Abstract
In this paper, we highlight the affinity between the genomes of key representatives of the Pooideae subfamily, revealed at the chromosomal level by genomic in situ hybridization (GISH). The analyses were conducted using labeled probes from each species to hybridize with chromosomes of every species used in this study based on a “round robin” rule. As a result, the whole chromosomes or chromosome regions were distinguished or variable types of signals were visualized to prove the different levels of the relationships between genomes used in this study. We observed the unexpected lack of signals in secondary constrictions of rye (RR) chromosomes probed by triticale (AABBRR) genomic DNA. We have also identified unlabeled chromosome regions, which point to species-specific sequences connected with disparate pathways of chromosome differentiation. Our results revealed a conservative character of coding sequence of 35S rDNA among selected species of the genera Aegilops, Brachypodium, Festuca, Hordeum, Lolium, Secale, and Triticum. In summary, we showed strong relationships in genomic DNA sequences between species which have been previously reported to be phylogenetically distant.
Keywords: Pooideae, Brachypodium, Cereals, Forage grasses, Comparative genomics, In situ hybridization
Introduction
The Poaceae family (Barnh., Gramineae Juss.) is the fourth largest group of plants on Earth and can be found in nearly all regions and climate zones. More than 10,000 species and 600 genera constitute this family, but only three subfamilies, namely Ehrhartoideae, Pooideae, and Panicoideae, play the principal role of providing the human race with nutrition and a source of renewable energy. The Pooideae subfamily includes some economically important cereals, such as bread wheat (Triticum aestivum L.), barley (Hordeum vulgare L.), rye (Secale cereale L.), and triticale (×Triticosecale Wittm.), as well as many lawn and pasture grasses, such as perennial ryegrass (Lolium perenne L.). The genome composition of the species within Pooideae differs in respect to their size, ploidy level, basic chromosome number, and repetitive sequences content. For example, bread wheat (Triticum aestivum L.; 17 Gb; 2n = 6x = 42 chromosomes) is an allopolyploid, composed of six sets of chromosomes belonging to A, B, and D genomes with high content of non-coding, repetitive DNA. Contrastingly, Brachypodium distachyon has a small genome (272 Mb; 2n = 2x = 10 chromosomes) and low amount of repetitive sequences (Vogel and Hill 2008; International Brachypodium Initiative (IBI) 2010), making it a good model for structural genomic studies of grasses. It was previously reported that B. distachyon is a comparative and functional genetic model and its potential relevance for research on cereals, on the basis of its physiological and genetic advantages (Draper et al. 2001; Vogel and Bragg 2009). For example, the high degree of genetic synteny between wheat, B. sylvaticum, and B. distachyon enabled the identification of the genes present in the Ph1 (Pairing homeologous 1) locus of wheat (Griffiths et al. 2006; Huo et al. 2009), demonstrating the potential application of the genus Brachypodium in comparative genomics. The phylogenetic relationships between B. distachyon and other cereals and grasses have been derived on the basis of the analysis of the internal transcribed spacer (ITS) and rDNA coding sequences (Hsiao et al. 1994), genomic markers, such as random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP) (Catalán et al. 1995), chloroplast restriction sites, and ndhF gene sequence (Kellogg 2001; Mochida and Shinozaki 2013). Those approaches showed that B. distachyon can be considered as a kind of missing link between rice (Oryza sativa L.) and temperate grasses.
Exploring the genomes of a wide range of plant species revealed the existence of highly conserved sequences and synteny in gene order. Comparative genomics can distinguish synteny, which means the presence of two or more loci on the same chromosome, and collinearity, which refers to the similarities in the physical order of loci (McCouch 2001). However, recently, the synteny is defined as the conservation of blocks of order within the sets of chromosomes which are being compared to each other. The first study of chromosome evolution and comparative genomics was performed between potato (Solanum tuberosum) and tomato (Lycopersicon esculentum) (Bonierbale et al. 1988). Initially, low copy RFLP markers and low resolution genetic maps were used to determine the phylogenetic relationships between Poaceae species, and established that most of the grass genomes are composed from 30 rice-independent linkage blocks (Gale and Devos 1998; Keller and Feuillet 2000). These methods enabled the macro-collinearity of grass genomes to be determined; however, they also overestimated the level of synteny. Recently, the development of additional genomic resources such as EST marker databases and whole genome sequences, combined with improved software, has enabled comparative genomic studies to reach a higher level of resolution, uncovering the micro-collinearity of sequences, which form the basis to establish a model of grass genome evolution (Devos 2005; Wei et al. 2007; Salse et al. 2008; Murat et al. 2014).
One of the main factors leading to chromosome evolution and speciation are changes in the amount and distribution of repetitive DNA sequences (Flavell et al. 1979; Cuadrado and Jouve 2002). It is hypothesized that these sequences play an important role in chromosome organization, stabilization of chromosome structure, recognition and segregation of chromosomes in mitosis and meiosis, and regulation of gene activity (Vershinin et al. 1995). Repetitive sequences account for up to 90 % of plant genomes (Heslop-Harrison 2000), and show diverse compositions in different genomes. Changes in repetitive sequences could be particularly useful in studies of genomic evolution. Some families of those sequences, e.g., Afa family (Nagaki et al. 1995), are present in more than one genus, which could be an evidence of their genomic relationship. Genomic in situ hybridization (GISH) can distinguish particular genomes at the chromosomal level. GISH can result in chromosome painting and show similarities of repetitive DNA distribution between related species. Moreover, the physical locations of conserved sequences can be visualized on chromosomes as well.
In this work, we compare both the model species (Brachypodium distachyon) as well as other important grasses, using genomic DNA probes. Our intention was to verify and classify cytologically visible similarities and differences in repetitive non-coding DNA sequences locations in given Pooideae species. A comparative approach using GISH, instead of comparison of selected sequences or specific chromosome regions, was performed to widen the understanding of the relationships within the Pooideae subfamily at the chromosomal level.
