plantsci.2006.07.019. [DOI] [Google Scholar]
- Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R. TaNAC2 a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J Exp Bot. 2012;63:2933–2946. doi: 10.1093/jxb/err462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M. NAC transcription factors NST1 and NST3 are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira FT, Schlögl PS, Camargo SR, Fernandez JH, De Rosa VE, Pompermayer P, Arruda P. SsNAC23 a member of the NAC domain protein family is associated with cold herbivory and water stress in sugarcane. Plant Sci. 2005;169:93–106. doi: 10.1016/j.plantsci.2005.03.008. [DOI] [Google Scholar]
- Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S. Genome-wide analysis of NAC transcription factor family in rice. Gene. 2010;465:30–44. doi: 10.1016/j.gene.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M, Sharoni AM, Satoh K, Karim MR, Harikrishna JA, Shimizu T, Sasaya T, Omura T, Haque MA, Hasan SM, Ahmad A, Kikuchi S. NAC transcription factor family genes are differentially expressed in rice during infections with Rice dwarf virus Rice black-streaked dwarf virus Rice grassy stunt virus Rice ragged stunt virus and Rice transitory yellowing virus. Front Plant Sci. 2015;6:676. doi: 10.3389/fpls.2015.00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct functionally diverse. Trends Plant Sci. 2005;10:79–87. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- Pinheiro GL, Marques CS, Costa MD, Reis PA, Alves MS, Carvalho CM, Fietto LG, Fontes EP. Complete inventory of soybean NAC transcription factors: sequence conservation and expression analysis uncover their distinct roles in stress response. Gene. 2009;444:10–23. doi: 10.1016/j.gene.2009.05.012. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2011;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S, Bahadur RP, Srivastava PS, Prasad M. Molecular cloning and characterization of a membrane associated NAC family gene SiNAC from foxtail millet [Setaria italica (L.) P. Beauv.] Mol Biotechnol. 2011;49:138–150. doi: 10.1007/s12033-011-9385-7. [DOI] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17:369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Rueda J, Realpe T, Mejia GI, Zapata E, Rozo JC, Ferro BE, Robledo J. Genotypic analysis of genes associated with independent resistance and cross-resistance to isoniazid and ethionamide in Mycobacterium tuberculosis clinical isolates. Antimicrob Agents CH. 2015;59:7805–7810. doi: 10.1128/AAC.01028-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samac DA, Jung HJG, Lamb JFS. Development of Alfalfa (Medicago sativa L.) as a feedstock for production of ethanol and other bioproducts. In: Minteer S, editor. Alcoholic Fuels. Boca Raton: CRC Press Taylor & Francis Group; 2006. pp. 79–98. [Google Scholar]
- Sanderson MA, Skinner RH, Barker DJ, Edwards GR, Tracy BF, Wedin DA. Plant species diversity and management of temperate forage and grazing land ecosystems. Crop Sci. 2004;44:1132–1144. doi: 10.2135/cropsci2004.1132. [DOI] [Google Scholar]
- Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2010;11:572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town CD. Annotating the genome of Medicago truncatula. Curr Opin Plant Biol. 2006;9:122–127. doi: 10.1016/j.pbi.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Tran LP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Zheng Y, Xin H, Fang L, Li S. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013;32:61–75. doi: 10.1007/s00299-012-1340-y. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wang C, Xue F, Zhang H, Ji W. Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiol Bioch. 2015;96:356–363. doi: 10.1016/j.plaphy.2015.08.013. [DOI] [PubMed] [Google Scholar]
- Yang SS, Tu ZJ, Cheung F, Xu WW, Lamb JAF, Jung HJG, Vance CP, Gronwald JW. Using RNA-Seq for gene identification, polymorphism detection and transcript profiling in two alfalfa genotypes with divergent cell wall composition in stems. BMC Genom. 2011;12:1–19. doi: 10.1186/1471-2164-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZT, Lu SJ, Wang MJ, Bi DL, Sun L, Zhou SF, Song ZT, Liu JX. A plasma membrane?ethered transcription factor NAC062/ANAC062/NTL6 mediates the unfolded protein response in Arabidopsis. Plant J. 2014;79:1033–1043. doi: 10.1111/tpj.12604. [DOI] [PubMed] [Google Scholar]
- Yoshii M, Yamazaki M, Rakwal R, Kishi Kaboshi M, Miyao A, Hirochika H. The NAC transcription factor RIM1 of rice is a new regulator of jasmonate signaling. Plant J. 2010;61:804–815. doi: 10.1111/j.1365-313X.2009.04107.x. [DOI] [PubMed] [Google Scholar]
- You J, Zong W, Hu H, Li X, Xiao J, Xiong L. A SNAC1-regulated protein phosphatase gene OsPP18 modulates drought and oxidative stress tolerance through ABA-independent reactive oxygen species scavenging in rice. Plant Physiol. 2014;114:251116. doi: 10.1104/pp.114.251116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye Z. SND1 a NAC domain transcription factor is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Nevo E, Sun D, Peng J. Phylogenetic analyses unravel the evolutionary history of NAC proteins in plants. Evolution. 2012;66:1833–1848. doi: 10.1111/j.1558-5646.2011.01553.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conserved motifs identified from members of the NAC proteins in Medicago truncatula (JPEG 45871 kb)
Venn diagram of shared expression MtNAC genes among cold stress freezing drought salt and ABA stresses (JPEG 18518 kb)
List of primers used in qRT-PCR (XLS 19 kb)
Detailed information of 97 Medicago truncatula NAC proteins (XLS 75 kb)