Materials and methods
Plant material
Ten species (Table 1) were selected for comparative GISH analysis: Aegilops speltoides Tausch., Aegilops tauschii Coss., Brachypodium distachyon L., Festuca pratensis L., Hordeum vulgare L., Lolium perenne L., Secale cereale L., Triticosecale Wittm., Triticum aestivum L., and Triticum urartu Tumanian ex Gandilyan. Brachypodium distachyon material was sourced from the University of Silesia in Katowice, Poland. Seeds of Ae. speltoides, S. cereale, T. aestivum, and T. urartu were kindly supplied for the study from the National Small Grains Germplasm Research Facility, National Small Grains Collection (Aberdeen, Idaho, USA). The remaining species were provided by a collection of the Institute of Plant Genetics, Polish Academy of Sciences in Poznan, Poland.
Table 1.
Origin, chromosome number, size, and constitution of genomes of the studied Pooideae species according to Bennett and Leitch (2012)
| No. | Species | Origin | Chromosome number (2n) | Genome size (1C; pg) | Genome constitution |
|---|---|---|---|---|---|
| 1 | Aegilops speltoides Tausch. | USDA (PI 542264) | 14 | 5.15 | SS |
| 2 | Aegilops tauschii Coss. | IPG PAS (D51) | 14 | 5.1 | DD |
| 3 | Brachypodium distachyon L. | US (Bd21) | 10 | 0.36 | BdBd |
| 4 | Festuca pratensis Huds. | IPG PAS (cv. ‘Skra’) | 14 | 2.23 | FpFp |
| 5 | Hordeum vulgare L. | IPG PAS (cv. ‘Georgia’) | 14 | 5.55 | HH |
| 6 | Lolium perenne L. | IPG PAS (cv. ‘Arka’) | 14 | 2.76 | LpLp |
| 7 | Secale cereale L. | USDA (PI 323382; cv. ‘Imperial’) | 14 | 8.28 | RR |
| 8 | Triticum aestivum L. | USDA (Cltr 14108; cv. ’Chinese Spring’ ) | 42 | 17.33 | AABBDD |
| 9 | Triticum urartu Tumanian ex Gandilyan | USDA (PI 428335) | 14 | 4.93 | AA |
| 10 | ×Triticosecale Wittm. | IPG PAS (cv. ‘Kitaro’) | 42 | 19.80 | AABBRR |
Chromosome preparation
Seeds of each species were germinated on filter paper in Petri dishes for 3–4 days in the dark. The root tips were immersed in ice-cooled water for 26 h, fixed in ethanol and acetic acid (3:1, v/v), and stored at −20 °C until required. Mitotic chromosome preparations were made from root tips digested in a mixture of enzymes, diluted with 0.01 M sodium citric buffer, containing 20 % (v/v) pectinase (Sigma), 1 % (w/v) cellulose (Calbiochem), and 1 % (w/v) cellulase ‘Onozuka R-10’ (Serva). Meristems were dissected from root tips, squashed in drops of 45 % acetic acid, and the good quality preparation was frozen (Hasterok et al. 2006).
DNA isolation and probe labeling
Genomic DNA (gDNA) from young leaves of all selected species were isolated with the C-TAB method (Doyle and Doyle 1990). DNA from all plants were obtained at the same stage. After the extraction of DNA, samples were labeled by nick translation with tetramethylrhodamine-5-dUTP (Sigma-Aldrich). The ribosomal sequences 35S rDNA and 5S rDNA were labeled with digoxigenin-11-dUTP by nick translation and with tetramethyl-rhodamine-5-dUTP (Sigma-Aldrich) by polymerase chain reaction (PCR), respectively (Kwiatek et al. 2016b).
In situ hybridization
The GISH procedures were performed according to the protocol of Kosmala et al. (2006) and Kwiatek et al. (2016a, b), with minor modifications. The GISH protocol was standardized by several repeats to ensure that the obtained results were comparable and reproducible. The hybridization mixture consisted of 50 % deionized formamide, 10 % dextran sulfate, 2 × SSC, 0.5 % SDS, as well as 100–120 ng/slide gDNA probe. The blocking DNA was not used. Several initial GISH experiments were carried out to reveal optimal specifications, such as probe concentrations and wash temperatures. After establishing optimal GISH conditions, we used only those conditions for all experiments described in this paper. Chromosome preparations and hybridization mixture were denatured together at 80 °C for 2 min and then hybridized overnight at 37 °C. The post-hybridization washes were performed, according to Heslop-Harrison (2000), at 42 °C in 2 × SSC buffer. Probes labeled with tetramethylrhodamine-5-dUTP were directly visualized. After the acquisition of images, selected slides were washed off and reprobed with a new set of probes (35S rDNA and 5S rDNA). The reprobing procedures were performed according to the protocol of Heslop-Harrison et al. (1992), with minor modifications. Immunodetection of the digoxigenated probe (35S rDNA) was performed using fluorescein isothiocyanate-conjugated anti-digoxigenin antibody (Sigma-Aldrich). Mitotic cells were examined with an Olympus XM10 CCD camera attached to an Olympus BX61 automatic epifluorescence microscope. Image processing was carried out using Olympus Cell-F imaging software and Micrografx Picture Publisher software.
Results
GISH analyses were performed in order to determine the relationships between total genomic DNA of key crops and forage species at the chromosomal level. The experiments were carried out using labeled probes from each species to hybridize with chromosomes of every species used in this study based on a “round robin” rule. Overall, 100 GISH experiments were used (10 probes × 10 species, Table 2). The experiments showed various types of signal locations, including: telomeric and centromeric regions, rDNA loci, subcentromeric and subtelomeric regions, as well as chromosome labeling or signals dispersed along whole chromosomes. Additionally, rDNA-FISH (ribosomal DNA fluorescent in situ hybridization) was carried out when GISH revealed rDNA-like signals.
Table 2.
Specification of comparative mapping of the Pooideae subfamily according to a “round robin” rule
| Species | B. distachyon | H. vulgare | S. cereale | T. aestivum | Ae. tauschii | Ae. speltoides | T. urartu | ×Triticosecale | F. pratensis | L. perenne |
|---|---|---|---|---|---|---|---|---|---|---|
| No. of rDNA loci | 35S 2 loci; 5S 2 loci | 35S 4 loci; 5S 8 loci | 35S 2 loci; 5S 4 loci | 35S 4 loci; 5S 12 loci | 35S 2 loci; 5S 4 loci | 35S 4 loci; 5S 2 loci | 35S 4 loci; 5S 4 loci | 35S 6 loci; 5S 10 loci | 35S 2 loci; 5S 2 loci | 35S 7 loci; 5S 2 loci |
| B. distachyon | Centromeric regions + 2 signals of 35S rDNA | 4 signals of 35S rDNA (NOR chromosomes 6 and 7) | Telomeric regions + 2 signals of 35S + 4 signals of 5S rDNA | Telomeric regions + 4 signals of 35S rDNA | Telomeric regions + 2 signals of 35S rDNA | 4 signals of 35S + 2 signals of 5S rDNA, centromeric regions | Telomeric regions + 4 signals of 35S rDNA | Telomeric regions + 6 signals of 35S + 10 signals of 5S rDNA | Telomeric regions + 2 signals of 35S rDNA | Telomeric regions + 7 signals of 35S rDNA |
| H. vulgare | 35S rDNA | Chromosome labeling + centromeric regions | Chromosome labeling + 2 pericentromeric signals in chromosomes without rDNA loci | Chromosome labeling, in some chromosomes lack of signals in centromeric and terminal regions | Labeling of chromosome segments | Chromosome labeling + pericentromeric signals | Chromosome labeling + 2 pericentromeric signals | Pericentromeric signals in 14 chromosomes of B genome | Chromosome labeling + 2 signals of 35S rDNA | Chromosome labeling + 7 signals of 35S rDNA |
| S. cereale | Strong dispersed signals + 2 signals of 35S + 2 signals of 5S rDNA | Centromeric regions | Chromosome labeling + telomeric regions | Chromosome labeling and signals in chromosomes without rDNA loci | Chromosome labeling + 6 terminal signals in chromosomes without rDNA loci | Chromosome labeling without terminal regions, 4–5 signals in chromosomes without rDNA loci | Chromosome labeling | Labeling of 14 chromosomes (genome R) | Weak chromosome labeling + 2 signals of 35S rDNA | Strong, dispersed signals |
| T. aestivum | 35S rDNA + 5S rDNA + telomeric regions + centromeric regions | Pericentromeric regions | Very weak chromosome labeling without NORs and telomeric regions | Chromosome labeling | Chromosome labeling | Chromosome labeling without terminal regions + pericentromeric signals | Chromosome labeling without terminal regions (1 arm in 2 chromosomes) | Labeling of 28 chromosomes (genomes A and B), very weak labeling of 14 R-genome chromosomes | Chromosome labeling + 2 signals of 35S rDNA | 7 signals of 35S rDNA |
| Ae. tauschii | 35S rDNA + telomeric regions + centromeric regions | Chromosome labeling | Chromosome labeling without telomeric regions | Chromosome labeling | Chromosome labeling + telomeric regions + signals in chromosomes without rDNA loci | Chromosome labeling without terminal regions | Chromosome labeling (8 chromosomes with stronger intensity) | Chromosome labeling, 14 chromosomes with stronger intensity (genome B) | Weak chromosome labeling + 2 signals of 35S rDNA | Chromosome labeling + 5 signals of 35S rDNA |
| Ae. speltoides | 2 signals of 35S + 2 signals of 5S rDNA + centromeric regions | Pericentromeric regions + centromeric regions | Chromosome labeling without terminal regions | 14 signals of 5S rDNA, centromeric regions in 14 chromosomes of B genome | Chromosome labeling + 2 terminal signals in 1 pair without rDNA loci | Chromosome labeling + pericentromeric regions | Chromosome labeling without terminal regions in 1 pair | Labeling of 14 chromosomes (genome B) | Chromosome labeling + 2 signals of 35S rDNA | Weak chromosome labeling + 7 signals of 35S rDNA |
| T. urartu | 2 signals of 35S rDNA | Chromosome labeling without terminal regions | Chromosome labeling without terminal regions | Chromosome labeling, 14 chromosomes with stronger intensity (genome A) | Chromosome labeling without terminal regions | Chromosome labeling without terminal regions | Chromosome labeling | Chromosome labeling (A genome - strong signals, B - weaker intensity, R - very weak labeling) | Weak chromosome labeling + 2 signals of 35S rDNA | Weak chromosome labeling + 7 signals of 35S rDNA |
| ×Triticosecale | 35S rDNA + 5S rDNA + strong dispersed signals | Pericentromeric regions | Chromosome labeling without 2 NORs + telomeric regions | Chromosome labeling without terminal regions, centromeric regions and 2 NORs | Chromosome labeling without terminal regions and NORs | Chromosome labeling of selected regions + pericentromeric signals | Chromosome labeling (6 chromosomes with stronger intensity; 2 chromosomes without labeling of terminal regions) | Chromosome labeling | 2 signals of 35S rDNA | Chromosome labeling |
| F. pratensis | 35S rDNA + centromeric regions | Chromosome labeling + centromeric regions | Chromosome labeling without terminal regions | Weak dispersed signals | Dispersed signals + telomeric regions (not in all chromosomes) | Chromosome labeling without terminal regions and interstitial block in chromosome 5 | Chromosome labeling | Chromosome labeling | Chromosome labeling | Chromosome labeling |
| L. perenne | 2 signals of 35S + 2 signals of 5S rDNA + telomeric regions | Chromosome labeling + 4 signals of 35S rDNA | Chromosome labeling without terminal regions + 2 signals of 35S rDNA | 4 signals of 35S rDNA + telomeric regions | 2 signals of 35S + 2 signals of 5S rDNA + telomeric regions + weak dispersed signals | 4 signals of 35S + 2 signals of 5S rDNA | 4 signals of 35S rDNA + terminal signals in 2 chromosomes + dispersed signals | Chromosome labeling + 6 signals of 35S rDNA | Strong, distinct signals | Chromosome labeling without terminal regions in 1 pair |
Columns = chromosome preparations; rows = gDNA probes
Brachypodium distachyon chromosomes
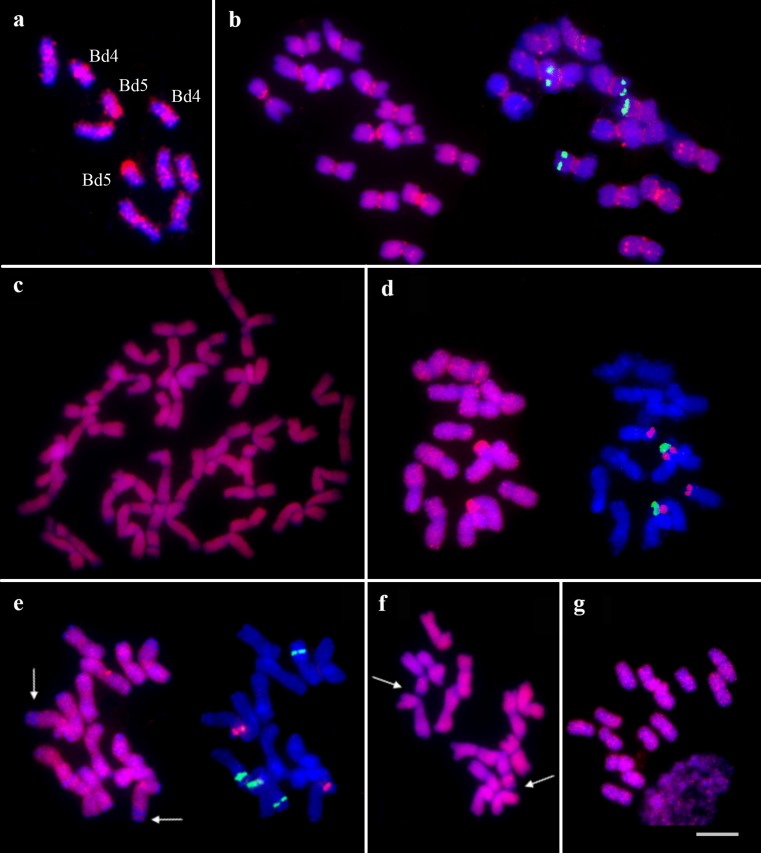
The GISH experiments on B. distachyon (2n = 2x = 10) chromosomes resulted in the 35S rDNA loci being identified using all probes of gDNA (genomic DNA) probes tested. The 5S rDNA loci were labeled by S. cereale, T. aestivum, Ae. speltoides (Fig. 1a), ×Triticosecale, and L. perenne gDNA probes only. Pericentromeric signals were observed when B. distachyon, T. aestivum, Ae. tauschii, Ae. speltoides (Fig. 1a), and F. pratensis gDNA probes were used. Rye and triticale probes also gave strong and clear signals dispersed in all B. distachyon chromosomes. Furthermore, B. distachyon telomeric regions were labeled when T. aestivum, Ae. tauschii, and L. perenne DNA were used as genomic probes.
Fig. 1.
Comparative GISH mapping within the Pooideae subfamily using gDNA (red) of selected species: a Ae. speltoides gDNA probe to B. distachyon chromosomes; b Ae. speltoides gDNA probe to H. vulgare chromosomes; c Ae. tauschii gDNA probe to T. aestivum chromosomes; d L. perenne gDNA probe to Ae. tauschii chromosomes; e F. pratensis gDNA probe to Ae. speltoides chromosomes; f Ae. speltoides gDNA probe to T. urartu chromosomes; g ×Triticosecale gDNA probe to L. perenne chromosomes. b, d, and e: on the right site are shown metaphase plates with 35S (green) and 5S rDNA (red). The white arrows indicate unlabeled regions in chromosomes. Scale bar = 5 μm
Hordeum vulgare chromosomes
All barley (H. vulgare; 2n = 2x = 14) chromosomes were labeled by H. vulgare, Ae. tauschii, F. pratensis, and L. perenne gDNA probes. By contrast, T. urartu probe marked every barley chromosome but left the telomeric regions unlabeled. Clear signals in the centromeric regions were observed when H. vulgare, S. cereale, Ae. speltoides (Fig. 1b), and F. pratensis gDNA probes were hybridized. In addition, pericentromeric regions were identified when T. aestivum, Ae. speltoides, and ×Triticosecale (Fig. 2b) gDNA probes were used. Hybridization using gDNA probes generated from B. distachyon and L. perenne resulted in four strong signals, which corresponded to the 35S rDNA loci.
Fig. 2.
Hybridization patterns for comparative mapping of gDNA (red): a B. distachyon gDNA probe to Ae. speltoides chromosomes showing centromeric regions, 5S and 35S rDNA; b ×Triticosecale gDNA probe to H. vulgare chromosomes: pericentromeric and peritelomeric signals; c L. perenne gDNA probe to F. pratensis chromosomes: other distinct, strong signals (white arrows); d S. cereale gDNA probe to T. urartu chromosomes: chromosome labeling and dispersed signals. Scale bar = 5 μm
Secale cereale chromosomes
All rye (S. cereale; 2n = 2x = 14) chromosomes were labeled by H. vulgare, S. cereale, and ×Triticosecale probes. The triticale probe did not hybridize with two 35S rDNA loci (Fig. 3c). The H. vulgare probe also identified an additional two signals located on the pericentromeric regions of the chromosomes lacking rDNA loci. Hybridizations with the rest of probes, except for B. distachyon probe, resulted in labeling of all the rye chromosome except for the telomeric regions. Similarly to triticale, the wheat probe also did not label two 35S rDNA loci on rye chromosomes. In contrast, B. distachyon gDNA probe gave signals in two 35S and four 5S rDNA loci, as well as in terminal regions. Finally, the L. perenne probe highlighted an additional two regions which corresponded to the 35S rDNA loci.
Fig. 3.
Labeling of all chromosomes using gDNA (red) of selected species beyond: a subtelomeric regions: T. urartu gDNA probe to Ae. speltoides chromosomes; b centromeric regions: H. vulgare gDNA probe to T. urartu chromosomes (the arrows indicate distinct pericentromeric signals); c NORs: triticale gDNA probe to S. cereale chromosomes (the arrows indicate NORs)
Triticum aestivum chromosomes
All wheat chromosomes (T. aestivum; 2n = 6x = 42) were labeled by T. aestivum, T. urartu, Ae. tauschii (Fig. 1c), and S. cereale gDNA probes, although the T. urartu probe resulted in 14 chromosomes of A genome being marked stronger. The rye gDNA probe showed strong and dispersed signals in most of the wheat chromosomes. Triticale and barley gDNA probes also labeled all chromosomes; however, the centromeric and telomeric regions as well as two 35S rDNA loci (only in the case of triticale probe) remained unlabeled. Contrastingly, the Ae. speltoides gDNA probe gave only 14 signals in 5S rDNA loci and 14 centromeric regions in B genome chromosomes. The B. distachyon probe resulted in labeled telomeric sites and four signals in 35S rDNA loci, while L. perenne yielded four 35S rDNA loci and telomeric regions. Finally, weak and dispersed signals were observed using F. pratensis gDNA probe.
Aegilops tauschii chromosomes
Aegilops tauschii (2n = 2x = 14) chromosomes were marked in GISH experiments using T. aestivum, Ae. tauschii, S. cereale, and Ae. speltoides gDNA probes. Additionally, Ae. tauschii probe hybridization resulted in strong telomeric signals and clear dispersed signals along all chromosomes. Six telomeric sites were detected using rye probe. The Ae. speltoides gDNA probe gave also two terminal signals in Ae. tauschii chromosomes. The ×Triticosecale and T. urartu gDNA probes also marked all chromosomes; however, the telomeric regions and two 35S rDNA loci (only in the case of triticale probe) remained unlabeled. Labeled segments of Ae. tauschii were observed when H. vulgare gDNA probe was used in the GISH experiments. In contrast, L. perenne (Fig. 1d) and F. pratensis gDNA probes resulted in weak dispersed sites observed in all chromosomes. Additionally, the L. perenne probe gave signals in two 35S and two 5S rDNA loci, and also labeled all telomeric regions (Fig. 1d). Two 35S rDNA loci and telomeric sites were also labeled when GISH was carried out using B. distachyon gDNA probe.
Aegilops speltoides chromosomes
The experiments with labeled DNA of Ae. tauschii, T. urartu (Fig. 3a), S. cereale, T. aestivum, and F. pratensis (Fig. 1e) revealed labeling of all Ae. speltoides (2n = 2x = 14) chromosomes with the exception of the telomeric regions, whereas gDNA probe of S. cereale revealed strong, distinct signals on several chromosomes. GISH with Ae. speltoides gDNA probe resulted in chromosome labeling and brighter signals in pericentromeric regions. Similar results for all chromosomes of Ae. speltoides were obtained when gDNA of ×Triticosecale, T. aestivum, and H. vulgare were used for GISH. What is more, the subtelomeric region of a pair of chromosomes possessing the 5S loci remained unlabeled when hybridization with F. pratensis gDNA (Fig. 1e) was performed. Four 35S and two 5S rDNA loci were obtained for the DNA of L. perenne and B. distachyon (Fig. 2a) and centromeric regions were highlighted using B. distachyon only.
Triticum urartu chromosomes
Labeled gDNA of five species, S. cereale (Fig. 2d), Ae tauschii, T. urartu, F. pratensis, and H. vulgare lead to chromosome labeling on their entire length. Additionally, distinct signals in the pericentromeric regions in two chromosomes and unlabeled centromeric regions in all chromosomes were observed in experiments using H. vulgare DNA (Fig. 3b). Chromosome painting with lack of signals in the terminal part of one pair of chromosomes was obtained for the DNA of Ae. speltoides (Fig. 1f), T. aestivum, and ×Triticosecale. Additionally, Ae. speltoides probe resulted in some strong, clear signals in pericentromeric regions (Fig. 1f). Brachypodium distachyon probe resulted in strong subtelomeric signals and gave four signals of 35S rDNA loci. The genomic DNA of L. perenne revealed only the 35S rDNA sequences.
×Triticosecale chromosomes
Hybridization with gDNA of B. distachyon and H. vulgare to chromosomes of triticale (×Triticosecale; 2n = 6x = 42) provided contrasting results to the remaining experiments, where chromosome labeling was observed. The gDNA of B. distachyon was mapped in telomeric sites and gave six signals of 35S rDNA loci and ten signals of 5S rDNA loci. The dispersed, strong signals located near the centromeric and telomeric regions in genome B were visible for the experiment with gDNA of H. vulgare. GISH with S. cereale probe enabled to distinguish the R genome chromosomes, while the DNA probe of Ae. speltoides visualized the B genome, and DNA of T. urartu the A genome chromosomes. What is more, the hybridization with DNA of T. aestivum enabled to identify A and B genome as well as R genome chromosomes in terms of negative discrimination (very weak labeling). Probing of homologous gDNA to triticale chromosomes resulted in entire chromosomes being labeled. Finally, probing with Ae. tauschii gDNA resulted in chromosome labeling, but with more intensity of the B genome of ×Triticosecale.
Festuca pratensis chromosomes
GISH with gDNA probes of selected species, with the exception of F. pratensis and L. perenne (Fig. 2c), revealed two signals of 35S rDNA in the karyotype of F. pratensis (2n = 2x = 14). Furthermore, in nearly all cases where 35S rDNA signals were detected, complete chromosome labeling was also observed; the exceptions were B. distachyon and ×Triticosecale. However, B. distachyon probe produced weak signals in telomeric regions. Chromosome labeling was clearly visible when F. pratensis gDNA was used to probe chromosomes of F. pratensis. Hybridization of L. perenne DNA to F. pratensis revealed strong, distinct signals in some chromosomes (Fig. 2c).
Lolium perenne chromosomes
The hybridization of genomic DNA of B. distachyon, H. vulgare, T. aestivum, Ae. tauschii, Ae. speltoides, and T. urartu to chromosomes of L. perenne (2n = 2x = 14) identified the 35S rDNA loci. What is more, the B. distachyon gDNA probe labeled the telomeric sites of L. perenne. Strong, dense signals were observed for S. cereale probe. Lolium perenne chromosomes probed with gDNA from H. vulgare, Ae. tauschii, T. urartu, and Ae. speltoides resulted in both 35S rDNA signals and chromosome labeling. Probing gDNA of ×Triticosecale (Fig. 1g), F. pratensis, and L. perenne resulted in the labeling of all chromosomes. GISH with gDNA of L. perenne to L. perenne chromosomes showed an unlabeled terminal part of a pair of chromosomes that lacked rDNA loci.
Discussion
In this study, we detected and described the hybridization patterns of various genomic DNA probes generated from key representatives of cereals, forage grasses, and B. distachyon on mitotic chromosomes of those species. The aim of this approach was to score the affinity of the given species by using gDNA as probes, without blocking DNA from the species chosen for chromosome spreads preparations. As a result, the whole chromosomes or chromosome regions were distinguished or variable types of signals were visualized to prove the different levels of the relationships between genomes used in this study.
A part of the presented results are either as they were expected or confirm the results reported in other papers. However, our study also reveals some novel insights, which require more specific analysis in the future. The most characteristic DNA motifs in the Pooideae subfamily are telomeric and rDNA sequences. Our results confirmed the previous study of Hasterok et al. (2004), who reported labeled pericentromeric regions and 35S rDNA loci on B. distachyon chromosomes using B. distachyon genomic probe. This phenomenon could be explained by the small content of repetitive sequences in this genome. Besides, Mayer et al. (2011) compared the gene order of B. distachyon with H. vulgare and reported that the number of conserved syntenic loci was high (14,422), reflecting a closer phylogenetic affinity. What is more, Ma et al. (2010) carried out GISH reciprocal experiments between B. distachyon and H. vulgare and observed mainly 35S ribosomal DNA loci. In the present study, the same type of affinity was observed in GISH runs with all of the gDNA probes, indicating that this high copy DNA sequence is conserved between analyzed grasses. Ribosomal DNA loci, which are essential for all organisms, can be detected at the cytogenetic level; however, homogenization operates very strongly in these regions in all Poaceae genomes (Sallares and Brown 1999). Signals observed in the centromeric regions of B. distachyon led to the assumption that this species is more closely related to Ae. speltoides and Ae. tauschii than with T. urartu. Moreover, we showed a certain degree of genomic relationship between B. distachyon and F. pratensis. This point of view could also be confirmed by the phylogenetic analysis of grass species based on the ndfH chloroplast gene sequence made by Mochida and Shinozaki (2013).
Considering their importance in agriculture, the most analyzed temperate cereal species in comparative genomics are barley and wheat, along with their relatives. Based on comparison to the previously constructed high-density physical marker map of wheat (Qi et al. 2004), Mayer et al. (2011) reported that barley contains an archetypal Triticeae genome. Most of the chromosome arms showed well-conserved synteny with previously reported chromosomal translocations (Qi et al. 2006). In the present work, we observed labeled pericentromeric regions on H. vulgare chromosomes using Ae. speltoides, T. aestivum, and ×Triticosecale gDNA probes. These observations are correlated with the results of Icsó et al. (2015), where gDNA of H. vulgare was used for the chromosome probing of T. aestivum. They reported that gDNA of barley produced hybridization bands mainly in the pericentromeric and intercalary chromosome regions of B genome chromosomes and it corresponded to the GAA FISH pattern. Surprisingly, we observed differences in the labeling of wheat chromosomes using barley genomic probe in comparison to chromosomes of wheat labeled with gDNA probes of its ancestral species. The three genomes of wheat share similar repetitive sequence types, with the D genome specific repetitive sequences being the most frequent (Nagaki et al. 1995; Jia et al. 2013). In addition, the existence of tandemly repeated sequence dpTa1 presented in 58 species of the Triticeae tribe have been reported (Vershinin et al. 1994). The occurrence of signals detected in phylogenetically distant species may indicate strong relationships between repetitive sequences in selected grasses. For example, the pSc119.2 sequence from rye is widely used in the identification of wheat chromosomes and its relatives (Cuadrado and Jouve 1994; Schneider et al. 2003, 2005; Wiśniewska et al. 2013).
On the other hand, we have observed an unexpected lack of signals in secondary constrictions of rye chromosomes probed by triticale genomic DNA. This may be connected with the differences in rDNA sequences between 1R chromosomes of rye and 1R chromosomes of triticale. In general, unlabeled chromosome regions can be related with species-specific sequences or disparate pathways of chromosome differentiation, which was exposed by the lack of labeled segments of Ae. speltoides chromosomes probed with F. pratensis gDNA. The most likely explanation for the unlabeled telomeric regions could be the fact that the Ae. speltoides genome carries subtelomeric sequences Spelt1 and Spelt52 (Salina et al. 2006, 2009). On the other hand, the lack of signals from the 5S chromosome segment of Ae. speltoides remains unknown but indicates that this region could have gone under structural changes during S genome evolution. What is more, we observed single signals in pericentromeric locations in some chromosomes of T. urartu probed with Ae. speltoides gDNA. Similarly, Belyayev et al. (2000) reported pericentromeric signals in the chromosomes of T. urartu and suggested that those regions are conserved in A and B genomes. Moreover, comparative GISH resulted in a lack of signals in the subtelomeric regions in the chromosomes of T. aestivum, Ae. speltoides, Ae. tauschii, and H. vulgare. This could be determined by the different constitution of subtelomeric regions. For example, the composition of the termini of S. cereale chromosomes is complex and, besides the basic telomeric sequence (T/A)1-4 G1-8, it also contains tandemly organized DNA families such as pSc119.2, pSc200, and pSc250 (Salina et al. 2009).
Our comparative analysis of genomic relationships within crucial Pooideae species revealed some intriguing results, such as unexpected chromosome labeling between distant species (e.g., ×Triticosecale vs. L. perenne), dispersed signals (e.g., L. perenne vs. F. pratensis), or labeling of selective chromosome regions (e.g., S. cereale vs. Ae. speltoides). The strong, distinct signals observed for L. perenne and F. pratensis can be useful for the karyotyping of F. pratensis chromosomes. Till now, only two pairs of chromosomes, 2F with 35S rDNA and 3F with 5S rDNA loci, can be easily identifiable (Jones et al. 2002; Thomas 1981).
In conclusion, our results are in parallel with the comparative studies made on model species and key representatives of the Pooideae subfamily. However, some results indicated unknown intergenomic dependencies that need further analysis. Moreover, reciprocal GISH analysis on Aegilops, Hordeum, and Triticum species could shed light on the chromosome structure and evolution within the Triticeae tribe.
Acknowledgments
The authors are grateful to Dylan Wyn Phillips from the Institute of Biological, Environmental and Rural Sciences, Aberystwyth University, Aberystwyth, Wales, United Kingdom for his invaluable advice and helpful comments on the manuscript.
Authors’ contributions
JM, MM, and MK conceived, designed, and performed the experiments, and wrote the paper. All authors analyzed the data.
Compliance with ethical standards
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no competing interests.
References
- Belyayev A, Raskina O, Korol A, Nevo E. Coevolution of A and B genomes in allotetraploid Triticum dicoccoides. Genome. 2000;43:1021–1026. doi: 10.1139/gen-43-6-1021. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ (2012) Plant DNA C-values database (release 6.0, December 2012). Available online at: http://data.kew.org/cvalues/
- Bonierbale MW, Plaisted RL, Tanksley SD. RFLP maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics. 1988;120:1095–1103. doi: 10.1093/genetics/120.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán P, Shi Y, Armstrong L, Draper J, Stace CA. Molecular phylogeny of the grass genus Brachypodium P. Beauv. based on RFLP and RAPD analysis. Bot J Linn Soc. 1995;117:263–280. [Google Scholar]
- Cuadrado A, Jouve N. Highly repetitive sequences in B chromosomes of Secale cereale revealed by fluorescence in situ hybridization. Genome. 1994;37:709–712. doi: 10.1139/g94-100. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Jouve N. Evolutionary trends of different repetitive DNA sequences during speciation in the genus Secale. J Hered. 2002;93:339–345. doi: 10.1093/jhered/93.5.339. [DOI] [PubMed] [Google Scholar]
- Devos KM. Updating the ‘crop circle’. Curr Opin Plant Biol. 2005;8:155–162. doi: 10.1016/j.pbi.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid total DNA preparation procedure for fresh plant tissue. Focus. 1990;12:13–15. [Google Scholar]
- Draper J, Mur LA, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, et al. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 2001;127:1539–1555. doi: 10.1104/pp.010196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell RB, O’Dell M, Smith DB. Repeated sequence DNA comparisons between Triticum and Aegilops species. Heredity. 1979;42:309–322. doi: 10.1038/hdy.1979.34. [DOI] [Google Scholar]
- Gale MD, Devos KM. Comparative genetics in the grasses. Proc Natl Acad Sci U S A. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, et al. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature. 2006;439:749–752. doi: 10.1038/nature04434. [DOI] [PubMed] [Google Scholar]
- Hasterok R, Draper J, Jenkins G. Laying the cytotaxonomic foundations of a new model grass, Brachypodium distachyon (L.) Beauv. Chromosome Res. 2004;12:397–403. doi: 10.1023/B:CHRO.0000034130.35983.99. [DOI] [PubMed] [Google Scholar]
- Hasterok R, Dulawa J, Jenkins G, Leggett M, Langdon T. Multi-substrate chromosome preparations for high throughput comparative FISH. BMC Biotechnol. 2006;6:20. doi: 10.1186/1472-6750-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison JS. Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. Plant Cell. 2000;12:617–636. doi: 10.1105/tpc.12.5.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Harrison GE, Leitch IJ. Reprobing of DNA:DNA in situ hybridization preparations. Trends Genet. 1992;8:372–373. doi: 10.1016/0168-9525(92)90287-E. [DOI] [PubMed] [Google Scholar]
- Hsiao C, Chatterton NJ, Asay KH, Jensen KB. Phylogenetic relationships of 10 grass species: an assessment of phylogenetic utility of the internal transcribed spacer region in nuclear ribosomal DNA in monocots. Genome. 1994;37:112–120. doi: 10.1139/g94-014. [DOI] [PubMed] [Google Scholar]
- Huo NX, Vogel JP, Lazo GR, You FM, Ma Y, McMahon S, et al. Structural characterization of Brachypodium genome and its syntenic relationship with rice and wheat. Plant Mol Biol. 2009;70:47–61. doi: 10.1007/s11103-009-9456-3. [DOI] [PubMed] [Google Scholar]
- Icsó D, Molnár-Láng M, Linc G. Constructing an alternative wheat karyotype using barley genomic DNA. J Appl Genet. 2015;56:45–48. doi: 10.1007/s13353-014-0230-0. [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative (IBI) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature. 2013;496:91–95. doi: 10.1038/nature12028. [DOI] [PubMed] [Google Scholar]
- Jones ES, Mahoney NL, Hayward MD, Armstead IP, Jones JG, Humphreys MO, et al. An enhanced molecular marker based genetic map of perennial ryegrass (Lolium perenne) reveals comparative relationships with other Poaceae genomes. Genome. 2002;45:282–295. doi: 10.1139/g01-144. [DOI] [PubMed] [Google Scholar]
- Keller B, Feuillet C. Colinearity and gene density in grass genomes. Trends Plant Sci. 2000;5:246–251. doi: 10.1016/S1360-1385(00)01629-0. [DOI] [PubMed] [Google Scholar]
- Kellogg EA. Evolutionary history of the grasses. Plant Physiol. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmala A, Zwierzykowski Z, Gąsior D, Rapacz M, Zwierzykowska E, Humphreys MW. GISH/FISH mapping of genes for freezing tolerance transferred from Festuca pratensis to Lolium multiflorum. Heredity (Edinb) 2006;96:243–251. doi: 10.1038/sj.hdy.6800787. [DOI] [PubMed] [Google Scholar]
- Kwiatek M, Belter J, Majka M, Wiśniewska H. Allocation of the S-genome chromosomes of Aegilops variabilis Eig. carrying powdery mildew resistance in triticale (× Triticosecale Wittmack) Protoplasma. 2016;253:329–343. doi: 10.1007/s00709-015-0813-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatek M, Majka M, Ślusarkiewicz-Jarzina A, Ponitka A, Pudelska H, Belter J, Wiśniewska H. Transmission of the Aegilops ovata chromosomes carrying gametocidal factors in hexaploid triticale (×Triticosecale Wittm.) hybrids. J Appl Genet. 2016;57:305–315. doi: 10.1007/s13353-015-0332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Vu GTH, Schubert V, Watanabe K, Stein N, Houben A, et al. Synteny between Brachypodium distachyon and Hordeum vulgare as revealed by FISH. Chromosome Res. 2010;18:841–850. doi: 10.1007/s10577-010-9166-3. [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Martis M, Hedley PE, Šimková H, Liu H, Morris JA, et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell. 2011;23:1249–1263. doi: 10.1105/tpc.110.082537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch SR. Genomics and synteny. Plant Physiol. 2001;125:152–155. doi: 10.1104/pp.125.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Shinozaki K. Unlocking Triticeae genomics to sustainably feed the future. Plant Cell Physiol. 2013;54:1931–1950. doi: 10.1093/pcp/pct163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat F, Zhang R, Guizard S, Flores R, Armero A, Pont C, et al. Shared subgenome dominance following polyploidization explains grass genome evolutionary plasticity from a seven protochromosome ancestor with 16K protogenes. Genome Biol Evol. 2014;6:12–33. doi: 10.1093/gbe/evt200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Tsujimoto H, Isono K, Sasakuma T. Molecular characterization of a tandem repeat, Afa family, and its distribution among Triticeae. Genome. 1995;38:479–486. doi: 10.1139/g95-063. [DOI] [PubMed] [Google Scholar]
- Qi LL, Echalier B, Chao S, Lazo GR, Butler GE, Anderson OD, et al. A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics. 2004;168:701–712. doi: 10.1534/genetics.104.034868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Friebe B, Gill BS. Complex genome rearrangements reveal evolutionary dynamics of pericentromeric regions in the Triticeae. Genome. 2006;49:1628–1639. doi: 10.1139/g06-123. [DOI] [PubMed] [Google Scholar]
- Salina EA, Lim KY, Badaeva ED, Shcherban AB, Adonina IG, Amosova AV, et al. Phylogenetic reconstruction of Aegilops section Sitopsis and the evolution of tandem repeats in the diploids and derived wheat polyploids. Genome. 2006;49:1023–1035. doi: 10.1139/G06-050. [DOI] [PubMed] [Google Scholar]
- Salina EA, Sergeeva EM, Adonina IG, Shcherban AB, Afonnikov DA, Belcram H, et al. Isolation and sequence analysis of the wheat B genome subtelomeric DNA. BMC Genomics. 2009;10:414. doi: 10.1186/1471-2164-10-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallares R, Brown TA. PCR-based analysis of the intergenic spacers of the Nor loci on the A genomes of Triticum diploids and polyploids. Genome. 1999;42:116–128. doi: 10.1139/gen-42-1-116. [DOI] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, et al. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell. 2008;20:11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Linc G, Molnár-Láng M, Graner A. Fluorescence in situ hybridization polymorphism using two repetitive DNA clones in different cultivars of wheat. Plant Breed. 2003;122:396–400. doi: 10.1046/j.1439-0523.2003.00891.x. [DOI] [Google Scholar]
- Schneider A, Linc G, Molnár I, Molnár-Láng M. Molecular cytogenetic characterization of Aegilops biuncialis and its use for the identification of 5 derived wheat–Aegilops biuncialis disomic addition lines. Genome. 2005;48:1070–1082. doi: 10.1139/g05-062. [DOI] [PubMed] [Google Scholar]
- Thomas HM. The Giemsa C-banding karyotypes of six Lolium species. Heredity. 1981;46:263–267. doi: 10.1038/hdy.1981.33. [DOI] [Google Scholar]
- Vershinin A, Svitashev S, Gummesson PO, Salomon B, von Bothmer R, Bryngelsson T. Characterization of a family of tandemly repeated DNA sequences in Triticeae. Theor Appl Genet. 1994;89:217–225. doi: 10.1007/BF00225145. [DOI] [PubMed] [Google Scholar]
- Vershinin AV, Schwarzacher T, Heslop-Harrison JS. The large-scale genomic organization of repetitive DNA families at the telomeres of rye chromosomes. Plant Cell. 1995;7:1823–1833. doi: 10.1105/tpc.7.11.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Bragg JN. Brachypodium distachyon, a new model for the Triticeae. In: Feuillet C, Muehlbauer GJ, editors. Genetics and genomics of the Triticeae. New York: Springer; 2009. pp. 427–449. [Google Scholar]
- Vogel J, Hill T. High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21-3. Plant Cell Rep. 2008;27:471–478. doi: 10.1007/s00299-007-0472-y. [DOI] [PubMed] [Google Scholar]
- Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E, et al. Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 2007;3:e123. doi: 10.1371/journal.pgen.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewska H, Kwiatek M, Kulak-Książczyk S, Apolinarska B. Introgression of A- and B-genome of tetraploid triticale chromatin into tetraploid rye. J Appl Genet. 2013;54:435–440. doi: 10.1007/s13353-013-0171-z. [DOI] [PMC free article] [PubMed] [Google Scholar]